Abstract
Sensitivity enhancement by isotope enrichment and DNP NMR enables detection of minor but biologically relevant species in native intact bone, including nucleic acids, choline from phospholipid headgroups, and histidinyl and hydroxylysyl groups. Labelled matrix from the aggressive osteosarcoma K7M2 cell line confirms the assignments of nucleic acid signals arising from purine, pyrimidine, ribose, and deoxyribose species. Detection of these species is an important and necessary step in elucidating the atomic level structural basis of their functions in intact tissue.
Towards elucidating their biological roles in intact tissue, DNP NMR reveals nucleic acids, and other important low abundance biomolecules in a complex biomaterial, bone, and in cancer extracellular matrix.
The extracellular matrix (ECM) is a complex, heterogeneous, and active, environment, playing structural roles, but also strongly influencing cell behaviour in both homeostasis and pathology. The difficulty of understanding the composition and regulation of the ECM parallels the situation within cells, but with the added complication that many molecular components are insoluble. Solid-state NMR (ssNMR) can contribute to a better understanding of the ECM through direct detection of sparse species, made possible by a strategy of isotopic enrichment of native biomaterials1 combined with rapidly evolving dynamic nuclear polarization (DNP) enhancement techniques.2 Briefly these are based on the transfer of the much greater electron spin polarization, excited by a microwave laser at the electron resonance frequency of an organic free radical dopant, to the (NMR active) nuclei of the molecules of interest, at low temperatures to maximize polarization. The conversion of electron to nuclear polarization results in NMR sensitivity enhancements of up to several orders of magnitude. Recently we have reported direct observation of important low abundance collagen post-translational modifications (PTMs) in intact native, and model, ECM.3 While we focussed on species derived from one specific amino acid (Lys) component of the ECM, it is also of great potential importance to investigate whether other sparse ECM species become observable with DNP enhancement. Non-protein minor species, besides low abundance amino acids, are of particular interest since they represent a realm in which NMR can provide atomic length scale molecular structural information complementing other current biophysical techniques. Incorporating non-protein molecules into the overall structural biology picture will broaden conceptions of which ECM components to investigate in further detail towards an integrated model of the biological and material properties of tissues.
Here, we report DNP-enhanced NMR on mouse bone1 enriched in a wide range of 13C, 15N-amino acids. Where necessary to distinguish between low abundance protein, and non-protein, signals, we confirmed amino acid type assignments by comparison with ECM produced by cells cultured on media supplemented with one or a few specific amino acids, using methods already described.4,5 Our previous analysis of bone focussed on the relatively abundant species observable with conventional ssNMR. The enhancement provided by DNP now reveals non-protein biomolecules biosynthesized from isotopically enriched amino acids and/or sugars, and their incorporation into bone. The preparation of stable isotope enriched biomaterials, and our DNP NMR methodology, is described in detail in the ESI,† Materials and methods sections.
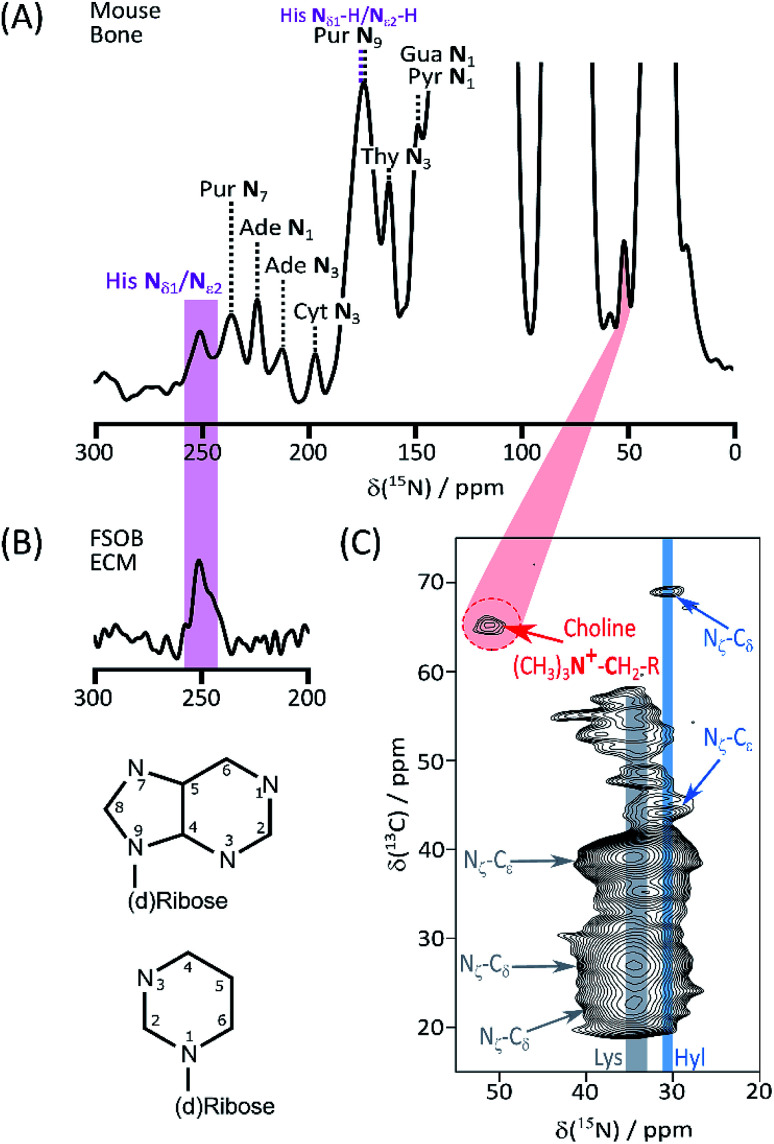
We begin our investigation on bone with a suite of ssNMR characterizations under DNP signal enhancement conditions. A full 1D 15N DNP NMR spectrum is shown in ESI Fig. S1.† Apart from the predictable major signals (from backbone and sidechain amides, and abundant Lys and Arg) the sensitivity enhancement of enrichment-DNP reveals a number of more minor resonances (Fig. 1A) from lower abundance species; the shifts of many of those at high spectral frequency coincide closely with the common (deoxy)nucleotide base ring nitrogens (Biological Magnetic Resonance Database, BMRB, as summarized in ESI Schemes S1† (DNA) and S2 (RNA)). Signals from the nitrogen atoms of protonated His imidazolium (ca. 180 ppm) and unprotonated imidazole (ca. 250 ppm) ring forms are also observed.6,7 The His assignments are reinforced by 15N data from model osteoblast (bone precursor) ECM selectively enriched in (U-13C6, 15N3)-His (His*) (Fig. 1B; see ESI† for preparation and comparison of 15N–13C correlation spectra, Fig. S2†). The low frequency region of the bone DARR-assisted 15N–13C correlation spectrum (Fig. 1C) displays a clear cross peak (65 ppm 13C, 52 ppm 15N) which we ascribe to the (CH3)3N̲+–C̲H2–CH2–O– correlation in the head groups of phosphatidylcholine (PC) lipids (13C assignments based on Kroon et al.;8 see ESI Fig. S3† for overlays with model compound spectra). The PC ethanolamine substructure is biosynthesized from a single serine unit so, in our bone material, all atoms will be concurrently labelled. Finally Fig. 1C also shows the distinctive Hyl Nζ–Cδ fingerprint correlations; the close correspondence with (U-13C6, 15N2)-Lys enriched ECM from vascular smooth muscle cells3 is shown in ESI Fig. S4,† proving that our labelling – DNP NMR combination readily detects the Hyl PTM even in this complex organic–inorganic composite native biomaterial, as well as in simpler native (skin) and model ECMs.3 Chemical shifts are summarized in Table 1.
Fig. 1. (A) High frequency region of the 15N DNP-NMR spectrum showing assignments of resolved base signals of nucleic acids, and ring His signals; (B) 15N spectrum of His* FSOb ECM highlighting correspondence with the signal from the imidazole form of the His ring; (C) DARR assisted 15N–13C correlation spectrum of labelled bone, transmitting magnetization along the Hyl (and Lys) sidechain to emphasize correlations between Hyl (as well as the intense Lys) Nζ and Cδ. Also highlighted is the signal from the (CH3)3N̲+–C̲H2–CH2–O– correlation from phosphatidylcholine headgroups. Atom numbering of the nucleic acid purine (top) and pyrimidine skeletons is also shown.
Chemical shifts of resolved signals from low abundance species detected in 13C, 15N enriched bone by DNP NMR.
| Amino acids/PTMs | ||||||
| Hyl | Cδ | Cε | Nζ | |||
| 69 | 44 | 31 | ||||
| His | Cγ | Cδ2 | Cε1 | Nδ1a | Nε2a | Nδ1/ε2b |
| 129 | 117 | 135 | 180 | 177 | 250 | |
| (Deoxy)nucleotide bases | ||||||
| N1 | N3 | N7 | N9 | |||
| Ade | 224 | 212 | 235c | 174c | ||
| Gua | 148 | 235c | 174c | |||
| Cyt | 148 | 197 | 197 | |||
| Thy/Ura | 162 | 148 | ||||
| Lipids | ||||||
| PC | –N̲(CH3)3+ | –C̲H2–N+ | ||||
| 52 | 65 | |||||
Protonated imidazolium form.
Unprotonated imidazole form.
Single unresolved signal.
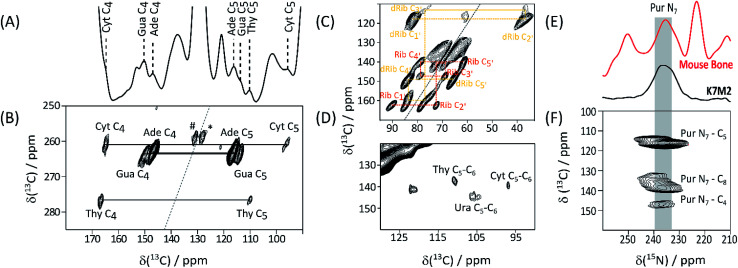
Detection of His, lipid components, and Hyl, in native biomaterials is not unexpected. In contrast nucleic acids in bone have been little discussed, although this tissue is a favourable source of forensic genetic material.9 Given this underrepresentation of bone in nucleic acid literature, we turned to ECM from the aggressively metastasizing K7M2 osteosarcoma cell line10 to substantiate these signals. A 13C DNP NMR spectrum of K7M2 ECM supplemented with (U-13C2, 15N)-Gly (Gly*) and (U-13C6)-glucose (Glc*) is shown in Fig. 2A; it shows a number of clear signals with shifts consistent with the common (deoxy)nucleotide bases11 (ESI Schemes S1 and S2†). Abundant nucleic acid in the K7M2 ECM is confirmed by conventional in situ propidium iodide staining and confocal fluorescence microscopy (ESI Fig. S5†). The biosynthetic routes to the bases (ESI Scheme S3†) dictates that all the base carbon atoms will be at least partially 13C enriched by Gly* and Glc*, although only the purine C4–C5–N7 substructure (from intact Gly*), and to some extent pyrimidine C4–C5–C6 from Glc* via Ser,12 will necessarily be concurrently labelled. (Deoxy)riboses are biosynthesized from Glc* and so all (deoxy)sugar rings will be concurrently 13C enriched. Fig. 2B details results of a through-bond (J-transmitted) INADEQUATE,13 which clearly shows the predicted purine C4–C5 and pyrimidine C4–C5 cross peaks. DARR14 (Fig. 2C) confirms these and indicates pyrimidine C6 signals in Cyt and Thy, and Ura from RNA. The presence of both DNA dRib, and RNA Rib, is confirmed by the low frequency part of the INADEQUATE spectrum (Fig. 2D). 15N DNP NMR shows a clear signal from purine N7's, corresponding closely to that of the putative purine N7's in bone (Fig. 2E). Also a 13C{15N} TEDOR15 (Fig. 2F) mapping short carbon-nitrogen distances, shows the expected purine N7–C5 and weaker N7–C4 correlations, but also purine N7–C8 cross peaks (the latter atom acquired from Glc* via N-formyl tetrahydrofolate). All DNA and RNA shifts measured in K7M2 ECM are summarized in Table 2.
Fig. 2. 13C DNP NMR spectra of Gly*, Glc* supplemented K7M2 ECM. (A) 13C CP-MAS; nucleotide base signals assigned by 2D experiments and database shift values (ESI Schemes S1 and S2†) are labelled; (B) high frequency region of INADEQUATE J-transmitted SQ-DQ correlation spectrum (265 ppm 13C DQ sweep width to avoid aliasing; #, * – correlations between lipid olefinic carbons); (C) low frequency region of the same spectrum tracing out correlations within ribose (Rib), and deoxyribose (dRib), rings; (D) high frequency region of the 13C–13C DARR correlation spectrum base; (E) 15N DNP NMR of Gly*Glc* K7M2 ECM compared with that of labelled bone, supporting the assignment of the 235 ppm signal as purine N7; (F) 13C{15N} TEDOR spectrum showing the expected purine N7–C5 and N7–C4, as well as the putative purine ring N7–C8, through-space correlations (15N sweep width 438 ppm to cover full 15N range).
Chemical shifts of resolved (deoxy)nucleotide signals in the K7M2 rat osteosarcoma cell line.
| C2 | C4 | C5 | C6 | C8 | N7 | |
|---|---|---|---|---|---|---|
| Ade | 153a | 147b,c | 116b,c | 153a | 139b,e | 236b,e |
| Gua | 153a | 150b,c | 114b,c | 167a | 136b,e | 239b,e |
| Cyt | 153a | 165c | 96c | 140d | ||
| Thy | 153a | 167c | 110c | 137d | ||
| Ura | 105d | 145d |
Signals observed in 1D spectrum although no SQ-DQ or DARR correlations observable; assignments are inferred from average shifts reported in BMRB.
Assignments to Ade and Gua are based on rank order of BMRB average shifts, and may be reversed.
From J-transmitted INADEQUATE SQ-DQ correlation.
From 13C–13C DARR correlation.
From 15N–13C TEDOR.
| C1′ | C2′ | C3′ | C4′ | C5′ | |
|---|---|---|---|---|---|
| dRib | 82 | 37 | 78 | 84 | 65 |
| Rib | 90 | 72 | 69 | 79 | 61 |
There are large number of catalogued nucleic acid shifts in the BMRB but as yet no robust equivalent of the chemical shift index16 of protein secondary structural elements, especially with respect to 15N. Nucleotide base 13C and 15N shifts are particularly sensitive to secondary structural features17–20 but linewidths and overlap in our 13C and 15N spectra currently preclude structural conclusions about K7M2 ECM. In principle though this work suggests that the potential of 15N shifts as indices of nucleic acid structure20,21 in whole tissues can be realized through several conformational “reporter” nitrogen atoms in the well resolved high frequency NMR window above 150 ppm. Nucleic acid in bone may be predominantly intracellular, unlike that of K7M2 where a higher extracellular proportion is expected, but it is interesting that these distinctive 15N signals can be observed in both systems. We envisage that detection by DNP-NMR in native tissue is an essential step in fulfilling this potential to characterize nucleic acid structure in the intact tissue and ECM environment.
NMR detection of nucleic acids in bone raises fresh questions about their biological role, and avenues to new insights. NMR detection is an essential first step in elucidating these roles at the molecular and atomic level in native materials. Extracellular roles e.g. signalling, of nucleic acids are likely to operate in K7M2 ECM, and potentially also in bone and bone diseases e.g. osteoporosis.22 Bone undergoes constant remodelling so a high degree of transcriptional, translational, and apoptotic activities are expected,23 leading to significant amounts of nucleic acid being produced in bone cells and released in the ECM. Once in the extracellular environment, some nucleic acids may have a different function in addition to their intracellular roles. For instance, nucleic acids have pronounced affinity for apatite mineral,24 and may participate in biomineralization,25 as calcium sequestering and mineral templating scaffolds, a role that is already proposed for structurally related polynucleotides like poly-ADP-ribose.4
Phospholipids26 are abundant in mineralized bone.27 They may represent the presence of matrix vesicles – extracellular structures ca. 100 nm in size, “pinched off” from the outer membranes of osteoblasts – which act as a nidus for initial calcium phosphate precipitation in normal28 and pathological29 biomineralization. The molecular mechanisms underlying the initial bioprecipitation events and processes remain to be elucidated, in particular the role of matrix vesicle phospholipids. Direct observation of the PC headgroup by NMR in healthy bone is a significant step in atomic level understanding of the relevant intermolecular and interionic interactions. In addition to matrix vesicles, of 40–100 nm sized exosomes are the basis of a pathway for cancer metastasis30 and therapy resistance31 through delivery of proteins, lipids, RNA and DNA to remote cells.32 The cargo carried by these vesicles may play unexpected roles if released directly in the extracellular space and allowed to interact with other components of the ECM.33 DNP-enhanced NMR is potential non-targeted method of better understanding the role played by these extracellular vesicles.
Hyl and its subsequent modifications are essential to the structure, properties, and functions, of collagen.34 The type I collagen heterotrimer contains 12 His residues. Both of those in each α1 (I) chain are two residues from those Lys residues which are sequence specifically δ-hydroxylated to Hyl, in a strongly conserved Lys-Gly-His-Arg tetrad.35 These His residues have been proposed to play a part in targeting lysyl hydroxylases and thereby the formation of the specific Lys and Hyl crosslinks36 essential to maintaining collagen and tissue structure. Some of the same His residues are not only hydroxylated to Hyl, but of these some are subsequently also enzymatically glycosylated to (Glc)GalHyl, an essential event in directing the as-yet poorly understood process of collagen fibril assembly. It has recently been proposed that these self same residues are also preferentially glycated in non-enzymatic, usually harmful, reactions with metabolic sugars.37 Finally there is evidence for a trivalent crosslink, histidinohydroxylysinonorleucine, between these same Lys/Hyl and His residues.38 Thus His and Lys/Hyl residues co-exist in crosslinking and glycosylation/glycation “hotspots” which may be key to better understanding the spatial relationships underlying orderly, and pathological, collagen assembly and structure. Detecting His, Lys and Hyl simultaneously, as enabled by isotopic enrichment and DNP-enhanced NMR, in native and model ECM, opens fertile new possibilities for studying these processes.
This communication demonstrates again3 the ability of ssNMR, combined with signal enhancement by DNP and targeted isotope labelling, to detect low abundance components in intact tissue that may have surprising biological roles, thus complementing other biophysical and molecular biology techniques in exploring diverse areas of matrix biology.
Conflicts of interest
There are no conflicts of interest. Animal procedures were unregulated in terms of the U. K. Animals (Scientific Procedures) Act 1986, but nevertheless were reviewed for compliance with institutional ethical guidelines by the Animal Welfare Ethical Review Board, University of Cambridge.
Supplementary Material
Acknowledgments
U. K. Medical Research Council (DGR, RR, RG75828), the Engineering and Physical Sciences Research Council PhD studentship (IG, AP), CSC Cambridge Scholarship for PhD studentship (RL), postdoctoral fellowship from the DAAD and Leibniz Association (WYC), Raymond and Beverly Sackler Fund for Physics of Medicine, University of Cambridge (AP), and support by iNEXT, grant number 653706, funded by the Horizon 2020 programme of the European Commission.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra03198g
Notes and references
- Wong V. W. C. Reid D. G. Chow W. Y. Rajan R. Green M. Brooks R. A. Duer M. J. J. Biomol. NMR. 2015;63:119. doi: 10.1007/s10858-015-9977-9. [DOI] [PubMed] [Google Scholar]
- Thankamony A. S. L. Wittmann J. J. Kaushik M. Corzilius B. Prog. Nucl. Magn. Reson. Spectrosc. 2017;102–3:120. doi: 10.1016/j.pnmrs.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Chow W. Y. Li R. Goldberga I. Reid D. G. Rajan R. Clark J. Oschkinat H. Duer M. J. Hayward R. Shanahan C. M. Chem. Commun. 2018;54:12570. doi: 10.1039/C8CC04960B. [DOI] [PubMed] [Google Scholar]
- Chow W. Y. Rajan R. Muller K. H. Reid D. G. Skepper J. N. Wong W. C. Brooks R. A. Green M. Bihan D. Farndale R. W. Slatter D. A. Shanahan C. M. Duer M. J. Science. 2014;344:742. doi: 10.1126/science.1248167. [DOI] [PubMed] [Google Scholar]
- Li R. Rajan R. Wong W. C. V. Reid D. G. Duer M. J. Somovilla V. J. Martinez-Saez N. Bernardes G. J. L. Hayward R. Shanahan C. M. Chem. Commun. 2017;53:13316. doi: 10.1039/C7CC06624D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. H. Hong M. J. Am. Chem. Soc. 2011;133:1534. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F. Schmidt-Rohr K. Hong M. J. Am. Chem. Soc. 2012;134:3703. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon P. A. Quinn D. M. Cordes E. H. Biochemistry. 1982;21:2745. doi: 10.1021/bi00540a026. [DOI] [PubMed] [Google Scholar]
- Kitayama T. Ogawa Y. Fujii K. Nakahara H. Mizuno N. Sekiguchi K. Kasai K. Yurino N. Yokoi T. Fukuma Y. Yamamoto K. Oki T. Asamura H. Fukushima H. Leg. Med. 2010;12:84. doi: 10.1016/j.legalmed.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Khanna C. Khan J. Nguyen P. Prehn J. Caylor J. Yeung C. Trepel J. Meltzer P. Helman L. Cancer Res. 2001;61:3750. [PubMed] [Google Scholar]
- Sergeyev I. V. Day L. A. Goldbourt A. McDermott A. E. J. Am. Chem. Soc. 2011;133:20208. doi: 10.1021/ja2043062. [DOI] [PubMed] [Google Scholar]
- Nelson D. L. and Cox H. M., Lehninger: Biochemistry, W. H. Freeman & Co., New York, 6th edn, 2013 [Google Scholar]
- Lesage A. Auger C. Caldarelli S. Emsley L. J. Am. Chem. Soc. 1997;119:7867. doi: 10.1021/ja971089k. [DOI] [Google Scholar]
- Takegoshi K. Nakamura S. Terao T. Chem. Phys. Lett. 2001;344:631. doi: 10.1016/S0009-2614(01)00791-6. [DOI] [Google Scholar]
- Jaroniec C. P. Filip C. Griffin R. G. J. Am. Chem. Soc. 2002;124:10728. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. Prog. Nucl. Magn. Reson. Spectrosc. 2011;58:62. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Borer P. N. LaPlante S. R. Zanatta N. Levy G. C. Nucleic Acids Res. 1988;16:2323. doi: 10.1093/nar/16.5.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlante S. R. Boudreau E. A. Zanatta N. Levy G. C. Borer P. N. Ashcroft J. Cowburn D. Biochemistry. 1988;27:7902. doi: 10.1021/bi00420a048. [DOI] [PubMed] [Google Scholar]
- Gaffney B. L. Goswami B. Jones R. A. J. Am. Chem. Soc. 1993;115:12607. doi: 10.1021/ja00079a059. [DOI] [Google Scholar]
- Xu X. P. Au-Yeung S. C. F. J. Phys. Chem. B. 2000;104:5641. doi: 10.1021/jp0007538. [DOI] [Google Scholar]
- Gao X. Jones R. A. J. Am. Chem. Soc. 1987;109:3169. doi: 10.1021/ja00244a064. [DOI] [Google Scholar]
- Yavropoulou M. P. Yovos J. G. J. Musculoskeletal Neuronal Interact. 2018;18:18. [PMC free article] [PubMed] [Google Scholar]
- Jilka R. L. Noble B. Weinstein R. S. Bone. 2013;54:264. doi: 10.1016/j.bone.2012.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreille O. M. Diegoli T. M. Irwin J. A. Coble M. D. Parsons T. J. Forensic Sci. Int.: Genet. 2007;1:191. doi: 10.1016/j.fsigen.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Coscas R. Bensussan M. Jacob M. P. Louedec L. Massy Z. Sadoine J. Daudon M. Chaussain C. Bazin D. Michel J. B. Atherosclerosis. 2017;259:60. doi: 10.1016/j.atherosclerosis.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Boskey A. L. Timchak D. M. Metab. Bone Dis. Relat. Res. 1983;5:81. doi: 10.1016/0221-8747(83)90006-1. [DOI] [PubMed] [Google Scholar]
- Haraszti R. A. Didiot M. C. Sapp E. Leszyk J. Shaffer S. A. Rockwell H. E. Gao F. Narain N. R. DiFiglia M. Kiebish M. A. Aronin N. Khvorova A. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub E. E. Biochim. Biophys. Acta. 2009;1790:1592. doi: 10.1016/j.bbagen.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. H. T. Camacho N. Aono H. Atherosclerosis. 1999;144:94. doi: 10.1016/S0021-9150(99)80363-3. [DOI] [Google Scholar]
- Williams C. Rodriguez-Barrueco R. Silva J. M. Zhang W. J. Hearn S. Elemento O. Paknejad N. Manova-Todorova K. Welte K. Bromberg J. Peinado H. Lyden D. Cell Res. 2014;24:766. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li I. Nabet B. Y. Mol. Cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerweij T. Perez-Lanzon M. Baglio S. R. J. Vis. Exp. 2018;135:e56932. doi: 10.3791/56932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L. Shen J. Tu C. Q. Hornicek F. Duan Z. F. Cancer Metastasis Rev. 2016;35:377. doi: 10.1007/s10555-016-9630-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J. Top. Curr. Chem. 2005;247:115. doi: 10.1007/b103821. [DOI] [Google Scholar]
- Rodriguez-Pascual F. Slatter D. A. Sci. Rep. 2016;6:37374. doi: 10.1038/srep37374. doi: 10.1038/srep37374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helseth D. L. Lechner J. H. Veis A. Biopolymers. 1979;18:3005. doi: 10.1002/bip.1979.360181208. [DOI] [Google Scholar]
- Hudson D. M. Archer M. King K. B. Eyre D. R. J. Biol. Chem. 2018;293:15620. doi: 10.1074/jbc.RA118.004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechanic G. L. Katz E. P. Henmi M. Noyes C. Yamauchi M. Biochemistry. 1987;26:3500. doi: 10.1021/bi00386a038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.