ABSTRACT
Longan (Dimocarpus longan) is a subtropical fruit tree best known for its nutritious fruit and regarded as a valuable tonic and traditional medicine since ancient times. A high-quality chromosome-scale genome assembly is valuable for functional genomic study and genetic improvement of longan. Here, we report a chromosome-level reference genome sequence for the longan cultivar JDB. The assembled genome is 455.5 Mb in size and anchored to fifteen chromosomes, representing a significant improvement in contiguity (contig N50 = 12.1 Mb, scaffold N50 = 29.5 Mb) over a previous draft assembly. A total of 40 420 protein-coding genes were predicted in the D. longan genome. Synteny analysis suggests that longan shares the widespread gamma event with core eudicots but has no other whole genome duplications. Comparative genomics showed that the D. longan genome experienced significant expansions of UDP-glucosyltransferase and phenylpropanoid biosynthesis-related gene families. Deep genome sequencing analysis of longan cultivars identified longan biogeography as a major contributing factor to its genetic diversity and revealed clear population admixture and introgression among cultivars of different geographic origins, suggesting a likely migration trajectory of longan that is confirmed by existing historical records. Finally, genome-wide association studies (GWAS) of longan cultivars identified quantitative trait loci (QTLs) for six different fruit quality traits and revealed a shared QTL that contained three genes for total soluble solids and seed weight. The chromosome-level reference genome assembly, annotation, and population genetic resources for D. longan will facilitate the molecular studies and breeding of desirable longan cultivars in the future.
Introduction
Longan (D. longan Lour.), also known as dragon’s eyeball and closely related to lychee, is a tropical/subtropical evergreen fruit tree in the Sapindaceae family with a diploid genome [1] (2n = 2x = 30). It is regarded as a valuable tonic and has traditionally been used as a medicinal plant with rich pharmaceutical effects from many plant parts, especially fruits, and it contributes to rural economic development in tropical and subtropical areas. The use of longan in traditional herbal remedies was recorded in the compendium of materia medica (Ben Cao Gang Mu in Chinese) by Li Shizhen, a famous traditional Chinese medicine expert during the Ming Dynasty, who called longan the king of fruits [2]. Given its high nutritional and economic value, longan is cultivated in many countries such as China, Australia, Thailand, and Vietnam. Among these countries, China has the highest production of longan with the largest cultivation area [3], including Guangdong, Guangxi, Fujian, Hainan, and other regions [4].
China has a long history of longan cultivation and use. Native to South China, longan has been cultivated in China for over 2000 years with rich germplasm resources [5]. Abundant wild resources are found in Yunnan and Hainan provinces, whence longan was introduced to South Asian countries such as Thailand [6]. A previous study based on the different pollen exine patterns of fourteen longan varieties supports Yunnan as the primary center of longan origin and Guangdong, Guangxi, and Hainan as the secondary centers [7]. To date, the population genetic structure of longan remains poorly characterized. Longan varieties have a rather ambiguous genetic background owing to the fact that they reproduce by both inbreeding and crossbreeding in the field. A previous analysis using ISSR (inter-simple sequence repeat) markers indicated that Thai and Vietnamese varieties have close genetic relationships [8]. However, the classification of longan varieties using these markers has been inconclusive because of differences in marker selection, number of varieties, and classification methods. A resolved population structure of longan varieties and a better understanding of its genetic diversity and migration history are essential for its conservation and breeding but are not yet available, as they require a large-scale phylogenomic study of longan varieties from around China and Southeast Asia.
Longan leaves, flowers, fruits, and seeds [9, 10] are rich in polyphenols with anti-cancer and anti-oxidant properties that are biosynthesized primarily through the shikimic acid, flavonoid, and phenylpropanoid pathways. The phenylpropanoid pathway is one of the most extensively investigated specialized metabolic pathways. Branches of the phenylpropanoid pathway produce metabolites such as flavonoids, hydroxycinnamic acid esters, hydroxycinnamic acid amides, and the precursors of lignin, lignans, and tannins. These phenolic natural products are key components of the nutritive, flavor, and medicinal properties of longan. Phenylpropanoids also play important roles in plant resistance to pathogen infection [11] either by acting as physical barriers against invasion or through chemical toxicity to herbivores and microbial pathogens. Therefore, these secondary metabolites are acquired traits that offer adaptive advantages to longan through evolution and can be exploited by humans as useful medicines. Despite the vital roles of phenylpropanoids in the nutrition and flavor of longan fruits, their biosynthetic pathways in longan remain uncharacterized because of limited genetic and genomic resources and the technical difficulty of genetic transformation, which impede the improvement of desired traits in longan fruits through molecular and genomic breeding.
Longan breeding, which relies mainly on sexual hybridization, typically targets two main traits: fruit size and sweetness [12]. Longan breeding is challenging and time-consuming because of its long juvenile period and the difficulty of genetic transformation. Marker-assisted selection (MAS) is an effective biotechnological tool that enables early selection of hybrid progenies at the seedling stage [13]. However, our knowledge about the genetic mapping of longan is limited. Guo et al. (2011) constructed a low-quality male and female genetic map consisting of 243 and 184 separate molecular markers [14]. Single nucleotide polymorphism (SNP) markers based on restriction site-associated DNA sequencing (RAD-seq) were developed for QTL identification using hybrid F1 progenies and their two parents as materials [13]. A high-quality reference genome sequence and information on the longan genetic background would significantly facilitate the investigation of genotype–phenotype associations of longan germplasms and thus expedite the longan breeding program. Although a draft genome sequence of the D. longan “HHZ” cultivar is available [9], the assembly is highly fragmented, composed of 51 392 contigs with a contig N50 of 26 kb.
Here, we produced a chromosome-level genome assembly for the D. longan JDB cultivar by combining Illumina paired-end (PE), PacBio single molecule real-time (SMRT), and high-throughput chromatin conformation capture (Hi-C) sequencing. We annotated the genome using ab initio prediction, homolog evidence, and multi-tissue transcriptomic data. In addition, we performed population genome sequencing from a collection of longan accessions, followed by an in-depth analysis of population structure using high-quality genetic variants. The analysis revealed the population genetic diversity of longan and demonstrated population admixture and introgression among cultivars from major longan growing areas. GWAS analysis revealed three genes that were associated with fruit total soluble solids and seed weight, suggesting parallel evolution in the evolutionary diversification of domesticated species. The genome assembly, annotations, and genetic variants are valuable for functional genomic studies as well as molecular breeding of D. longan to improve yield and fruit quality and exploit its medicinal properties.
Results
Genome assembly and annotation
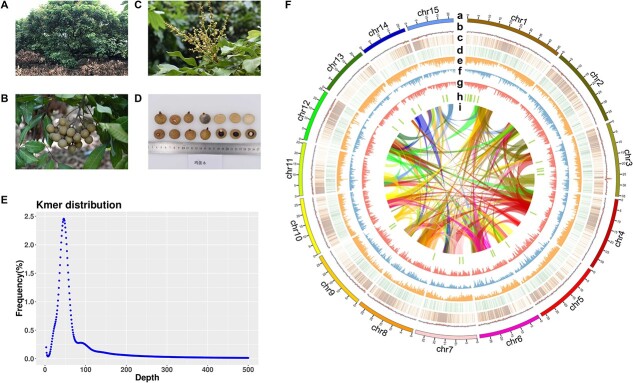
The D. longan cultivar “JDB” originated in Fujian and is planted in the Longan Germplasm Repository of Guangdong Province (Figure 1A-1D); fresh young leaves were collected for genomic DNA isolation and sequencing. To construct a chromosome-level reference genome for D. longan, a total of 184.4 Gb PacBio SMRT reads (~415× coverage), 25.3 Gb (~56× coverage) Illumina PE reads, and 57.6 Gb (~127× coverage) Hi-C Illumina read pairs were generated (Supplementary Table 1). We estimated the genome size of D. longan cultivar JDB as 474.98 Mb with a heterozygosity rate of 0.36% via the distribution of k-mer frequency using Illumina PE reads (Figure 1E). PacBio SMRT reads were used to assemble the D. longan genome with Canu15, followed by polishing contigs with Pilon16 using Illumina PE reads, yielding a draft genome assembly of 455.5 Mb (Table 1). Next, Hi-C paired-end reads were used to anchor the contigs to chromosomes with 3D-DNA [17]. The final D. longan JDB genome assembly of 455.5 Mb covers 95.90% of the estimated genome size (474.98 Mb), and 98.7% of the assembled sequences are anchored onto 15 chromosomes (Figure 1F; Supplementary Figure 1) with contig and scaffold N50s of 12.1 Mb and 29.6 Mb, respectively (Table 1). Thus, this longan genome assembly represents a significant improvement over the highly fragmented D. longan HHZ genome assembly (contig N50: 0.026 Mb) released previously [9]. Genome completeness was assessed using the plant dataset of the Benchmarking Universal Single Copy Orthologs (BUSCO) database v1.22 [18] with e-value <1e−5. BUSCO evaluation revealed the 98.1% completeness of our D. longan genome assembly (88.4% single copy, 9.7% duplicated copy, 1.1% fragmented, and 0.8% missing) (Table 1, Supplementary Table 2).
Figure 1.
Chromosome-level genome assembly of longan (Dimocarpus longan Lour.). (A–D): Photos of tree (A), flower (B), fruit cluster (C), and fruit section (D) of the longan cultivar JDB. (E) Kmer frequency distribution analysis for the JDB genome based on Illumina paired-end reads. (F) Overview of the D. longan genome. Tracks a to i: chromosomes, GC-content, density of Gypsy LTRs, density of Copia LTRs, density of protein-coding genes, SNP density, Indel density, distribution of secondary metabolic gene clusters (predicted using plantiSMASH), and syntenic blocks (colored ribbons). The density statistics were calculated within genomic windows 150 kb in size.
Table 1.
Statistics for Dimocarpus longan JDB genome assembly and annotations
| Statistics | D. longan JDB (this study) | D. longan Honghezi 9 | |
|---|---|---|---|
| Contig | Total number of contigs | 250 | 51 392 |
| Assembly size (Mb) | 455.5 | 471.9 | |
| Contig N50 (Mb) | 12.1 | 0.026 | |
| Contig N90 (Mb) | 1.8 | 0.006 | |
| Largest Contig (Mb) | 31.1 | 0.17 | |
| Scaffold | Total number of scaffolds | 90 | 17 367 |
| Assembly size (Mb) | 455.5 | 495.3 | |
| Scaffold N50 (Mb) | 29.6 | 0.57 | |
| Scaffold N90 (Mb) | 22.3 | 0.12 | |
| Largest scaffold (Mb) | 46.6 | 6.9 | |
| Annotation | Number of genes | 40 420 | 31 007 |
| Repeat content (%) | 41.7 | 52.9 | |
| Number of ncRNAs | 2555 | NA | |
| BUSCO (%) | 98.1% | 94% | |
| GC content (%) | 43.9 | 33.7 |
We next performed genome annotations using the BRAKER2 pipeline, combining evidence from ab initio prediction, protein homologs, and multi-tissue (root, shoot, leaf, and fruit) transcriptome sequencing data. The genome annotation pipeline predicted a total of 40 420 protein-coding genes and 2555 non-coding RNAs for D. longan (Table 1). The longan genome has an overall guanine-cytosine (GC) content of 34% and a gene density of 89 genes per Mb (Supplementary Table 2). About 89.0% of the genes were annotated with the non-redundant protein sequence database (Nr), and 84.6% of the genes were annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG) terms (Supplementary Table 3). Repetitive elements make up 41.7% of the D. longan genome, 54.9% and 25.4% of which are long terminal repeat retrotransposons (LTRs) and DNA transposons, respectively. Two major LTR subtypes, LTR-Copia (179.64 Mb) and LTR-Gypsy (66.18 Mb), represent 8.55% and 15.53% of the longan genome, respectively (Supplementary Table 4).
Comparative genomics and synteny analysis
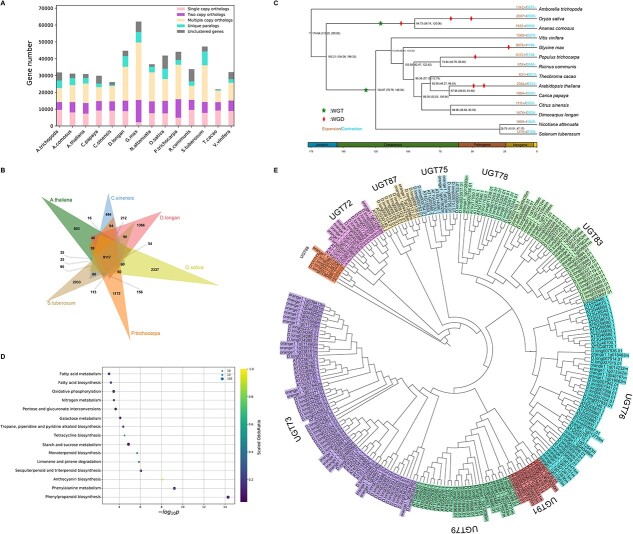
Next, we performed intraspecies synteny analysis of the D. longan genome to investigate its genome evolutionary history. Intraspecies syntenic gene pairs in D. longan were identified using MCScanX, which supported the presence of a whole genome triplication (WGT) event in the longan genome (Supplementary Figure 2A). The synonymous substitution rate (Ks) distribution for syntenic gene pairs also suggested that the D. longan genome had experienced the WGT (Supplementary Figure 2B). In addition, the 1:1 ratio of syntenic blocks between longan and grape (Vitis vinifera) and the 1:2 ratio of syntenic blocks between longan and poplar (Populus trichocarpa) indicated that longan had only the WGT (γ) event and did not have other whole genome duplications (WGDs) (Supplementary Figure 2C). To reveal the genome evolution and divergence of longan, we performed phylogenomic analysis of longan and thirteen representative plant species, including eight Rosids (Citrus sinensis, Carica papaya, Arabidopsis thaliana, Theobroma cacao, P. trichocarpa, Ricinus communis, Glycine max, V. vinifera), two Solanaceae (Solanum tuberosum, Nicotiana attenuata), one Poaceae (Oryza sativa), and a basal angiosperm (Amborella trichopoda). Orthogroup (gene family) identification revealed that these plants shared 7530 orthogroups, 137 of which are single-copy groups (Figure 2A; Supplementary Table 5). In particular, we identified 1366 orthogroups unique to D. longan compared to A. thaliana, C. sinensis, S. tuberosum, and P. trichocarpa (Figure 2B). The multiple sequence alignments of 137 single-copy orthologs in 14 species were concatenated and used for phylogeny construction and divergence time estimation using MCMCTree calibrated with fossil records (Figure 2C). Among the thirteen species, longan was phylogenetically closest to C. sinensis. The two species shared a last common ancestor at around 67 million years ago (Mya) that diverged from the asterids (N. attenuata, S. tuberosum) at around 125 Mya (Figure 2C).
Figure 2.
Phylogenomic genomics of Dimocarpus longan. (A) Summary of gene family clustering of D. longan and 13 related species. Single copy orthologs: 1-copy genes in an ortholog group. Multiple copy orthologs: multiple genes in an ortholog group. Unique orthologs: species-specific genes. Other orthologs: the rest of the clustered genes. Unclustered genes: number of genes outside of cluster. (B) Comparison of orthogroups (gene families) among six angiosperm species, D. longan (longan), A. thaliana (Arabidopsis), C. sinensis (citrus), S. tuberosum (potato), P. trichocarpa (poplar), and O. sativa (rice). (C) Phylogenetic relationships and divergence time estimates (with confidence intervals). The numbers of gene family expansions and contractions are indicated by red and blue numbers, respectively. (D) Bubble plot summarizing the most significantly enriched KEGG terms associated with expanded gene families in D. longan. The x-axis is the log10 transformed p-value. The size of the bubble is scaled to the number of genes. The color scale represents the scale of odds ratio in observed versus expected (genomic background) numbers of genes annotated with specific KEGG terms. (E) A phylogenetic tree of UGTs (UDP-glucosyltransferases) in three angiosperms, including D. longan.
Expanded gene families related to phenylpropanoid biosynthesis and UDP-glucosyltransferases
Gene family contraction and expansion are the evolutionary forces that drive rapid speciation and result in the diversification of plants [9]. Gene family analysis suggested that the longan genome has 1474 expanded and 2424 contracted gene families (Figure 2C) compared with the common ancestor of C. sinensis and D. longan. KEGG enrichment showed that the expanded gene families were significantly enriched (P < 0.05) in “phenylpropanoid biosynthesis”, “phenylalanine metabolism”, “anthocyanin”, “sesquiterpenoid and triterpenoid biosynthesis”, and “monoterpenoid biosynthesis” (Figure 2D). The 97 expanded longan phenylpropanoid biosynthesis genes were classified into seven gene families: phenylalanine ammonia-lyase (PAL, 5 members), peroxidase (POD, 38 members), O-methyltransferase (OMT, 3 members), glycosyl hydrolase family 1 (GH1, 26 members), aldehyde dehydrogenase family (ADH, 18 members), AMP-binding enzyme (4 members), and beta-galactosidase (BGL, 3 members) (Supplementary Table 6), probably involved in the biosynthesis of various lignin precursors. Lignin is a major component of some plant cell walls and is likely to have been involved in longan speciation [19]. Structural lignins provide physical barriers against pathogen infection and mechanical support for plant growth and the long-distance transport of water and nutrients [20]. Key enzymes such as PAL, POD, and PPO of the phenylpropanoid and lignin pathways are involved in these processes. Catalyzing the first step in the phenylpropanoid biosynthetic pathway, PALs were expressed at higher levels in roots, leaves, and stems but not in green fruits (Supplementary Figure 3), consistent with a previous report [9]. In addition, 28 of the 38 PODs showed differential expression in four major tissues (leaves, stems, roots, and fruits) (Supplementary Figure 4). The phenylpropanoid pathway is a conserved biosynthetic pathway in plants that leads to the production of various phenylpropanoid secondary metabolites such as phenolics, lignins, and flavonoids. The longan expansion of genes encoding these biosynthetic enzymes suggests that this pathway has probably evolved to generate diverse types of phenylpropanoids in longan through natural and artificial selection. A previous transcriptome-based study contrasted with our findings, suggesting that structural genes in phenylpropanoid pathways showed contraction instead of expansion [9]. However, we argue that a high-quality genome assembly and annotation is essential for the correct and complete characterization of gene families, producing results that are more trustworthy than transcriptome analysis alone.
Plants have evolved exquisite mechanisms for the biosynthesis of phenylpropanoids through acylation, methylation, glycosylation, and hydroxylation [21]. Most of the compounds synthesized by the phenylpropanoid pathway can be glycosylated by UDP-glucosyltransferases (UGTs). UGTs are key glycosylation enzymes that stabilize and enhance the solubility of small metabolites to maintain intracellular homeostasis [22]. For example, UGTs glycosylate volatile benzenoids/phenylpropanoids, the monoterpene linalool, and a strawberry aroma 4-hydroxy-2,5-di-methyl-3(2H)-furanone. InterPro (IPR) protein domain enrichment analysis showed that the expanded longan gene families are significantly enriched in IPR domains such as UGTs and cytochrome P450s (Supplementary Figure 5). The longan genome encodes more (215) UGTs (Supplementary Table 7) than those of Arabidopsis (107), C. sinensis (135), C. grandis (145), and V. vinifera (181) but fewer than that of apple (241) [ 23]. UGTs also participate in plant development, growth, and defense responses. Phenylpropanoid metabolism plays important roles in resistance to pathogen infection through the UGTs21. A phylogenetic tree was constructed using UGT protein sequences from longan and other plants, including Arabidopsis and Citrus (Figure 2E), and 115 expanded UGTs were divided into ten groups. Five groups (A, D, E, G, and L) expanded more than the others, although the numbers of genes in these groups varied widely among species [23]. In longan, groups A, D, H, I, and L expanded more than the other groups. For example, the numbers of longan UGTs in group D (31 UGTs, UGT73) and group I (19 UGTs, UGT83) were significantly increased compared with those in Arabidopsis and Citrus. A group D member, UGT73C7, mediated the redirection of phenylpropanoid metabolism to hydroxycinnamic acids (HCAs) and coumarin biosynthesis under biotic stress, resulting in SNC1-dependent Arabidopsis immunity [24]. In group I, the number of UGTs was highest compared with other fruits such as peach (5 UGTs), apple (11 UGTs), and grapevine (14 UGTs). UGT83A1 (GSA1) was required for metabolite reprogramming under abiotic stress through the redirection of metabolic flux from lignin biosynthesis to flavonoid biosynthesis and the accumulation of flavonoid glycosides, which coordinately confer high crop productivity and enhanced abiotic stress tolerance [25]. A total of 115 expanded UGT genes were unevenly distributed on the 15 chromosomes of the longan genome (Supplementary Figure 6). Chromosome 15 contained the most UGT genes (24), followed by 20 on Chromosome 13. Many of these UGT genes form clusters on several chromosomes (Supplementary Figure 6), suggesting that they are probably derived from tandem duplications of ancient UGT genes during longan evolution. Transcriptomic profiling suggested that 96 UGTs were differentially expressed in longan, with four (accounting for 4.2%), fourteen (14.6%), and ten (10.4%) UGTs uniquely expressed in leaves, roots, and fruit, respectively (Supplementary Figure 7, Supplementary Table 8).
Population structure, migration, and genetic admixture of longan cultivars
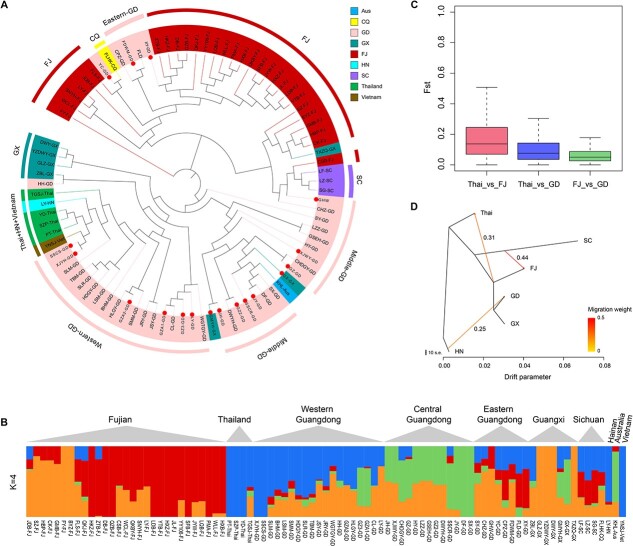
To understand longan genomic dynamics across its current distribution range in southern China and southeast Asian countries, we performed whole-genome resequencing analysis of 87 accessions (Supplementary Table 9) from five southern provinces in China (Guangdong, Fujian, Guangxi, Sichuan, and Hainan) and three other countries (Thailand, Vietnam, and Australia) with an average sequencing depth of 50×. Read mapping to the longan reference genome and variant detection yielded 29 730 132 single nucleotide polymorphisms (SNPs). After filtering, 11 421 213 high-quality SNP loci (minor allele frequency > 5%) were used for subsequent population genetic analyses. Although Guangdong borders on Fujian, the climate of the two provinces differs greatly during the longan growing season. After generations of planting and screening, different cultivation areas have formed their own longan variety characteristics and types. Using the genetic variant data, we analyzed the population structure within these longan cultivars using phylogenomic analysis and principal component analysis. Phylogenomic analysis clustered 87 longan accessions into relatively distinct domestic Guangdong and Fujian groups after the removal of artificial breeding populations (Figure 3A). Three Sichuan cultivars were next to the Fujian group and distant from the Guangdong group. Notably, two Guangdong cultivars, FLD and CPZ, were clustered with the Fujian group, probably because they come from eastern Guangdong, adjacent to Fujian. Guangdong cultivars are divided into two subgroups, the “Shixai” (SX)-centered group and the “Chuliang” (CL)-centered group from central and western Guangdong, respectively (Figure 3A); these are also the two main cultivars widely grown in Guangdong and Guangxi. Consistent with the phylogenetic tree, the principal component analysis showed that the Guangdong and Fujian cultivars were grouped separately overall, and the Thai and Vietnamese populations were distant from the Chinese populations if artificial breeding cultivars were removed (Supplementary Figure 8).
Figure 3.
Population structure and admixture analysis of Dimocarpus longan. (A) A neighbor-joining phylogenetic tree of all D. longan individuals was constructed using SNPs. The artificial breeding individual was marked with red dots inside. Colors represent different geographic groups. (B) A biogeographical ancestry (admixture) analysis of D. longan accessions with four ancestral clusters colored differently in the heatmap, in which each column represents a longan sample. (C) Distribution of Fst values (a measure of genetic differentiation) between longan populations from Thailand (Thai), Fujian (FJ), and Guangdong (GD). (D) Maximum-likelihood tree and migration events among seven groups of D. longan. The migration events are colored according to their weight.
To investigate the genetic background of longan from various regions, we performed a biogeographical ancestry (admixture) analysis based on high-quality SNPs and tested it with ancestral group values (k) ranging from 2 to 8. With a choice of four ancestral groups (k = 4), which gave the smallest cross-validation error (Supplementary Figure 9), the admixture analysis revealed a distinct genetic structure within longan accessions of different geographic origins. Longan cultivars from Fujian are primarily composed of two ancestral groups, whereas Guangdong, Guangxi, and Sichuan cultivars contain fractions of all four ancestral groups, indicating their more complex ancestral backgrounds (Figure 3B). The more similar ancestry composition between eastern Guangdong and Fujian cultivars is consistent with the geographic closeness of the two growing regions, suggesting a common ancestral origin or a possible exchange of cultivars between the two regions. By contrast, Thai and Vietnamese cultivars have a simple composition overall, with one predominant ancestral group, most likely shared with western Guangdong (Figure 3B). Thai cultivars were genetically more related to western Guangdong cultivars (Figure 3A) but distant from Fujian cultivars. Consistent with this result, we also detected stronger genetic differentiation (measured as the Fst value) between Thailand and Fujian than between Thailand and Guangdong (Figure 3C). Notably, the Australian cultivar has a genetic background that resembles the middle Guangdong cultivar, suggesting that it is a possible cultivar of middle-Guangdong origin that has recently been introduced into Australia.
With the diverse ancestral backgrounds of these longan cultivars, we were curious about the migration history of longan germplasms and therefore investigated potential gene flows among different growing areas due to such migration using TreeMix analysis. Given its reported origin in China, many wild longan resources are present in the Yunnan and Hainan provinces of China [6]. Therefore, the Hainan cultivar was used as an outgroup in this analysis. The TreeMix analysis detected a migration event directed from Hainan to Guangdong. The highest gene flow (migration weight 0.44) was observed between Sichuan and Fujian (Figure 3D). Gene flows were also detected from the Fujian, Guangdong, and Guangxi populations to Thailand (migration weight 0.31) (Figure 3D). The detection of gene flows was consistent with historical records of longan migration. Longan was first cultivated in the “Ling-nan” district of China, including Guangdong, Guangxi, and Hainan, about 2000 years ago, recorded by the painting of “San Fu Huang”. According to historical records, longan was not successfully moved to northern China-Shaanxi Province but was successfully introduced to Sichuan and then to Fujian, with suitable climate conditions (Yang Fu, “Chronicles of the South”, 1st century A.D.). Taken together, our analysis results broadly matched historical records that there was gene flow from Hainan wild germplasms to Guangdong, then a strong flow from Sichuan to Fujian, and finally gene flow from China to Thailand.
GWAS for longan fruit quality traits
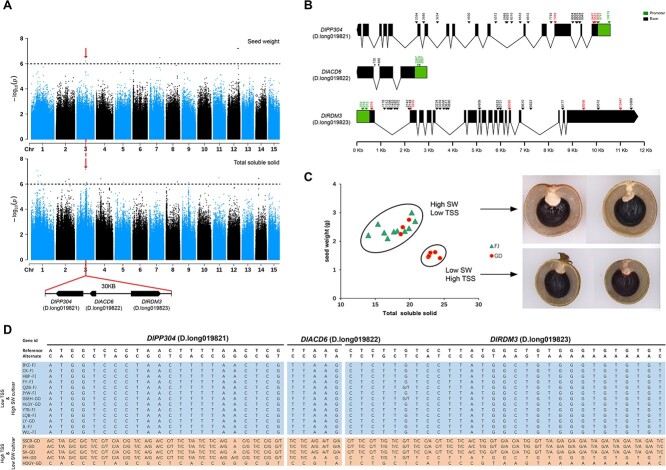
We next performed GWAS for fruit qualities to uncover QTLs underlying these traits. Phenotyping data, including pericarp thickness, pulp thickness, fruit horizontal diameter, total soluble solids, edible rate, and seed weight, were collected from 80 longan cultivars at the Longan Germplasm Repository of Guangdong Province that were genotyped in this study. To validate the reliability of the phenotype data, we calculated the Pearson correlation coefficient. Consistent with a previous study of longan [26], the coefficient of variation of single fruit weight and seed weight was the largest, whereas the coefficient of variation of fruit weight, pericarp thickness, and edible rate was the smallest (Supplementary Figure 10). GWAS analysis using 11 421 213 biallelic SNPs identified ten candidate QTLs that were significantly associated with six longan traits (p-value <1e−6) (Supplementary Table 11), associating 12 503, 7834, 11 184, 6348, 46 182, and 23 523 SNPs with pericarp thickness, pulp thickness, fruit horizontal diameter, edible rate, seed weight, and total soluble solids, respectively (Supplementary Figure 11). Genomic regions associated with seed weight contained several genes (Supplementary Table 9) related to seed development, such as UBQ10, UBIQ1, THI4, CBL10, and CTPS2. Specifically, a genomic region 0.94–0.99 Mb on Chromosome 6 contained three tandemly duplicated DlBGL42 genes encoding β–glucosidase (Supplementary Table 12), consistent with reports that seed size is controlled by Clbg1 (watermelon β–glucosidase) via decreased ABA content [27].
Fruit sweetness is generally measured by total soluble solids content [28]. For a long time, high total soluble solids and edible rate have been key longan traits selected in longan breeding [14]. Interestingly, GWAS identified a genomic region located on Chromosome 3 (at ~18.37–18.45 Mb) that harbored QTLs shared between total soluble solids and seed weight (Figure 4A). A SNP variance analysis of this region found polymorphic site mutations of ~37 kb at approximately 18.379 to 18.416 Mb, harboring the three successive genes DlPP304 (D. long019821.01), DlACD6 (D. long019822.01), and DlRDM3 (D. long019823.01), which encode a mitochondrial pentatricopeptide repeat-containing protein, an accelerated cell death protein, and an RNA-directed DNA methylation protein, respectively (Figure 4B). The co-localization of trait-linked loci usually reflects significant positive correlations among different traits [29]. Sugar content is a fruit characteristic that varies among cultivars and contributes to the distinctive flavor profile of longan varieties. There are reports that sugar contents are affected by fruit size and seed weight in sapodilla [30], and soluble sugar accumulation affects Arabidopsis seed size [31].
Figure 4.
GWAS mapping of seed weight and total soluble solids in longan fruit. (A) Top: Manhattan plot summarizing the GWAS results for seed weight based on analysis performed with a randomly down-sampled SNP call set. Middle: Manhattan plot of GWAS results for total soluble solids. Dotted lines represent the Bonferroni significance threshold. Red vertical lines represent the overlapping region of the two traits as highlighted below. Bottom: close-up of highlighted regions with three genes in the vicinity of the GWAS peak; (B) Diagram of three genes with respect to reference sequences and the haplotypes observed in samples collected from China. Black solid arrows indicate synonymous SNPs, and red solid arrows indicate non-synonymous SNPs specific to varieties with high total soluble solids (TSS) and low SW (seed weight); (C) Correlations of SW and TSS; (D) SNPs within the three genes for the most extreme TSS and SW phenotypes at each end of the high-type and low-type distributions (orange mutation is heterozygous to homozygous; blue: non-mutated) (see Supplementary Table 11 for all samples).
We next analyzed seed weight and total soluble solids phenotypes in 80 longan cultivars (Supplementary Figure 12). We observed that a group of ten Fujian and three Guangdong varieties displayed low soluble solids but bigger and heavier seeds, whereas another group of five Guangdong varieties had high soluble solids but smaller and lighter seeds (Figure 4C; Supplementary Figure 10). Examination of SNPs in the DlPP304, DlACD6, and DlRDM3 regions suggested that varieties with high soluble solids and low seed weight contained a larger number of variants than those with low soluble solids and high seed weight (Figure 4D, Supplementary Table 11). In particular, we found that the HDGY-GD variety, which contained mostly homozygous variants at DlPP304, DlACD6, and DlRDM3 gene regions, had the highest total soluble solids and the lowest seed weight among the cultivars (Figure 4D). Interestingly, most of the SNPs within the three genes were heterozygous in the group with high total soluble solids, hinting that sugar content is likely to be associated with heterosis, as high total soluble solids has long been a desired trait in longan breeding programs through hybridization. Previous studies have found a large proportion of the yield advantage of hybrids over their parents of elite inbred varieties [32, 33]. For most of the heterosis-related loci identified, dominance or incomplete dominance of heterozygous loci plays an important role. However, this hypothesis remains to be validated in further experiments. In summary, our GWAS analysis revealed a list of QTLs and candidate genes for further validation by functional genomics or forward genetics approaches.
Discussion
Longan is one of the most economically important fruit trees in China and Southeast Asia. However, molecular breeding of longan cultivars has been slow because of a lack of genomic and biotechnological resources. Using a combination of single-molecule (PacBio) and Hi-C sequencing data, we successfully produced a chromosome-level longan genome assembly, followed by protein-coding gene annotations. This genome assembly for longan represents a significant improvement over a current draft reference genome based on short-read sequencing data [9]. The improvement mainly stems from assembly of contigs using PacBio long reads, followed by scaffolding based on chromatin interaction information given by Hi-C data. As longan is primarily an outbreeding tree species, it typically has a high heterozygosity rate, which presents a challenge to the contiguous assembly of its diploid genome. The JDB cultivar used for de novo genome assembly has a relatively low heterozygosity rate of 0.3%, probably due to repeated inbreeding. Together, these two factors facilitated the highly contiguous assembly of the genome. The repetitive content (41%) in our genome assembly is less than that reported previously for an assembly (52%) based on Illumina short reads by Lin et al. [9], indicating that long read sequencing has helped to resolve the ambiguous placement and assembly of repetitive regions that are typically prone to mis-assembly from short reads. The high BUSCO score (98%) shows that completeness of the present longan assembly is also improved compared with that of Lin et al. [9], although we admit that there is still much to do to obtain a complete longan genome sequence in the future. In short, we present a high-quality, chromosome-scale genome assembly of longan, the best longan genome assembly reported to date.
Using this reference genome, we investigated the evolution of the longan genome and showed that it shared a whole genome triplication and lacked additional WGDs. Lin et al. [9] performed 4DTv analysis of their longan genome assembly and found that longan had an ancient genome duplication event, but no information was given with respect to whether the event occurred in related plants. By contrast, we conducted inter-species analysis using the longan and grape genomes and showed that longan and grape had a 1:1 syntenic relationship, confirming that the ancient duplication event in longan [9] was actually a whole genome triplication (γ event) shared with grape and other core eudicots. Given that the poplar genome underwent a post-γ WGD, the 1:1 and 1:2 syntenic relationships between longan and grape and between longan and poplar suggested the lack of a WGD following the divergence of grape and longan and of longan and poplar. Both Lin et al. [9] and our study identified a peak in Ks or 4DTv distribution for longan paralogs, indicating that many gene duplications have occurred recently in the longan genome, probably resulting from the activities of transposable elements.
The delightful taste and rich nutrition of longan fruits are among many unique traits of this plant. Longan fruit are rich in polyphenolic compounds. Genomic analysis of longan can reveal critical information about how the plant evolved such a unique chemical reservoir. We detected significant gene family expansions in biosynthetic genes of phenylpropanoids, terpenoids (sesquiterpenoids and triterpenoids), etc., compared with related plant lineages, suggesting that the longan genome has evolved towards innovation of these secondary metabolites through natural or artificial selection. By contrast, Lin et al. [9] analyzed their draft longan genome assembly and also showed that the longan genome had experienced gene family expansions and contractions. However, no functional terms related to specialized metabolism were found to be enriched in the expanded or contracted gene families they identified, although they did show enrichment of terms related to general biological processes such as cellular component organization. These different enrichment results may be explained by several factors, such as an improved genome assembly and annotations, the selection of plant genomes for comparison, and the functional enrichment analysis tools used. Our analysis also identified the expansion of UGT gene families, which are essential for the diversification of structural features and control the final oxidation, hydroxylation, and glycosylation steps to yield secondary metabolites in longan. The exact roles of UGTs in the formation and modification of specific longan natural products that give longan fruits their unique taste and nutrition remain to be determined.
Longan is widely grown in China and many Southeast Asian countries where longan cultivars were initially introduced from China, the origin of longan with the largest cultivation area in the world. Chinese longan consists mainly of the Guangdong population, the Fujian population, the Guangxi population, and so forth. Exchange of longan germplasms among different growing regions probably led to genetic introgression. Because longan can undergo both inbreeding and outbreeding in the natural environment, its genetic background and population structure are rather complex and await elucidation. The 87 cultivated varieties in this study were mostly collected from mainland China and a few neighboring South Asian countries; they have excellent characteristics and are often used as breeding parents, but most evolved through long-term natural hybridization. The evolutionary model of natural hybridization is unclear, resulting in the unclear genetic background of germplasm resources. As a result, confusion among names of longan varieties is not uncommon, such that different varieties have the same name, or the same variety has different names. Combining a high-quality genome assembly and population genomic sequencing of longan varieties, we have resolved the complex genetic background of longan, revealed its population admixture model, and deduced possible migration routes of longan consistent with historical records.
The high-quality SNP molecular markers generated in this study enabled the discovery of multiple QTLs strongly associated with useful traits such as seed weight and soluble solids, providing excellent resources for future molecular breeding. A QTL locus containing the three successive genes DIPP304, DIACD6, and DIRDM3 was significantly associated with total soluble solids and seed weight. Literature shows that the homologs of these genes have been reported to function in sugar content and fruit ripening, seed set, fruit abscission, and so forth [34–37]. The PPR (pentatricopeptide repeat) gene affecting sugar content in plants is likely to be involved in regulating cellular carbon metabolism. For example, the mitochondrial PPR protein SLO2 is required for carbon energy balance in Arabidopsis, and sugar contents were much lower in the slo2 mutants [34]. Thus, the longan DlPP304 gene encoding a PPR gene homolog probably plays similar roles in regulating the carbon energy balance that affects soluble solid contents in fruits. As for DlACD6, its Arabidopsis homolog AtACD6 (ACCELERATED CELL DEATH 6) is a transmembrane ankyrin repeat protein that modulates the activity of pattern recognition receptors (PRRs) associated with hybrid necrosis [38]. A single amino acid insertion of ACD6 was sufficient to convert the reference Col-0 allele into a necrosis-inducing allele [36], resulting in reduced seed set. Therefore, based on the literature and GWAS results in our study, we speculate that genetic variation in DlACD6 probably contributes to variable seed characteristics such as seed weight among longan cultivars. DlRDM3 is a protein homolog of the RNA-directed DNA methylation (RDM) family. Repression of the RDM pathway is reportedly required for fruit abscission in C. sinensis [37]. Taken together, these findings pave the way for research through the integration of genomic, transcriptomic, and metabolomic data to provide abundant promising candidates for future improvement of desired longan traits.
Materials and methods
Germplasm genetic resources
A 30-year-old D. longan tree cultivar named JDB from the Longan Germplasm Repository of Guangdong Province (which belongs to the Institute of Fruit Tree Research at the Guangdong Academy of Agricultural Sciences in China) was used for genome sequencing and de novo assembly. Eighty-six additional D. longan cultivars (Supplementary Table 9) from the Longan Germplasm Repository of Guangdong Province were collected for genome resequencing.
DNA and RNA isolation
The longan cultivar JDB was planted in the Longan Germplasm Repository of Guangdong Province. Fresh and healthy young leaves were collected, cleaned, and used for genomic DNA isolation and sequencing. Genomic DNA was extracted from young fresh leaves of D. longan using the modified cetyltrimethylammonium bromide (CTAB) method [39]. The concentration and purity of the extracted DNA were assessed using a Nanodrop 2000 spectrophotometer (Thermo, MA, USA) and a Qubit 3.0 system (Thermo, CA, USA), and the integrity of the DNA was measured using pulsed-field electrophoresis with 0.8% agarose gel. In addition, fresh leaves and other tissues (roots, shoots, and young fruits) of the JDB cultivar were collected for RNA isolation and transcriptome sequencing. Total RNA was isolated with the RNAprep Pure Plant Kit (Tiangen Biotech) according to the manufacturer’s instructions. The integrity and quantity of extracted RNA were analyzed on an Agilent 2100 Bioanalyzer. For each tissue, three biological replicates were prepared for sequencing.
Genome and transcriptome sequencing
DNA sequencing libraries were constructed and sequenced on the Illumina NovaSeq 6000 platform at 50× depth according to the manufacturer’s protocols (Illumina). To generate long-read sequencing reads for D. longan, DNA libraries were prepared for PacBio SMRT sequencing following the PacBio standard protocols and sequenced on the Sequel platform. In brief, genomic DNA was randomly sheared to an average size of 20 kb using a g-Tube (Covaris). The sheared gDNA was end-repaired using polishing enzymes. After purification, a 20-kb insert SMRTbell library was constructed according to the PacBio standard protocol with the BluePippin size-selection system (Sage Science), and sequences were generated on the PacBio Sequel (9 cells) and PacBio RS II (1 cell) platforms by Biomarker Technologies. Raw subreads were filtered based on read quality (≥0.8) and read length (≥1000 bp). For chromosome-level genome scaffolding, Hi-C libraries were prepared from fresh leaves and sequenced on the Illumina HiSeq X Ten platform. DNA was digested with HindIII enzyme, and the ligated DNA was sheared into sizes of 200–400 bp. The resulting libraries were sequenced using the Illumina NovaSeq 6000 platform. For transcriptome sequencing, RNA sequencing (RNA-seq) libraries were constructed using the True-Seq kit (Illumina, CA) and sequenced using the Illumina HiSeq X Ten platform. Illumina raw reads were trimmed using Trimmomatic (v0.39) [40] with parameters “LEADING: 10 TRAILING:10 SLIDINGWINDOW:3:20 MINLEN:36” to remove adapter sequences and low-quality reads, yielding a total of ~77.7 Gb clean RNA-seq data from four tissues.
Genome assembly and evaluation
To estimate the genome size and heterozygosity level of D. longan, cleaned Illumina PE reads were used for k-mer spectrum analysis with GenomeScope (v2.0) [41] based on 21-mer statistics. PacBio SMRT reads were used for de novo genome assembly with the Canu pipeline (V1.9) [15] using the parameters “correctedErrorRate=0.045 corMhapSensitivity=normal ‘batOptions=-dg 3 -db 3 -dr 1 -ca 500 -cp 50”. Alternative haplotig sequences were removed using purge_dups [42] with default settings, and only primary contigs were retained for downstream analysis. To correct base-pair-level errors in raw assembly sequences, two rounds of polishing were conducted using high-quality Illumina DNA reads with Pilon (v1.23) [16]. The longan contigs were further anchored to chromosomes using 3D-DNA [17] based on the Hi-C contact map, followed by manual correction using Juicebox (v1.11.08) [43] to fix assembly errors. The completeness of the genome assembly was assessed by BUSCO v1.2218 using the 2121 eudicotyledons_odb10 single-copy genes. PacBio sequence reads and Illumina DNA reads were aligned to the genome sequences using minimap2 [44] and BWA [45], respectively.
Repetitive element annotation
We used a combination of the de novo repeat library and homology-based strategies to identify repetitive structures. TransposonPSI [46] was used to identify transposable elements. The GenomeTools suite [47] (LTR harvest and LTR digest) was used to annotate LTR-RTs with protein HMMs from the Pfam database. Then, a de novo repeat library of the longan genome was built using RepeatModeler [48], and each of the three repeat libraries was classified with Repeat_Classifier. Subsequently, the non-redundant repeat library was analyzed using BLASTx to search the transposase database (evalue = 1e−10) and non-redundant plant protein database (evalue = 1e−10) to remove protein-coding genes. Then, the de novo repeat library was used to discover and mask the assembled genome with RepeatMasker [49] with the “-xsmall -excln” parameter.
Prediction and annotation of protein-coding genes
For gene structure annotations, the RNA-seq data from four different tissues were aligned to the repeat-soft masked genome using STAR [50], which generates intron hints for gene structure annotation. The structural annotation of protein-coding genes was performed using BRAKER2 [51] by combining the aligned results from ab initio predictions, homologous protein mapping, and RNA-seq mapping to produce the final gene prediction. Genes with protein length < 120 amino acids and expression level < 0.5 TPM were removed. Predicted genes were assigned functions by performing BLAST searches against the NCBI non-redundant protein database with an e-value threshold of 1e−10. In addition, a comprehensive annotation was also performed using InterProScan (5.36–75.0) [52].
Comparative genomics analysis
Putative orthologs were identified using protein sequences from two monocots, ten eudicots, Amborella trichopoda, and longan. Only the longest protein sequence was selected as representative of each gene. Orthogroups were inferred with OrthoFinder (v2.4.1) [53]. The species tree was used as a starting tree to estimate species divergence times using MCMCTree in the paml package (v4.9) [54]. Speciation event dates for Ananas comosus-O. sativa (102–120 MYA), P. trichocarpa-R. communis (70–86 MYA), A. thaliana-Carica papaya (63–82 MYA), and G. max-C. sinensis (98–117 MYA) obtained from TimeTree (www.timetree.org) were used to calibrate the divergence time estimates. We conducted two independent MCMCTree runs using the following settings: burnin = 20 000, sampfreq = 30, and nsample = 20 000.
The ortholog count table and phylogenetic tree topology inferred from OrthoFinder were provided to CAFÉ (v4.2) [55], which identifies significant expansion or contraction in each gene family across species using a random birth and death model to estimate the size of each family at each ancestral node. Among expanded gene families, longan genes enriched with IPR002213 (UDP-glucuronosyl/UDP-glucosyltransferase) and IPR036396 (Cytochrome P450 superfamily) and their ortholog CDS sequences from the A. thaliana and C. sinensis genomes were retrieved. Multiple sequence alignment was performed using MUSCLE (v3.8.1551) [56] software. IQ-TREE was used to construct a maximum likelihood tree with parameters “-m MF”. The tree file was loaded into the Interactive Tree of Life (iTOL) web server for tree visualization and figure preparation [57].
Transcriptomic analysis
After removing adapters and trimming low-quality bases, RNA-seq reads were mapped to the longan reference genome using STAR [50] with parameters “--alignIntronMax 6000 –align IntronMin 50” and then using the RSEM tool [58] for transcript quantification. Outliers among the individual experimental samples were verified based on the Pearson correlation coefficient (r2 ≥ 0.85). Differential expression analysis was performed using the DESeq2 package [59]. Genes were differentially expressed between two conditions if the adjusted p-value was <0.01 and fold change was >1.
Genetic variation detection
Population resequencing reads were mapped to the chromosome-level genome assembly of longan using BWA [45] with default parameters. Alignments for each sample were processed by removing duplicate reads using SAMtools [60]. The mpileup function in SAMtools was used to generate mpileup files for each sample. BCFtools [61] was used to identify SNPs and small indels. The SNPs were filtered with criteria of read mapping quality (MQ) > 40, minimum coverage > 10, base quality > 30, and genotype missing rate < 20% (of all samples). The high quality SNPs were used in further analysis.
Population structure analysis
The vcf-format SNP sets were transformed into binary ped format using PLINK [62]. To estimate individual admixture assuming different numbers of clusters, the population structure and ancestry were investigated using ADMIXTURE [63] based on all SNPs. A linkage disequilibrium pruning step was performed with PLINK [62] using the parameters “--indep-pairwise 50 10 0.1”. We analyzed the number (K) of ancestral clusters ranging from 2 to 8 and found that the cross-validation error was smallest at K = 4. We also inferred a population-level phylogeny across all groups using the maximum likelihood approach implemented in TreeMix [64]. For this analysis, samples with a genotype missing rate > 20% were filtered out. SNPs with an imputation info score < 0.8, minor allele frequency (MAF) < 5%, and significant deviation (p < 10e–4) from Hardy–Weinberg equilibrium (HWE) were also removed.
GWAS analysis
Phenotypic data from the D. longan cultivars in the Longan Germplasm Repository of Guangdong Province, including pericarp thickness, pulp thickness, fruit horizontal diameter, total soluble solids, edible rate, and seed weight, were measured in 2020 and 2021. Fifty mature fruits per cultivar were harvested for phenotypic measurement from five forty-year-old longan trees that had even canopy sizes and fruit loads. To ensure the statistical power of the analysis, we used SNPs with MAF > 5% to represent the genotyping information for each GWAS. GEMMA (version 0.98.1) was used to carry out the GWAS analyses with a mixed linear model (MLM) [65]. Furthermore, to minimize false positives in GWAS, population structure was taken into account using a kinship matrix that was estimated using PLINK [62]. The genome-wide significance threshold was set to 1e−6.
Acknowledgement
This project is supported by the Key-Area Research and Development Program of Guangdong Province (2020B020220006) and the Guangdong Provincial Crops Germplasm Nursery Construction and Resources Collection, Preservation, Identification & Evaluation Foundation. In addition, LG is supported by the National Natural Science Foundation of China (31970317) and a faculty startup package from Peking University Institute of Advanced Agricultural Sciences. The authors would also like to thank anonymous reviewers for their comments and suggestions to improve this manuscript.
Author contributions
Project design and oversight: LG, JL, and WQ; Sample collection and curation: DG and SH; Conducting experiment and data analysis: JW, ZL, and LG; Result interpretation: LG, JW, JL, BL, and WQ; Figure and table preparation: LG, JW, and LZ; Manuscript writing and revision: LG, JW, BL, and WQ; Funding: JL and LG; All authors have read and approved the final version of this manuscript.
Data availability
The genome sequencing data, RNA-seq data, Hi-C data, and genome assembly of D. longan generated in this study have been deposited in the Genome Sequence Archive (GSA) database at the National Genomics Data Center of China National Center for Bioinformation under accession number CRA004281.
Conflict of interest statement
The authors declare no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
REFERENCES
- 1. Zhang YF, BoBin L, Ying Wet al. The chromosomes observation of several rare germplasm in litchi and Longan. Acta Horticulture Sinica. 2010;37:1991–4. [Google Scholar]
- 2. Tang YY, He X-M, Sun Jet al. Polyphenols and alkaloids in byproducts of longan fruits (Dimocarpus Longan Lour.) and their bioactivities. Molecules. 2019;24:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altendorf S. Minor Tropical Fruits: Mainstreaming a Niche Market Food and Agriculture Organization of the United Nations. 2018;67–74.
- 4. Chen YH, Sun J, Lin Het al. Salicylic acid reduces the incidence of Phomopsis longanae chi infection in harvested longan fruit by affecting the energy status and respiratory metabolism. Postharvest Biol Technol. 2020;160:111035. [Google Scholar]
- 5. Zheng SQ, Wei XQ, Jiang JMet al. Actual state and corresponding strategy on longan breeding in China. Fujian Fruits. 2010;4:35–40. [Google Scholar]
- 6. Menzel CM, Waite GK. Litchi and Longan: Botany, Production. And Uses. Trowbridge: Cromwell Press; 2005. [Google Scholar]
- 7. Ke GW, Wang CC, Tang ZF. Palynological studies on the origin of longan cultivation. Horticultural Plant Journal. 1994;4:323–8. [Google Scholar]
- 8. Zhu JH, Pan L-M, Qin S-Qet al. Analysis on genetic relations in different ecotypes of Longan (Dimocarpus longan Lour.) germplasm resources by ISSR markers. Journal of Plant Genetic Resources. 2013;14:65–9. [Google Scholar]
- 9. Lin YL, Min J, Lai Ret al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience. 2017;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Guo DL, Han DMet al. A comprehensive insight into the metabolic landscape of fruit pulp, peel, and seed in two longan (Dimocarpus longan Lour.) varieties. Int J Food Prop. 2020;23:1527–39. [Google Scholar]
- 11. Dixon RA, Achnine L, Kota Pet al. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol Plant Pathol. 2002;3:371–90. [DOI] [PubMed] [Google Scholar]
- 12. Zheng S, et al. Fruit scientific research in new China in the past 70 years: longan. J Fruit Sci. 2019;36:1414–20. [Google Scholar]
- 13. Jue DW, Liu L, Sang Xet al. SNP-based high-density genetic map construction and candidate gene identification for fruit quality traits of Dimocarpus longan Lour. Sci Hortic. 2021;284:110086. [Google Scholar]
- 14. Guo YS, Zhao Y, Liu C. QTLs analysis of several traits in Longan. Biotechnol & Biotechnol Equip. 2011;25:2203–9. [Google Scholar]
- 15. Koren S, Walenz BP, Berlin Ket al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker BJ, Abeel T, Shea Tet al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;19:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudchenko O, Batra SS, Omer ADet al. De novo assembly of the Aedes aegypti genome using hi-C yields chromosome-length scaffolds. Science. 2017;356:92–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simão FA, Waterhouse RM, Ioannidis Pet al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- 19. Neutelings G. Lignin variability in plant cell walls: contribution of new models. Plant Sci. 2011;181:379–86. [DOI] [PubMed] [Google Scholar]
- 20. Zhao S, Zhao L, Liu Fet al. NARROW AND ROLLED LEAF 2 regulates LEAF shape, male fertility, and seed size in rice. J Integr Plant Biol. 2016;58:983–96. [DOI] [PubMed] [Google Scholar]
- 21. Yuan L, Grotewold E. Plant specialized metabolism. Plant Sci. 2020;298:110579. [DOI] [PubMed] [Google Scholar]
- 22. Ross J, Li Y, Lim EKet al. Higher plant glycosyltransferases. Genome Biol. 2001;2:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu B, Liu XH, Xu Ket al. Genome-wide characterization, evolution and expression profiling of UDP-glycosyltransferase family in pomelo (Citrus grandis) fruit. BMC Plant Biol. 2020;20:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang XX, Wang Y, Lin JSet al. The novel pathogen-responsive glycosyltransferase UGT73C7 mediates the redirection of phenylpropanoid metabolism and promotes SNC1-dependent Arabidopsis immunity. Plant J. 2021;107:149–65. [DOI] [PubMed] [Google Scholar]
- 25. Dong NQ, Sun Y, Guo Tet al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun. 2020;11:2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen XP, et al. Diverse seed characteristics of Longan germplasms. Fujian Journal of Agricultural Sciences. 2016;31:1318–22. [Google Scholar]
- 27. Wang Y, Wang J, Guo Set al. CRISPR/Cas9-mediated mutagenesis of ClBG1 decreased seed size and promoted seed germination in watermelon. Hortic Res. 2021;8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shuai L, Gu LT, Liu WHet al. Characteristic analysis of sugar returning in the fruit of different Longan cultivars. Chinese Journal of Tropical Crops. 2016;37:1900–7. [Google Scholar]
- 29. Ben Chaim A, Paran I, Grube RCet al. QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor Appl Genet. 2001;102:1016–28. [Google Scholar]
- 30. Setiawan E. Sugar content as affected by fruits size and seed number of sapodilia in madura. Agrovigor. 2016;9:105–10. [Google Scholar]
- 31. Meng LS, Xu MK, Li Det al. Soluble sugar accumulation can influence seed size via AN3-YDA gene cascade. J Agric Food Chem. 2017;65:4121–32. [DOI] [PubMed] [Google Scholar]
- 32. Huang X, Yang S, Gong Jet al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat Commun. 2015;6:6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang X, Yang S, Gong Jet al. Genomic architecture of heterosis for yield traits in rice. Nature. 2016;537:629–33. [DOI] [PubMed] [Google Scholar]
- 34. Zhu Q, Dugardeyn J, Zhang Cet al. SLO2, a mitochondrial pentatricopeptide repeat protein affecting several RNA editing sites, is required for energy metabolism. Plant J. 2012;71:836–49. [DOI] [PubMed] [Google Scholar]
- 35. Subburaj S, Tu L, Lee Ket al. A genome-wide analysis of the Pentatricopeptide repeat (PPR) gene family and PPR-derived markers for flesh color in watermelon (Citrullus lanatus). Genes (Basel). 2020;11:1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Todesco M, Kim S-T, Chae Eet al. Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet. 2014;10:e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sabbione A, Daurelio L, Vegetti Aet al. Genome-wide analysis of AGO, DCL and RDR gene families reveals RNA-directed DNA methylation is involved in fruit abscission in Citrus sinensis. BMC Plant Biol. 2019;19:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Świadek M, Proost S, Sieh Det al. Novel allelic variants in ACD6 cause hybrid necrosis in local collection of Arabidopsis thaliana. New Phytol. 2017;213:900–15. [DOI] [PubMed] [Google Scholar]
- 39. Allen GC, Flores-Vergara MA, Krasynanski Set al. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006;1:2320–5. [DOI] [PubMed] [Google Scholar]
- 40. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ranallo-Benavidez TR, Jaron KS, Schatz MC. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11:1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan D, McCarthy SA, Wood Jet al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Durand NC, Robinson JT, Shamim MSet al. Juicebox provides a visualization system for hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hass B. Transposon PSI: An Application of PSI-Blast to Mine (Retro-) Transposon ORF Homologies. Cambridge, MA, USA: Broad Institute; 2010. [Google Scholar]
- 47. Gremme G, Steinbiss S, Kurtz S. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform. 2013;10:645–56. [DOI] [PubMed] [Google Scholar]
- 48. Smit A, Hubley R. RepeatModeler open-1.0. Available athttp://www.repeatmasker.org (2015).
- 49. Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015. http://www.repeatmasker.org (2013).
- 50. Dobin A, Davis CA, Schlesinger Fet al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hoff KJ, Lomsadze A, Borodovsky Met al. Whole-genome annotation with BRAKER. Methods Mol Biol. 2019;1962:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Quevillon E, Silventoinen V, Pillai Set al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. [DOI] [PubMed] [Google Scholar]
- 55. Han MV, Thomas GWC, Lugo-Martinez Jet al. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30:1987–97. [DOI] [PubMed] [Google Scholar]
- 56. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Danecek P, Marshall J, Danecek Pet al. HTSlib: C library for reading/writing high-throughput sequencing data. GigaScience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chang CC. Data management and summary statistics with PLINK. Methods Mol Biol. 2020;2090:49–65. [DOI] [PubMed] [Google Scholar]
- 63. Zhou H, Alexander D, Lange K. A quasi-Newton acceleration for high-dimensional optimization algorithms. Stat Comput. 2011;21:261–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequencing data, RNA-seq data, Hi-C data, and genome assembly of D. longan generated in this study have been deposited in the Genome Sequence Archive (GSA) database at the National Genomics Data Center of China National Center for Bioinformation under accession number CRA004281.