Abstract
Background
Meningiomas are the most common primary intracranial tumor in adults. Clinical care is currently guided by the World Health Organization (WHO) grade assigned to meningiomas, a 3-tiered grading system based on histopathology features, as well as extent of surgical resection. Clinical behavior, however, often fails to conform to the WHO grade. Additional prognostic information is needed to optimize patient management.
Methods
We evaluated whether chromosomal copy-number data improved prediction of time-to-recurrence for patients with meningioma who were treated with surgery, relative to the WHO schema. The models were developed using Cox proportional hazards, random survival forest, and gradient boosting in a discovery cohort of 527 meningioma patients and validated in 2 independent cohorts of 172 meningioma patients characterized by orthogonal genomic platforms.
Results
We developed a 3-tiered grading scheme (Integrated Grades 1-3), which incorporated mitotic count and loss of chromosome 1p, 3p, 4, 6, 10, 14q, 18, 19, or CDKN2A. 32% of meningiomas reclassified to either a lower-risk or higher-risk Integrated Grade compared to their assigned WHO grade. The Integrated Grade more accurately identified meningioma patients at risk for recurrence, relative to the WHO grade, as determined by time-dependent area under the curve, average precision, and the Brier score.
Conclusion
We propose a molecularly integrated grading scheme for meningiomas that significantly improves upon the current WHO grading system in prediction of progression-free survival. This framework can be broadly adopted by clinicians with relative ease using widely available genomic technologies and presents an advance in the care of meningioma patients.
Keywords: meningioma, copy-number alterations
Key Points.
Aggressive meningiomas have recurrent copy-number alterations.
Incorporating copy-number alterations into a grading scheme identifies high-risk tumors.
Importance of the Study.
Meningiomas are among the most common brain tumors. Grading based on histopathology often fails to predict tumor behavior. Incomplete tumor sampling, interobserver variability in histology assessment, and ambiguous histologic features all pose challenges and lead to uncertainty in patient management. We show that a grading scheme that incorporates mitotic index and multiple high-risk copy-number alterations allows for identification of patients at risk for tumor recurrence, despite complete tumor resection, and in some cases, despite benign-appearing histopathology (WHO grade I). A critical aspect of our approach is that the features are transparent, and can be assessed by a number of genomic platforms that have proliferated across medical centers. The work described here lays a foundation for more effective grading of meningiomas, genomically informed clinical trials, and improved patient management.
Meningioma is the most common primary intracranial tumor in the United States with approximately 35 000 new cases every year,1 and an estimated population prevalence of approximately 1 in every 100 adult persons over the age of 45.2 Given the large number of cases, a reliable and accessible predictor of clinical outcome after diagnosis is needed. At present, the World Health Organization (WHO) places meningioma into 1 of the 3 grades based on histopathology, and is the primary measure used to predict outcome and guide postsurgical treatment decision making.3 It is well recognized, however, that the behavior of a number of meningiomas does not conform to their assigned WHO grade, with some histologically benign meningiomas developing repeated recurrences despite aggressive treatment while other higher-grade meningiomas remain stable after surgical resection.
The limitations of WHO grading result in part from interobserver variability in histological assessment,4 the potential for under-sampling of a tumor type with known histologic and molecular heterogeneity,5–7 and the possibility that malignant potential may not be uniformly reflected in assessment of histologic features.8 Molecular characterization may represent a means to overcome these concerns. Molecular-based approaches, including methylation profiling and DNA and RNA sequencing,9–12 have shown the ability to predict behavior; however, these methodologies rely on platforms limited to select centers while the majority of meningiomas are treated outside of large, academic institutions.13,14 Copy-number variations (CNVs), previously described in higher-grade meningioma,15–18 are an attractive option given the wide availability of the technology required, even in resource-limited settings. In an attempt to devise an improved prognostically relevant grading scheme that is broadly accessible, we evaluated 699 meningiomas with detailed clinical, imaging, histologic, and molecular annotation, to formulate a molecularly integrated grade that is simple to apply, transparent, scalable, and accurate in long-term prediction of clinical behavior.
Methods
Patients
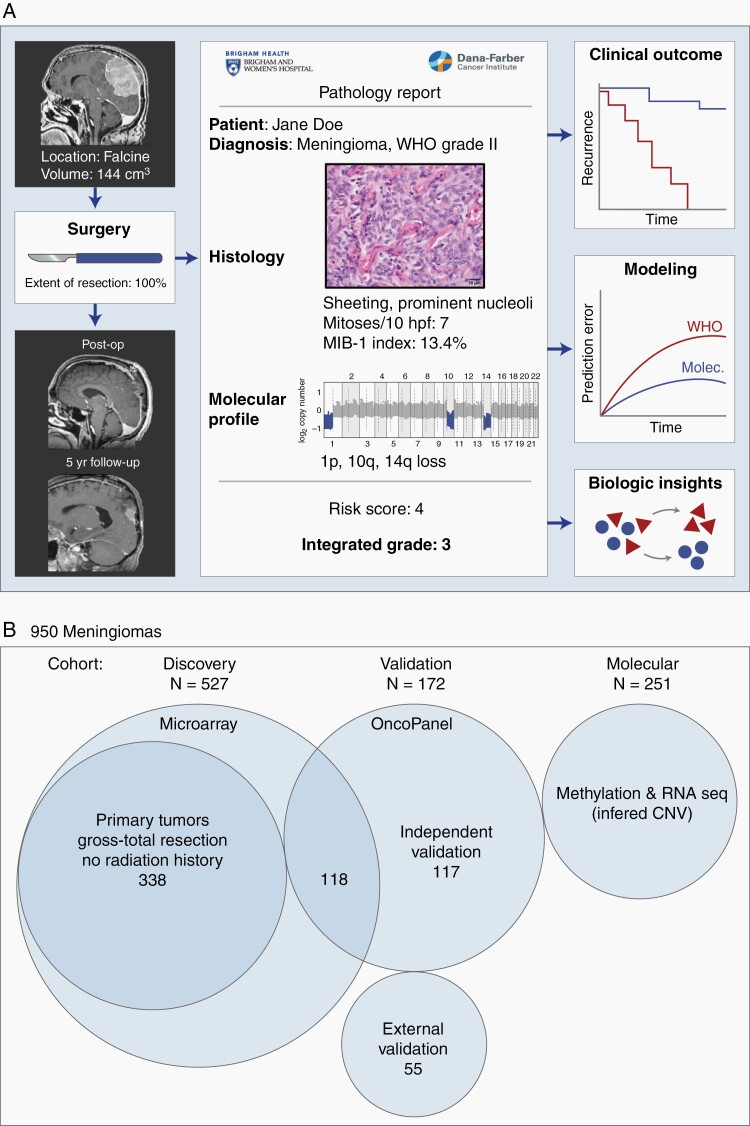
We examined 527 meningiomas resected from unique patients evaluated at the Brigham and Women’s Hospital (BWH) from 2003 to 2019 as the discovery cohort. An additional 172 patients with surgically resected meningioma, including 117 from BWH and 55 from the University of Toronto, were examined as independent validation cohorts. Of the BWH discovery cohort, 81% of cases were prospectively analyzed as part of standard clinical testing in a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory and reported in the clinical record, which was initiated in 2012. This study was approved by the institutional review boards of BWH and University of Toronto.
Clinical Annotation
Clinical history, tumor location, and radiographic recurrence were assessed. Preoperative and postoperative MRIs underwent volumetric contouring to define the extent of resection, with gross total resection (GTR) defined by no residual enhancing nodular tumor on imaging, and all others classified as subtotal resection (STR), modeled upon the RANO meningioma response assessment imaging criteria.19 All follow-up MRIs were independently reviewed by 3 authors (W.L.B., J.D., and S.T.) to evaluate for recurrence. This included evaluation of the MRI images, the radiology reports, and the clinical chart. When personal review of imaging and the radiology reports demonstrated discordance for progression/recurrence, the authors discussed these inconsistent cases to achieve consensus. Simpson grade of resection was not available, as it was not routinely recorded in the operative note or patient chart. Anatomic location was categorized as midline skull base, lateral skull base, lateral convexity, falcine/parasagittal, or other, which included intraventricular and spinal meningiomas.
Pathology
Histopathologic review of all tumors was performed by board-certified neuropathologists and tumor grade was abstracted from the pathology reports issued by the BWH Neuropathology division according to the WHO Classification of Tumors (2007 and 2016). Cases with unclear grading were re-reviewed and assigned a grade according to the criteria from the WHO Classification of Tumors 2016 (by S.S.).20 Cases with brain invasion were also assigned to WHO grade II after re-review. Atypical features, mitoses per 10 high-powered fields (HPF), and MIB-1 proliferative index were recorded. Mitotic count was recorded from the clinical record when available or performed in cases where specific information was absent. Following the WHO guidelines, we assess mitotic count by visual inspection of H&E stained sections with ≥4 mitoses per/10 HPF (of 0.16 mm2) used to define atypical meningioma (WHO grade 2) and ≥20 mitoses/10 HPF used to define anaplastic meningioma (WHO grade 3). MIB-1 index was assessed using a semi-quantitative approach by quantifying MIB-1 positive labeling in ~1000 cells. In a minority of cases where the index was reported as a range, we list the mid-point of the range for statistical analysis.
Genomic Characterization
Whole-genome microarray analysis for DNA copy-number profiling was available for 527 tumors from the discovery cohort and 83 samples from the BWH validation cohort. Targeted mutational profiling of 227-447 cancer-associated genes (OncoPanel, versions 1-3)21,22 was available for 118 samples from the discovery cohort and all 117 of the BWH validation set. The external validation cohort underwent methylation-based copy-number profiling at the University of Toronto.23 Using previously described methods,24–26 we derived broad CNVs from the OncoPanel dataset and publicly available methylation and RNAseq datasets. The fraction (proportion of the length) of the genome altered was estimated for each sample among these datasets (additional details in the Supplementary Appendix).
Outcome
The primary outcome assessed was progression-free survival, defined as time from surgery until radiographic recurrence on follow-up imaging in cases of GTR, or time until tumor progression in cases of STR.
Model Design, Validation
To develop a grading score reflective only of intrinsic tumor factors and to exclude the potential confounding influence of varying treatment on subsequent recurrence, we first modeled recurrence in 338 patients with primary, previously untreated meningiomas with GTR in the discovery cohort. We investigated covariates that define the WHO grading scheme (mitotic index, the presence of atypical features, and brain invasion), as well as MIB-1 proliferative index and chromosome arm-level and CDKN2A CNVs. Each feature was individually evaluated for association with time-to-recurrence using the log-rank test. Covariates were further investigated using Cox LASSO regularization,27 random survival forests,28 and gradient boosting.29 Shared high-risk features identified across selection methods were chosen for iterative model building to construct an Integrated Grade with the best predictive performance for tumor recurrence. Features were added sequentially into the model until addition of no more features improve model performance, as assessed using the Brier score. The Integrated Grade was compared to WHO grade for predicting tumor recurrence using time-dependent receiver operator curves (ROC), time-dependent average precision (AP) curves, and Brier curves. The Integrated Grade was internally validated in the discovery cohort using cross-validation techniques (10 000 bootstrap resampling and leave-one-out-cross-validation) and externally validated in 2 independent cohorts.
Using all of the discovery cohort, which included tumors with varied treatment histories (Supplementary Table S1), the Integrated Grade was included along with extent of resection, tumor size, and tumor status (primary vs recurrent) into a Cox proportional hazards model to generate a nomogram for recurrence risk. A decision curve analysis was performed to evaluate the clinical utility of the nomogram.30
Additional details of genomic characterization, model design, and assessment are described in the Supplementary Appendix.
Results
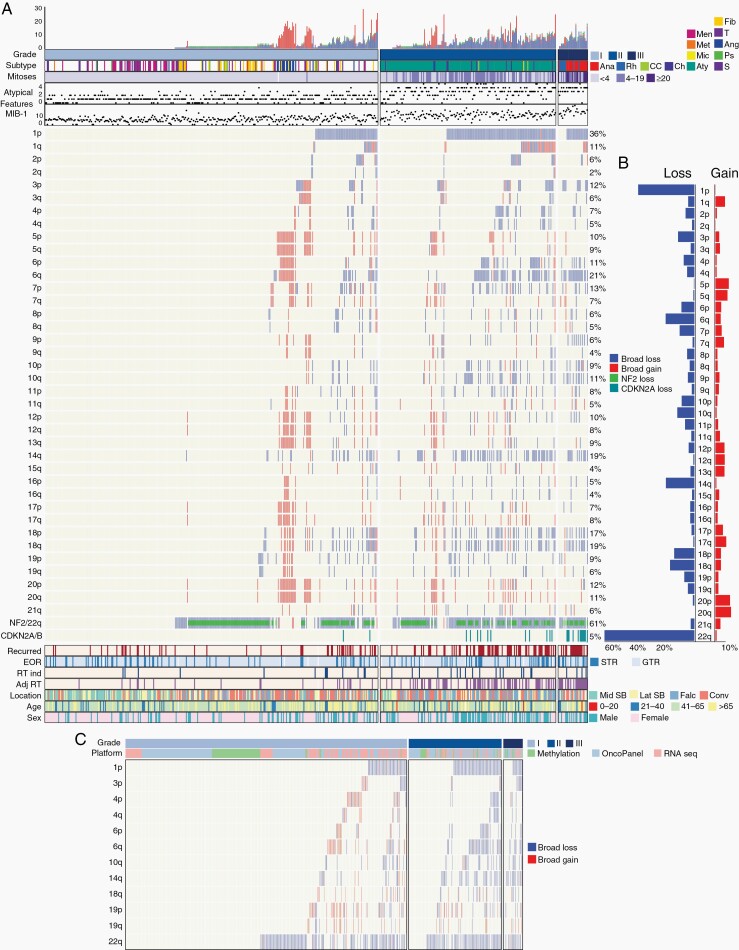
The discovery cohort included 527 meningiomas (326 WHO grade I, 172 WHO grade II, 29 WHO grade III; Figure 1). The cohort was 67% female, with a mean age of 57 (range 9-90) years; there were 132 recurrences across a mean follow-up of 43 months (Supplementary Table S1). Broad CNVs were absent in 27% of tumors and monosomy 22 was present alone in 18% of tumors, followed by frequent loss of chromosomes 1p (36%), 14q (19%), 6p/q (17%), 18p/q (16%), 10p/q (10%), and gain of 20p/q (11%), 5p/q (10%), 7p/q (6%), 17p/q (8%), and 12p/q (7%) (Figure 2A, B). Chromosomal gains were largely restricted to angiomatous and microcystic meningiomas, a subset of which also exhibited chromosome 1p loss (Figure 2A).16 Of 34 patients with radiation-induced meningiomas, frequent broad CNVs were present with 71% exhibiting 1p loss (Supplementary Figure S2). Loss of 3p, encompassing BAP1 and PBRM1, was seen in 5/9 cases with rhabdoid histology.31,32 WHO grade III meningiomas demonstrated significantly greater genome disruption than WHO grade II meningiomas (P = .027) with a trend toward more arm-level CNVs (8.3 vs 6.7, P = 06; Supplementary Figure S3A).31
Fig. 1.
Study overview and description of patient cohorts. (A) A clinical cohort of meningiomas were evaluated for patient demographics, histologic and molecular features, preoperative and postoperative tumor volume, and clinical follow-up including recurrence and death. (B) Description of 950 meningiomas analyzed. The discovery cohort consisted of 527 tumor samples from unique patients, with 338 primary tumors without history of treatment or prior radiotherapy. Validation was performed on 2 independent cohorts of 117 patients and 55 patients. Copy number was additionally derived from a molecular cohort of publicly available methylation and RNAseq datasets.
Fig. 2.
Copy-number profile of meningiomas. (A) Distribution of copy-number alterations, histologic features, treatment factors, and demographic features across 527 meningiomas in the discovery cohort. Tumor subtypes were annotated as: Anaplastic (Ana), Rhabdoid (Rh), Clear Cell (CC), Chordoid (Ch), Atypical (Aty), Microcystic (Mic), Metaplastic (Met), Meningothelial (Men), Transitional (T), Angiomatous (Ang), Psammomatous (Ps), Secretory (S), and Fibroblastic (F). Mitoses were classified as <4, 4-19, or >20. (B) Frequency of copy-number gains and losses across each chromosomal arm among the discovery cohort. (C) Inferred copy-number alterations in high-risk chromosomes of interest among 510 meningiomas with methylation, mutational (OncoPanel), or RNAseq data.
We inferred CNVs from 509 meningiomas with DNA mutation panel sequencing (OncoPanel, n = 258), methylation profiling (n = 95), or bulk RNAseq (n = 156) (Figure 2C). The broad high-risk CNVs extrapolated from OncoPanel were 96% concordant with microarray-derived CNVs in tumors evaluated by both methods (n = 118, Supplementary Figure S3B), demonstrating the reliability of CNV inference from other genomic platforms.
Development of a Molecularly Based Integrated Grade
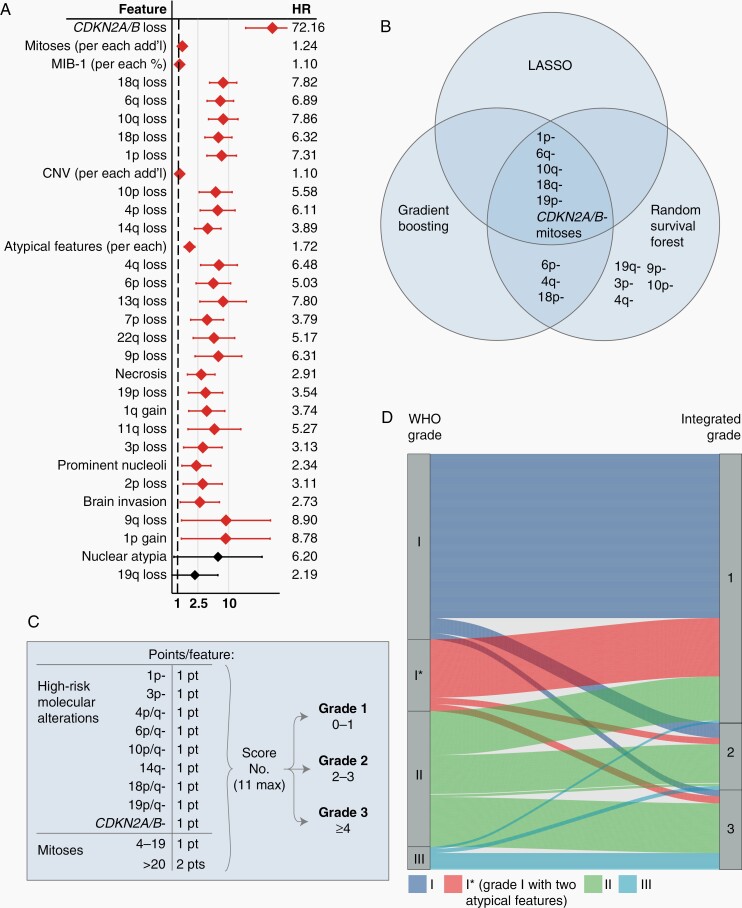
With the aim of developing a grading scheme that is prognostically relevant and that reflects tumor-intrinsic factors, we devised an Integrated Grade (see Methods; Figure 3A, B), which accounts for mitotic count, focal hemizygous or homozygous loss of CDKN2A, and loss of 1p, 3p, 4p/q, 6p/q, 10p/q, 14q, 18p/q, and 19p/q. Several high-risk chromosomes, including 4, 6, 10, 18, and 19, exhibited synchronous loss of the short and long arms when altered; therefore, we assigned loss of either arm of these chromosomes as 1 point. While mitotic count and the MIB-1 proliferative index were both associated with recurrence, they were significantly correlated (R = 0.76, P < .001), and mitotic count exhibited less variance (Supplementary Figure S3C). We assigned 1 point for the presence of any of the above chromosomal losses, as well as for CDKN2A loss and a mitotic count of 4-19; 2 points were assigned for mitotic count >20. Tumors are divided into three Integrated Grades based on their point score: Integrated Grade 1 (0-1 pt), Integrated Grade 2 (2-3 pts), and Integrated Grade 3 (>4 pts) (Figure 3C).
Fig. 3.
Development of a molecularly Integrated Grade. (A) Univariate Cox proportional hazards analysis evaluating tumor-intrinsic, histologic, and molecular features and their associated hazard risk for tumor recurrence across 338 patients with primary meningiomas who underwent GTR without prior radiation. Specific hazard ratios are listed with error bars representing 95% CI. Features are ranked in order from top-down as most significant to least significant, with those with P < .05 denoted by red confidence interval bars. (B) Features most significantly associated with meningioma recurrence, as identified by LASSO, random survival forest, and gradient boosting methods. (C) Integrated Grading scheme: points are assigned for the presence of listed molecular features or elevated mitoses, with the resultant sum categorizing tumors into 1 of the 3 Grades with increasing recurrence risk. (D) Association of WHO grade and Integrated Grade for 527 meningiomas, with 87% concordance between WHO grade I and Integrated Grade 1, 31% concordance between WHO grade II and Integrated Grade 2, and 72% concordance between WHO grade III and Integrated Grade 3. The majority of WHO grade III tumors that reclassified into a different Integrated Grade were rhabdoid meningiomas, while 19/20 anaplastic meningiomas remained Integrated Grade 3. Abbreviation: GTR, gross total resection.
Association Between WHO Grade and Integrated Grade
While WHO grade was significantly associated with Integrated Grade (chi-squared test, P < .001), 32% of cases were reclassified, mostly among the WHO grade II meningiomas (Figure 3D). WHO grade I tumors with or without 2 atypical features distributed similarly across Integrated Grades. Among WHO grade III tumors, rhabdoid meningiomas are frequently reclassified into lower-risk Integrated Grades, with 3/9 as Integrated Grade 1 and 4/9 designated as Integrated Grade 2, consistent with the challenge in grading this histologic meningioma subtype33 and a low incidence of recurrent events such as loss of 1p. In contrast, 19/20 WHO grade III anaplastic meningiomas remained Integrated Grade 3.
Integrated Grade and Clinical Outcome
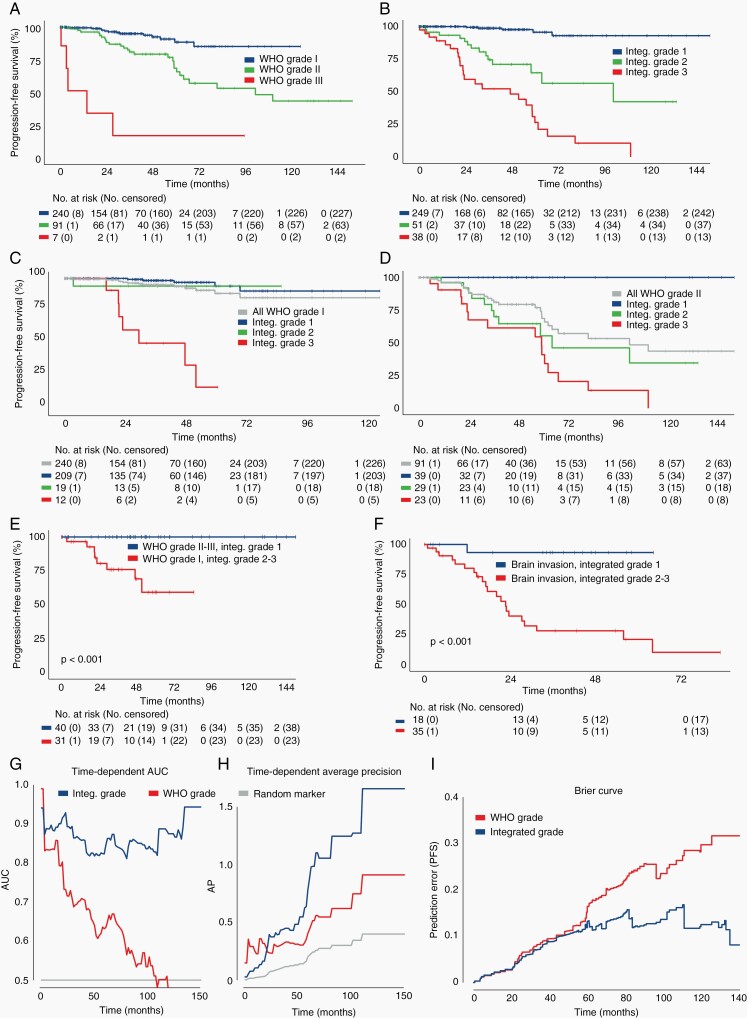
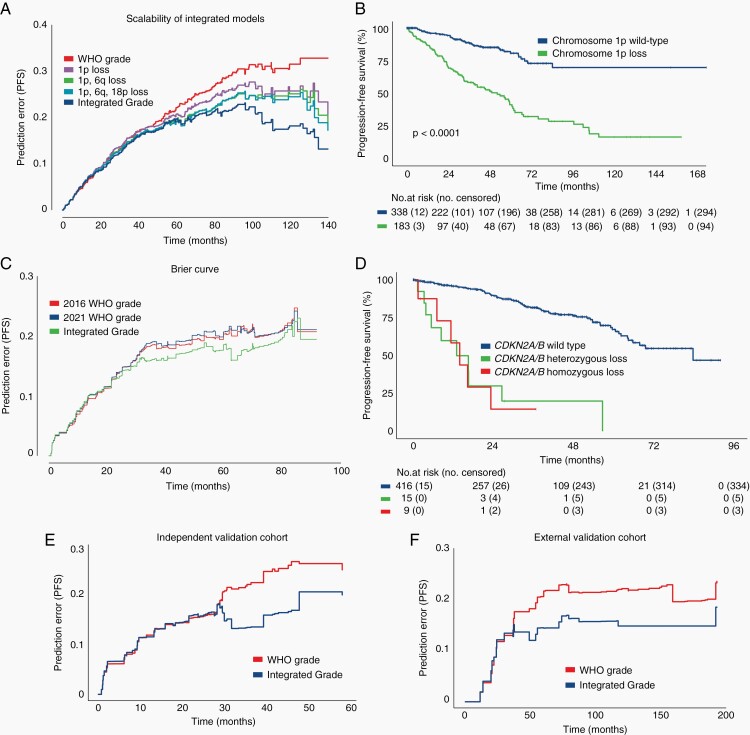
In the subset of 338 primary non-irradiated meningiomas with GTR, tumors demonstrated distinct progression-free survival curves as stratified by either WHO grade or Integrated Grade (P < .001; Figure 4A, B). The application of Integrated Grade further differentiated recurrence within WHO grade I or II meningiomas (Figure 4C, D). WHO grade I meningiomas designated Integrated Grade 2 or 3 fared significantly worse than WHO grade II-III meningiomas with Integrated Grade 1 (P < .001; Figure 4E). Integrated Grade also predicted recurrence more reliably than the presence of brain invasion, a feature sufficient for WHO grade II designation in the current schema, as meningiomas with brain invasion and Integrated Grade 1 fared significantly better than those with brain invasion but Integrated Grade 2-3 (P < .001; Figure 4F). Taken together, the Integrated Grading scheme significantly improved the ability to predict recurrence risk compared to the WHO grade, as evaluated by time-dependent (ROC) area under the curve (5-year AUC 0.823 vs 0.632), AP (5-year AP 0.781 vs 0.405), and Brier score (0.098 vs 0.180) (Figure 4G–I), even when restricted to the prospectively collected cases alone (5-year AUC 0.795 vs 0.624; 5-year AP 0.638 vs 0.341; Supplementary Figure S4A, B). Notably, the predictive capacity of the Integrated Grade compared to WHO grade strengthened with follow-up time.
Fig. 4.
Evaluation of Integrated Grade as a predictor of meningioma behavior. (A, B) Progression-free survival of 338 patients with primary meningiomas after GTR, stratified by (A) WHO grade or (B) Integrated Grade. (C, D) Progression-free survival stratified by Integrated Grade among (C) WHO grade I and (D) WHO grade II tumors. (E) Progression-free survival of discrepantly graded meningiomas (P < .01, log-rank test). (F) Progression-free survival among patients with brain invasion on histopathologic analysis, stratified by Integrated Grade 1 vs Integrated Grades 2-3. (G) Time-dependent area under the curve (AUC) of a ROC for Integrated Grade and WHO grade (Integrated Grade: 5-year AUC 0.823, 95% CI 0.724-0.91 vs WHO grade: 5-year AUC 0.632, 95% CI 0.521-0.737). (H) Time-dependent average precision for Integrated Grade and WHO grade (Integrated Grade: 5-year AP 0.781, 95% CI 0.485-1.0; vs WHO grade: 5-year AP 0.405, 95% CI 0.258-0.586). (I) Brier prediction curve for progression-free survival comparing Integrated Grade to WHO grade in the 338 patients (Brier score: 0.098 vs 0.180). Abbreviation: GTR, gross total resection.
Across the full discovery cohort, WHO grade and Integrated Grade stratified patients into differing progression-free outcome curves (Supplementary Figure S4C, D), and the Integrated Grade continued to outperform the WHO grade in predicting recurrence (Brier score 0.157 vs 0.233; Figure 5A). The addition of individual molecular features into the grading scheme incrementally strengthened the predictive performance for recurrence over WHO grade (Figure 5A). The presence of 1p loss as a single feature conferred a significantly higher risk of recurrence compared to tumors with intact 1p (Figure 5B). With regard to CDKN2A/B, meningiomas with either homozygous or heterozygous loss had similar rates of recurrence (Figure 5B). Of the 244 patients with available mutational data for TERTp coverage, the Integrated Grade outperformed the new 2021 WHO grading scheme, which incorporates TERTp mutations and CDKN2A/B homozygous loss into the definition of WHO grade 3 (Brier score: 2016 WHO 0.163 vs 2021 WHO 0.166 vs Integrated Grade 0.151; Figure 5C).34 We validated the performance of the Integrated Grade in 2 independent cohorts, 1 of the 117 patients (Brier score 0.178 vs 0.214) and another of 55 patients (Brier score 0.143 vs 0.181) (Figure 5D, E). In each of these cohorts, we found that the Integrated Grade was superior to WHO grade in predicting recurrence, similarly to the discovery cohort, most notable over longer-term follow-up. In addition, the Integrated Grade was superior to WHO grade in assessing overall survival on long-term follow-up (Supplementary Figure S4E).
Fig. 5.
Validation of integrated grade among multiple patient cohorts. (A) Brier prediction curve for progression-free survival among the 527 patients with varied treatment histories in the discovery cohort (Brier score: 0.157 vs 0.233). (B) Progression-free survival of 527 patients stratified by chromosome 1p status. (C) Progression-free survival of 527 patients stratified by CDKN2A/B status, showing no difference in outcome between patients with homozygous or heterozygous loss. (D) Brier prediction curve for progression-free survival among 244 patients for which mutational data was available in addition to copy-number data. Brier scores for this cohort were determined by the assigned 2016 WHO grading scheme, the new 2021 WHO grading scheme, and the Integrated Grading scheme (Brier score; 2016 WHO 0.163 vs 2021 WHO 0.166 vs Integrated Grade 0.151). (E) Brier prediction curve for progression-free survival calculated for Integrated Grade and WHO grade among the 117-patient internal validation cohort (Brier score 0.178 vs 0.214). (F) Brier prediction curve for progression-free survival calculated for WHO grade and Integrated Grade for the 55-patient external validation cohort (Brier score 0.133 vs 0.181).
Mutations
Only a small number of mutations were observed that are associated with aggressive behavior, such as in SMARCB1 (n = 7), SMARCE1 (n = 1), TERT promoter (TERTp, n = 6), and BAP1 (n = 2). Of the 6 cases with TERTp mutation (3 WHO grade I, 3 WHO grade II), 3 recurred; the 3 TERTp-mutated tumors that did not recur were Integrated Grade 1, while the 3 TERTp-mutated tumors that recurred were Integrated Grade 2-3 (Supplementary Figure S4F).
Impact of Treatment
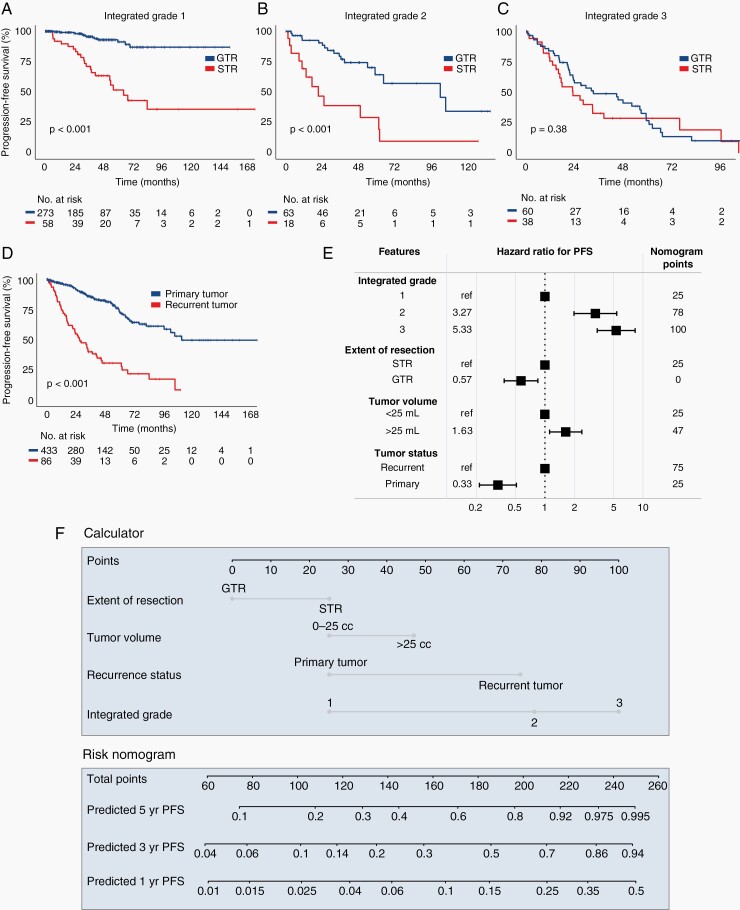
The treatment received significantly influences the clinical course of meningiomas, with surgery playing a central role. GTR decreased recurrence risk for Integrated Grade 1 and Integrated Grade 2 meningiomas (P < .001); however, GTR did not significantly delay recurrence in Integrated Grade 3 tumors (P = .38) (Figure 6A–C). In de novo higher-grade meningiomas, either by Integrated or WHO grading, receipt of adjuvant radiation did not prolong progression-free survival (Supplementary Figure S5A–D). Recurrent tumors had a much higher likelihood of subsequent recurrence than primary tumors (P < .001; Figure 6D).
Fig. 6.
Impact of treatment and development of nomogram for tumor recurrence. Progression-free survival of 527 meningioma patients in the discovery cohort, stratified by (A–C) extent of resection status (GTR vs STR) for each Integrated Grade, and (D) primary vs recurrent tumor status. (E) Multivariate Cox proportional hazards model incorporating Integrated Grade with extent of resection, tumor volume, and primary vs recurrent tumor status. (F) Nomogram for predicting 1-, 3-, and 5-year recurrence risk based on the four listed parameters. Abbreviation: GTR, gross total resection; STR, subtotal resection.
Multivariate Risk Model
The Integrated Grade, primary vs recurrent status, tumor volume >25 mL, and extent of resection were all significantly associated with hazard of recurrence, but the Integrated Grade had the greatest impact (Figure 6E) and most predictive power (Supplementary Figure S6A). Previously described risk factors, such as age, male sex, and tumor location, were not independently associated with recurrence after accounting for Integrated Grade (Supplementary Figure S6B).18 Additionally, the receipt of early adjuvant radiation after surgery was did not influence recurrence rate after accounting for the Integrated Grade in this cohort (Supplementary Figure S6C). We constructed a nomogram based on the multivariate model which can be applied to individual patient scenarios (Figure 6F). Decision curve analysis was performed to assess the clinical utility of this model, with the multivariate model rendering a net benefit exceeding null models among a wide range of threshold probabilities (Supplementary Figure S6D).30
Discussion
Since the publication of the seminal monograph Meningioma: Their classification, regional behavior, life history, and surgical end results by Harvey Cushing and Loise Eisenhardt in 1938, the grading of meningiomas has been based on histopathologic features, the interpretation of which may differ across pathologists and inconsistently reflects tumor behavior. The development of a prognostically relevant grading scheme with combined histologic and molecular features, as exists for other central nervous system tumors,35–38 remains inchoate for meningioma.
We present an alternative to the WHO grading scheme for meningioma with improved accuracy in predicting tumor behavior. We incorporated mitotic index, a proven reliable predictor of tumor behavior,39 as well as genomic markers significantly associated with recurrence, all of which have been previously observed in malignant meningiomas.15,18,40 Our combinatorial risk stratification scheme accommodates the variable genomic profiles observed in aggressive meningiomas. The ability of CNVs to predict tumor behavior has been debated, thereby, limiting the use of CNVs in prognostication despite the relative technical ease of accessing this information.9,10,17,18 While the underlying biologic drivers of aggressive meningiomas still remain poorly understood, copy-number alterations appear to occur early in progression and before clinical detection, and offer predictive value most notable over long-term follow-up compared to WHO grade.16
The Integrated Grade varied from the WHO grade in a substantial portion of patients (ie, ~one-third), and could therefore provide immediate and meaningful clinical benefit. For example, the administration of adjuvant radiation for WHO grade II and some grade I meningiomas, especially in the setting of STR, remains controversial and predominantly clinician-dependent.41 Given the excellent clinical course of Integrated Grade 1 meningiomas, independent of their WHO grade and brain invasiveness, deferring the potential long-term adverse sequelae of upfront radiation may be safe.42,43 Residual tumor in those meningiomas without copy-number alterations or with only monosomy 22 may continue to exhibit indolent growth with observation. Conversely, the presence of multiple high-risk copy-number alterations in the presence of grade I or grade I with atypical features histology should alert clinicians to maintain close surveillance for tumor recurrence over long follow-up, even after GTR, and raise consideration for adjuvant radiotherapy or clinical trial enrollment. While the biological imprint of meningiomas plays a vital role in tumor behavior, it does not supplant the impact of surgery: a genomically stable meningioma (Integrated Grade 1) will have the best chance for long-term progression-free survival after complete excision. Given the high incidence of meningiomas globally, the framework we present would influence the care of thousands of meningioma patients worldwide.1,44
One concern for the integration of genomic data into a grading scheme is the feasibility of implementation in academic and community settings. Accessibility and ease of interpretation are critical. Copy-number profiling is widely available in standard practice because of its demonstrated utility in the diagnosis of other tumors (eg, 1p/19q status in gliomas).35 The observation that the summation of high-risk molecular features incrementally strengthened the ability to predict recurrence allows for selective application of available genomic resources, ranging from targeted fluorescence in situ hybridization (FISH) to whole-genome assays. Furthermore, the accurate inference of broad chromosomal loss or gain from a number of genomic platforms, independent of proprietary technologies, increases the appeal.11,45 The binary read-out of copy-number alterations and the transparency offered by a point-based risk stratification system further enhances the ease of interpretation.
This analysis harbors several limitations. Mutational coverage in this study was incomplete, including those associated with prognosis such as in TERTp, BAP1, and SMARCE1.31,46–50 While these mutations are relatively rare, the Integrated Grade was able to stratify outcome amongst TERT promoter-mutated cases. Additionally, there was a sparsity of certain WHO high-grade histologic subtypes (clear cell, chondroid, rhabdoid, and papillary). It is possible these subtypes may have other important genomic features driving aggressive tumor behavior not captured by our Integrated Grading scheme, such as SMARCE1 mutations in clear cell meningiomas. Simpson grade, the most precise measure for extent of tumor resection, was unavailable. Hence, we adopted volumetric measurements based on quantitative imaging analysis as an indicator of tumor resection to accommodate for subjectivity and inter-surgeon variability in reporting,51 acknowledging that the failure to remove tumor-involved bone and dura remains an important consideration for evaluating recurrence risk. Additionally, methylation signatures were not available for our institutional cohort, limiting comparison to previously described methylation classification schemes.9,10,12 Broad access to copy-number-based profiling, made available by divergent genomic platforms that have proliferated across medical centers and commercial labs, will expand the number of patients who may benefit from molecular characterization beyond WHO grade at institutions without ready access to methylation platforms. Copy-number profiling has been integrated into the workflow at our institution, and the largely prospective collection of these cases strengthens the validity of our findings. Despite the need for assessment of larger cohorts of meningioma with rare mutations and of uncommon histologic subtypes, we show that mitotic index and copy-number profile can alone appraise tumor behavior with satisfactory results and significantly improve upon existing methods.
Taken together, the modular nature of the proposed risk-stratified integrated grading scheme lends itself to future refinement with incorporation of additional axes of genomic data as scientific discovery advances and has immediate relevance to management of meningioma patients. We envision that the Integrated Grade can enhance the clinical care of meningioma patients and aid in the design of future prospective clinical trial
Supplementary Material
Contributor Information
Joseph Driver, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Samantha E Hoffman, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA; Harvard-MIT Program in Health Science Technology, MD-PhD Program, Harvard Medical School, Boston, Massachusetts, USA.
Sherwin Tavakol, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Eleanor Woodward, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Eduardo A Maury, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA; Harvard-MIT Program in Health Science Technology, MD-PhD Program, Harvard Medical School, Boston, Massachusetts, USA; Bioinformatics and Integrative Genomics Program, Harvard Medical School, Boston, Massachusetts, USA.
Varun Bhave, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Noah F Greenwald, Cancer Biology Program, Stanford University School of Medicine, Stanford, California, USA.
Farshad Nassiri, Department of Neurosurgery, University of Toronto, Toronto, Ontario, Canada.
Kenneth Aldape, National Cancer Institute, Bethesda, Maryland, USA.
Gelareh Zadeh, Department of Neurosurgery, University of Toronto, Toronto, Ontario, Canada.
Abrar Choudhury, Departments of Radiation Oncology and Neurological Surgery, University of California San Francisco, San Francisco, California, USA.
Harish N Vasudevan, Departments of Radiation Oncology and Neurological Surgery, University of California San Francisco, San Francisco, California, USA.
Stephen T Magill, Departments of Radiation Oncology and Neurological Surgery, University of California San Francisco, San Francisco, California, USA.
David R Raleigh, Departments of Radiation Oncology and Neurological Surgery, University of California San Francisco, San Francisco, California, USA.
Malak Abedalthagafi, King Fahad Medical City and King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia.
Ayal A Aizer, Department of Radiation Oncology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Brian M Alexander, Department of Radiation Oncology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Keith L Ligon, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
David A Reardon, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
Patrick Y Wen, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
Ossama Al-Mefty, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Azra H Ligon, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Adrian M Dubuc, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Rameen Beroukhim, Center for Neuro-Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA; Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Elizabeth B Claus, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA; Yale School of Public Health, New Haven, Connecticut, USA.
Ian F Dunn, Department of Neurosurgery, Oklahoma University Medical Center, Oklahoma City, Oklahoma, USA.
Sandro Santagata, Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Wenya Linda Bi, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts, USA; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
Funding
This work has been supported by the Brigham Research Institute Precision Medicine Award (W.L.B.), the Brain Science Foundation (S.S.), the Jared Branfman Sunflowers for Life Fund (S.S.), the National Institute of General Medical Sciences to the Harvard/MIT MD-PhD program (E.A.M., T32GM007753), and the Biomedical Informatics and Data Science Research Training Program (E.A.M., T15LM007098).
Conflict of interest statement. None of the authors have any pertinent conflicts of interest to disclose in relation to the work presented in this manuscript.
Authorship statement. Data collection and analysis: J.D., S.E.H., S.T., E.W., E.A.M., V.B., N.F.G., F.N., G.Z., R.B., E.B.C., S.S., and W.L.B. Interpretation of analysis: all authors. Drafting of the initial manuscript: J.D., S.S., and W.L.B. Reviewing, editing, and approval of final manuscript: all authors.
References
- 1. Ostrom QT, Cioffi G, Gittleman H, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21(Suppl 5):v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vernooij MW, Ikram MA, Tanghe HL, et al. Incidental findings on brain MRI in the general population. N Engl J Med. 2007;357(18):1821–1828. [DOI] [PubMed] [Google Scholar]
- 3. Louis DN, Ohgaki H, Wiestler OD, et al. WHO Classification of Tumours of the Central Nervous System. Revised 4th ed. IARC; 2016. [Google Scholar]
- 4. Rogers CL, Perry A, Pugh S, et al. Pathology concordance levels for meningioma classification and grading in NRG Oncology RTOG Trial 0539. Neuro Oncol. 2016;18(4):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sayagués JM, Tabernero MD, Maíllo A, et al. Intratumoral patterns of clonal evolution in meningiomas as defined by multicolor interphase fluorescence in situ hybridization (FISH): is there a relationship between histopathologically benign and atypical/anaplastic lesions? J Mol Diagn. 2004;6(4):316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abedalthagafi MS, Bi WL, Merrill PH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015;208(6):345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juratli TA, Thiede C, Koerner MVA, et al. Intratumoral heterogeneity and TERT promoter mutations in progressive/higher-grade meningiomas. Oncotarget. 2017;8(65):109228–109237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jenkinson MD, Santarius T, Zadeh G, Aldape KD. Atypical meningioma-is it time to standardize surgical sampling techniques? Neuro Oncol. 2017;19(3):453–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 10. Nassiri F, Mamatjan Y, Suppiah S, et al. ; International Consortium on Meningiomas. DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019;21(7):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel AJ, Wan Y-W, Al-Ouran R, et al. Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proc Natl Acad Sci U S A. 2019;116(43):21715–21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vasudevan HN, Braunstein SE, Phillips JJ, et al. Comprehensive molecular profiling identifies FOXM1 as a key transcription factor for meningioma proliferation. Cell Rep. 2018;22(13):3672–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu P, Du XL, Zhu JJ, Esquenazi Y. Improved survival of glioblastoma patients treated at academic and high-volume facilities: a hospital-based study from the National Cancer Database. J Neurosurg. 2019;132(2):491–502. [DOI] [PubMed] [Google Scholar]
- 14. Curry WT Jr, Barker FG 2nd. Racial, ethnic and socioeconomic disparities in the treatment of brain tumors. J Neurooncol. 2009;93(1):25–39. [DOI] [PubMed] [Google Scholar]
- 15. Al-Mefty O, Kadri PA, Pravdenkova S, Sawyer JR, Stangeby C, Husain M. Malignant progression in meningioma: documentation of a series and analysis of cytogenetic findings. J Neurosurg. 2004;101(2):210–218. [DOI] [PubMed] [Google Scholar]
- 16. Bi WL, Greenwald NF, Abedalthagafi M, et al. Genomic landscape of high-grade meningiomas. NPJ Genom Med. 2017;2:525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro Oncol. 2016;18(2):269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Domingues PH, Sousa P, Otero Á, et al. Proposal for a new risk stratification classification for meningioma based on patient age, WHO tumor grade, size, localization, and karyotype. Neuro Oncol. 2014;16(5):735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang RY, Bi WL, Weller M, et al. Proposed response assessment and endpoints for meningioma clinical trials: report from the Response Assessment in Neuro-Oncology Working Group. Neuro Oncol. 2019;21(1):26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Louis D, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. Lyon, France: International Agency for Research on Cancer; 2016:408. [Google Scholar]
- 21. Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141(6):751–758. [DOI] [PubMed] [Google Scholar]
- 23. conumee: enhanced copy-number variation analysis using Illumina 450k methylation arrays. R package 0.99.
- 24. Hovestadt V, Zapatka M. conumee: enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 1.9.0. http://bioconductor.org/packages/conumee/.
- 25. Serin Harmanci A, Harmanci AO, Zhou X. CaSpER identifies and visualizes CNV events by integrative analysis of single-cell or bulk RNA-sequencing data. Nat Commun. 2020;11(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bi WL, Greenwald NF, Ramkissoon SH, et al. Clinical identification of oncogenic drivers and copy-number alterations in pituitary tumors. Endocrinology. 2017;158(7):2284–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishwaran H, Kogalur UB. Consistency of random survival forests. Stat Probab Lett. 2010;80(13-14):1056–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hothorn T, Buehlmann P, Kneib T, Schmid M, Hofner B. Model-based Boosting 2.0. J Mach Learn Res. 2010;11:2109–2113. [Google Scholar]
- 30. Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19(4):535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams EA, Wakimoto H, Shankar GM, et al. Frequent inactivating mutations of the PBAF complex gene PBRM1 in meningioma with papillary features. Acta Neuropathol. 2020;140(1):89–93. [DOI] [PubMed] [Google Scholar]
- 33. Vaubel RA, Chen SG, Raleigh DR, et al. Meningiomas with rhabdoid features lacking other histologic features of malignancy: a study of 44 cases and review of the literature. J Neuropathol Exp Neurol. 2016;75(1):44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Louis DN, Perry A, Wesseling P, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pajtler KW, Witt H, Sill M, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27(5):728–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Northcott PA, Korshunov A, Witt H, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 39. Perry A, Stafford SL, Scheithauer BW, Suman VJ, Lohse CM. Meningioma grading: an analysis of histologic parameters. Am J Surg Pathol. 1997;21(12):1455–1465. [DOI] [PubMed] [Google Scholar]
- 40. Boström J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001;159(2):661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogers L, Barani I, Chamberlain M, et al. Meningiomas: knowledge base, treatment outcomes, and uncertainties: a RANO review. J Neurosurg. 2015;122(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weber DC, Ares C, Villa S, et al. Adjuvant postoperative high-dose radiotherapy for atypical and malignant meningioma: a phase-II parallel non-randomized and observation study (EORTC 22042-26042). Radiother Oncol. 2018;128(2):260–265. [DOI] [PubMed] [Google Scholar]
- 43. Rogers L, Zhang P, Vogelbaum MA, et al. Intermediate-risk meningioma: initial outcomes from NRG Oncology RTOG 0539. J Neurosurg. 2018;129(1):35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leece R, Xu J, Ostrom QT, Chen Y, Kruchko C, Barnholtz-Sloan JS. Global incidence of malignant brain and other central nervous system tumors by histology, 2003-2007. Neuro Oncol. 2017;19(11):1553–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Collord G, Tarpey P, Kurbatova N, et al. An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep. 2018;8(1):13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016;108(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45(3):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gauchotte G, Peyre M, Pouget C, et al. Prognostic value of histopathological features and loss of H3K27me3 immunolabeling in anaplastic meningioma: a multicenter retrospective study. J Neuropathol Exp Neurol. 2020;79(7):754–762. [DOI] [PubMed] [Google Scholar]
- 51. Schwartz TH, McDermott MW. The Simpson grade: abandon the scale but preserve the message. J Neurosurg. 2020;135:1–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.