Abstract
STUDY QUESTION
Can additional genetic variants for circulating anti-Müllerian hormone (AMH) levels be identified through a genome-wide association study (GWAS) meta-analysis including a large sample of premenopausal women?
SUMMARY ANSWER
We identified four loci associated with AMH levels at P < 5 × 10−8: the previously reported MCM8 locus and three novel signals in or near AMH, TEX41 and CDCA7.
WHAT IS KNOWN ALREADY
AMH is expressed by antral stage ovarian follicles in women, and variation in age-specific circulating AMH levels has been associated with disease outcomes. However, the physiological mechanisms underlying these AMH-disease associations are largely unknown.
STUDY DESIGN, SIZE, DURATION
We performed a GWAS meta-analysis in which we combined summary statistics of a previous AMH GWAS with GWAS data from 3705 additional women from three different cohorts.
PARTICIPANTS/MATERIALS, SETTING, METHODS
In total, we included data from 7049 premenopausal female participants of European ancestry. The median age of study participants ranged from 15.3 to 48 years across cohorts. Circulating AMH levels were measured in either serum or plasma samples using different ELISA assays. Study-specific analyses were adjusted for age at blood collection and population stratification, and summary statistics were meta-analysed using a standard error-weighted approach. Subsequently, we functionally annotated GWAS variants that reached genome-wide significance (P < 5 × 10−8). We also performed a gene-based GWAS, pathway analysis and linkage disequilibrium score regression and Mendelian randomization (MR) analyses.
MAIN RESULTS AND THE ROLE OF CHANCE
We identified four loci associated with AMH levels at P < 5 × 10−8: the previously reported MCM8 locus and three novel signals in or near AMH, TEX41 and CDCA7. The strongest signal was a missense variant in the AMH gene (rs10417628). Most prioritized genes at the other three identified loci were involved in cell cycle regulation. Genetic correlation analyses indicated a strong positive correlation among single nucleotide polymorphisms for AMH levels and for age at menopause (rg = 0.82, FDR = 0.003). Exploratory two-sample MR analyses did not support causal effects of AMH on breast cancer or polycystic ovary syndrome risk, but should be interpreted with caution as they may be underpowered and the validity of genetic instruments could not be extensively explored.
LARGE SCALE DATA
The full AMH GWAS summary statistics will made available after publication through the GWAS catalog (https://www.ebi.ac.uk/gwas/).
LIMITATIONS, REASONS FOR CAUTION
Whilst this study doubled the sample size of the most recent GWAS, the statistical power is still relatively low. As a result, we may still lack power to identify more genetic variants for AMH and to determine causal effects of AMH on, for example, breast cancer. Also, follow-up studies are needed to investigate whether the signal for the AMH gene is caused by reduced AMH detection by certain assays instead of actual lower circulating AMH levels.
WIDER IMPLICATIONS OF THE FINDINGS
Genes mapped to the MCM8, TEX41 and CDCA7 loci are involved in the cell cycle and processes such as DNA replication and apoptosis. The mechanism underlying their associations with AMH may affect the size of the ovarian follicle pool. Altogether, our results provide more insight into the biology of AMH and, accordingly, the biological processes involved in ovarian ageing.
STUDY FUNDING/COMPETING INTEREST(S)
Nurses’ Health Study and Nurses’ Health Study II were supported by research grants from the National Institutes of Health (CA172726, CA186107, CA50385, CA87969, CA49449, CA67262, CA178949). The UK Medical Research Council and Wellcome (217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the listed authors, who will serve as guarantors for the contents of this article. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). Funding for the collection of genotype and phenotype data used here was provided by the British Heart Foundation (SP/07/008/24066), Wellcome (WT092830M and WT08806) and UK Medical Research Council (G1001357). M.C.B., A.L.G.S. and D.A.L. work in a unit that is funded by the University of Bristol and UK Medical Research Council (MC_UU_00011/6). M.C.B.’s contribution to this work was funded by a UK Medical Research Council Skills Development Fellowship (MR/P014054/1) and D.A.L. is a National Institute of Health Research Senior Investigator (NF-0616-10102). A.L.G.S. was supported by the study of Dynamic longitudinal exposome trajectories in cardiovascular and metabolic non-communicable diseases (H2020-SC1-2019-Single-Stage-RTD, project ID 874739). The Doetinchem Cohort Study was financially supported by the Ministry of Health, Welfare and Sports of the Netherlands. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Ansh Labs performed the AMH measurements for the Doetinchem Cohort Study free of charge. Ansh Labs was not involved in the data analysis, interpretation or reporting, nor was it financially involved in any aspect of the study. R.M.G.V. was funded by the Honours Track of MSc Epidemiology, University Medical Center Utrecht with a grant from the Netherlands Organization for Scientific Research (NWO) (022.005.021). The Study of Women's Health Across the Nation (SWAN) has grant support from the National Institutes of Health (NIH), DHHS, through the National Institute on Aging (NIA), the National Institute of Nursing Research (NINR) and the NIH Office of Research on Women’s Health (ORWH) (U01NR004061; U01AG012505, U01AG012535, U01AG012531, U01AG012539, U01AG012546, U01AG012553, U01AG012554, U01AG012495). The SWAN Genomic Analyses and SWAN Legacy have grant support from the NIA (U01AG017719). The Generations Study was funded by Breast Cancer Now and the Institute of Cancer Research (ICR). The ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent official views of the funders. The Sister Study was funded by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (Z01-ES044005 to D.P.S.); the AMH assays were supported by the Avon Foundation (02-2012-065 to H.B. Nichols and D.P.S.). The breast cancer genome-wide association analyses were supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the ‘Ministère de l’Économie, de la Science et de l’Innovation du Québec’ through Genome Québec and grant PSR-SIIRI-701, The National Institutes of Health (U19 CA148065, X01HG007492), Cancer Research UK (C1287/A10118, C1287/A16563, C1287/A10710) and The European Union (HEALTH-F2-2009-223175 and H2020 633784 and 634935). All studies and funders are listed in Michailidou et al. (Nature, 2017). F.J.M.B. has received fees and grant support from Merck Serono and Ferring BV. D.A.L. has received financial support from several national and international government and charitable funders as well as from Medtronic Ltd and Roche Diagnostics for research that is unrelated to this study. N.S. is scientific consultant for Ansh Laboratories. The other authors declare no competing interests.
Keywords: anti-Müllerian hormone, AMH, genome-wide association analysis, ovarian ageing, reproductive ageing
Introduction
Anti-Müllerian hormone (AMH) is generally known for its function in sexual differentiation, during which AMH signalling is essential for the regression of internal female reproductive organs in male embryos (Jost, 1947). In women, AMH is expressed by granulosa cells of primary ovarian follicles, and AMH expression continues until the antral stage (Weenen et al., 2004). AMH becomes undetectable after menopause, when the ovarian reserve is depleted, and AMH can therefore be used as a marker for reproductive ageing (Finkelstein et al., 2020).
Variation in age-specific circulating AMH levels has been associated with the occurrence of several non-communicable diseases, including breast cancer (Ge et al., 2018). In addition, it has been suggested that AMH may be involved in the pathogenesis of polycystic ovary syndrome (PCOS) (Homburg and Crawford, 2014). Gaining more insight into genetic variation and biological mechanisms underlying inter-individual variation in AMH expression through genome-wide association studies (GWASs) could provide new clues regarding postnatal functions of AMH, and possibly, the aetiologies of non-communicable diseases associated with AMH levels.
Previous GWASs on circulating AMH levels included either a mixture of male and female adolescents (Perry et al., 2016), a very small study population (Schuh-Huerta et al., 2012a), or women of late reproductive age (Ruth et al., 2019) in whom AMH levels are generally very low. Of the previous GWASs, only the largest (n = 3344) identified a single genetic variant for AMH levels in premenopausal women, at chromosome 20 (rs16991615) (Ruth et al., 2019), which is also associated with natural age at menopause (He et al., 2009; Stolk et al., 2009). As the sample sizes of previous GWASs were relatively small, a larger GWAS meta-analysis might lead to detection of more AMH variation loci. Moreover, as most variation in AMH levels in women is observed at ages 20–40 years (de Kat et al., 2016), the inclusion of younger women will increase power to identify additional loci. Therefore, we aimed to identify additional genetic variants for AMH through a GWAS meta-analysis including 7049 premenopausal female participants. For that, we combined summary statistics from the largest previous AMH GWAS meta-analysis (Ruth et al., 2019) with GWAS data from 3705 additional women of early and middle reproductive age from three different cohorts.
Materials and methods
Study population
We included data from 7049 premenopausal female participants (median age ranged from 15.3 to 48 years across cohorts; Table I) of European ancestry. In addition to the data from the AMH GWAS meta-analysis by Ruth et al. (2019) (n = 3344), we included data from the Doetinchem Cohort Study (Verschuren et al., 2008; Picavet et al., 2017) (n = 2084), the Study of Women’s Health Across the Nation (SWAN) (Sowers et al., 2000) (www.swanstudy.org) (n = 425) and data from adolescent daughters of the Avon Longitudinal Study of Parents and Children (ALSPAC) (www.bristol.ac.uk/alspac/) (Boyd et al., 2013) (n = 1196). The GWAS by Ruth et al. included data from the Generations Study (Swerdlow et al., 2011), Sister Study (Nichols et al., 2015), Nurses’ Health Study (Hankinson et al., 1995), Nurses’ Health Study II (Eliassen et al., 2006) and ALSPAC mothers (Fraser et al., 2013). For the current study, we requested summary statistics excluding data from ALSPAC mothers, as we wanted to analyse data from the ALSPAC mothers separately to investigate potential bias due to first-degree relatedness. More details about participating studies and the definitions used for the assessment of menopausal status are described in the Supplementary Materials and Methods and Supplementary Table SI. All studies received ethical approval from an institutional ethics committee.
Table I.
Distributions of AMH and age per participating study.
| Study | n | AMH, pmol/l (median (IQR)) | Age at blood collection, years (median (IQR)) |
|---|---|---|---|
| Studies contributing to summary statistics GWAS Ruth et al. (2019)a | |||
| Generations Study | 379 | 3.9 (0.8, 11.7) | 44 (40, 48) |
| Sister Study | 438 | 1.2 (0.1, 6.0) | 48 (45, 51) |
| Nurses’ Health Studies | 642 | 6.1 (2.0, 13.9) | 44 (41, 47) |
| Additional studies | |||
| Doetinchem Cohort Study | 2084 | 10.9 (2.9, 25.6) | 37.2 (31.2, 42.9) |
| ALSPAC mothers | 1885 | 2.0 (0.4, 5.2) | 46 (44, 49) |
| ALSPAC daughters | 1196 | 26.1 (18.2, 39.8) | 15.3 (15.3, 15.5) |
| SWAN | 425 | 1.1 (0.2, 3.3) | 47.3 (45.3, 49.3) |
| Total | 7049 | ||
In the original study ALSPAC mothers were included as well, but in the current analyses, summary statistics from ALSPAC were included separately to assess potential genomic inflation due to inclusion of both ALSPAC mothers and daughters. Therefore, we treated the ALSPAC mothers as individual study.
AMH, anti-Müllerian hormone; GWAS: genome-wide association analysis; SWAN, Study of Women’s Health Across the Nation.
AMH measurements
Included studies measured AMH in either serum or plasma using different manual AMH ELISA assays. The picoAMH assay (Ansh Labs, Webster, TX) was used to measure circulating AMH levels in the Generations Study, both Nurses’ Health Studies, the Doetinchem Cohort Study and SWAN. For the Sister Study, the ultrasensitive AMH ELISA (Ansh Labs, Webster, TX, USA) was used, and for ALSPAC, AMH was measured using the Gen II AMH ELISA (Beckman Coulter UK Ltd, High Wycombe, UK). Antibodies differ between the Ansh Labs and Beckman Coulter assays (Moolhuijsen and Visser, 2020), but detailed information about these differences in antibodies is not available. The methodology for handling AMH measurements below the assay limit of detection (LOD) also differed across studies. A detailed overview of these study-specific details has been included in Supplementary Table SI. Across studies, the percentage of measurements under the assay-specific LODs ranged from 0% to 24.2%.
Genotyping and imputation
Extensive details on genotyping and imputation procedures for each participating study are presented in Supplementary Table SII. Briefly, samples of the Generations Study, Sister Study and most samples of the Nurses’ Health Studies, were genotyped using the OncoArray array (Ruth et al., 2019). The remaining 225 samples of the Nurses’ Health Studies were genotyped using Illumina HumanHap550 and HumanHap610 arrays (Ruth et al., 2019). Samples of the Doetinchem Cohort Study were genotyped using the Illumina Infinium Global Screening Array-24 Kit (Illumina Inc., San Diego, CA, USA). For genotyping of samples from ALSPAC mothers and daughters, the Illumina Human660W-Quad array and Illumina HumanHap550 quad genome-wide SNP genotyping platform were used, respectively. SWAN participants were genotyped using the Illumina Multi-Ethnic Global Array (MEGA A1). All participating studies performed sample and SNP QC prior to imputation, which was done using the Haplotype Reference Consortium (HRC) panel version r1.1 2016 (Supplementary Table SII).
Association analyses
All studies converted AMH concentrations to pmol/l using 1 pg/ml = 0.00714 pmol/l. As AMH levels are not normally distributed, AMH measurements were transformed using rank-based inverse normal transformation in all studies, as described previously (Ruth et al., 2019).
In all studies, linear models were fitted, assuming additive SNP effects, adjusted for age at blood collection (years) using different software tools (Marchini et al., 2007; Zhou and Stephens, 2012; Zhan et al., 2016) (Supplementary Materials and methods). Analyses were further adjusted for population stratification by inclusion of either 10 principal components (ALSPAC, SWAN) or a kinship matrix (Generations Study, Sister Study, Nurses’ Health Studies, Doetinchem Cohort Study). In addition, we included summary statistics of the meta-analysis of the Generations Study, Sister Study and Nurses’ Health Studies, which was performed using METAL (Willer et al., 2010), as described elsewhere (Ruth et al., 2019). Separate association analyses were conducted for the ALSPAC mothers and daughters, because of the large differences in both age and AMH distributions between these groups (Supplementary Materials and methods).
Prior to meta-analysis, we performed file-level and meta-level QC on all summary statistics files to clean and check the data, and to identify potential study-specific problems. File-level and meta-level QC were conducted using the R package EasyQC (v9.2; www.uni-regensburg.de/medizin/epidemiologie-praeventivmedizin/genetische-epidemiologie/software/), following a previously published protocol (Winkler et al., 2014) (Supplementary Materials and methods). No study-specific issues were identified through these QC procedures (Supplementary Figs S1, S2, S3, S4 and S5). In addition, we sought to confirm that inclusion of ALSPAC mothers and daughters as separate cohorts would not result in inflation of effect estimates due to first-degree relatedness (411 mother–daughter pairs were present in the ALSPAC data; 5.8% of the total study population). We checked this through meta-analysing only data of the two ALSPAC cohorts and both checking the corresponding QQ plot and calculating λ. Given the absence of genomic inflation (λ = 1.01, QQ plot in Supplementary Fig. S6), we included summary statistics of both ALSPAC mothers and daughters in the meta-analysis to increase statistical power.
We performed an inverse variance weighted (IVW) meta-analysis using METAL (version 2011-03-25; http://csg.sph.umich.edu/abecasis/metal/) (Willer et al., 2010). Genomic control was applied for all included studies. SNPs with a minor allele frequency (MAF) < 1% and/or poor imputation quality (info score < 0.4 or r2 < 0.3, depending on which metric was provided) were excluded. As a result, 8 298 138 autosomal SNPs were included in this AMH GWAS meta-analysis. To assess whether observed effect estimates were homogeneous across studies, we additionally performed a heterogeneity analysis in METAL (Willer et al., 2010).
To identify lead and secondary SNPs within genome-wide significant associated loci, we performed an approximate conditional and joint association analysis (Yang et al., 2012). We used Genome-wide Complex Trait Analysis (GCTA) (Yang et al., 2011) (version 1.93.1f beta; https://cnsgenomics.com/software/gcta/) to run a stepwise model selection procedure to select independently associated SNPs (cojo-slct) using the summary-level data. We estimated linkage disequilibrium (LD) between SNPs using data of 4059 unrelated participants from the EPIC-NL cohort (Beulens et al., 2010) as the LD reference panel.
Because of the strong correlation between AMH and age, and the difference in both the AMH and age distributions in the ALSPAC daughters compared to the other included cohorts, we performed a sensitivity analysis in which we excluded the ALSPAC daughters. Furthermore, this sensitivity analysis served as an additional check that inclusion of both ALSPAC mothers and daughters did not cause identification of false-positive hits.
Gene-based genome-wide association analysis
We performed a gene-based genome-wide association analysis using the MAGMA (de Leeuw et al., 2015) implementation (v1.08) in the online Functional Mapping and Annotation of Genome-wide Association Studies (FUMA) platform (Watanabe et al., 2017) (https://fuma.ctglab.nl/) (parameter settings are listed in Supplementary Table SIII). For this analysis, SNPs located in gene bodies were aggregated to 18 896 protein coding genes (Ensembl build 92). MAGMA tests the joint association of all SNPs in each gene with inverse normal transformed AMH levels using a multiple linear regression approach, which takes LD between SNPs into account (de Leeuw et al., 2015). FUMA considered genes to be significantly associated with circulating AMH levels if P < 2.65 × 10−6 (Bonferroni corrected P-value; 0.05/18 896).
Functional annotation using FUMA
FUMA is an integrative web-based platform that uses 18 biological resources and can be used to functionally annotate lead variants from GWAS, and to prioritize the most likely causal SNPs and genes (Watanabe et al., 2017). We used the SNP2GENE process integrated into FUMA (v1.3.6a) (Watanabe et al., 2017) for the characterization of genomic loci and functional gene mapping (parameter settings are listed in Supplementary Table SIII). We included SNPs identified in our approximate conditional and joint analysis as predefined lead SNPs for the characterization of genomic risk loci. SNPs that were in LD with these lead SNPs (r2 > 0.6) within a 500 kb window based on the 1000G Phase 3 European reference panel population in FUMA and a GWAS meta-analysis P-value < 0.05 were selected as candidate SNPs. Non-GWAS-tagged SNPs from the 1000G Phase 3 European reference that met these LD and distance criteria were also selected as candidate SNPs. Candidate SNPs were annotated based on Combined Annotation Dependent Depletion scores (Kircher et al., 2014), Regulome DB scores (Boyle et al., 2012) and chromatin states (Kundaje et al., 2015) (Supplementary Table SIII). Positional mapping, eQTL mapping and chromatin interaction mapping were used to map SNPs to genes (Supplementary Table SIII). For chromatin states, eQTL mapping and chromatin interaction mapping, we only selected tissues and cell types that are most likely to be involved in AMH expression and signalling (Supplementary Table SIII).
Pathway analysis using DEPICT
We used the hypothesis-free pathway analysis tool DEPICT (v1; https://github.com/perslab/depict) (Pers et al., 2015) to prioritize the most likely causal genes at associated loci, to highlight gene sets enriched in genes within associated loci, and to identify tissues/cell types that are implicated by the associated loci. For these analyses, we included all suggestive significant SNPs (P < 5 × 10−6), which were clumped at LD r2 < 0.1 and a physical distance of 500 kb using PLINK v.1.9 as part of the DEPICT pipeline.
Linkage disequilibrium score regression
We estimated SNP heritability using the LD Hub web interface (v1.9.3; http://ldsc.broadinstitute.org/) (Zheng et al., 2017) for LD score regression (Bulik-Sullivan et al., 2015). In addition, we used LD Hub to estimate SNP-based genetic correlations between AMH and phenotypes that have been associated with AMH in observational studies. These genetic correlation analyses make use of GWAS summary statistics for all SNPs to estimate genetic covariance among SNPs for two traits (Bulik-Sullivan et al., 2015). Included phenotypes comprised reproductive traits, hormones (leptin), anthropometric traits, blood lipids, glycemic traits, metabolites, cardiometabolic traits, cancer, autoimmune diseases, bone mineral density, ageing and smoking behaviour. Of the 597 UK Biobank traits in the LD Hub database, we only included traits that corresponded to these phenotype categories, resulting in 345 comparisons. To correct for multiple testing, we calculated false discovery rates (FDR), using the p.adjust function in R (R package ‘stats’) (R Core Team, 2017). FDR adjusted P-values < 0.05 were considered to be significant.
Mendelian randomization
In observational studies, AMH has been associated with breast cancer (Ge et al., 2018), and PCOS (Laven et al., 2004), amongst other diseases. As the exact function of AMH in the aetiology of these diseases is unclear, and actual AMH levels are associated with predicted future age at menopause and current menopausal status, causality of these AMH-disease associations remains to be determined. Mendelian randomization (MR) is a specific instrumental variable analysis method that may provide evidence for causality of observational associations (Davey Smith and Ebrahim, 2005). Briefly, SNPs that are robustly associated with AMH can be used as instrumental variables to estimate the causal effect of AMH on other traits and/or disease outcome.
Because our AMH GWAS meta-analysis only included women, and previous research suggests that genetic variants for inter-individual differences in AMH levels differ between males and females (Perry et al., 2016), we performed MR analyses for the female-specific outcomes breast cancer and PCOS only. We performed two-sample MR analyses (Pierce and Burgess, 2013) using the R package TwoSampleMR (version 0.5.1) (Hemani et al., 2018). We included identified lead SNPs as genetic instrumental variables for AMH. For the outcomes, we included summary statistics from recent large-scale GWASs for breast cancer (n = 228 951; 122 977 cases) (Michailidou et al., 2017) and PCOS (n = 113 238; 10 074 cases) (Day et al., 2018). Wald ratio estimates were calculated for individual SNPs and a random effects IVW meta-analysis approach was used to combine these estimates. To assess the strength of included genetic variants for AMH, we calculated F-statistics corresponding to the IVW analyses, using the proportion of variance in AMH explained by the genetic variants, the sample size of the outcome GWASs and the number of variants included (Burgess and Thompson, 2015). We compared the overall MR estimate (i.e. IVW) to SNP-specific MR estimates (i.e. Wald ratio) since inconsistent estimates are indicative of horizontal pleiotropy, which is a violation of the MR assumptions (Hemani et al., 2018). In addition, we tested for heterogeneity in causal effects amongst the genetic variants using Cochrane’s Q statistics and performed leave-one-out sensitivity analyses to assess the potential effect of outlying variants.
Post-hoc analysis: assay-specific AMH levels across rs10417628 genotype categories
A recent case report suggested that the amino acid substitution corresponding to rs10417628 (AMH locus) reduces AMH detection by the picoAMH assay from Ansh Labs without influencing AMH bioactivity (Hoyos et al., 2020). We sought to verify this finding in a subsample of the Doetinchem Cohort Study, for which AMH was measured using both the picoAMH assay and the less sensitive Gen II assay from Beckman Coulter. In addition, we requested median AMH levels across rs10417628 genotype categories from both ALSPAC mothers and daughters, because ALSPAC also used the AMH Gen II assay from Beckman Coulter.
Results
Descriptive statistics on age and AMH levels of the study participants included in this GWAS meta-analysis are presented per study in Table I. Median AMH ranged from 1.1 pmol/l in SWAN to 26.1 pmol/l in ALSPAC daughters. Median age ranged from 15.3 years in ALSPAC daughters to 48 years in the Sister Study.
Genome-wide association analysis
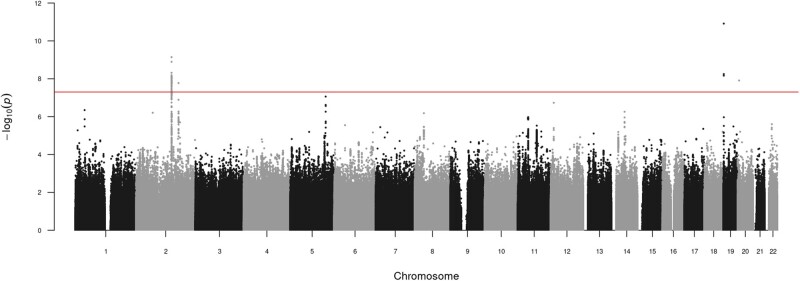
We identified four genome-wide significant lead SNPs (P < 5 × 10−8) for inverse normally transformed AMH, in four loci (Table II, Fig. 1 and Supplementary Figs S7 and S8). Approximate conditional and joint analysis did not reveal secondary signals. In addition to the previously reported locus on chromosome 20 (rs16991615, nearest gene: MCM8), we identified one locus on chromosome 19 (nearest gene: AMH) and two loci on chromosome 2 (nearest genes: TEX41 and CDCA7). The strongest signal was rs10417628 on chromosome 19, which is physically located in the AMH gene (β = −0.34, SE = 0.05, P = 1.2 × 10−11) (Supplementary Fig. S8A). Combined, the four lead SNPs explained 1.47% of the variance in AMH levels.
Table II.
Loci significantly associated (P < 5 × 10−8) with inverse normally transformed AMH in women.
| Nearest gene | SNP | Chr | Pos | EA | OA | EAF | n | Imputation quality | Effect (SE) | P | Direction | P het | Percentage of variance in AMH explained |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMH | rs10417628 | 19 | 2 251 817 | T | C | 0.02 | 7049 | 0.83 | −0.34 (0.05) | 1.2 × 10−11 | −−−−− | 0.14 | 0.50% |
| TEX41 | rs13009019 | 2 | 145 670 572 | A | G | 0.69 | 7049 | 0.95 | −0.09 (0.01) | 7.2 × 10−10 | −−−−− | 0.24 | 0.35% |
| MCM8 | rs16991615 | 20 | 5 948 227 | A | G | 0.07 | 7049 | 0.99 | 0.16 (0.03) | 1.2 × 10−8 | ++−++ | 0.0009 | 0.30% |
| CDCA7 | rs11683493 | 2 | 174 259 325 | T | C | 0.57 | 7049 | 0.97 | −0.08 (0.01) | 1.7 × 10−8 | −−−−− | 0.03 | 0.32% |
Definition of columns: nearest gene, nearest gene identified using DEPICT tool (Subjects and Methods); SNP, genetic variant identified as lead SNP; Chr, chromosome; Pos, base pair position genomic build GRCh37; EA, effect allele; OA, other allele; EAF, effect allele frequency; n, number of samples contributing to estimate; imputation quality, mean imputation quality over the included studies; effect (SE); effect size and corresponding standard error; P, P-value; Direction, direction of effect for previous GWAS, ALSPAC mothers, ALSPAC daughters, Doetinchem Cohort Study and SWAN, respectively; Phet, P-value for heterogeneity of effect across studies; AMH, anti-Müllerian hormone; SWAN, Study of Women’s Health Across the Nation.
Figure 1.
Manhattan plot of genome-wide association results for inverse normally transformed AMH in women. Results are association results from meta-analysis of inverse normally transformed AMH in 7049 women of European ancestry. Individual studies adjusted analyses for age at AMH measurement and population stratification (through kinship matrix or 10 principal components). AMH, anti-Müllerian hormone.
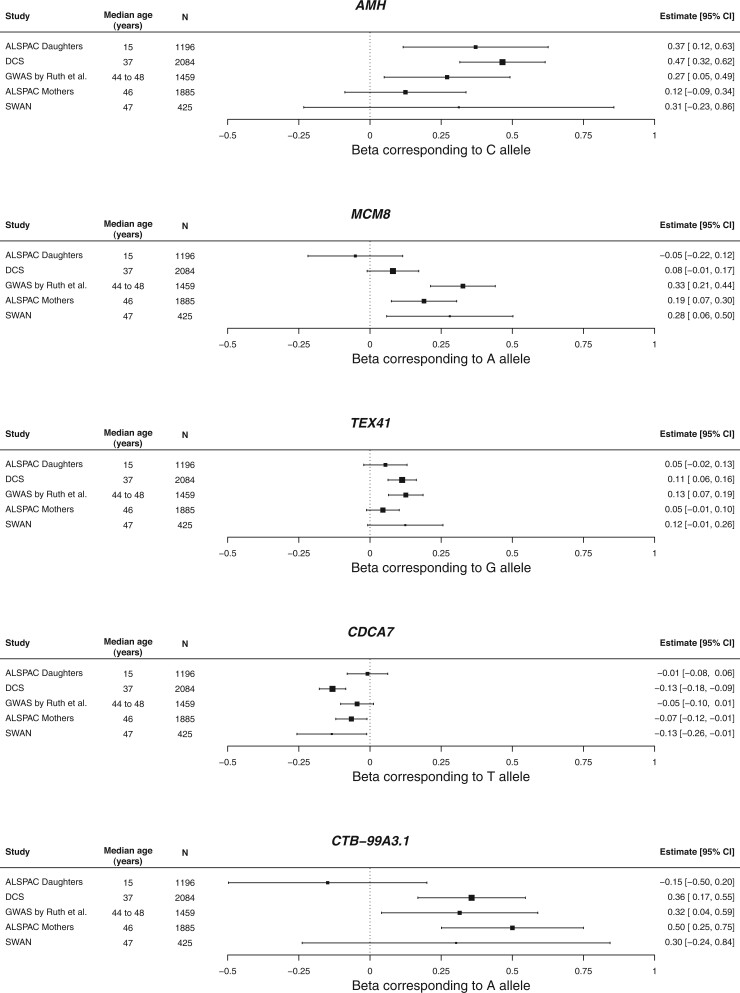
We observed no systematic deviation of associations in the ALSPAC adolescents from the associations in women of middle reproductive age (Fig. 2). Only for a locus at chromosome 5, the association in the ALSPAC adolescents was radically different than in the other cohorts. For the MCM8 locus, estimates were weaker in both the ALSPAC adolescents and women in the Doetinchem Cohort Study compared to the remaining studies.
Figure 2.
Effect estimates for the five SNPs that reached genome-wide significance in the main and sensitivity analyses across included studies.
In the sensitivity analysis excluding ALSPAC daughters, all four loci from the main analysis remained genome-wide significant and beta estimates were very similar to those from the main analysis. However, an additional locus at chromosome 5 (rs116090962, nearest gene: CTB-99A3.1) was identified (β = 0.38, SE = 0.07, P = 6.0 × 10−9) and P-values corresponding to the MCM8, and CDCA7 loci were lower in this sensitivity analysis compared to the main analysis (Supplementary Table SIV).
Gene-based genome-wide association analysis
Gene-based genome-wide association analysis, which tested associations between 18 896 protein coding genes and inverse normal transformed AMH, highlighted the following two significant genes: AMH and BMP4 (Supplementary Fig. S9).
Functional annotation using FUMA
Through the SNP2GENE process, FUMA identified 82 candidate SNPs that were in LD with the four identified lead SNPs (Supplementary Table SV). These candidate SNPs were used for the prioritization of genes.
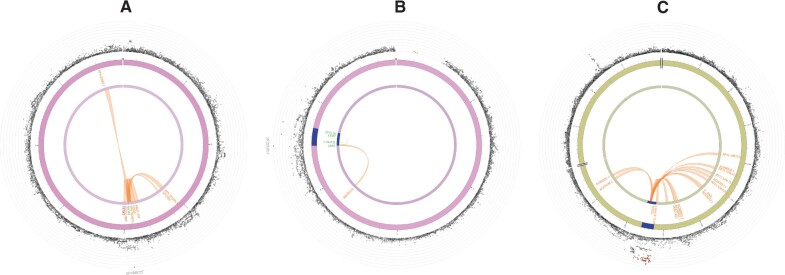
In total, 12 genes were mapped to the locus of the previously identified SNP on chromosome 20 (rs16991615) (Supplementary Table SVI), of which MCM8 and CRLS1 were prioritized based on eQTL mapping (Fig. 3A). CRLS1 was the only gene prioritized based on both eQTL mapping and chromatin interactions. However, as rs16991615 is a missense variant located in exon 9 of the MCM8 gene, and this was the only SNP identified for this locus, MCM8 is the most likely gene causing this signal.
Figure 3.
Circos plots for the genome-wide significant loci for inverse normally transformed AMH in women. Circos plots are presented for each of the identified loci: MCM8 (A), AMH (B), TEX41 and CDCA7 (C). The outer layer represents the Manhattan plot. The second (including genomic positions) and third layers represent the chromosome ring; genomic risk loci are depicted in blue. Only genes mapped by either eQTLs or chromatin mapping are plotted. Genes only mapped by eQTLs are green; genes only mapped by chromatin interactions are orange; and genes mapped by both have a red colour. Orange-coloured lines represent chromatin interactions, green-coloured lines are eQTL links. Plots were created using the FUMA platform (Watanabe et al., 2017). AMH, anti-Müllerian hormone; FUMA, Functional Mapping and Annotation of Genome-wide Association Studies.
For the locus on chromosome 19, three candidate SNPs (rs10417628, rs12462821, rs7247495) were identified. The lead SNP in this locus (rs10417628) is also a missense variant, located in exon 5 of the AMH gene, making this the most likely causal gene at this locus. The other two variants were located in intronic and intronic non-coding RNA regions. Based on the used parameter settings, FUMA mapped eight genes to the AMH locus (Supplementary Table SVI), of which four were highlighted by eQTL mapping (AMH, C19orf35, SPPL2B and LSM7) and one was highlighted through chromatin interactions (ABHD17A) (Fig. 3B).
Most of the candidate SNPs were identified for the locus on chromosome 2 near TEX41. All 77 variants were located in either intronic or exonic long non-coding RNA regions. Of the 15 genes mapped to this locus (Supplementary Table SVI), no genes were prioritized based on eQTL mapping, but several genes were prioritized based on chromosome interactions, including ZEB2-AS1 (Fig. 3C). In the other locus on chromosome 2, for which CDCA7 is the nearest gene, no additional candidate SNPs were identified, and no genes were mapped to this locus.
Pathway analysis using DEPICT
Using the DEPICT tool, 188 suggestive associated SNPs (P < 5 × 10−6) were clumped at LD r2 < 0.1 and a physical distance of 500 kb, resulting in 24 clumps and 18 loci as input for the enrichment analyses (Supplementary Table SVII). The top three prioritized gene sets were ‘URI1 PPI subnetwork’, ‘NFYB PPI subnetwork’ and ‘nuclear inner membrane’ (Supplementary Table SVIII). ‘Induced Pluripotent Stem Cells’ was identified as the highest prioritized cell type (Supplementary Table SIX). However, none of these enrichments were statistically significant (FDR values for all comparisons > 0.05).
DEPICT prioritized nine genes at FDR < 0.05 as most likely causal genes (Supplementary Table SX). Of the genome-wide associated loci, only MCM8 (rs16991615) and CDCA7 (rs11683493) were prioritized at this FDR threshold. AMH and BMP4 were also prioritized by DEPICT, but FDR values were > 0.20.
Linkage disequilibrium score regression
We used LD score regression implemented in LD Hub to calculate SNP heritability for AMH based on the meta-analysis summary statistics. Total SNP heritability (hg2) on the observed scale was estimated to be 15% (SE = 7%). We additionally performed genetic correlations analyses between AMH and 345 traits on LD Hub. After correction for multiple testing, AMH was only significantly correlated with age at menopause (rg = 0.82, SE = 0.19, FDR = 0.003) (Supplementary Table SXI).
Mendelian randomization analyses
IVW MR estimates did not indicate a causal effect of circulating AMH on breast cancer risk (ORIVW = 1.00, 95% CI: 0.74–1.36; Table III and Supplementary Fig. S10). Results from the single SNP analysis including the variant in the AMH locus also did not support a causal association with breast cancer (ORIVW = 0.99, 95% CI = 0.87–1.12), whereas analyses for the remaining variants suggested a risk decreasing effect of the SNPs in the TEX41 and CDCA7 loci and a risk increasing effect of the variant in the MCM8 locus (Table III). In agreement with these findings, a formal heterogeneity test for the IVW estimate indicated heterogeneity in causal effects amongst the four genetic variants (2.13 × 10−11), although the interpretation of this heterogeneity P-value is limited due to the small number of included SNPs. Leave-one-out sensitivity analyses supported the outlying effect of rs16991615 (MCM8 locus) (Supplementary Fig. S11).
Table III.
Mendelian randomization estimates for causal effects of circulating AMH on breast cancer and PCOS risk.
| Outcome | Method | Odds ratio | 95% CI | P |
|---|---|---|---|---|
| Breast cancer | IVW | 1.00 | 0.74–1.36 | 0.98 |
| Wald ratio estimate for rs10417628 (AMH) | 0.99 | 0.87–1.12 | 0.85 | |
| Wald ratio estimate for rs13009019 (TEX41) | 0.84 | 0.72–0.97 | 0.02 | |
| Wald ratio estimate for rs16991615 (MCM8) | 1.60 | 1.37–1.87 | 1.79 × 10−9 | |
| Wald ratio estimate for rs11683493 (CDCA7) | 0.76 | 0.65–0.89 | 9.41 × 10−4 | |
| PCOS | IVW | 1.29 | 0.85–1.95 | 0.23 |
| Wald ratio estimate for rs10417628 (AMH) | 1.27 | 0.64–2.56 | 0.49 | |
| Wald ratio estimate for rs13009019 (TEX41) | 1.66 | 0.80–3.45 | 0.18 | |
| Wald ratio estimate for rs16991615 (MCM8) | 1.75 | 0.83–3.69 | 0.14 | |
| Wald ratio estimate for rs11683493 (CDCA7) | 0.66 | 0.29–1.50 | 0.32 | |
Odds ratio and 95% CI are per 1 unit increase in inverse normally transformed AMH.
AMH, anti-Müllerian hormone; PCOS, polycystic ovary syndrome; IVW, inverse variance weighted; MR, Mendelian randomization.
For PCOS, the IVW MR estimate did not provide evidence for a causal effect either (ORIVW = 1.29, 95% CI = 0.85–1.95; Table III and Supplementary Fig. S12), although the wide confidence interval indicated a considerable degree of uncertainty around this estimate. Single SNP analyses resulted in a similar effect estimate for the variant in the AMH locus (ORIVW = 1.27, 95% CI = 0.64–2.56), risk increasing effects for the SNPs in the TEX41 and MCM8 loci, and a risk decreasing effect of the variant in the CDCA7 locus (Table III). The heterogeneity test did not suggest heterogeneous effects of the individual SNPs (P = 0.30), most likely because of the high uncertainty in individual SNP estimates, but again interpretation of this P-value is limited with only four SNPs. Leave-one-out sensitivity analyses indicated rs11683493 (CDCA7 locus) affected the IVW estimate most, and that exclusion of this variant resulted in a positive association (ORIVW = 1.53, 95% CI = 1.01–2.33) (Supplementary Fig. S13).
Post-hoc analysis: assay-specific AMH levels across rs10417628 genotype categories
For 1518 of the 2084 included Doetinchem Cohort Study participants, both picoAMH assay and AMH Gen II assay measurements were available. In general, median AMH levels measured using the Gen II assay were less different between women homozygous for the reference allele (C allele) and heterozygous women (median AMH levelshomozygousrefallele = 953.0 pg/ml, IQR: 428.0—1999.0; median AMH levelsheterozygous = 848.0 pg/ml, IQR: 509.0—1310.0), compared to AMH levels measured using the picoAMH assay (median AMH levelshomozygousrefallele = 1485.9 pg/ml, IQR: 704.4–3150.0; median AMH levelsheterozygous = 811.0 pg/ml, IQR: 462.3–1480.9). Only one woman was estimated to be homozygous for the effect allele (dosage T allele = 1.9, age at measurement = 28.3 years). Whereas AMH levels for this woman were undetectable using the picoAMH assay, circulating AMH levels were detected using the less sensitive Gen II assay (318 pg/ml).
For ALSPAC, which also used the Gen II assay to measure AMH, the distribution of AMH levels was similar across adult women homozygous for the reference allele and heterozygous women as well. However, in the ALSPAC daughters, median AMH levels were clearly higher in adolescents homozygous for the reference allele compared to heterozygous adolescents. Among the ALSPAC participants, only one adolescent was homozygous for the T allele, but her AMH levels could not be shared due to disclosure risk.
Discussion
We identified four loci for circulating AMH levels in women of European ancestry. In addition to confirming a previously reported signal in the MCM8 locus, we discovered three new signals in and near the AMH, TEX41 and CDCA7 genes. In total, 35 genes were prioritized for these loci based on physical position, eQTL mapping and chromatin interactions, but pathway analyses did not reveal enrichments of gene-sets, tissues or cell types for genes annotated to suggestive associated SNPs. Genetic correlation analyses supported a shared genetic architecture between AMH levels and age at menopause. Exploratory MR analyses did not provide strong evidence of a causal effect of circulating AMH on breast cancer or PCOS.
We confirmed the association between rs16991615 and circulating AMH levels, previously reported by Ruth et al. (2019). This SNP is a missense variant located in exon 9 of the MCM8 gene, rendering MCM8 the most likely causal gene at this locus. In humans, MCM8 plays a role in in homologous recombination, which is critical for DNA repair (Park et al., 2013). Previous studies have linked MCM8 deficiency to premature ovarian failure and infertility, but also to cancer development (Michailidou et al., 2017; Griffin and Trakselis, 2019; Lutzmann et al., 2019). Associations between rs16991615 and age at menopause (Day et al., 2015) and number of ovarian follicles (Schuh-Huerta et al., 2012b) have also been reported, which suggests that this locus is associated with circulating AMH levels because of its influence on the number of antral follicles.
Our GWAS study is the first AMH GWAS that has identified a missense SNP (rs10417628) in the AMH gene in women. A previous AMH GWAS including adolescents from ALSPAC identified three SNPs in the AMH gene that were only significantly associated with AMH levels in male adolescents, and of which one (rs2385821) was in moderate LD with our lead SNP rs10417628 (R2 = 0.55) (Perry et al., 2016). However, approximate conditional and joint analyses suggested that these variants represent the same signal at the AMH locus. Although identification of a genetic variant in the gene encoding for AMH itself suggests that we revealed an actual signal for circulating AMH concentrations, a recent case report suggests that the amino acid substitution corresponding to rs10417628 reduces AMH detection by the picoAMH assay from Ansh Labs without influencing AMH bioactivity (Hoyos et al., 2020). We sought to verify this finding in a subsample of the Doetinchem Cohort Study, for which AMH was measured using both the picoAMH and the less sensitive Gen II assay from Beckman Coulter. In addition, we requested median AMH levels across genotype categories for ALSPAC, which also used this Beckman Coulter assay to measure AMH. In line with the finding of Hoyos et al., our post-hoc analyses indicated that AMH levels were undetectable using the picoAMH, but not using the Gen II assay, for the only homozygous woman in the Doetinchem Cohort Study. Moreover, median AMH levels were less different between women homozygous for the reference allele and heterozygous women in both the Doetinchem Cohort Study and the adult ALSPAC participants. In contrast, we did observe a clear difference in median AMH levels between ALSPAC adolescents homozygous for the reference allele and heterozygous adolescents. Because of this limited and inconsistent evidence, and the lack of publicly available information on the antibodies and conformational epitopes of the Gen II assay, we do not want to draw any definite conclusions about this yet.
For the associated loci on chromosome 2, it is more challenging to assign possible causal genes, as TEX41 is a long non-coding RNA and the SNP in the CDCA7 locus was located in an intergenic region. Gene mapping based on chromatin interactions with TEX41 highlighted several genes, including the long non-coding RNA ZEB2-AS1 (ZEB2 antisense RNA 1). ZEB2-AS1 up-regulates expression of the protein ZEB2 (Beltran et al., 2008). ZEB2 (also known as SIP1) inhibits signal transduction in TGF-β and BMP signalling through interaction with ligand-activated SMAD proteins (Postigo, 2003; Conidi et al., 2011). Among other BMP proteins, BMP4 has been reported to regulate AMH expression through activation of SMAD proteins (Estienne et al., 2015; Pierre et al., 2016). Based on identification of BMP4 in our gene-based association analysis and its prioritization by DEPICT, we hypothesize that BMP4-induced AMH expression may be regulated by ZEB2 interaction. However, fundamental laboratory research is needed to prove this hypothesis.
CDCA7 (also known as JPO1) is a direct target gene of the transcription factor MYC and is involved in apoptosis (Gill et al., 2013). Functional annotation did not map any genes to this locus and thus the mechanism through which this locus affects circulating AMH levels remains to be elucidated. Based on the involvement of CDCA7 in apoptosis, it may be possible that this gene affects the number of antral follicles, which are the main producers of AMH in women (Weenen et al., 2004). Ideally, future studies should explore whether the observed genetic associations may merely reflect the size of the ovarian reserve, by adjusting analyses for antral follicle count. Such analyses would also show whether we can actually use the identified variants as instruments for circulating AMH levels itself or for the quantity of antral follicles in MR analyses.
We did not find support for a causal effect of circulating AMH levels on breast cancer and PCOS risk in our exploratory MR analyses. To be valid genetic instrumental variables for MR, SNPs have to fulfil the following three criteria (Lawlor et al., 2008): (i) SNPs have to be associated with the circulating AMH levels; (ii) SNPs cannot be associated with confounders of the studied AMH-outcome associations and (iii) SNPs cannot influence the outcomes through mechanisms that do not involve circulating AMH levels. Because rs10417628 in the AMH gene potentially reflects AMH detection instead of AMH expression, analyses including this variant should be interpreted with caution. However, leave-one-out analyses excluding this variant did not affect IVW MR estimates. Based on the function of genes mapped to the loci on chromosomes 20 and 2, it is likely that these variants affect breast cancer and PCOS risk through mechanisms independent of AMH (e.g. DNA replication and apoptosis), in particular the MCM8 locus, which has also been identified in breast cancer GWAS (Michailidou et al., 2017). Due to the limited number of identified lead SNPs, it was not possible to assess whether our results were indeed biased by horizontal pleiotropy. Furthermore, weak instrument bias may still have biased MR results towards the null, since the F statistics may be overestimated in this GWAS (853.9 for breast cancer, and 422.4 for PCOS). Consequently, we should be cautious about excluding a causal effect of AMH on the studied outcomes.
Initially, we decided to analyse both the ALSPAC adolescents and adult women together to increase the total number of included participants, and consequently statistical power. Previous research suggests that AMH levels in females rise during puberty, until the mid to late twenties, and after that, decreases until menopause (Kelsey et al., 2011; de Kat et al., 2016); that is AMH levels are heterogeneous across different reproductive phases. Based on these observations and the differences in both age and AMH distributions between the ALSPAC adolescents and other participants, we performed a sensitivity analysis excluding the adolescents from ALSPAC. This analysis revealed an additional locus on chromosome 5 (rs116090962, nearest gene: CTB-99A3.1), although we could not find clues for its association with circulating AMH levels in adult women only, nor with AMH levels in general. Study-specific betas revealed an opposite effect for the MCM8 locus and a minimal effect for the CDCA7 locus in adolescents compared to effects in adult women. However, we observed no systematic deviation of SNP-AMH associations in the adolescents from associations in the adult women. Nevertheless, our observation that the P-values corresponding to the MCM8, CDCA7 and CTB-99A3.1 loci were lower in the analyses excluding the ALSPAC adolescents suggests that including these samples increased heterogeneity, rather than statistical power, even though our conclusions remained the same. A larger GWAS including older adolescents and a larger proportion of females aged 20–40, would be required to reveal potential gene–age interactions explain variation in AMH expression.
The main strengths of the current GWAS meta-analysis are its size, which is twice the size of the previous GWAS meta-analysis, and its larger proportion of women of early-reproductive age. Given the function of AMH in ovarian follicle development, circulating levels and variation in AMH levels decrease with age. As a result, statistical power to identify genetic variants for circulating AMH increases if younger women are included. Still, our sample size remains relatively small for a GWAS, and future larger studies may lead to the detection of additional variants for circulating AMH levels. This is supported by our chip heritability estimate of 15% (SE = 7%), which indicates that there are likely more SNPs that contribute to variability in AMH levels. Identification of additional genetic variants will also facilitate increased power to identify pathways and tissues enriched for genes involved in AMH expression. A second limitation of this study is the potential overlap in participants between the current AMH GWAS and the GWAS for breast cancer (Michailidou et al., 2017) (maximum n = 1459; 20.7% of current study) and PCOS (Day et al., 2018) (maximum n = 225; 3.2% of current study). Overlap in participants in two-sample MR analyses may bias effect estimates and inflate Type 1 error rates (Burgess et al., 2016).
In conclusion, we replicated the previously reported association with the MCM8 locus and identified three novel loci for circulating AMH levels in women, including the AMH locus. The strongest signal in this locus possibly affects AMH detection by specific assays rather than AMH bioactivity, but further research is required to confirm this hypothesis. Genes mapped to the MCM8, TEX41 and CDCA7 loci are involved in the cell cycle and processes like DNA replication and apoptosis. The mechanism underlying their associations with AMH may affect the size of the ovarian follicle pool. MR analyses did not support a causal effect of AMH on breast cancer or PCOS, but these finding should be interpreted with caution because we could not robustly explore how valid the instruments were and because of the considerable uncertainty in effect estimates.
Data availability
The data underlying this article are available in the NHGRI-EBI Catalog of human genome-wide association studies, at https://www.ebi.ac.uk/gwas/. Code underlying the analyses for this article are available via GitHub, at https://github.com/reneemgverdiesen/AMH_GWAS.
Supplementary Material
Acknowledgements
The Generations Study thank Breast Cancer Now and the Institute of Cancer Research for support and funding of the Generations Study, and the study participants, study staff, and the doctors, nurses and other healthcare staff and data providers who have contributed to the study. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. We would like to thank the participants and staff of the Nurses’ Health Study and the Nurses’ Health Study II.
For the Doetinchem Cohort Study, we thank Dr HSJ Picavet, A Blokstra MSc and Petra Vissink for their crucial role in data collection and data management.
For the ALSPAC Cohort, we are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
SWAN thanks the study staff at each site and all the women who participated in SWAN. Clinical Centers: University of Michigan, Ann Arbor—Siobán Harlow, PI 2011–present, MaryFran Sowers, PI 1994–2011; Massachusetts General Hospital, Boston, MA—Joel Finkelstein, PI 1999–present; Robert Neer, PI 1994–1999; Rush University, Rush University Medical Center, Chicago, IL—Howard Kravitz, PI 2009–present; Lynda Powell, PI 1994–2009; University of California, Davis/Kaiser—Ellen Gold, PI; University of California, Los Angeles—Gail Greendale, PI; Albert Einstein College of Medicine, Bronx, NY—Carol Derby, PI 2011–present, Rachel Wildman, PI 2010–2011; Nanette Santoro, PI 2004–2010; University of Medicine and Dentistry—New Jersey Medical School, Newark—Gerson Weiss, PI 1994–2004; and the University of Pittsburgh, Pittsburgh, PA—Karen Matthews, PI. NIH Program Office: National Institute on Aging, Bethesda, MD—Chhanda Dutta 2016–present; Winifred Rossi 2012–2016; Sherry Sherman 1994–2012; Marcia Ory 1994–2001; National Institute of Nursing Research, Bethesda, MD—Program Officers. Central Laboratory: University of Michigan, Ann Arbor—Daniel McConnell (Central Ligand Assay Satellite Services). SWAN Repository: University of Michigan, Ann Arbor—Siobán Harlow 2013–present; Dan McConnell 2011–2013; MaryFran Sowers 2000–2011. Coordinating Center: University of Pittsburgh, Pittsburgh, PA—Maria Mori Brooks, PI 2012—present; Kim Sutton-Tyrrell, PI 2001–2012; New England Research Institutes, Watertown, MA—Sonja McKinlay, PI 1995–2001. Steering Committee: Susan Johnson, Current Chair, Chris Gallagher, Former Chair.
The Sister Study acknowledges the contributions of Jack A. Taylor, MD, PhD, Molecular & Genetic Epidemiology Group, Epidemiology Branch, National Institute of Environmental Health Sciences, Durham, NC who secured initial funding for the Sister Study GWAS, and Hazel B. Nichols, PhD, Associate Professor, Department of Epidemiology, Gillings School of Global Public Health who was the PI of the grant that allowed the AMH assays for the case-control study of AMH and breast cancer in premenopausal women.
Authors’ roles
R.M.G.V., Y.T.v.d.S., C.H.v.G. and N.C.O.M. were involved in the initial study design. W.M.M.V. and F.J.M.B. (Doetinchem Cohort Study), D.A.L. (ALSPAC), A.H.E. (Nurses’ Health Studies), P.K. (Nurses’ Health Studies), D.P.S. (Sister Study), S.D.H. (SWAN) and A.J.S. (Generations Study) were involved in acquisition of (part of) the data. R.M.G.V., M.C.B., A.L.G.S., J.A.S. and K.S.R. carried out analyses. RMGV drafted the article. All authors (including NS, MJS and AM) gave input on the initial study design and analyses, were involved in the interpretation of the results, revising and approving the manuscript.
Funding
Nurses’ Health Study and Nurses’ Health Study II were supported by research grants from the National Institutes of Health (CA172726, CA186107, CA50385, CA87969, CA49449, CA67262, CA178949). The UK Medical Research Council and Wellcome (217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the listed authors, who will serve as guarantors for the contents of this article. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). Funding for the collection of genotype and phenotype data used here was provided by the British Heart Foundation (SP/07/008/24066), Wellcome (WT092830M and WT08806) and UK Medical Research Council (G1001357). M.C.B., A.L.G.S. and D.A.L. work in a unit that was funded by the University of Bristol and UK Medical Research Council (MC_UU_00011/6). M.C.B.’s contribution to this work was funded by a UK Medical Research Council Skills Development Fellowship (MR/P014054/1) and D.A.L. is a National Institute of Health Research Senior Investigator (NF-0616-10102). A.L.G.S. was supported by the study of Dynamic longitudinal exposome trajectories in cardiovascular and metabolic non-communicable diseases (H2020-SC1-2019-Single-Stage-RTD, project ID 874739). The Doetinchem Cohort Study was financially supported by the Ministry of Health, Welfare and Sports of the Netherlands. The funder had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Ansh Labs performed the AMH measurements for the Doetinchem Cohort Study free of charge. Ansh Labs was not involved in the data analysis, interpretation or reporting, nor was it financially involved in any aspect of the study. R.M.G.V. was funded by the Honours Track of MSc Epidemiology, University Medical Center Utrecht with a grant from the Netherlands Organization for Scientific Research (NWO) (022.005.021). The Study of Women’s Health Across the Nation (SWAN) has grant support from the National Institutes of Health (NIH), DHHS, through the National Institute on Aging (NIA), the National Institute of Nursing Research (NINR) and the NIH Office of Research on Women’s Health (ORWH) (U01NR004061; U01AG012505, U01AG012535, U01AG012531, U01AG012539, U01AG012546, U01AG012553, U01AG012554, U01AG012495). The SWAN Genomic Analyses and SWAN Legacy have grant support from the NIA (U01AG017719). The Generations Study was funded by Breast Cancer Now and the Institute of Cancer Research (ICR). The ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent official views of the funders. The Sister Study was funded by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (Z01-ES044005 to D.P.S.); the AMH assays were supported by the Avon Foundation (02-2012-065 to H.B. Nichols and D.P.S.). The breast cancer genome-wide association analyses were supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the ‘Ministère de l’Économie, de la Science et de l’Innovation du Québec’ through Genome Québec and grant PSR-SIIRI-701, The National Institutes of Health (U19 CA148065, X01HG007492), Cancer Research UK (C1287/A10118, C1287/A16563, C1287/A10710) and The European Union (HEALTH-F2-2009-223175 and H2020 633784 and 634935). All studies and funders are listed in Michailidou et al. (Nature, 2017).
Conflict of interest
F.J.M.B. has received fees and grant support from Merck Serono and Ferring BV. D.A.L. has received financial support from several national and international government and charitable funders as well as from Medtronic Ltd and Roche Diagnostics for research that is unrelated to this study. N.S. is scientific consultant for Ansh Laboratories. The other authors declare no competing interests.
References
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG.. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 2008;22:756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beulens JW, Monninkhof EM, Verschuren WM, van der Schouw YT, Smit J, Ocke MC, Jansen EH, van Dieren S, Grobbee DE, Peeters PH. et al. Cohort profile: the EPIC-NL study. Int J Epidemiol 2010;39:1170–1178. [DOI] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Smith GD.. Cohort profile: the ‘children of the 90s’–the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 2013;42:111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012;22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JR, Patterson N, Robinson EB. et al. ; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM. et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Davies NM, Thompson SG.. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson SG.. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. Boca Raton, Florida, USA: CRC Press, 2015. [Google Scholar]
- Conidi A, Cazzola S, Beets K, Coddens K, Collart C, Cornelis F, Cox L, Joke D, Dobreva MP, Dries R. et al. Few Smad proteins and many Smad-interacting proteins yield multiple functions and action modes in TGFbeta/BMP signaling in vivo. Cytokine Growth Factor Rev 2011;22:287–300. [DOI] [PubMed] [Google Scholar]
- Davey Smith G, Ebrahim S.. What can Mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ 2005;330:1076–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, Kraft P, Lin N, Huang H, Broer L. et al. ; 23andMe Research Team. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet 2018;14:e1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Ruth KS, Thompson DJ, Lunetta KL, Pervjakova N, Chasman DI, Stolk L, Finucane HK, Sulem P, Bulik-Sullivan B. et al. ; LifeLines Cohort Study. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet 2015;47:1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kat AC, van der Schouw YT, Eijkemans MJ, Herber-Gast GC, Visser JA, Verschuren WM, Broekmans FJ.. Back to the basics of ovarian aging: a population-based study on longitudinal anti-Mullerian hormone decline. BMC Med 2016;14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, Posthuma D.. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliassen AH, Missmer SA, Tworoger SS, Spiegelman D, Barbieri RL, Dowsett M, Hankinson SE.. Endogenous steroid hormone concentrations and risk of breast cancer among premenopausal women. J Natl Cancer Inst 2006;98:1406–1415. [DOI] [PubMed] [Google Scholar]
- Estienne A, Pierre A, di Clemente N, Picard JY, Jarrier P, Mansanet C, Monniaux D, Fabre S.. Anti-Mullerian hormone regulation by the bone morphogenetic proteins in the sheep ovary: deciphering a direct regulatory pathway. Endocrinology 2015;156:301–313. [DOI] [PubMed] [Google Scholar]
- Finkelstein JS, Lee H, Karlamangla A, Neer RM, Sluss PM, Burnett-Bowie SM, Darakananda K, Donahoe PK, Harlow SD, Prizand SH. et al. Antimullerian hormone and impending menopause in late reproductive age: the study of women’s health across the nation. J Clin Endocrinol Metab 2020;105:e1862–e1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Smith GD, Henderson J, Macleod J, Molloy L, Ness A. et al. Cohort profile: the Avon longitudinal study of parents and children: ALSPAC mothers cohort. Int J Epidemiol 2013;42:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Clendenen TV, Afanasyeva Y, Koenig KL, Agnoli C, Brinton LA, Dorgan JF, Eliassen AH, Falk RT, Hallmans G. et al. Circulating anti-Mullerian hormone and breast cancer risk: a study in ten prospective cohorts. Int J Cancer 2018;142:2215–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RM, Gabor TV, Couzens AL, Scheid MP.. The MYC-associated protein CDCA7 is phosphorylated by AKT to regulate MYC-dependent apoptosis and transformation. Mol Cell Biol 2013;33:498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin WC, Trakselis MA.. The MCM8/9 complex: a recent recruit to the roster of helicases involved in genome maintenance. DNA Repair (Amst) 2019;76:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson SE, Willett WC, Manson JE, Hunter DJ, Colditz GA, Stampfer MJ, Longcope C, Speizer FE.. Alcohol, height, and adiposity in relation to estrogen and prolactin levels in postmenopausal women. J Natl Cancer Inst 1995;87:1297–1302. [DOI] [PubMed] [Google Scholar]
- He C, Kraft P, Chen C, Buring JE, Paré G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI.. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet 2009;41:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homburg R, Crawford G.. The role of AMH in anovulation associated with PCOS: a hypothesis. Hum Reprod 2014;29:1117–1121. [DOI] [PubMed] [Google Scholar]
- Hoyos LR, Visser JA, McLuskey A, Chazenbalk GD, Grogan TR, Dumesic DA.. Loss of anti-Müllerian hormone (AMH) immunoactivity due to a homozygous AMH gene variant rs10417628 in a woman with classical polycystic ovary syndrome (PCOS). Hum Reprod 2020;35:2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost A. The age factor in the castration of male rabbit fetuses. Proc Soc Exp Biol Med 1947;66:302. [DOI] [PubMed] [Google Scholar]
- Kelsey TW, Wright P, Nelson SM, Anderson RA, Wallace WH.. A validated model of serum anti-Mullerian hormone from conception to menopause. PLoS One 2011;6:e22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ. et al. ; Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laven JS, Mulders AG, Visser JA, Themmen AP, De Jong FH, Fauser BC.. Anti-Mullerian hormone serum concentrations in normoovulatory and anovulatory women of reproductive age. J Clin Endocrinol Metab 2004;89:318–323. [DOI] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163. [DOI] [PubMed] [Google Scholar]
- Lutzmann M, Bernex F, da Costa de Jesus C, Hodroj D, Marty C, Plo I, Vainchenker W, Tosolini M, Forichon L, Bret C. et al. MCM8- and MCM9 deficiencies cause lifelong increased hematopoietic DNA damage driving p53-dependent myeloid tumors. Cell Rep 2019;28:2851–2865.e4. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P.. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–913. [DOI] [PubMed] [Google Scholar]
- Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, Lemacon A, Soucy P, Glubb D, Rostamianfar A. et al. ; ConFab/AOCS Investigators. Association analysis identifies 65 new breast cancer risk loci. Nature 2017;551:92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolhuijsen LME, Visser JA.. Anti-Müllerian hormone and ovarian reserve: update on assessing ovarian function. J Clin Endocrinol Metab 2020;105:3361–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols HB, Baird DD, Stanczyk FZ, Steiner AZ, Troester MA, Whitworth KW, Sandler DP.. Anti-Mullerian hormone concentrations in premenopausal women and breast cancer risk. Cancer Prev Res (Phila) 2015;8:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Long DT, Lee KY, Abbas T, Shibata E, Negishi M, Luo Y, Schimenti JC, Gambus A, Walter JC. et al. The MCM8-MCM9 complex promotes RAD51 recruitment at DNA damage sites to facilitate homologous recombination. Mol Cell Biol 2013;33:1632–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JR, McMahon G, Day FR, Ring SM, Nelson SM, Lawlor DA.. Genome-wide association study identifies common and low-frequency variants at the AMH gene locus that strongly predict serum AMH levels in males. Hum Mol Genet 2016;25:382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T. et al. ; Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 2015;6:5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picavet HSJ, Blokstra A, Spijkerman AMW, Verschuren WMM.. Cohort profile update: the Doetinchem Cohort Study 1987–2017: lifestyle, health and chronic diseases in a life course and ageing perspective. Int J Epidemiol 2017;46:1751–1751g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre A, Estienne A, Racine C, Picard JY, Fanchin R, Lahoz B, Alabart JL, Folch J, Jarrier P, Fabre S. et al. The bone morphogenetic protein 15 up-regulates the anti-Mullerian hormone receptor expression in granulosa cells. J Clin Endocrinol Metab 2016;101:2602–2611. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Burgess S.. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013;178:1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. EMBO J 2003;22:2443–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2017.
- Ruth KS, Soares ALG, Borges MC, Eliassen AH, Hankinson SE, Jones ME, Kraft P, Nichols HB, Sandler DP, Schoemaker MJ. et al. Genome-wide association study of anti-Mullerian hormone levels in pre-menopausal women of late reproductive age and relationship with genetic determinants of reproductive lifespan. Hum Mol Genet 2019;28:1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh-Huerta SM, Johnson NA, Rosen MP, Sternfeld B, Cedars MI, Reijo Pera RA.. Genetic markers of ovarian follicle number and menopause in women of multiple ethnicities. Hum Genet 2012a;131:1709–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh-Huerta SM, Johnson NA, Rosen MP, Sternfeld B, Cedars MI, Reijo Pera RA.. Genetic variants and environmental factors associated with hormonal markers of ovarian reserve in Caucasian and African American women. Hum Reprod 2012b;27:594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers MFR, Crawford SL, Sternfeld B, Morganstein D, Gold EB, Greendale GA, Evans DA, Neer R, Matthews KA, Sherman S.. SWAN: a multicenter, multiethnic, community-based cohort study of women and the menopausal transition. In: Lobo RA, Kelsey J and Marcus R (eds.). Menopause: Biology and Pathobiology. San Diego, CA: Academic Press, 2000, 175–188. [Google Scholar]
- Stolk L, Zhai G, van Meurs JB, Verbiest MM, Visser JA, Estrada K, Rivadeneira F, Williams FM, Cherkas L, Deloukas P. et al. Loci at chromosomes 13, 19 and 20 influence age at natural menopause. Nat Genet 2009;41:645–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow AJ, Jones ME, Schoemaker MJ, Hemming J, Thomas D, Williamson J, Ashworth A.. The Breakthrough Generations Study: design of a long-term UK cohort study to investigate breast cancer aetiology. Br J Cancer 2011;105:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschuren WM, Blokstra A, Picavet HS, Smit HA.. Cohort profile: the Doetinchem Cohort Study. Int J Epidemiol 2008;37:1236–1241. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weenen C, Laven JS, von Bergh AR, Cranfield M, Groome NP, Visser JA, Kramer P, Fauser BC, Themmen AP.. Anti‐Müllerian hormone expression pattern in the human ovary: potential implications for initial and cyclic follicle recruitment. Mol Hum Reprod 2004;10:77–83. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR.. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE. et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 2014;9:1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ. et al. ; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–375.S1–S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM.. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ.. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics 2016;32:1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Pourcain BS. et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017;33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M.. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012;44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the NHGRI-EBI Catalog of human genome-wide association studies, at https://www.ebi.ac.uk/gwas/. Code underlying the analyses for this article are available via GitHub, at https://github.com/reneemgverdiesen/AMH_GWAS.