ABSTRACT
Background
Human milk oligosaccharides (HMOs) are an abundant class of compounds found in human milk and have been linked to the development of the infant, and specifically the brain, immune system, and gut microbiome.
Objectives
Advanced analytical methods were used to obtain relative quantitation of many structures in approximately 2000 samples from over 1000 mothers in urban, semirural, and rural sites across geographically diverse countries.
Methods
LC-MS−based analytical methods were used to profile the compounds with broad structural coverage and quantitative information. The profiles revealed their structural heterogeneity and their potential biological roles. Comparisons of HMO compositions were made between mothers of different age groups, lactation periods, infant sexes, and residing geographical locations.
Results
A common behavior found among all sites was a decrease in HMO abundances during lactation until approximately postnatal month 6, where they remained relatively constant. The greatest variations in structural abundances were associated with the presence of α(1,2)-fucosylated species. Genomic analyses of the mothers were not performed; instead, milk was phenotyped according to the abundances of α(1,2)-fucosylated structures. Mothers from the South American sites tended to have higher proportions of phenotypic secretors [mothers with relatively high concentrations of α(1,2)-fucosylated structures] in their populations compared to the rest of the globe, with Bolivia at ∼100% secretors, Peru at ∼97%, Brazil at ∼90%, and Argentina at ∼85%. Conversely, the cohort sampled in Africa manifested the lowest proportion of secretors (South Africa ∼ 63%, the Gambia ∼ 64%, and Malawi ∼ 75%). Furthermore, we compared total abundances of HMOs in secretors compared with nonsecretors and found that nonsecretors have lower abundances of HMOs compared to secretors, regardless of geographical location. We also observed compositional differences of the 50+ most abundant HMOs between milk types and geographical locations.
Conclusions
This study represents the largest structural HMO study to date and reveals the general behavior of HMOs during lactation among different populations.
Keywords: human milk oligosaccharides, oligosaccharides, mass spectrometry, glycans, breast milk, carbohydrates, lactose, secretor, FUT2
Introduction
Human milk for infants is a manifestation of a highly adapted, dynamic, and personalized process of human postnatal development. Human milk is dynamic in the sense that time-dependent changes occur in the presence and concentration of milk bioactives within and across mothers and personalized in the sense that maternal genotype, health status, and environmental exposures, including diet, can impact milk compositions (1–5).
Human milk oligosaccharides (HMOs) are among the most abundant and diverse components of breast milk, with hundreds of unique structures identified to date (6–9). HMOs serve as nutrients for highly adapted, early bacterial colonizers of the infant gut, including specific strains of bifidobacteria endowed with suites of gene-encoded proteins dedicated to the import and utilization of HMOs (10–14). While the prebiotic effect is believed to be a major function, a small fraction of HMOs are absorbed in the small intestine and detectable in plasma (15) and urine (16), suggesting the potential for direct effects on host physiology, including immunomodulation (17–19) and brain development (20).
HMOs are assembled by glycosyltransferases to form either branched or linear structures. HMOs generally consist of a lactose [glucose and galactose (Gal)] core, with variable combinations of N-acetylglucosamine (GlcNAc), and can be further bound to monosaccharides, including sialic acids [N-acetylneuraminic acid (Neu5Ac)] and fucose (Fuc) (21). The process yields an extensive number of oligosaccharides that in many cases are unique to human milk (22).
Variations in HMOs are greatest among secretor genotypes. Secretors are individuals with a functioning FUT2 gene encoding α(1,2)-fucosyltransferase that attaches fucose via an α(1,2)-linkage to terminal Gal residues, thereby producing blood antigens into secreted fluids (e.g., sweat, tears, semen, and milk) (21, 23–25). Nonsecretors have diminished ability to produce ABH or Lewis b antigens [Fucα1,2Galβ1,3(Fucα1,4)GlcNAcβ] due to mutations in the FUT2 gene (21, 23–25). Genetic studies have documented variations in the prevalences of the wild-type and mutant FUT2 alleles around the world (26–28). The prevailing existence of different genotypes in populations shows that there are unique advantages to individuals. This notion is consistent with evidence suggesting a protective effect against, for example, otitis media (29) and autoimmune diseases (30, 31) for secretors and against viral diarrhea for nonsecretors (32). While different alterations in the FUT2 gene among various populations determine the secretor status of the mother, the milk and its function is guided by the abundances of HMO structures. Thus, classifying the milk phenotype—for example, the amount of sialylation and fucosylation—is a more direct approach in surveying the differences between mothers’ milks and correlating infant health outcomes. Advanced analytical methods now make it possible to accurately determine the phenotypic secretor status by directly quantitating the abundances of α(1,2)-fucosylated structures present in the breast milk (23). For the purpose of this study, we refer to milk that corresponds to high amounts of α(1,2)-fucosylated structures as S+ milk and milk corresponding to low abundances as S− milk. HMOs can therefore be used to type the milk according to the presence or absence of α(1,2)-fucosylated structures, regardless of the genotype (8, 23).
MS−based analytical methods have enabled more rapid and precise characterization of human milk, including the capability to simultaneously determine the abundances of hundreds of distinct carbohydrate structures. However, determination of HMOs is largely limited to studies of cohorts living in a small number of sites or geographic locales (33–35). In this report, we describe the results of a cross-sectional analysis that quantified the abundances of HMOs to determine the natural variations during lactation among various sites involving mothers from different ethnic groups. Milk samples were obtained and analyzed from over 1000 mothers living in 15 countries, encompassing 6 continents, and representing a diverse set of ecological and cultural backgrounds.
Methods
Sources of breast milk samples
Breast milk samples (N = 2234) were collected from mothers (N = 1090) in 16 global sites, including urban, rural, and semirural communities in 15 sites spanning Africa, Eurasia, the Americas, and Australia. Samples and resulting data were collected from different studies, as detailed in the Supplemental Text. However, the quality controls and analytical methods for each sample remained the same. A detailed summary of sample information, including country, population, collection procedures, sample size, and infant age, is provided in Table 1 and Supplemental Table 1. All infants were delivered full term. Written informed consent was obtained from all parents/guardians prior to study enrollment. Samples were collected using standardized protocols for all populations, as detailed in the Supplemental Methods. Details on the ethical approval identifiers, trial registration information, and participant inclusion/exclusion criteria for each study site can also be found in the Supplemental Methods.
TABLE 1.
Summary of successful milk collections as a function of location and lactation month1
| Postpartum lactation month | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | −1 | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 21 | 24 | 26 | Total |
| Argentina | — | — | 1 | 1 | — | 2 | 1 | 3 | 2 | — | 3 | 3 | 2 | 2 | — | — | — | — | — | — | — | — | — | — | 20 |
| Bangladesh | — | 15 | 15 | 14 | 12 | 12 | 12 | 12 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 92 |
| Bolivia | — | 1 | 1 | 3 | 7 | 4 | 1 | 6 | 4 | 2 | 5 | 3 | 2 | 1 | 7 | 2 | 3 | 2 | 3 | 1 | 2 | — | — | 1 | 61 |
| Boston, MA | — | — | — | 2 | 1 | 4 | 2 | 2 | 2 | 1 | 1 | — | 4 | 1 | — | — | — | — | — | — | — | — | — | — | 20 |
| Brazil | — | — | 22 | 18 | 14 | 20 | 18 | 17 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 109 |
| Davis, CA | 8 | 59 | 40 | 31 | 37 | 38 | — | 30 | — | — | — | — | 1 | 11 | — | — | — | — | — | — | — | — | — | — | 255 |
| Gambia | — | — | 33 | — | — | 33 | 33 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 99 |
| India | — | — | 42 | 42 | 42 | 42 | 40 | 39 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 247 |
| Malawi | — | — | 73 | — | 84 | — | — | 652 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 809 |
| Namibia | — | — | 3 | 2 | — | 1 | — | — | 1 | 2 | — | — | — | — | 1 | 1 | — | — | — | 1 | — | — | — | — | 12 |
| Nepal | — | — | 1 | 3 | — | 3 | 2 | 4 | 4 | 2 | 3 | 2 | — | — | 1 | — | — | — | — | — | — | 1 | 2 | — | 28 |
| Perth | — | 28 | 29 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 57 |
| Peru | — | 35 | 36 | 33 | 34 | 32 | 31 | 32 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 233 |
| Philippines | — | 5 | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | — | — | — | — | — | — | — | — | — | — | 18 |
| Poland | — | — | 1 | 2 | 1 | 4 | 2 | 2 | 4 | 1 | — | 3 | — | 2 | 1 | — | — | — | — | — | — | — | — | — | 23 |
| South Africa | — | — | 26 | 26 | 27 | 26 | 25 | 26 | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | 151 |
| Total | 8 | 143 | 324 | 178 | 261 | 223 | 169 | 816 | 19 | 10 | 13 | 12 | 11 | 18 | 10 | 3 | 3 | 2 | 3 | 2 | 2 | 1 | 2 | 1 | 2234 |
Values presented correspond to the number of samples collected as a function of geographical location and month of lactation.
HMO extraction and MS analysis
HMOs were extracted from breast milk samples using previously reported methods (7, 8, 36, 37). Briefly, entire milk samples were aliquoted into 96-well plates, diluted, and then defatted via centrifugation. Proteins were precipitated with ethanol, and the resulting glycans were reduced with sodium borohydride (Sigma-Aldrich). Solid-phase extraction was performed on graphitized carbon cartridges (Glygen) to remove lactose and salts. After the solvent was evaporated, purified HMOs were reconstituted and diluted prior to analysis. Standard solutions were made of HMO in water, with concentrations ranging from 0.05–0.2 mg/mL.
Extracted HMOs were analyzed on a nano–HPLC–time of flight (TOF)–MS. The HPLC unit (model series 1200, Agilent Technologies) utilizes a capillary pump for sample loading (4 μL/min) and a nano pump for analyte separation (0.3 μL/min). Loading and separation were performed on a microfluidic chip packed with porous graphitized carbon via enrichment and analytical columns, respectively, using a binary gradient of solvent A [3% acetonitrile (ACN) in 0.1% formic acid] and solvent B (90% ACN in 0.1% formic acid). This system was coupled to an Agilent 6220 series TOF MS. Detection was performed in the positive mode, and calibration was achieved with a dual nebulizer electrospray source with calibrant ions with mass-to-charge (m/z) values ranging from 118.086 to 2721.895.
Structural annotation of HMOs
Data were collected using Agilent MassHunter Workstation Data Acquisition software (B.02.01) and analyzed using Agilent MassHunter Qualitative Analysis software B.03.01 and B.06.00. The “Find Compound by Molecular Feature” function was used to extract ion abundances to within 20 ppm of theoretical HMO masses. Individual HMOs were identified by accurate mass, retention time, and elution order, as defined in previously developed HMO libraries (7, 8). An in-house software program was used to align peaks due to a minor retention time shift (37). HMOs were grouped into classes as follows: fucosylated HMOs (any structure with Fuc), sialylated HMOs (any structure with Neu5Ac), undecorated HMOs (neither Fuc nor Neu5Ac present), and fucosylated plus sialylated HMOs (both Fuc and Neu5Ac present). Relative abundances (%) were calculated by normalizing class and individual compound abundances to the total HMO abundance in each breast milk sample. Compounds that were not identified in individual samples but were present in at least 50% of all samples were given an (Limit of Detection)/2 abundance.
Classification of milk secretor phenotype as S+ and S− based on HMO abundances
The phenotypic secretor status was determined following our previously published method (23). Structures with known α(1,2)-Fuc linkages were identified by matching exact masses and retention times to previously developed annotated HMO libraries (7, 8). The abundances of the most abundant α(1,2)-fucosylated structures—namely, 2′-fucosyllactose (2′FL), lactodifucotetraose (LDFT), difucosyllacto-N-hexaose a (DFLNHa), and trifucosyllacto-N-hexaose (TFLNH)—were summed and normalized to the total HMO abundances in a given sample, so that a relative α(1,2)-fucosylation value could be determined. The secretor status was assigned based on a previously established and validated threshold of 6% (23). If this value exceeded the threshold. the mother was deemed a secretor (S+); conversely, if the value fell below this threshold, the mother was deemed a nonsecretor (S−). The numbers of mothers producing S+ and S− milk in each location were determined, and the proportion of S− mothers was calculated for each location. If a mother provided multiple samples from different postpartum time points, her secretor status was determined based on the secretor status determination in the majority of her samples. If there was no majority milk type, she was excluded from the statistical analysis (1 mother from Davis, CA; 2 mothers from Malawi; and 3 mothers from Perth, Australia).
Statistical analyses
Mann-Whitney U tests were used to determine the differences between absolute and relative abundances of the HMO classes. Furthermore, the data were grouped to show how secretor status, geographical location, age, sex, and lactation month affected the HMO profiles. An alpha correction of 0.05 was used for the statistical analysis. Differences were determined when all samples from all time points were combined (S+ n = 1709, S− n = 524) and when samples were split by location, including all time points (Argentina: S+ n = 17, S− n = 3; Bolivia: S+ n = 52, S− n = 0; Bangladesh: S+ n = 72, S− n = 20; Boston, MA: S+ n = 15, S− n = 5; Brazil: S+ n = 97, S− n = 12; Davis, CA: S+ n = 194, S− n = 58; Gambia: S+ n = 63, S− n = 36; India: S+ n = 166, S− n = 81; Malawi: S+ n = 601, S− n = 208; Namibia: S+ n = 10, S− n = 2; Nepal: S+ n = 20, S− n = 8; Australia: S+ n = 42, S− n = 15; Peru: S+ n = 227, S− n = 8; Philippines: S+ n = 13, S− n = 5; Poland: S+ n = 18, S− n = 5; and South Africa: S+ n = 93, S− n = 58). Due to the vast changes in milk composition throughout lactation, samples were binned based on lactation month; therefore, milk samples collected from a single mother at different time points were treated as independent samples.
Results
HMO-based classification of milk into S+ and S− phenotypes
Over 2000 breast milk samples were collected from 1090 mothers in 15 geographical sites. The samples obtained from 6 continents were analyzed under 1 protocol, allowing direct comparison of abundances by classes and individual structures (Table 1). Using nano–HPLC–quadruple TOF (qTOF)–MS, we identified 60 structures that were common to most samples. However, the total number of unique structures varied for each mother, with the average count of nearly 100 structures in a single mother. The abundances for these structures varied widely, spanning 4 orders of magnitude.
HMOs containing Lewis b structures [α(1,2)-fucose], a feature of the secretor genotype (homozygous or heterozygous for the functional FUT2 allele) (23, 25, 38), were most variable between mothers. We defined milk rich in Lewis b structures (2′FL, LDFT, DFLNHa, and TFLNH) that were consistently represented in samples collected within and across the different geographic sites S+ milk, belonging to a secretor mother. Milk containing a total relative abundance of <6% of these 4 Lewis b structures was defined as S- milk, thus belonging to a nonsecretor mother. This criterion was developed previously and has been validated with genomic data (23). Phenotyping the milk addresses what the infant receives, while genotyping the mother does not necessarily translate to HMO abundances.
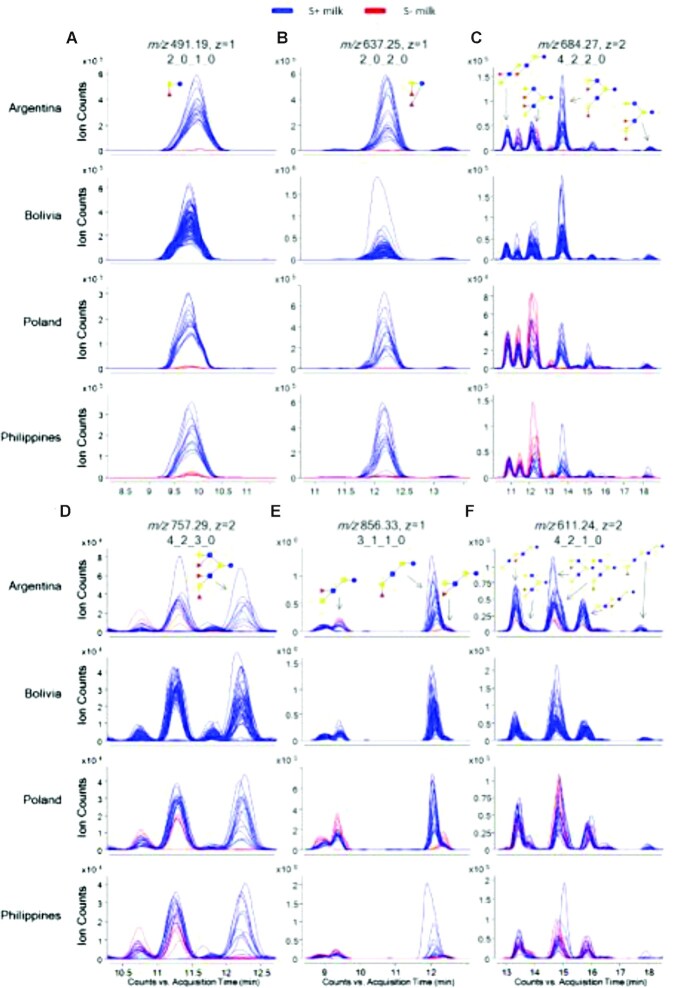
Figure 1 shows extracted ion chromatograms of the most abundant α(1,2)-fucosylated compounds in human milk: namely, 2′FL and LDFT (Figure 1A and B). These compounds were consistently higher in S+ milk (blue) compared to S− milk (red, near baseline) across all sites (P < 0.05). Five other α(1,2)-Fuc-containing structures—namely, lacto-N-fucopentaose (LNFP) I, lacto-N-difucohexaose I, monofucosyllacto-N-hexaose I (MFLNH I), isomer I fucosyl-paralacto-N-hexaose, and difucosyllacto-N-hexaose c—were much less ubiquitous and, when detected, were present at much lower abundances than the 4 structures used for determination of secretor status (Table 2). Conversely, there were structures, particularly α(1,3)- and α(1,4)-fucosylated HMOs, that were significantly higher in S− milk (Figure 1C–F; Table 2). Three isomers with composition 3Hex:1HexNAc:1Fuc and having α(1,3/4)-Fuc linkages were produced in higher abundances in mothers with S− milk (Figure 1E).
FIGURE 1.
EICs displaying differences in abundances of HMO markers with α(1–2)-linked Fuc between mothers with S+ (–) and S− (–) milk from different locations around the world. Locations were chosen to represent different areas with both S+ and S− milk producers. Bolivia was also chosen to display EICs of 100% S+ milk producers. Monosaccharide compositions of structures are given as Hex_HexNAc_Fuc_Neu5Ac and represented as glucose ( ), galactose (
), galactose ( ), N-acetylglucosamine (
), N-acetylglucosamine ( ), and fucose (
), and fucose ( ). (A) EIC of 2’FL with m/z 491.19. (B) EIC of LDFT with m/z 637.25. (C) EIC of isomers DFpLNH II, DFLNHb, DFLNHa, and DFLNHc with m/z 684.27. (D) EIC of TFLNH with m/z 757.29. (E) EIC of isomers LNFP I, II, and III with m/z 856.33. (F) EIC of isomers MFpLNH IV, 4120a, MFLNH III, MFLNH I, IFLNH III, and IFLNH I with m/z 611.24. DFLNH, difucosyllacto-N-hexaose; DFpLNH II, difucosyl-parap-lacto-N-hexaose; EIC, extracted ion chromatograms; Fuc, fucose; HMO, human milk oligosaccharide; IFLNH I, isomer I fucosyl-para-lacto-N-hexaose; IFLNH III, isomer III fucosyl-para-lacto-N-hexaose; LDFT, lactodifucotetraose; LNFP, lacto-N-fucopentaose; MFLNH, monofucosyllacto-N-hexaose; MFpLNH IV, fucosyl-para-lacto-N-hexaose; m/z, mass-to-charge; Neu5Ac, N-acetylneuraminic acid; p-LNH, para-lacto-N-hexaose; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures; S-LNH, monosialyllacto-N-hexaose; TFLNH, trifucosyllacto-N-hexaose; 2’FL, 2’-fucosyllactose.
). (A) EIC of 2’FL with m/z 491.19. (B) EIC of LDFT with m/z 637.25. (C) EIC of isomers DFpLNH II, DFLNHb, DFLNHa, and DFLNHc with m/z 684.27. (D) EIC of TFLNH with m/z 757.29. (E) EIC of isomers LNFP I, II, and III with m/z 856.33. (F) EIC of isomers MFpLNH IV, 4120a, MFLNH III, MFLNH I, IFLNH III, and IFLNH I with m/z 611.24. DFLNH, difucosyllacto-N-hexaose; DFpLNH II, difucosyl-parap-lacto-N-hexaose; EIC, extracted ion chromatograms; Fuc, fucose; HMO, human milk oligosaccharide; IFLNH I, isomer I fucosyl-para-lacto-N-hexaose; IFLNH III, isomer III fucosyl-para-lacto-N-hexaose; LDFT, lactodifucotetraose; LNFP, lacto-N-fucopentaose; MFLNH, monofucosyllacto-N-hexaose; MFpLNH IV, fucosyl-para-lacto-N-hexaose; m/z, mass-to-charge; Neu5Ac, N-acetylneuraminic acid; p-LNH, para-lacto-N-hexaose; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures; S-LNH, monosialyllacto-N-hexaose; TFLNH, trifucosyllacto-N-hexaose; 2’FL, 2’-fucosyllactose.
TABLE 2.
The 60 most common HMO structures in S+ and S− milks across all study sites1
| Neutral mass (Da) | Composition (Hex_HexNAc_Fuc_Neu5Ac) | HMO | S− type milk mean | S+ type milk mean | P value |
|---|---|---|---|---|---|
| 490.19 | 2010 | 2′FL | 0.002 ± 0.005 | 0.07 ± 0.04 | <0.0001 |
| 636.24 | 2020 | LDFT | 0.001 ± 0.003 | 0.04 ± 0.03 | <0.0001 |
| 709.26 | 3100 | LNT + LNnT | 0.2 ± 0.09 | 0.1 ± 0.06 | <0.0001 |
| 855.32 | 3110 | LNFP II | 0.04 ± 0.03 | 0.02 ± 0.02 | <0.0001 |
| 855.32 | 3110 | LNFP I + LNFP III | 0.02 ± 0.02 | 0.06 ± 0.04 | <0.0001 |
| 1074.39 | 4200 | LNH | 0.009 ± 0.01 | 0.009 ± 0.007 | 0.06 |
| 1074.39 | 4200 | LNnH | 0.009 ± 0.01 | 0.01 ± 0.01 | <0.0001 |
| 1074.39 | 4200 | p-LNH | 0.004 ± 0.007 | 0.004 ± 0.005 | <0.0001 |
| 1220.45 | 4210 | MFpLNH IV | 0.02 ± 0.01 | 0.02 ± 0.009 | 0.1 |
| 1220.45 | 4210 | 412Oa | 0.007 ± 0.01 | 0.003 ± 0.006 | <0.0001 |
| 1220.45 | 4210 | MFLNH III + MFLNH I | 0.03 ± 0.02 | 0.02 ± 0.01 | 0.02 |
| 1220.45 | 4210 | IFLNH III | 0.008 ± 0.006 | 0.01 ± 0.006 | <0.0001 |
| 1220.45 | 4210 | IFLNH I | 0.001 ± 0.003 | 0.004 ± 0.004 | <0.0001 |
| 1366.51 | 4220 | DFpLNH II | 0.01 ± 0.007 | 0.01 ± 0.006 | <0.0001 |
| 1366.51 | 4220 | DFLNHb | 0.02 ± 0.01 | 0.01 ± 0.007 | <0.0001 |
| 1366.51 | 4220 | DFLNHa | 0.001 ± 0.002 | 0.01 ± 0.01 | <0.0001 |
| 1512.57 | 4230 | TFLNH | 0.004 ± 0.004 | 0.006 ± 0.005 | <0.0001 |
| 1585.58 | 5310 | 5130a | 0.006 ± 0.006 | 0.004 ± 0.003 | <0.0001 |
| 1585.58 | 5310n | F-LNO | 0.004 ± 0.003 | 0.003 ± 0.002 | 0.002 |
| 1731.64 | 5320 | DFLNO I | 0.007 ± 0.005 | 0.003 ± 0.003 | <0.0001 |
| 1731.64 | 5320 | DFLNnO II | 0.004 ± 0.005 | 0.003 ± 0.003 | 0.9 |
| 1731.64 | 5320 | 5230a + DFLNnO I/DFLNO II | 0.004 ± 0.003 | 0.006 ± 0.004 | <0.0001 |
| 635.22 | 2001 | 6'SL | 0.003 ± 0.003 | 0.002 ± 0.003 | <0.0001 |
| 635.22 | 2001 | 3'SL | 0.01 ± 0.008 | 0.01 ± 0.007 | 0.4 |
| 1000.36 | 3101 | LSTc + LSTb | 0.03 ± 0.02 | 0.03 ± 0.01 | <0.0001 |
| 1000.36 | 3101 | LSTa | 0.003 ± 0.002 | 0.002 ± 0.002 | <0.0001 |
| 1365.49 | 4201 | S-LNH | 0.003 ± 0.004 | 0.002 ± 0.002 | <0.0001 |
| 1365.49 | 4201 | 4021a + S-LNnH II | 0.004 ± 0.005 | 0.006 ± 0.005 | <0.0001 |
| 490.19 | 2010 | %2'FL | 0.4 ± 0.9 | 11.1 ± 4.6 | <0.0001 |
| 636.24 | 2020 | %LDFT | 0.2 ± 0.5 | 5.8 ± 5.1 | <0.0001 |
| 709.26 | 3100 | %LNT + LNnT | 30.3 ± 10 | 21.7 ± 5.9 | <0.0001 |
| 855.32 | 3110 | %LNFP II | 6.1 ± 5 | 2.9 ± 2.6 | <0.0001 |
| 855.32 | 3110 | %LNFP I + LNFP III | 3.8 ± 2.5 | 8.2 ± 5 | <0.0001 |
| 1074.39 | 4200 | %LNH | 1.5 ± 1.6 | 1.2 ± 0.9 | 0.5 |
| 1074.39 | 4200 | %LNnH | 1.4 ± 1.8 | 1.7 ± 1.4 | <0.0001 |
| 1074.39 | 4200 | %p-LNH | 0.7 ± 1.2 | 0.6 ± 0.7 | <0.0001 |
| 1220.45 | 4210 | %MFpLNH IV | 2.8 ± 1.5 | 2.3 ± 1.2 | <0.0001 |
| 1220.45 | 4210 | %412Oa | 1.1 ± 1.7 | 0.6 ± 1 | <0.0001 |
| 1220.45 | 4210 | %MFLNH III + MFLNH I | 4.3 ± 2.6 | 3.3 ± 1.8 | <0.0001 |
| 1220.45 | 4210 | %IFLNH III | 1.3 ± 0.9 | 1.4 ± 0.9 | 0.007 |
| 1220.45 | 4210 | %IFLNH I | 0.2 ± 0.5 | 0.5 ± 0.6 | <0.0001 |
| 1366.51 | 4220 | %DFpLNH II | 2.2 ± 1.3 | 1.5 ± 0.8 | <0.0001 |
| 1366.51 | 4220 | %DFLNHb | 3.3 ± 2.6 | 1.6 ± 1.2 | <0.0001 |
| 1366.51 | 4220 | %DFLNHa | 0.2 ± 0.3 | 2 ± 1.8 | <0.0001 |
| 1512.57 | 4230 | %TFLNH | 0.7 ± 0.6 | 0.9 ± 0.7 | <0.0001 |
| 1585.58 | 5310 | %5130a | 0.9 ± 0.8 | 0.5 ± 0.4 | <0.0001 |
| 1585.58 | 5310 | %F-LNO | 0.6 ± 0.4 | 0.4 ± 0.3 | <0.0001 |
| 1731.64 | 5320 | %DFLNO I | 1.1 ± 0.7 | 0.5 ± 0.4 | <0.0001 |
| 1731.64 | 5320 | %DFLNnO II | 0.7 ± 0.7 | 0.5 ± 0.4 | 0.0007 |
| 1731.64 | 5320 | %5230a + DFLNnO I/DFLNO II | 0.7 ± 0.5 | 0.8 ± 0.4 | <0.0001 |
| 635.22 | 2001 | %6'SL | 0.5 ± 0.5 | 0.3 ± 0.4 | <0.0001 |
| 635.22 | 2001 | %3'SL | 2.6 ± 1.7 | 2.2 ± 1.4 | <0.0001 |
| 1000.36 | 3101 | %LSTc + LSTb | 5.2 ± 2 | 3.8 ± 1.6 | <0.0001 |
| 1000.36 | 3101 | %LSTa | 0.5 ± 0.3 | 0.3 ± 0.3 | <0.0001 |
| 1365.49 | 4201 | %S-LNH | 0.5 ± 0.5 | 0.3 ± 0.3 | <0.0001 |
| 1365.49 | 4201 | %4021a + S-LNnH II | 0.6 ± 0.7 | 0.8 ± 0.6 | <0.0001 |
Values are presented as mean abundance ± SD. All data collected were used for this analysis, including data from the same mother at different time points of lactation. The monosaccharide composition is represented by a 4-digit code (Hex_HexNAc_Fuc_Neu5Ac). Common HMO abbreviations were used to name oligosaccharides. Oligosaccharides with 2 compound names are isomers that were difficult to resolve chromatographically, and the data presented are the sum of their combined abundances. An oligosaccharide name proceeding a percentage symbol indicates that the values presented are mean relative abundances. P values were obtaining using Mann-Whitney U tests. DFLNH, difucosyllacto-N-hexaose; DFLNnO, difucosyllacto-N-neooctaose; DFLNO, difucosyllacto-N-octaose; DFpLNH II, difucosyl-para-lacto-N-hexaose II; F-LNO, monofucosyllacto-N-octaose; Fuc, fucose; HMO, human milk oligosaccharide; IFLNH I, isomer I fucosyl-para-lacto-N-hexaose; IFLNH III, isomer III fucosyl-para-lacto-N-hexaose; LDFT, lactodifucotetraose; LNFP, lacto-N-fucopentaose; LNH, lacto-N-hexaose; LNnH, lacto-N-neohexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; LSTa, Sialyllacto-N-tetraose a; LSTb, sialyllacto-N-tetraose b; LSTc, sialyllacto-N-neotetraose; MFLNH, monofucosyllacto-N-hexaose; MFpLNH IV, fucosyl-para-lacto-N-hexaose; Neu5Ac, N-acetylneuraminic acid; p-LNH, para-lacto-N-hexaose; S-LNH, monosialyllacto-N-hexaose; S-LNnH II, monosialyllacto-N-neohexaose II; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures; TFLNH, trifucosyllacto-N-hexaose; 2’FL, 2’-fucosyllactose; 3'SL: 3'-sialyllactose, 6'SL: 6'-sialyllactose.
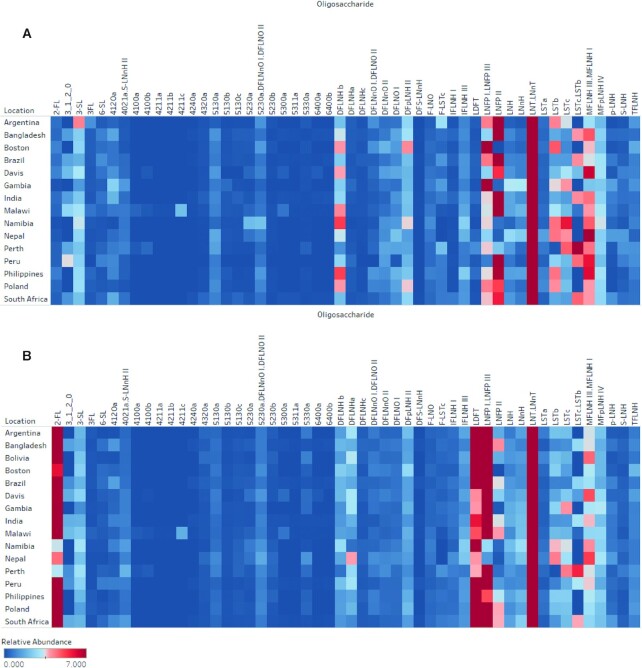
The relative abundances of the 60 most common structures were shown for S+ and S− milk in Figure 2 (see Table 2). The abundance of each HMO was normalized to the total abundances of the selected group, which made up approximately 97% of all abundances. For mothers that produced S+ milk, the most abundant HMOs were lacto-N-tetraose/lacto-N-neotetraose (LNT/LNnT). These 2 compounds are isomers and were difficult to resolve chromatographically, and the data presented are the sum of their combined abundances. At all sites, mothers with S− milk had higher abundances of LNFP II, suggesting this HMO is a potential marker of S− milk (P < 0.0001; Mann-Whitney U test). Other HMOs, including MFLNH I, MFLNH III, and difucosyl-para-lacto-N-hexaose II, were also higher in S− milk, achieving statistical significance (P < 0.05; Mann-Whitney U test) at all sites except Brazil (P < 0.38; Mann-Whitney U test).
FIGURE 2.
Heat map of relative abundances of the most common (60) HMOs across 15 geographically diverse sites. Comparison of abundances from mothers who are (A) S− producers and (B) S+ producers. HMO abundance values correspond to HPLC-qTOF MS spectral abundance, normalized to the mean of the total abundance of counts from each sample. HMOs that were not baseline separated (resolution > 1.5) were grouped together and labeled accordingly. DFLNH, difucosyllacto-N-hexaose; DFLNnO, difucosyllacto-N-neooctaose; DFS-LNnH, difucosylmonosialyllacto-N-neohexaose; DFpLNH II, difucosyl-parap-lacto-N-hexaose; DFS-LNnH, Difucosylmonosialyllacto-N-neohexaose; F-LNO, monofucosyllacto-N-octaose; F-LSTc, Monofucosylmonosialyllacto-N-neotetraose; HMO, human milk oligosaccharide; IFLNH I, isomer I fucosyl-para-lacto-N-hexaose; IFLNH III, isomer III fucosyl-para-lacto-N-hexaose; LDFT, lactodifucotetraose; LNFP, lacto-N-fucopentaose; LNH, lacto-N-hexaose; LNnH, lacto-N-neohexaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; LST, sialyllacto-N-tetraose; MFLNH, monofucosyllacto-N-hexaose; MFpLNH IV, fucosyl-para-lacto-N-hexaose; qTOF, quadruple time of flight; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures; S-LNnH II, Monosialyllacto-N-neohexaose II; 2’FL, 2’-fucosyllactose; 3′FL, 3′-fucosyllactose; 3′SL, 3'SL: 3'-sialyllactose, 6'SL: 6'-sialyllactose.
Total HMO abundances between sites and during lactation
The total abundances of HMOs between mothers from various sites were compared as a function of months postpartum. Figure 3 compares total HMOs from mothers living in Brazil; Bangladesh; Davis, CA; Peru; India; and South Africa sampled between postpartum months 1–6. These sites were selected because they provided the most extensive longitudinal sampling. The highest abundances were observed at month 1 and decreased uniformly thereafter. Each site showed similar behavior and similar decreases in total abundances, with some variations in months 3 and 4. For example, in both the Peru and India sites, total abundances decreased at month 3 and remained nearly constant thereafter, while in the other sites HMO abundances dropped uniformly between months 1 and 6. When comparing total absolute abundances in secretors between months 1 and 6, statistical significance was achieved across all selected sites, with higher significance achieved in sites with more sampling (Figure 3B). Similarly, statistical significance was similarly achieved when comparing abundances in nonsecretors between months 1 and 6, where there were sufficient numbers of samples. HMO abundances were also compared between S+ and S− milk across these sites by averaging the means of all time points within 1 site. In all counties except Brazil, S+ milk possessed higher abundances of HMOs (P < 0.0001; Mann-Whitney U test).
FIGURE 3.
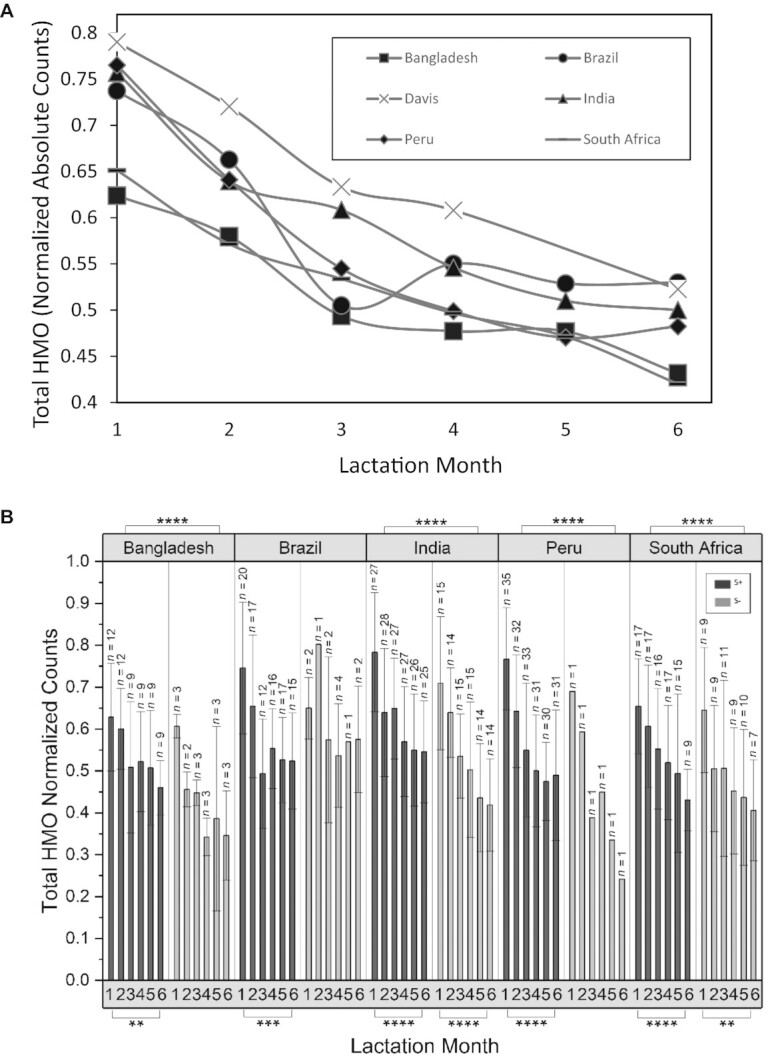
(A) Mean changes in HMO concentrations in breastmilk samples collected monthly during the first 6 months postpartum at sites with extensive longitudinal sampling. (B) Total HMO abundances as a function of location, lactation month, and secretor status. HMO abundance values correspond to HPLC-qTOF MS spectral abundance, normalized to the mean of the total abundance of ion counts from each sample. N values correspond to the number of samples. Error bars represent the SD. P values were obtaining using Mann-Whitney U tests with an α correction of 0.05. HMO, human milk oligosaccharide; qTOF, quadruple time of flight. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ****P ≤ 0.0001.
Total HMO abundances and maternal age
Variations in HMOs by maternal age were examined at sites with large numbers of samples [Malawi; Davis, CA; India; Peru]. The milk samples were binned into age groups of <20, 21–30, 31–40, and 41–50 years. Total absolute abundances of HMOs were not different between the age groups (Supplemental Figure 1) or between S+ and S− milk (data not shown). There were also no maternal age–related differences in total fucosylation, total sialylation, or levels of 2′FL (the latter among S+ mothers).
Total HMO abundances and infant sex
HMOs were compared to determine whether the sex of the infant affected total HMO abundances. Comparisons were made within sites (Supplemental Figure 2). We found no statistically significant differences in absolute HMO abundances in milk from mothers of male compared to female infants. Similarly, total fucosylation and total sialylation yielded no differences based on the sex of the offspring (not shown).
Variations in HMO subtypes across study sites and secretor status
Fucosylated HMOs
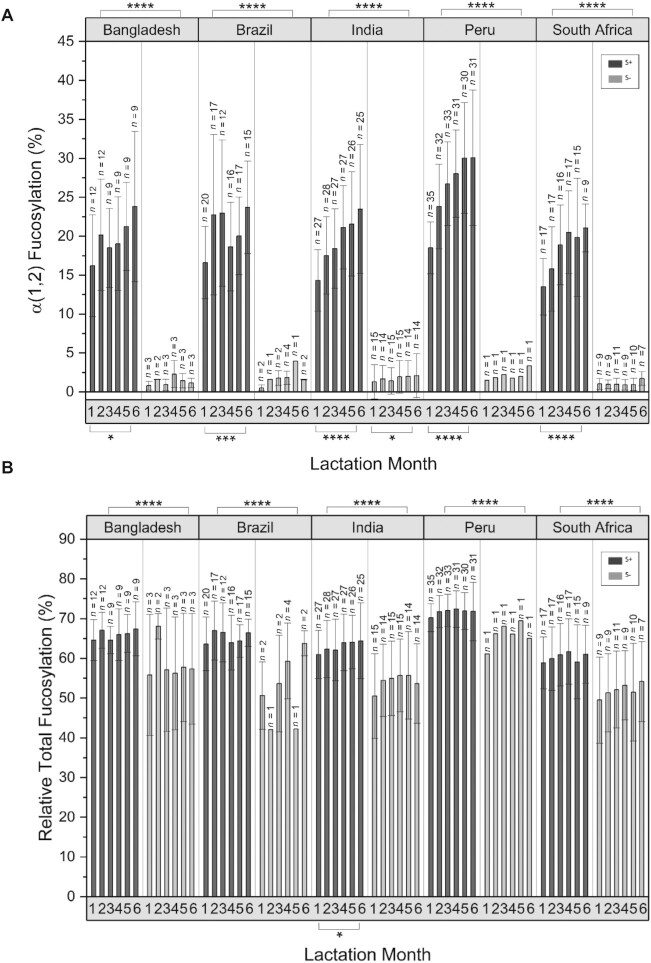
Fucosylated oligosaccharides comprised the largest and most abundant groups of HMO structures. We first compared the total abundances of α(1–2)-fucosylated HMOs in milk collected monthly during the first 6 months from mothers at study sites with longitudinal sampling: namely, Bangladesh, Bolivia, India, Peru, and South Africa (Figure 4A; Table 3). The proportions of α(1,2)-fucosylated structures in secretors increased significantly between postnatal months 1–6, with statistical significance across all selected sites (P < 0.05 to P < 0.0001; Mann-Whitney U test). However, for S− milk, only the site with large sample size (India) achieved statistical significance. S+ milk had consistently higher abundances of α(1,2)-fucosylated HMOs (ranging from 15%–20% of total HMOs) compared to S− milk (<5% of total HMOs; P < 0.0001; Mann-Whitney U test). Interestingly, S− milk, while containing very low abundances of α(1,2)-fucosylated HMOs, at times had non-0 abundances slightly above the baseline (P < 0.5; Mann-Whitney U test). As 2′FL is the most abundant of these structures, it increased over time as expected (Supplemental Figure 3). Total fucosylation (oligosaccharides that contain fucose regardless of the linkage) increased between months 1 and 6 in all sites; however, significance was only achieved in the India site, and only for S+ milk (P < 0.05; Mann-Whitney U test; Figure 4B). Total fucosylation differed slightly between S+ and S− milk (Figure 4B). In general, S− milk had low abundances of fucosylated structures compared to S+ milk, reflecting the deficit in α(1,2)-fucosylated HMOs, but partially compensated by increases in (1,3/4)-fucose linkages.
FIGURE 4.
Variations in fucosylated HMOs during lactation between geographical sites. (A) Mean relative abundance of total α(1,2)-fucose-containing HMOs in breastmilk samples as a function of lactation month (child's postnatal age) at sites with extensive longitudinal sampling. (B) Summed mean relative abundance of all fucose containing HMOs in the same samples (percentages of total fucosylation). S− type producers ( ) and S+ type producers (
) and S+ type producers ( ). HMO abundance values correspond to HPLC-qTOF MS spectral abundance, normalized to the mean of the total abundance of counts from each sample. Samples from multiple time points provided by a single mother, as well as samples which were provided at only 1 time point, were included in this analysis. N values correspond to the number of samples. Error bars represent the SD. P values were obtaining using Mann-Whitney U tests with an α correction of 0.05. HMO, human milk oligosaccharide; qTOF, quadruple time of flight; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ****P ≤ 0.0001.
). HMO abundance values correspond to HPLC-qTOF MS spectral abundance, normalized to the mean of the total abundance of counts from each sample. Samples from multiple time points provided by a single mother, as well as samples which were provided at only 1 time point, were included in this analysis. N values correspond to the number of samples. Error bars represent the SD. P values were obtaining using Mann-Whitney U tests with an α correction of 0.05. HMO, human milk oligosaccharide; qTOF, quadruple time of flight; S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ****P ≤ 0.0001.
TABLE 3.
Mean levels of total HMO, ɑ(1,2)-fucosylated HMOs, total fucosylation, total sialylation, and total undecorated HMOs by study site and secretor status1
| Site location | GDP per capita | Total HMO, normalized counts | Total fucosylation, % | Total sialylation, % | Total undecorated, % | α(1,2)-fucosylated, % | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B+ | B- | B+ | B- | B+ | B- | B+ | B- | B+ | B | |||
| Argentina | Namqom | 14,508 | 0.71 ± 0.14 | 0.37 ± 0.07 | 69.04 ± 4.69 | 64.66 ± 5.62 | 10.40 ± 2.62 | 24.16 ± 2.86 | 23.47 ± 5.03 | 22.44 ± 5.07 | 24.48 ± 4.94 | 2.22 ± 0.53 |
| Bolivia | Amazonian Lowlands | 3351 | 0.61 ± 0.13 | 0 ± 0 | 68.90 ± 6.39 | 0 ± 0 | 11.14 ± 2.69 | 0 ± 0 | 23.20 ± 6.39 | 0 ± 0 | 24.12 ± 8.87 | 0 ± 0 |
| Brazil | Fortaleza, Ceará | 9881 | 0.59 ± 0.16 | 0.59 ± 0.12 | 65.18 ± 6.19 | 54.80 ± 10.01 | 11.74 ± 3.64 | 14.76 ± 5.66 | 27.68 ± 6.64 | 36.94 ± 11.55 | 20.51 ± 7.32 | 1.72 ± 1.01 |
| Peru | Loreto | 6723 | 0.61 ± 0.19 | 0.48 ± 0.17 | 71.26 ± 4.79 | 64.15 ± 5.57 | 13.63 ± 3.22 | 15.99 ± 3.46 | 20.35 ± 4.92 | 26.74 ± 6.41 | 24.64 ± 7.65 | 1.98 ± 0.68 |
| Australia | Perth | 53,831 | 0.98 ± 0.24 | 0.94 ± 0.16 | 63.24 ± 4.58 | 58.02 ± 10.77 | 29.43 ± 6.73 | 34.17 ± 6.98 | 21.42 ± 6.08 | 26.32 ± 10.71 | 10.84 ± 5.06 | 2.61 ± 0.82 |
| Bangladesh | Mirpur, Dhaka | 1564 | 0.58 ± 0.15 | 0.47 ± 0.15 | 65.30 ± 5.42 | 56.16 ± 12.40 | 11.44 ± 2.95 | 15.17 ± 3.19 | 27.76 ± 5.79 | 34.31 ± 12.66 | 18.45 ± 7.10 | 1.28 ± 0.90 |
| India | Vellore, Tamil Nadu | 1980 | 0.62 ± 0.16 | 0.54 ± 0.16 | 62.94 ± 7.43 | 54.15 ± 9.78 | 10.78 ± 2.87 | 15.13 ± 4.15 | 29.92 ± 7.18 | 36.37 ± 9.16 | 19.32 ± 6.49 | 1.17 ± 2.07 |
| Nepal | Nubri Valley | 900 | 0.64 ± 0.13 | 0.63 ± 0.24 | 62.23 ± 6.64 | 49.66 ± 19.50 | 14.31 ± 2.88 | 19.19 ± 4.04 | 28.42 ± 6.12 | 38.71 ± 19.37 | 12.82 ± 3.68 | 0.99 ± 0.37 |
| Philippines | Cebu | 2982 | 0.62 ± 0.09 | 0.51 ± 0.11 | 64.11 ± 9.35 | 60.90 ± 5.89 | 11.54 ± 2.34 | 16.11 ± 4.35 | 27.65 ± 9.94 | 28.74 ± 6.34 | 20.09 ± 5.37 | 2.24 ± 0.36 |
| Gambia | 673 | 0.58 ± 0.18 | 0.55 ± 0.17 | 53.63 ± 8.49 | 42.60 ± 16.81 | 13.85 ± 5.07 | 16.34 ± 5.76 | 36.62 ± 7.68 | 45.96 ± 16.20 | 18.49 ± 65.85 | 1.69 ± 1.47 | |
| Malawi | Mangochi district | 357 | 0.72 ± 0.20 | 0.66 ± 0.20 | 62.01 ± 9.91 | 54.22 ± 16.59 | 12.21 ± 3.37 | 14.31 ± 5.61 | 29.92 ± 9.32 | 36.37 ± 15.11 | 20.54 ± 7.20 | 1.32 ± 0.74 |
| Namibia | Omuhonga Basin | 5516e | 0.52 ± 0.25 | 0.47 ± 0.02 | 63.63 ± 10.08 | 57.52 ± 1.67 | 17.92 ± 4.0 | 25.43 ± 1.54 | 26.07 ± 8.41 | 26.93 ± 0.67 | 15.10 ± 10.04 | 1.11 ± 0.03 |
| South Africa | Thohoyandou, Limpopo Province | 6120 | 0.56 ± 0.15 | 0.51 ± 0.18 | 60.15 ± 7.71 | 50.95 ± 10.45 | 13.16 ± 3.26 | 15.20 ± 3.08 | 31.32 ± 7.69 | 39.58 ± 9.85 | 17.88 ± 5.84 | 1.03 ± 0.74 |
| Poland | Beskid Wyspowy Mtns | 13,871 | 0.70 ± 0.22 | 0.54 ± 009 | 63.58 ± 6.60 | 51.46 ± 6.98 | 10.69 ± 3.66 | 17.35 ± 6.01 | 29.13 ± 7.25 | 36.68 ± 9.3 | 18.86 ± 5.06 | 1.20 ± 0.29 |
| United States | Boston, Massachusetts | 59,939 | 0.76 ± 0.25 | 0.58 ± 0.27 | 62.44 ± 3.88 | 58.95 ± 4.89 | 12.73 ± 3.47 | 16.45 ± 2.88 | 29.40 ± 5.35 | 30.87 ± 5.70 | 18.41 ± 6.77 | 2.87 ± 0.97 |
| United States | Davis, California | 59,939 | 0.72 ± 0.24 | 0.65 ± 0.24 | 64.00 ± 5.86 | 59.92 ± 9.23 | 13.57 ± 4.40 | 19.41 ± 6.36 | 27.50 ± 5.98 | 29.10 ± 9.19 | 15.44 ± 7.19 | 2.11 ± 1.52 |
GDP values for each country are representative of the year of collection. Values are presented as means ± SDs. All data collected were used for this analysis, including data from the same mother at different time points of lactation. P values were obtaining using Mann-Whitney U tests. GDP, gross domestic product; HMO, human milk oligosaccharide.
Variations in fucosylated structures were observed between milk from different countries within each secretor phenotype (Figure 2). Mothers with S+ milk from Nepal had significantly higher levels of DFLNHa (P < 0.0002; Mann-Whitney U test) and lower levels of 2′FL (P < 0.0001; both S+, secretor markers) compared to the other sites. Human milk obtained from mothers residing in the Gambia and Boston, MA, sites had high relative abundances of LNFP I and LNFP III compared to all other sites, and low abundances of LNFP II. Milk from mothers residing in Perth, Australia, had relatively high abundances of sialyllacto-N-neotetraose (LSTc) and sialyllacto-N-tetraose b (LSTb) compared to other sites. Interestingly, 2′FL was not generally the most abundant in S+ milk among all countries. Australia; Boston, MA; and Namibia all had S+ milk with the most abundant α(1–2)-fucosylated compound being LDFT (P = 0.005; Mann-Whitney U test).
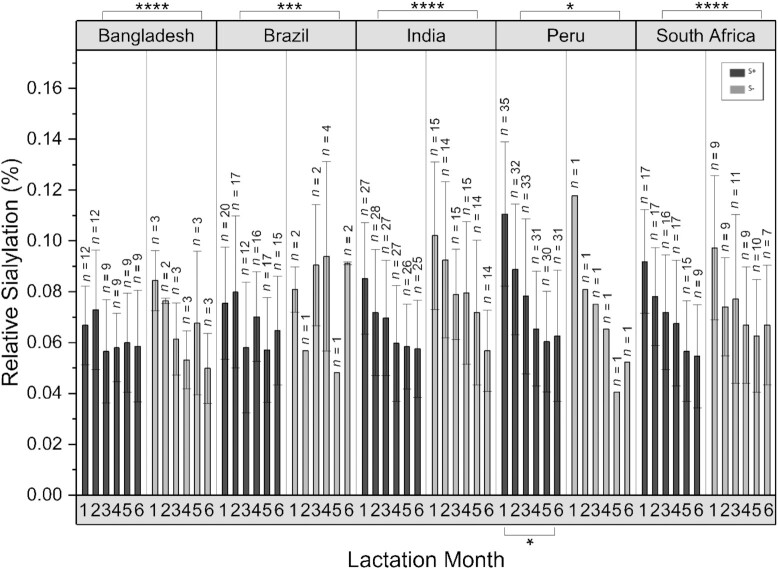
Sialylated HMOs
Sialylated oligosaccharides generally had much lower abundances in human breast milk than fucosylated species. The mean abundances of sialylated HMOs in samples collected over the first postnatal 6 months from different study sites are shown in Figure 5 and Table 3. Total abundances of sialylated HMOs were relatively constant throughout lactation for both S+ and S− phenotypes. Likewise, the relative abundances of sialylated HMOs were comparable across study sites with extensive sampling (Bangladesh, Bolivia, India, Peru, and South Africa), averaging 12.3% ± 3.4 of the total HMOs in S− milk and 15.2% ± 3.8 in S+ milk (P < 0.05-P < 0.0001; Mann-Whitney U test). The most abundant sialylated HMOs when considering all samples were the combined group of LSTc + LSTb (in both S+ and S− samples; Figure 2). Note, however, that the values from Peru for S− milk were obtained from samples provided by a single mother, due to the very low prevalence of S− milk in this population. In populations with greater frequencies of S− milk, there was a greater relative abundance of sialylated structures in S− milk compared to S+ milk. In samples from India, Bangladesh, and South Africa, the absolute abundances of sialylated HMOs in month 1 were nearly 15% greater in S− compared to S+ milk.
FIGURE 5.
Mean relative abundances of total sialic acid (Neu5Ac)–containing HMOs (percentage of total sialylation) in breastmilk samples as a function of lactation month (child's postnatal age) at sites with extensive longitudinal sampling. Samples from multiple time points provided by a single mother, as well as samples from multiple mothers with 1 time point, were all included in this analysis. HMO abundances corresponded to HPLC-qTOF MS abundances, normalized to the mean of the total ion counts from each sample. Error bars represent the SD. P values were obtained using Mann-Whitney U tests with an α correction of 0.05. HMO, human milk oligosaccharide; Neu5Ac, N-acetylneuraminic acid; qTOF, quadruple time of flight. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, ****P ≤ 0.0001.
It is noteworthy that the sialylated HMO DSLNT, which has been shown to be protective against necrotizing enterocolitis in an animal model (39) and in a human preterm infant cohort (40), was often found in low abundance and was not 1 of the 60 HMOs found in all sites.
Undecorated (nonfucosylated and nonsialylated) HMOs
The total abundances of undecorated HMOs (lacking both fucose and sialic acid) in milk from mothers at the various study sites were compared in Supplemental Figure 4 and Table 3. Undecorated HMOs were lower in S+ milk compared to S− milk (P = 0.002; Mann-Whitney U test) (41). Between sites, overall relative abundances of undecorated HMOs were comparable (31.6 ± 8.9%), with the exception of Peru, which exhibited significantly lower levels (20.5 ± 5.2%; P = <0.0001) due to the higher amounts of fucosylation and diminished numbers of S− milk. Across all samples, the most abundant undecorated HMO was the LNT/LNnT group (Figure 2).
Global variations of S+ and S− milk
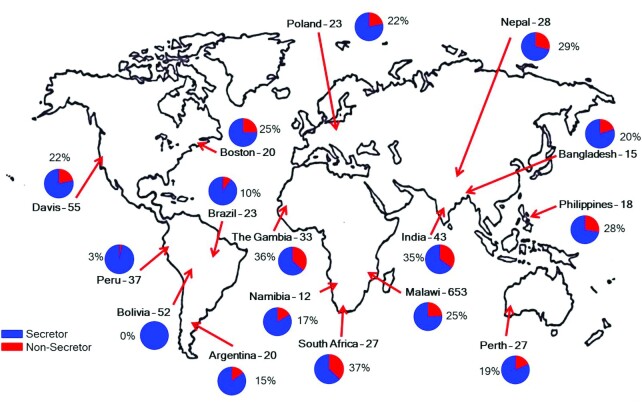
With the samples from different locations and the ability to distinguish S+ and S− milk, we determined the distribution of the 2 phenotypes globally. The sites are located on the map, along with the fractions of S+ (blue) and S− (red) milk (Figure 6). The fraction of the mothers that produced S− milk was lower than that of the mothers that produced S+ at every geographical site. High proportions of S− milk were found in Africa (37% in South Africa and 36% in the Gambia). These values contrast with other sites in Africa, including Namibia (17%) and Malawi (25%), that had lower proportions of S− milk. Other sites with high levels of S− milk included those in India (35%) and Nepal (29%). Bangladesh was lower, with only 20% S− mothers. The US sites [Davis, CA (22%), and Boston, MA (25%)], were similar to the Poland (22%) and Australian (19%) sites. Low values were found in the South American sites of Bolivia (0%), Peru (3%), and Brazil (10%). The samples from these sites were obtained from indigenous populations. The other South American site, Argentina (15%), included nonindigenous populations.
FIGURE 6.
Proportion of samples tested from each study location that were S+ type (i.e., secretor mothers; ( ) and S− type (nonsecretor mothers; (
) and S− type (nonsecretor mothers; ( ). Labels are presented as “country–number of mothers.” If samples from multiple time points were provided by the same mother, the secretor status determination was concluded based on the majority of her samples. If there was no majority milk type, she was excluded from the statistical analysis (1 mother from Davis, CA; 2 mothers from Malawi; and 3 mothers from Perth, Australia). S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures.
). Labels are presented as “country–number of mothers.” If samples from multiple time points were provided by the same mother, the secretor status determination was concluded based on the majority of her samples. If there was no majority milk type, she was excluded from the statistical analysis (1 mother from Davis, CA; 2 mothers from Malawi; and 3 mothers from Perth, Australia). S− milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; S+ milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures.
Discussion
The analyses of the HMO compositions of human milk samples collected from 16 sites around the world using a sensitive HPLC-qTOF-MS-based approach provided the most extensive data set reported to date. While the samples were collected from different studies, the analytical method for each sample was the same. Unfortunately, absolute quantitation of total HMO and specific structures was not possible due to the low availability of standards in the earliest analysis.
This study significantly expands on our previous work at a single site in the Gambia (23). The large number of samples from various geographical locations allowed us to further explore factors that may affect HMO production during lactation. Factors such as the age of the mother and the sex of the infant do not affect HMO abundances. The sex of the infant had been previously reported to affect milk compositions, suggesting that some components of human milk may be tailored to sex-differentiated developmental priorities (42). However, examination of milk among all populations and within each site yielded no significant variations in total HMO abundances based on the sex of a mother's child. Because neither dietary data nor maternal nutritional status data were available for this study, we were not able to determine the extents to which these factors might influence abundances of individual structures, nor levels of fucosylation and sialylation. However, the comparison of per capita gross domestic products (GDPs) was obtained by comparing total abundances with published GDPs. Interestingly, we observed some correlation between per capita GDPs and total levels of HMOs from mothers living at the different sites (Supplemental Figure 5). As shown, countries with a high GDP per capita tended to have milk with the most abundant levels of HMOs. Likewise, mothers from sites with a low GDP per capita tended to have lower levels of HMOs. Among countries with low GDPs, there was a common minimum level of HMOs, while the trend towards higher abundances of HMOs did not appear to manifest until significantly higher GDPs were obtained. It would be difficult to make conclusions regarding GDPs and HMO production, as the amounts of HMOs fed to the infant can vary depending on local feeding practices. Furthermore, the sample size, though large in totality, is still small at the local level and cannot fully represent the respective nation. However, we encourage further studies on societal effects on milk production.
Genomic analysis has been previously used to obtain global distributions of secretors and nonsecretors (43). While this approach obtains the genotypic status, the phenotype—that is, the actual abundances of different structures and structural types in the milk—are those that affect infant health and developmental outcomes (44). Hence, the concentrations of HMOs are crucial to understanding the health outcomes of infants, providing information of distinct nutritional components that align with the secretor status. Additionally, among secretors there are large variations in the abundances of fucosylated and sialylated species that are important contributors to infant development. The concentrations can then be used to phenotype milk based on the abundances of specific structures—namely, α(1,2)-fucosylated species—without genomic data. For the purposes of this study, we therefore refer to milk that corresponds to high amounts of α(1,2)-fucosylated structures as S+ milk, and milk corresponding to low abundances as S− milk.
The fractions of mothers who produced S− milk were nearly 40% in West and South Africa and South Asia, while nearly 0% in parts of Latin America. The fraction of nonsecretor mothers (and hence S− milk) is often cited to be ∼20%. However, this number is based primarily on studies of European and Euro-American mothers, which is consistent with our own results for sites in Europe and the United States. The US sites (Davis, CA; Boston, MA) and Poland (the sole European site) had S− milk in proportions similar to those previously reported (34). The rarity of S− milk among indigenous populations in South America suggests either founder effects during human migration into Beringia or selection from pathogen pressure. Many infectious diseases, including cholera, whose severity is associated with blood type, can have devastating effects on populations (17). As previously discussed, blood typing is similar to human milk in that the presence or absence of α(1,2)-fucosylated structures is the key determinant. A study of the cholera outbreak in Peru in 1991 found that those with blood type O, which occurs at a very high frequency among South American indigenous populations, had more severe symptoms and were 8 times more likely to be hospitalized, emphasizing the relevance of glycosylation on infectious diseases (45).
A distinguishing feature of humans and primates compared to other mammals is the high level of fucosylated structures, with humans having the highest abundances (46). Fucosylated structures, or the presence of at least 1 fucose, increased in relative abundances throughout lactation. Previous studies similarly noted that fucosylation increased throughout lactation for the first 6 months, regardless of secretor status (41). Fucosylation was generally higher in S+ milk. As a consequence of higher fucosylation in S+ milk, the amount of undecorated HMOs was lower in S+ milk compared to S− milk, consistent with earlier findings (41). In contrast, sialylation is the lowest among humans but is significantly greater in bovine and porcine milk (47). In this study, though sialylated oligosaccharides in the human milk samples were still low, they were relatively higher in S− compared to S+ milk. Previous studies have also reported the higher abundances of sialylated HMOs in nonsecretor (S−) milk (41).
The structural variety of HMOs and their different relative abundances have the potential to endow human milk from different mothers with distinct functional properties, including modulating its effect on the developing microbiota and its effects on infection/colonization with enteropathogens (48). Members of the infant gut microbiota have specific glycosyl hydrolases and glycan-binding proteins that either promote the fitness of specific community members or block the host from infection. For example, HMOs promote the growth of Bifidobacterium longum subsp. infantis, a gut symbiont that is richly endowed with a suite of genes specifically adapted to import and utilize HMOs. The HMO composition of breastmilk thus has the potential to influence the fitness of strain-level variants of this and other related bifidobacterial species and to shape a program of normal postnatal community development (succession) that has been identified in healthy individuals and that is impaired in those with undernutrition (13, 49). Preclinical studies in gnotobiotic animals and clinical studies of the effects of repairing microbial community immaturity in children with acute malnutrition support the notion that healthy development of the microbiota is causally linked to healthy growth (13, 49) Similarly, HMOs with α(1–2)-linked Fuc (S+ milk) are associated with decreased incidences and severity of diarrhea caused by Campylobacter jejuni and enteropathogenic Escheria coli (50, 51): enteropathogens that are ubiquitous in many low-income countries where childhood undernutrition is prevalent (52). S− milk is enriched in HMOs that bind to Helicobacter pylori and enteropathogenic E. coli, preventing their attachment to gut epithelial cells (53). Individuals homozygous for FUT2 mutations (nonsecretors) also show resistance to norovirus infection (54, 55) or, considering this relationship from the viewpoint of the pathogen, both rotavirus and norovirus (2 of the most common causes of viral diarrhea in infants) appear to prefer the secretor host (32). These mothers could in turn provide protection to their infant by delivering S− milk. Determining infant infectious disease risks should ideally include consideration of the secretor statuses of both the mother and the infant to address such key questions as whether S+ milk is particularly beneficial for the nonsecretor infant and vice versa.
The global results suggest that the secretor status and the complement of HMOs produced by a mother during lactation are influenced in part by adaptations shaped by ancestral nutritional and disease ecologies experienced by diverse human populations. Indeed, immunofactors in breastmilk are associated with subsistence practices that affect nutritional intakes and pathogen exposures of diverse, traditional societies and demonstrate the importance of considering populations within their contemporary, historical, and prehistorical contexts (45). The “first-step” findings described here highlight the importance of multipopulation studies to better characterize the relationships among maternal characteristics, HMO compositions, early gut community development, the products of microbial metabolism of these HMOs, and measures of infant health status (46). Delineation of these relationships, along with those mediated by other key constituents of breastmilk, such as secreted immunoglobulins and antibacterial proteins, will help guide the design of future prebiotic approaches based on purified milk components (or synthetically produced mimetics) and/or synbiotics (prebiotics combined with probiotic microorganisms) that promote healthy gut community development, healthy growth of infants, and even healthy immune and inflammatory responses over a lifetime.
Supplementary Material
Acknowledgments
We thank all of the mothers who participated and the communities who allowed the research and contributed samples. We also thank all the fieldworkers, staff and researchers, clinical staff, and laboratory technicians who collected the samples and data, and field assistant/translator John Jakurama.
We are grateful to the Principal Investigators (PIs) and their colleagues who oversaw the studies at Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and Enteric Disease (MAL-ED) sites [Tahmeed Ahmed (icddr, b, Bangladesh), Aldo Lima (Universidade Federal do Ceará, Brazil), Margaret Kosek (Johns Hopkins School of Public Health, Iquitos, Peru), Gagandeep Kang (Christian Medical College, Vellore, India), and Pascale Bessong (University of Venda, South Africa)]. We are grateful to the Interantional Lipid-Based Nutrient Supplement (iLNS-DYAD)-Malawi PIs and study team [Ken Maleta (University of Malawi College of Medicine), Per Ashorn (University of Tampere School of Medicine, Finland), Robin Bernstein (University of Colorado), and Jennifer T Smilowitz and Kathryn Dewey (University of California, Davis)]. We thank Andrzej Galbarzyck for critical organizing at the Mogielica Human Ecology Study Site (Poland). Milk samples from Nepal were collected among ethnic Tibetan mothers in the northern Gorkha district (Jhangchuk Sangmo, Nyima Sangmo, and Tsewang Palton were Tibetan field assistants; Dr Diki Bista was co-PI); milk samples from the Philippines were collected from mothers in rural and urban Cebu, Philippines (Office of Population Studies, University of San Carlos). The Argentine samples were from Qom mothers (Qom field assistants and Fundación ECO).
The authors’ responsibilities were as follows – LDK, MM, EAQ, AB, KH, JTS, AMZ, MJB, JIG, MAU, CV, JBG: oversaw sample collection and provided samples; LDW, SMT, JCCD: prepared samples; AV, LDW, SMT, JCCD: analyzed human milk oligosaccharide data; AV, JCCD: performed statistical analyses on human milk oligosaccharide data and infant disease metadata; CBL: designed the study and oversaw the analyses; AV, CBL: interpreted results; AV, CBL, JCCD, MJB, JIG: wrote the manuscript together with; and all authors: read and approved the final manuscript.
Notes
This work was supported by grants from the Bill and Melinda Gates Foundation (to JCCD and CBL), the NIH (grants AT007079 to DAM, HD061923 to CBL, R01AG024119 to MG and HK, and AT008759 to DAM), the Peter J. Shields Endowed Chair in Dairy Food Science (to DAM), and NSF DDIG 1233270 to MM. The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) Project was carried out as a collaborative project supported by the Bill & Melinda Gates Foundation, the Foundation for the National Institutes of Health, and the Fogarty International Center. The iLiNS-DYAD study was funded by the Bill & Melinda Gates Foundation and FANTA-2 project, AED, antiepileptic drugs. The Early Nutrition and Immune Development (ENID) trial was supported by the UK Medical Research Council (MRC; MC-A760-5QX00) and the UK Department for International Development (DFID), under the MRC/DFID Concordat agreement.
Author disclosures: CBL, DAM, and JBG are cofounders of Evolve Biosystems, Inc., a company focused on diet-based manipulation of the gut microbiota. JIG is a founder of Matatu, a company focused on microbial-based manipulation for improvement of health. All other authors report no conflicts of interest.
Supplemental Text, Supplemental Table 1, Supplemental Methods, and Supplemental Figures 1–5 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Abbreviations used: ACN, acetonitrile; DFLNHa, difucosyllacto-N-hexaose a; Fuc, fucose; Gal, galactose; GDP, gross domestic product; GlcNAc, N-acetylglucosamine; Hex, Hexose, HexNAc, N-Acetylhexosamine; HMO, human milk oligosaccharide; LDFT, lactodifucotetraose; LNFP, lacto-N-fucopentaose; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; LSTb, sialyllacto-N-tetraose b; LSTc, sialyllacto-N-neotetraose; Neu5Ac, N-acetylneuraminic acid; qTOF, quadruple time of flight; nonsecretor phenotype, (S−) milk, milk that corresponds to low amounts of α(1,2)-fucosylated structures; secretor phenotype, (S+) milk, milk that corresponds to high amounts of α(1,2)-fucosylated structures; TFLNH, trifucosyllacto-N-hexaose; TOF, time of flight; 2′FL, 2′-fucosyllactose.
Contributor Information
Anita Vinjamuri, Department of Chemistry, University of California, Davis, CA, USA; Foods for Health Institute, University of California, Davis, CA, USA.
Jasmine C C Davis, Department of Chemistry, University of California, Davis, CA, USA; Foods for Health Institute, University of California, Davis, CA, USA.
Sarah M Totten, Department of Chemistry, University of California, Davis, CA, USA; Foods for Health Institute, University of California, Davis, CA, USA.
Lauren D Wu, Department of Chemistry, University of California, Davis, CA, USA; Foods for Health Institute, University of California, Davis, CA, USA.
Laura D Klein, Department of Human Evolutionary Biology, Harvard University, Cambridge, MA, USA.
Melanie Martin, Department of Anthropology, University of Washington, Seattle, WA, USA.
E A Quinn, Department of Anthropology, Washington University in St. Louis, St. Louis, MO, USA.
Brooke Scelza, Department of Anthropology, University of California Los Angeles, Los Angeles, CA, USA.
Alicia Breakey, Wildwood School, Los Angeles, CA, USA.
Michael Gurven, Department of Anthropology, University of California Santa Barbara, Santa Barbara, CA, USA.
Grazyna Jasienska, Department of Environmental Health, Faculty of Health Sciences, Jagiellonian University Medical College, Krakow, Poland.
Hillard Kaplan, Health Economics and Anthropology, Chapman University, Orange, CA, USA.
Claudia Valeggia, Department of Anthropology, Yale University, New Haven, CT, USA.
Katie Hinde, School of Human Evolution and Social Change, Arizona State University, Tempe, AZ, USA.
Jennifer T Smilowitz, Foods for Health Institute, University of California, Davis, CA, USA; Department of Food Science and Technology, University of California, Davis, CA, USA.
Robin M Bernstein, Department of Anthropology, University of Colorado, Boulder, CO, USA; Institute of Behavioral Science, University of Colorado, Boulder, CO, USA.
Angela M Zivkovic, Foods for Health Institute, University of California, Davis, CA, USA; Department of Nutrition, University of California, Davis, CA, USA.
Michael J Barratt, Edison Family Center for Genome Sciences and Systems Biology, Washington University School of Medicine, St. Louis MO, USA; Center for Gut Microbiome and Nutrition Research, Washington University School of Medicine, MO, USA.
Jeffrey I Gordon, Edison Family Center for Genome Sciences and Systems Biology, Washington University School of Medicine, St. Louis MO, USA; Center for Gut Microbiome and Nutrition Research, Washington University School of Medicine, MO, USA.
Mark A Underwood, Foods for Health Institute, University of California, Davis, CA, USA; Department of Pediatrics, University of California, Davis, CA, USA.
David A Mills, Foods for Health Institute, University of California, Davis, CA, USA; Department of Food Science and Technology, University of California, Davis, CA, USA.
J Bruce German, Foods for Health Institute, University of California, Davis, CA, USA; Department of Food Science and Technology, University of California, Davis, CA, USA.
Carlito B Lebrilla, Department of Chemistry, University of California, Davis, CA, USA; Foods for Health Institute, University of California, Davis, CA, USA.
Data Availability
Data that support the findings of this study are available upon request from the corresponding author (CBL).
References
- 1. Ballard O, Morrow A. Human milk composition nutrients and bioactive factors. Pediatr Clin North Am. 2013;60(1):49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dorea JG. Selenium and breast-feeding. Br J Nutr. 2002;88(5):443–61. [DOI] [PubMed] [Google Scholar]
- 3. Innis SM. Impact of maternal diet on human milk composition and neurological development of infants. Am J Clin Nutr. 2014;99(3):734S–41S. [DOI] [PubMed] [Google Scholar]
- 4. Picciano MF. Nutrient composition of human milk. Pediatr Clin North Am. 2001;48(1):53–67. [DOI] [PubMed] [Google Scholar]
- 5. Hinde K, Milligan LA. Primate milk: Proximate mechanisms and ultimate perspectives. Evol Anthropol. 2011;20(1):9–23. [DOI] [PubMed] [Google Scholar]
- 6. Ruhaak LR, Lebrilla CB. Analysis and role of oligosaccharides in milk. BMB Rep. 2012;45(8):442–51. [DOI] [PubMed] [Google Scholar]
- 7. Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10(2):856–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9(8):4138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newburg DS, Neubauer SH. Handbook of milk composition. Academic Press, Inc: 1995; p. 919. [Google Scholar]
- 10. Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71(6):1589–96. [DOI] [PubMed] [Google Scholar]
- 11. Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: Mechanisms and implications. Microbiology. 2013;159(Pt_4):649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. Glycoprofiling of bifodobaterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55(22):8914–9. [DOI] [PubMed] [Google Scholar]
- 13. Gehrig JL, Venkatesh S, Chang H-W, Hibberd MC, Kung VL, Cheng J, Chen RY, Subramanian S, Cowardin CA, Meier MFet al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019:365(6449):eaau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinde K, Lewis ZT. Mother's littlest helpers. Science. 2015;348(6242):1427–8. [DOI] [PubMed] [Google Scholar]
- 15. Ruhaak LR, Stroble C, Underwood MA, Lebrilla CB. Detection of milk oligosaccharides in plasma of infants. Anal Bioanal Chem. 2014;406(24):5775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Leoz MLA, Wu S, Strum JS, Ninonuevo MR, Gaerlan SC, Mirmiran M, German JB, Mills DA, Lebrilla CB, Underwood MA. A quantitative and comprehensive method to analyze human milk oligosaccharide structures in the urine and feces of infants. Anal Bioanal Chem. 2013;405(12):4089–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Coppa GV, Zampini L, Galeazzi T, Facinelli B, Ferrante L, Capretti R, Orazio G. Human milk oligosaccharides inhibit the adhesion to caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr Res. 2006;59(3):377–82. [DOI] [PubMed] [Google Scholar]
- 18. Hong P, Ninonuevo MR, Lee B, Lebrilla C, Bode L. Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br J Nutr. 2008;101(4):482–6. [Google Scholar]
- 19. Eiwegger T, Stahl B, Haidl P, Schmitt J, Boehm G, Dehlink E, Urbanek R, Szépfalusi Z. Prebiotic oligosaccharides: In vitro evidence for gastrointestinal epithelial transfer and immunomodulatory properties. Pediatr Allergy Immunol. 2010;21(8):1179–88. [DOI] [PubMed] [Google Scholar]
- 20. Wang B. Sialic acid is an essential nutrient for brain development and cognition. Annu Rev Nutr. 2009;29(1):177–222. [DOI] [PubMed] [Google Scholar]
- 21. Kunz C, Rudloff S. Biological functions of oligosaccharides in human milk. Acta Paediatr. 1993;82(12):903–12. [DOI] [PubMed] [Google Scholar]
- 22. Urashima T, Kitaoka M, Asakuma S, Messer M. Milk oligosaccharides. In: McSweeney P, Fox P, editors. Advanced dairy chemistry: Lactose, water, salts, and minor constituents. New York, NY: Springer Science; 2009:295–348. [Google Scholar]
- 23. Totten SM, Zivkovic AM, Wu S, Ngyuen U, Freeman SL, Ruhaak RL, Darboe MK, German JB, Prentice AM, Lebrilla CB. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res. 2012;11(12):6124–33. [DOI] [PubMed] [Google Scholar]
- 24. Brooks SA, Dwek MV, Schumacher U. Functional and molecular glycobiology. Oxford, UK: BIOS Scientific Publishers Ltd; 2002. [Google Scholar]
- 25. Thurl S, Henker J, Siegel M, Tovar K, Sawatzki G. Detection of four human milk groups with respect to Lewis blood group dependent oligosaccharides. Glycoconjugate J. 1997:14(7):795–9. [Google Scholar]
- 26. Mottram L, Wiklund G, Larson G, Qadri F, Svennerholm A-M. FUT2 non-secretor status is associated with altered susceptibility to symptomatic enterotoxigenic Escherichia coli infection in Bangladeshis. Sci Rep. 2017;7(1):10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Koda Y, Soejima M, Pang H, Schlaphoff T, du Toit ED, Kimura H. Extensive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum Genet. 1998;103(2):204–10. [Google Scholar]
- 28. Kudo T, Iwasaki H, Nishihara S, Shinya N, Ando T, Narimatsu I, Narimatsu H. Molecular genetic analysis of the human Lewis histo-blood group system: II. Secretor gene inactivation by a novel single missense mutation A385T in Japanese nonsecretor individuals. J Biol Chem. 1996;271(16):9830–7. [DOI] [PubMed] [Google Scholar]
- 29. Santos-Cortez RLP, Chiong CM, Frank DN, Ryan AF, Giese APJ, Bootpetch Roberts T, Daly KA, Steritz MJ, Szeremeta W, Pedro Met al. FUT2 variants confer susceptibility to familial otitis media. Am J Hum Genet. 2018;103(5):679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smyth DJ, Cooper JD, Howson JMM, Clarke P, Downes K, Mistry T, Stevens H, Walker NM, Todd JA. FUT2 nonsecretor status links type 1 diabetes susceptibility and resistance to infection. Diabetes. 2011;60(11):3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ye BD, Kim BM, Jung S, Lee HS, Hong M, Kim K, Moon JW, Baek J, Oh EH, Hwang SWet al. Association of FUT2 and ABO with Crohn's disease in Koreans. J Gastroenterol Hepatol. 2020;35(1):104–9. [DOI] [PubMed] [Google Scholar]
- 32. Rossouw E, Brauer M, Meyer P, du Plessis NM, Avenant T, Mans J. Virus etiology, diversity and clinical characteristics in South African children hospitalised with gastroenteritis. Viruses. 2021;13(2):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erney RM, Malone WT, Skelding MB, Marcon AA, Kleman-Leyer KM, O'Ryan ML, Ruiz-Palacios G, Hilty MD, Pickering LK, Prieto PA. Variability of human milk neutral oligosaccharides in a diverse population. J Pediatr Gastroenterol Nutr. 2000;30(2):181–92. [DOI] [PubMed] [Google Scholar]
- 34. McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SEet al. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr. 2017;105(5):1086–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plows JF, Berger PK, Jones RB, Alderete TL, Yonemitsu C, Najera JA, Khwajazada S, Bode L, Goran MI. Longitudinal changes in human milk oligosaccharides (HMOs) over the course of 24 months of lactation. J Nutr. 2021;151(4):876–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm Ret al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54(20):7471–80. [DOI] [PubMed] [Google Scholar]
- 37. Totten SM, Wu LD, Parker EA, Davis JCC, Hua S, Stroble C, Ruhaak LR, Smilowitz JT, German JB, Lebrilla CB. Rapid-throughput glycomics applied to human milk oligosaccharide profiling for large human studies. Anal Bioanal Chem. 2014;406(30):7925–35. [DOI] [PubMed] [Google Scholar]
- 38. Kumazaki T, Yoshida A. Biochemical evidences that secretor gene, Se, is a structural gene, coding a specific fucosyltransferase. Proc Natl Acad Sci. 1984;81(13):4193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang W, He-Yang J, Zhuang W, Liu J, Zhou X. Causative role of mast cell and mast cell-regulatory function of disialyllacto-N-tetraose in necrotizing enterocolitis. Int Immunopharmacol. 2021;96:107597. [DOI] [PubMed] [Google Scholar]
- 40. Masi AC, Embleton ND, Lamb CA, Young G, Granger CL, Najera J, Smith DP, Hoffman KL, Petrosino JF, Bode Let al. Human milk oligosaccharide DSLNT and gut microbiome in preterm infants predicts necrotising enterocolitis. Gut. 2021;70(12):2273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu G, Davis JCC, Goonatilleke E, Smilowitz JT, German JB, Lebrilla CB. Absolute quantitation of human milk oligosaccharides reveals phenotypic variations during lactation. J Nutr. 2017;147(1):117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galante L, Milan AM, Reynolds CM, Cameron-Smith D, Vickers MH, Pundir S. Sex-specific human milk composition: The role of infant sex in determining early life nutrition nutrients [Internet]. Basel; 2018. Available from: http://europepmc.org/abstract/MED/30200404 [Google Scholar]
- 43. Arrouzet CJ, Ellis K, Kambhampati A, Chen Y, Steele M, Lopman B. Population-level human secretor status is associated with genogroup 2 type 4 norovirus predominance. J Infect Dis. 2020;221(11):1855–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klein LD, Huang J, Quinn EA, Martin MA, Breakey AA, Gurven M, Kaplan H, Valeggia C, Jasienska G, Scelza Bet al. Variation among populations in the immune protein composition of mother's milk reflects subsistence pattern. Evol Med Public Health. 2018;2018(1):230–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Swerdlow DL, Mintz ED, Rodriguez M, Tejada E, Ocampo C, Espejo L, Barrett TJ, Petzelt J, Bean NH, Seminario Let al. Severe life-threatening cholera associated with blood group O in Peru: implications for the Latin American epidemic. J Infect Dis. . 1994;170:(2):468–72. [DOI] [PubMed] [Google Scholar]
- 46. Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, Gagneux P, German JB, Lebrilla CB. Evolutionary glycomics: Characterization of milk oligosaccharides in primates. J Proteome Res. 2011;10(4):1548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tao N, DePeters EJ, German JB, Grimm R, Lebrilla CB. Variations in bovine milk oligosaccharides during early and middle lactation stages analyzed by high-performance liquid chromatography-chip/mass spectrometry. J Dairy Sci. 2009;92(7):2991–3001. [DOI] [PubMed] [Google Scholar]
- 48. Underwood MA. Impact of probiotics on necrotizing enterocolitis. Semin Perinatol. 2017;41(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raman AS, Gehrig JL, Venkatesh S, Chang H.-W., Hibberd MC, Subramanian S, Kang G, Bessong PO, Lima AAM, Kosek MNet al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science. 2019;365(6449):eaau4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newburg DS, Ruiz-Palacios G, Altaye M, Chaturvedi P, Meinzen-Derr J, de Lourdes Guerrero M, Morrow AL.. Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology. 2004;14(3):253–63. [DOI] [PubMed] [Google Scholar]
- 51. Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. Human milk oligosaccharide blood group epitopes and innate immune protection against campylobacter and calcivirus diarrhea in breastfed infants. In: Pickering LK, Morrow AL, Ruiz-Palacios GM, Schanler RJ, editors. Protecting infants through human milk. Springer; Boston; 2004;443–6. [Google Scholar]
- 52. Platts-Mills JA, Liu J, Rogawski ET, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama Ret al. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: A reanalysis of the MAL-ED cohort study. Lancet Glob Health. 2018;6(12):e1309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simon PM, Goode PL, Mobasseri A, Zopf D. Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect Immun. 1997;65(2):750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thorven M, Grahn A, Hedlund KO, Johansson H, Wahlfrid C, Larson G, Svensson L. A homozygous nonsense mutation (428G→A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J Virol. 2005;79(24):15351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larsson MM, Rydell GEP, Grahn A, Rodríguez-Díaz J, Åkerlind B, Hutson AM, Estes MK, Larson G, Svensson L. Antibody prevalence and titer to norovirus (genogroup II) correlate with secretor (FUT2) but not with ABO phenotype or Lewis (FUT3) genotype. J Infect Dis. 2006;194(10):1422–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available upon request from the corresponding author (CBL).