Abstract
The interocular distance, or orbital telorism, is a distinctive craniofacial trait that also serves as a clinically informative measure. While its extremes, hypo- and hypertelorism, have been linked to monogenic disorders and are often syndromic, little is known about the genetic determinants of interocular distance within the general population. We derived orbital telorism measures from cranial magnetic resonance imaging by calculating the distance between the eyeballs’ centre of gravity, which showed a good reproducibility with an intraclass correlation coefficient of 0.991 (95% confidence interval 0.985–0.994). Heritability estimates were 76% (standard error = 12%) with a family-based method (N = 364) and 39% (standard error = 2.4%) with a single nucleotide polymorphism-based method (N = 34 130) and were unaffected by adjustment for height (model II) and intracranial volume (model III) or head width (model IV). Genome-wide association studies in 34 130 European individuals identified 56 significantly associated genomic loci (P < 5 × 10−8) across four different models of which 46 were novel for facial morphology, and overall these findings replicated in an independent sample (N = 10 115) with telorism-related horizontal facial distance measures. Genes located nearby these 56 identified genetic loci were 4.9-fold enriched for Mendelian hypotelorism and hypertelorism genes, underlining their biological relevance. This study provides novel insights into the genetic architecture underlying interocular distance in particular, and the face in general, and explores its potential for applications in a clinical setting.
Introduction
Orbital telorism, the distance between the eyes, is a craniofacial trait that varies between individuals as a result of growth processes of the skull and brain. Outside the medical field, the eye-to-eye distance marks a prominent feature of human facial variation. In a clinical setting, it is primarily known for its two extremes, i.e. hypo- and hypertelorism, respectively, a decreased and an increased interocular distance. These two extremes are considered dysmorphic features linked to specific genetic syndromes or disorders (1).
The embryonic development of the face—with cell migration towards and fusion in the midline—plays a crucial role in variations of orbital telorism, mostly thought to be controlled by the Sonic hedgehog (Shh) signalling pathway on a molecular level (2). More specifically, Shh initiates expression of Nkx2.1, which is necessary for ventral forebrain development (3). Problems during this embryonic process can not only lead to hypo- and hypertelorism, but also various other midline anomalies such as cleft lip and palate (4,5). Postnatally, the interorbital area continues to grow and develop, primarily during the first three years of life continuing until early adulthood, resulting in an increase of the interorbital distance (6,7).
Although some monogenic Mendelian disorders have been related to hypo- and hypertelorism, little is known about common genetic factors leading to variations in orbital telorism in the general population. Quantitative orbital traits seem to be moderately to highly heritable, with estimates ranging from 34 to 72% (8,9), which suggests that a large part of interindividual variation can be explained by genetic factors. Whole-exome sequencing and genome-wide association studies (GWAS) have indeed been performed to study the genetics underlying facial morphology including the interorbital distance, yet so far few associated genetic loci have been identified for interorbital distance (10–19). A GWAS specifically focusing on orbital telorism has not been carried out before.
To further elucidate genetic influences on orbital telorism in adult humans, we measured interorbital distance from magnetic resonance imaging (MRI) scans in 34 130 European individuals. We then estimated the heritability of this trait using different methods and performed GWAS with replication in an independent sample from the largest facial morphology GWAS thus far (N = 10 115) (18), followed by further exploration of the biological and clinical relevance of the identified genetic loci.
Results
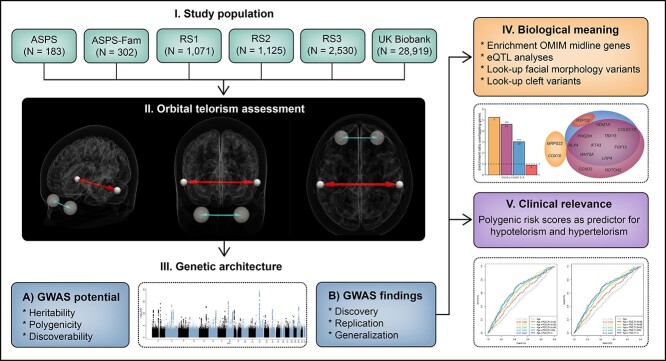
A schematic overview of the study design is shown in Figure 1. In short, we included 34 130 participants from four population-based studies, namely the UK Biobank (20) (N = 28 919), the Austrian Stroke Prevention Study (ASPS) (N = 183), the ASPS Family Study (ASPS-Fam) (21,22) (N = 302) and the Rotterdam Study (23,24) (N = 4726). Descriptive information about these studies, including population characteristics, genotyping and imputation information, and MRI acquisition and analysis parameters are presented in Supplementary Material, Table S1.
Figure 1.
Study design. Figure showing the design of the study. ASPS, Austrian Stroke Prevention Study; ASPS-Fam, Austrian Stroke Prevention Family Study; GWAS, genome-wide association study; OMIM, Online Mendelian Inheritance of Man database; RS, Rotterdam Study.
Repeatability, reproducibility and correlations
First, we assessed the reliability of our MRI-derived measures for orbital telorism and head width. Repeatability scans were made in a subset of the Rotterdam Study (N = 85), which showed good repeatabilities for these two measures, i.e. an intraclass correlation coefficient (ICC) of 0.991 [95% confidence interval (CI) 0.985–0.994] for orbital telorism and 0.982 (95% CI 0.972–0.989) for head width (Supplementary Material, Figs S1 and S2). In addition, a good correlation was shown between orbital telorism and manual measurements of interpupillary distance in another subset of the Rotterdam Study (N = 316), with an ICC of 0.837 (95% CI 0.800–0.867) (Supplementary Material, Fig. S3).
Since orbital telorism is a trait reflecting growth of the human body, we then explored the added value of orbital telorism beyond other anthropometric traits. To do so, we performed a correlation analysis between orbital telorism and potential confounders including important anthropometric traits within the UK Biobank sample. This showed that orbital telorism is moderately correlated with height (r = 0.51), intracranial volume (r = 0.30) and head width (r = 0.51) (Supplementary Material, Fig. S4), and may therefore reflect growth processes independent of these related anthropometric traits.
Heritability, polygenicity and discoverability
To determine the overall contribution of genetic influences on orbital telorism, we then estimated the heritability, polygenicity and discoverability. For the heritability estimation, we used two different methods, i.e. family-based heritability and single nucleotide polymorphism (SNP)-based heritability, providing, respectively, upper and lower bounds for the true heritability (25). Using a family-based approach, the heritability in ASPS-Fam was estimated to be 0.76 [standard error (SE) 0.12]. The SNP heritability estimate based on GWAS summary statistics was 0.39 (SE 0.02). These estimates were unaffected upon adjustment for height and intracranial volume or head width (Table 1). The polygenicity, i.e. the proportion of common SNPs from the reference panel involved in orbital telorism, was estimated to be 8.41 × 10−4, and the discoverability, i.e. the mean strength of association, was estimated at 1.80 × 10−4. The polygenicity and discoverability are comparable to those of height, and orbital telorism seems less polygenic and more discoverable than education and intelligence, and more polygenic and less discoverable than lipid traits (Supplementary Material, Fig. S5).
Table 1.
Heritability estimates
| Model | Family-based heritability (N = 364), h2 (SE) | SNP-based heritability (N = 34 130), h2 (SE) |
|---|---|---|
| 1 (age and sex) | 0.76 (0.12) | 0.39 (0.02) |
| 2 (model 1 + height) | 0.75 (0.12) | 0.39 (0.02) |
| 3 (model 2 + intracranial volume) | 0.74 (0.13) | 0.40 (0.02) |
| 4 (model 2 + head width) | 0.82 (0.12) | 0.40 (0.03) |
h2, narrow-sense heritability; N, sample size; SE, standard error.
GWAS and replication
Subsequently, we aimed to detect which specific genetic factors have an influence on variability in orbital telorism. Using a GWAS approach, we first analysed each cohort separately and subsequently meta-analysed the results (N = 34 130). We tested four models, adjusting for age, sex, principal components and study-specific covariates (model I), and additionally for height (model II) and intracranial volume (model III) or head width (model IV). On a genome-wide significant level (P < 5 × 10−8), we identified 56 genetic loci across the four different models, of which 23 genetic loci were consistent across all models (Fig. 2A; Supplementary Material, Table S2). However, none of the 78 genetic lead variants showed significant differences in effect sizes across the four models (Supplementary Material, Table S3). After additional multiple testing adjustments (P < 3.3 × 10−8), 55 loci with 73 lead variants remained genome-wide significant (Supplementary Material, Tables S2 and S3). In all models, genomic inflation was observed (λ = 1.2), although this could primarily be attributed to polygenicity rather than population stratification, with linkage disequilibrium (LD) score intercept values ranging from 1.02 to 1.03 (Supplementary Material, Table S4).
Figure 2.
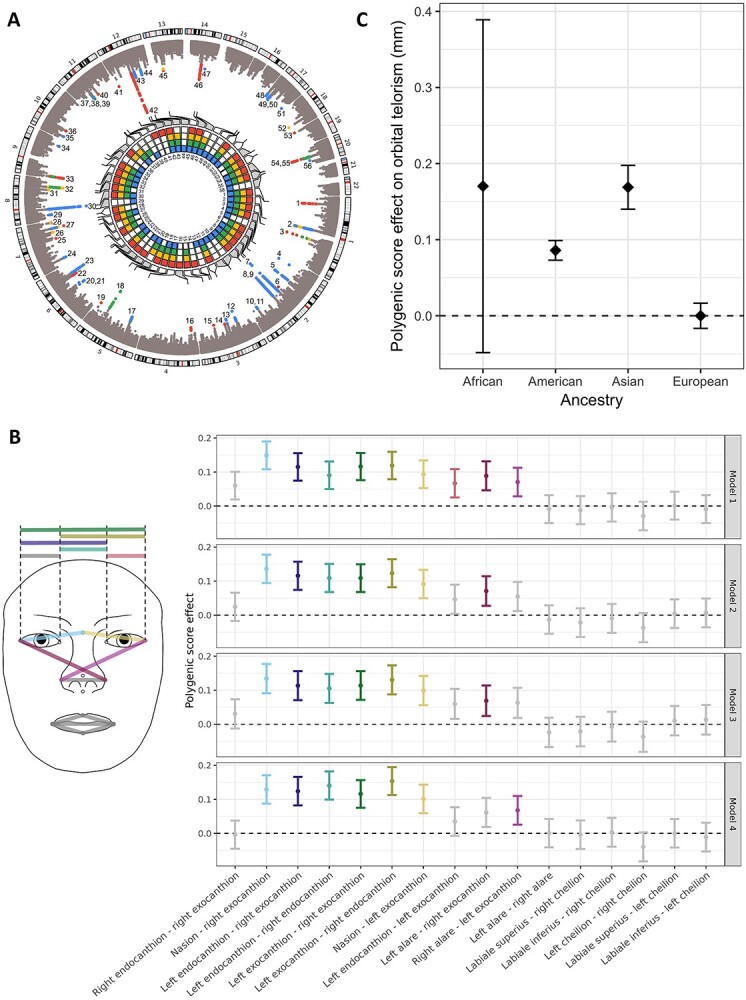
Genetic findings: discovery, replication and generalization. (A) Discovery. Circos plot showing the genome-wide association study findings across different models. The outer layer is a Manhattan plot, with genome-wide significant variants depicted in colour, with colours corresponding to the model with the lowest P-value. The inner layer is a heatmap showing for each genetic locus the model(s) in which the genetic locus has been identified. The colours corresponding to each model are red (model I, adjusted for age and sex), yellow (model II, adjusted for age, sex and height), green (model III, adjusted for age, sex, height and intracranial volume) and blue (model IV, adjusted for age, sex, height and head width). (B) Replication using polygenic scores. Plot showing the overall contribution of genetic lead variants to facial shape measures in the genome-wide association study by Xiong et al. (18) across the different models. Point estimates with their 95% confidence intervals are shown, with coloured error bars depicting facial morphological distances surviving the Bonferroni multiple testing threshold (P < 0.05/16). (C) Generalization. Bar plot showing the overall differences in orbital telorism in millimetres expected due to the allele frequency differences of the identified genetic lead variants across ancestries, in combination with their corresponding effect sizes. The null line corresponds to the average polygenic score effect of the investigated data of European samples.
As a replication dataset, we used the discovery data from a previously published GWAS on facial shape in 10 115 European individuals (18). In this dataset, 64 of the 78 lead genetic variants identified in the current study were available, of which four replicated after adjusting for the number of genetic variants tested (P < 0.05/64) and one variant (rs1371044) after additional correction for the 16 proxy phenotypes (P < 4.9 × 10−5) (Supplementary Material, Table S5). However, across the different models, polygenic scores (PGSs) of the available genetic lead variants were significantly associated with 9 out of the 10 horizontal eye distance measures (Fig. 2B;Supplementary Material, Table S6), also after excluding genetic variants that replicated individually (Supplementary Material, Fig. S6). Since the Rotterdam Study was present in both the facial shape and orbital telorism GWAS, we also performed the replication analysis excluding the Rotterdam Study sample from the current orbital telorism GWAS meta-analysis, which did not change the results substantially (Supplementary Material, Fig. S7).
Non-European ancestry differences
Since orbital telorism not only differs between individuals within ancestries but also across ancestries (26,27), we hypothesized that the ethnic differences in orbital telorism could partially be explained by differences in allele frequencies of genetic variants for orbital telorism. Based on the 1000 Genome allele frequencies, we estimated that the allele frequency differences between ancestries of the identified model I lead variants would result in a +0.17 mm difference in orbital telorism for African ancestry individuals, +0.09 mm for American ancestry individuals and +0.17 mm for Asian ancestry individuals, compared with European ancestry individuals (Supplementary Material, Table S7; Fig. 2C).
Impact on craniofacial morphology
To disentangle the telorism-specific and general craniofacial effects, we investigated the associations of genetic factors for other craniofacial traits with orbital telorism. First, we performed a look-up of 219 independently associated lead genetic variants identified in previous facial morphology GWAS (10–15,17–19,28,29). We found that 26 of these 219 signals were also significantly related to orbital telorism after multiple testing adjustments (Supplementary Material, Table S8). Although these were not restricted to eye features (i.e. left eye to nasion distance, right eye to nasion distance, shape of forehead and eye area), we did observe a higher proportion of significant signals for telorism-related features compared with nose, mouth and chin features [eye and forehead 28.1% (9/32); nose 6.0% (5/83); mouth 5.4% (2/37); chin 12.7% (7/55); global or other 22.7% (10/44)].
Since previous studies have shown genetic overlap of cleft lip and palate with facial morphology (30,31), we also performed a look-up of genetic variants identified for non-syndromic cleft lip with or without cleft palate and cleft palate alone. From the total of 53 independent genetic variants for these traits, 3 were also associated with orbital telorism after correcting for multiple testing (Supplementary Material, Table S9).
Enrichment and functionality
We further hypothesized that genes linked to extremes of telorism, i.e. hypo- and hypertelorism, would also be relevant for normal variation in telorism. Therefore, we assessed whether there was enrichment for genes previously associated with Mendelian disorders accompanied by hypo- or hypertelorism. Indeed, there was a 5.3 times enrichment for Mendelian hypotelorism genes, albeit non-significant (Ngenes = 2, P = 0.069), and a significant 4.6 times enrichment for Mendelian hypertelorism genes (Ngenes = 11, P = 9.2 × 10−8) compared with the rest of the genome (Fig. 3A; Supplementary Material, Tables S10 and S11). Only one additional gene (RSPO2) was identified by including cleft and midline genes (Fig. 3B), resulting in an enrichment ratio of 3.0 for cleft genes (Ngenes = 10, P = 5.2 × 10−4) and 0.89 for midline genes (Ngenes = 1, P = 1).
Figure 3.
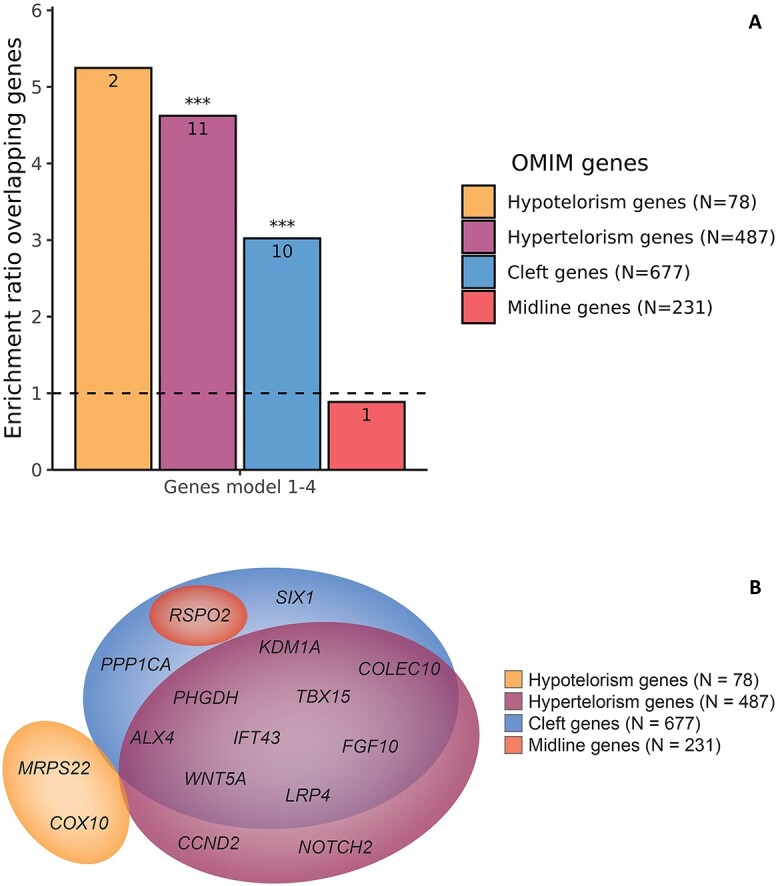
Enrichment and functional annotation. (A) Bar plot showing the enrichment ratio of the genes in or nearby the identified loci for Online Mendelian Inheritance of Man database (OMIM) genes associated to disorders with midline defects, compared with the rest of the genome. In each bar, the number of overlapping genes is shown and above the bar the significance is shown: non-significant (ns); *P < 0.05; **P < 0.01; ***P < 0.05/5. (B) Venn diagram showing the genes close to the identified lead variants or genetic variants in linkage disequilibrium (<10 kb) associated with hypotelorism, hypertelorism, clefts and midline defects in OMIM and their in-between overlap.
To further explore which genes could potentially be the target genes, we performed expression quantitative trait loci (eQTL) analyses. Genes for which eQTLs were present were also significantly enriched for Mendelian hypertelorism (4.7-fold, P = 1.9 × 10−12), cleft (3.2-fold, P = 9.8 × 10−7) and midline (3.2-fold, P = 6.3 × 10−3) genes (Supplementary Material, Tables S10 and S11). In total, eQTLs were found for 22 genes associated with OMIM midline anomalies, of which five (CCND2, FGFR1, GBA, LRP4, RSPO2) in brain tissue. For example, we found that CCND2, involved in the cell cycle G1/S transition, is not only a gene nearby an identified locus, but genome-wide significant variants were also eQTLs for CCND2 in brain cerebrum and cerebellar tissue. Mutations in this gene are known to cause Megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome 3, for which, hypertelorism is one of the clinical features. No additional Mendelian hypotelorism and hypertelorism genes were identified in an osteoclast-like cell culture eQTL study.
Application
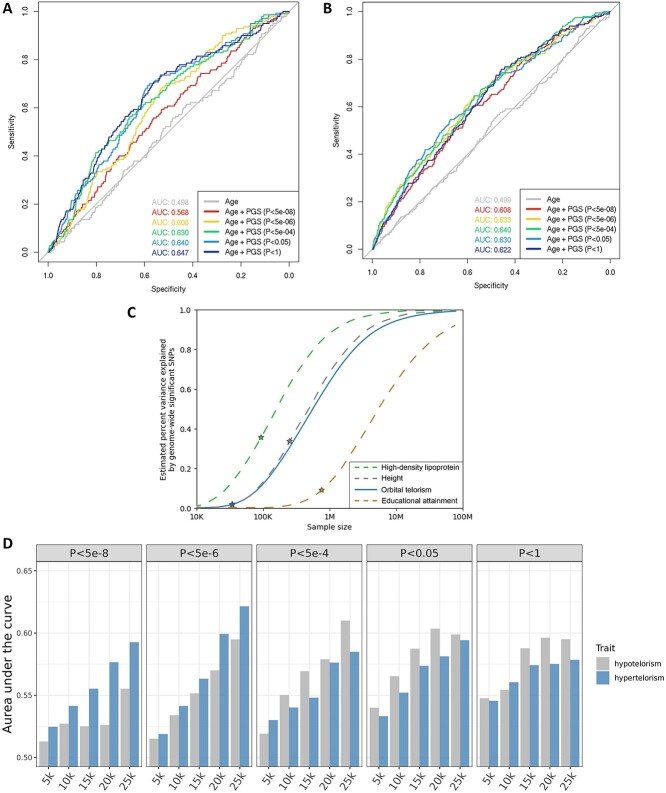
Next, we assessed the applicability of the genetic findings in an independent dataset of the UK Biobank (N = 7580). Polygenic scores summarizing the effects of the identified genetic variant at multiple P-value thresholds explained up to 2.7% of the variation in orbital telorism values (Supplementary Material, Table S12). Compared with basic prediction models for hypotelorism and hypertelorism including age, the area under the curve (AUC) increased from 0.498 to 0.647 for hypotelorism and from 0.499 to 0.640 for hypertelorism (Fig. 4A and B). Compared with a basic prediction model with age and height, the AUC increased from 0.614 to 0.679 for hypotelorism and from 0.587 to 0.670 for hypertelorism (Supplementary Material, Fig. S8A and B); and compared with a basic prediction model with age, height and intracranial volume, the AUC increased from 0.667 to 0.708 for hypotelorism and from 0.607 to 0.687 for hypertelorism (Supplementary Material, Fig. S8C and D). Given that genome-wide significant variants explain only 1.8% of the orbital telorism heritability (Fig. 4C), we explored to what extent larger sample sizes of future studies will result in an increase in this explained variance. For example, we estimated that a 3-fold increase in the sample size, from the current 34 k to 100 k individuals, will lead to an ~7-fold increase in explained variance (Fig. 4C). Similarly, we observed a positive relation between sample sizes of the discovery GWAS and discriminative ability of PGSs for hypo- and hypertelorism in an independent sample (Fig. 4D).
Figure 4.
Potential for clinical application. (A) Receiver operating characteristic (ROC) curve for the discriminative value of polygenic scores for hypotelorism, as shown by the area under the curve (AUC) for polygenic scores with different P-value thresholds for the inclusion of genetic variants. (B) ROC curve for the discriminative value of polygenic scores for hypertelorism, as shown by the AUC for polygenic scores with different P-value thresholds for the inclusion of genetic variants. (C) Power plot showing the power of the current genome-wide association studies for orbital telorism in comparison to high-density lipoprotein levels (64), height (65) and educational attainment (66). The lines show the expected fraction of heritability explained as a function of the sample size. The stars denote the fraction of heritability explained by genome-wide significant variants at the current sample size. (D) Bar plots showing the average AUC for hypotelorism and hypertelorism using telorism polygenic scores with different genome-wide association study sample sizes and P-value thresholds. The x-axis shows the discovery sample sizes of the telorism genome-wide association studies. The panels show the results for polygenic scores at different P-value thresholds for the inclusion of genetic variants.
Discussion
In this study, we elucidated the genetic architecture of orbital telorism and identified 78 genome-wide significantly associated lead genetic variants located at 56 genetic loci across four different models. These loci were enriched for genes known to cause hypo- and hypertelorism in Mendelian syndromes, underlining the biological importance of the identified loci.
To our knowledge, this is the largest GWAS on orbital telorism so far, with a three-time increase of the sample size compared with previous efforts investigating facial morphology (18). To exploit available population imaging data, we developed an automated tool to derive orbital telorism measures from brain MRI scans, which showed very good repeatability and correlated well with manual interpupillary distance measures, which is a proxy of orbital telorism. In fact, it has long been acknowledged that radiographic measurements are more reliable than manual measurements (6). Thus, our MRI-derived automated telorism measure yields promise for future studies incorporating the increasingly available large-scale imaging datasets. Not only will this allow researchers to study orbital telorism in larger samples, but also other MRI-derived craniofacial traits. Importantly, the field is moving from simple craniofacial measurements to more comprehensive craniofacial traits, which resembles the complex nature of craniofacial morphologies.
Our results confirm that orbital telorism is a heritable trait, with estimates ranging from 0.74 to 0.82 using family-based heritability methods, and 0.39 to 0.40 using SNP-based heritability methods. This is in line with heritability estimates for other facial morphological traits, which are known to be highly genetically determined (32). We additionally estimated the polygenicity, i.e. the proportion of causally associated genetic variants from the total number of variants in a reference panel, and the discoverability, i.e. the causal effect size variance (33). We observed polygenicity and discoverability estimates that were similar to those of height (33), a trait that has proven to be a very successful trait for GWAS discoveries. These findings therefore highlight the potential of orbital telorism to further elucidate the genetic underpinnings of facial morphology.
As orbital telorism reflects specific growth processes of the human body, it is expected to be correlated with more global growth measures such as height or intracranial volume as a proxy of head size. Although in clinical practice hypo- and hypertelorism are usually determined irrespective of other anthropometric traits, we were interested whether genetic factors related to orbital telorism are independent of general growth factors for the human body and head. We therefore created four different models, not only adjusting for age and sex, but also for height, and either intracranial volume or head width—which can also be useful for future research since the results of each model can be of interest depending on the research question at hand. Interestingly, however, even after adjusting for height and either intracranial volume or head width, the effects of the identified lead variants did not change significantly. This suggests that genetic factors related to orbital telorism are largely independent of more global growth factors related to the growth of the body and head.
Not only were the GWAS results similar across the four different models, our overall findings also replicated despite notable differences in the replication sample compared with the discovery sample, i.e. (1) measures were derived from three-dimensional facial images instead of MRI scans; (2) identical measurements of orbital telorism were not available and (3) other adjustments were made, i.e. body mass index and a generalized procrustes analysis (GPA) in order to remove variation due to scaling, shifting and rotation instead of adjustments for height, intracranial volume and head width. The replications of our findings were restricted to eye width-related measures and did not extend to width metrics of the lower half of the face. This indicates that overall, the identified genetic variants are specifically associated with eye width-related measures.
Previous studies have shown differences in interpupillary or eye-to-eye distance depending on ethnicity (26,27,34). Although we were not able to include non-European samples in our discovery set, we did explore the generalizability of the findings to other ethnicities attributable to differences in allele frequencies. In accordance with phenotypic studies (26,27,34), individuals with African or Asian ancestries were on average predicted to have higher orbital telorism values than individuals with European ancestries based on allele frequency differences. It should, however, be noted that this is a hypothesis unverified in non-European samples with data on orbital telorism. More diverse samples could provide more insight into the biology underlying these ancestral differences and may reveal novel genetic loci contributing to variation in orbital telorism, but few samples are available.
Traditionally, midline traits including telorism are thought to be mainly influenced by the Shh pathway (2). However, only one nearby gene (WNT5A) overlapped with the KEGG (35) Hedgehog signalling pathway, and one additional eQTL gene (WNT16). This suggests that a variety of gene types and pathways may play a role in subtle variations in telorism in a general population. Nonetheless, the biological relevance of the identified loci was illustrated by the enrichment of nearby genes for Mendelian syndromes associated with hypotelorism or hypertelorism, mainly driven by hypertelorism genes. For example, a common (minor allele frequency = 0.165) exonic lead variant (rs10494217) in the T-Box Transcription Factor 15 gene (TBX15) in LD with a genetic lead variant (r2 = 0.993) was significantly associated with orbital telorism (Pmodel1 = 1.87 × 10−8) with predicted deleterious effects (CADD score 22.7). TBX15 is a gene coding for a transcription factor that regulates the development of the skull. Mutations in this gene are associated with Cousin syndrome, an autosomal recessive Mendelian syndrome accompanied by facial dysmorphologies including hypertelorism and cleft palate. Another common (minor allele frequency = 0.331) 3′-UTR variant with a CADD score of 14.52 (rs1374961) located in the Fibroblast Growth Factor 10 gene (FGF10) was in LD with another lead variant (r2 = 0.607). The protein encoded by this gene is involved in various biological processes including embryonic development of the brain. Mutations in this gene can cause lacrimoauriculodentodigital syndrome, with hypertelorism as one of its clinical features. This indicates that these genes from various pathways influence orbital telorism across the entire spectrum, from normal to extreme.
Since facial morphological features are known to be phenotypically and genetically correlated (18), we investigated the genetic overlap with other craniofacial traits. Indeed, variants previously associated with facial morphology were also associated with telorism in our sample, suggesting that these genetic factors affect multiple facial morphological features. Previous studies have also shown an overlap between the genetics of non-syndromic cleft lip with or without cleft palate and normal variations in facial shape morphology (30,31). Since orbital telorism and clefts are both facial midline phenotypes, we hypothesized that our findings would be enriched for variants and genes associated with clefts and midline biology in general. Although only few of the previously identified variants for cleft lip and/or palate were associated with telorism after multiple testing, the majority of the overlapping OMIM genes for hypertelorism were also known for their association with clefts. One additional overlapping OMIM gene was identified by broadening the search to clefts and midline biology in general, namely the R-Spondin 2 gene (RSPO2), which enhances Wnt signalling through ubiquitin E3 ligase inhibition. Mutations in this gene are known to be associated with Tetraamelia syndrome 2. This clinical syndrome is characterized by partial or complete absence of the limbs and bilateral lung agenesis, but is also accompanied by facial dysmorphological features including facial clefts. Genetic variants in LD (r2 > 0.97) with the genetic lead variant rs62537502 are in eQTL with RSPO2 in the brain cortex, pointing towards a potential important role of this gene in normal variations in orbital telorism. Although this entails only one of the two main tissues thought to be involved, i.e. brain and bone tissue, it is important to realize that bone tissue is usually not present in well-established eQTL databases, including GTEx v8. We did look into another smaller eQTL dataset established using osteoclast-like cell cultures (N = 158) (36), but none of the significant eQTLs was linked to genes known for Mendelian disorders associated with hypotelorism or hypertelorism. Despite this, a similar enrichment of OMIM midline genes was seen for eQTL genes compared to the enrichment of nearby genes, suggesting a substantial involvement of genes that are not exclusively expressed in bone tissue.
GWAS findings are increasingly being used in clinical practice (37). For telorism, PGSs could potentially be used in the clinic to discriminate monogenic from polygenic hypotelorism or hypertelorism. Therefore, we tested whether PGSs of our findings showed a good discriminative ability for hypotelorism and hypertelorism in a general population. Although PGSs had a substantially higher AUC compared with a basic model including age, AUC estimates were moderate. Thus, while the results look promising, PGSs for telorism need to perform better before they become useful for clinical applications. However, with the current sample size only a fraction of the heritability is explained, which will increase in future GWAS with more power. Using random subsets of the UK Biobank sample, we also observed a positive relation between discovery GWAS sample sizes and discriminative abilities of PGSs in an independent sample. With the increasing availability and samples sizes of biobanks, polygenic risk scores could distinguish monogenic from polygenic telorism anomalies at a level that is helpful for clinicians.
In conclusion, our study provides insights into the genetics underlying subtle variations of orbital telorism in a general population. Follow-up analyses revealed an enrichment for genes associated with Mendelian disorders associated with midline anomalies. GWAS with larger sample sizes and studies in clinical populations are needed to improve the applicability of telorism GWAS findings in a clinical setting.
Materials and Methods
Study population
As discovery samples, we used data from the ASPS, the ASPS-Fam (21,22), the Rotterdam Study (24) and the UK Biobank (20). An independent subset of the UK Biobank was used to assess the applicability of the genetic findings. An overview of the population characteristics of the discovery and application samples is provided in Supplementary Material, Table S1.
Telorism and head width assessment
MRI scanners from different manufacturers and with different characteristics such as field strength were used to calculate telorism in the discovery samples and the UK Biobank application sample (Supplementary Material, Table S1). Calculations were performed on T2-weighted sequences, and if not available on T1-weighted sequences. In short, advanced normalization tools registration (38) was performed between the native and standard space (MNI). Then, the eyeball mask manually derived from the MNI template was registered to the native space using the metrics from the initial registration. The centre of gravity of the eyeballs was used to calculate the distance between the x, y and z values. The distances were manually checked if the distances, eye mask coordinates or mean voxel intensity, were >2.5 standard deviations away from the mean. Similarly, a mask containing fiducial points on the skull of the MNI template was also registered to native space. Head width was measured using points within the MNI template located at [68.3, −30.0, −19.7] and [−69.1, −30.7, −19.7], which was translated to the subject space using the obtained deformation file. The distance between the subject space points was obtained using the centre of gravity function.
In 85 individuals from the Rotterdam Study, a repeatability MRI scan was made. We used these scans to assess the ICC for orbital telorism and head width. In addition, for a subset of the Rotterdam Study measurements of interpupillary distance derived from eye examinations were available (N = 316), which we compared with our orbital telorism measurements since they should highly correlate.
Heritability, polygenicity and discoverability
To determine the overall contribution of common genetic variants on variability in telorism, we calculated the heritability of telorism using two different methods. In the ASPS-Fam sample, we performed a variance-components linkage analysis using the SOLAR software (39), which provides a family-based heritability estimate. Using the GWAS summary statistics, we estimated the SNP heritability using the LD score regression (LDSC) software (40). In both analyses, we adjusted for age and sex (model I), and additionally for height (model II), intracranial volume (model III) and head width (model IV). To estimate the polygenicity and discoverability of orbital telorism, we used the MiXeR software (http://github.com/precimed/mixer) (33).
Genome-wide association studies
Information regarding the genotyping and imputation procedures, as well as association testing is provided in Supplementary Material, Table S1. GWAS were performed using linear regression under an additive model. We adjusted for principal components (if needed), study centre (if applicable), genotyping array (if applicable) and—similar to the heritability analyses—for age and sex (model I), and additionally for height (model II), height and intracranial volume (model III), and height and head width (model IV). Genetic variants were filtered on study level using imputation quality (INFO or r2) * number of individuals * minor allele frequency > 5. A meta-analysis of the results was performed in METAL (41), using an inverse-variance weighted fixed-effects model with a SE analysis scheme. After meta-analysis, genetic variants were additionally filtered using the lowest imputation quality (INFO or r2) * number of individuals * minor allele frequency > 100. To assess differences in estimates between the models, we performed Z-tests. For the main analysis, we used P < 5 × 10−8 as P-value threshold for significance. We also performed 10 000 permutations in the UK Biobank sample to calculate the number of independent tests across the four different models. This resulted in a P-value threshold of P < 3.3 × 10−8 (5 × 10−8/1.5) to additionally adjust for multiple testing. The LDSC software (40) was used to calculate the LDSC intercept and ratio. These values can be used to assess whether genomic inflation (λ > 1) can be attributed to a trait’s polygenicity. When the LDSC intercept is close to one or the LDSC ratio close to zero, the genomic inflation is thought to be caused by polygenicity rather than population stratification.
Replication
We replicated our results in a GWAS on facial shape phenotypes (18). The assessment of facial shape measures in this dataset, including proxies of orbital telorism, has been described previously (18). In short, three-dimensional facial photographs were used to extract 78 Eucledian distance variables from all possible combinations between 13 facial landmarks. To remove variation due to shifting, rotation and scaling, GPA was performed. In addition to the GPA scaling of the phenotypes, GWAS analyses were adjusted for age, sex, body mass index and if needed familial relationships and principal components. This is most comparable to our model III and IV, with body mass index as a proxy for height and GPA as a proxy for intracranial volume or head width. For the replication analysis in this facial GWAS data set, we focused on facial width variables and variables highly correlated (r2 > 0.6) with these facial width distances. The eight available facial width phenotypes were left alare–right alare; right cheilion–left cheilion; left endocanthion–left exocanthion; left endocanthion–right endocanthion; left endocanthion–right exocanthion; left exocanthion–right endocanthion; left exocanthion–right exocanthion; right endocanthion–right exocanthion. The eight highly correlated phenotypes were labiale inferius–left cheilion; labiale inferius–right cheilion; labiale superius–left cheilion; labiale superius–right cheilion; left alare–right exocanthion; nasion–left exocanthion; nasion–right exocanthion; and right alare–left exocanthion. The combined association of the available genetic lead variants with these facial shape metrics were assessed using the inverse-variance weighting method implemented in the gtx package in R.
To validate the replication results of the combined associations of the lead variants, we performed two sensitivity analyses. First, we were interested whether our results were driven by the genetic variants that replicated individually after multiple testing adjustments (P < 0.05/64), which we therefore excluded in the first sensitivity analysis. Second, since the Rotterdam Study was included in both the discovery and replication sample, we performed another sensitivity analysis excluding the Rotterdam Study from the discovery GWAS meta-analysis. We then extracted new lead variants based on this GWAS meta-analysis excluding the Rotterdam Study and repeated the same analysis.
Generalization
Orbital telorism is known to differ across ancestries (26,27). Therefore, we were interested to see if these differences could partially be explained by the differences in allele frequencies of the identified genetic lead variants. We extracted the 1000 Genomes allele frequencies of the identified genetic lead variants using HaploReg version 4.1 (42). Based on the differences between the GWAS allele frequencies and the 1000 Genomes allele frequencies, we calculated the overall difference in the telorism population averages across ancestries due to these genetic variants. To obtain this overall population difference, we took the difference in allele frequency between 1000 Genomes and the GWAS, and multiplied this number by the GWAS effect estimate.
Impact on craniofacial morphology
To assess whether the identified genetic variants are general craniofacial variants or specific telorism variants, we performed a look-up of variants in other craniofacial GWAS (10–15,17–19,28,29). In addition, since previous studies have shown a link between the genetics of cleft lip and palate with craniofacial morphology (30), we also performed a look-up of significant variants in previous GWAS investigating cleft lip or palate (43–57). To calculate the number of independent genetic variants tested, we clumped the genetic variants using PLINK 1.9 (58) with a LD r2 threshold of 0.8 and a clumping window of 250 kb. The P-value adjusted for multiple tested was defined as 0.05 divided by the number of independent genetic variants.
Enrichment and functional annotation
To investigate enrichment of the identified genes for hypo- and hypertelorism and midline defects, we extracted genes associated with clinical syndromes manifesting with those symptoms using the OMIM database (59). For hypotelorism, we used the search term ‘hypotelorism’ OR ‘closely spaced eyes’; for hypertelorism, we used the search term ‘hypertelorism’ OR ‘widely spaced eyes’ OR ‘increased interocular distance’. Additionally, we investigated the enrichment for genes and midline abnormalities by adding respectively ‘cleft’ and ‘midline’ to the search term. Subsequently, we took these genes overlapping with OMIM forward to additional eQTL analyses, in order to assess which genes might be causally related to telorism. For these different searches, overlapping genes were manually curated to prevent any false positive matches. We extracted eQTLs using Genotype-Tissue Expression version 8 within the Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) software (60), and an eQTL study of human osteoclast-like cell cultures differentiated from peripheral blood mononuclear cells available via the GEnetic Factors for OSteoporosis Consortium (N = 158) (36).
Application
In an independent sample of participants of the UK Biobank, i.e. the fifth imaging data release, we tested the applicability of the genetic findings for clinical use. We created weighted PGSs of the model I findings using PRSice-2 (61) with the default clumping parameters. P-value thresholds were set at P < 5 × 10−8, P < 5 × 10−6, P < 5 × 10−4, P < 0.05 and P < 1. We then tested the predictive ability for hypo- and hypertelorism of these different PGSs. Hypo- and hypertelorism were defined as more than respectively two standard deviations below and above the sex-specific mean. As baseline models, we created prediction models (1) only including age, (2) including age and height and (3) including age, height and intracranial volume. Receiver operating characteristic curves with AUC were created and calculated using the pROC R package (62). The MiXeR software (63) was used to create a GWAS power plot, comparing orbital telorism with high-density lipoprotein (64), height (65) and educational attainment (66). To investigate the relation between GWAS sample size and discriminative abilities of PGSs based on these GWAS results, we first created 10 random subsets of 5, 10, 15, 20 and 25 k individuals from the UK Biobank. In these 50 random subsets of individuals, we then performed GWAS on telorism as described before. In the independent sample of the UK Biobank, we then created PGSs based on these GWAS results and calculated the average AUC for hypo- and hypertelorism.
Abbreviations
- ASPS
Austrian Stroke Prevention Study
- AUC
area under the curve
- CI
confidence interval
- eQTL
expression quantitative trait loci
- GPA
Generalized Procrustes Analysis
- GWAS
genome-wide association study
- ICC
intraclass correlation coefficient
- LD
linkage disequilibrium
- LDSC
linkage disequilibrium score regression
- MRI
magnetic resonance imaging
- PGS
polygenic score
- ROC
receiver operation characteristic
- Shh
Sonic hedgehog
- SNP
single nucleotide polymorphism
Supplementary Material
Acknowledgements
This research has been conducted using the UK Biobank Resource under Application Number 23509.
The authors thank the staff and the participants for their valuable contributions to ASPS and ASPS-Fam. We thank Birgit Reinhart for her long-term administrative commitment, Elfi Hofer for the technical assistance at creating the DNA bank, Ing. Johann Semmler and Anita Harb for DNA sequencing and DNA analyses by TaqMan assays and Irmgard Poelzl for supervising the quality management processes after ISO9001 at the biobanking and DNA analyses.
The Medical University of Graz and the Steiermärkische Krankenanstaltengesellschaft support the databank of the ASPS and ASPS-Fam.
The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, MSc, and Carolina Medina-Gomez, MSc, for their help in creating the GWAS database, and Karol Estrada, PhD, Yurii Aulchenko, PhD, and Carolina Medina-Gomez, MSc, for the creation and analysis of imputed data. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists. Conflict of Interest statement. None declared.
Contributor Information
Maria J Knol, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Mikolaj A Pawlak, Department of Neurology and Cerebrovascular Disorders, Poznan University of Medical Sciences Poznan, Poland; Department of Clinical Genetics, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Sander Lamballais, Department of Clinical Genetics, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Natalie Terzikhan, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Edith Hofer, Department of Neurology, Clinical Division of Neurogeriatrics, Medical University Graz, Auenbruggerplatz 22 8036 Graz, Austria; Institute of Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 22 8036 Graz, Austria.
Ziyi Xiong, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands; Department of Genetic Identification, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Caroline C W Klaver, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands; Department of Ophthalmology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Lukas Pirpamer, Department of Neurology, Clinical Division of Neurogeriatrics, Medical University Graz, Auenbruggerplatz 22 8036 Graz, Austria.
Meike W Vernooij, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
M Arfan Ikram, Department of Epidemiology, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Reinhold Schmidt, Department of Neurology, Clinical Division of Neurogeriatrics, Medical University Graz, Auenbruggerplatz 22 8036 Graz, Austria.
Manfred Kayser, Department of Genetic Identification, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Tavia E Evans, Department of Clinical Genetics, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Hieab H H Adams, Department of Clinical Genetics, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands; Department of Radiology and Nuclear Medicine, Erasmus MC University Medical Center Rotterdam, Rotterdam 3015 CE, The Netherlands.
Data and code availability
The genome-wide summary statistics that support the findings of this study will be made available via the NHGRI-EBI GWAS Catalog website (https://www.ebi.ac.uk/gwas/downloads/summarystatistics). The code to derive orbital telorism from structural MRI images is available at https://github.com/Neuroguide/orbitaltelorism.
Funding
ZonMW (grant number 916.19.151 to H.H.H.A.); H2020 (grant 730897 to M.A.P.) “Transnational Access Program for a Pan-European Network of HPC Research Infrastructures and Laboratories for scientific computing.
References
- 1. Dollfus, H. and Verloes, A. (2004) Dysmorphology and the orbital region: a practical clinical approach. Surv. Ophthalmol., 49, 547–561. [DOI] [PubMed] [Google Scholar]
- 2. Hu, D. and Helms, J.A. (1999) The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development, 126, 4873–4884. [DOI] [PubMed] [Google Scholar]
- 3. Echelard, Y., Epstein, D.J., St-Jacques, B., Shen, L., Mohler, J., McMahon, J.A. and McMahon, A.P. (1993) Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell, 75, 1417–1430. [DOI] [PubMed] [Google Scholar]
- 4. Weinberg, S.M., Leslie, E.J., Hecht, J.T., Wehby, G.L., Deleyiannis, F.W.B., Moreno, L.M., Christensen, K. and Marazita, M.L. (2017) Hypertelorism and orofacial clefting revisited: an anthropometric investigation. Cleft Palate Craniofac. J., 54, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Converse, McCarthy, J.G. and Wood-Smith, D. (1975) Orbital hypotelorism. Pathogenesis, associated facio-cerebral anomalies, surgical correction. Plast. Reconstr. Surg., 56, 389–394. [PubMed] [Google Scholar]
- 6. Cohen, M.M., Jr., Richieri-Costa, A., Guion-Almeida, M.L. and Saavedra, D. (1995) Hypertelorism: interorbital growth, measurements, and pathogenetic considerations. Int. J. Oral Maxillofac. Surg., 24, 387–395. [DOI] [PubMed] [Google Scholar]
- 7. Ford, E.H.R. (1958) Growth of the human cranial base. Am. J. Orthod., 44, 498–506. [Google Scholar]
- 8. Im, S.W., Kim, H.J., Lee, M.K., Yi, J.H., Jargal, G., Sung, J., Cho, S.I. and Kim, J.I. (2010) Genome-wide linkage analysis for ocular and nasal anthropometric traits in a Mongolian population. Exp. Mol. Med., 42, 799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raposo-do-Amaral, C.M., Krieger, H., Cabello, P.H. and Beiguelman, B. (1989) Heritability of quantitative orbital traits. Hum. Biol., 61, 551–557. [PubMed] [Google Scholar]
- 10. Paternoster, L., Zhurov, A.I., Toma, A.M., Kemp, J.P., St Pourcain, B., Timpson, N.J., McMahon, G., McArdle, W., Ring, S.M., Smith, G.D.et al. (2012) Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am. J. Hum. Genet., 90, 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu, F., van derLijn, F., Schurmann, C., Zhu, G., Chakravarty, M.M., Hysi, P.G., Wollstein, A., Lao, O., deBruijne, M., Ikram, M.A.et al. (2012) A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet., 8, e1002932–e1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaffer, J.R., Orlova, E., Lee, M.K., Leslie, E.J., Raffensperger, Z.D., Heike, C.L., Cunningham, M.L., Hecht, J.T., Kau, C.H., Nidey, N.L.et al. (2016) Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genet., 12, e1006149–e1006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee, M.K., Shaffer, J.R., Leslie, E.J., Orlova, E., Carlson, J.C., Feingold, E., Marazita, M.L. and Weinberg, S.M. (2017) Genome-wide association study of facial morphology reveals novel associations with FREM1 and PARK2. PLoS One, 12, e0176566–e0176566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cha, S., Lim, J.E., Park, A.Y., Do, J.-H., Lee, S.W., Shin, C., Cho, N.H., Kang, J.-O., Nam, J.M., Kim, J.-S.et al. (2018) Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics, 19, 481–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiao, L., Yang, Y., Fu, P., Hu, S., Zhou, H., Peng, S., Tan, J., Lu, Y., Lou, H., Lu, D.et al. (2018) Genome-wide variants of Eurasian facial shape differentiation and a prospective model of DNA based face prediction. J. Genet. Genomics, 45, 419–432. [DOI] [PubMed] [Google Scholar]
- 16. Wu, W., Zhai, G., Xu, Z., Hou, B., Liu, D., Liu, T., Liu, W. and Ren, F. (2019) Whole-exome sequencing identified four loci influencing craniofacial morphology in northern Han Chinese. Hum. Genet., 138, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Claes, P., Roosenboom, J., White, J.D., Swigut, T., Sero, D., Li, J., Lee, M.K., Zaidi, A., Mattern, B.C., Liebowitz, C.et al. (2018) Genome-wide mapping of global-to-local genetic effects on human facial shape. Nat. Genet., 50, 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong, Z., Dankova, G., Howe, L.J., Lee, M.K., Hysi, P.G., deJong, M.A., Zhu, G., Adhikari, K., Li, D., Li, Y.et al. (2019) Novel genetic loci affecting facial shape variation in humans. elife, 8, e49898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White, J.D., Indencleef, K., Naqvi, S., Eller, R.J., Hoskens, H., Roosenboom, J., Lee, M.K., Li, J., Mohammed, J., Richmond, S.et al. (2021) Insights into the genetic architecture of the human face. Nat. Genet., 53, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sudlow, C., Gallacher, J., Allen, N., Beral, V., Burton, P., Danesh, J., Downey, P., Elliott, P., Green, J., Landray, M.et al. (2015) UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med., 12, e1001779–e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt, R., Lechner, H., Fazekas, F., Niederkorn, K., Reinhart, B., Grieshofer, P., Horner, S., Offenbacher, H., Koch, M. and Eber, B. (1994) Assessment of cerebrovascular risk profiles in healthy persons: definition of research goals and the Austrian stroke prevention study (ASPS). Neuroepidemiology, 13, 308–313. [DOI] [PubMed] [Google Scholar]
- 22. Schmidt, R., Fazekas, F., Kapeller, P., Schmidt, H. and Hartung, H.P. (1999) MRI white matter hyperintensities: three-year follow-up of the Austrian stroke prevention study. Neurology, 53, 132–139. [DOI] [PubMed] [Google Scholar]
- 23. Ikram, M.A., Brusselle, G., Ghanbari, M., Goedegebure, A., Ikram, M.K., Kavousi, M., Kieboom, B.C.T., Klaver, C.C.W., deKnegt, R.J., Luik, A.I.et al. (2020) Objectives, design and main findings until 2020 from the Rotterdam Study. Eur. J. Epidemiol., 35, 483–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ikram, M.A., Brusselle, G.G.O., Murad, S.D., vanDuijn, C.M., Franco, O.H., Goedegebure, A., Klaver, C.C.W., Nijsten, T.E.C., Peeters, R.P., Stricker, B.H.et al. (2017) The Rotterdam Study: 2018 update on objectives, design and main results. Eur. J. Epidemiol., 32, 807–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayhew, A.J. and Meyre, D. (2017) Assessing the heritability of complex traits in humans: methodological challenges and opportunities. Curr. Genomics, 18, 332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pryor, H.B. (1969) Objective measurement of interpupillary distance. Pediatrics, 44, 973–977. [PubMed] [Google Scholar]
- 27. Pivnick, E.K., Rivas, M.L., Tolley, E.A., Smith, S.D. and Presbury, G.J. (1999) Interpupillary distance in a normal black population. Clin. Genet., 55, 182–191. [DOI] [PubMed] [Google Scholar]
- 28. Pickrell, J.K., Berisa, T., Liu, J.Z., Ségurel, L., Tung, J.Y. and Hinds, D.A. (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet., 48, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adhikari, K., Fuentes-Guajardo, M., Quinto-Sánchez, M., Mendoza-Revilla, J., Camilo Chacón-Duque, J., Acuña-Alonzo, V., Jaramillo, C., Arias, W., Lozano, R.B., Pérez, G.M.et al. (2016) A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat. Commun., 7, 11616–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howe, L.J., Lee, M.K., Sharp, G.C., Davey Smith, G., St Pourcain, B., Shaffer, J.R., Ludwig, K.U., Mangold, E., Marazita, M.L., Feingold, E.et al. (2018) Investigating the shared genetics of non-syndromic cleft lip/palate and facial morphology. PLoS Genet., 14, e1007501–e1007501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Indencleef, K., Roosenboom, J., Hoskens, H., White, J.D., Shriver, M.D., Richmond, S., Peeters, H., Feingold, E., Marazita, M.L., Shaffer, J.R.et al. (2018) Six NSCL/P loci show associations with normal-range craniofacial variation. Front. Genet., 9, 502–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richmond, S., Howe, L.J., Lewis, S., Stergiakouli, E. and Zhurov, A. (2018) Facial genetics: a brief overview. Front. Genet., 9, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holland, D., Frei, O., Desikan, R., Fan, C.-C., Shadrin, A.A., Smeland, O.B., Sundar, V.S., Thompson, P., Andreassen, O.A. and Dale, A.M. (2020) Beyond SNP heritability: polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet., 16, e1008612–e1008612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farkas, L.G., Katic, M.J., Forrest, C.R., Alt, K.W., Bagic, I., Baltadjiev, G., Cunha, E., Cvicelová, M., Davies, S., Erasmus, I.et al. (2005) International anthropometric study of facial morphology in various ethnic groups/races. J. Craniofac. Surg., 16, 615–646. [DOI] [PubMed] [Google Scholar]
- 35. Kanehisa, M. and Goto, S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mullin, B.H., Tickner, J., Zhu, K., Kenny, J., Mullin, S., Brown, S.J., Dudbridge, F., Pavlos, N.J., Mocarski, E.S., Walsh, J.P.et al. (2020) Characterisation of genetic regulatory effects for osteoporosis risk variants in human osteoclasts. Genome Biol., 21, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Torkamani, A., Wineinger, N.E. and Topol, E.J. (2018) The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet., 19, 581–590. [DOI] [PubMed] [Google Scholar]
- 38. Avants, B.B., Tustison, N.J., Song, G., Cook, P.A., Klein, A. and Gee, J.C. (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 54, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Almasy, L. and Blangero, J. (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet., 62, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bulik-Sullivan, B.K., Loh, P.R., Finucane, H.K., Ripke, S., Yang, J., Schizophrenia Working Group of the Psychiatric Genomics, C, Patterson, N., Daly, M.J., Price, A.L. and Neale, B.M. (2015) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet., 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Willer, C.J., Li, Y. and Abecasis, G.R. (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26, 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ward, L.D. and Kellis, M. (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res., 44, D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Birnbaum, S., Ludwig, K.U., Reutter, H., Herms, S., Steffens, M., Rubini, M., Baluardo, C., Ferrian, M., Almeida de Assis, N., Alblas, M.A.et al. (2009) Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet., 41, 473–477. [DOI] [PubMed] [Google Scholar]
- 44. Grant, S.F.A., Wang, K., Zhang, H., Glaberson, W., Annaiah, K., Kim, C.E., Bradfield, J.P., Glessner, J.T., Thomas, K.A., Garris, M.et al. (2009) A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediatr., 155, 909–913. [DOI] [PubMed] [Google Scholar]
- 45. Mangold, E., Ludwig, K.U., Birnbaum, S., Baluardo, C., Ferrian, M., Herms, S., Reutter, H., deAssis, N.A., Chawa, T.A., Mattheisen, M.et al. (2010) Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet., 42, 24–26. [DOI] [PubMed] [Google Scholar]
- 46. Beaty, T.H., Murray, J.C., Marazita, M.L., Munger, R.G., Ruczinski, I., Hetmanski, J.B., Liang, K.Y., Wu, T., Murray, T., Fallin, M.D.et al. (2010) A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet., 42, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ludwig, K.U., Mangold, E., Herms, S., Nowak, S., Reutter, H., Paul, A., Becker, J., Herberz, R., AlChawa, T., Nasser, E.et al. (2012) Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet., 44, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun, Y., Huang, Y., Yin, A., Pan, Y., Wang, Y., Wang, C., Du, Y., Wang, M., Lan, F., Hu, Z.et al. (2015) Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun., 6, 6414–6414. [DOI] [PubMed] [Google Scholar]
- 49. Ludwig, K.U., Ahmed, S.T., Böhmer, A.C., Sangani, N.B., Varghese, S., Klamt, J., Schuenke, H., Gültepe, P., Hofmann, A., Rubini, M.et al. (2016) Meta-analysis reveals genome-wide significance at 15q13 for nonsyndromic Clefting of both the lip and the palate, and functional analyses implicate GREM1 as a plausible causative gene. PLoS Genet., 12, e1005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leslie, E.J., Carlson, J.C., Shaffer, J.R., Feingold, E., Wehby, G., Laurie, C.A., Jain, D., Laurie, C.C., Doheny, K.F., McHenry, T.et al. (2016) A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet., 25, 2862–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leslie, E.J., Liu, H., Carlson, J.C., Shaffer, J.R., Feingold, E., Wehby, G., Laurie, C.A., Jain, D., Laurie, C.C., Doheny, K.F.et al. (2016) A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am. J. Hum. Genet., 98, 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leslie, E.J., Carlson, J.C., Shaffer, J.R., Butali, A., Buxó, C.J., Castilla, E.E., Christensen, K., Deleyiannis, F.W., Leigh Field, L., Hecht, J.T.et al. (2017) Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet., 136, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ludwig, K.U., Böhmer, A.C., Bowes, J., Nikolic, M., Ishorst, N., Wyatt, N., Hammond, N.L., Gölz, L., Thieme, F., Barth, S.et al. (2017) Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip ± cleft palate and cleft palate only. Hum. Mol. Genet., 26, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu, Y., Zuo, X., He, M., Gao, J., Fu, Y., Qin, C., Meng, L., Wang, W., Song, Y., Cheng, Y.et al. (2017) Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun., 8, 14364–14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Butali, A., Mossey, P.A., Adeyemo, W.L., Eshete, M.A., Gowans, L.J.J., Busch, T.D., Jain, D., Yu, W., Huan, L., Laurie, C.A.et al. (2019) Genomic analyses in African populations identify novel risk loci for cleft palate. Hum. Mol. Genet., 28, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carlson, J.C., Anand, D., Butali, A., Buxo, C.J., Christensen, K., Deleyiannis, F., Hecht, J.T., Moreno, L.M., Orioli, I.M., Padilla, C.et al. (2019) A systematic genetic analysis and visualization of phenotypic heterogeneity among orofacial cleft GWAS signals. Genet. Epidemiol., 43, 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Rooij, I.A., Ludwig, K.U., Welzenbach, J., Ishorst, N., Thonissen, M., Galesloot, T.E., Ongkosuwito, E., Bergé, S.J., Aldhorae, K., Rojas-Martinez, A.et al. (2019) Non-syndromic cleft lip with or without cleft palate: genome-wide association study in Europeans identifies a suggestive risk locus at 16p12.1 and supports SH3PXD2A as a Clefting susceptibility gene. Genes (Basel), 10, 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A., Bender, D., Maller, J., Sklar, P., deBakker, P.I., Daly, M.J. and Sham, P.C. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Amberger, J.S., Bocchini, C.A., Schiettecatte, F., Scott, A.F. and Hamosh, A. (2015) OMIM.org: online Mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res., 43, D789–D798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Watanabe, K., Taskesen, E., vanBochoven, A. and Posthuma, D. (2017) Functional mapping and annotation of genetic associations with FUMA. Nat. Commun., 8, 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choi, S.W. and O'Reilly, P.F. (2019) PRSice-2: polygenic risk score software for biobank-scale data. Giga Science, 8, giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J.C. and Müller, M. (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics, 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Frei, O., Holland, D., Smeland, O.B., Shadrin, A.A., Fan, C.C., Maeland, S., O'Connell, K.S., Wang, Y., Djurovic, S., Thompson, W.K.et al. (2019) Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun., 10, 2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Willer, C.J., Schmidt, E.M., Sengupta, S., Peloso, G.M., Gustafsson, S., Kanoni, S., Ganna, A., Chen, J., Buchkovich, M.L., Mora, S.et al. (2013) Discovery and refinement of loci associated with lipid levels. Nat. Genet., 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wood, A.R., Esko, T., Yang, J., Vedantam, S., Pers, T.H., Gustafsson, S., Chu, A.Y., Estrada, K., Luan, J., Kutalik, Z.et al. (2014) Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet., 46, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee, J.J., Wedow, R., Okbay, A., Kong, E., Maghzian, O., Zacher, M., Nguyen-Viet, T.A., Bowers, P., Sidorenko, J., Karlsson Linnér, R.et al. (2018) Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet., 50, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome-wide summary statistics that support the findings of this study will be made available via the NHGRI-EBI GWAS Catalog website (https://www.ebi.ac.uk/gwas/downloads/summarystatistics). The code to derive orbital telorism from structural MRI images is available at https://github.com/Neuroguide/orbitaltelorism.