Summary
The cyclic pyrimidines cCMP and cUMP have been reported in multiple organisms and cell types. As opposed to the cyclic nucleotides cAMP and cGMP, which are second messenger molecules with well-established regulatory roles across all domains of life, the biological role of cyclic pyrimidines remained unclear. Here we report that cCMP and cUMP are second messengers functioning in bacterial immunity against viruses. We discovered a family of bacterial pyrimidine cyclase enzymes that specifically synthesize cCMP and cUMP following phage infection, and demonstrate that these molecules activate immune effectors that execute an antiviral response. A crystal structure of a uridylate cyclase enzyme from this family explains the molecular mechanism of selectivity for pyrimidines as cyclization substrates. Defense systems encoding pyrimidine cyclases, denoted here “Pycsar” for Pyrimidine Cyclase Systems for Antiphage Resistance, are widespread in prokaryotes. Our results assign clear biological function to cCMP and cUMP as immunity signaling molecules in bacteria.
Introduction
Cyclic nucleotide monophosphates are ubiquitous signaling molecules that function as second messengers in all domains of life. 3′,5′-cyclic adenosine monophosphate (cAMP) is the most common cyclic nucleotide with widespread roles in both prokaryotic and eukaryotic signaling. In bacteria, cAMP signaling controls gene expression and is essential for growth and adaptation to glucose starvation (Green et al., 2014; Pastan and Perlman, 1970). In eukaryotic cells, cAMP activates protein kinase signaling, allowing cells to respond to external stimuli. Human and animal cells additionally use 3′,5′-cyclic guanosine monophosphate (cGMP) as a second messenger, which regulates vasodilation through relaxation of smooth-muscle tissue and also functions within the vertebrate visual system (Lincoln, 1989; Stryer, 1986). Growing evidence suggests that cGMP serves as a second messenger also in bacteria (Linder, 2010; Rauch et al., 2008; Ryu et al., 2015).
cAMP and cGMP are generated by adenylate and guanylate cyclase enzymes that convert ATP and GTP into cyclized products. cAMP/cGMP cyclases comprise distinct enzyme classes, with class III adenylate/guanylate cyclases exhibiting the most widespread organismal distribution. Class III cyclase enzymes function as obligate dimers (Khannpnavar et al., 2020). The cyclase active site is composed of residues contributed from each enzyme protomer that together coordinate a divalent metal cation-dependent reaction and catalyze synthesis of cAMP and cGMP in response to upstream stimuli (Steegborn, 2014).
In contrast to the well-established functions of cAMP and cGMP, the existence and possible functional importance of cyclic pyrimidine nucleotides, 3′,5′-cyclic cytosine monophosphate (cCMP) and 3′,5′-cyclic uridine monophosphate (cUMP), remains controversial (Gaion and Krishna, 1979; Seifert, 2015). The presence of cCMP in mammalian tissues was reported more than three decades ago (Newton et al., 1984), and advancements in mass spectrometry methods have enabled detection of cCMP and cUMP in multiple animal cells (Bähre et al., 2014, 2015). Although cCMP/cUMP have been hypothesized to be involved in diverse processes including development and apoptotic cell death (Seifert, 2017), to date it is not known if cCMP/cUMP have specific biological roles, or whether these molecules are by-products of weak, non-specific activity of adenylate and guanylate cyclases.
In this study we report a novel family of bacterial pyrimidine cyclases that synthesize cCMP and cUMP as the primary products. By determining the crystal structure of a uridylate cyclase from this family, we identify a unique active-site configuration that explains selection of UTP as the primary substrate for cyclization. We further show that cCMP/cUMP cyclases function as part of a family of bacterial anti-phage defense systems, and that production of cyclic pyrimidine molecules is specifically triggered by phage infection. The cCMP and cUMP molecules then activate effector proteins that execute abortive infection through membrane impairment or depletion of cellular NAD+. Our results show that cytidylate and uridylate cyclases are widespread in bacteria and archaea, and establish a biological function of cyclic pyrimidines as second messengers mediating bacterial immunity.
Results
A defense system that produces cCMP
We analyzed ~40,000 microbial genomes for uncharacterized genes potentially involved in anti-phage defense and identified a family of genes with an annotated adenylate cyclase protein domain. Analysis of this gene family revealed 967 bacterial and archaeal genes with frequent genomic localization next to known defense systems such as CRISPR-Cas and restriction modification, a trait common to many phage-resistance systems (Figure S1A; Table S1; Methods) (Doron et al., 2018). As adenylate cyclase proteins have important housekeeping regulatory roles in all domains of life (Hanoune and Defer, 2001), we were intrigued by their possible roles in phage defense. The predicted cyclase genes are usually found in an operon together with an additional gene (Figure 1A), and we therefore cloned such two-gene operons, one from Escherichia coli 303145 and the other from E. coli E831, into the laboratory E. coli strain MG1655. Challenging the transformed bacteria with a panel of 12 phages showed that both operons provided substantial defense against multiple phages (Figure 1B; Figure S1B). Mutation in the predicted active site of the cyclase domain abolished defense, suggesting that the nucleotide cyclase activity is essential for the defensive capacity of the system (Figure 1C). Expression of the cyclase gene alone did not protect from infection, indicating that the second gene in the operon is also essential for defense against phage (Figure 1C).
Figure 1. The Pycsar defense system encodes a cytidylate cyclase.
(A) A two-gene system encoding a protein with a nucleotide cyclase domain and a protein with transmembrane (TM) helices from Escherichia coli E831. The gene IDs in the IMG database (Chen et al., 2019) are shown above the genes. (B) Pycsar systems defend against phages. Systems from two E. coli strains were cloned, together with the native promoter regions, and transformed into E. coli MG1655. Fold defense was measured using serial dilution plaque assays, comparing the efficiency of plating of phages on the Pycsar-containing strain to that of the control strain that lacks the system. Data represent an average of three replicates (see detailed data in Figure S1B). (C) Mutation in the active site of the predicted cyclase domain (D102A, predicted to abolish metal binding), or deletion of the TM-domain gene, abolishes the defensive activity of Pycsar from E. coli E831. Data represent plaque-forming units per ml (PFU/ml) of T5 phages infecting control cells, cells expressing the WT Pycsar from E. coli E831, and cells expressing mutated Pycsar. Shown is the average of three replicates, with individual data points overlaid. (D) Thin-layer chromatography analysis of nucleotide second messenger synthesis by the E. coli E831 cyclase (EcPycC). N, all four NTPs; Pi, inorganic phosphate. Data are representative of 3 independent experiments. (E) HPLC analysis of chemical standards compared to the product of EcPycC. (F) Chemical structure of 3′,5′ cyclic cytidine monophosphate (cCMP). (G) MSMS fragmentation spectra of a cCMP standard (bottom) and the cyclase product (top). (H) Concentrations of cCMP in lysates extracted from EcPycC-expressing cells or control GFP-expressing cells, that were infected by T5 phage, as measured by LC-MSMS with synthesized cCMP standard. X-axis represents minutes post infection, with zero representing non-infected cells. Cells were infected by phage T5 at an MOI of 2 at 37°C. EcPycC expression was induced by 0.2% arabinose. Bar graphs represent the average of two biological replicates, with individual data points overlaid.
We next expressed and purified the predicted adenylate cyclase from E. coli E831 and analyzed its enzymatic activity by thin-layer chromatography. Incubation of the purified protein with radiolabeled ATP resulted in no significant production of cAMP, counter to the hypothesized enzymatic activity that was predicted by homology-based protein annotation (Figure 1D). To test if the protein uses other nucleotides as substrates, we incubated it with radiolabeled GTP, CTP and UTP. Remarkably, the protein efficiently converted CTP into a new product that migrated as a single species (Figure 1D). Enzymatic activity was dependent on the presence of Mn2+ and occurred over a wide pH range from 7.0–9.5 (Figure S2A,B). Liquid chromatography followed by tandem mass spectrometry (LC-MSMS) analysis compared to a chemically synthesized standard showed that the product of the enzymatic activity is 3′,5′ cyclic cytidine monophosphate (cCMP) (Figure 1E-1G; Figure S2C). These results demonstrate that this E. coli defensive gene codes for a cytidylate cyclase, an enzyme which, to our knowledge, was previously hypothesized (Seifert, 2015) but never identified. We therefore named the defense system “Pycsar” for Pyrimidine Cyclase System for Antiphage Resistance, and denoted the cyclase domain-containing gene pycC.
To confirm that this enzyme synthesizes cCMP also in vivo, we used LC-MSMS to examine production of cCMP in E. coli MG1655 cells expressing the PycC cyclase protein from E. coli E831. We could not detect cCMP above the background level in lysates derived from exponentially growing cells (Figure 1H). However, in lysates derived from phage-infected cells we detected substantial levels of cCMP (Figure 1H). The cCMP molecule begun accumulating between 15 and 30 min after initial infection by phage T5 but did not accumulate in infected control cells lacking the cyclase, indicating that cCMP is indeed produced by the cyclase and suggesting that cCMP production in vivo is stimulated by phage infection (Figure 1H).
A family of pyrimidine cyclases that mediate phage resistance
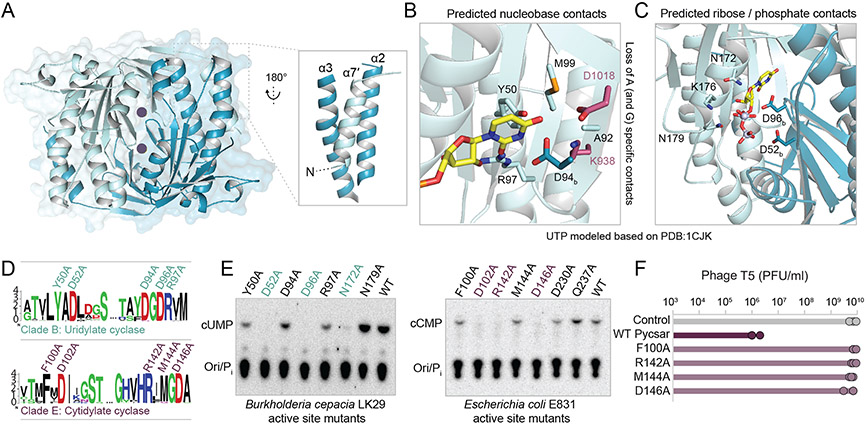
We next constructed a phylogenetic tree using multiple sequence alignment of the cyclase domain in the 967 bacterial and archaeal pycC genes. The tree topology showed that these proteins form five major clades, with the cCMP cyclases verified in this study found in clade E (Figure 2A). Closely related sequences from the same clade were frequently derived from bacteria and archaea of diverse phyla, suggesting horizontal gene transfer between phylogenetically distant organisms. To examine the substrate specificity of enzymes in this family of nucleotide cyclase enzymes, we selected two protein representatives from each clade, expressed and purified them, and incubated them with radiolabeled nucleotides. Surprisingly, proteins from clades A through D did not use CTP as substrate, but rather showed strong uridylate cyclase activities, converting UTP to 3′,5′ cyclic uridine monophosphate (cUMP) (Figure 2B-2E; Figure S3A,B). These are the first enzymes reported to generate cUMP as the primary product (Seifert, 2015). We tested a Pycsar system from Xanthomonas perforans, encoding a PycC cyclase from clade B, and found that it provided defense against phage T7 (Figure 2F).
Figure 2. Diversity of pyrimidine cyclases in prokaryotic genomes.
(A) Phylogenetic tree of proteins with pyrimidine cyclase domains. Only non-redundant sequences were used to build the tree (Table S1). The percentage of genes that are located near known defense genes is indicated for each clade. Bootstrap values are indicated for major clades. The outermost ring indicates the type of effector gene that is associated with the cyclase gene; Upper left, color code for the effector types. TM stands for transmembrane domain. Pie charts represent the phylum distribution of cyclase-containing prokaryotes in each of the clades on the tree. The common domain organization of the Pycsar system in each clade is presented to the right of the pie chart. Genes for which cyclase activity was demonstrated in vitro are marked by grey wedges, with U marking cUMP production and C marking cCMP. (B) Thin-layer chromatography analysis of cyclase proteins from two representatives of each clade. Pi, inorganic phosphate; Data are representative of 3 independent experiments. (C) HPLC analysis of chemical standards compared to the enzymatic product of the BcPycC cyclase. (D) Chemical structure of 3′,5′ cyclic uridine monophosphate (cUMP). (E) MSMS fragmentation spectra of a cUMP standard (bottom) and the BcPycC cyclase product (top). (F) A Pycsar system from Xanthomonas perforans GEV1001, when cloned into E. coli MG1655, defends against phage T7. Data represent plaque-forming units per ml (PFU/ml) of T7 phage infecting control cells and system-expressing cells. Shown is the average of three replicates, with individual data points overlaid.
Some of the tested proteins, and specifically the ones from clade D, also showed minor guanylate cyclase activities (Figure 2B; Figure S3C), in addition to producing cUMP as the major product. Adenylate cyclase activities were very low in all tested proteins, suggesting a strong selection against cAMP production that might interfere with cAMP-dependent cellular regulatory processes (Figure 2B, S2C).
The domain organization of the pyrimidine cyclases varied between clades. In clades B, C and D, the nucleotide cyclase domain was the only identifiable domain within the protein sequence, whereas clade E cyclases also contained an AGS-C domain, which was previously found to be fused to signal-producing proteins in CBASS antiphage systems and was hypothesized to serve as a phage recognition domain (Burroughs et al., 2015; Millman et al., 2020a). Structural prediction of the AGS-C domain suggests that it adopts an immunoglobulin-like fold (Figure S3D), supporting the hypothesis that it might be used in phage detection. A deletion of the AGS-C domain of EcPycC rendered the system inactive against phages (Figure S3E). PycC proteins from clade A contained two consecutive nucleotide cyclase domains, as opposed to single cyclase domains in other clades (Figure 2A).
Uridylate cyclase structure reveals the molecular basis for pyrimidine cyclization
To understand the molecular principles defining the substrate specificity of pyrimidine-specific cyclase enzymes, we determined a 1.5 Å crystal structure of a clade B uridylate cyclase from Burkholderia cepacia LK29 (BcPycC) (Figure 3A; Table S1). The BcPycC crystal structure reveals a two-fold symmetric homodimeric enzyme with three main α-helices wrapped around a central anti-parallel β-sheet. BcPycC shares clear structural homology with class III adenylate and guanylate cyclase proteins conserved in prokaryotic and eukaryotic signaling systems (Steegborn, 2014). Unlike most class III enzymes, BcPycC has an additional alpha helix (α7′) at the N-terminus which makes contacts to α-helices 2 and 3 of the opposing protomer (Figure 3A).
Figure 3. Structural and functional analysis of pyrimidine cyclases.
(A) Crystal structure of a uridylate cyclase from Burkholderia cepacia LK29. Individual protomers of the homodimer are depicted in light and dark blue. Active site locations are denoted by purple circles. (B) Zoom-in cutaway of the nucleotide binding pocket in the cUMP cyclase structure. UTP modeled into the apo protein active site highlights potential amino acid interactions (subscript b denotes opposing monomer). D1018 and K938 are residues essential for adenylate selectivity in mammalian adenylate cyclases, and are modeled here from superposition of BcPycC with a mammalian soluble adenylate cyclase (PDB 1CJK). (C) Zoom-in cutaway of predicted phosphate and ribose interactions in the cUMP cyclase structure. (D) Conserved motifs in cUMP and cCMP cyclase of clade B and clade E, respectively. The residues mutated in B. cepacia LK29 and E. coli E831 PycC cyclases are indicated above. (E) Thin-layer chromatography analysis of pyrimidine cyclase mutants of Burkholderia cepacia LK29 (left) and Escherichia coli E831 (right). The mutations in the cyclase proteins are indicated above. Marked in colors are mutations that completely abolished cUMP/cCMP production. (F) Plating efficiency of phage T5 on control cells, E. coli E831 Pycsar-expressing cells, and strains mutated in the cyclase protein. Data represent plaque-forming units per ml (PFU/ml), average of three replicates with individual data points overlaid.
Analysis of the BcPycC active site reveals that each protomer in the homodimeric enzyme contains catalytic aspartate residues, D52 and D96, required for divalent metal ion coordination, and a conserved asparagine at position N172 predicted to contact the ribose ester oxygen (Figure 3B-3D; Figure S4A). Substitutions at either of these sites abolished cUMP synthesis (Figure 3E), supporting that BcPycC uses the same highly conserved catalytic mechanism shared by all class III cyclase enzymes (Linder, 2005; Tesmer et al., 1999). Using the structure of a mammalian class III adenylate cyclase in complex with an ATP analog as a guide (Tesmer et al., 1999), we modeled a UTP nucleotide into the BcPycC active site to identify residues involved in pyrimidine selection (Figure 3B,C). Class III adenylate and guanylate cyclases control nucleobase selectivity through base-specific contacts with a pair of highly conserved lysine and aspartate residues (adenylate cyclases) or cysteine and glutamate residues (guanylate cyclases) (Linder, 2005; Steegborn, 2014). Notably, the BcPycC nucleotide binding pocket lacks analogous side chains positioned for purine nucleotide selection. Instead, in BcPycC residues Y50, D94, and R97 protrude into the nucleotide binding pocket and sterically occlude the space required to coordinate ATP or GTP (Figure S4B). BcPycC residues Y50 and R97 are conserved in cUMP synthases from clade B. Likewise, cCMP synthases (clade E) including EcPycC are predicted to contain F and R residues at similar structural positions, suggesting a role for these amino acids in PycC pyrimidine base selection (Figure S4C,D). Using alanine scanning mutagenesis, we next tested substitutions to each BcPycC and EcPycC active site residue. BcPycC and EcPycC nucleobase pocket mutations reduced activity, but most of them did not completely abolish cCMP/cUMP synthesis in vitro (Figure 3E). In vivo experiments, however, showed that nucleobase pocket mutations abolished defense in bacteria (Figure 3F). Together, these results explain how Pycsar cyclases have evolved for pyrimidine specificity.
The structure of a Pseudomonas aeruginosa cyclase from clade A of the PycC phylogenetic tree was previously determined, and it was observed that the protein lacks adenylate or guanylate cyclase activities (PDB 6YII) (Linder et al., 2020). We purified this P. aeruginosa protein (renamed here PaPycC) and demonstrated that PaPycC is an active uridylate cyclase (Figure 2B). Structural comparison of the clade A PaPycC and the clade B BcPycC shows that the α7′ helix we identified at the N-terminus of BcPycC is also present in the PaPycC structure, suggesting that this helix may be a hallmark for PycC pyrimidine cyclases. Whereas BcPycC comprised a homodimer, PaPycC is encoded as a single chain with the two consecutive nucleotide cyclase domains folding into a pseudo-heterodimeric structure (Linder et al., 2020). In PaPycC, the C-terminal cyclase domain contributes the metal binding residues and the N-terminal cyclase domain presumably provides the binding pocket for nucleobase selective residues (Linder et al., 2020). Such tandem-domain configurations on a single protein chain are common in mammalian class III adenylate cyclase enzymes, although in these enzymes the architecture of the two domains is reversed as compared to PaPycC (Steegborn, 2014). A previous biochemical analysis of PaPycC showed that it can bind ATP as determined by isothermal titration calorimetry, and the PaPycC structure was solved together in complex with ATP (Linder et al., 2020). However, the configuration of ATP bound within PaPycC was shown to be incompatible with ATP hydrolysis or cAMP formation (Figure S4E) (Linder et al., 2020). Analysis of the predicted UTP binding pocket within PaPycC shows that many residues are conserved with BcPycC including the BcPycC Y50 and R97 residues predicted to sterically occlude purine bases (Figure S4E-G). These results further support that the active site architecture of PycC enzymes selectively prevents ATP/GTP usage in order to specify pyrimidine nucleotide selection.
cCMP and cUMP activate effector genes that mediate abortive infection
Our results suggest that phage infection stimulates Pycsar cyclases to produce cCMP and cUMP. To understand the roles of these molecules in mediating the bacterial response to infection, we next analyzed the operons in which these cyclases are embedded. These operons typically contain at least one additional gene, which is essential for defense from phage (Figures 1; Figure 2). In some cases, the additional gene contains an N-terminal domain annotated as “cyclic nucleotide-binding domain” (pfam accession PF00027), which is common in proteins that bind, and are regulated by, cyclic nucleotides such as cAMP and cGMP (Gomelsky and Galperin, 2013) (Figure S5A, Table S1). This suggests that the second protein binds the cyclic nucleotide produced by the cyclase in response to phage infection and serves as the effector of the Pycsar system. In the majority of cases, the effector gene encodes an additional domain, either a domain with one to four transmembrane helices (57%) or a Toll/interleukin-1 receptor (TIR) domain (30%), and we denote these effector genes pycTM and pycTIR, respectively (Figure 2A). Transmembrane-helix domains and TIR domains are frequently found in bacterial defense systems such as CBASS (Cohen et al., 2019; Millman et al., 2020a; Morehouse et al., 2020) and retrons (Millman et al., 2020b), where, once activated, they are responsible for inflicting cell death or growth arrest before the phage is able to replicate, a process termed abortive infection (Lopatina et al., 2020).
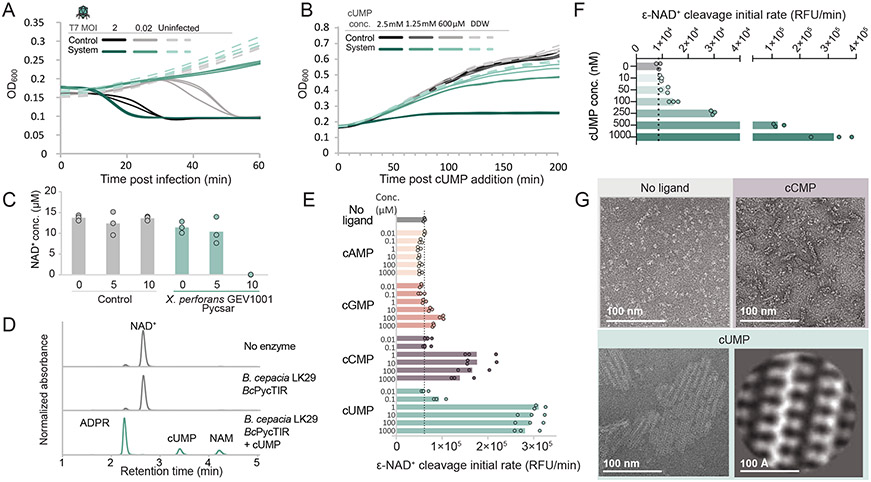
To test whether the systems that encode pyrimidine cyclases defend by triggering abortive infection, we performed infection experiments in liquid culture with cells encoding the Pycsar system from E. coli E831, which comprises an effector protein with three predicted transmembrane helices and a cytidylate cyclase (Figure 1). At multiplicity of infection (MOI) of 0.02, in which ~2% of bacteria are infected by the initial phage inoculum, the culture of the wild-type cells collapsed due to phage propagation, whereas the culture of the cells containing the Pycsar system survived (Figure 4A). Conversely, at an MOI of 2, in which almost all bacteria are infected by the initial phage inoculum, the culture containing the Pycsar operon entered a state of growth stasis (Figure 4A). These results are consistent with a phenotype of abortive infection, in which growth arrest of infected bacteria eliminates infected cells from the bacterial population and protects the culture from the spread of the phage (Lopatina et al., 2020).
Figure 4. cCMP mediates abortive infection in Pycsar.
(A) Growth curves of cells expressing the Pycsar system from E. coli E831 (purple) and control cells (black) with and without infection by phage T5 at multiplicity of infection (MOI) of 2 or 0.02. Results of three experiments are presented as individual curves. (B) Growth curves of Pycsar-expressing cells (purple) and control cells (black) with and without the addition of different concentrations of cCMP to the medium, or double distilled water (DDW) control. Results of three experiments are presented as individual curves. (C) Fluorescence microscopy images of E. coli MG1655 cells expressing the Pycsar system from E. coli E831 (lower panel), or control cells lacking the system (upper panel). Shown are DNA (blue) and membrane (red) stains. Images were captured at 60 min after the addition of 250 μM cCMP to the medium. Arrows point to abnormal membrane protrusions. Representative images from a single replicate out of three independent replicates are shown.
To directly determine if the effector protein is activated by cCMP to impose a toxic effect on the cell, we incubated cells expressing the E. coli E831 Pycsar in media supplemented with chemically synthesized cCMP. Although cCMP is a polar molecule that is not expected to efficiently diffuse through cell membranes, we reasoned that at high enough concentrations in the growth medium some cCMP molecules will be able to reach the interior of the cell and activate the effector. Indeed, Pycsar-expressing cells showed severe growth arrest in the presence of 250 μM cCMP (Figure 4B). cCMP did not cause growth arrest in control cells that did not express the defense system, suggesting that the toxicity is inflicted by the system following exposure to cCMP (Figure 4B). Addition of chemically synthesized cUMP to the growth medium did not cause toxicity, suggesting that the E. coli E831 PycTM effector is specific to the cCMP molecule produced by its cognate PycC cyclase (Figure S5B). Growth arrest was associated with the appearance of abnormal membrane protrusions in some of the cells, suggesting that the transmembrane-spanning effector exerts its toxicity by impairing membrane integrity, as suggested for transmembrane-spanning effectors of the CBASS (Millman et al., 2020a) and retron abortive infection defense systems (Millman et al., 2020b) (Figure 4C).
The second-most abundant effector gene in Pycsar systems, which encodes a TIR domain (pycTIR), is associated with uridylate cyclases (Figure 2). The cyclase-TIR Pycsar system from X. perforans also demonstrated an abortive infection phenotype in liquid culture infection experiments (Figure 5A), and the addition of high concentration cUMP to the medium resulted in growth arrest while the addition of cCMP to the cells did not (Figures 5B, S5C). TIR-domain effectors in microbial defense systems are known to serve as NADases that deplete the cell of NAD+ once they receive a signal indicative of phage infection, leading to abortive infection (Morehouse et al., 2020). Indeed, NAD+ was completely depleted from cells expressing the X. perforans Pycsar during infection by phage T7 (Figure 5C). We purified the TIR-domain effector protein from Burkholderia cepacia LK29 (BcPycTIR) which is found in the same defense operon as the crystallized BcPycC uridylate cyclase (Figure 2, Figure 5). Incubation of BcPycTIR with cUMP triggered robust NADase activity as observed by high-performance liquid chromatography (Figure 5D). BcPycTIR activation generated the expected NAD+ degradation products, ADP-ribose and nicotinamide (Figure 5D). We next tested the cNMP-specificity of this TIR effector protein using a fluorescent NAD+ analog. BcPycTIR exhibited strong selectivity for cyclic pyrimidines with ~50–100 nM cUMP sufficient for robust NADase activation (Figure 5E,F). Conversely, cAMP, even when added at millimolar concentrations, did not activate BcPycTIR (Figure 5E).
Figure 5. cUMP activates Pycsar TIR effectors that execute abortive infection.
(A) Growth curves of cells expressing the cyclase-TIR Pycsar system from Xanthomonas perforans GEV1001 (green) and control cells (black) with and without infection by phage T7 at multiplicity of infection (MOI) of 2 or 0.02. Results of three experiments are presented as individual curves. (B) Growth curves of XpPycsar-expressing cells (green) and control cells (black) with and without the addition of cUMP to the medium. Results of three experiments are presented as individual curves. (C) Concentrations of NAD+ in lysates extracted from XpPycsar expressing cells and control cells, infected by phage T7, as measured by LC-MSMS with synthesized NAD+ standard. X-axis represents minutes post infection, with zero representing non-infected cells. Cells were infected by phage T7 at an MOI of 2 at 37°C. Bar graphs represent the average of three biological replicates, with individual data points overlaid. (D) HPLC analysis of effector-mediated NAD+ cleavage by BcPycTIR. The TIR-domain effector cleaves NAD+ into the products ADP-ribose (ADPR) and nicotinamide (NAM), and the activity is strictly dependent on the presence of the cUMP molecule. cUMP was added at a concentration of 250 nM and incubated for 1 hr with the TIR effector from B. cepacia LK29 at 500 nM in the presence of 500 μM NAD+. Data are representative of 3 independent experiments. (E) Analysis of NAD+ cleavage activity of BcPycTIR using the fluorescent substrate ε-NAD+ at varying concentration of cNMPs. Data are average of three biological replicates with individual data points overlaid. (F) NAD+ cleavage activity of BcPycTIR in response to cUMP at finer resolution of concentrations span as compared to panel E. (G) Electron microscopy analysis of BcPycTIR filament formation in the presence of 50 μM cCMP, cUMP, or without a ligand. The 2D class average of the effector when activated by cUMP is presented at the bottom right. Scale bar for 2D class average image is 100 Å.
Activation of TIR NADase domains is known to occur through protein oligomerization (Jiang et al., 2020; Morehouse et al., 2020; Sporny et al., 2019). To determine the mechanism of cNMP-mediated effector activation we next used negative stain electron microscopy to image BcPycTIR in the presence of cUMP. Remarkably, BcPycTIR recognition of cUMP resulted in the formation of large 100–300 nm two-dimensional sheets (Figure 5G). 2D class averages revealed that the BcPycTIR sheets are constructed from interlocking individual filaments resembling a zipper-like configuration. cCMP, a weak activator of BcPycTIR NADase activity, induced formation of short flexible filaments (Figure 5G). These results explain how cyclic pyrimidine signaling drives BcPycTIR NADase activation and further support that protein oligomerization is a common mechanism controlling effector function in anti-phage defense (Lowey et al., 2020; Morehouse et al., 2020).
Phages can evolve to escape Pycsar-mediated defense
To determine whether phages can evolve to evade Pycsar systems, we attempted to isolate phage mutants that could overcome the defense conferred by the E. coli E831 cytidylate cyclase and its cognate effector. We were able to isolate 17 such mutants of phage T5 that partially escaped defense, and sequenced the full genome of each of these mutants. In almost all of these escapee phages (16 of 17) we identified a missense point mutation within the gene D20-21, encoding the pb8 protein, the major capsid protein precursor of T5 (Figure 6A,B; Table S2). Pb8 is the main component of the T5 head, forming an icosahedral capsid shell that contains 775 pb8 protomers (Huet et al., 2019). In all cases the point mutations resulted in an amino acid substitution, and in most cases the mutation in pb8 was the only mutation differing between the mutant and WT phage, indicating that single amino acid alterations in the T5 major capsid protein enable the phage to escape defense (Table S2).
Figure 6. Phage escape from Pycsar-mediated defense.
(A) Representative phage mutants capable of escaping defense by the Pycsar system of E. coli E831. Shown are 10-fold serial dilution plaque assays, comparing the plating efficiency of WT and mutant phages on E. coli MG1655 cells that contain the system and a control strain that lacks the system and contains an empty vector instead. Mutations of all escape mutant phages are detailed in Table S2. (B) Positions of mutations within the T5 Pb8 protein. (C) Concentrations of cCMP in cell lysates extracted from cells infected by WT or mutant T5 phages, as measured by LC-MSMS with synthesized cCMP standard. X-axis represents minutes post infection, with zero representing non-infected cells. Cells were infected at an MOI of 2 at 37°C. Bar graphs represent the average of three biological replicates, with individual data points overlaid. EcPycC expression was induced by 0.02% arabinose. (D) A competition fitness assay between WT and mutant T5 phage on both EcPycsar-expressing and control cells. Y-axis represents the fraction of each strain out of the total phage mixture based on sequenced DNA reads. X-axis represents the infection passage, with 0 representing the mixture prior to first passage. In each passage, phage lysate was taken and used for infection of a fresh bacterial culture.
We next measured production of cCMP in cells expressing the EcPycC cyclase during infection by mutant phages. Mutant phages elicited substantially less cCMP production at 40 min following phage infection as compared to WT T5 phage (Figure 6C). These results suggest that the cytidylate cyclase is less efficiently activated by the mutant phages, leading to reduced cCMP production, hence allowing phage escape from defense.
Overexpression of pb8 in cells expressing the Pycsar system did not elicit effector-mediated toxicity, suggesting that activation of the cyclase may necessitate another factor in addition to the pb8 capsid precursor (Figure S6A). We were also unable to demonstrate binding between pb8 and the cytidylate cyclase, and co-incubation of purified EcPycC with pb8 did not increase its cyclase activity rate in vitro (Figure S6B-D). These results imply that the interaction between the two proteins is either indirect or, again, necessitates another factor in addition to pb8.
To examine whether the escape mutations in pb8 confer a fitness cost to the phage we performed competition assays between WT and mutant T5 strains. For this, we infected a liquid culture of bacteria with a mixture of WT and mutant T5 phages and measured the ratio between WT and mutant phages following several infection cycles (Figure 6D). As expected, the mutant phage rapidly took over the phage mixture when infecting cells expressing Pycsar, because the system protects from the WT phage but not from the mutant (Figure 6D). However, when infecting cells not expressing the Pycsar system, the mutant phage was almost eliminated from the phage mixture after a single passage (Figure 6D). The results replicated for other phage mutants (Figure S6E,F). These results suggest that mutations conferring resistance to Pycsar-mediated defense also result in a severe fitness cost to the phage and are thus expected to be selected against in natural settings in which the majority of bacterial hosts do not encode the defense system.
Discussion
After decades of debate on whether cCMP and cUMP have distinct biological roles (Seifert, 2015, 2017; Seifert et al., 2011), our results show that these molecules are essential second messengers within Pycsar defense systems, establishing a function for cyclic pyrimidines in bacterial immunity. Our data show high specificity for cyclic pyrimidines in Pycsar systems. The PycC cyclases have strong preference to either produce cCMP or cUMP with almost no adenylate cyclase activity; and while PycTIR effectors are robustly activated by 50–100 nM of cUMP, they are not responsive to cAMP even at millimolar concentrations. It is likely that this strong substrate selectivity has evolved to prevent interference between Pycsar defense signaling and housekeeping regulation by cAMP.
Enzymes of the PycC family reported here were recently predicted to be involved in phage defense based on their close proximity to known defense genes, and in particular genes typically associated with CBASS systems (cap genes) (Millman et al., 2020a), leading to the prediction that they would produce immune signaling molecules. However, based on sequence annotation these enzymes were incorrectly predicted to synthesize cAMP or cGMP derivatives. We show that the cCMP and cUMP produced by PycC proteins in response to phage infection act as signaling molecules that mediate cell death. This discovery joins the recent identification of immune mechanisms such as type III CRISPR-Cas, CBASS, and Thoeris, all of which rely on the production of a unique signaling compound that activates downstream effectors to prevent phage propagation within an infected cell (Cohen et al., 2019; Kazlauskiene et al., 2017; Ofir et al., 2021). These mechanistically diverse systems, utilizing a variety of signaling molecules and effectors, represent a conserved immunological principle in which the production of a second messenger mediates the immune response. The enzymatic production of the signaling molecule rapidly amplifies the signal upon phage recognition, and presumably accelerates the execution of the immune response (Lopatina et al., 2020).
cCMP and cUMP have been detected in human cells, and were previously implicated in early embryonic development (cCMP) (Chan, 1987) and promotion of apoptotic cell death (cUMP) (Beckert et al., 2014). Recent studies have demonstrated conserved immune functions for signaling molecules between bacteria and eukaryotes: cyclic GMP–AMP (cGAMP) is produced both by the cGAS-STING antiviral pathway in animals as well as by CBASS antiviral systems in bacteria (Bernheim and Sorek, 2020; Cohen et al., 2019; Morehouse et al., 2020; Whiteley et al., 2019), and TIR immune proteins generate similar signaling molecules in both bacteria and plants (Ofir et al., 2021). It is therefore intriguing to speculate that cCMP and cUMP may function as immune signaling molecules in eukaryotes as well. We envision that our identification of the unique composition of the active site in PycC pyrimidine cyclases, which determines the strong specificity to CTP or UTP as substrates, may allow future discovery of pyrimidine cyclase enzymes in other organisms including eukaryotes, and would ultimately extend our understanding on the biological roles of cCMP and cUMP.
Limitations of the study
An important open question is to understand how the Pycsar system senses phage replication. We found that in vivo, the Pycsar system produces cyclic nucleotides only in response to phage infection. However, purified PycC enzymes produce the cyclic signaling molecules even without phage-derived stimulus. One explanation for these findings is that recognition of phage replication in vivo results in PycC clustering and activation of cyclic nucleotide synthesis due to a sharp increase in local enzyme concentration. PycC activity in vitro in the absence of phage recognition could therefore be explained by the elevated concentration of highly purified protein. Alternatively, similar to sensors that function as guards in plant innate immunity pathways and detect pathogens indirectly (Dodds and Rathjen, 2010), PycC enzymes may be inhibited by a cellular co-factor in vivo that is depleted during phage replication. Finally, discovery of the Pycsar system provides a new opportunity to explain how cyclase enzymes discriminate between purine and pyrimidine bases. Our analysis of the apo structure of BcPycC reveals an active-site configuration associated with pyrimidine selection. Determining the structure of PycC in complex with pyrimidine nucleotide substrates and defining the mechanism of cCMP/cUMP recognition by downstream Pycsar receptors will be important goals for future studies.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contacts, Rotem Sorek (rotem.sorek@weizmann.ac.il) and Philip J. Kranzusch (philip_kranzusch@dfci.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Crystal structure of BcPycC was deposited at the protein data base under identifier PDB: 7R65, and is publicly available as of the date of publication.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains and phages
E. coli strain MG1655 (ATCC 47076) was grown in MMB (LB + 0.1 mM MnCl2 + 5 mM MgCl2, with or without 0.5% agar) at 37°C or room temperature (RT). Whenever applicable, media were supplemented with ampicillin (100 μg ml−1), to ensure the maintenance of plasmids. Infection was performed in MMB media at 37°C or room temperature as detailed in each section. E. coli BL21-DE3 was grown in MDG (0.5% glucose, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 0.25% aspartic acid, 100 mg ml–1 ampicillin, 34 mg ml–1 chloramphenicol, and trace metals). Overnight MDG cultures were used to inoculate cultures of M9ZB media (0.5% glycerol, 1% Cas-amino Acids, 47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, 85.6 mM NaCl, 2 mM MgSO4, 100 mg ml–1 ampicillin, 34 mg ml–1 chloramphenicol, and trace metals, as described previously (Zhou et al., 2018)), which were grown and harvested for each experiment. Phages used in this study are listed in Table S3 and in the Key resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli K-12 MG1655 | American Type Culture Collection (ATCC) | ATCC 47076 |
| E. coli BL21-DE3 RIL | Agilent | Cat#230245 |
| Phage Lambda-vir | U. Qimron | N/A |
| Phage SECPhi17 | Doron et al., 2018 | N/A |
| Phage SECphi18 | Doron et al., 2018 | N/A |
| Phage SECphi27 | Doron et al., 2018 | N/A |
| Phage SECphi6 | Millman et al, 2020 | N/A |
| Phage SECphi4 | Millman et al, 2020 | N/A |
| Phage P1 | U. Qimron | N/A |
| Phage T2 | German Collection of Microorganisms and Cell Cultures GmbH (DSMZ) | DSM 16352 |
| Phage T4 | U. Qimron | N/A |
| Phage T5 | U. Qimron | N/A |
| Phage T6 | German Collection of Microorganisms and Cell Cultures GmbH (DSMZ) | DSM 4622 |
| Phage T7 | U. Qimron | N/A |
| Phage T5 – pb8 mutant n.1 | This paper | N/A |
| Phage T5 – pb8 mutant n.4 | This paper | N/A |
| Phage T5 – pb8 mutant n.9 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| [ɑ-32P] ATP | Perkin Elmer | Cat#BLU003H250UC |
| [ɑ-32P] GTP | Perkin Elmer | Cat#BLU006H250UC |
| [ɑ-32P] UTP | Perkin Elmer | Cat#BLU007H250UC |
| [ɑ-32P] CTP | Perkin Elmer | Cat#BLU008H250UC |
| NTPs (ATP, GTP, UTP, CTP) | New England Biolabs | Cat#N0450S |
| 3′,5′-cCMP | Biolog | Cat#C 001 |
| 3′,5′-cUMP | Biolog | Cat#U 001 |
| 3′,5′-cAMP | Biolog | Cat#A001S |
| 3′,5′-cGMP | Biolog | Cat#G001 |
| Nicotinamide adenine dinucleotide (NAD+) | Sigma-Aldrich | Cat#N0632 |
| ε-NAD+ Nicotinamide 1,N6-ethenoadenine dinucleotide (ε-NAD+) | Sigma-Aldrich | Cat#N2630 |
| Adenosine 5′-diphosphoribose (ADPR) | Sigma-Aldrich | Cat#A0752 |
| Nicotinamide (NAM) | Sigma-Aldrich | Cat#N3376 |
| membrane stain FM1-43 | Thermo Fisher Scientific | Cat#T35356 |
| DNA stain DAPI | Sigma-Aldrich | Cat#D9542 |
| Ni-NTA Agarose | Qiagen | Cat#30250 |
| HiLoad 16/600 Superdex 75 PG | GE Healthcare | Cat#28989333 |
| HiLoad 16/600 Superdex 200 PG | GE Healthcare | Cat#28989336 |
| Zorbax Bonus-RP HPLC Column | Agilent | Cat#863668-901 |
| Silica Gel-60 F254 glass-backed TLC plate | MilliporeSigma | Cat#M1057290001 |
| Alkaline Phosphatase, Calf Intestinal (CIP) | New England Biolabs | Cat#M0290S |
| SYPRO Orange Fluorescent Dye | ThermoFisher | Cat#S6650 |
| HEPES | VWR | Cat#97061-824 |
| Tris base | VWR | Cat#97062-416 |
| PEG 3350 | Sigma-Aldrich | Cat#202444 |
| Ammonium acetate | ThermoFisher | Cat#A639 |
| Imidazole | VWR | Cat#97065-016 |
| Tris[−2carboxyethyl] phosphine hydrochloride (TCEP) | GoldBio | Cat#TCEP50 |
| Dithiothreitol (DTT) | GoldBio | Cat#DTT25 |
| EcPycC recombinant protein, and mutants as described | This paper | N/A |
| BcPycC recombinant protein, and mutants as described | This paper | N/A |
| PaPycC recombinant protein | This paper | N/A |
| RsPycC recombinant protein | This paper | N/A |
| RsmPycC recombinant protein | This paper | N/A |
| TpPycC recombinant protein | This paper | N/A |
| GmPycC recombinant protein | This paper | N/A |
| PtPycC recombinant protein | This paper | N/A |
| MePycC recombinant protein | This paper | N/A |
| SaPycC recombinant protein | This paper | N/A |
| Ec303145PycC recombinant protein | This paper | N/A |
| BcPycTIR recombinant protein | This paper | N/A |
| Critical Commercial Assays | ||
| DNeasy blood and tissue kit | Qiagen | Cat #69504 |
| Nextera DNA Library Prep Kit | Illumina | Cat#15027865, Cat#15027866 |
| RNase A (within QIAprep Spin Miniprep Kit) | Qiagen | Cat# 27106 |
| DNase-I | Merck | Cat#11284932001 |
| Deposited Data | ||
| Crystal structure of BcPycC cUMP synthase | This paper | PDB: 7R65 |
| Oligonucleotides | ||
| Primers, see Table S4 | This paper | N/A |
| Recombinant DNA | ||
| pBbS8k-RFP | Lee et al., 2011 | Addgene Cat#35276 |
| pSG1 | Doron et al., 2018 | N/A |
| pBAD-GFP | Thermo Fisher Scientific | Cat #43001 |
| E. coli E831 Pycsar | Genscript Corp. | N/A |
| E. coli E831 Pycsar cyclase mutant F100A | Genscript Corp. | N/A |
| E. coli E831 Pycsar cyclase mutant R142A | Genscript Corp. | N/A |
| E. coli E831 Pycsar cyclase mutant M144A | Genscript Corp. | N/A |
| E. coli E831 Pycsar cyclase mutant D146A | Genscript Corp. | N/A |
| E. coli E831 Pycsar delta AGS-C | Genscript Corp. | N/A |
| E. coli 303145 Pycsar | Genscript Corp. | N/A |
| X. perforans GEV1001 Pycsar | Genscript Corp. | N/A |
| pBbS8k-pb8 | This paper | N/A |
| pET16-6 × His-SUMO2 | Zhou et al., 2018 | N/A |
| pET16-6 × His-MBP-SUMO2 | This paper | N/A |
| Software and Algorithms | ||
| The Integrated Microbial Genomes (IMG) | Chen et al., 2019 | https://img.jgi.doe.gov/m/ |
| MMseqs2 | Steinegger and Söding, 2017 | https://github.com/soedinglab/mmseqs2 |
| RNAfold WebServer | Lorenz et al., 2011 | http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi |
| BLASTclust from the BLAST NCBI package | Altschul et al., 1990 | https://ftp.ncbi.nlm.nih.gov/blast/executables/legacy.NOTSUPPORTED/2.2.26/ |
| WAR web server | Torarinsson and Lindgreen, 2008 | http://genome.ku.dk/resources/war |
| HHpred | Soding et al., 2005 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
| FastTree | Price et al., 2009 | http://microbesonline.org/fasttree |
| iTOL | Letunic and Bork, 2016 | https://itol.embl.de/ |
| Breseq (version 0.29.0) | Deatherage and Barrick, 2014 | http://barricklab.org/breseq |
| Prism v9.0d | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| Pymol v2.4.0 | Schrödinger, LLC | https://pymol.org/2/ |
| Coot v0.8.9.3-pre | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix v1.18.2-3874 | Liebschner et al., 2019 | https://www.phenix-online.org/ |
| Image J | NIH | https://imagej-nih-gov.ezp-prod1.hul.harvard.edu/ij/index.html |
| ImageQuant TL v8.2.0 | GE Healthcare | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/molecular-imaging-for-proteins/imaging-software/imagequant-tl-8-2-image-analysis-software-p-09518 |
| Jalview v2.11.1.4 | Waterhouse et al., 2009 | http://www.jalview.org |
| MAFFT v7 | Katoh et al., 2019 | https://mafft.cbrc.jp/alignment/server/ |
METHOD DETAILS
Detection of cNMP cyclase systems in defense islands
Protein sequences of all genes in 38,167 bacterial and archaeal genomes were downloaded from the Integrated Microbial Genomes (IMG) database (Chen et al., 2019) in October 2017 and clustered into protein families as described in (Millman et al., 2020a). Defense scores were calculated as previously described (Doron et al., 2018), recording the fraction of genes in each cluster that have known defense genes in their genomic environment spanning 10 genes upstream and downstream the inspected gene. For each cluster an average number of known defense genes in the genomic environment of all members was also calculated. Clusters where the most common pfam annotation was PF00211, with average protein size >200 aa, defense score >0.35 and average number of known defense genes in the environment >0.8 were included in the family of cyclase genes studied here.
Plasmid and strain construction
E. coli E831, E. coli 303145 and X. perforans GEV1001 Pycsar systems used in this study were synthesized by Genscript Corp. and cloned into the pSG1 plasmid (Doron et al., 2018) with their native promoters, or into the pBAD plasmid (Thermofisher, cat. #43001) (Table S4) as previously described (Bernheim et al., 2021; Doron et al., 2018). Mutants of the cyclase gene of E. coli E831 were also synthesized and cloned by Genscript. All synthesized sequences are presented in Table S4. A pBAD plasmid containing the cCMP cyclase of E. coli E831 was cloned using primers NT148 and NT149 listed in Table S4.
Plaque assays
Phages were propagated by picking a single phage plaque into a liquid culture of E. coli MG1655 grown at 37°C to OD600 of 0.3 in MMB medium until culture collapse. The culture was then centrifuged for 10 min at 3200×g and the supernatant was filtered through a 0.2 μm filter to remove remaining bacteria and bacterial debris. Lysate titer was determined using the small drop plaque assay method as described in (Mazzocco et al., 2009).
Plaque assays were performed as previously as described in (Mazzocco et al., 2009). Bacteria (E. coli MG1655 with the pSG1 plasmid containing the Pycsar system from E. coli E831 and E. coli 303145 or the pBAD plasmid with the system from X. perforans GEV1001) and negative control (E. coli MG1655 with an empty pSG1 vector or pBAD-GFP) were grown overnight at 37°C. Then 300 μl of the bacterial culture was mixed with 30 ml melted MMB agar (LB + 0.1 mM MnCl2 + 5 mM MgCl2 + 0.5% agar, with or without 0.2% arabinose, as described in Table S4) and left to dry for 1 h at room temperature. 10-fold serial dilutions in MMB were performed for each of the tested phages and 10 μl drops were put on the bacterial layer. Plates were incubated overnight at room temperature. Plaque forming units (PFUs) were determined by counting the derived plaques after overnight incubation.
Infection in liquid culture
Overnight cultures of bacteria harboring the Pycsar system (E. coli MG1655 with a pSG1 plasmid containing the system from E. coli E831 or a pBAD plasmid containing the system from X. perforans GEV1001) or negative control (E. coli MG1655 with a an empty pSG1 plasmid or a pBAD-GFP plasmid) were diluted 1:100 in MMB medium. For pBAD containing cultures, 0.2% arabinose was added. Cells were incubated at 37°C while shaking at 200 rpm until early log phase (OD600 0.3). 180 μl of the bacterial culture were transferred into wells in a 96-well plate containing 20 μl of phage lysate for a final MOI of 2 or 0.02, or phage buffer (50 mM Tris-HCl pH 7.4, 100 mM MgCl2, 10 mM NaCl) for uninfected control. Infections were performed in triplicates from overnight cultures prepared from three separate colonies. Plates were incubated at 37°C with shaking in a TECAN Infinite200 plate reader and OD600 was followed with measurement every 10 min.
cCMP/cUMP supplementation assays
Overnight cultures of bacteria harboring the system (E. coli MG1655 with a pBAD plasmid containing the system from E. coli E831 or a pBAD plasmid containing the system from X. perforans GEV1001) or negative control (E. coli MG1655 with a pBAD-GFP plasmid) were diluted 1:100 in MMB medium with 0.2% arabinose. Cells were incubated at 37°C while shaking at 200 rpm until early log phase (OD600 0.3). 180 μl of the bacterial culture were transferred into wells in a 96-well plate containing 20 μl of different concentrations of 3′,5′-cCMP, 3′,5′-cUMP or ultrapure water as a control. Plates were incubated at 37°C with shaking in a TECAN Infinite200 plate reader and OD600 was followed with measurement every 10 min.
Cell lysate preparation
Overnight cultures of E. coli harboring the Pycsar systems and negative controls were diluted 1:100 in 250 ml MMB medium (with or without arabinose, as described in Table S4) and grown at 37°C (250 rpm) until reaching OD600 of 0.3. The cultures containing the cyclase from E. coli E831 were infected by T5, and cells containing the Pycsar system from X. perforans GEV1001 were infected by T7 at a final MOI of 2. Following the addition of phage, at 15, 30, and 40 min post infection for T5- (WT or mutant) infected cells, or at 5 and 10 min post infection for T7-infected cells (plus an uninfected control sample). 50 ml samples were taken and centrifuged for 5 min at 3200×g. The culture pellets were flash frozen using dry ice and ethanol. The pellets were re-suspended in 600 μl of 100 mM phosphate buffer at pH 8 and supplemented with 4 mg ml−1 lysozyme. The samples were then transferred to a FastPrep Lysing Matrix B 2 ml tube (MP Biomedicals cat. #116911100) and lysed using a FastPrep bead beater for 40 s at 6 m s−1 (two cycles). Tubes were then centrifuged at 4°C for 15 min at 15,000×g. Supernatant was transferred to Amicon Ultra-0.5 Centrifugal Filter Unit 3 kDa (Merck Millipore cat. #UFC500396) and centrifuged for 45 min at 4°C at 12,000×g. Filtrate was taken and used for LC-MSMS analysis.
Quantification of nucleotides by HPLC-MSMS
Cell lysates were prepared as described above and analyzed by LC-MSMS. Quantification of nucleotides was carried out using an Acquity I-class UPLC system coupled to Xevo TQ-S triple quadrupole mass spectrometer (both Waters, US). The UPLC was performed using an Atlantis Premier BEH C18 AX column with the dimension of 2.1 × 100 mm and particle size of 1.7 μm (Waters). Mobile phase A was 20 mM ammonium formate at pH 3 and acetonitrile was mobile phase B. The flow rate was kept at 300 μl min−1 consisting of a 2 min hold at 2% B, followed by linear gradient increase to 100% B during 5 min. The column temperature was set at 25°C and an injection volume of 1 μl. An electrospray ionization interface was used as ionization source. Analysis was performed in positive ionization mode. Metabolites were detected using multiple-reaction monitoring, using argon as the collision gas. Quantification was made using standard curve in 0–10 μM concentration range. 15N5-adenosine 5′-monophosphate (Sigma) was added to standards and samples as internal standard (0.5 μM). TargetLynx (Waters) was used for data analysis.
Mass spectrometry of in vitro reactions
20 μl reactions containing purified EcPycC or BcPycC with 500 μM CTP or UTP were incubated at 37°C overnight in buffer (50 mM CAPSO pH 9.4, 100 mM KCl, 10 mM MgCl2, 1 mM MnCl2), heat inactivated at 95°C for 2 min, diluted to 200 μl with RNase-free water and filtered through 10 kDa cutoff Amicon™ Ultra centrifuge filters (Millipore). Sample analysis by high-resolution LC-MSMS was carried out by MS-Omics (Vedbæk).
Thermal denaturation assay
EcPycC (wildtype or D102A mutant) was added at a final concentration of 20 μM with 3× SYPRO Orange Dye in a buffer of 100 mM KCl, 20 mM HEPES-KOH pH 7.5. Samples were heated from 20 to 95°C over the course of 2 h using a qPCR CFX96 thermocycler (Biorad) and fluorescence in the HEX channel was measured every 0.5°C. The first derivative of each fluorescence curve was calculated, and the melting temperature was identified as the peak of each derivative curve.
Isolation of mutant phages
To isolate mutant phages that escape defense, phages were plated on bacteria expressing the Pycsar system (E. coli E831) using the double-layer plaque assay (Mazzocco et al., 2009). For this, 100 μl of bacterial cells grown in MMB to an OD600 of 0.3 were mixed with 100 μl of T5 phage, and left at room temperature for 10 min. 5 ml of pre-melted 0.3% MMB were added and the mixture was poured onto a bottom layer of 1.1% MMB. The double layer plates were incubated overnight at 37°C (mutants #1–10) or room temperature (mutants #11–17) and single plaques were picked into 90 μl phage buffer.
For mutants #10 and #17, instead of picking single plaques, the entire top layer was scraped into 2 ml of phage buffer to enrich for phages that escape Pycsar defense. Phages were left for 1 h at room temperature during which the phages were mixed several times by vortex to release them from the agar into the phage buffer. The phages were centrifuged at 3200×g for 10 min to remove agar and bacterial cells, and the supernatant was transferred to a new tube.
In order to test the phages for the ability to escape from the defense system, the small drop plaque assay was used (Mazzocco et al., 2009). 300 ml of an overnight culture of either E. coli E831 or E. coli MG1655 with plasmid pSG1 were mixed with 30 ml melted MMB 0.3% agar and left to dry for 1 h at room temperature. 10-fold serial dilutions in phage buffer were performed for the ancestor T5 phage (WT phage used for the original double layer plaque assay) and the T5 phage mutants #1–17 isolated from plaques formed on the strain expressing the defense genes. 10 μl drops of the phage were put on the bacterial layer. The plates were incubated overnight at 37°C or room temperature for mutants #1–10 and #11–17 respectively.
Amplification of mutant phages
Isolated phages for which there was decreased defense compared to the ancestor phage were further propagated by picking a single plaque formed on E. coli MG1655 containing the E. coli E831 Pycsar system in the small drop plaque assay into a liquid culture of the same strain, which was grown at 37°C in MMB with shaking at 200 rpm to an OD600 of 0.3. The phages were incubated with the bacteria at 37°C with shaking at 200 rpm for 3 h, and then an additional 9 ml of bacterial culture grown to OD600 of 0.3 in MMB was added, and incubated for 3 additional hours in the same conditions. The lysates were then centrifuged at 3200×g for 10 min and the supernatant was filtered through a 0.2 μm filter to remove remaining bacteria. Phage titer was then checked using the small drop plaque assay on the negative control strain as described above.
Sequencing and genome analysis of phage mutants
High titer phage lysates (>107 PFU/ml) of the ancestor phage and isolated phage mutants were used for DNA extraction. 0.5 ml of phage lysate was treated with DNase-I (Merck cat #11284932001) added to a final concentration of 20 μg ml−1 and incubated at 37°C for 1 h to remove bacterial DNA. DNA was extracted using the QIAGEN DNeasy blood and tissue kit (cat. #69504) starting from a proteinase-K treatment to degrade the phage capsids. Libraries were prepared for Illumina sequencing using a modified Nextera protocol as previously described (Baym et al., 2015). Following sequencing on Illumina NextSeq500, reads were aligned to the T5 phage reference genome (GenBank accession number: AY543070) and mutations compared to the reference genome were identified using Breseq (version 0.34.1) with default parameters (Deatherage and Barrick, 2014). Only mutations that occurred in the isolated mutants, but not in the ancestor phage, were considered. Silent mutations within protein coding regions were disregarded as well.
Microscopy of infected cells
E. coli MG1655 cells that contain an inducible pBAD plasmid containing the E. coli E831-derived Pycsar or a negative control containing a pBAD-GFP plasmids were grown in MMB medium at 37°C with 0.2% arabinose. When growth reached an OD600 of 0.3, 250μM of cCMP was added to the medium. Sixty min following the addition of cCMP, 500 μl of the samples were centrifuged at 10,000×g for 2 min at 25°C and resuspended in 5 μl of 1× phosphate-buffered saline (PBS), supplemented with 1 μg ml−1 membrane stain FM4-64 (Thermo Fisher Scientific T-13320) and 2 μg ml−1 DNA stain 4,6-diamidino2-phenylindole (DAPI) (Sigma-Aldrich D9542-5MG). Cells were visualized and photographed using an Axioplan2 microscope (ZEISS) equipped with ORCA Flash 4.0 camera (HAMAMATSU). System control and image processing were carried out using Zen software version 2.0 (Zeiss).
Phylogenetic analysis
To generate the phylogenetic tree in Figure 2A the ‘clusthash’ option of MMseqs2 (Steinegger and Söding, 2017) (release 6-f5a1c) was used to remove protein redundancies (using the ‘--min-seq-id 0.9’ parameter). Sequences that were shorter than 200 amino acids were also removed and HHsearch was used to identify the cyclase domain in each protein sequence. Eight sequences of diguanylate cyclase were added (IMG ID: 2503740511, 2689262325, 2688711941, 2688713579, 2661427045, 2661426508, 2688078042, 2688079262) and were used as an outgroup. Sequences of the cyclase domain were aligned using clustal-omega v.1.2.4 with default parameters (Sievers and Higgins, 2018). FastTree was used to generate a tree from the multiple-sequence alignment using default parameters (Price et al., 2009). iTOL was used for tree visualization (Letunic and Bork, 2019). To generate the sequence motifs of clades B and E, sequences of each clade were aligned using clustal-omega v.1.2.4 with default parameters (Sievers and Higgins, 2018). Weblogo3 (version 2.8.2) was used to generate the motifs for each clade (Crooks et al., 2004).
Fitness assay of mutant pb8 phages
Mutant and WT T5 phages were mixed in a target ratio of approximately 1:1. Overnight cultures of bacteria harboring the defense system (E. coli MG1655 with a pSG1 plasmid containing the system from E. coli E831 or a negative control strain with an empty vector) were diluted 1:100 in MMB medium. Cells were incubated at 37°C while shaking at 200 rpm until early log phase (OD600 0.3). System-expressing and control cells were infected with the phage lysate for a final MOI of 0.2, and incubated for 3 h at 37°C while shaking at 200 rpm. Cultures were then centrifuged at 3200×g for 5 min and phage-containing supernatant was filtered through a 0.2 μm filter to get rid of remaining bacteria. Phage titer was then checked using the small drop plaque assay on the negative control strain as described above. This phage lysate was subsequently used for the infection of a fresh culture of bacteria at an MOI of 0.2 (second passage). The same process was carried out for the third passage. In each of the passages, 0.5 ml of the phage lysate was kept for DNA extraction and sequencing, as detailed above.
Protein expression and purification
BL21-RIL E. coli (Agilent) were transformed with plasmid DNA encoding 6×His-SUMO2-tagged gene expression constructs and plated on MDG-agar plates (0.5% glucose, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4, 2 mM MgSO4, 0.25% aspartic acid, 100 mg ml−1 ampicillin, 34 mg ml−1 chloramphenicol, and trace metals), then colonies were used to inoculate overnight MDG liquid cultures (same components minus the agar). Overnight MDG cultures were used to inoculate cultures of M9ZB media (0.5% glycerol, 1% Cas-amino Acids, 47.8 mM Na2HPO4, 22 mM KH2PO4, 18.7 mM NH4Cl, 85.6 mM NaCl, 2 mM MgSO4, 100 mg ml−1 ampicillin, 34 mg ml−1 chloramphenicol, and trace metals), which were grown and harvested in each experiment as described below. For BcPycTIR expression, M9ZB media was supplemented with 10 mM nicotinamide (NAM, Sigma). Selenomethionine-labeled protein for structural determination was prepared as previously described (Eaglesham et al., 2019).
Recombinant cyclase enzymes and BcPycTIR were purified as previously described (Morehouse et al., 2020; Zhou et al., 2018). Briefly, large scale cultures (1–2 liters) were grown for ~5 h at 37°C, then induced with IPTG overnight at 16°C with 230 rpm shaking. Cell pellets were harvested, washed with PBS, and flash frozen in liquid N2 prior to short-term storage at −80°C. Cells were resuspended and sonicated in lysis buffer (20 mM HEPES-KOH pH 7.5, 400 mM NaCl, 30 mM imidazole, 10% glycerol, supplemented with 1 mM DTT) and purified using Ni-NTA resin (Qiagen). Resin-bound protein was washed with lysis buffer supplemented to 1 M NaCl and eluted with lysis buffer supplemented to 300 mM imidazole. The eluent was dialyzed into 20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM DTT overnight (16–20 h) while removing the SUMO2 tag with recombinant human SENP2 protease (D364–L589, M497A). Proteins were concentrated using a 10K-cutoff concentrator (Millipore) and either passed back over Ni-NTA resin to remove cleaved 6×His-SUMO2 and uncleaved protein or further purified by size-exclusion chromatography on a 16/600 Superdex 75 column using the same dialysis buffer system. Proteins were concentrated to >10 mg ml−1, flash frozen with liquid nitrogen, and stored at −80°C until needed. 10% glycerol was included in the final storage buffer as a cryoprotectant for those proteins which could not be concentrated >10 mg ml−1.
Crystallization and structure determination
BcPycC was crystallized using the hanging drop vapor diffusion method at 18°C. Protein was thawed on ice before dilution into 25 mM HEPES-KOH pH 7.5 with 75 mM KCl and 1 mM TCEP bringing the final protein concentration before setting crystallization trays to 8 mg ml−1. Drops were set by mixing 1:1 protein and reservoir solution in 2 μl drops over a 350 μl reservoir in 15-well Easy-Xtal trays (Qiagen) following sparse screening to identify initial crystal hits. BcPycC crystallized in less than 24 h in 0.1 M ammonium acetate (NH4CH3CO2), 0.1 M HEPES-KOH pH 7.5, and 29% PEG-3350. Crystals were harvested and cryoprotected in reservoir solution supplemented with 15% ethylene glycol before plunge freezing in liquid N2.
X-ray data collection was carried out at the Northeastern Collaborative Access Team beamline 24-ID-E (P30 GM124165), fixed at 12.66 keV and used an Eiger detector (S10OD021527) and the Argonne National Laboratory Advanced Photon Source (DE-AC02-06CH11357). X-ray crystallography data were processed with XDS and AIMLESS (Kabsch, 2010) using the SSRL autoxds script (A. Gonzalez, Stanford SSRL). Experimental phase information was determined using data collected from selenomethionine-substituted crystals. 16 anomalous sites (from 4 protein copies in the ASU) were identified with HySS and an initial map was produced using SOLVE/RESOLVE in Phenix (Liebschner et al., 2019). Model building was performed using Coot (Emsley and Cowtan, 2004), then refined in Phenix. Statistics were analyzed as described in Table S5 (Chen et al., 2010; Karplus and Diederichs, 2012; Weiss, 2001).
Thin-layer chromatography analysis of cyclic nucleotide products
Purified cyclases were mixed at a final concentration of 5 μM with 100 μM NTPs and trace radiolabeled α−32P-NTP in 10 μL reactions with 100 mM KCl, 10 mM MgCl2, 1 mM MnCl2, and 50 mM Tris-HCl pH 8.5. These reactions were incubated for 2 h at 37°C and then terminated by treatment with 1 μl of 5 units μl−1 Calf Intestinal Phosphatase (CIP, New England Biolabs) for 1 h at 37°C. 1 μl of each reaction was spotted on glass-backed silica-coated thin layer chromatography (TLC) plates (Millipore) and developed for one hour in a solvent system composed of 11:7:2 1-propanol:conc. NH4OH:water as previously described (Whiteley et al., 2019). TLC plates were dried and then processed by exposing with a phosphor screen followed by imaging with an Amersham Typhoon IP (Cytiva). For optimization experiments reactions were left at 37°C for 25 min then terminated at 85°C for 2 min, and CIP treated for 30 min at 37°C before spotting on TLC.
High-performance liquid chromatography analysis of enzymatic reactions
Cyclic nucleotide synthesis was carried out in 125 μl reactions with 5 μM purified cyclase and 500 μM ATP, CTP, GTP, and UTP in 1× reaction buffer (100 mM KCl, 10 mM MgCl2, 1 mM MnCl2, 50 mM Tris-HCl pH 8.5), then incubated overnight at 37°C. Reactions were filtered through 10 kDa cutoff Amicon™ Ultra centrifuge filters (Millipore) and injected onto an Agilent 1260 infinity series HPLC. Comparison of major peak products to cyclic nucleotide standards (cAMP [Biolog A 001 S], cGMP [Biolog G 001], cCMP [Biolog C 001], and cUMP [Biolog U 001]) was carried out using a C18 column (Agilent Zorbax Bonus-RP 4.6×150 mm, 3.5-micron). The column was heated to 40°C and run at 1 ml min−1 with a mobile phase of 50 mM NaH2PO4 (pH 6.8 with NaOH) supplemented with 3% acetonitrile. Absorbance signals were monitored at 254 nm.
NADase reactions were carried out in a similar manner with 500 μM NAD+ (Sigma), 500 nM BcPycTIR, and 0–1mM cNMP ligands. Reactions were incubated for 1 h at 25°C and then filtered prior to HPLC analysis.
Fluorescence plate reader analysis of NADase activity
A plate reader assay was devised to monitor the NADase activity of purified BcPycTIR. Briefly, 30 μl reactions were built in white walled clear bottom 96-well plates. The reactions consisted of buffer (20 mM HEPES-KOH pH 7.5, 100 mM KCl), 1 μM BcPycTIR, a range of 10 nM to 1 mM synthetic cyclic nucleotide ligands, and 500 μM of the fluorescent NAD+ analog ε-NAD+. Fluorescent measurements (300 nm excitation/410 nm emission) of each well were taken in kinetic mode every 1 min for a total of 60 min in a Synergy H1 plate reader (BioTek). Initial rates were calculated based on linear fits of the first 3 min of the reaction.
Negative stain electron microscopy and processing
50 μM (~1.66 mg ml−1) BcPycTIR was incubated with an equimolar ratio of cyclic nucleotide in gel filtration buffer (20 mM HEPES-KOH pH 7.5, 150 mM KCl, 1 mM TCEP) on ice for ~5 min. Mixtures were then serially diluted to a final concentration of 0.15 mg ml−1 protein with gel filtration buffer supplemented with the appropriate cyclic nucleotide at 50 μM. 3 μl of each sample were applied to a glow-discharged (30 s, 30 mA) 400-mesh carbon-coated grid, followed by a 30 s absorption step and side blotting to remove bulk solution. The grid was immediately stained twice with a 1.5% uranyl formate solution, with side blotting after each step.
Electron micrographs (30 micrographs for each condition) were collected using an FEI Tecnai T12 TEM operated at 120 keV equipped with a Gatan 4K × 4K CCD camera at a nominal magnification of 67,000×, corresponding to a calibrated pixel size of 1.68 Å, and a target defocus of −1.5 μm. Image processing was performed in RELION-3.1 (Zivanov et al., 2020). CTF estimation was performed with CTFFIND4.1 (Rohou and Grigorieff, 2015). Particles were either picked manually or using Gautomatch (Kai Zhang, https://www.mrc-lmb.cam.ac.uk/kzhang/) followed by manual curation. Particles were extracted using a 148 pixel box size and subjected to 2D reference-free classification using a circular 225 Å diameter mask.
Pb8 binding assay
An in vitro pull-down experiment was designed to assess the ability of purified phage T5 pb8 protein to form a stable interaction with EcPycC. An N-terminal 6×His-SUMO2-tagged construct of pb8 was purified as described above for the cyclases and BcPycTIR. 0.5 μM EcPycC ‘bait’ protein which contained an N-terminal 6×His-MBP-SUMO2 tag was incubated (30 min, 4°C) with 100 μl of 50% slurry (50 μl packed beads) amylose resin (New England Biolabs) diluted in 400 μl of binding buffer (20 mM HEPES-KOH pH 7.5, 100 mM KCl, 1 mM TCEP, 0.01% NP-40 detergent). 4 μM (8× [‘bait’]) untagged pb8 protein (‘prey’) was added to the mixture and incubated for an additional 1 h at 4°C. Resin was pelleted at 4°C and 500 × g for 3 min and the supernatant was removed by aspiration. The beads were washed, pelleted, and resuspended in 1 ml of binding buffer a total of 4 times before ‘elution’ was achieved by heating the beads at 95°C for 2 min in 50 μl of SDS-PAGE loading dye. Eluted samples were loaded onto a 10% SDS-PAGE gel and visualized with Coomassie staining.
QUANTIFICATION AND STATISTICAL ANALYSIS
The average of triplicates is shown throughout with individual points overlaid, unless stated otherwise.
Supplementary Material
Figure S1. A two-gene defense system found in defense islands (related to Figure 1A,B). (A) Representative instances of a two-gene system encoding a gene with nucleotide cyclase domain (pink) and a gene with transmembrane (TM) domains (purple), in their genomic neighborhoods. Genes known to be involved in defense are shown in yellow. RM, restriction modification; Gabija and Zorya are a recently described defense systems (Doron et al., 2018). The bacterial species and the accession of the relevant genomic scaffold in the Integrated Microbial Genomes (IMG) database (Chen et al., 2019) are indicated on the left. (B) Bacteria expressing WT Pycsar systems from E. coli 303145 or E. coli E831, as well as the system from E. coli E831 with point mutations in the cyclase or deletion of the TM effector (“cyclase only”), and a negative control that contains an empty vector, were grown on agar plates in room temperature. Tenfold serial dilutions of the phage lysate were dropped on the plates. Data represent plaque-forming units per milliliter for phages tested in this study. Each bar graph represents average of three replicates, with individual data points overlaid. The data for T5-infected WT and mutated E. coli E831 Pycsar are also presented in Figures 1C and 3F.
Figure S2. In vitro characterization of EcPycC and BcPycC cyclase catalytic activities (related to Figures 1D,1E, 2C). (A) Titration of divalent cation concentrations affects the in vitro activity of the EcPycC (top) and BcPycC (bottom) cyclases. Magnesium and manganese concentrations are in mM. (B) Cyclases display optimal activity at a broad range of neutral to basic pH. (C) HPLC traces for EcPycC in vitro reactions with individual NTPs showing no significant cyclase activity for non-cognate substrates. Chemical standards were used to confirm reaction products. (D) Thermofluor stability assays of WT and mutant EcPycC. Assays were performed in the following conditions: 3× SYPRO orange fluorescent dye, 20 μM enzyme, 20 mM HEPES pH 7.5, 100 mM KCl buffer. 20 μL reaction volume. Measured at 20-95 °C at 0.5 °C increments every 30 seconds. Average of three independent experiments is shown.
Figure S4. Sequence alignment and structural comparisons of PycC cyclases (related to Figure 3). (A) Representation of BcPycC UTP binding pocket highlighting proposed amino acid contacts. A “b” subscript indicates residue contacts from the opposing monomer in the dimeric active site. (B) ATPαS modeled (transparent grey) into the BcPycC binding pocket would cause significant clashing interactions with active site residues. UTP (yellow) modeled based on alignment with ATPαS in 1CJK (C) Homology model of the CTP binding pocket in EcPycC prepared with the Phyre2 prediction server (Kelley et al., 2015) using the crystal structure of BcPycC cUMP synthase as a template and CTP (yellow) modeled based on alignment with ATPαS in 1CJK. (D) Structure guided sequence alignment of nucleotide cyclases. Structure of BcPycC of clade B was used to seed sequence alignment with Promals3D. For the two-domain cyclases PaPycC of clade A and human soluble adenylate cyclase, both N- and C-term cyclase domains are depicted. Blue asterisks indicate catalytic residues. Purple asterisks indicate residues known to be important for selection of adenosine base in most adenylate cyclases. Residues in red were mutated in this study. Purple shading depicts sequence conservation. The cyclases identified as most similar to BcPycC by a Dali search (Holm, 2020) include photoactivated adenylyl cyclase (PDB 5X4V), P. aeruginosa CyaB adenylate cyclase (PDB 3R5G), and the cyanobacterial guanylate cyclase Cya2 (PDB 20W1). (E) PaPycC (PDB 6YII) binds ATP (yellow) in a configuration that differs significantly from the extended ATPαS binding configuration in 1CJK (grey), and is not compatible for cyclization. (F) The non-productive ATP conformation observed in the PaPycC structure can be potentially accommodated in the BcPycC binding pocket but notably in both PaPycC and BcPycC active sites steric clashes would prevent the extended and cyclization competent configuration. (G) Superposition of PaPycC and BcPycC nucleotide binding pockets (ATP removed for clarity).
Figure S3. Biochemical analysis of cyclic pyrimidine mononucleotide phosphate synthases (related to Figure 2). (A) HPLC separation of primary cyclic nucleotide products generated by representative uridylate cyclases from various clades of the PycC phylogeny. (B) HPLC analysis displaying cCMP synthesis activity for an additional clade E cyclase. (C) Some synthases can produce minor quantities of cGMP in vitro. All reactions contained 500 μM NTPs and 5 μM enzyme. For panels A through C, HPLC of standard molecule is shown in grey. (D) The AGS-C domain of EcPycC (residues 318–450) shares predicted structural homology to immunoglobulin fold protein domains. Prediction was carried out with AlphaFold2 Google collaborative notebook with default parameters (Jumper et al., 2021). (E) Bacteria expressing the WT Pycsar system from E. coli E831, the same system with EcPycC in which the AGS-C domain was deleted, and a negative control that contains an empty vector, were grown on agar plates in room temperature. Tenfold serial dilutions of phage lysates (T5 and P1) were dropped on the plates. Data represent plaque-forming units per milliliter of the tested phages. Each bar graph represents average of three replicates, with individual data points overlaid.
Figure S5. Pycsar effector specificity to cyclic nucleotides (related to Figures 4B and 5B). (A) Sequence alignment of cyclic nucleotide binding domains (CNBD, pfam accession PF00027) of PycTIR proteins and previously studied CNBDs. Sequences were aligned using MAFFT and Promals3D. Presented are CNBDs from PaPycTIR, AhPycTIR, BcPycTIR and XpPycTIR, E. coli cAMP receptor protein (CRP), H. sapiens PRKAR1α cAMP-dependent protein kinase type I-α regulatory subunit (PKA), H. sapiens PRKG1 cGMP-dependent protein kinase 1 (PKG), H. sapiens cGMP-gated cation channel alpha-1 (CNGA1), and H. sapiens hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4). (B-C) Non-cognate cyclic nucleotides are not toxic to Pycsar-expressing cells. (B) Growth curves of E. coli MG1655 cells expressing the Pycsar system from E. coli E831 (purple) and control cells (black) with and without the addition of the non-cognate ligand cUMP to the medium at various concentrations. (C) Growth curves of E. coli MG1655 cells expressing the Pycsar system from X. perforans GEV1001 (blue) and control cells (black) with and without the addition of the non-cognate ligand cCMP to the medium at various concentrations. For both panels B and C, results of three experiments are presented as individual curves.
Table S3. Phages used in this study, related to figure 1.
Table S5. Summary of crystallography statistics, related to figure 3.
Figure S6. Phages mutated in the Pb8 protein (related to Figure 6). (A) Growth curves of E. coli MG1655 cells expressing the Pycsar system from E. coli E831, that also contain a vector harboring the pb8 protein under an inducible promoter (purple) and control cells (black) containing the same pb8-containing construct but not the defense system. Results of three experiments are presented as individual curves. X-axis represents minutes post arabinose induction. (B) In vitro pull-down assay using MBP-tagged proteins demonstrating lack of interaction between T5 pb8 and the E. coli E831 PycC cyclase. EcPycC was expressed with an N-terminal 6×His–MBP–SUMO2 tag and the resulting full-length protein has a predicted molecular weight of ~105 kDa. The lower molecular weight bands correspond to truncated 6×His–MBP species co-purified during Ni-NTA purification. (C,D) The impact of pb8 T5 phage capsid protein on cyclase activity of EcPycC was assessed via thin-layer chromatography. Reactions contained 1 μM EcPycC and 0, 1, or 5 μM purified pb8 in buffer containing 50 mM Tris pH 8.5, 1 mM MnCl2, 100 mM KCl, 500 μM CTP, and trace α−32P-CTP. Reactions were incubated for the indicated times at 37°C, quenched at 85°C for 2 min, cooled on ice for 2 min, then treated with CIP for 30 min at 37°C to digest uncyclized CTP. (C) A representative TLC image. (D) The percent fractional signal associated with cCMP production from three independent experiments was quantified and average values are plotted, with individual data points overlaid. (E,F) A competition fitness assay between WT and mutant T5 phages (mutant n.1 and mutant n.4 for panels C and D, respectively) on both Pycsar-expressing and control cells. Y-axis represents the fraction of each strain out of the total phage mixture based on sequenced DNA reads. X-axis represents the infection passage, with 0 representing the mixture prior to first passage. In each passage, phage lysate was taken and used for infection of a fresh bacterial culture.
Table S1. Pycsar systems identified in this study (provided as an Excel file), related to figure 2.
Table S2. Mutant phages isolated in this study (provided as an Excel file), related to figure 6.
Table S4. Synthesized sequences and primers used in this study (provided as an Excel file), related to STAR methods.
Acknowledgements
We thank the Sorek and Kranzusch laboratory members for comments on the manuscript and fruitful discussion, and the Molecular Electron Microscopy Suite at Harvard Medical School. We thank Brianna Duncan-Lowey for her assistance in predicting AGS-C homology to immunoglobulin folds. R.S. was supported, in part, by the European Research Council (grant ERC-CoG 681203), the Ernest and Bonnie Beutler Research Program of Excellence in Genomic Medicine, the German Research Council (DFG) priority program SPP 2330 (grant SO 1611/2), the Minerva Foundation with funding from the Federal German Ministry for Education and Research, the Knell Family Center for Microbiology, the Yotam project and the Weizmann Institute Sustainability And Energy Research (SAERI) initiative, and the Dr. Barry Sherman Institute for Medicinal Chemistry. P.J.K. was supported by grants from the Pew Biomedical Scholars Program, Mark Foundation for Cancer Research, The Mathers Foundation, the Parker Institute for Cancer Immunotherapy, and a Burroughs Wellcome Fund PATH award. B.R.M. was supported as a Ruth L. Kirschstein NRSA Postdoctoral Fellow NIH F32GM133063. A.M. was supported by a fellowship from the Ariane de Rothschild Women Doctoral Program and, in part, by the Israeli Council for Higher Education via the Weizmann Data Science Research Center, and by a research grant from Madame Olga Klein-Astrachan. S.S. was supported by grants from the Vallee Scholars Program and a Packard fellowship.
Footnotes
Declaration of Interests
R.S. is a scientific cofounder and advisor of BiomX, Pantheon Bioscience and Ecophage.
References
- Bähre H, Danker KY, Stasch JP, Kaever V, and Seifert R (2014). Nucleotidyl cyclase activity of soluble guanylyl cyclase in intact cells. Biochem. Biophys. Res. Commun 443, 1195–1199. [DOI] [PubMed] [Google Scholar]
- Bähre H, Hartwig C, Munder A, Wolter S, Stelzer T, Schirmer B, Beckert U, Frank DW, Tümmler B, Kaever V, et al. (2015). cCMP and cUMP occur in vivo. Biochem. Biophys. Res. Commun 460, 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, and Kishony R (2015). Inexpensive Multiplexed Library Preparation for Megabase-Sized Genomes. PLoS One 10, e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckert U, Grundmann M, Wolter S, Schwede F, Rehmann H, Kaever V, Kostenis E, and Seifert R (2014). cNMP-AMs mimic and dissect bacterial nucleotidyl cyclase toxin effects. Biochem. Biophys. Res. Commun 451, 497–502. [DOI] [PubMed] [Google Scholar]
- Bernheim A, and Sorek R (2020). The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol 18, 113–119. [DOI] [PubMed] [Google Scholar]
- Bernheim A, Millman A, Ofir G, Meitav G, Avraham C, Shomar H, Rosenberg MM, Tal N, Melamed S, Amitai G, et al. (2021). Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schäffer DE, Iyer LM, and Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 43, 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PJ (1987). The effect of cyclic cytidine 3′,5′-monophosphate (cCMP) on the in vitro development, hatching and attachment of the mouse blastocyst. Experientia 43, 929–930. [DOI] [PubMed] [Google Scholar]
- Chen IMA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, et al. (2019). IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, Doron S, Amitai G, and Sorek R (2019). Cyclic GMP–AMP signalling protects bacteria against viral infection. Nature 574, 691–695. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE, and Barrick JE (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol 1151, 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, and Rathjen JP (2010). Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet 2010 118 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, and Sorek R (2018). Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglesham JB, Pan Y, Kupper TS, and Kranzusch PJ (2019). Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS–STING signalling. Nature 566, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Gaion RM, and Krishna G (1979). Cytidylate cyclase: The product isolated by the method of Cech and Ignarro is not cytidine 3′,5′-monophosphate. Biochem. Biophys. Res. Commun 86, 105–111. [DOI] [PubMed] [Google Scholar]
- Gomelsky M, and Galperin MY (2013). Bacterial second messengers, cGMP and c-di-GMP, in a quest for regulatory dominance. EMBO J. 32, 2421–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Stapleton MR, Smith LJ, Artymiuk PJ, Kahramanoglou C, Hunt DM, and Buxton RS (2014). Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: Adaptation for different ecological niches. Curr. Opin. Microbiol 18, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoune J, and Defer N (2001). Regulation and role of adenylyl cyclase isoforms. Annu. Rev. Pharmacol. Toxicol 41, 145–174. [DOI] [PubMed] [Google Scholar]
- Holm L (2020). Using Dali for Protein Structure Comparison. Methods Mol. Biol 2112, 29–42. [DOI] [PubMed] [Google Scholar]
- Huet A, Duda RL, Boulanger P, and Conway JF (2019). Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A 116, 21037–21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu T, Lee CH, Chang Q, Yang J, and Zhang Z (2020). The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature 588, 658–663. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, and Diederichs K (2012). Linking crystallographic model and data quality. Science 336, 1030–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, and Siksnys V (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khannpnavar B, Mehta V, Qi C, and Korkhov V (2020). Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol 63, 34–41. [DOI] [PubMed] [Google Scholar]
- Letunic I, and Bork P (2019). Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln TM (1989). Cyclic GMP and mechanisms of vasodilation. Pharmacol. Ther 41, 479–502. [DOI] [PubMed] [Google Scholar]
- Linder JU (2005). Substrate selection by class III adenylyl cyclases and guanylyl cyclases. IUBMB Life 57, 797–803. [DOI] [PubMed] [Google Scholar]
- Linder JU (2010). cGMP production in bacteria. Mol. Cell. Biochem 334, 215–219. [DOI] [PubMed] [Google Scholar]
- Linder J, Hupfeld E, Weyand M, Steegborn C, and Moniot S (2020). Crystal structure of a class III adenylyl cyclase-like ATP-binding protein from Pseudomonas aeruginosa. J. Struct. Biol 211. [DOI] [PubMed] [Google Scholar]
- Lopatina A, Tal N, and Sorek R (2020). Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol 7, 371–384. [DOI] [PubMed] [Google Scholar]
- Lowey B, Whiteley AT, Keszei AFA, Morehouse BR, Mathews IT, Antine SP, Cabrera VJ, Kashin D, Niemann P, Jain M, et al. (2020). CBASS Immunity Uses CARF-Related Effectors to Sense 3′–5′- and 2′–5′-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 182, 38–49.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzocco A, Waddell TE, Lingohr E, and Johnson RP (2009). Enumeration of bacteriophages using the small drop plaque assay system. Methods Mol. Biol 501, 81–85. [DOI] [PubMed] [Google Scholar]
- Millman A, Melamed S, Amitai G, and Sorek R (2020a). Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol 5, 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman A, Bernheim A, Stokar-Avihail A, Fedorenko T, Voichek M, Leavitt A, Oppenheimer-Shaanan Y, and Sorek R (2020b). Bacterial Retrons Function In Anti-Phage Defense. Cell 183, 1551–1561.e12. [DOI] [PubMed] [Google Scholar]
- Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, Shao S, Sorek R, and Kranzusch PJ (2020). STING cyclic dinucleotide sensing originated in bacteria. Nature 586, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RP, Salih SG, Salvage BJ, and Kingston EE (1984). Extraction, purification and identification of cytidine 3’,5’-cyclic monophosphate from rat tissues. Biochem. J 221, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, Tal N, Malheiro DBA, Malitsky S, Amitai G, et al. (2021). Antiviral activity of bacterial TIR domains via signaling molecules that trigger cell death. BioRxiv 2021.01.06.425286. [DOI] [PubMed] [Google Scholar]
- Pastan I, and Perlman R (1970). Cyclic adenosine monophosphate in bacteria. Science 169, 339–344. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP (2009). Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Leipelt M, Russwurm M, and Steegborn C (2008). Crystal structure of the guanylyl cyclase Cya2. Proc. Natl. Acad. Sci. U. S. A 105, 15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Youn H, Kang IH, and Gomelsky M (2015). Identification of bacterial guanylate cyclases. Proteins Struct. Funct. Bioinforma 83, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert R (2015). cCMP and cUMP: Emerging second messengers. Trends Biochem. Sci 40, 8–15. [DOI] [PubMed] [Google Scholar]
- Seifert R (2017). cCMP and cUMP across the tree of life: From cCMP and cUMP generators to cCMP- and cUMP-regulated cell functions. In Handbook of Experimental Pharmacology, (Springer New York LLC), pp. 3–23. [DOI] [PubMed] [Google Scholar]
- Seifert R, Beste K, Burhenne H, Voigt U, Wolter S, Hammerschmidt A, Reinecke D, Sandner P, Pich A, Schwede F, et al. (2011). Cyclic CMP and cyclic UMP: new (old) second messengers. BMC Pharmacol. 11, O34. [Google Scholar]
- Sievers F, and Higgins DG (2018). Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporny M, Guez-Haddad J, Lebendiker M, Ulisse V, Volf A, Mim C, Isupov MN, and Opatowsky Y (2019). Structural Evidence for an Octameric Ring Arrangement of SARM1. J. Mol. Biol 431, 3591–3605. [DOI] [PubMed] [Google Scholar]
- Steegborn C (2014). Structure, mechanism, and regulation of soluble adenylyl cyclases - similarities and differences to transmembrane adenylyl cyclases. Biochim. Biophys. Acta 1842, 2535–2547. [DOI] [PubMed] [Google Scholar]
- Steinegger M, and Söding J (2017). MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol 35, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Stryer L (1986). Cyclic GMP cascade of vision. Annu. Rev. Neurosci VOL. 9, 87–119. [DOI] [PubMed] [Google Scholar]
- Tesmer JJG, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, and Sprang SR (1999). Two-metal-ion catalysis in adenylyl cyclase. Science 285, 756–760. [DOI] [PubMed] [Google Scholar]
- Weiss MS (2001). Global indicators of X-ray data quality. J. Appl. Crystallogr 34, 130–135. [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC, Morehouse BR, Lowey B, Nieminen EA, Danilchanka O, King DS, Lee ASY, Mekalanos JJ, et al. (2019). Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Whiteley AT, de Oliveira Mann CC, Morehouse BR, Nowak RP, Fischer ES, Gray NS, Mekalanos JJ, and Kranzusch PJ (2018). Structure of the Human cGAS–DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 174, 300–311.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, and Scheres SHW (2020). Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A two-gene defense system found in defense islands (related to Figure 1A,B). (A) Representative instances of a two-gene system encoding a gene with nucleotide cyclase domain (pink) and a gene with transmembrane (TM) domains (purple), in their genomic neighborhoods. Genes known to be involved in defense are shown in yellow. RM, restriction modification; Gabija and Zorya are a recently described defense systems (Doron et al., 2018). The bacterial species and the accession of the relevant genomic scaffold in the Integrated Microbial Genomes (IMG) database (Chen et al., 2019) are indicated on the left. (B) Bacteria expressing WT Pycsar systems from E. coli 303145 or E. coli E831, as well as the system from E. coli E831 with point mutations in the cyclase or deletion of the TM effector (“cyclase only”), and a negative control that contains an empty vector, were grown on agar plates in room temperature. Tenfold serial dilutions of the phage lysate were dropped on the plates. Data represent plaque-forming units per milliliter for phages tested in this study. Each bar graph represents average of three replicates, with individual data points overlaid. The data for T5-infected WT and mutated E. coli E831 Pycsar are also presented in Figures 1C and 3F.
Figure S2. In vitro characterization of EcPycC and BcPycC cyclase catalytic activities (related to Figures 1D,1E, 2C). (A) Titration of divalent cation concentrations affects the in vitro activity of the EcPycC (top) and BcPycC (bottom) cyclases. Magnesium and manganese concentrations are in mM. (B) Cyclases display optimal activity at a broad range of neutral to basic pH. (C) HPLC traces for EcPycC in vitro reactions with individual NTPs showing no significant cyclase activity for non-cognate substrates. Chemical standards were used to confirm reaction products. (D) Thermofluor stability assays of WT and mutant EcPycC. Assays were performed in the following conditions: 3× SYPRO orange fluorescent dye, 20 μM enzyme, 20 mM HEPES pH 7.5, 100 mM KCl buffer. 20 μL reaction volume. Measured at 20-95 °C at 0.5 °C increments every 30 seconds. Average of three independent experiments is shown.
Figure S4. Sequence alignment and structural comparisons of PycC cyclases (related to Figure 3). (A) Representation of BcPycC UTP binding pocket highlighting proposed amino acid contacts. A “b” subscript indicates residue contacts from the opposing monomer in the dimeric active site. (B) ATPαS modeled (transparent grey) into the BcPycC binding pocket would cause significant clashing interactions with active site residues. UTP (yellow) modeled based on alignment with ATPαS in 1CJK (C) Homology model of the CTP binding pocket in EcPycC prepared with the Phyre2 prediction server (Kelley et al., 2015) using the crystal structure of BcPycC cUMP synthase as a template and CTP (yellow) modeled based on alignment with ATPαS in 1CJK. (D) Structure guided sequence alignment of nucleotide cyclases. Structure of BcPycC of clade B was used to seed sequence alignment with Promals3D. For the two-domain cyclases PaPycC of clade A and human soluble adenylate cyclase, both N- and C-term cyclase domains are depicted. Blue asterisks indicate catalytic residues. Purple asterisks indicate residues known to be important for selection of adenosine base in most adenylate cyclases. Residues in red were mutated in this study. Purple shading depicts sequence conservation. The cyclases identified as most similar to BcPycC by a Dali search (Holm, 2020) include photoactivated adenylyl cyclase (PDB 5X4V), P. aeruginosa CyaB adenylate cyclase (PDB 3R5G), and the cyanobacterial guanylate cyclase Cya2 (PDB 20W1). (E) PaPycC (PDB 6YII) binds ATP (yellow) in a configuration that differs significantly from the extended ATPαS binding configuration in 1CJK (grey), and is not compatible for cyclization. (F) The non-productive ATP conformation observed in the PaPycC structure can be potentially accommodated in the BcPycC binding pocket but notably in both PaPycC and BcPycC active sites steric clashes would prevent the extended and cyclization competent configuration. (G) Superposition of PaPycC and BcPycC nucleotide binding pockets (ATP removed for clarity).
Figure S3. Biochemical analysis of cyclic pyrimidine mononucleotide phosphate synthases (related to Figure 2). (A) HPLC separation of primary cyclic nucleotide products generated by representative uridylate cyclases from various clades of the PycC phylogeny. (B) HPLC analysis displaying cCMP synthesis activity for an additional clade E cyclase. (C) Some synthases can produce minor quantities of cGMP in vitro. All reactions contained 500 μM NTPs and 5 μM enzyme. For panels A through C, HPLC of standard molecule is shown in grey. (D) The AGS-C domain of EcPycC (residues 318–450) shares predicted structural homology to immunoglobulin fold protein domains. Prediction was carried out with AlphaFold2 Google collaborative notebook with default parameters (Jumper et al., 2021). (E) Bacteria expressing the WT Pycsar system from E. coli E831, the same system with EcPycC in which the AGS-C domain was deleted, and a negative control that contains an empty vector, were grown on agar plates in room temperature. Tenfold serial dilutions of phage lysates (T5 and P1) were dropped on the plates. Data represent plaque-forming units per milliliter of the tested phages. Each bar graph represents average of three replicates, with individual data points overlaid.
Figure S5. Pycsar effector specificity to cyclic nucleotides (related to Figures 4B and 5B). (A) Sequence alignment of cyclic nucleotide binding domains (CNBD, pfam accession PF00027) of PycTIR proteins and previously studied CNBDs. Sequences were aligned using MAFFT and Promals3D. Presented are CNBDs from PaPycTIR, AhPycTIR, BcPycTIR and XpPycTIR, E. coli cAMP receptor protein (CRP), H. sapiens PRKAR1α cAMP-dependent protein kinase type I-α regulatory subunit (PKA), H. sapiens PRKG1 cGMP-dependent protein kinase 1 (PKG), H. sapiens cGMP-gated cation channel alpha-1 (CNGA1), and H. sapiens hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4). (B-C) Non-cognate cyclic nucleotides are not toxic to Pycsar-expressing cells. (B) Growth curves of E. coli MG1655 cells expressing the Pycsar system from E. coli E831 (purple) and control cells (black) with and without the addition of the non-cognate ligand cUMP to the medium at various concentrations. (C) Growth curves of E. coli MG1655 cells expressing the Pycsar system from X. perforans GEV1001 (blue) and control cells (black) with and without the addition of the non-cognate ligand cCMP to the medium at various concentrations. For both panels B and C, results of three experiments are presented as individual curves.
Table S3. Phages used in this study, related to figure 1.
Table S5. Summary of crystallography statistics, related to figure 3.
Figure S6. Phages mutated in the Pb8 protein (related to Figure 6). (A) Growth curves of E. coli MG1655 cells expressing the Pycsar system from E. coli E831, that also contain a vector harboring the pb8 protein under an inducible promoter (purple) and control cells (black) containing the same pb8-containing construct but not the defense system. Results of three experiments are presented as individual curves. X-axis represents minutes post arabinose induction. (B) In vitro pull-down assay using MBP-tagged proteins demonstrating lack of interaction between T5 pb8 and the E. coli E831 PycC cyclase. EcPycC was expressed with an N-terminal 6×His–MBP–SUMO2 tag and the resulting full-length protein has a predicted molecular weight of ~105 kDa. The lower molecular weight bands correspond to truncated 6×His–MBP species co-purified during Ni-NTA purification. (C,D) The impact of pb8 T5 phage capsid protein on cyclase activity of EcPycC was assessed via thin-layer chromatography. Reactions contained 1 μM EcPycC and 0, 1, or 5 μM purified pb8 in buffer containing 50 mM Tris pH 8.5, 1 mM MnCl2, 100 mM KCl, 500 μM CTP, and trace α−32P-CTP. Reactions were incubated for the indicated times at 37°C, quenched at 85°C for 2 min, cooled on ice for 2 min, then treated with CIP for 30 min at 37°C to digest uncyclized CTP. (C) A representative TLC image. (D) The percent fractional signal associated with cCMP production from three independent experiments was quantified and average values are plotted, with individual data points overlaid. (E,F) A competition fitness assay between WT and mutant T5 phages (mutant n.1 and mutant n.4 for panels C and D, respectively) on both Pycsar-expressing and control cells. Y-axis represents the fraction of each strain out of the total phage mixture based on sequenced DNA reads. X-axis represents the infection passage, with 0 representing the mixture prior to first passage. In each passage, phage lysate was taken and used for infection of a fresh bacterial culture.
Table S1. Pycsar systems identified in this study (provided as an Excel file), related to figure 2.
Table S2. Mutant phages isolated in this study (provided as an Excel file), related to figure 6.
Table S4. Synthesized sequences and primers used in this study (provided as an Excel file), related to STAR methods.
Data Availability Statement
Crystal structure of BcPycC was deposited at the protein data base under identifier PDB: 7R65, and is publicly available as of the date of publication.