Abstract
Increased levels of oxidative stress and inflammation are the underlying mechanisms behind the aging process and age-related diseases. The purpose of our research is to explore whether epigallocatechin gallate (EGCG) can extend replicative life span by preventing the oxidative stress and inflammatory effects of WI-38 fibroblasts and the involved mechanisms in vitro. WI-38 cells were treated with different concentrations of EGCG (0, 25, 50 and 100 μM) at population doubling (PD) 25. At late-stage cells, we determined the age-associated genes with signaling through transcriptome sequencing. The expression profile of the targets in WI-38 fibroblasts was confirmed by bioinformatics analysis, qPCR and western blot. We found that EGCG markedly decreased reactive oxygen species (ROS), and inflammation factors, tumor necrosis factor-α (TNF-α), interleukin (IL)-6, and significantly increased cell proliferation at PD 35 and 45. EGCG treatments significantly decreased p53 and retinoblastoma (Rb) expressions, markedly increased p-Rb and E2F2 expressions as well as antioxidant enzymes and superoxide dismutase (SOD) 1 and SOD2 content, and obviously decreased the expressions of inflammation factors IL-32, TNF-α expressions at PD 45 WI-38 cells. Moreover, the effects were changed by EGCG treatment by p53 siRNA or overexpression. These findings in our studies reveal that EGCG treatments improved senescence and enhanced the replicative life span through alleviating oxidative stress and inflammation in WI-38 fibroblasts.
Increased levels of oxidative stress and inflammation are the underlying mechanisms behind the aging process and age-related diseases.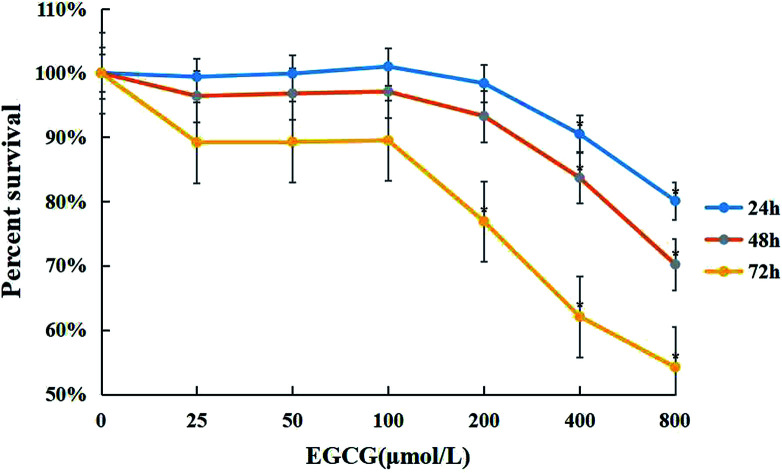
Introduction
Increasing evidence reports that elevated oxidative stress and inflammation are associated with aging and aging-related diseases, including obesity, diabetes, hypertension, dyslipidemia and cardiovascular disease.1,2 Oxidative stress is often accompanied by inflammation, and the release of inflammatory factors aggravates the degree of oxidative stress. The interactions between oxidative stress and inflammation can damage body cells and tissues, further affecting human lifespan.3,4
In the early 1960s, the concept that primary cells isolated from mammalian tissues can undergo only a finite number of divisions when grown in culture was established by Hayflick.5,6 Cells lose their replicative potential and they are termed and viewed as senescent cells during cells cultivation. Cellular senescence has been shown to reflect some features of aging in vivo, while replicative senescence has been confirmed to play an essential role in tumor inhibition.7 Therefore, senescence cells represent a valid model for studying mammalian cellular aging and understanding the mechanisms of cellular aging, which will provide insight into organismal aging. Oxidative stress and chronic low-grade inflammatory responses are associated with the increase of population doublings in WI-38 fibroblasts. The accumulation of oxidative stress and inflammation factors within cells is a common feature of aging.
Catechins are the predominant form of tea polyphenols and mainly consist of epigallocatechin gallate (EGCG), epigallocatechin (EGC), epicatechin gallate (ECG), and epicatechin (EC).8 Epidemiological studies suggest that the beneficial health effects of diets rich in green tea are provided by members of this family such as EGCG. The effects of EGCG include anti-inflammatory, anti-oxidative, anti-cancer, hypolipemic, anti-hyperglycemic and modulating metabolic activity of a cell.9 EGCG mediates its effects by altering various targets that encompass the cell membrane and intracellular proteins, cell signaling molecules and miRNA profile. In addition, our previous studies demonstrated that rats treated with EGCG exhibited a significantly longer lifetime that was accompanied by a reduction in oxidative stress and inflammation by activating the proteins involved in the regulation of longevity FOXO3a and SIRT1.10 Furthermore, studies in various model organisms indicate a lifespan extension by green tea and EGCG treatment in both invertebrates and mammalian species.11–13 However, the underlying mechanisms by which EGCG prolongs life span caused by alleviating inflammation and oxidative stress remain unclear. In the current study, we investigated the protective effects and mechanisms of EGCG in the development of oxidative stress and inflammation caused by increased population doublings in WI-38 fibroblasts.
Materials and methods
Materials
Dulbecco's Modified Eagle Medium (DMEM) was obtained from HyClone (Logan, Utah, USA); fetal bovine serum (FBS) was obtained from (PAA Laboratories, Pasching, Austria); EGCG and dimethyl sulfoxide (DMSO) for cell experiments were obtained from Sigma-Aldrich (St. Louis, MO, USA); 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) for cytotoxicity was obtained from MP Biomedicals (MP, CA, USA); β-actin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). p53 siRNA (h) was purchased from Cell Signaling Technology (Danvers, MA, USA).
Cell culture and treatment
WI-38 human diploid fibroblast cell line was obtained from the Chinese Academy of Science (Shanghai, China). The cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic at 37 °C in an atmosphere containing 95% air and 5% CO2.5 The WI-38 cells were treated with different concentrations of EGCG (25, 50 and 100 μM) or without EGCG (control) at PD 25.
Cell viability
The cell viability was assessed using MTT assay as previously described.14
Senescence-associated β-galactosidase staining
Staining for β-galactosidase (SA-β-gal) activity was performed as described by Dimri et al.15 Briefly, WI-38 cells were washed with PBS, fixed in 4% formaldehyde for 30 min at room temperature and washed again with PBS three times. Cells were then stained with SA-β-gal staining solution containing 1 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) overnight at 37 °C (in the absence of CO2). Four hundred cells were counted each time, and the number of positive cells was calculated as a percentage of the whole cells.16
Cell proliferation
Cell proliferation was estimated using the EdU incorporation assay. Briefly, cells were cultured in 6-well plates and exposed to 50 mM EdU (Ribobio, Guangzhou, China) for 4 h at 37 °C. The cells were then fixed in 4% formaldehyde for 30 min at room temperature and permeabilized in 0.5% Triton X-100 for 10 min. Cells were washed with PBS, and each well was incubated with 400 mL 1XApolloH reaction mixture for 30 min. The fluorescence was red and the excitation was from the a 550 nm laser beam.17
Oxidative stress and inflammatory factors in WI-38 cells
ROS was assayed using commercial kits (Beyotime, Shanghai, China) using the dichloro-dihydrofluorescein diacetate (DCFH-DA) method, or the data from 10 000 single cell events were collected using a flow cytometer (BD LSRFortessa) within 20 s for each measurement. TNF-α and IL-6 were tested with an enzyme-linked immunosorbent assay (ELISA) using commercial kits (Boster, Wuhan, China), according to the manufacturers' instructions.
Cell transfection
WI-38 cells were treated with different concentrations of EGCG (25, 50 and 100 μM) at PD 45. The p53 siRNA (10 μM, Beyotime Biotechnology, China) was mixed with a transfection reagent Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in a buffer as well as the negative control siRNA (NCsiRNA), followed by incubation for 20 min at room temperature. We replaced the normal culture after overnight transfection and did not add antibiotics to the media during transfection as this would have caused cell death. After 12 h the WI-38 cells were collected for measurement of the key proteins by western blot.
WI-38 cells were transfected with the pEXP-RB-Mam-EGFP-p53 recombinant plasmid (RiboBio, Guangzhou, China) using the Lipofectamine 2000 for 48 h. Then, the cells were determined of p53 and key proteins by western blot.
RNA extraction and transcriptome sequencing
Total RNAs were extracted from WI-38 fibroblasts using TRIzol (Invitrogen, Carlsbad, CA, USA) and treated with RNase-free DNase to remove any residual genomic DNA. RNA purity was checked using the kaiaoK5500® spectrophotometer (Kaiao, Beijing, China). RNA integrity and concentration were assessed using the RNA Nano 6000 Assay Kit from the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). A total of 2 μg RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (#E7530L, NEB, USA), following the manufacturer's recommendations and index codes were added to attribute sequences to each sample.
Quantitative PCR
Total RNAs was isolated from cells with the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA was reverse transcribed to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed with the SYBR Green PCR Master Mix and a 7500 FAST Real-time PCR System (Applied Biosystems). All primers were purchased by Sheng Gong.
Western blotting analysis
The p53 (Cell Signaling Technology), Rb (Santa Cruz Biotechnology), p-Rb (Santa Cruz Biotechnology), E2F2 (Abcam), SOD1 (Santa Cruz Biotechnology), SOD2 (Santa Cruz Biotechnology), IL-32 (Boster), TNF-α (Santa Cruz Biotechnology) proteins were determined by the western blot analysis as previously described.18
Statistical analysis
All data analysis was performed with the Statistical Package of Social Sciences (SPSS) 18.0 software. Results were presented as mean ± standard deviation. Statistical analyses were conducted by the one-way ANOVA, pairwise comparisons using the Dunnett's test, and P < 0.05 was considered to be statistically significant.
Results
Effects of EGCG on the viability of WI-38 fibroblasts
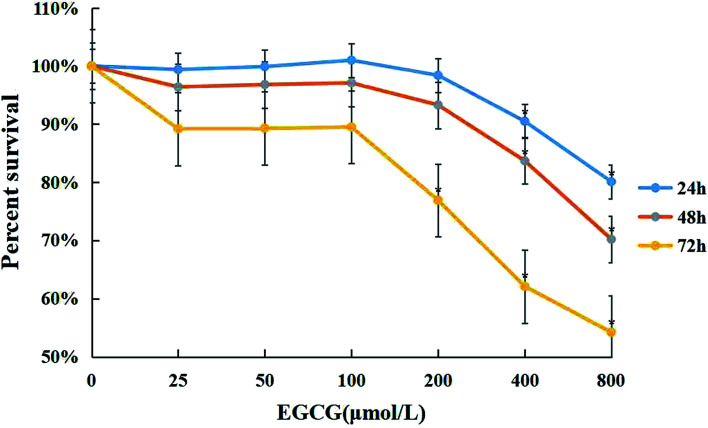
WI-38 cells were treated with 0–800 μM of EGCG for 24, 48 and 72 h to assess cell viability by the MTT assay (Fig. 1). Cytotoxicity was observed when the concentration of EGCG reached 400 μM for 24 h in the WI-38 cells. Therefore, the EGCG concentration was safe within 25–100 μM in the WI-38 cells in the present study.
Fig. 1. The cytotoxic effects of EGCG in WI-38 fibroblasts. The effects of EGCG (0, 25, 50, 100, 200, 400, and 800 μM) for 24, 48 and 72 h with the MTT assay. The experiments were repeated 3 times. Data are presented as means ± SD (n = 3). *P < 0.05 compared with the control group.
Aging is associated with ROS, inflammation factors and proliferative capacity in the WI-38 fibroblasts
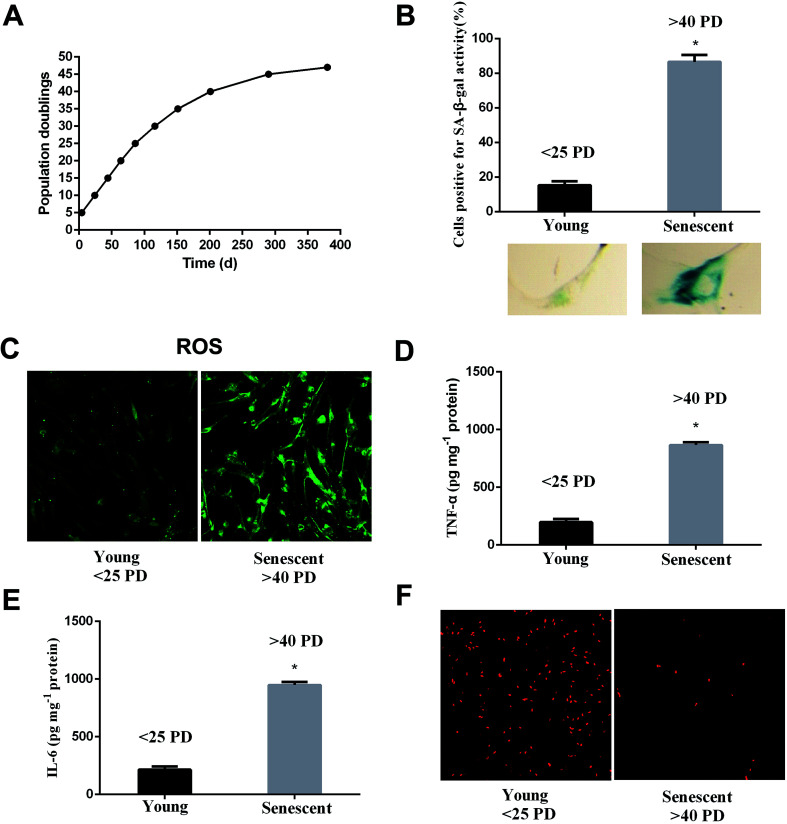
WI-38 fibroblasts were grown as described in the proliferation curve (Fig. 2A). The two stages of WI-38 cell life were investigated: young (<25 PD) and senescent (>40 PD). SA β-gal and EdU staining were used to assess the aging state at both early and late passages of the WI-38 fibroblasts. The number of cells that indicated positive staining for the senescence-associated β-galactosidase was higher in the senescence cells as compared to that of the young ones (Fig. 2B, P < 0.05). WI-38 cells exhibited morphological changes as they became larger and appeared in irregular shapes when entering replicative senescence. Moreover, the ROS and inflammation factor levels markedly increased (Fig. 2C–E, P < 0.05), and the percentage of cells that continued to incorporate EdU in aging cells was significantly lowered in comparison with that in young cells (Fig. 2F).
Fig. 2. Aging is associated with increased ROS, inflammation factors and decreased proliferation. (A) Proliferation curve of WI-38 fibroblasts. (B) Early-passage cells (<25 PD) were defined as young because only 10% were positive for SA-β-Gal staining. Late-passage cells (>40 PD) were defined as senescent cells because more than 90% of the cells were positive for SA-β-Gal staining. (C) The ROS levels at early and late passages. (D) The TNF-α levels at early and late passages. (E) The IL-6 levels at early and late passages. (F) Cell proliferation shows the characteristic EdU staining. Data are presented as means ± SD (n = 3). *P < 0.05 compared with young.
EGCG extended lifespan in WI-38 fibroblasts
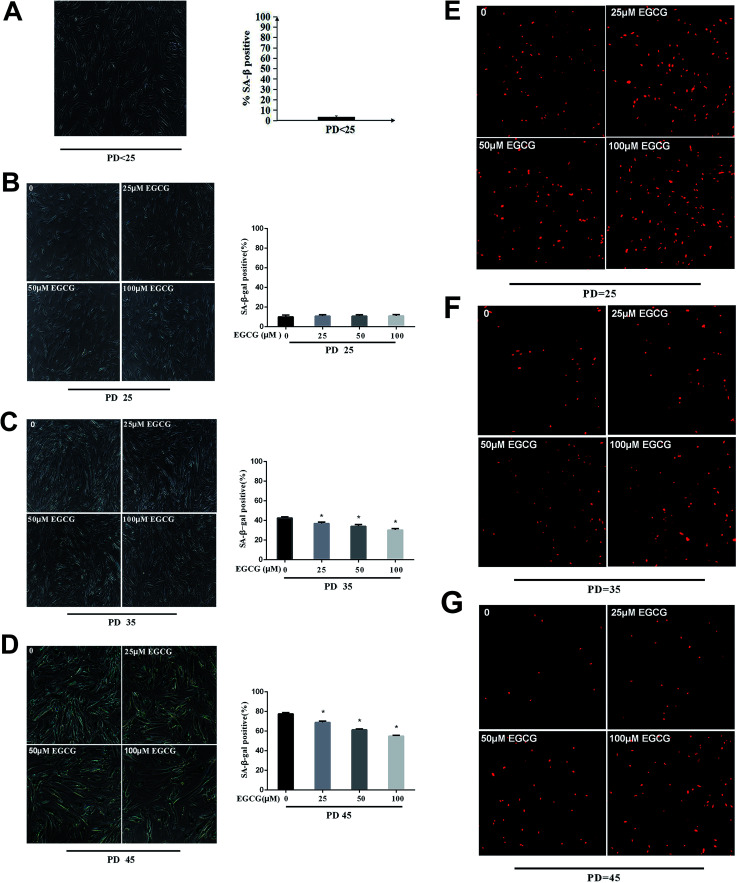
We examined the life span of cells grown in the presence or absence of EGCG at each stages of WI-38 fibroblasts. The number of cells positive for senescence-associated beta-galactosidase (SA β-gal) at the early stages did not change (Fig. 3A and B, P > 0.05), but markedly decreased by EGCG at PD 35 and 45 (Fig. 3C and D, P < 0.05). In addition, the percentage of cells that continued to incorporate EdU in the EGCG-treated cultures was increased relative to that in control cultures at PD 35, 45 of WI-38 fibroblasts (Fig. 3E–G). These findings are consistent with the increased cell proliferation at late passage in the EGCG-treated cells compared to that in the control cells.
Fig. 3. The senescent state was dynamically monitored in WI-38 fibroblasts. (A) The senescent state at PD < 25 of WI-38. (B) The senescent state at PD 25 of WI-38. (C) The senescent state at PD 35 of WI-38. (D) The senescent state at PD 45 of WI-38. The proliferation with EdU staining at PD 25 (E), 35 (F), 45 (G) of WI-38 fibroblasts. Data are presented as means ± SD (n = 3). *P < 0.05 compared with the control group.
EGCG-attenuated oxidative stress and inflammation in WI-38 fibroblasts
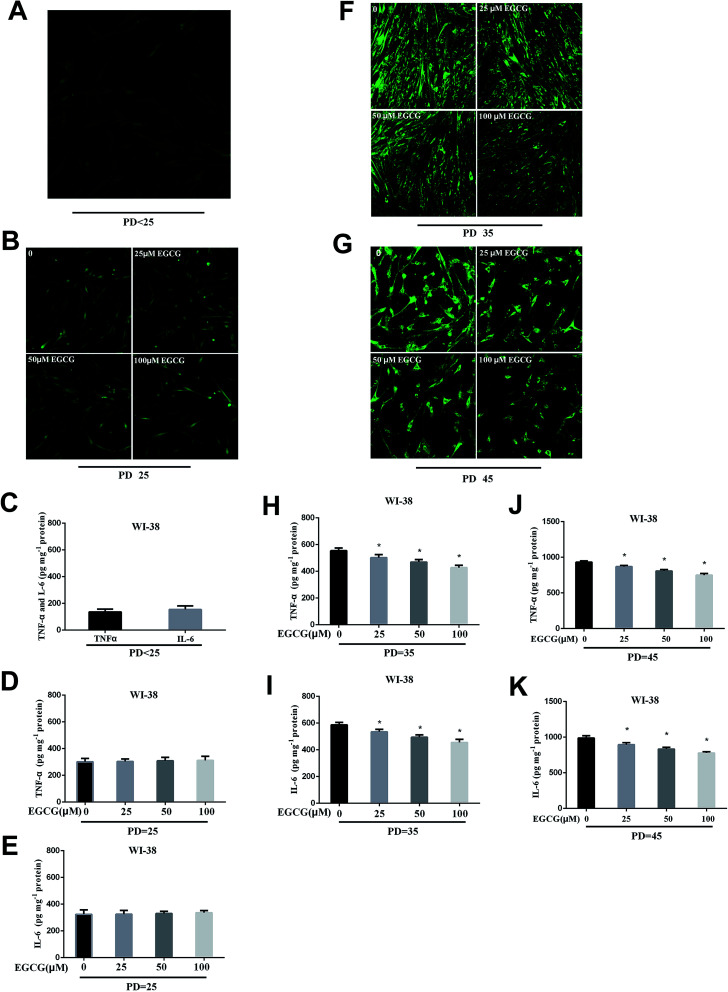
We dynamically monitored the ROS levels and inflammation factors in the presence or absence of EGCG at each stages of WI-38 fibroblasts. At the early stages of WI-38 fibroblasts, the ROS (Fig. 4A and B) and inflammation factor levels (Fig. 4C–E, P > 0.05) did not change. With the increase in cell population doublings, the ROS and inflammation factor levels clearly enhanced in the control group, and EGCG treatments markedly decreased the levels of ROS (Fig. 4F and G), TNF-α and IL-6 (Fig. 4H–K, P < 0.05) at PD 35 and 45 of WI-38 fibroblasts.
Fig. 4. The ROS and inflammation factors levels were dynamically measured in WI-38 fibroblasts. The ROS (A and B), TNF-α and IL-6 levels (C–E) at the early passage of WI-38 fibroblasts. The ROS levels (F) at PD 35 of WI-38 fibroblasts. The ROS levels (G) at PD 45 of WI-38 fibroblasts. The TNF-α (H) and IL-6 (I) levels at PD 35 of WI-38 fibroblasts. The TNF-α (J) and IL-6 (K) levels at PD 45 of WI-38 fibroblasts. Data are presented as means ± SD (n = 3). *P < 0.05 compared with the control group.
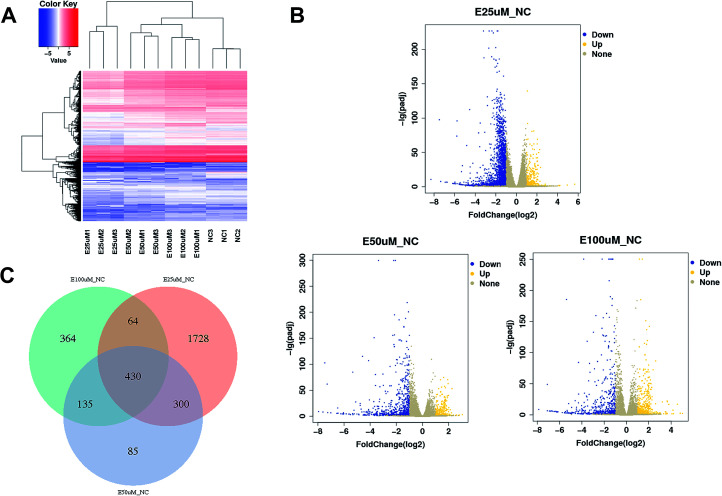
Identifying differentially expressed genes
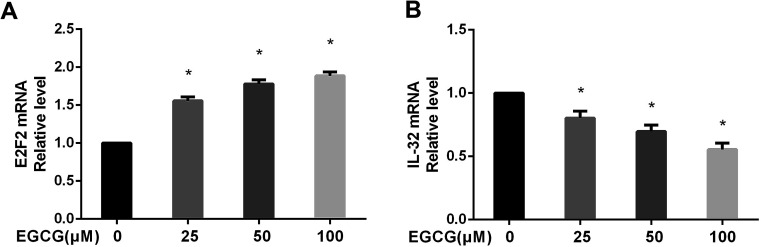
The gene expression profiles of WI-38 fibroblasts (PD 45) of the control with different EGCG concentrations were compared. Different expressions of mRNAs between the WI-38 cells treated without EGCG and treated with EGCG were identified through log 2 fold change, which was calculated as the ratio between two group means using data before column-wise normalization was applied with a threshold set for up- and down-regulated genes of |log 2 fold change| ≥ 1.5 with P value < 0.05. Finally, hierarchical clustering was performed to show distinguishable mRNA expression profiling among the samples (Fig. 5A–C). To validate our expression data obtained by RNA sequencing, the differential expression patterns of 21 unigenes were verified by qPCR. Among them, E2F2 transcription factor was found to be the most significantly upregulated, strongly correlated with the inflammation factor IL-32, and qPCR confirmed that EGCG dose-dependently up-regulated E2F2 expression (Fig. 6A) and downregulated IL-32 expression in WI-38 fibroblasts (Fig. 6B).
Fig. 5. mRNA expression profiling among samples. Heatmap (A), volcano plots (B) and Venn plot (C) reflect mRNA differential expression among samples.
Fig. 6. The E2F2 and IL-32 mRNA levels were verified in WI-38 fibroblasts. The E2F2 (A) and IL-32 (B) mRNA levels were determined by real time-PCR at PD 45 of WI-38 fibroblasts. Data are presented as means ± SD (n = 3). *P < 0.05 compared with the control group.
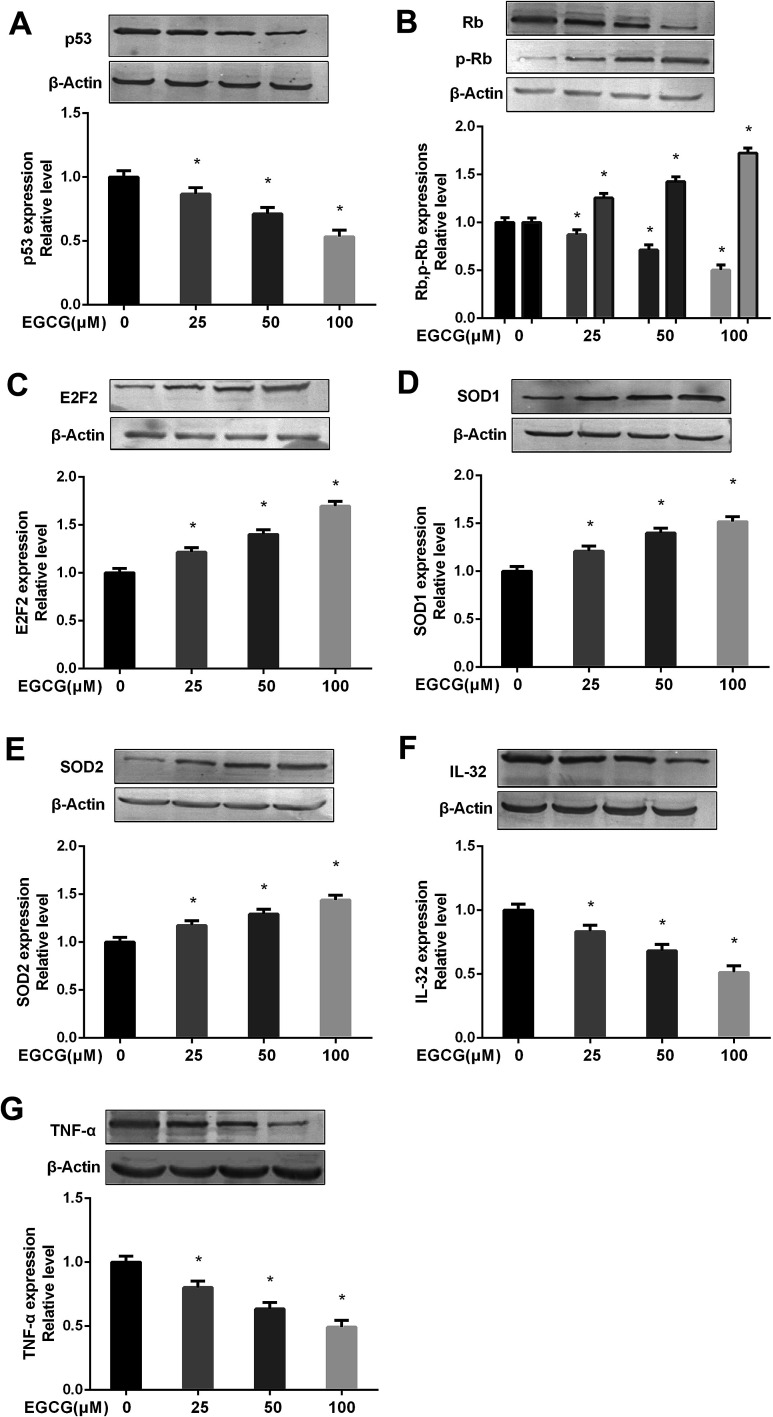
Effects of EGCG on the key proteins involved in cell proliferation, oxidative stress and inflammation in WI-38 cells
We measured the key protein expressions to determine the mechanism of EGCG on relieving the cell proliferation, oxidative stress and inflammation. The results showed that EGCG treatments significantly decreased p53, Rb expression, markedly increased p-Rb, E2F2 expressions (Fig. 7A–C, P < 0.05) and antioxidant enzymes, SOD1 and SOD2 (Fig. 7D and E, P < 0.05), and obviously decreased inflammation factors IL-32, TNF-α expressions (Fig. 7F and G, P < 0.05) at PD 45 WI-38 cells.
Fig. 7. Effects of EGCG on key proteins involved in cell proliferation, oxidative stress and inflammation at PD 45 of WI-38 cells. The p53 (A), (p)Rb (B), E2F2 (C), SOD1 (D), SOD2 (E), IL-32 (F) and TNF-α (G) protein levels were determined by western blot method at PD 45 of WI-38 fibroblasts. The experiments were repeated 3 times. Data are presented as means ± SD (n = 3). *P < 0.05 compared with the control group.
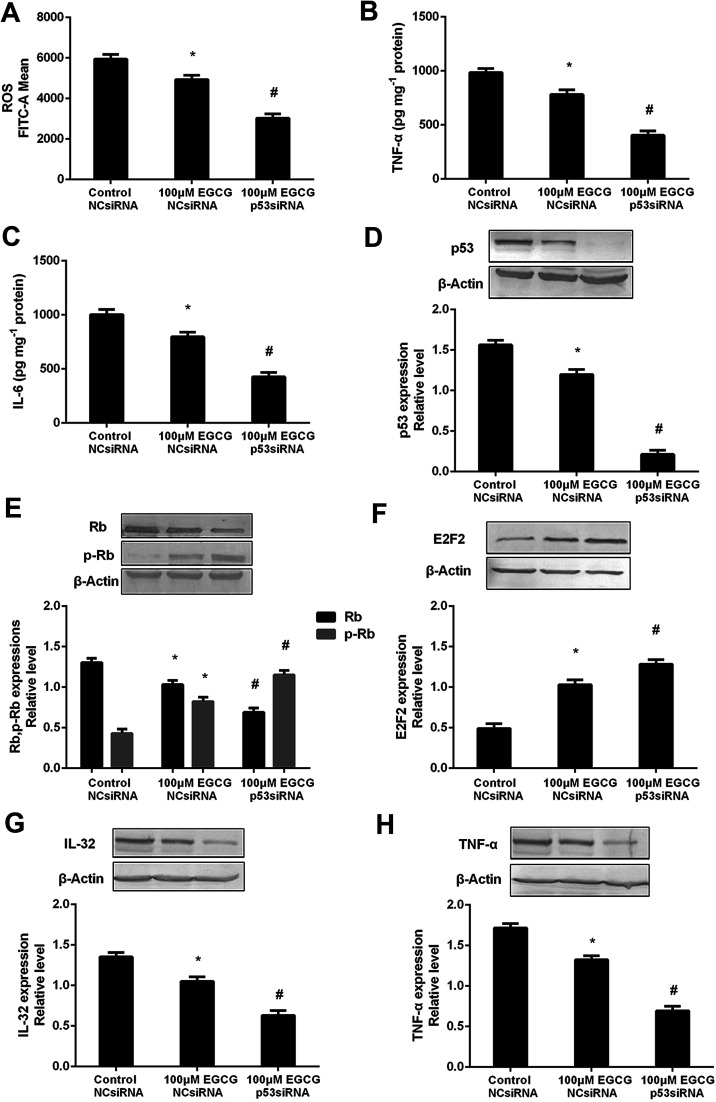
The p53 pathway contributed to EGCG-protection against aging in WI-38 cells
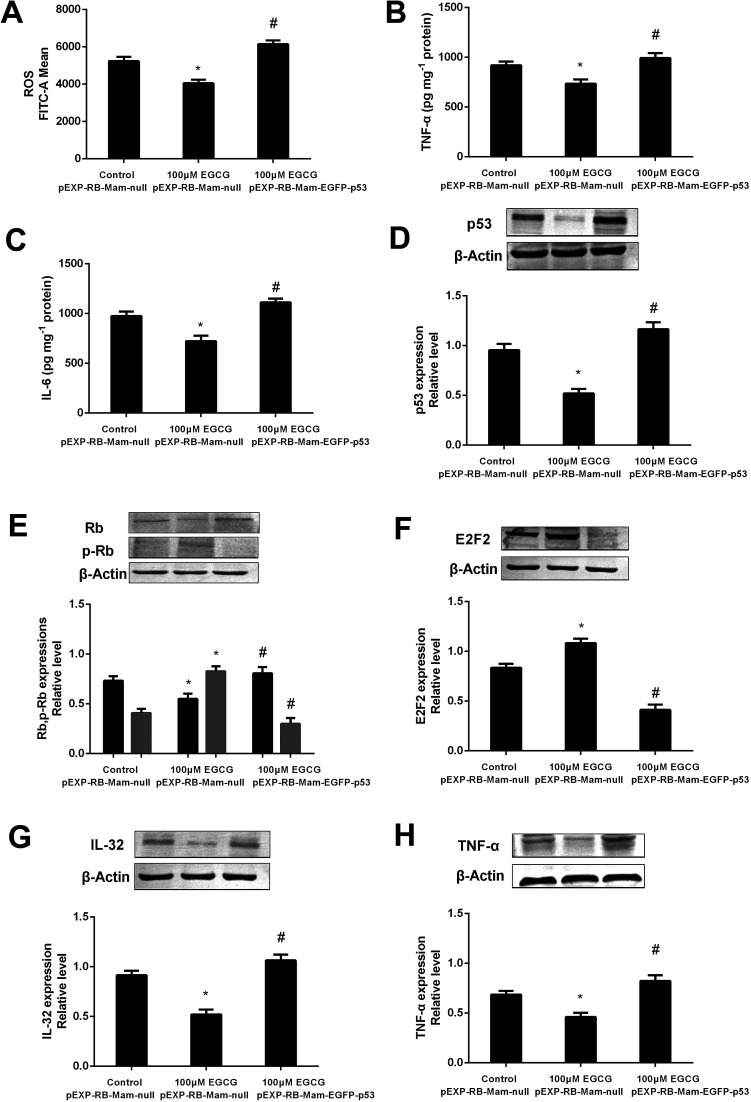
To prove that the function of p53 in mediating the EGCG improvement of aging, both the presence or absence of p53 siRNA and pEXP-RB-Mam-EGFP-p53 were executed at late stage of WI-38 cells. EGCG significantly lowered ROS, TNF-α and IL-6 levels (Fig. 8A–C, P < 0.05), the effects of which were markedly repressed by p53 siRNA. The effects of EGCG on p53, Rb, p-Rb, E2F2, IL-32 and TNF-α expressions (Fig. 8D–H, P < 0.05) were significantly changed in the presence of p53 siRNA in WI-38 cells. The ROS, TNF-α and IL-6 levels were significantly increased by EGCG after p53 overexpression (Fig. 9A–C, P < 0.05). The effects of EGCG on p53, Rb, p-Rb, E2F2, IL-32 and TNF-α expressions (Fig. 9D–H, P < 0.05) were significantly changed in the presence of pEXP-RB-Mam-EGFP-p53 in human embryonic fibroblasts. These results clearly demonstrated that EGCG protected against oxidative stress or inflammation factor-induced aging via the p53 pathway in WI-38 human embryonic fibroblasts.
Fig. 8. p53 mediated EGCG improvement of cell senescence in WI-38 cells. The ROS (A), TNF-α (B) and IL-6 (C) levels, and p53 (D), (p)Rb (E), E2F2 (F), IL-32 (G) and TNF-α (H) protein expressions after transfected with p53 siRNA at PD 45 of WI-38 cells. The experiments were repeated 3 times. Data are presented as means ± SD (n = 3). * Compared with control + NCsiRNA, P < 0.05; # compared with EGCG + NCsiRNA group, P < 0.05.
Fig. 9. The p53 pathway involved into EGCG-regulation of cell lifespan in WI-38 cells. The ROS (A), TNF-α (B) and IL-6 (C) levels, and p53 (D), (p)Rb (E), E2F2 (F), IL-32 (G) and TNF-α (H) protein expressions after transfected with pEXP-RB-Mam-EGFP-p53 at PD 45 of WI-38 cells. The experiments were repeated 3 times. Data are presented as means ± SD (n = 3). * Compared with pEXP-RB-Mam-null, P < 0.05; # compared with 100 μM EGCG pEXP-RB-Mam-null, P < 0.05.
Discussion
EGCG, which accounts for up to 50% of the catechins, has been found to possess versatile biological functions for the prevention or treatment of diabetes, cancer, heart diseases etc.19 Previous studies examining the cell-associated activities and effects of EGCG on gene transcription and regulation mainly focused on the 10–100 μM concentration range.20,21 However, due to poor bioavailability, the absorption and metabolism of EGCG in the intestine being critical for its bioactivities and health benefits, there are number of challenges in developing EGCG into therapeutic agent.
EGCG may improve the longevity effects of green tea in various species. In C. elegans, catechin modulated an energy-intensive stress response and repaired system that resulted in reduced body length and an enhanced lifespan.22 In Drosophila melanogaster, EGCG extended the life span via an induction of endogenous antioxidant enzymes.12 The data indicated that a reduction in glucose metabolism contributes to the better fitness and the extended lifespan in EGCG-treated Drosophila melanogaster.23 Furthermore, in mice receiving tea polyphenols from the 13th month of life onward, life span was significantly longer than control animals.13 Although senescence is a multicausal complex process, inflammation and oxidative stress are reputed to be significant factors affecting the aging progress and species longevity, which are associated with hyperlipidemia, diabetes and cardiovascular disease to injure body organisms and cells. However, little is known about its mechanism of EGCG on prolonging longevity by ameliorating inflammation and oxidative stress in vitro.
WI-38 human embryonic fibroblasts were grown as described in the proliferation curve. The two stages of cells life span were investigated: young (PD < 25) and senescent (PD > 40). In addition, WI-38 fibroblasts showed morphological changes as they became larger and exhibited irregular shapes when entering replicative senescence. Moreover, the ROS and inflammation factors levels were significantly augmented, and proliferative capacity was obviously decreased in senescent WI-38 fibroblasts. In this study, WI-38 fibroblasts were treated with 0, 25, 50 and 100 μM concentrations of EGCG simultaneously at population doubling (PD) 25. The difference in life span between groups was analyzed to determine whether EGCG could prolong the cell replicative life span. We dynamically monitored the senescence state, ROS, inflammatory factors and proliferative capacity in WI-38 fibroblasts throughout their replicative life span. As expected, our results suggest that the number of cells positive for senescence-associated beta-galactosidase (SA beta-gal) activity at high population doublings were reduced by EGCG. In addition, the percentage of cells that continued to incorporate EdU in the EGCG-treated cultures was increased in senescent WI-38 fibroblasts. These findings are consistent with decline in the SA beta-gal activity and increased proliferation at PD 35 and 45 in the EGCG-treated cultures relative to the control cultures.
In addition, it has been reported that rats with acetic acid-induced colitis showed an increase in the activity of SOD in the EGCG-treated group when compared with that in the placebo or control group.24 The results showed that EGCG prevented DON-induced cytotoxicity in a dose–response manner through anti-inflammatory and anti-oxidative in HT-29 cells.25 Furthermore, EGCG could be an effective approach to decrease inflammation and oxidative stress induced by UA.26 EGCG attenuated microcystin-LR induced oxidative stress and inflammation in human umbilical vein endothelial cells.27 These results collectively further suggest that EGCG can relieve the situation of oxidative stress and inflammation both in vivo and in vitro.
We postulated that the alleviation of endogenous oxidative and inflammation production through EGCG treatments might reduce the endogenous oxidative and inflammation responses, and that these might underlie the increased replicative life span observed in EGCG-treated cultures. In the current study, we measured the levels of oxidative stress and inflammation in the presence or absence of EGCG at each stages of the WI-38 fibroblasts. As we expected, EGCG treatments showed decreased levels of oxidative stress and inflammation factors relative to controls at PD 35 and 45. Therefore, we propose that EGCG exerts the effects of prolonging replicative life span by mitigating the oxidative stress and inflammation in WI-38 fibroblasts.
With senescent-passage cells, we determined the age-associated effects with signaling through gene sequencing. The study was conducted by using the methods of bioinformatics, including hierarchical clustering, gene enrichment, KEGG analysis and correlational analyses. The effects of EGCG on the expression profile of some interesting genes were confirmed by qPCR. Among the EGCG-regulated genes, we identified several modules and numerous significant genes that were associated with cell cycle, cell proliferation and inflammation-related functions, which then identified the transcription factor E2F2 as highly up-regulated and strongly correlated with the inflammation factor IL-32. Our results confirmed that E2F2 mRNA level increased and that the IL-32 mRNA level was lowered with EGCG treatments by qPCR at PD 45 of WI-38 cells. We found that E2F2 involved in cell cycle and cell proliferation was modulated by p53/Rb. p53 is a critical and complex player in the regulation of apoptosis, cell cycle, senescence and longevity.28 Rb is found at aging in its active, hypophosphorylated form, which binds to the E2F protein family members to repress their transcriptional targets.29 Researches have also linked the p53 and RB1 pathways with the E2F transcription factor and pRb-family proteins, clearly playing key roles in both cell proliferation and differentiation.30,31 The results from our study indicated that the effects of EGCG were associated with the inhibition of the p53/Rb signaling pathway, which resulted in increased p-Rb, E2F2 and antioxidant enzymes, SOD expressions in senescent WI-38 cells. The increasing evidence demonstrates that interleukin (IL)-32 is a proinflammatory cytokine, inducing IL-1α, IL-1β, IL-6, IL-8, and tumor necrosis factor (TNF)-α via nuclear factor (NF)-κB and p38MAPK signal transduction pathways.32 We have found that IL-32 and TNF-α are involved in inflammation, a conclusion consistent with the decline in the IL-32 and TNF-α protein expressions. In addition, the effects of ROS, inflammatory factors levels, and p53, Rb, p-Rb, E2F2, IL-32 and TNF-α expressions were obviously changed by EGCG after treated with p53 siRNA or pEXP-RB-Mam-EGFP-p53 in WI-38 fibroblasts.
Conclusion
Our studies reported that EGCG improved the cell replicative lifespan in a dose-dependent manner, which inhibited ROS, inflammation factors and enhanced the proliferation capacity of WI-38 cells, thereby preventing or delaying a potential cellular and tissue dysfunction through the attenuation of the oxidative stress, inflammation signaling activation and the modulation of cell proliferation.
Conflicts of interest
The authors declare that they have no conflict of interests.
Supplementary Material
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81372997, 81673152).
References
- Khansari N. Shakiba Y. Mahmoudi M. Recent Pat. Inflammation Allergy Drug Discovery. 2009;3:73–80. doi: 10.2174/187221309787158371. [DOI] [PubMed] [Google Scholar]
- Zhou J. Ouedraogo M. Qu F. Duez P. Phytother. Res. 2013;27:1745–1755. doi: 10.1002/ptr.4942. [DOI] [PubMed] [Google Scholar]
- Tsubota K. Nippon Ganka Gakkai Zasshi. 2007;111:193–205. [PubMed] [Google Scholar]
- Sampayo J. N. Gill M. S. Lithgow G. J. Biochem. Soc. Trans. 2003;31:1305–1307. doi: 10.1042/bst0311305. [DOI] [PubMed] [Google Scholar]
- Hayflick L. Moorhead P. S. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hayflick L. Exp. Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Campisi J. Science. 2005;309:886–887. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- Orso D. Floriano L. Ribeiro L. C. Bandeira N. M. G. Prestes O. D. Zanella R. Food Anal. Methods. 2016;9:1638–1653. doi: 10.1007/s12161-015-0339-8. [DOI] [Google Scholar]
- Legeay S. Rodier M. Fillon L. Faure S. Clere N. Nutrients. 2015;7:5443–5468. doi: 10.3390/nu7075230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y. Na L. Feng R. Gong L. Zhao Y. Li Q. Li Y. Sun C. Aging Cell. 2013;12:1041–1049. doi: 10.1111/acel.12133. [DOI] [PubMed] [Google Scholar]
- Bonillaramirez L. Jimenezdelrio M. Velezpardo C. Gene. 2013;512:355. doi: 10.1016/j.gene.2012.09.120. [DOI] [PubMed] [Google Scholar]
- Li Y. M. Chan H. Y. Huang Y. Chen Z. Y. Mol. Nutr. Food Res. 2007;51:546–554. doi: 10.1002/mnfr.200600238. [DOI] [PubMed] [Google Scholar]
- Kitani K. Osawa T. Yokozawa T. Biogerontology. 2007;8:567–573. doi: 10.1007/s10522-007-9100-z. [DOI] [PubMed] [Google Scholar]
- Gerlier D. Thomasset N. J. Immunol. Methods. 1986;94:57–63. doi: 10.1016/0022-1759(86)90215-2. [DOI] [PubMed] [Google Scholar]
- Dimri G. P. Lee X. Basile G. Acosta M. Scott G. Roskelley C. Medrano E. E. Linskens M. Rubelj I. Pereira-Smith O. et al. . Proc. Natl. Acad. Sci. U. S. A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed E. K. Rogowska-Wrzesinska A. Roepstorff P. Bulteau A. L. Friguet B. Aging Cell. 2010;9:252–272. doi: 10.1111/j.1474-9726.2010.00555.x. [DOI] [PubMed] [Google Scholar]
- Sun L. Chen L. Sun L. Pan J. Yu L. Han L. Yang Z. Luo Y. Ran Y. Mol. Cell. Proteomics. 2013;12:395–406. doi: 10.1074/mcp.M112.020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Yuan H. Zhang C. Guan Y. Wu Y. Ling F. Niu Y. Li Y. Diabetes Res. Clin. Pract. 2018;142:363–373. doi: 10.1016/j.diabres.2018.06.017. [DOI] [PubMed] [Google Scholar]
- Cheng J. Wu F. H. Wang P. Ke J. P. Wan X. C. Qiu M. H. Bao G. H. J. Agric. Food Chem. 2018;66:7948–7957. doi: 10.1021/acs.jafc.8b02266. [DOI] [PubMed] [Google Scholar]
- Khan N. Afaq F. Saleem M. Ahmad N. Mukhtar H. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- Intra J. Kuo S. M. Chem.-Biol. Interact. 2007;169:91–99. doi: 10.1016/j.cbi.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul N. Pietsch K. Menzel R. Sturzenbaum S. R. Steinberg C. E. Mech. Ageing Dev. 2009;130:477–486. doi: 10.1016/j.mad.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Wagner A. E. Piegholdt S. Rabe D. Baenas N. Schloesser A. Eggersdorfer M. Stocker A. Rimbach G. Oncotarget. 2015;6:30568–30578. doi: 10.18632/oncotarget.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z. H. Chen C. Xiao S. D. Biomed. Pharmacother. 2008;62:189–196. doi: 10.1016/j.biopha.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Kalaiselvi P. Rajashree K. Bharathi Priya L. Padma V. V. Food Chem. Toxicol. 2013;56:110–118. doi: 10.1016/j.fct.2013.01.042. [DOI] [PubMed] [Google Scholar]
- Xie H. Sun J. Chen Y. Zong M. Li S. Wang Y. Oxid. Med. Cell. Longevity. 2015;2015:214836. doi: 10.1155/2015/214836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. Deng H. Pan H. Xu Y. Zhang M. Chemosphere. 2017;168:25–31. doi: 10.1016/j.chemosphere.2016.10.037. [DOI] [PubMed] [Google Scholar]
- Feng Z. Lin M. Wu R. Genes Cancer. 2011;2:443–452. doi: 10.1177/1947601911410223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M. Nåñez S. Heard E. Narita M. Lin A. W. Hearn S. A. Spector D. L. Hannon G. J. Lowe S. W. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- Kailong L. Du X. Yani H. Lin Z. Jvrong Y. Ruihua S. Lin C. Biocell. 2007;31:213–223. [PubMed] [Google Scholar]
- Dyson N. Genes Dev. 1998;12(15):2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- Noldpetry C. A. Nold M. F. Zepp J. A. Kim S. H. Voelkel N. F. Dinarello C. A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3883–3888. doi: 10.1073/pnas.0813334106. [DOI] [PMC free article] [PubMed] [Google Scholar]