Abstract
Populations of the entomopathogenic fungus Batkoa major were analyzed using sequences of four genomic regions and evaluated in relation to their genetic diversity, insect hosts and collection site. This entomophthoralean pathogen killed numerous insect species from 23 families and five orders in two remote locations during 2019. The host list of this biotrophic pathogen contains flies, true bugs, butterflies and moths, beetles, and barkflies. Among the infected bugs (Order Hemiptera), the spotted lanternfly (Lycorma delicatula) is a new invasive planthopper pest of various woody plants that was introduced to the USA from Eastern Asia. A high degree of clonality occurred in the studied populations and high gene flow was revealed using four molecular loci for the analysis of population structure. We did not detect any segregation in the population regarding host affiliation (by family or order), or collection site. This is the first description of population structure of a biotrophic fungus-generalist in the entomopathogenic Order Entomophthorales. This analysis aimed to better understand the potential populations of entomopathogen-generalists infecting emerging invasive hosts in new ecosystems.
Introduction
Species in the Entomophthorales are predominantly arthropod pathogens, serving important ecological roles ranging from modifying host behavior to regulating population dynamics [1–4]. However, host range among entomophthoralean species is poorly understood, complicated by limited information on species identities of both fungal pathogens and arthropod hosts. Moreover, advances in sequencing technologies have revealed the presence of several species complexes, resulting in what was once considered a species with multiple hosts in fact being several cryptic species with distinctive host specificities (e.g., the Entomophaga aulicae species complex [5], and the Entomophthora muscae species complex [6]). Additional complications arise when entomophthoralean-arthropod species combinations tested in the lab demonstrate pathogenicity, although field studies often reveal a narrower host range (i.e., ecological host range) than the lab host range (physiological host range [7]). Therefore, to understand the dynamics of diseases caused by entomophthoralean fungi in arthropod populations, it is critically important to identify the spectrum of potential arthropod hosts.
Batkoa is an excellent example of an entomophthoralean genus whose recent phylogenetic revision [8, 9] allows for careful comparisons with arthropod hosts. The genus was first described in 1989 [10] and now includes ten species [11]. Although at one point divided across six genera, recent phylogenetic studies provide parallel evidence that Batkoa is a single and distinct genus [9, 12]. Across all Batkoa species, the host range of B. major is among the best documented, with several reported host associations; in a worldwide compendium of entomophthoralean species, Bałazy [2] lists B. major as occurring in North and South America, as well as in Europe and Asia, and that it was “infecting several insect species of different orders,” including a ptilodactylid (Coleoptera), tipulids (Diptera), aphids (Hemiptera), and an ichneumonid (Hymenoptera). In 2018, B. major was also found alongside Beauveria bassiana (another entomopathogenic fungal species), co-infecting populations of the invasive spotted lanternfly (Lycorma delicatula, Fulgoridae, Hemiptera) [13]. This invasive fulgorid planthopper is only distantly related to native insects in the area of the co-epizootic (i.e., there are no native species in the same family, the Fulgoridae, in this area). At the time of the 2018 epizootics, B. major had only been cited from North America one time since its description in 1888 [8].
This study began with the goal of identifying the native reservoir hosts for B. major, a poorly known pathogen causing epizootics in outbreak populations of a new invasive insect. Based on trends in host range in the Entomophthorales, it was assumed that there would be few native host species and that these would predominantly belong to the order Hemiptera. We present results of a survey of naturally occurring infections in northeastern US forests that was conducted to identify hosts of B. major. We hypothesized that B. major in sampled locations is genetically diverse, but that it is not a species complex and that its populations are mostly clonal. Subsequent analyses investigated the genetic diversity and population structure of B. major to evaluate the potential for host specific clones and gene flow among collection sites.
Materials and methods
Sample collection and fungal isolation
Native insect populations were sampled in a mixed hardwood forest near Ithaca, New York. On nine days between 19 June and 14 September 2019, cadavers of insects killed by entomophthoralean fungi were collected in Danby State Forest, Tompkins County, New York. Collections were made along the Abbott Loop hiking trail between 42°18’56.3"N, 76°29’42.2"W and 42°17’44.9"N, 76°29’10.8"W. Native insects killed by entomophthoralean fungi were also collected along the borders of the Angora Fruit Farm (40°21’30.6”N, 75°53’00.4”W), Berks County Parks and Recreation, Pennsylvania on 19 September 2019, near Ailanthus altissima (tree of heaven; the preferred host tree for L. delicatula) and in the adjacent hardwoods. At both sites, all sides of leaves, twigs, and branches from the ground to 2.5 m were carefully surveyed for dead insects. Arthropod cadavers were placed in 29 ml clear plastic cups containing 5 ml of 1.5% water agar and were transported to the laboratory at 4°C. Collected insects were from low density populations of native species. Collection trips were made within 24–48 hours after rainfall and collections took place over a period of two to three hours per site. The two sample sites are approximately 220 km from each other.
In the laboratory cadavers that were sporulating or were ready to sporulate were moved to high humidity enclosures at room temperature. Each cadaver was separately covered with the base of a 60 mm petri dish containing malt extract agar (MEA; 30 g malt extract, 20 g agar, 1 L distilled water) to allow “ascending” conidia to be collected on the MEA [14]. After approximately 6 hours, petri dishes with conidia were removed. Cadavers that had not yet sporulated were left under high humidity at 20°C overnight and were irregularly checked for sporulation for a total of 48 h. After conidia had been collected, the body of each insect was stored at -20°C and subsequently examined to morphologically identify arthropod host species.
All MEA plates with conidia were maintained at 20°C. Once conidia had begun to germinate, thin sections of MEA containing hyphae were excised and placed in 35 mm petri dishes containing 1.5 ml 95% Grace’s insect medium (Lonza, Walkersville, MD) and 5% fetal bovine serum (Life Technologies, Grand Island, NY). Once hyphal growth was evident, hyphae were transferred to egg yolk Sabouraud maltose agar (EYSMA [14]) in 100 mm petri dishes. When cultures were mature, they were frozen in 10% glycerol in 2 ml cryotubes at -80°C, using a CoolCell Freezing System (Corning, NY) and deposited in the ARSEF culture collection (Table 1).
Table 1. Insect hosts collected in Pennsylvania and New York State, infected by Batkoa major in 2019–2020 (S1 File).
| Collection | Host | ARSEF # (HLBio#) | GenBank accession # | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Site | Date | Order | Family or Suborder | Species | ITS1 | ITS2 | 28S | RPB2 | |
| AFF | 9/10 2018 | Hemiptera | Fulgoridae | Lycorma delicatula | Bat13769 | OL335101 | OL335159 | OL332699 | OL624704 |
| DSF | 6/27/2019 | Diptera | Rhagionidae | * | 14421 (Bat42) | OL335043 | OL335102 | OL332659 | OL624643 |
| DSF | 6/27/2019 | Hemiptera | Cixiidae | Cixius sp. | 14420 (Bat73) | OL335044 | OL335103 | OL332659 | OL624644 |
| DSF | 6/27/2019 | Coleoptera | Elateridae | Athous brightwelli | 14426 (Bat83) | OL335045 | OL335104 | OL332659 | OL624645 |
| DSF | 6/27/2019 | Coleoptera | Cantharidae | Rhagonycha fraxini | (Bat86) | OL335046 | OL335105 | OL332637 | OL624646 |
| DSF | 6/27/2019 | Diptera | Dolichopodidae | Gymnopterus sp. | (Bat88) | OL335047 | OL335106 | OL332638 | OL624647 |
| DSF | 6/27/2019 | Coleoptera | Elateridae | Athous brightwelli | 14444 (Bat90) | OL335048 | OL335107 | OL332639 | OL624648 |
| DSF | 6/27/2019 | Coleoptera | Tenebrionidae | Isomira sericea | (Bat91) | OL335049 | - | OL332640 | OL624649 |
| DSF | 7/10/2019 | Diptera | Lauxaniidae | Homoneura inserta | (Bat120) | OL335050 | OL335108 | OL332641 | OL624650 |
| DSF | 7/10/2019 | Diptera | Sciaridae | * | (Bat121) | - | - | OL332642 | - |
| DSF | 7/10/2019 | Diptera | Rhagionidae | Symphoromyia sp. | (Bat123) | OL335051 | OL335109 | OL332643 | OL624651 |
| DSF | 7/10/2019 | Diptera | Rhagionidae | * | (Bat124) | OL335052 | OL335110 | OL332644 | OL624652 |
| DSF | 7/10/2019 | Diptera | Rhagionidae | * | 14427 (Bat132) | OL335053 | OL335111 | OL332645 | OL624653 |
| DSF | 7/10/2019 | Diptera | Rhagionidae | * | 14430 (Bat156) | OL335054 | OL335112 | OL332646 | OL624654 |
| DSF | 8/1/2019 | Lepidoptera | Blastobasidae | * | 14431 (Bat160) | OL335055 | OL335113 | OL332647 | OL624655 |
| DSF | 8/1/2019 | Lepidoptera | Tineidae | Dryadaula sp. | 14448 (Bat163) | OL335056 | - | OL332648 | OL624656 |
| DSF | 8/1/2019 | Lepidoptera | Erebidae | Lophocampa caryae | 14435 (Bat164) | OL335057 | OL335114 | OL332649 | OL624657 |
| DSF | 8/1/2019 | Lepidoptera | Crambidae | Eudonia sp. | 14437 (Bat165) | OL335058 | OL335115 | OL332650 | OL624658 |
| DSF | 8/1/2019 | Lepidoptera | Blastobasidae | * | 14428 (Bat167) | OL335059 | - | OL332651 | OL624659 |
| DSF | 8/1/2019 | Diptera | Rhagionidae | * | 14432 (Bat173) | OL335060 | - | OL332652 | OL624660 |
| DSF | 8/1/2019 | Diptera | Dolichopodidae | Thrypticus sp. | 14436 (Bat174) | OL335061 | - | OL332653 | OL624661 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | (179) | OL335062 | OL335116 | OL332654 | OL624662 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | 14425 (Bat181) | OL335063 | OL335117 | OL332655 | OL624663 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | (Bat183) | OL335064 | OL335118 | OL332656 | OL624664 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | 14433 (Bat184) | OL335065 | OL335119 | OL332657 | OL624665 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | 14438 (Bat186) | OL335066 | OL335120 | OL332658 | OL624666 |
| DSF | 8/1/2019 | Diptera | Sciaridae | * | 14449 (Bat189) | OL335067 | OL335121 | OL332659 | - |
| DSF | 8/7/2019 | Coleoptera | Cantharidae | Rhagonycha sp. | 14434 (Bat204) | OL335068 | OL335122 | OL332660 | OL624667 |
| DSF | 8/7/2019 | Diptera | Sciaridae | * | 14439 (Bat205) | OL335069 | OL335123 | OL332661 | OL624668 |
| DSF | 8/7/2019 | Coleoptera | Cantharidae | Rhagonycha sp. | (Bat207) | OL335070 | OL335124 | OL332662 | OL624669 |
| DSF | 8/7/2019 | Hemiptera | Cixiidae | Cixius sp. | (Bat209) | OL335071 | OL335125 | OL332663 | OL624670 |
| DSF | 8/7/2019 | Lepidoptera | Blastobasidae | * | 14424 (Bat210) | OL335072 | - | OL332664 | OL624671 |
| DSF | 8/7/2019 | Coleoptera | Cantharidae | Rhagonycha sp. | (Bat217) | OL335073 | OL335126 | OL332665 | OL624672 |
| DSF | 8/7/2019 | Hemiptera | Derbidae | Apache degeeri | 14457 (Bat220) | OL335074 | OL335127 | OL332666 | OL624673 |
| DSF | 8/7/2019 | Lepidoptera | Oecophoridae | Fabiola edithella | 14440 (Bat221) | OL335075 | OL335128 | OL332667 | OL624674 |
| DSF | 8/15/2019 | Hemiptera | Cicadellidae | * | 14441 (Bat222) | OL335076 | OL335129 | OL332668 | OL624675 |
| DSF | 8/15/2019 | Lepidoptera | Erebidae | Lymantria dispar | (Bat228) | OL335077 | OL335130 | OL332669 | OL624676 |
| DSF | 8/15/2019 | Hemiptera | Cicadellidae | * | 14423 (Bat241) | OL335078 | OL335131 | OL332670 | OL624677 |
| DSF | 8/15/2019 | Hemiptera | Cixiidae | Cixius sp. | 14422 (Bat242) | - | OL335132 | OL332671 | OL624678 |
| DSF | 8/15/2019 | Diptera | Anthomyiidae | * | (Bat249) | OL335079 | OL335133 | OL332672 | OL624679 |
| DSF | 9/4/2019 | Diptera | Heleomyzidae | Tephrochlamys rufiventris | (Bat269) | OL335080 | OL335134 | OL332673 | OL624680 |
| DSF | 9/4/2019 | Psocoptera | Amphipsocidae | Polypsocus corruptus | (Bat270) | - | OL335135 | OL332674 | OL624681 |
| DSF | 9/4/2019 | Diptera | Sciaridae | * | 14450 (Bat271) | OL335081 | OL335136 | OL332675 | OL624682 |
| DSF | 9/4/2019 | Diptera | Psychodidae | * | 14451 (Bat273) | OL335082 | OL335137 | OL332676 | OL624683 |
| DSF | 9/4/2019 | Diptera | Dolichopodidae | Gymnopterus sp. | (Bat275) | - | - | OL332677 | - |
| DSF | 9/4/2019 | Hemiptera | Cixiidae | Cixius sp. | (Bat277) | - | - | - | OL624684 |
| DSF | 9/4/2019 | Hemiptera | Achilidae | * | (Bat287) | OL335083 | OL335138 | OL332678 | OL624685 |
| DSF | 9/4/2019 | Hemiptera | Achilidae | * | (Bat288) | OL335084 | OL335139 | OL332679 | OL624686 |
| DSF | 9/4/2019 | Hemiptera | Achilidae | * | (Bat291) | OL335085 | OL335140 | OL332680 | OL624687 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | (Bat297) | OL335086 | OL335141 | OL332681 | OL624688 |
| DSF | 9/14/2019 | Hemiptera | Cixiidae | Cixius sp. | 14458 (Bat300) | OL335087 | OL335142 | OL332682 | OL624689 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | (Bat301) | OL335088 | OL335143 | OL332683 | OL624690 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | 14452 (Bat304) | OL335089 | OL335144 | OL332684 | OL624691 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | 14453 (Bat309) | OL335090 | OL335145 | OL332685 | OL624692 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | 14429 (Bat320) | OL335091 | OL335146 | OL332686 | OL624693 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | 14454 (Bat321) | OL335092 | OL335147 | OL332687 | OL624694 |
| DSF | 9/14/2019 | Lepidoptera | Geometridae | Lambdina fiscellaria | (Bat322) | - | OL335148 | OL332688 | OL624695 |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | 14455 (Bat323) | - | OL335149 | OL332689 | - |
| DSF | 9/14/2019 | Diptera | Sciaridae | * | (Bat324) | OL335093 | OL335150 | OL332690 | OL624696 |
| DSF | 9/14/2019 | Hemiptera | Derbidae | Apache degeeri | 14459 (Bat326) | OL335094 | OL335151 | OL332691 | OL624697 |
| AFF | 9/19/2019 | Diptera | Sciaridae | * | (Bat327) | OL335095 | OL335152 | OL332692 | OL624698 |
| AFF | 9/19/2019 | Diptera | Lauxaniidae | * | (Bat332) | OL335096 | OL335153 | OL332693 | - |
| AFF | 9/19/2019 | Diptera | Dolichopodidae | Medetera sp. | (Bat333) | - | OL335154 | OL332694 | OL624699 |
| AFF | 9/19/2019 | Diptera | * | * | (Bat334) | OL335097 | OL335155 | OL332695 | OL624700 |
| AFF | 9/19/2019 | Diptera | Milichiidae | Madiza glabra | (Bat335) | OL335098 | OL335156 | OL332696 | OL624701 |
| AFF | 9/19/2019 | Psocoptera | Psocomorpha | * | (Bat340) | OL335099 | OL335157 | OL332697 | OL624702 |
| AFF | 8/31/2020 | Hemiptera | Fulgoridae | Lycorma delicatula | (Bat565) | OL335100 | OL335158 | OL332698 | OL624703 |
AFF—Angora Fruit Farm, Berks County Parks and Recreation, Pennsylvania, DSF–Danby State Forest, Tompkins County, New York.–missing data,
*—not identified to the family/suborder, genus, or species level.
DNA extraction and amplification
Fungal tissues from in vitro growth were transferred to lysis buffer and beaten with 0.5 g of 0.7 mm diameter zirconia beads at 4800 rpm for 1 min. DNA extraction and PCR were performed as described in Hajek et al. [15]. PCR was performed on 4 loci: 28S (large subunit of structural ribosomal RNA (rRNA) of eukaryotic cytoplasmic ribosomes), ITS1 and ITS2 (large subunit of structural ribosomal RNA (rRNA) of eukaryotic cytoplasmic ribosomes), and RPB2(large subunit of structural ribosomal RNA (rRNA) of eukaryotic cytoplasmic ribosomes). 28S amplification used forward primer LR0R [16] and reverse primer LR5 [17]. ITS1 amplification used forward primer ITS5 [18] and reverse primer 5.8S [17]. ITS2 amplification used forward primer ITS3 [18] and reverse primer ITS4sub: 5’-TGGAGCAAGTACAAACAACACT-3’. RPB2 amplification used forward primer BatRPB2f: 5’- ACCCTCAGAAACCTCTCGTC-3’ and reverse primer BatRPB2r: 5’- CAAACCGAGCCAGCAATTTG-3’.
PCR conditions for 28S were initial denaturation for 5 min at 95°C followed by 6 cycles of denaturation for 1 min at 95°C, annealing at 58°C for 1 min that decreased by 1°C for each cycle, and extension for 1.5 minutes at 72°C. The 6 cycles were followed by 30 cycles of denaturation for 30 sec at 95°C, annealing at 52°C for 1 min, and extension for 1 min at 72°C. The final step was extension at 72°C for 10 sec. PCR conditions for ITSI and ITSII were an initial denaturation for 5 min at 94°C followed by 35 cycles of denaturation at 94°C for 45 sec, annealing at 55°C for 50 sec, and extension at 72°C for 1 min. The final step was extension at 72°C for 10 min. PCR conditions for RPB2 were an initial denaturation at 95°C for 4 min followed by 34 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 1 min, a ramp that increased the temperature at a rate of 0.3°C/sec for 1.23 min from 50 to 72°C, and extension for 1 min at 72°C. The final step was extension for 10 min at 72°C [19]. To check whether the PCR products were viable, products underwent agarose gel electrophoresis in 1x TAE buffer and were visualized with ACCURIS SmartDoc (Accuris, New Jersey, USA). Successful products were purified by combining 8μL of product with 2μL of a master mix (1.6μL molecular water, 0.2μL 10X PCR buffer, 0.1μL SAP enzyme, and 0.1μL EXO enzyme) at 37°C for 35 min followed by deactivating the enzymes at 90°C for 13 min. Purified PCR products were sequenced by Genewiz LLC (South Plainfield, New Jersey, USA). Sequences were edited, assembled, aligned, and searched using Geneious software v. 8.1.8 (Biomatters Ltd).
Phylogenetic reconstruction and analysis of population structure
A single FASTA file was prepared from each of the four loci used to identify B. major: ITS1 (N = 39), ITS2 (N = 54), 28S (N = 66), and RPB2 (N = 62). Each FASTA file was aligned using MAFFT version 7 (using default parameters with a scoring matrix of 1PAM/ κ = 2 for closely related sequences and was imported into R version 4.0.2 [20] for Maximum Likelihood inference and building a phylogenetic tree [21]. All analyses were conducted using the adegenet and poppr packages [22, 23]. Fungal samples in each FASTA file were further labelled according to the geographic location and arthropod host from which they were collected (accounting for host order, family, and species).
To infer the number of genetic clusters across our data set, and to evaluate the utility of arthropod host as a predictor of population structure in B. major, alignments of each locus were subjected to a Discriminant Analysis of Principle Components (DAPC) [24–26]. An additional DAPC was performed retaining only one sample per genotype per population using the clonecorrect command in poppr [23]. Population differentiation was further analyzed by calculating FST according to B. major host order and family using hierfstat [27].
Results
In 2019, we collected a total of 213 insects that appeared to have been killed by entomophthoralean fungi. Most entomophthoralean species are difficult to isolate so collections resulted in a total of 67 samples of B. major (cultures plus fungus directly from cadavers) that could be used for molecular analysis.
Morphological characterization
Our morphological observations of B. major from the cadavers of host insects do not differ from previously published records [2]. Diameters of conidia, size of conidial papillae, and the number of nuclei in conidia are typical for the species (Fig 1).
Fig 1. Micromorphology of Batkoa major.
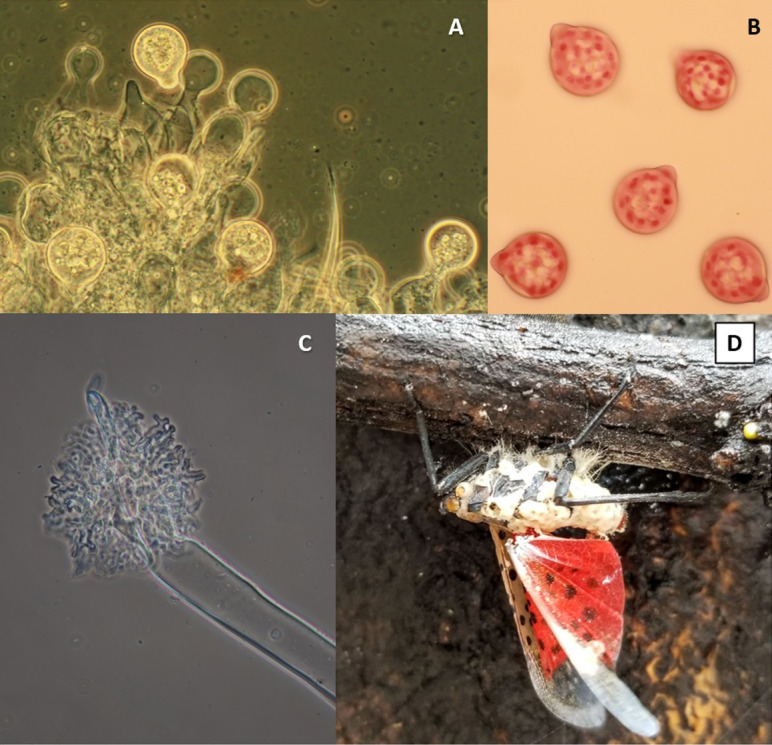
A. Conidiophores with conidia. Conidia average 41.5 μm wide x 49.3 μm long. B. Multinucleate conidia (nuclei stained with aceto-orcein). C. Distal end of rhizoid with holdfast. D. Cadaver of the spotted lanternfly attached to a twig by rhizoids (Photo by E.H. Clifton).
Genetic polymorphism reveals the lack of host specificity
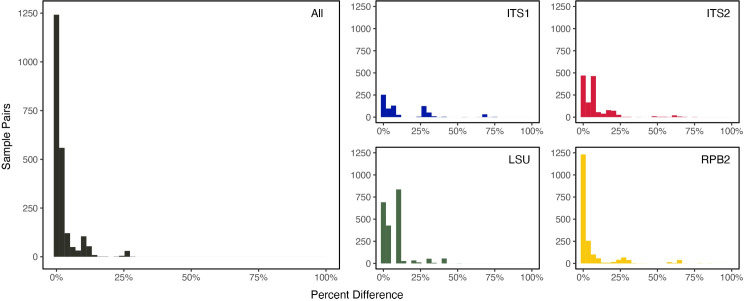
Genotype comparisons and phylogenetic reconstructions suggests that these B. major populations consist of numerous genotypes. Alignments of four loci (28S, ITS1, ITS2 and RPB2) of B. major (S2 File) reveal a high degree of genetic polymorphism among individuals (Fig 2).
Fig 2. Percent genetic difference among pairwise comparisons of all specimens.
Approximately half of all specimen pairs are identical for any given locus, as well as when all four loci are combined, suggesting that asexuality is an important aspect of the B. major life history. The least degree of polymorphism occurs in RPB2, whereas the most occurs in ITS1, in contrast to observations in other fungal species [28, 29].
The combined alignments consisted of 3,201 positions (N = 67 specimens), with each locus presenting a varied degree of sites: 28S had a length of 1,067 bp with 20 polymorphic positions (N = 66), ITS1 had a length of 911 bp with 174 polymorphic positions (N = 39), ITS2 had a length of 639 bp with 58 polymorphic positions (N = 54), and RPB2 had a length of 584 bp with 29 polymorphic positions (N = 62). Although we were unable to amplify and sequence the entire ITS region, and thus combine ITS1 and ITS2 sequences, we estimate the total length of the ITS region is greater than 1,600 nucleotides.
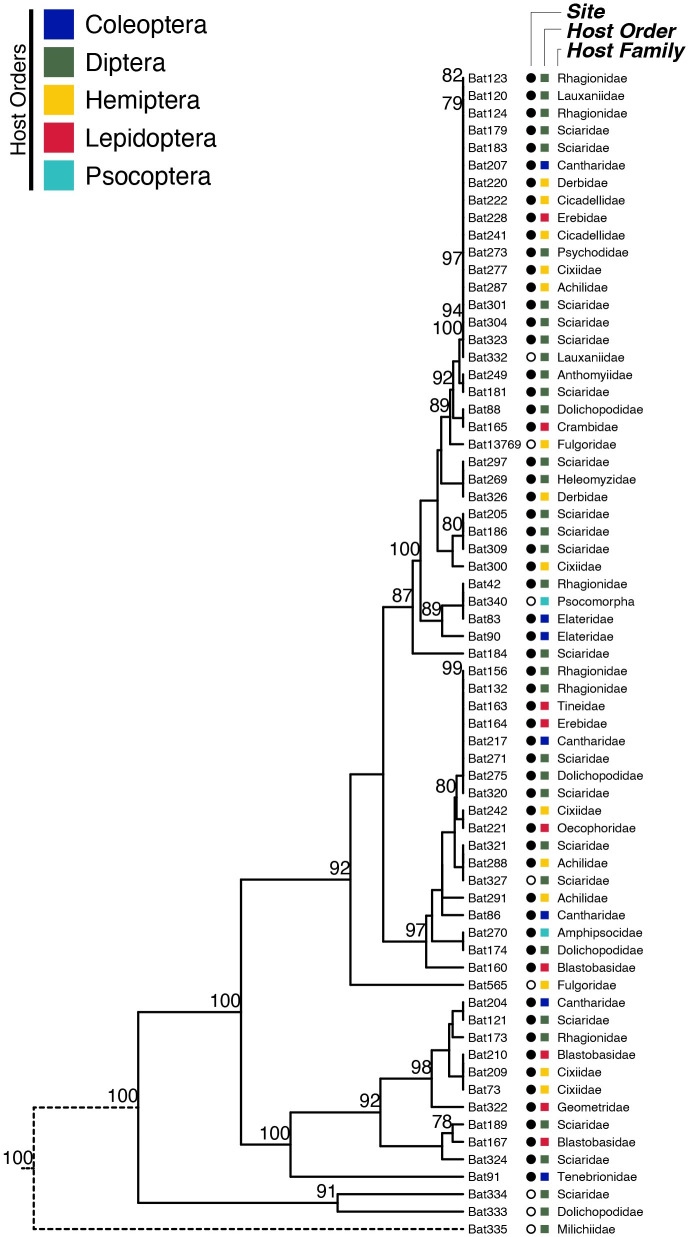
Notably, genetic polymorphism does not correspond to arthropod host (Fig 3). For example, several clonal sample (i.e., those with branch lengths of zero in Fig 3) were collected from arthropod hosts distantly related to each other and belonging to different orders. Even when genetically dissimilar specimens are compared, several host orders or families are represented in a single clade.
Fig 3. Dendrogram of all B. major isolates.
Each specimen is labelled according to the order and family of the arthropod host from which it was sampled. Population origin is also specified, with an open circle for the Angora Fruit Farm population and a closed circle for the Danby State Forest population. Nodes whose bootstrap support was greater than 75 are labelled accordingly. A dotted black line was drawn to depict the outermost tree branch to indicate that it was shortened for aesthetic purposes.
Mapping hosts on the B. major phylogenetic tree demonstrated that hosts from the same family can be infected by pathogens that have different genotypes. There is no visible grouping of the hosts with particular clades of the pathogen, either on the single locus trees (S1 Fig) or on the combined four-locus tree (Fig 3). Major host orders and families are located on the B. major phylogenetic tree randomly. Slightly more than half of the infected insects belong to order Diptera (35 out of 67 samples). Among individual families, the most samples were from the Sciaridae (dark-winged fungus gnats; 19 out of 67 samples). Insect orders represented less frequently in the current screen are (in descending order): Hemiptera (N = 14 samples), Lepidoptera (N = 9), Coleoptera (N = 7), Psocoptera (N = 2).
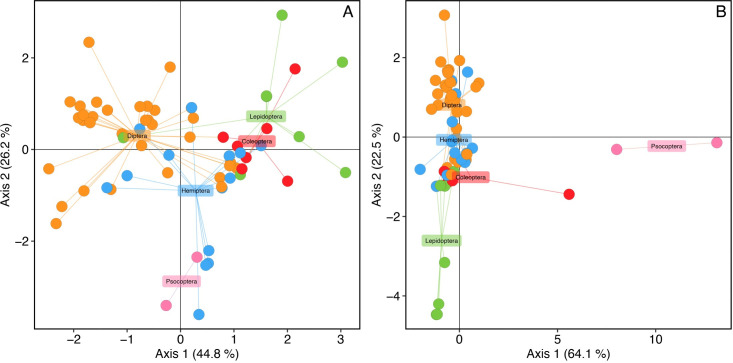
Our DAPC analysis corroborates a lack of host specificity in B. major, illustrating a high degree of genotypic overlap when grouping samples by host order. The first (44.2% [64.1% clone corrected]) and second (26.2% [22.5% clone corrected]) principal components capture most of the genetic variation among samples and reveal no apparent clustering of individuals in correlation to their arthropod host (including when only a single specimen per genotype is retained in the analysis). An exception was the two samples from the Order Psocoptera that clustered together in the clone-corrected analysis (Fig 4).
Fig 4. DAPC analysis for the combined ITS1, ITS2, LSU and RPB2.
Data in full (A) and clone-corrected (B). Specimens are labeled according to host order. Axis 1 explained 44.8% (64.1% for clone-corrected data) and axis 2 explained 26.2% (22.5% for clone-corrected data) of the genetic variation among individuals.
High gene flow within and between populations of B. major
Genotypes of B. major identified with either ITS1, ITS2, 28S or RPB2 do not cluster according to collection site, in addition to not clustering by host order or family (Figs 3 and 4). Despite differences in sample size from our two collection sites, hosts were infected (often belonging to different arthropod orders and families) at each location by the same genotype, despite being separated by 220 km. Moreover, genetically distinct fungal specimens from the same population do not cluster phylogenetically, illustrating a broader pattern of high gene flow among genotypes of B. major.
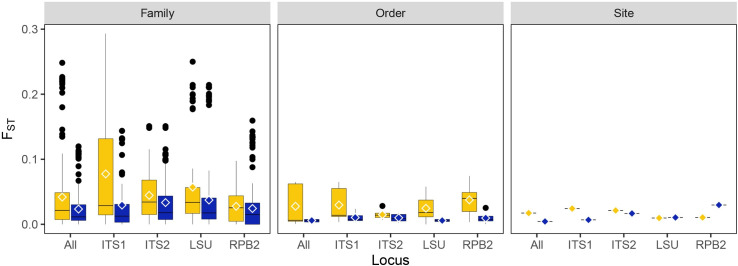
The lack of population structure by arthropod host is further supported by FST values calculated between host order and family, in addition to values calculated between the two collection sites (Fig 5). In all cases, median FST is 0.05–0.035 in all pairwise comparisons of host orders and families, with the maximum FST values approximately 0.20 when comparing host family by the 28S locus.
Fig 5. FST calculations for each locus and all four combined.
FST calculated between host order, host family, and collection site (syn. population). Yellow–all data, blue–clone corrected data. Mean FST is illustrated by an open rhombus.
Discussion
Entomophthoralean species have several different modes of host range diversity. Batkoa major, B. apiculata, Zoophthora radicans, and Conidiobolus thromboides have broader host ranges that include hosts in different insect orders. Then, there are some species of Entomophthorales with an intermediate type of host specificity, only infecting insects within the same insect order. For example, studies of physiological host range demonstrated that all three species in the Entomophaga aulicae species complex infect only species of Lepidoptera [30, 31]. Finally, there are highly specialized species, like Strongwellsea magna and S. castrans, that, even with extensive study, are known only from host species within individual families of Diptera [32]. The group of species with broad host ranges seems to be the smallest group within this fungal order [2]. Theory has suggested that more host specific parasites can lead to greater survival success [33].
Contrary to expectations, we found that the native fungal entomopathogen B. major has a diverse host range, including native insects in five insect orders. Why this fungal species has not been reported more previously is not known although one possibility is the difficulty of sampling insects in forested locations. Nonetheless, our findings agree with those for other fungal pathogens with broad host ranges, which are generally held to be more likely to form symbioses with novel hosts in invasive contexts [34, 35].
Invasive L. delicatula is thus a competent host for this native pathogen that is a generalist and which we found in abundance during a fall epizootic in L. delicatula [13] or all season long in different native host species. The well-known idiom ‘jack of all trades and master of none’ has been used to suggest that generalists would be less successful than specialists [33]. The alternate opinion that changes the idiom to ‘jack of all trades and master of all’ [36] is more consistent with results from the present study where B. major was found all season long infecting a diversity of hosts, although prevalence was not high in these lower density populations. Woolhouse et al. [37] suggest that conditions predisposing pathogens to generalism include high levels of genetic diversity as well as ample opportunities for cross-species transmission. While we found clonality for some groups of B. major, we found numerous clones including multiple samples from different hosts and gene flow occurred among some of them. We see some similarity in the population structure between B. major (our observations) and E. muscae [38]. In addition, our predominant collecting site was a native forest in New York during summer and the native insect fauna provided a diversity of hosts. Clones also existed within each of two clades of the entomophthoralean E. muscae infecting two species of flies [39].
Unusually rDNA sequence length significantly contributes to the polymorphism in our samples, possibly also due to higher substitution rates compared to other entomophthoralean fungi [9]. Curiously, the total length of the ITS region in B. major exceeds 1600 bp, which is quite an unusual feature compared to most fungal species. However, long ITS is also characteristic of other entomophthoralean species, e.g. for E. muscae [40] and Zoophthora species [41]. Also, genomes in some fungi contain multiple ITS copies [42]. Therefore, high population diversity in the B. major population recorded for the ITS1 and ITS2 regions might significantly reflect random ITS copies rather than real genetic diversity. The longer that the length of the ITS region is, the larger the number of mutations that might occur and therefore the larger number of different ITS copies that might be amplified and sequenced, which can be reflected as population diversity for that genomic region. In contrast, high numbers of identical copies suggest a high degree of clonality in the population, i.e., identical copies with different placement on the phylogenetic tree. It seems unlikely that we have randomly sampled the same ITS copy. This fact might be a good indication that the copies in B. major are homogenized by concerted evolution and sexual processes are occurring.
Interestingly, FST values indicate that not only is there high gene flow among populations, but also that there is high gene flow within populations vis-à-vis arthropod host. More specifically, our data not only indicate a lack of host specificity, but also that genotypes of B. major readily exchange genetic information (i.e., undergo sexual reproduction) with other genotypes infecting phylogenetically distant arthropod hosts.
Our finding of the same genotype of B. major at collection sites 220 km apart suggests that long-distance movement of B. major genotypes is occurring. Longer distance dispersal of B. major could be accomplished via dispersal of infected hosts. Insects can disperse longer distances and infected migratory aphids are known to provide dispersal of entomophthoralean fungi [43]. Most entomophthoralean fungi actively eject asexual spores which can result in airborne dispersal of these pathogens [1]. Such dispersal has been documented during epizootics for several species [44–46] and long distance dispersal has been modelled for Entomophaga maimaiga based on spread of this introduced pathogen [47].
Generalist pathogens are thought to potentially experience trade-offs in that they are not as well adapted to all the hosts that they infect [37]. While Bufford et al. [35] found that taxonomic similarity of co-evolved hosts with novel hosts was more important than contact opportunity, our study did not find any such patterns. In the present study, it could be possible that fitness could differ when B. major infects the invasive L. delicatula versus the diverse native hosts that were infected. Similar observations were made for the efficiency of E. muscae infecting even closely related muscoid species at the same location [48]. As opposed to the present study, the clones in E. muscae were associated with host species. For the fungal clavicipitacean genus Metarhizium, clones occurred within different species; however, because isolates came from soil samples, host relationships are not known [49].
Yet, even if individual fitness was decreased when B. major infected the novel invasive L. delicatula, being a generalist allowed B. major to take advantage of an outbreak population of an invasive host and we did not find native specialist pathogens responding to these outbreak invasive populations.
Conclusion
The studied populations of B. major can infect various hosts in the same location. Analysis of molecular data supports the hypothesis of the clonal nature of the studied population. This can serve as a good example of a genetically diverse population of a pathogen-generalist with a certain amount of gene flow between its members. Use of a broad host range enabled B. major to switch to infection of the spotted lanternfly, a new invasive pest in the USA, which only appeared in Pennsylvania in 2014.
Supporting information
(PDF)
Molecular data on Batkoa major.
(XLSX)
Batkoa major alignment.
(TXT)
Acknowledgments
We thank David Harris and Sarah Stefanik for collection, isolation, and identification of fungi. We thank Jim Liebherr, Tyler Hagerty, Charles Bartlett, Jason Dombroskie for identification of some fungal-killed insects. We thank reviewers for their helpful comments.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
USDA NIFA SCRI 2019-51181-30014. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pell JK, Eilenberg J, Hajek AE, Steinkraus DC. Biology, ecology and pest management potential of Entomophthorales. In: Magan N, Butt TM, Jackson C, editors. Fungi as biocontrol agents: progress, problems and potential. Wallingford, Oxon: CABI International; 2001. pp. 71–153. Available from: https://repository.rothamsted.ac.uk/item/892x9/biology-ecology-and-pest-management-potential-of-entomophthorales [Google Scholar]
- 2.Entomophthorales Bałazy S. Flora of Poland. Fungi (Mycota). Vol. 24. Polish Academy of Science; 1993. [Google Scholar]
- 3.Keller S. Arthropod-pathogenic Entomophthorales of Switzerland. I. Conidiobolus, Entomophaga and Entomophthora. Sydowia. 1987;40: 122–167. [Google Scholar]
- 4.Evans HC. Mycopathogens of insects of epigeal and aerial habitats. Wilding N, Collins NM, Hammond PM, Webber JF, editors. Insect-fungus interactions. 1989; pp. 205–239. Available from:https://scholar.google.com/scholar_lookup?title=Mycopathogens+of+insects+of+epigeal+and+aerial+habitats&author=Evans%2C+H.C.&publication_year=1989 [Google Scholar]
- 5.Walsh SRA, Tyrrell D, Humber RA, Silver JC. DNA restriction fragment length polymorphisms in the rDNA repeat unit of Entomophaga. Exp Mycol. 1990;14: 381–392. [Google Scholar]
- 6.Jensen AB, Thomsen L, Eilenberg J. Value of host range, morphological, and genetic characteristics within the Entomophthora muscae species complex. Mycol Res. 2006;110(8): 941–950. [DOI] [PubMed] [Google Scholar]
- 7.Hajek AE, Butler L. Predicting the host range of entomopathogenic fungi. In: Follett PA, Duan JJ, editors. Nontarget effects of biological control. Boston, MA: Springer US; 2000. pp. 263–276. Available from: 10.1007/978-1-4615-4577-4_15 [DOI] [Google Scholar]
- 8.Gryganskyi AP, Humber RA, Smith ME, Hodge K, Huang B, Voigt K, et al. Phylogenetic lineages in Entomophthoromycota. Persoonia Mol Phylogeny Evol Fungi. 2013;30: 94–105. doi: 10.3767/003158513X666330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gryganskyi AP, Humber RA, Smith ME, Miadlikowska J, Wu S, Voigt K, et al. Molecular phylogeny of the Entomophthoromycota. Mol Phylogenet Evol. 2012;65(2): 682–94. doi: 10.1016/j.ympev.2012.07.026 [DOI] [PubMed] [Google Scholar]
- 10.Humber RA. Synopsis of a revised classification for the Entomophthorales (Zygomycotina). Mycotaxon. 1989;34: 441–460. [Google Scholar]
- 11.Bensch K. Mycobank [Internet]. 2018. Available from: https://www.mycobank.org/page/Home
- 12.Hodge KT, Hajek AE, Gryganskyi A. The first entomophthoralean killing millipedes, Arthrophaga myriapodina n. gen. n. sp., causes climbing before host death. J Invertebr Pathol. 2017;149: 135–140. doi: 10.1016/j.jip.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 13.Clifton EH, Castrillo LA, Gryganskyi A, Hajek AE. A pair of native fungal pathogens drives decline of a new invasive herbivore. Proc Natl Acad Sci. 2019;116(19): 9178–9180. doi: 10.1073/pnas.1903579116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajek AE, Papierok B, Eilenberg J. Methods for study of the Entomophthorales. In: Lacey LA, editor. Manual of techniques in invertebrate pathology. 2nd ed. Amsterdam: Academic Press; 2012. pp. 285–316. [Google Scholar]
- 15.Hajek AE, Gryganskyi A, Bittner T, Liebherr JK, Liebherr JH, Jensen AB, et al. Phylogenetic placement of two species known only from resting spores: Zoophthora independentia sp. nov. and Z. porteri comb nov. (Entomophthorales: Entomophthoraceae). J Invertebr Pathol. 2016;140: 68–74. doi: 10.1016/j.jip.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 16.Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98(6): 625–34. [Google Scholar]
- 17.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8): 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 19.Matheny PB. PCR primers to amplify and sequence rpb2 (RNA polymerase II second largest subunit) in the Basidiomycota (Fungi). 2006. Available from: https://www2.clarku.edu/faculty/dhibbett/downloads2%20syllabi%20proposals%20etc/rpb2%20primers.pdf
- 20.R Core Team. R: A language and environment for statistical computing [Internet]. R Foundation for Statistical Computing. 2014. [cited 2021 Jun 23]. Available from: http://www.R-project.org/ [Google Scholar]
- 21.Nixon KC. Phylogeny. In: Levin SA, editor. Encyclopedia of biodiversity (Second Edition). Waltham: Academic Press, 2001. pp. 16–23. Available from: https://www.sciencedirect.com/science/article/pii/B9780123847195001088 [Google Scholar]
- 22.Jombart T, Ahmed I. adegenet 1.3–1: new tools for the analysis of genome-wide SNP data. Bioinforma Oxf Engl. 2011;27(21): 3070–3071. doi: 10.1093/bioinformatics/btr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamvar ZN, Tabima JF, Grünwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2: e281. doi: 10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2): 945–959. doi: 10.1093/genetics/155.2.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11(1): 94. doi: 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grünwald NJ, Goss EM. Evolution and population genetics of exotic and re-emerging pathogens: novel tools and approaches. Annu Rev Phytopathol. 2011;49: 249–267. doi: 10.1146/annurev-phyto-072910-095246 [DOI] [PubMed] [Google Scholar]
- 27.Goudet J. hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5(1): 184–186. [Google Scholar]
- 28.Gryganskyi AP, Lee SC, Litvintseva AP, Smith ME, Bonito G, Porter TM, et al. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLOS ONE. 2010;5(12): e15273. doi: 10.1371/journal.pone.0015273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X-L, Li Q, Peng W-H, Zhou J, Cao X-L, Wang D, et al. Intra- and inter-isolate variation of ribosomal and protein-coding genes in Pleurotus: implications for molecular identification and phylogeny on fungal groups. BMC Microbiol. 2017;17(1): 139. doi: 10.1186/s12866-017-1046-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soper RS, Shimazu M, Humber RA, Ramos ME, Hajek AE. Isolation and characterization of Entomophaga maimaiga sp. nov., a fungal pathogen of gypsy moth, Lymantria dispar, from Japan. J Invertebr Pathol. 1988;51(3): 229–241. [Google Scholar]
- 31.Hajek AE, Butler L, Wheeler MM. Laboratory bioassays testing the host range of the gypsy moth fungal pathogen Entomophaga maimaiga. Biol Control. 1995;4(5): 530–544. [Google Scholar]
- 32.Eilenberg J, Jensen AB. Strong host specialization in fungus genus Strongwellsea (Entomophthorales). J Invertebr Pathol. 2018;157: 112–116. doi: 10.1016/j.jip.2018.08.007 [DOI] [PubMed] [Google Scholar]
- 33.MacArthur RH. Geographical ecology: patterns in the distribution of species. Princeton University Press; 1984. 296 pp. [Google Scholar]
- 34.Barrett LG, Heil M. Unifying concepts and mechanisms in the specificity of plant-enemy interactions. Trends Plant Sci. 2012;17(5): 282–292. doi: 10.1016/j.tplants.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 35.Bufford JL, Hulme PE, Sikes BA, Cooper JA, Johnston PR, Duncan RP. Taxonomic similarity, more than contact opportunity, explains novel plant–pathogen associations between native and alien taxa. New Phytol. 2016;212(3): 657–667. doi: 10.1111/nph.14077 [DOI] [PubMed] [Google Scholar]
- 36.Remold S. Understanding specialism when the Jack of all trades can be the master of all. Proc Biol Sci. 2012;279(1749): 4861–4869. doi: 10.1098/rspb.2012.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292(5519): 1109–1112. doi: 10.1126/science.1059026 [DOI] [PubMed] [Google Scholar]
- 38.Lihme M, Jensen AB, Rosendahl S. Local scale population genetic structure of Entomophthora muscae epidemics. Fungal Ecol. 2009;2(2): 81–86. [Google Scholar]
- 39.Gryganskyi AP, Humber RA, Stajich JE, Mullens B, Anishchenko IM, Vilgalys R. Sequential utilization of hosts from different fly families by genetically distinct, sympatric populations within the Entomophthora muscae species complex. PLOS ONE. 2013;8(8): e71168. doi: 10.1371/journal.pone.0071168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter D, Powell MJ, et al. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia. 2006;98(6): 860–871. doi: 10.3852/mycologia.98.6.860 [DOI] [PubMed] [Google Scholar]
- 41.Chen M-J, Lin Y, Huang B. Taxonomic position of Zoophthora radicans and its related species and analysis of nrDNA ITS of the isolates. Mycosystema. 2019;38: 1653–1660. [Google Scholar]
- 42.Rush TA, Golan J, McTaggart A, Kane C, Schneider RW, Aime MC. Variation in the internal transcribed spacer region of Phakopsora pachyrhizi and implications for molecular diagnostic assays. Plant Dis. 2019;103(9): 2237–2245. doi: 10.1094/PDIS-08-18-1426-RE [DOI] [PubMed] [Google Scholar]
- 43.Feng M-G, Chen C, Chen B. Wide dispersal of aphid-pathogenic Entomophthorales among aphids relies upon migratory alates. Environ Microbiol. 2004;6(5): 510–516. doi: 10.1111/j.1462-2920.2004.00594.x [DOI] [PubMed] [Google Scholar]
- 44.Hemmati F, Pell JK, McCartney HA, Deadman ML. Airborne concentrations of conidia of Erynia neoaphidis above cereal fields. Mycol Res. 2001;105(4): 485–489. [Google Scholar]
- 45.Steinkraus DC, Hollingsworth RG, Boys GO. Aerial spores of Neozygites fresenii (Entomophthorales: Neozygitaceae): density, periodicity, and potential role in cotton aphid (Homoptera: Aphididae) epizootics. Environ Entomol. 1996; 25: 48–57. [Google Scholar]
- 46.Elkinton JS, Bittner TD, Pasquarella VJ, Boettner GH, Liebhold AM, Gould JR, et al. Relating aerial deposition of Entomophaga maimaiga conidia (Zoopagomycota: Entomophthorales) to mortality of gypsy moth (Lepidoptera: Erebidae) larvae and nearby defoliation. Environ Entomol. 2019;48(5): 1214–1222. doi: 10.1093/ee/nvz091 [DOI] [PubMed] [Google Scholar]
- 47.Dwyer G, Elkinton JS, Hajek AE. Spatial scale and the spread of a fungal pathogen of gypsy moth. Am Nat. 1998;152(3): 485–494. doi: 10.1086/286185 [DOI] [PubMed] [Google Scholar]
- 48.Mullens B, Rodriguez J, Meyer J. An epizootiological study of Entomophthora muscae in muscoid fly populations on Southern California poultry facilities, with emphasis on Musca domestica. Hilgardia. 1987;55(3): 1–41. [Google Scholar]
- 49.Rehner SA. Genetic structure of Metarhizium species in western USA: Finite populations composed of divergent clonal lineages with limited evidence for recent recombination. J Invertebr Pathol. 2020;177: 107491. doi: 10.1016/j.jip.2020.107491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Molecular data on Batkoa major.
(XLSX)
Batkoa major alignment.
(TXT)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.