Abstract
The endoplasmic reticulum (ER)-located ATP/ADP-antiporter (ER-ANT1) occurs specifically in vascular plants. Structurally different transporters mediate energy provision to the ER, but the cellular function of ER-ANT1 is still unknown. Arabidopsis (Arabidopsis thaliana) mutants lacking ER-ANT1 (er-ant1 plants) exhibit a photorespiratory phenotype accompanied by high glycine levels and stunted growth, pointing to an inhibition of glycine decarboxylase (GDC). To reveal whether it is possible to suppress this marked phenotype, we exploited the power of a forward genetic screen. Absence of a so far uncharacterized member of the HaloAcid Dehalogenase (HAD)-like hydrolase family strongly suppressed the dwarf phenotype of er-ant1 plants. Localization studies suggested that the corresponding protein locates to chloroplasts, and activity assays showed that the enzyme dephosphorylates, with high substrate affinity, the B6 vitamer pyridoxal 5′-phosphate (PLP). Additional physiological experiments identified imbalances in vitamin B6 homeostasis in er-ant1 mutants. Our data suggest that impaired chloroplast metabolism, but not decreased GDC activity, causes the er-ant1 mutant dwarf phenotype. We present a hypothesis, setting transport of PLP by ER-ANT1 and chloroplastic PLP dephosphorylation in the cellular context. With the identification of this HAD-type PLP phosphatase, we also provide insight into B6 vitamer homeostasis.
Knocking out the activity of a pyridoxal 5′-phosphate phosphatase suppresses the dwarf phenotype of mutants lacking an ATP/ADP transporter and provides insight into vitamin B6 homeostasis.
Introduction
Nucleotides represent building blocks of DNA and RNA may activate metabolic precursors during polymer production and act as cofactors in enzymatic reactions or as second messengers in signal cascades (Roux and Steinebrunner, 2007; Geigenberger et al., 2010; Möhlmann et al., 2014). Among the different nucleotides, ATP plays a central role because it is the major energy currency of the cell. It acts as cosubstrate in a multitude of different reactions and thus is required in almost every cell compartment. However, in most eukaryotes, ATP regeneration from ADP and phosphate mainly takes place in mitochondria. Consequently, ATP has to be transported from the site of its production to the compartment where needed (Klingenberg, 1989). ADP/ATP carriers (AACs), the most abundant transporters in the inner mitochondrial membrane, catalyze the export of ATP, supplying the cytosol and, indirectly, other organelles with mitochondrial energy (Millar and Heazlewood, 2003). The concomitant ADP import provides one substrate for oxidative phosphorylation, whereas the second substrate (inorganic phosphate) enters the mitochondrion via specific phosphate carriers.
Besides mitochondrial energy metabolism, plants can regenerate high amounts of ATP in chloroplasts during photosynthesis. Therefore, chloroplasts appear energetically quite independent since photosynthetic ATP provides the energy for anabolic processes in the stroma. Heterotrophic plastids, however, rely on ATP uptake from the cytosol and the same holds true for chloroplasts in the night or during phases of limited photosynthetic activity (Tjaden et al., 1998; Reiser et al., 2004; Reinhold et al., 2007). The corresponding import is catalyzed by members of the major facilitator superfamily, the so-called nucleoside triphosphate transporters (NTTs) (Winkler and Neuhaus, 1999; Haferkamp et al., 2011). Interestingly, NTT-mediated ATP uptake occurs in exchange with ADP and phosphate, and thus energy provision to the plastid enables the subsequent export of the two products of energy consumption (Trentmann et al., 2008).
The endoplasmic reticulum (ER) harbors several energy-demanding processes such as protein folding, protein maturation, and protein quality control, but lacks the capacity for ATP regeneration (Guillén and Hirschberg, 1995). Therefore, this organelle essentially relies on the ATP uptake from the cytosol (Mayinger and Meyer, 1993). Although substantial ATP passage across the ER membrane was demonstrated as early as 1992 (Clairmont et al., 1992), the molecular nature of the corresponding facilitator protein remained unknown for a long time.
In 2008, an ATP/ADP antiporter was identified in the ER of Arabidopsis (Arabidopsis thaliana) (ER-ANT1) and thus could be considered as the first promising candidate. Structurally, this protein represents a homolog to the mitochondrial AACs and catalyzes, when recombinantly expressed in Escherichia coli, the exchange of ATP and ADP (Leroch et al., 2008). The corresponding gene shows increased expression in organs and tissues with high protein folding activity in the ER, and loss-of-function mutants (er-ant1) exhibit decreased transcription of ATP-dependent ER enzymes, as well as less lipid and storage proteins in seeds (Leroch et al., 2008). These observations tempted us to speculate that ER-ANT1 proteins, which have been identified at the molecular level in the representative mono and dicotyledoneous plants, rice (Oryza sativa) and Arabidopsis, respectively, fulfill functions in energy metabolism and ATP dependent processes in the ER lumen (Leroch et al., 2008; Zhang et al., 2016). However, several later observations suggest that ER-ANT1 does not represent the universal long-sought major ATP supplier of the ER because energy provision to the lumen is required in all eukaryotes, but ER-ANT1 homologs are restricted to vascular plants (Haferkamp and Schmitz-Esser, 2012; Hoffmann et al., 2013). Thus, even in the absence of ER-ANT1 sufficient ATP supply to the ER is guaranteed, and is accordingly mediated by an alternative transport system (Leroch et al., 2008; Hoffmann et al., 2013).
In fact, recent studies identified ATP/ADP exchanger in the ER membrane (AXER) as the major ATP/ADP exchanger in this organelle in mammalian cells (Klein et al., 2018). The corresponding protein belongs to the solute carrier family and is not an AAC-type protein. It was shown that this ATP/ADP exchanger is of high relevance for the ER energy metabolism and homologs to AXER exist in all eukaryotes, including plants (Klein et al., 2018). Thus, since AXER catalyzes the main ATP provision to the ER the exact physiological function of ER-ANT1 is unknown. Moreover, from an evolutionary point of view it is hard to envision why a mitochondrial AAC homolog was recruited to the ER to deliver ATP (given the subsequently established function of AXER) and, in addition, it is unclear why solely vascular plants would possess two different types of ATP/ADP transporters.
However, independent of the exact physiological role of ER-ANT1, its importance for the plant is beyond doubt (Leroch et al., 2008). This is, because Arabidopsis er-ant1 knock-out lines (er-ant1) exhibit a dwarf phenotype and decreased chlorophyll levels when grown at ambient CO2 concentrations (Leroch et al., 2008). Moreover, er-ant1 mutants accumulate the photorespiratory key-intermediate glycine, are impaired in CO2 fixation, and exhibit a higher CO2 compensation point than wild-types (wts; Hoffmann et al., 2013), which are typical characteristics for many photorespiration mutants (Bauwe et al., 2010). Furthermore, while er-ant1 Arabidopsis and rice mutants suffer severely under ambient air, they are rather unaffected under conditions of high CO2 availability (Hoffmann et al., 2013; Zhang et al., 2016), which is also consistent with other photorespiratory mutants (Bauwe et al., 2010).
Thus, the current data suggest that ER-ANT1 may at least contribute to the energy metabolism of the ER and fulfills a specific role in vascular plants that is somehow connected to photorespiration. However, although the different enzymatic steps of the photorespiratory pathway are distributed among several cellular compartments, none are associated with the ER membrane or its lumen (Timm et al., 2016). Therefore, the link between ER-mediated adenine nucleotide transport, catalyzed by ER-ANT1, and the photorespiratory phenotype of the loss-of-function mutant remains enigmatic.
To shed light on the plant-specific function of ER-ANT1 and how its absence might affect photorespiration we conducted a suppressor screen of er-ant1. Alleviation of the growth defect of the ER-ANT1 loss-of-function mutant could be achieved by knocking out the activity of a further protein, namely a putative HaloAcid Dehalogenase (HAD)-like hydrolase. Interestingly, the latter protein specifically acts on pyridoxal 5′-phosphate (PLP), identifying the HAD-like hydrolase as a PLP phosphatase (PLPP). Further studies demonstrate that changes in the concentration of PLP are causative for the er-ant1 phenotype and localization experiments revealed that this protein locates in chloroplasts. The analyses conducted lead to the hypothesis that ER-ANT1 influences subcellular B6 vitamer levels, connecting an ER-located PLP reserve to the rest of the cell. In addition, we provide evidence for an active dephosphorylation of PLP in chloroplasts which contributes to vitamin B6 homeostasis.
Results
Suppressor mutants of er-ant1 are phenotypically diverse
To gain further insights into the properties of er-ant1 mutants, we conducted mutagenesis of er-ant1 plants with ethyl methanesulfonate (EMS). The resulting M1 plants were grown in pools of ∼500 plants to produce the M2 generation of seeds. The progeny was screened for putative suppressors of the er-ant1 phenotype (termed ser-ant1). For this, M2 seeds were cultivated at ambient air on soil for 3 weeks and then screened visually for plants substantially larger than er-ant1. This strategy led to the identification of several putative ser-ant1 mutants. False positives were excluded by confirmation of the er-ant1 T-DNA insertion and after self-pollination of the isolated M2 ser-ant1 mutants, we used 13 lines of the M3 generation for further characterization and mapping.
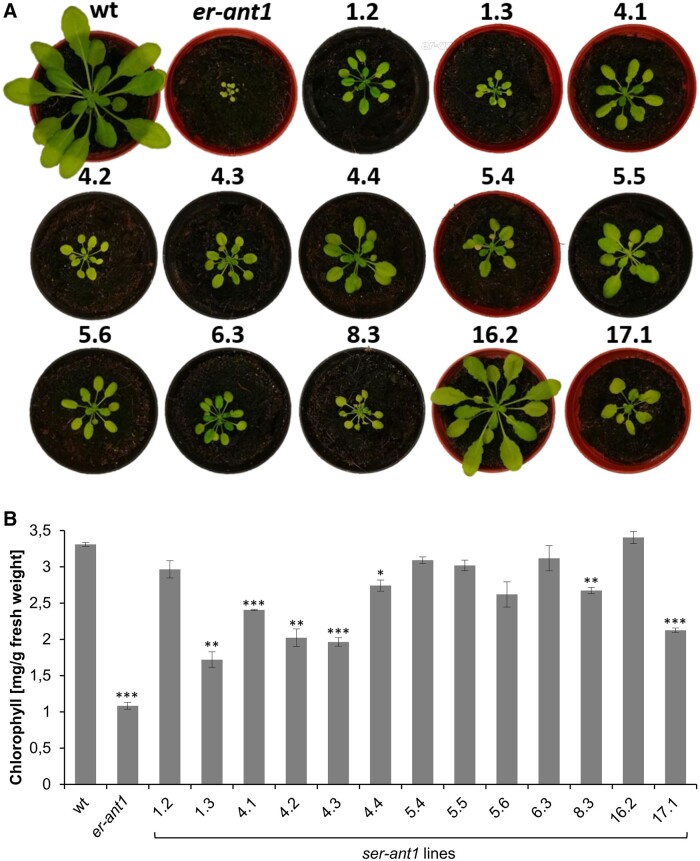
During visual inspection of these M3 plants, it became directly evident that the individual ser-ant1 lines differ in terms of rosette size, coloration, and shape of leaves (Figure 1A). This heterogeneous appearance is suggestive of genetic variations among the different lines. Although all selected M3 ser-ant1 mutants were larger, appeared greener, and developed more leaves than er-ant1 mother plants, they did not reach the wt level. Solely line 16.2 was almost as large and as green as wt, but showed a more pronounced leaf serration. Line 1.3 was only slightly larger than er-ant1 plants, whereas the remaining lines exhibited intermediate sizes. The young leaves in the center of the rosette of several ser-ant1 lines were of normal green coloration, while older leaves appeared comparatively pale. The young leaves of line 6.3 appeared even darker green than those of wt. Line 5.4 had strikingly yellow cotyledons, whereas line 4.2 was generally very pale and, in this context, highly resembled the phenotype of er-ant1 mutants.
Figure 1.
Suppression of the er-ant1 phenotype by ser-ant1 mutations. A, Suppressor mutants selected for further characterization and mapping. Photographs show representative 28-d-old third-generation (M3) plants for each selected mutant line, plants were grown on soil at ambient air. Images were digitally extracted for comparison. B, Total chlorophyll content in leaves of er-ant1 and M3 ser-ant1 plants relative to the wt. Mean values of three individual replicates ± se. Asterisks indicate the significance level between wt and mutant plants according to a Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
To analyze whether the apparent suppression of chlorosis in the different lines becomes reflected by an increase in chlorophyll per fresh weight (FW), we quantified this pigment in the ser-ant1 lines, er-ant1, and the wt (Figure 1B). Generally, the chlorophyll content of er-ant1 is low and makes up only about a third of wt levels. Notably, all tested ser-ant1 lines show higher chlorophyll contents when compared with er-ant1. They reached at least 50% of the wt level and five of the selected 13 lines even approach it (≥90% of wt). Interestingly, the chlorophyll content of the smallest line 1.3 represents the lowest among the ser-ant1 mutants, while the chlorophyll level of line 16.2 is nearly identical to that of wts. Therefore, suppression of the er-ant1 growth defect is generally accompanied by an increase in chlorophyll. However, even though their chlorophyll content was almost fully recovered many plants were still substantially smaller than the wt.
Suppressor mutants exhibit better photosynthetic performance than er-ant1
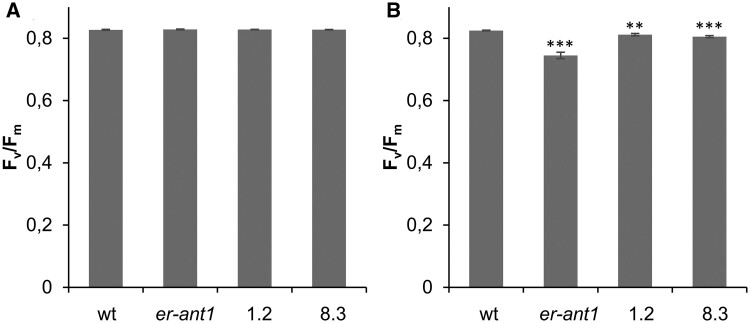
Previous analyses revealed that er-ant1 is substantially impaired in photosynthesis (Hoffmann et al., 2013). Its limited photosynthetic capacity is presumably causative, or at least contributes to the growth retardation typical for this mutant plant. To check whether suppressor lines not only contain more chlorophyll but also show higher photosynthetic capacities than er-ant1, we determined their maximum quantum yield of photosystem II (PSII), represented by (Fv/Fm). For this, we selected two suppressor lines (no. 8.3 and no. 1.2) exhibiting almost complete recovery of the chlorophyll contents, namely 80%–90% of the wt level. We detected no significant differences between the Fv/Fm values of the wt, er-ant1, and the two suppressor lines when plants were cultivated under high CO2 (Figure 2A). Their Fv/Fm values were at ∼0.83 (Figure 2A) which is indicative of unstressed plants (Demmig and Björkman, 1987).
Figure 2.
Maximum quantum yield of PSII (Fv/Fm) of wt, er-ant1, and two selected M3 ser-ant1 plants. A, Fv/Fm at 2,000 parts per million (ppm) CO2. B, Fv/Fm at ambient air. Data represent mean values of seven individual replicates, ±se. Asterisks indicate the significance level between wt and mutant plants according to a Student’s t test (**P < 0.01, ***P < 0.001).
Exposure to ambient CO2 conditions for 5 d did not alter the maximum quantum yield of PSII in the wt. However, a significant decrease of the maximal quantum yield became apparent in er-ant1 mutants (Fv/Fm ∼0.74; Figure 2B) whereas the two suppressor lines were less affected (Fv/Fm ∼0.8). The improved growth of the suppressor lines thus can be attributed to their increased chlorophyll contents and higher photosynthetic capacities when compared with er-ant1.
Suppressor mutants still accumulate glycine
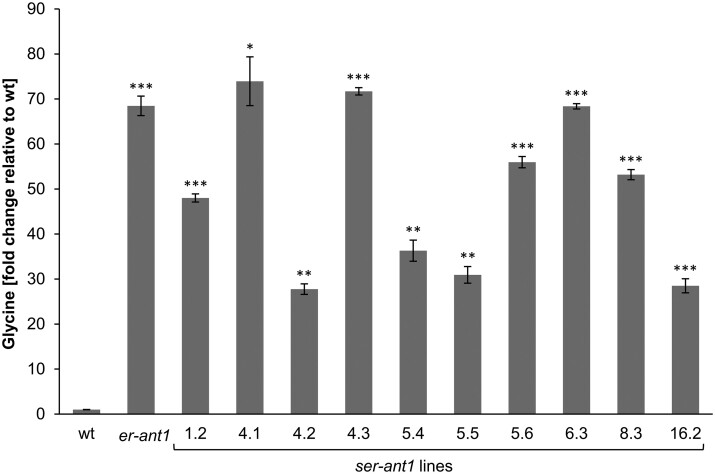
Another striking characteristic of er-ant1 is its marked glycine accumulation in leaves (Hoffmann et al., 2013). Since some ser-ant1 lines show restored growth and chlorophyll contents, as well as improved photosynthesis, we were interested in whether they accumulate less glycine than er-ant1 mutants. Interestingly, not only did er-ant1 plants contain approximately about 70-fold more glycine than the wt plants, but all tested ser-ant1 lines also accumulated substantial amounts of this amino acid. Their glycine concentration was at least 30-fold higher than in wt plants and several plants even contained as much glycine as in the original er-ant1 mutant (Figure 3).
Figure 3.
Glycine content of leaves from er-ant1 and ser-ant1 mutants relative to the wt level. Plants were cultivated at ambient CO2 conditions. The glycine content of the wt was set to 1. Data represent mean values of five individual replicates ± se. Asterisks indicate the significance between the glycine levels of wt and mutant plants according to a Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
Identification of ser-ant1 loci
To identify the suppression causing mutations in individual ser-ant1 lines, we used the mapping-by-sequencing approach (James et al., 2013). To this end, the homozygous ser-ant1 lines were backcrossed (BC1) individually with the parental er-ant1 T-DNA insertion line that was used for generating the EMS suppressor mutant collection. The fact that all individuals of the BC1-F1 progeny displayed the dwarf er-ant1 phenotype, whereas the BC1-F2 progeny segregated in a 1:3 ratio of ser-ant1- to er-ant1 phenotypes suggests that the suppressor mutations are recessive. After self-pollination, the BC1-F2 segregating populations (∼500 plants per line) were analyzed for the ser-ant1 phenotype. Subsequently, two bulked pools of plants per line displaying either the ser-ant1- or the er-ant1 phenotype were created, each consisting of 50 individuals. Pooled genomic DNA from each bulk was sequenced with average depths in the range of 41× to 56×. The fragmented sequences were mapped to the reference genome (Arabidopsis TAIR10) and the identified EMS mutation variants were analyzed (and filtered) to exclude noncausative mutations. By analyzing the allelic variant frequencies (AFs) it became clear that nine ser-ant1 lines carry the suppression causing mutation in a specific region at chromosome 2. Interestingly, seven of these lines possessed the highest AFs for variants in the genetic information of At2g33255 (Supplemental Table S1). Therefore, we considered mutations in this gene as the most promising candidate suppressors. In line 6.3, the mutation resulted in a premature stop codon, which suggests that the alleviation of the er-ant1 phenotype is due to activity loss of the corresponding protein (Supplemental Figure S1). To rule out that the identified single-nucleotide polymorphism (SNP) in At2g33255 is a false-positive, the mutation was checked by Sanger-sequencing of gDNA from individuals of the 6.3 M3 generation (Supplemental Figure S2).
Confirmation of the candidate suppressor mutations in At2g33255 by reverse genetics
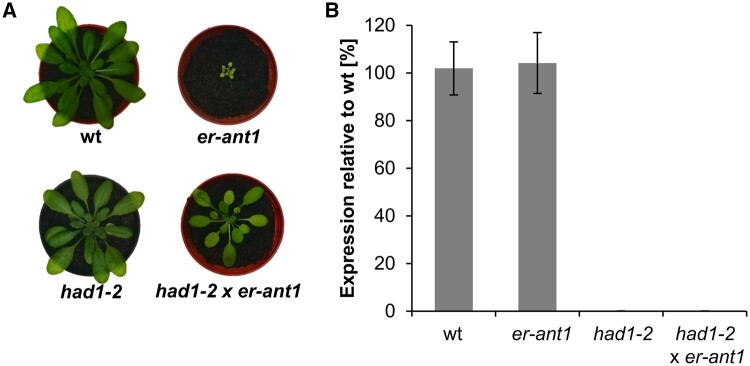
We used reverse genetics to investigate whether loss of function of the At2g33255 encoded protein is causative for the observed phenotype suppression. For this, three individual At2g33255 T-DNA insertion lines were ordered and by the help of reverse transcription-quantitative PCR (RT-qPCR) we verified that two of them (termed had1-2 and had1-3, see below) represent true loss-of-function (knock out) mutants, whereas one (had1-1) still contained almost 50% of the wt transcript level (Supplemental Figure S3A). Morphologically, had1-2 and had1-3 were indistinguishable from wt plants (Supplemental Figure S3B). We used the had1-2 line for crossing with er-ant1. When cultivated under ambient CO2 levels, the segregating F2 progeny exhibited either the had1-2 (wt like) phenotype, the original er-ant1 (dwarf) phenotype, or an intermediate size (semi-size) phenotype (Figure 4A). Using a T-DNA screen confirmed that exclusively the semi-sized plants were homozygous for T-DNA insertions in both the er-ant1 and the At2g33255 gene. Subsequently, homozygous F3-plants were analyzed for expression of the At2g33255 gene by RT-qPCR (Figure 4B). The absence of the corresponding transcript demonstrates that the semi-sized plants represent true had1-2 x er-ant1 double knockouts (Figure 4B). This observation demonstrates that absence of the At2g33255 transcript alleviates the growth defect of er-ant1. To further confirm that the absence of HAD1 activity is causative for alleviating the dwarf phenotype, we reintroduced the active HAD1 by expression of the wt HAD1 gene under either the constitutive ubiquitin promotor or the endogenous HAD1 promotor, named ubiq::HAD1 x had1-2 x er-ant1 and promHAD1::HAD1 x had1-2 x er-ant1, respectively (Supplemental Figure S4). Several homozygous ubiq::HAD1 x had1-2 x er-ant1- and promHAD1::HAD1 x had1-2 x er-ant1 triple mutant lines exhibited increased HAD1 mRNA, appeared morphologically markedly smaller and showed pale leaves (Supplemental Figure S4, A and B), as is typical for the original er-ant1 line (Figure 4A; Leroch et al., 2008).
Figure 4.
Absence of the At2g33255 transcript suppresses the growth defect of er-ant1. A, Phenotypic comparison of 4 weeks old plants grown at ambient CO2. Images were digitally extracted for comparison. B, At2g33255 gene expression analyzed by RT-qPCR. RNA for cDNA synthesis was extracted from leaves of 3 weeks old plants. Data were normalized to the SAND (At2g28390) housekeeping gene. Data represent mean values of three biological replicates ± se.
At2g33255 encodes a PLPP
Although the T-DNA insertion in At2g33255 was proven as causative to suppress dwarf growth of er-ant1 mutants (Figure 4A) the physiological background for this finding was completely unclear, particularly because the function of the corresponding enzyme is unknown. The protein encoded by At2g33255 is annotated as a member of the HAD-like superfamily (HADSF). Generally, the HADSF comprises proteins with a conserved α/β-domain, which shares similarity with the Rossmann fold and is termed “hydrolase fold” (Burroughs et al., 2006). Interestingly, different from the implication of its name, the HADSF consists predominantly of phosphatases (∼79%) and ATPases (∼20%), whereas dehalogenases as well as phosphonatases, phosphomutases, and phosphomannomutases represent the minority of the protein superfamily members (Koonin and Tatusov, 1994; Kuznetsova et al., 2015).
Within the HADSF the protein encoded by At2g33255 belongs to the HAD subfamily IA. Representatives of this subfamily are characterized by a small α-helical cap between loop 1 and loop 2 of the core domain. In Arabidopsis, the HADSF subfamily IA is formed by 19 proteins (IPR006439) (Krishnakumar et al., 2014; Mitchell et al., 2018) and 7 of these proteins have been characterized as phosphatases (Caparrós-Martín et al., 2013). Therefore, it appeared likely that At2g33255 also encodes a phosphatase. Phosphatases are generally not very substrate-specific and often accept various phosphorylated substrates. However, a previous screen by others using dl-glycerol-3-phosphate, d-fructose-6-phosphate, d-fructose-1-phosphate, d-fructose-1,6-bisphosphate, d-glucose-6-phosphate, α-d-glucose-1-phosphate, 2-deoxy-d-glucose-6-phosphate, d-mannose-6-phosphate, or α-d-mannose-1-phosphate failed to detect any phosphatase activity of the At2g33255 gene product (Caparrós-Martín et al., 2013).
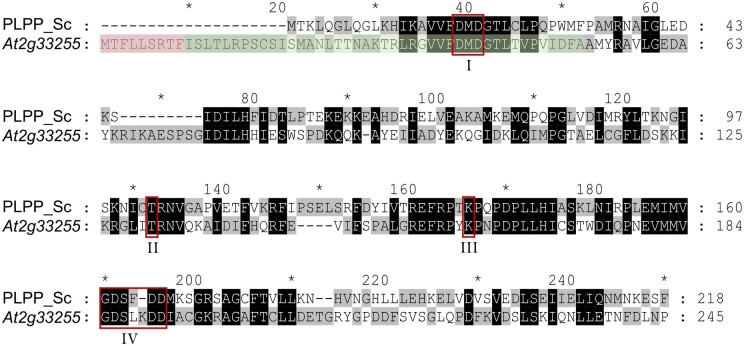
To get an idea about its possible function, we used its predicted amino acid sequence for NCBI Blast searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against nonplant genomes. Interestingly, we identified high similarities to the YOR131C-like HADSF members from Saccharomyces cerevisiae (baker’s yeast). YOR131C was identified as a PLPP (Kuznetsova et al., 2015). Due to the considerable amino acid similarity of the predicted protein sequence of At2g33255 to YOR131C (44%; Figure 5), we hypothesized that it might also exhibit hydrolytic activity against PLP.
Figure 5.
ClustalW alignment of the amino acid sequences of the PLPP from S. cerevisiae (PLPP_Sc; YOR131C) and the At2g33255 gene product. Residues identical among the two sequences or with similar properties are indicated by black shading. Residues not identical are highlighted by different shading (gray/white). Red boxes mark conserved HADSF motifs I–IV (I: DxD; II, T/S; III: K/R; IV: E/DD, GDxxxD, or GDxxxxD). The putative mitochondrial targeting sequence (according to TargetP) is shaded in red. The predicted chloroplast targeting sequence (according to ChloroP) comprises also the green shaded residues. Dashes represent introduced gaps for alignment improvement. Numbers at the right indicate amino acid positions. The amino acid sequence alignment also demonstrates that the At2g33255 encoded protein is N-terminally extended by 20 residues when compared with the cytosolically located PLPP YOR131C from S. cerevisiae. Such extensions often act as targeting sequences signifying the import of proteins into mitochondria or chloroplasts. Notably, the At2g33255 encoded protein does not share substantial sequence similarity (25%) to a recently identified chloroplastic PLP phosphatase from N. tabacum, named NtPLPP1 (ShuoHao et al., 2019; Supplemental Figure S5).
Subcellular localization of the At2g33255 encoded protein
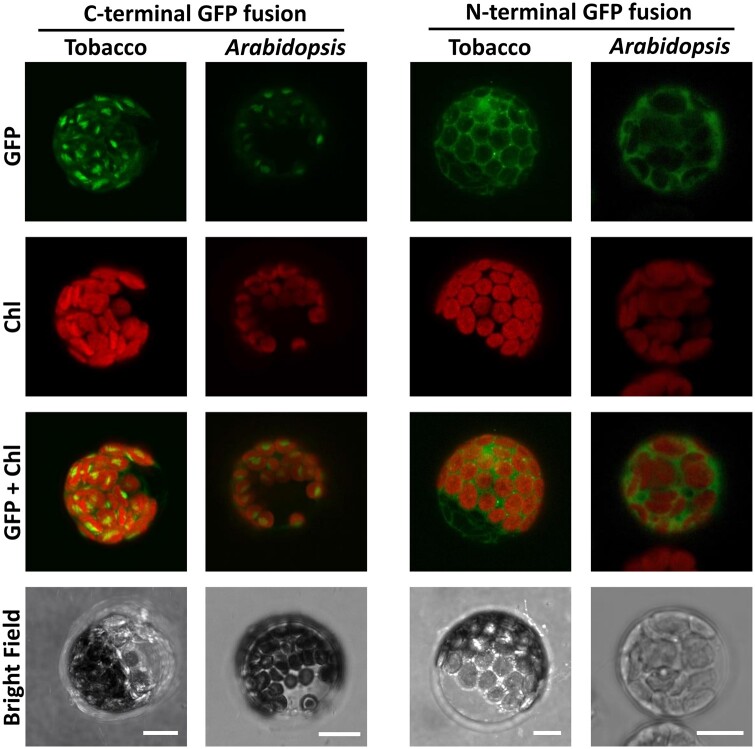
The N-terminal extension of the amino acid sequence of At2g33255, when compared with the yeast homolog (Figure 5), points to a mitochondrial or plastidic targeting of the encoded protein. This putative localization is supported by in silico prediction programs, however, with quite moderate reliability scores. Thus, to clarify the ambiguous localization we transiently expressed the At2g33255 sequence in either Nicotiana benthamiana or Arabidopsis protoplasts as a translational fusion to green fluorescent protein (GFP), either at the N or the C terminus. Protoplast transformation with the C-terminally attached marker protein resulted in weak green fluorescence surrounding the position of the chloroplast. The strongest signal intensity, however, was detectable in the center of the organelle (Figure 6) indicating accumulation of the GFP fusion protein in the stroma. With GFP attached to the N-terminus, the fluorescence signal of the fusion protein was distributed throughout the cell but not detectable in chloroplasts (Figure 6). This observation suggests that the N-terminally attached GFP masks the chloroplast targeting signal of the HAD-type hydrolase and by this hinders its entry into the organelle.
Figure 6.
Localization of the HAD-type hydrolase in N. benthamiana and Arabidopsis protoplasts. Pictures were taken with a confocal laser-scanning microscope 16 h after transient transformation. Scale bars = 10 μm. Chl, chlorophyll fluorescence.
The uncharacterized HAD-type hydrolase from Arabidopsis acts as a PLPP
The sequence similarities between the yeast PLPP (YOR131C) and the HAD-type hydrolase are (Figure 5) indicative of a related enzymatic activity. To gain insights into the biochemical properties of the HAD-type hydrolase from Arabidopsis, the protein was heterologously expressed in E. coli, purified to apparent homogeneity via immobilized metal affinity chromatography (IMAC; Supplemental Figure S6) and subsequently used in enzymatic activity tests.
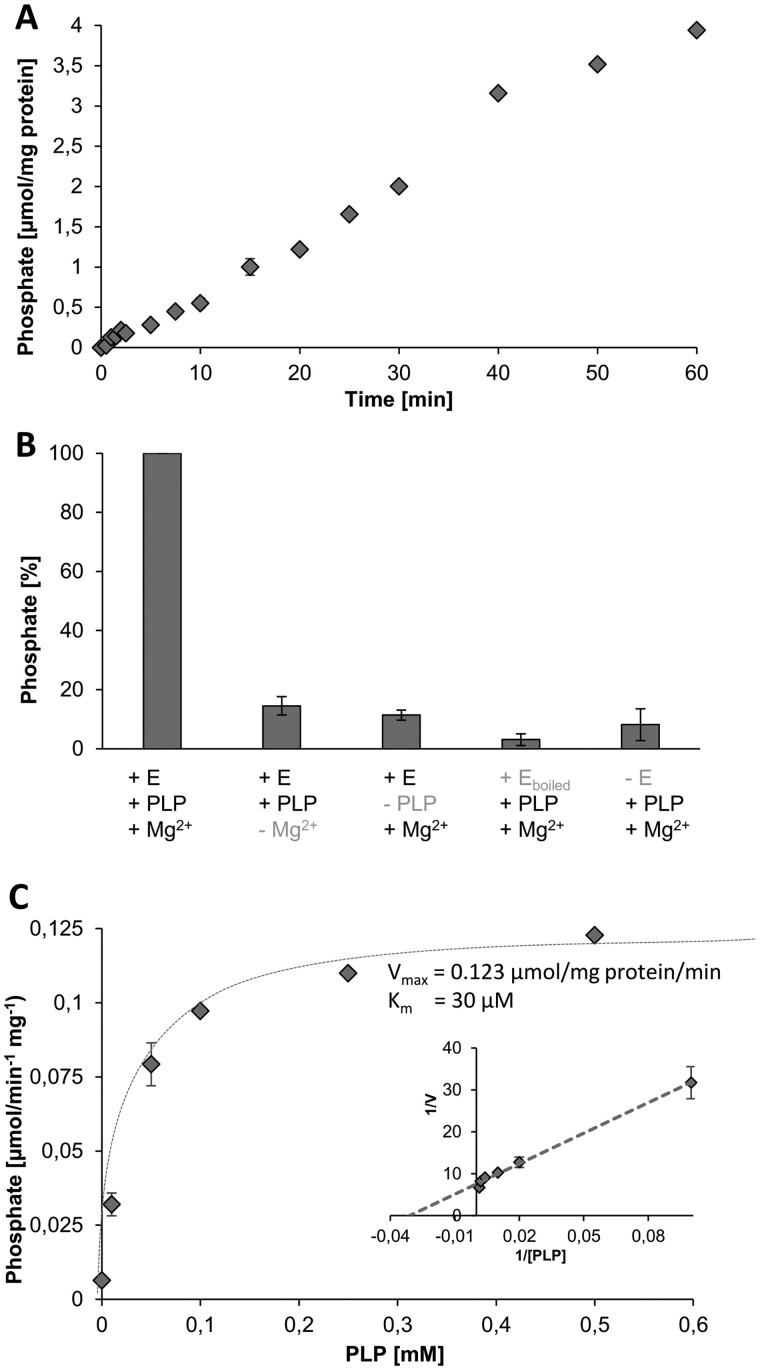
Interestingly, in the presence of PLP, we observed measurable phosphate release. The generation of free phosphate increased nearly linearly with time and reached values of about 3.9 µmol mg protein−1 h−1 (Figure 7A). Omitting magnesium (Mg2+) from the test medium substantially decreased phosphate release (Figure 7B) and no enzymatic activity was detectable when PLP was absent (Figure 7B). Similarly, use of heat-denatured enzyme or lack of enzyme in the test only led to background levels of phosphate release (Figure 7B). These observations demonstrate that (1) phosphate release depends on the activity of the recombinant protein, (2) phosphate is released from PLP, and (3) the enzyme activity depends markedly upon the presence of Mg2+, which is a typical feature of HADSF-type phosphatases (Kuznetsova et al., 2015). Because of the PLPP activity, we term the HAD-type hydrolase At2g33255 from now on AtPLPP1.
Figure 7.
Enzyme assays verified the PLPP function of the At2g33255 encoded protein. A, Time course of phosphate release from PLP (0.2 mM). B, Phosphate release is dependent on the active enzyme (+E), presence of PLP (0.2 mM), and Mg2+. Each assay time was 30 min. C, Michaelis–Menten curve indicating substrate saturation of enzyme activity, the inset shows a corresponding Lineweaver–Burk analysis indicating an apparent Km for PLP of 30 µM and a Vmax of 0.123 µmol mg protein−1 min−1. Each assay time was 30 min. Shown are mean values of four replicates ± se.
A previous study failed to detect any hydrolytic activity of the At2g33255 encoded enzyme with nine different substrates (Caparrós-Martín et al., 2013), pointing to a notable substrate specificity of the protein. However, it is important to mention that PLP was not among the previously tested compounds (Caparrós-Martín et al., 2013). The fact that we could observe phosphate release from PLP shows that the recombinant enzyme is active, which is an important prerequisite for the investigation of its substrate preference. Therefore, we checked whether the AtPLPP1 exhibits hydrolytic activity toward a range of other phosphorylated compounds (Table 1). Six of the 13 tested metabolites did not serve as substrates. All other compounds tested showed ˂10% of the phosphate release observed when using PLP as substrate (Table 1), with the exception of ATP (30.5%). These observations support the conclusion that At2g33255 acts as a AtPLPP1. We also analyzed the rate of phosphate release as a function of the PLP concentration allowing us to determine an apparent Km value of 30 µM, and a Vmax of the purified protein of 0.123 µmol mg protein−1 min−1 (Figure 7C, inset therein).
Table 1.
Enzymatic activity of At2g33255 on different putative substrates
| Tested substrate | Phosphate release as % |
|---|---|
| 6-phosphogluconic acid | n.d. |
| d-Fructose-6-phosphate | n.d. |
| d-Fructose-1,6-bisphosphate | n.d. |
| d-Glucose-6-phosphate | 3.5 |
| α-d-glucose-1-phosphate | n.d. |
| Glyceraldehyde 3-phosphate | n.d. |
| 3-Phosphoglyceric acid | 6.5 |
| Dihydroxyacetone-phosphate | 9.0 |
| Phosphoenolpyruvate | n.d. |
| Adenosine triphosphate | 30.5 |
| Adenosine diphosphate | 1.8 |
| Adenosine monophosphate | 3.2 |
| PLP | 100.0 |
Notes: Assays were performed with 1 mM substrate and 6.25 µg of the recombinant phosphatase per 0.5 mL. A colorimetric change of the assay mix indicated release of phosphate after 30 min of incubation at 23°C. n.d., not detectable ≤ 1%.
Indications for a perturbed B6 vitamer metabolism in er-ant1
The group of B6 vitamins is composed of six vitamers: pyridoxal (PL), pyridoxine (PN) and pyridoxamine (PM), and their phosphorylated forms PLP, PNP, and PM 5′-phosphate (PMP) (Fitzpatrick et al., 2007; Hellmann and Mooney, 2010). PLP, acting as an essential coenzyme for a wide range of enzymes, can be biosynthesized de novo in the cytosol (Tambasco-Studart et al., 2005). Salvage pathways allow for interconversion of the different vitamers (Parra et al., 2018) and are distributed throughout the cellular organelles (Rueschhoff et al 2012; Colinas et al., 2016).
Here, the localization studies suggest that AtPLPP1 is targeted to the chloroplast (Figure 6). Consequently, absence of this enzyme would decrease the rate of PLP hydrolysis in the stroma and by this could increase PLP availability in this organelle. The observed suppression of dwarfism by activity loss of the AtPLPP1 might thus point to a perturbed cellular B6 vitamer metabolism in er-ant1 mutant plants that is alleviated by PLP availability.
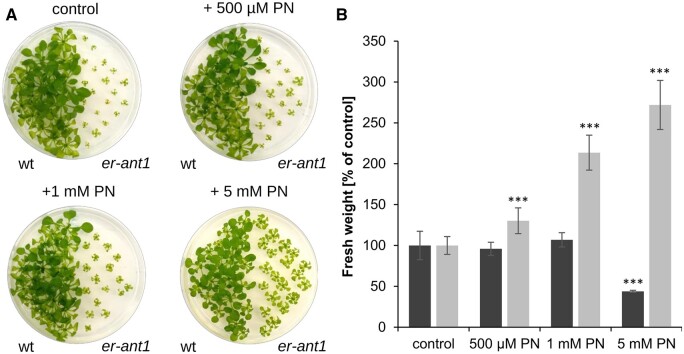
If this hypothesis is true, exogenously supplied B6 vitamers might be beneficial for mutant development. To test this assumption, we cultivated er-ant1 and wt seedlings on agar plates supplemented with increasing concentrations of the B6 vitamer PN. In this context, it is important to note that solely nonphosphorylated B6 vitamers can enter the cell (Rueschhoff et al., 2012) and that PN supplements B6 vitamin metabolism since it can act as a precursor for PLP generation (Rueschhoff et al., 2012). The feeding assay revealed that the growth of wt seedlings remained unaltered by PN additions up to 1 mM (Figure 8A). However, the highest PN concentration used (5 mM) had a negative impact on growth of the wt seedlings and led to a substantial decrease in biomass (˂50% of the corresponding control, 0 mM PN, Figure 8B). In contrast, er-ant1 seedlings grew much better in the presence of PN supplementation. Their size and biomass increased with rising PN concentrations (Figure 8, A and B) and the highest PN concentration tested resulted in almost three-fold higher biomass of the er-ant1 seedlings when compared with the corresponding control (0 mM PN; Figure 8B). This stimulatory effect of PN feeding on seedling growth is thus indicative for limiting levels of the essential vitamin B6 in er-ant1 plants.
Figure 8.
Effect of PN feeding on growth of er-ant1 and wt plants. Twenty plants were cultivated on agar plates (0.5 MS including GA5 vitamins, 0.8% agar) at ambient CO2 conditions (10-h light/14-h dark) for 25 d. A, wts and er-ant1 plants on agar plates without additional PN (control) and with different concentrations of additional PN. B, FW of er-ant1 (light gray) and corresponding wt plants (dark gray) grown on agar plates containing different concentrations of PN. The FW is given as a percentage of the respective control FW. Shown are mean values of at least five individual plates ± se. Asterisks indicate the significance level between wt and mutant plants according to a Student’s t test (***P < 0.001).
Except for glycine levels, the simultaneous loss of AtPLPP1 function largely restores the perturbed amino acid metabolism of er-ant1 mutants
Although alleviated in growth, the ser-ant1 mutants accumulate high amounts of glycine (Figure 3). The glycine content of line 6.3, with the premature stop codon in the At2g33255 gene, was even identical to that of er-ant1 (Figure 3). To check whether this is also true for the had1-2 x er-ant1 double knock-out mutant, we quantified its glycine concentration and compared it with that of the wt and the two single knock-out plants. Moreover, we additionally determined the concentrations of all amino acids already known to be markedly altered in er-ant1 when compared with the wt (Hoffmann et al., 2013). The amino acid content in had1-2 and had1-3 generally resembled those of the wt (Table 2). As previously reported, besides its high glycine accumulation er-ant1 plants exhibit particularly high arginine (Arg) content, whereas the levels of aspartate (Asp), glutamate (Glu), and alanine (Ala) are substantially lower than present in wt leaves (Table 2). Interestingly, the absence of AtPLPP1 in er-ant1 (had1-2 x er-ant1 suppressor mutant plants) did not markedly reduce the glycine accumulation but restored the remaining amino acid concentrations to almost wt levels (Table 2).
Table 2.
Altered amino acid contents of had1-2, had1-3, er-ant1, and had1-2 x er-ant1 mutants when compared with wt plants
| Amino acid | had1-2 (%) | had1-3 (%) | er-ant1 (%) | had1-2 x er-ant1 (%) |
|---|---|---|---|---|
| Asp | 95 | 98 | 12*** | 93* |
| Glu | 104 | 93 | 24*** | 82* |
| Glycine | 158 | 102 | 6,847*** | 6363*** |
| Ala | 115 | 104 | 20*** | 96 |
| Arg | 109 | 122 | 847*** | 124 |
Notes: Plants were grown at 2,000 ppm CO2 for 4 weeks and subsequently shifted to ambient air for 5 d. Samples were taken on the fifth day, 5 h after onset of light. Shown are the mean values of four individual replicates. The obtained concentrations were normalized to the respective wt concentration (set to 100%). Asterisks indicate the significance levels between wt and mutants according to a Student’s t test.
P < 0.05,
P < 0.001.
Arg can be taken as an indicator for the plant nitrogen status (Winter et al., 2015). It appeared remarkable that er-ant1 plants contained ∼8.5 times more Arg when compared with wt, while the corresponding levels in mutant lines had1-2 and had1-3, as well as in the double mutant had1-2 x er-ant1 resembled wt concentrations (Table 2).
Insights into vitamin B6 metabolism of the different mutant plants
The data of this study led us to the conclusion that the HAD-type hydrolase acts as a PLPP in chloroplasts of Arabidopsis and that er-ant1 mutants might suffer from imbalanced B6 vitamer homeostasis (Figures 7 and 8). To identify possible changes in B6 vitamer contents, we determined the levels of PMP, PLP, PM, PN, and PL in wt-, single- and double-knockout plants (Figure 9, A and B).
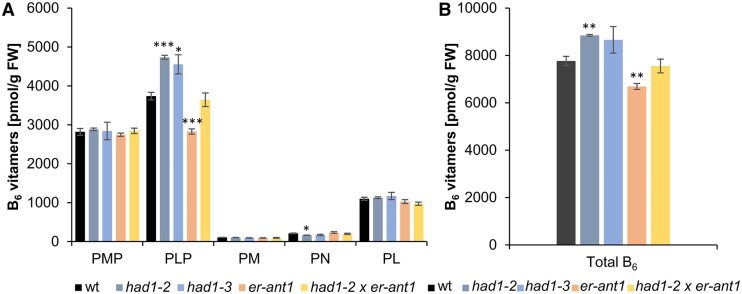
Figure 9.
Analysis of the B6 vitamer contents in the different plant lines. A, Analysis of the individual B6 vitamers PMP, PLP, PM, PN, and PL profile of the lines as indicated. B, Total vitamer contents (sum of PMP, PLP, PM, PN, and PL). Plants were grown at high CO2 (2,000 ppm) for 33 d to avoid pleiotropic effects on B6 vitamers in dwarf type er-ant1 plants. Data represent the mean of four replicates per line that was used for two extractions each. SEs are given. Statistical relevance was determined by a Student’s t test using the wt of the respective growth condition as a reference: *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005.
The levels of the low abundant B6 vitamers PM and PN range between 100 and 250 pmols g Fw−1 and are similar in all five plant lines analyzed (Figure 9A). Assuming that the newly identified AtPLPP1 dephosphorylates PLP it is not surprising that both HAD loss-of-function lines, had1-2, and had1-3, contained significantly increased PLP levels when compared with wt (Figure 9A). The corresponding levels of PL and PMP in both had1 mutants closely resembled wt concentrations (Figure 9A). er-ant1 mutants showed decreased PLP levels when compared with wts, while both PL and PMP concentrations were again similar to the corresponding concentrations found in wt (Figure 9A). The sum of all B6 vitamers in had1-2 plants is slightly increased, but decreased in er-ant1 plants when compared with the corresponding levels in wt (Figure 9B).
Discussion
Although ER-ANT1 was biochemically characterized more than a decade ago (Leroch et al., 2008) and although corresponding loss-of-function plants have been analyzed in detail (Hoffmann et al., 2013), the exact cellular role of this transporter remains elusive. It is particularly unclear how impaired ER-ANT1 activity in the membrane of the ER causes a defect in photorespiration, since a connection between these two processes was not evident. Thus, to gain deeper insights into the metabolism of the er-ant1 mutant and to enlighten the possible link between ER-ANT1 and photorespiration, we made use of the power of an EMS-based forward-genetic screen. By this approach, we identified a promising candidate mutation suppressing the marked growth defect of er-ant1 (Figure 1 and Supplemental Table S1). The corresponding gene variant led to a premature stop codon in a putative HAD-type hydrolase (Supplemental Figures S1 and S2), which suggests that loss of this enzyme activity suppresses the er-ant1 phenotype (Figures 1 and 3). In fact, subsequent reverse genetics confirmed that loss of the HAD-type phosphatase is causative for suppression of the dwarf growth typical for er-ant1 mutants (Figure 4 and Supplemental Figure S4, A and B).
Despite the positive outcome of the EMS screen, it was initially unclear why the missing activity of a putative HAD-type hydrolase exhibits suppressor function, since its subcellular localization and biological function were unknown. However, GFP-based studies on both N. benthamiana and Arabidopsis protoplasts were indicative for its location in the chloroplast stroma (Figure 6), which concurs with the presence of a putative organellar targeting-sequence extension at the N-terminal end of this protein, and which is absent from the structural homolog YOR131C from baker’s yeast (Figure 5). The affiliation of the enzyme to the HAD-type hydrolase superfamily suggested that it acts as a phosphatase, since most members of this protein group exhibit phosphatase activity (Burroughs et al., 2006). Moreover, the considerable amino acid sequence similarity of this protein to the PLPP YOR131C from baker’s yeast (44%; Figure 5; Kuznetsova et al., 2015) strongly supported the assumption of a related biochemical function in Arabidopsis chloroplasts. This assumption was supported by the positive effect of the PLP precursor PN on growth of er-ant1 mutants (Figure 8) suggesting that PLP was lacking in the latter mutants.
Accordingly, enzyme assays revealed that this HAD-type phosphatase is a functional and PLP-specific phosphatase (Table 1). The marked Mg2+ dependency of this enzyme (Figure 7B) supports to some degree its stromal location as the concentration of Mg2+ is high in chloroplasts when compared with all other plant cell organelles (Karley and White, 2009). However, Mg2+-dependent enzymes are not restricted to chloroplasts and we cannot rule out that a fraction of this novel enzyme locates outside the chloroplast, as corresponding GFP signals might be too weak to be detected.
That the newly identified protein is not only functional in enzyme activity tests (Figure 7, A and B) but also in planta was then confirmed by analyzing the B6 vitamer levels in both wt and loss-of-function plants. These analyses showed that the two independent had1 loss-of-function lines (had1-2 and had1-3) exhibit significantly increased PLP concentrations when compared with wts (Figure 9A). Therefore, it can be concluded that this enzyme hydrolyzes PLP not only in vitro but also in planta, and that its activity in chloroplasts contributes to the control of cellular PLP levels and with this to B6 vitamer homeostasis. Accordingly, we assign the name PLPP1 to this protein from Arabidopsis (AtPLPP1). In this context, it is important to mention that a recent study identified an alternative PLPP in chloroplasts of Nicotiana tabacum (NtPLPP), which similar to AtPLPP1 exhibits marked Mg2+ dependency (ShuoHao et al., 2019). Thus, it is likely that the presence of chloroplast-located PLPP enzymes extends to other vascular plants.
The recombinant AtPLPP1 exhibits an apparent Km for PLP of 30 µM (Figure 7C), which represents ∼15 times higher substrate affinity to that reported for NtPLPP (0.45 mM, see ShuoHao et al., 2019). A phylogenetic analysis indicates that Arabidopsis harbors a homolog to NtPLPP (ShuoHao et al., 2019), which, however, has so far not been characterized. Thus, it remains elusive whether two independent chloroplastic PLPPs are active in Arabidopsis. An answer to this question not only depends upon the respective substrate affinity but also upon the level of the enzyme in chloroplasts which is unknown. In any case, the markedly high affinity of AtPLPP1 for PLP is fully in line with the low concentration of this vitamer in Arabidopsis (Fitzpatrick et al., 2007; González et al., 2007).
It has been shown previously that the dwarf growth of er-ant1 mutants corresponds to a photorespiratory phenotype (Hoffmann et al., 2013). This phenotype is apparently caused by an inhibition of the activity of glycine decarboxylase (GDC; Hoffmann et al., 2013), representing an essential enzyme complex involved in photorespiration and locating to the mitochondrial matrix (Bauwe et al., 2010; Timm et al., 2012). Since er-ant1 plants exhibit a high accumulation of glycine, when compared with wts (Table 2; Hoffmann et al., 2013) and because glycine accumulation can be prevented under conditions of high CO2 (Hoffmann et al., 2013), which also abolishes the dwarf growth of er-ant1, it was speculated that the considerable glycine accumulation on the mutant line might cause impaired plant development (Hoffmann et al., 2013). However, here in this study, given that the suppressor mutant had1-2 x er-ant1 contains nearly identical glycine levels as observed in er-ant1 mother plants (64- and 68-fold increase, respectively; Table 2), we can now conclude that such high glycine levels in Arabidopsis are not the direct cause for the er-ant1 dwarf phenotype. This statement is consistent with observations on transgenic GDC knock-down dosage potato mutants exhibiting even up to 100-fold higher glycine levels as present in the wt, but are still viable and able to produce tubers (Heineke et al., 2001). So far it is unclear whether had1-2 x er-ant1 mutants show less unfolded protein response (UPR), which is a typical characteristic of er-ant1 loss-of-function mutants as revealed in both Arabidopsis and rice (Hoffmann et al., 2013; Zhang et al., 2016). Interstingly, to the best of our knowledge modified PLP levels have not been connected to UPR in the ER. However, the had1-2 x er-ant1 mutants might serve as a starting point to conduct future corresponding experiments.
Thus, the metabolic origin for the phenotypic peculiarity of the dwarf growth of er-ant1 must be due to something else. Quantification of amino acids known to be markedly altered in er-ant1 (Hoffmann et al., 2013) revealed that the cellular concentrations of Asp, Glu, and Ala, while strongly decreased in leaves of er-ant1, approach wt levels in had1-2 x er-ant1 plants (Table 2;Hoffmann et al., 2013). The biosynthesis of all three amino acids starts in chloroplasts and is dependent upon the activity of the enzymes Glu/2-oxo-glutarate amino transferase (GOGAT), as well as on Ala and Asp aminotransferases (Reyes-Prieto and Moustafa, 2012). Although all three enzymes use specific substrates, they share the conserved amino acid transferase property which is dependent upon the coenzyme PLP (Eliot and Kirsch, 2004). Therefore, given that (1) AtPLPP1 resides in chloroplasts (Figure 6), that (2) er-ant1 plants suffer from low PLP levels (Figure 9A), and that (3) had1 loss-of-function mutants show increased PLP levels (Figure 9A) while had1-2 x er-ant1 plants exhibit PLP as well as Asp, Glu, Arg, and Ala concentrations similar to wts (Figure 9A; Table 2), we propose that the restoration of the stromal PLP levels are causative for the marked suppression of the er-ant1 dwarf phenotype.
It is worth to mention that the decrease of Asp, Glu, and Ala levels in er-ant1 mutants is specific and is not simply due to the sequestration of in the high pool of glycine (Table 2). This is because the had1-2 x er-ant1 suppressor plants exhibit similarly increased glycine levels as observed in er-ant1, but show Asp, Glu, and Ala concentrations similar to wt (Table 2). Moreover, er-ant1 plants also exhibit markedly increased levels of Arg (Table 2) and the latter amino acid is generally a specific indicator for a high cellular availability (Sato and Yanagisawa, 2014; Winter et al., 2015). In fact, our assumption that limited amino acid transferase activity due to PLP deprivation in the stroma is causative for the er-ant1 dwarf phenotype under ambient CO2 is further supported by the observation that these mutants grow like the wt under high CO2 conditions (Hoffmann et al., 2013).
It is known that a high metabolic flux through the photorespiratory pathway (which is suppressed under high CO2) leads to a remarkable release of at the mitochondrial GDC (Douce et al., 2001). The chloroplastic enzyme couple of glutamine synthase and Glu synthase (GOGAT) is mandatory for reassimilation of which is released during photorespiration (Keys, 2006). The low GDC activity in er-ant1 plants, which is ∼40% of that in wts, is not caused by a decrease of the corresponding protein level (Hoffmann et al., 2013). Thus, it was speculated in a latter study that, besides an inhibitory glutathionylation of GDC, the GDC activity in er-ant1 is limited by further unknown factors (Hoffmann et al., 2013). Given that low PLP levels limit chloroplast located PLP dependent reactions (Table 2), we propose that the GDC activity in er-ant1 mitochondria is also inhibited by insufficient availability of PLP in corresponding mesophyll cells. This assumption is supported by the fact that GDC belongs to the most abundant enzymes in plant cells, representing about one-third of all mitochondrial proteins (Oliver et al., 1990), and that therefore its demand for PLP is relatively high.
Since had1-2 x er-ant1 suppressor mutants develop similarly to wt plants (Figures 1 and 4A) but still contain high glycine levels, the question arises: why does a stromal increase in PLP not rescue the GDC activity in mitochondria? An answer to this might be that although the absence of the newly identified AtPLPP1 leads to increased stromal PLP levels (Figure 9A) this vitamer is most likely not able to pass the chloroplast envelope (Rueschhoff et al., 2012). Thus, even if a PLP carrier protein exists in plant mitochondrial membranes, as is the case in mitochondria of yeast and mammals (Lui et al., 1981, 1982; Whittaker et al., 2015; Parra et al., 2018), the restored stromal PLP levels in had1-2 x er-ant1 plants do not communicate with the surrounding cytosol.
A hypothesis of how ER-ANT1 might affect the PLP availability in plant cells
The data above show that metabolic disturbances in the chloroplast caused by impaired PLP homeostasis in er-ant1 mutants can be suppressed by knocking out AtPLPP1. Thus, as a final question, we have to ask: how might a carrier protein residing in the ER membrane affect plant PLP homeostasis?
For a hypothesis setting ER-ANT1 into the scene of PLP homeostasis, we first have to recall that the ER of vascular plants harbors many specific functions in, for example lipid metabolism, cell-to-cell communication, or osmotolerance (Galili et al., 1998; Barton et al., 2011; Corso et al., 2018), which are absent in other eukaryotes. Moreover, the plant ER lumen harbors an impressive number of solutes, in particular solutes of low concentrations, which fulfill essential functions in signaling and stress responses typical for and restricted to plant cells (Helliwell, 2006; Friml and Jones, 2010; Wulfetange et al., 2011; Ju et al., 2012; Oda et al., 2016). Accordingly, the presence of ER-ANT1, which is solely found in vascular plants but not in other eukaryotes (Hoffmann et al., 2013; Zhang et al., 2016), is in line with the specific functions of the plant ER.
Given that ER-ANT1 is acting as a counter exchanger (Leroch et al., 2008) and in line with all the data above we propose that a fraction of cellular PLP in vascular plants locates to the ER lumen and is exported via ER-ANT1, in counter exchange with cytosolic ATP. Thus, in er-ant1 mutants, a low cytosolic PLP level prevents its uptake into mitochondria, leading to an impaired GDC activity (Hoffmann et al., 2013; Table 2). As another result of this PLP scavenging, the cytosol will probably also contain low PL levels, which subsequently limit stromal PLP synthesis by chloroplast located enzyme SOS4 kinase (Rueschhoff et al., 2012). The observation that ATP import into E. coli cells expressing ER-ANT1 is not inhibited by PLP (Leroch et al., 2008) does not argue against our assumption since it is unknown whether PLP is able to pass the outer bacterial membrane, or whether a specific orientation of ER-ANT1 in the E. coli membrane prevents access of PLP to the binding site.
We are fully aware that this hypothesis needs further experimental evaluation, but we are also fully aware that upcoming analyses which are from now on possible on had1-2 x er-ant1 plants as well as on the other so far not further analyzed suppressor mutants (Figures 1 and 3) will in future allow to clarify why the absence of ER-ANT1 activity causes the marked phenotype in corresponding loss-of-function mutants.
Materials and methods
Plant material and growth conditions
Studies were performed with Arabidopsis (A. thaliana) ecotype Columbia-0 wt plants, T-DNA insertion mutants with an insertion in At5g17400 (er-ant1: SALK_043626) (Leroch et al., 2008), EMS mutagenized er-ant1 plants of the M3-generation and T-DNA insertion mutants with an insertion in At2g33255 (plpp: SALK_041216C verified with primer pairs—plpp_LP: TCAAAGCAGAGAGCCCTAGTG, plpp_RP: GCTATGGAGAAAGGGTTGACC; plpp-2: SAIL_871_G11 verified with primer pairs—plpp-2_LP: TTGCATACACCAGATTTTGCTC, plpp-2_ RP: ATCAGACGAAAACCCAAATCC). Before germination, seeds were incubated for 2 d in the dark at 4°C on standardized ED73 soil (Weigel and Glazebrook, 2002). If not stated otherwise, plants were grown diurnally for 10 h at 22°C and 120 μmol photons m−2 s−1 light intensity followed by 14 h at 18°C without light. Plants were either grown at ambient CO2 levels (∼400 ppm) or high CO2 levels (2,000 ppm) in Fitotron plant climate chambers (Weiss Technik, Reiskirchen, Germany). For cultivation in sterile culture, seeds were surface sterilized (5 min 70% EtOH, 10 min 5% sodium hypochlorite for 10 min followed by washing three times with ddH2O) and subsequently stratified in water for 2 d at 4°C in the dark, before transfer to sterile 0.5 MS agar plates (0.2203% MS salts incl. vitamins, 0.5% [w/v] sucrose, 0.05% [w/v] MES, pH 5.7 [KOH], 0.8% [w/v] plant agar).
EMS mutagenesis and suppressor screen and construction of a mapping population
About 125,000 er-ant1 seeds were mutagenized by incubation in 0.4% (vol/vol) EMS solution for 8 h as previously described (Kim et al., 2006). The resulting M1 plants were grown and allowed to self-pollinate in pools of ∼500 plants at 2,000 ppm CO2 on soil. M2 seeds of the individual pools were grown at ambient CO2 conditions on soil. After 3 weeks, individual suppressors of the er-ant1 phenotype (ser-ant1) M2 plants were allowed to self-pollinate. The er-ant1 background of the M3 generation was ensured by verifying the presence of the T-DNA-insertion in At5g17400 (er-ant1) by PCR (Leroch et al., 2008).
BSA and NGS
To identify mutations causative for the suppression, bulked segregant analysis (BSA) was combined with whole-genome next-generation sequencing (NGS), similar to previously described by Song et al. (2017). For the generation of mapping populations, M3 mutants were backcrossed with individuals of the parental er-ant1 line. The resulting F1 generations were allowed to self-pollinate. Subsequently, the individual segregating F2 generations were used for mapping. For each investigated suppressor line, gDNA was extracted and pooled from rosette leaves of 50 plants with a ser-ant1 phenotype (positive pool) and from 50 plants showing the er-ant1 phenotype (negative pool). gDNA extraction was performed as previously described (Pallotta et al., 2000). The pooled gDNA samples of 13 mapping populations and pooled gDNA of 50 original er-ant1 mutant plants were used for NGS. The whole-genome resequencing and bioinformatic data analysis of the raw NGS data were provided by Novogene (Beijing, China). Briefly, the genomic DNA was randomly sheared into fragments of ∼350bp and subjected to library construction using the “Illumina TruSeq Library Construction Kit” (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After library quality control, 150-bp paired-end sequencing was performed on the “Illumina HiSeq” platform (Illumina). Image files were firstly transformed to sequence reads by base calling with CASAVA software (version 1.8; Illumina). The sequencing quality was assessed using FastQC Phred score evaluation. Adapters as well as low-quality reads were removed. The effective sequencing data were aligned to the reference sequence TAIR10 (ftp://ftp.Arabidopsis.org/home/tair/Genes/TAIR10_genome_release/) through BWA software and duplicates were removed with SAMtools (Li et al., 2009). To evaluate the similarity between each sample and the reference genome, mapping statistics such as mapping rate, average sequencing depth, and coverage were evaluated. SNPs were detected with SAMtools. To reduce the error rate in SNP detection, results were filtered, so that only SNPs were taken into account with more than four supporting reads and a mapping quality >20. Candidate SNPs were identified by SNP ratio mapping and the application of Candi-SNP (Etherington et al., 2014) and were verified via Sanger sequencing.
RNA isolation
Rosette leaves were harvested and ground in liquid nitrogen. Total RNA was isolated from 50 mg of the triturated material using the “NucleoSpin RNA Plant Kit” according to the manufacturer’s instructions (Macherey & Nagel, Düren, Germany). The RNA concentration was determined by photometric measurement at a nanodrop machine (Peqlab, Erlangen, Germany).
RT-qPCR
From total RNA, mRNA was transcribed into cDNA with reverse transcriptase (Verso-Kit, Thermo Fisher Scientific, Karlsruhe, Germany). For determining the expression of AtPLPP1, “PerfeCTa SYBR Green” was used according to the manufacturer’s instructions (Quantabio, Gaithersburg, MD, USA) with the following primer combination: PLPP_qRT_fwd: GAGTCCTGATAAACAGCAGAAGGC and PLPP_qRT_rev: AGCCACATAGTTCAGCAGTACCAG. For normalization, SAND (At2g28390) was used as a housekeeping gene (SAND_qRT_fwd: AACTCTATGCAGCATTTGATCCACT, SAND_qRT_rev: TGATTGCATATCTTTATCGCCATC). Amplification and fluorescence detection were performed using a MyiQ iCycler (Bio-Rad, Munich, Germany). Relative quantification was performed using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Chlorophyll quantification and photosynthetic measurements
Rosette leaves for chlorophyll determination were harvested and ground in liquid nitrogen. Extinctions of leaf extracts at the red absorption maxima of Chla (∼663 nm) and Chlb (∼646 nm) were measured and total chlorophyll content was determined (Porra, 2002). To investigate PSII performance via PAM analysis, plants were dark adapted for 15 min prior to exposure to actinic light. The effective PSII quantum yield, Y(II), was measured and calculated according to Genty et al. (1989).
Quantification of amino acids
Rosette leaves were harvested and ground in liquid nitrogen. An aliquot of 500 µL water was added to 100 mg of triturated material and the mixture was shaken for 15 min at 99°C (600 rpm). The starch and cell debris were sedimented by centrifugation at 20,000×g. For the measurement, 20 μL of the sample supernatant was mixed with 60 μL borate buffer (0.2 M boric acid, pH 8.8) and then derivatized with 20 μL AQC (fluorescent reagent 6-aminoquinolyl-N-hydroxy succinimidyl carbamate; Watrex, Prague, Czech Republic). The quantification of the amino acids was carried out with appropriate standard curves in an HPLC system consisting of a P680 HPLC Dionex pump, ASI 100 Dionex autosampler, RF 2000 fluorescence detector, UCI 50 Dionex interface, and a UVD 170 U Dionex UV detector. Nucleodur cc 250/4 100-5 c18ec served as the HPLC column. Aliquots of 25 μL were heated to 37°C on an autosampler and 20 μL were applied to the column by a pump, with a flow rate of 1 mL/min. The chromatograms were evaluated using the software Chromeleon version 6.7 (ThermoFisher, Waltham, MA, USA).
Determination of vitamin B6
Vitamin B6 extraction was performed and analyzed as described previously (Colinas et al., 2016) with the following changes: two separate extractions were performed, one with 16 volumes (extraction 1) and one with 8 volumes (extraction 2) of 50 mM ammonium acetate (pH 4.0), respectively. A 50-μL injection volume was used for a single run per extract. The calculations of PMP, PLP, and PL were based on extraction 1 while the calculations of PM and PN were based on extraction 2.
Heterologous expression of AtPLPP1 in E. coli
Heterologous AtPLPP1 expression was performed using the isopropyl-β-d-thiogalactopyranoside-inducible T7 RNA polymerase pET-vector/Rosetta 2 (DE3) expression system (Merck Biosciences, Novagen, Darmstadt, Germany). For this, the coding sequence of AtPLPP1 was amplified from Arabidopsis leaf cDNA with the following primer combination: PLPP_NdeI_fwd: NNNCATATGACTTTCCTCTTATCAAGAACATTTATTTC, PLPP_BamHI_rev NNN GGATCCCTACGGGTTCAGGTCGAAGTTCGTCTCC, allowing in-frame-insertion (via the NdeI and BamHI restriction sites) with the His-tag sequence of the pET16b vector. The correctness of the expression construct was verified by Sanger sequencing. Rosetta 2 cells containing the expression construct were cultivated in 50 mL standard YT media (0.8% [w/v] peptone, 0.5% [w/v] yeast extract, 0.25% [w/v] NaCl, pH 7.0) at 37°C aerobically overnight. From this culture, a new culture was started under the same condition. At an optical density of 0.2 at 600 nm, growth of the culture was continued aerobically at 15°C. At OD600 = 0.5, recombinant protein synthesis was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside, and the cells were harvested the next day by centrifugation (5 min; 5,000 × g). Cell disruption, protein purification, and immunostaining were performed as previously described (Trentmann et al., 2007). However, in contrast to Trentmann et al. (2007), the soluble fraction was used for protein enrichment via IMAC.
Phosphatase assay
The biochemical characterization of the purified HAD-type phosphatase AtPLPP1 was assayed as described previously (Sussman and Avron, 1981). The reaction mixture used contained 20 mM Tris–HCl pH 7.0, 5 mM MgCl2, and the tested substrates (in the given concentrations) in a total volume of 0.5 mL at 23°C. Approximately 6.25 μg/0.5 mL of purified protein was used in each enzymatic reaction. Reactions were stopped by adding perchloric acid to the reaction mixture (4.5% [v/v]). Subsequently, 200 µL of the mixture was used for phosphate determination using a standardized colorimetric assay (Ames, 1966).
GFP localization studies
To analyze the subcellular localization of AtPLPP1 in Arabidopsis and N. benthamiana, AtPLPP1–GFP fusion constructs were generated using the Gateway destination vectors pK7FWG2 (Karimi et al., 2002) for GFP attached to the C-terminus of AtPLPP1 and pk7WGF2 (Karimi et al., 2002) for an N-terminal fusion construct. For this, cDNA of AtPLPP1 was amplified omitting the stop codon via PCR using PLPP-specific primers (PLPP_fwd: ggggacaagtttgtacaaaaaagcaggcttaATGACTTTCCTCTTATCAAGAACATTTATTTCTCT CAC, PLPP_rev_+stop: ggggaccactttgtacaagaaagctgggttCTACGGGTTCAGGTCGAAGTT PLPP_rev_-stop: ggggaccactttgtacaagaaagctgggttCGGGTTCAGGTCGAAGTTCG), and including the attB1 and attB2 sites (lower case letters of the sequences). The amplified sequence was cloned into the entry vector pDONRZEO (Invitrogen) via the BP reaction and subsequently by an LR reaction into the respective destination vector. Protoplasts from mesophyll cells were isolated and transiently transformed according to Yoo et al. (2007). For imaging, a Leica TCS SP5 II confocal laser-scanning microscope was used (Objective: HCX PL APO lambda blue 63.0×1.20 WATER UV; Excitation wavelength laser: 488 nm Argon laser; Filter bandwidth emission: 495–540 nm [GFP], 651–704 nm [chloroplasts autofluorescence]).
Statistical analyses
Leaves or total rosettes from individual plants of the same age were used in physiological studies and acted as replicates. To determine the significance level of the mean of two sample sets (wt data or er-ant1 data were taken as reference) one-tailed Student’s t tests were used. Prior to the statistical analysis, normal distribution of data and equality of the variances were analyzed.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: At5g17400 (ER-ANT1), At2g33255 (AtPLPP1), YOR131C (PLPP_Sc from baker’s yeast), and At2g28330 (SAND housekeeping gene). A0A1S3ZWH3_TOBAC (NtPLPP1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. SNP induced changes in the At2g33255 gene product.
Supplemental Figure S2. Confirmation of the candidate SNP in At2g33255 of ser-ant1 line 6.3.
Supplemental Figure S3. Analysis of At2g33255 T-DNA insertion lines.
Supplemental Figure S4. Analysis of promHAD1::HAD1 x had1-2 x er-ant1 and ubiq::HAD1 x had1-2 x erant1 lines.
Supplemental Figure S5. ClustalW alignment of the amino acid sequences of the PLPP from N. tabacum (NtPLPP1) and the At2g33255 gene product.
Supplemental Figure S6. Purification of the HAD-type hydrolase, At2g33255, heterologously expressed in E. coli Rosetta cells.
Supplemental Table S1. SNP in the coding sequence of At2g33255 from different er-ant1 suppressor lines.
Supplementary Material
Acknowledgments
We are grateful to Jochen Wartenberg for excellent photography (Supplemental Figure S4).
Funding
Work in the lab of H.E.N. was financially supported by the Deutsche Forschungsgemeinschaft (DFG, Project NE 418/19-1). Cooperation between H.E.N., D.L., and T.K. was funded by the DFG in the frame of the TRR175 (projects B03, C05. and C01). T.B.F. would like to gratefully acknowledge funding from the Swiss National Science Foundation (Grants 31003A-141117/1 and 310030-192466).
Conflict of interest statement. None declared.
Contributor Information
Jacqueline Altensell, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
Ruth Wartenberg, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
Ilka Haferkamp, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
Sebastian Hassler, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
Vanessa Scherer, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
Priscille Steensma, Department of Botany and Plant Biology, University of Geneva, Geneva 1211, Switzerland.
Teresa B Fitzpatrick, Department of Botany and Plant Biology, University of Geneva, Geneva 1211, Switzerland.
Anurag Sharma, Copenhagen Plant Science Center, University of Copenhagen, Frederiksberg 1871, Denmark.
Omar Sandoval-Ibañez, Copenhagen Plant Science Center, University of Copenhagen, Frederiksberg 1871, Denmark.
Mathias Pribil, Copenhagen Plant Science Center, University of Copenhagen, Frederiksberg 1871, Denmark.
Martin Lehmann, Department of Biology I, Ludwig-Maximilians University of Munich, Planegg-Martinsried 82152, Germany.
Dario Leister, Department of Biology I, Ludwig-Maximilians University of Munich, Planegg-Martinsried 82152, Germany.
Tatjana Kleine, Department of Biology I, Ludwig-Maximilians University of Munich, Planegg-Martinsried 82152, Germany.
H Ekkehard Neuhaus, Department of Plant Physiology, University of Kaiserslautern, Kaiserslautern 67653, Germany.
T.K. performed the ethyl methanesulfonate (EMS) treatment of seeds. EMS studies were started by S.H. and finalized by J.A. T.K. supported J.A. in next-generation sequencing and corresponding data analyses. A.S., O.S.I., and J.A. performed and M.P. supervised the photosynthesis studies. V.S., J.A., and I.H. conducted functional prediction and analysis of the phosphatase, J.A. performed analyses of single and double mutants, and R.W. generated and analyzed the triple mutants. P.S. and T.B.F. conducted vitamer quantification. J.A., I.H., M.L., D.L., T.B.F., and H.E.N. discussed the data. I.H., J.A., and H.E.N. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: H. Ekkehard Neuhaus (neuhaus@rhrk.uni-kl.de).
References
- Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatases. In Neufeld EF, Ginsburg V, eds, Methods in Enzymology, Vol VIII. Academic Press, New York, NY [Google Scholar]
- Barton DA, Cole L, Collings DA, Liu DYT, Smith PMC, Day DA, Overall RL (2011) Cell-to-cell transport via the lumen of the endoplasmic reticulum. Plant J 66: 806–817 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L (2006) Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol 361: 1003–1034 [DOI] [PubMed] [Google Scholar]
- Caparrós-Martín JA, McCarthy-Suárez I, Culiáñez-Macià FA (2013) HAD hydrolase function unveiled by substrate screening: Enzymatic characterization of Arabidopsis thaliana subclass I phosphosugar phosphatase AtSgpp. Planta 237: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairmont CA, De Maio A, Hirschberg CB (1992) Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. J Biol Chem 267: 3983–3990 [PubMed] [Google Scholar]
- Colinas M, Eisenhut M, Tohge T, Pesquera M, Fernie AR, Weber APM, Fitzpatrick TB (2016) Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. Plant Cell 28: 439–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corso M, Doccula FG, de Melo JRF, Costa A, Verbruggen N (2018) Endoplasmic reticulum-localized CCX2 is required for osmotolerance by regulating ER and cytosolic Ca2+ dynamics in Arabidopsis. Proc Natl Acad Sci USA 115: 3966–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig B, Björkman O (1987) Comparison of the effect of excessive light on chlorophyll fluorescence (77K) and photon yield of O2 evolution in leaves of higher plants. Planta 171: 171–184 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F (2001) The glycine decarboxylase system: A fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF (2004) Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Ann Rev Biochem 73: 383–415 [DOI] [PubMed] [Google Scholar]
- Etherington GJ, Monaghan J, ZipfelC, MacLean D (2014) Mapping mutations in plant genomes with the userfriendly web application CandiSNP. Plant Meth 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Kappes B, Macheroux P, Tews I, Raschle T (2007) Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem J 407: 1–13 [DOI] [PubMed] [Google Scholar]
- Friml J, Jones AR (2010) Endoplasmic Reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G, Sengupta-Gopalan C, Ceriotti A (1998) The endoplasmic reticulum of plant cells and its role in protein maturation and biogenesis of oil bodies. Plant Mol Biol 38: 1–29 [PubMed] [Google Scholar]
- Geigenberger P, Riewe D, Fernie AR (2010) The central regulation of plant physiology by adenylates. Trends Plant Sci 15: 98–105 [DOI] [PubMed] [Google Scholar]
- González E, Danehower D, Daub ME (2007) Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol 145: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén E, Hirschberg CB (1995) Transport of adenosine triphosphate into the endoplasmatic reticulum proteoliposomes. Biochemistry 34: 5472–5476 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Fernie AR, Neuhaus HE (2011) Adenine nucleotide transport in plants: much more than a mitochondrial issue. Trends Plant Sci 16: 507–515 [DOI] [PubMed] [Google Scholar]
- Haferkamp I, Schmitz-Esser S (2012) The plant mitochondrial carrier family: Functional and evolutionary aspects. Front Plant Sci 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke D, Bykova N, Gardestrom P, Bauwe H (2001) Metabolic response of potato plants to an antisense reduction of the P-protein of glycine decarboxylase. Planta 212: 880–887 [DOI] [PubMed] [Google Scholar]
- Helliwell C (2006) The ER and plant hormones. In Robinson DG, ed., The Plant Endoplasmic Reticulum. Springer, Berlin, Heidelberg, pp 233–249 [Google Scholar]
- Hellmann H, Mooney S (2010) Vitamin B6: a molecule for human health? Molecules 15: 442–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Plocharski B, Haferkamp I, Leroch M, Ewald R, Bauwe H, Riemer J, Herrmann JM, Neuhaus HE (2013) From endoplasmic reticulum to mitochondria: Absence of the Arabidopsis ATP antiporter Endoplasmic Reticulum Adenylate Transporter1 perturbs photorespiration. Plant Cell 25: 2647–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James GV, Patel V, Nordström KJV, Klasen JR, Salomé PA, Weigel D, Schneeberger K (2013) User guide for mapping-by-sequencing in Arabidopsis. Genome Biol 14: R61–R61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al. (2012) CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci USA 109: 19486–19491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Karley AJ, White PJ (2009) Moving cationic minerals to edible tissues: Potassium, magnesium, calcium. Curr Opin Plant Biol 12: 291–298 [DOI] [PubMed] [Google Scholar]
- Keys A (2006) The re-assimilation of ammonia produced by photorespiration and the nitrogen economy of C3 higher plants. Photosynth Res 87: 165–175 [DOI] [PubMed] [Google Scholar]
- Kim Y, Schumaker KS, Zhu JK (2006) EMS mutagenesis of Arabidopsis. Methods Mol Biol 323: 101–103 [DOI] [PubMed] [Google Scholar]
- Klein MC, Zimmermann K, Schorr S, Landini M, Klemens PAW, Altensell J, Jung M, Krause E, Nguyen D, Helms V, et al. (2018) AXER is an ATP/ADP exchanger in the membrane of the endoplasmic reticulum. Nat Commun 9: 3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M (1989) Molecular aspects of adenine nucleotide carrier from mitochondria. Arch Biochem Biophys 270: 1–14 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Tatusov RL (1994) Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity: application of an iterative approach to database search. J Mol Biol 244: 125–132 [DOI] [PubMed] [Google Scholar]
- Krishnakumar V, Hanlon MR, Contrino S, Ferlanti ES, Karamycheva S, Kim M, Rosen BD, Cheng C-Y, Moreira W, Mock SA, et al. (2014) Araport: the Arabidopsis information portal. Nucl Acids Res 43: D1003–D1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova E, Nocek B, Brown G, Makarova KS, Flick R, Wolf YI, Khusnutdinova A, Evdokimova E, Jin K, Tan K, et al. (2015) Functional diversity of haloacid dehalogenase superfamily phosphatases from Saccharomyces cerevisiae: Biochemical, structural, and evolutionary insights. J Biol Chem 290: 18678–18698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroch M, Neuhaus HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J (2008) Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 20: 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lui A, Lumeng L, Li TK (1981) Metabolism of vitamin B6 in rat liver mitochondria. J Biol Chem 256: 6041–6046 [PubMed] [Google Scholar]
- Lui A, Lumeng L, Li TK (1982) Transport of pyridoxine and pyridoxal 5'-phosphate in isolated rat liver mitochondria. J Biol Chem 257: 14903–14906 [PubMed] [Google Scholar]
- Mayinger P, Meyer DI (1993) An ATP transporter is required for protein translocation into the yeast endoplasmic reticulum. EMBO J 12: 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL (2003) Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol 131: 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, et al. (2018) InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47: D351–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlmann T, Steinebrunner I, Neuhaus HE (2014) Nucleotides and nucleosides: transport, metabolism and signaling function of extracellular ATP. In Lüttge U, Beyschlag W, Cushman JC, eds, Prog. Bot. Vol 75. Springer, Heidelberg, Germany, pp 119–144 [Google Scholar]
- Oda K, Kamiya T, Shikanai Y, Shigenobu S, Yamaguchi K, Fujiwara T (2016) The Arabidopsis Mg transporter, MRS2-4, is essential for Mg homeostasis under both low and high Mg conditions. Plant Cell Physiol 57: 754–763 [DOI] [PubMed] [Google Scholar]
- Oliver DJ, Neuburger M, Bourguignon J, Douce R (1990) Interaction between the component enzymes of the glycine decarboxylase multienzyme complex. Plant Physiol 94: 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta MA, Graham RD, Langridge P, Sparrow DHB, Barker SJ (2000) RFLP mapping of manganese efficiency in barley. Theor Appl Genet 101: 1100–1108 [Google Scholar]
- Parra M, Stahl S, Hellmann H (2018) Vitamin B6 and its role in cell metabolism and physiology. Cells 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Reinhold T, Alawady A, Grimm B, Beran KC, Jahns P, Conrath U, Bauer J, Reiser J, Melzer M, Jeblick W, et al. (2007) Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J 50: 293–304 [DOI] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE (2004) Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis thaliana. Plant Physiol 136: 3524–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A (2012) Plastid-localized amino acid biosynthetic pathways of Plantae are predominantly composed of non-cyanobacterial enzymes. Sci Rep 2: 955–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux SJ, Steinebrunner I (2007) Extracellular ATP: an unexpected role as a signaler in plants. Trends Plant Sci 12: 522–527 [DOI] [PubMed] [Google Scholar]
- Rueschhoff E, Gillikin J, Sederoff H, Daub M (2012) The SOS4 pyridoxal kinase is required for maintenance of vitamin B6-mediated processes in chloroplasts. Plant Physiol Biochem 63C: 281–291 [DOI] [PubMed] [Google Scholar]
- Sato S, Yanagisawa S (2014) Characterization of metabolic states of Arabidopsis thaliana under diverse carbon and nitrogen nutrient conditions via targeted metabolomic analysis. Plant Cell Physiol 55: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShuoHao H, Jing L, Jie Z, JianYun Z, LongQuan H (2019) Identification and characterization of a pyridoxal 5'-phosphate phosphatase in tobacco plants. Plant Sci 278: 88–95 [DOI] [PubMed] [Google Scholar]
- Song J, Li Z, Liu Z, Guo Y, Qiu LJ (2017) Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front Plant Sci 8: 919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman I, Avron M (1981) Characterization and partial purification of dl-glycerol-1-phosphatase from Dunaliella salina. Biochim Biophys Acta 661: 199–204 [DOI] [PubMed] [Google Scholar]
- Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102: 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm S, Florian A, Fernie AR, Bauwe H (2016) The regulatory interplay between photorespiration and photosynthesis. J Exp Bot 67: 2923–2929 [DOI] [PubMed] [Google Scholar]
- Timm S, Mielewczik M, Florian A, Frankenbach S, Dreissen A, Hocken N, Fernie AR, Walter A, Bauwe H (2012) High-to-low CO2 acclimation reveals plasticity of the photorespiratory pathway and indicates regulatory links to cellular metabolism of Arabidopsis. PLoS ONE 7: e42809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden J, Möhlmann T, Kampfenkel K, Henrichs G, Neuhaus HE (1998) Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum) tuber morphology, yield and composition of tuber starch. Plant J 16: 531–540 [Google Scholar]
- Trentmann O, Horn M, van Scheltinga AC, Neuhaus HE, Haferkamp I (2007) Enlightening energy parasitism by analysis of an ATP/ADP transporter from chlamydiae. PLoS Biol 5: e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentmann O, Jung B, Neuhaus HE, Haferkamp I (2008) Non-mitochondrial ATP/ADP transporters accept phosphate as third substrate. J Biol Chem 283: 36486–36493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis. In A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, NY: [Database] [Google Scholar]
- Whittaker MM, Penmatsa A, Whittaker JW (2015) The Mtm1p carrier and pyridoxal 5'-phosphate cofactor trafficking in yeast mitochondria. Arch Biochem Biophys 568: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler HH, Neuhaus HE (1999) Non-mitochondrial ATP transport. Trends Biochem Sci 24: 64–68 [DOI] [PubMed] [Google Scholar]
- Winter G, Todd CD, Trovato M, Forlani G, Funck D (2015) Physiological implications of arginine metabolism in plants. Front Plant Sci 6: 534–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T (2011) The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol 156: 1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zheng X, Ke S, Zhu H, Liu F, Zhang Z, Peng X, Guo L, Zeng R, Hou P, et al. (2016) ER-localized adenine nucleotide transporter ER-ANT1: An integrator of energy and stress signaling in rice. Plant Mol Biol 92: 701–715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.