Abstract
It is important to control CRISPR/Cas9 when sufficient editing is obtained. In the current study, rational engineering of guide RNAs (gRNAs) is performed to develop small-molecule-responsive CRISPR/Cas9. For our purpose, the sequence of gRNAs are modified to introduce ligand binding sites based on the rational design of ligand–RNA pairs. Using short target sequences, we demonstrate that the engineered RNA provides an excellent scaffold for binding small molecule ligands. Although the ‘stem–loop 1’ variants of gRNA induced variable cleavage activity for different target sequences, all ‘stem–loop 3’ variants are well tolerated for CRISPR/Cas9. We further demonstrate that this specific ligand–RNA interaction can be utilized for functional control of CRISPR/Cas9 in vitro and in human cells. Moreover, chemogenetic control of gene editing in human cells transfected with all-in-one plasmids encoding Cas9 and designer gRNAs is demonstrated. The strategy may become a general approach for generating switchable RNA or DNA for controlling other biological processes.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is the basis of a revolutionary system for biological studies (1–4). CRISPR systems contain two components: a guide RNA (gRNA) and a CRISPR-associated endonuclease (Cas) (1,2). CRISPR/Cas9 is currently the most powerful gene editing toolkit (5,6). However, it is associated with a variety of adverse effects, some of which may arise from the excessive activities (7,8). Efforts were taken to develop Cas9 variants with improved specificity through protein engineering (9–12). On-demand activation of CRISPR/Cas9 was also developed to reduce the possibility of prolonged activities (13–15). However, once the chemically masked CRISPR/Cas9 is activated in cells, the events of gene editing are still difficult to restrain (16,17). The need of safe gene editing has created the demand for developing CRISPR inhibitors (18,19). The most investigated CRISPR/Cas9 inhibitors are the phage-derived anti-CRISPR proteins (20–23). However, it remains challenging to develop the use of either anti-CRISPR or small-molecule inhibitor of Cas9 due to the limited target specificity (24).
For CRISPR/Cas9, the gRNA forms a functional complex with the Cas9 (25,26). The function of gRNAs is determined by their specific secondary and tertiary structures (25). These higher order structures are important for the function of CRISPR/Cas9. We therefore consider to develop a RNA-targeting strategy by utilizing ligand–RNA interactions, which may be potentially achieved using covalent labeling of either the ligand or RNA (27,28). However, there were concerns about inevitable effects of chemical modifications on RNA folding, activity, and safety as compared with natural RNAs in living cells. It is also not easy to genetically encode any chemical modification into RNAs. Recently, a number of mismatch binding ligands (MBLs) were developed to target noncanonical base-pairs in nucleic acids (29–32). These compounds usually contain aromatic components with ability to form complementary hydrogen bonds with the nucleobase (33,34). One type of such aromatic moieties is a 1,8-naphthyridine ring containing two imino groups, which favors guanine as a binding partner (35,36). MBLs were demonstrated to induce nucleotide flip-out in nucleic acids (37,38). As a result, we envision that the development of small-molecule-responsive CRISPR/Cas9 can benefit from the sequence modification of gRNAs, which may provide specific ligand-binding sites.
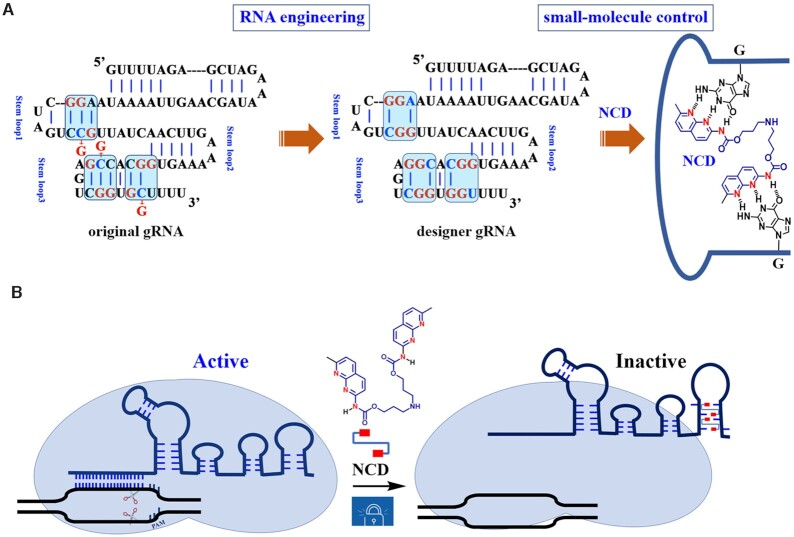
In the current study, rational engineering of gRNAs is performed to create and develop small-molecule-responsive CRISPR/Cas9. For our purpose, the sequence of gRNAs are modified to introduce ligand binding sites based on the rational design of ligand–RNA pairs (Figure 1A). We focus on a naphthyridine carbamate dimer (NCD), which consists of two units of a naphthyridine carbamate connected by a flexible linker (Figure 1A). Using short target sequences, we demonstrate that NCD recognizes MBL-binding units in those engineered RNA stem–loops. We further demonstrate that this specific ligand–RNA interaction can be utilized for functional control of CRISPR/Cas9 in vitro and in human cells (Figure 1B). Moreover, chemogenetic control of gene editing in human cells transfected with all-in-one plasmids encoding Cas9 and designer gRNAs is demonstrated. The strategy presented here may become a general approach for generating switchable RNA or DNA for controlling other biological processes.
Figure 1.
Schematic illustration of the design and workflow. (A) Engineering guide RNA to make it responsive to small molecules. The gRNA scaffold contains multiple well-defined structural motifs, including a repeat:anti-repeat duplex and three stem–loops. NCD and its hydrogen-bonding pattern to the introduced G-G mismatch sites are demonstrated. The structural unit of N-acyl-2-amino-1,8-naphthyridine contains three hydrogen-bonding groups that are fully complementary to that of G. (B) Illustration of interference of Cas9-mediated DNA cleavage by RNA-binding small molecules. The Cas9 complex with designer gRNAs still retains wild-type levels of activity, while the exposure of this complex to MBLs leads to evident inhibition of DNA cutting activities. The protospacer adjacent motif (PAM) is indicated. Red rectangles: 2-amino-1,8-naphthyridine.
MATERIALS AND METHODS
Materials
The oligonucleotides at HPLC purity were obtained from TaKaRa company (Dalian, China). Cas9 nuclease, Streptococcus pyogenes (product# M0646), Bst DNA pol, Large Fragment (product# M0275), T4 DNA Ligase (product# M0202S), Ribonucleotide solution mix (NTPs, product# N0450) and deoxy-ribonucleoside triphosphates (dNTPs, product# N0446) were purchased from New England Biolabs (Ipswich, MA, USA). Transcript Aid T7 High Yield Transcription kit (product# K0441) and Lipofectamine 3000 transfection agent (product# L3000015) were purchased from Thermo Fisher Scientific Inc. Pyrobest™ DNA Polymerase (product# R005A) and PrimeSTAR HS DNA Polymerase (product#R010A) was purchased from TaKaRa Shuzo Co. Ltd. (Tokyo, Japan). DNeasy Blood & Tissue kit (product# 69504) was purchased from Qiagen (Germany). The nucleic acid stains Super GelRed (NO.: S-2001) was purchased from US Everbright Inc. (Suzhou, China). The pH was measured with Mettler Toledo, FE20-Five Easy™ pH (Mettler Toledo, Switzerland). The concentration of nucleic acids was quantified by NanoDrop 2000c (Thermo Scientific, USA). The gel electrophoresis was run in a temperature-controlled vertical electrophoretic apparatus (DYCZ-22A, Liuyi Instrument Factory, Beijing, China). Gel Imaging was performed using Pharos FX Molecular imager (Bio-Rad, USA). The Discovery Series Quantity One 1D Analysis Software Version 4.6.9 was used to determine the percentage of indel formation. The UV melting studies were performed on a Jasco-810 spectropolarimeter (Jasco, Easton, MD, USA) equipped with a Peltier temperature controller.
Synthesis
The NCD and Z-NCTS were synthesized according to previous reports (39).
Construction of designer gRNAs with MBL-binding units
In vitro transcription reactions were performed to prepare each sgRNA and tracrRNA. Point mutations were introduced at nts 41, 66 and 76, respectively (GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT to GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCGGTTATCAACTTGAAAAAGTGGCACGGAGTCGGTGGTTTT). An overlap extension is performed to assemble smaller DNA fragments into a larger sequence, which was used for making each sgRNA and tracrRNA (15). For sgRNA preparation, the forward primer was designed to contain T7 promoter, variable guide sequence, the first 31 nt of the conserved region of the sgRNA scaffold (sg-SLX4IP-F, sg-HPRT1-F, sg-GFP-F or sg-HBEGF-F). And the reverse primer was designed to contain the reverse complement of the conserved region of the sgRNA scaffold (sgRNA-R, sgRNA-S1a-R, sgRNA-S1b-R, sgRNA-S1c-R, sgRNA-S2a-R, sgRNA-S2b-R, sgRNA-S2c-R or sgRNA-S3-R in Supplementary Table S1). Pyrobest™ DNA Polymerase was used for overlap extension reaction. Assembled template (300 ng) was used as a substrate for in vitro transcription by T7 RNA polymerase, using the Transcript Aid T7 High Yield Transcription kit following the manufacturer's instructions. Resulting transcription reactions were treated with DNase I, and RNA was purified using the NaOAc/phenol/chloroform method.
For tracrRNA preparation, the forward primer was designed to contain T7 promoter, the first 46 nt of the tracrRNA scaffold (tracrRNA-F). And a 66 nt primer was designed to contain the reverse complement of the tracrRNA scaffold (tracrRNA-R, tracrRNA-S1a-R, tracrRNA-S1b-R, tracrRNA-S1c-R, tracrRNA-S2a-R, tracrRNA-S2b-R, tracrRNA-S2c-R or tracrRNA-S3-R in Supplementary Table S1). For overlap extension and in vitro transcription reaction, the conditions were the same as the above sgRNA protocol.
Ligand control of designer sgRNAs for CRISPR/Cas9
In vitro Cas9 cleavage assay was performed in 1 × NEBuffer™ 3.1, which contained 100 mM NaCl, 50 mM Tris–HCl, 10 mM MgCl2 and 100 μg/ml BSA at pH 7.9 @ 25°C. Target GFP fragments were PCR amplified from pEGFP-C1 vector (Clontech) using t-GFP-F and t-GFP-R as primers. Other target DNA fragments were PCR amplified from genomic DNA (HeLa cells) using the following primers: t-SLX4IP-F and t-SLX4IP-R for t-SLX4IP; t-HPRT1-F and t-HPRT1-R for t-HPRT1; t-HBEGF-F and t-HBEGF-R for t-HBEGF. Briefly, original or designer sgRNAs (50 ng) were incubated with each MBL at various concentrations in the presence of Cas9 (0.2 μM), in 1 × NEBuffer™ 3.1 buffer at room temperature for 30 min. The Cas9-mediated DNA cleavage reactions were started by rapid mixing of equal volumes of the sgRNA/Cas9 preparation with a solution containing target DNA fragments (50 ng) in 1 × NEBuffer™ 3.1 buffer, and subjected to an incubation at 37°C for different incubation times. Reactions were quenched by adding SDS containing loading dye and loaded onto a 1.5% agarose gel containing 1.5 × Super GelRed for visualization (100 V, 1.5 h). The GeneRuler 100-bp DNA Ladder was used as DNA size marker. After electrophoresis, in gel targets were analyzed using a Pharos FX Molecular imager (Bio-Rad, USA).
Ligand control of designer tracrRNAs for CRISPR/Cas9
In vitro Cas9 cleavage assay was performed in 1 × NEBuffer™ 3.1. For the 2-part gRNA study, crRNA and each designer tracrRNA were annealed at room temperature for 10 min to form the gRNA complexes. For the MBL incubation step, similar reactions were set up using 15 ng of each crRNA and 25 ng of tracrRNA instead of 50 ng of each sgRNA. For Cas9-mediated DNA cleavage reactions, the conditions were the same as the above sgRNA protocol. The original tracrRNAs were used in control reactions.
Ligand control of designer sgRNAs in a stable Cas9-expressing cell line
One day prior to transfection, 4 × 105 HeLa-OC cells per well were seeded into a 6-well plate containing 0.5 ml medium. HeLa-OC cells were transfected with 2.5 μg each sgRNA using a standard transfection protocol. Each sgRNA was transfected as biological triplicates in three separate wells and processed independently. In brief, each sgRNA was mixed in a 125-μl DMEM. Thereafter, an equal volume of DMEM with Lipofectamine 3000 transfection agent (5.0 μl) was added and incubated for 15 min. The complex was subsequently added dropwise to the cells. After an incubation time of 4 h, the medium was changed to complete medium supplemented with appropriate concentrations of NCD. Cells were further cultured at 37°C in 5% CO2 for an additional 24 h.
Cell cultures were washed with PBS prior to DNA extraction. The genomic DNA was extracted using Qiagen DNeasy Blood and Tissue Kit following the manufacturer's protocol. PCR across the target site in different genes was run using the according amplicon primer set. All PCRs were performed using PrimeSTAR HS DNA Polymerase. PCR was performed in solution with a final volume of 20 μl, containing 0.5 μM of each target region amplification primer. The cycling conditions are as following: initial denaturation at 94°C for 15 s; 35 cycles consisting of 10 s of denaturation at 98°C, 1 min of annealing and extension at 68°C. The amplified DNA products were purified via Zymo Research DNA Clean and Concentrator Kit.
The cleavage activity of CRISPR/Cas9 at endogenous loci was quantified using T7E1 assay. All subsequent steps were performed independently for the triplicates. 100 ng of PCR product carrying the target loci was heated to 98°C and slowly cooled down to let DNA reanneal. Annealed DNA was digested with T7 endonuclease I (NEB) for 30 min at 37°C. Reaction was quenched by adding SDS containing loading dye and loaded onto a 1.5% agarose gel containing 1.5 × Super GelRed for visualization (100 V, 1.5 h). The percentage of cleavage was quantified using a Pharos FX Molecular imager (Bio-Rad, USA) running Image Lab™ 4.1. The percentage of indel formation = 100 × (1 – (1 – fraction cleaved)0.5).
Ligand-control of the hybrid system in human cells
PX165 (pSpCas9) was obtained from Addgene (plasmid# 48137) (5). One day prior to transfection, 4 × 105 HeLa cells per well were seeded into a six-well plate containing 0.5 ml medium. HeLa cells were transfected with 2.5 μg PX165 plasmid using a standard transfection protocol. In brief, PX165 plasmid was mixed in a 125-μl DMEM. Thereafter, an equal volume of DMEM with Lipofectamine 3000 transfection agent (5.0 μl) was added and incubated for 15 min. The complex was subsequently added dropwise to the cells. After 4-h of incubation, cells were treated with Lipofectamine 3000 loaded with endogenous gene targeting sgRNAs (original or designer) for 4 h. The sgRNA dose was 2.5 μg/well each sgRNA. After an incubation time of 4 h, the medium was changed to complete medium supplemented with appropriate concentrations of NCD. Cells were further cultured at 37°C in 5% CO2 for an additional 24 h. The target editing efficiency and inhibition thereof were quantified by T7E1 assay. Data were normalized on samples without NCD treatment.
Construction of designer plasmids with MBL-binding units
PX459 (pSpCas9(BB)-2A-Puro V2.0) was obtained from Addgene (Plasmid #62988) (5). A starting PX459 plasmid was used to construct the designer plasmid with MBL-binding units (GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT was changed to GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACGGAGTCGGTGGTTTT). Point mutations were created through overlap extension PCR of the region between unique restriction sites (PciI/SnaBI) of PX459, and two primer pairs [forward primer in upstream region (S2c-up-F), reverse primer in upstream region (S2c-up-R); forward primer in downstream region (S2c-down-F), reverse primer in downstream region (S2c-down-R)] were used. The two purified fragments were used in the overlap extension PCR to produce the fused fragment containing MBL-binding units. The expected sizes for amplicons were as follows: 376 bp for S2c-up (upstream region); 403 bp for S2c-down (downstream region); 749 bp for the fused fragment. Assembled PCR products were amplified with Pyrobest™ DNA Polymerase using forward and reverse oligonucleotides (S2c-up-F and S2c-down-R) with homology upstream or downstream of the PciI and SnaBI restriction sites, respectively. PX459 plasmid was digested with PciI/SnaBI to remove the original sgRNA cassette. The assembled PCR products and the digested plasmids were assembled with the in-fusion cloning method to produce the target plasmid PX459-S2c. Constructed plasmids were sequenced to confirm the replaced strand region using the primer (5′-CTTTTGCTGGCCTTTTGCTCA-3′).
Forward and reverse oligonucleotides containing the guide sequences for SLX4IP and HBEGF with appropriate overhangs were annealed by mixing 100 pmol of each pair and then incubated at 90°C for 5 min and slowly ramped to room temperature. The PX459 or PX459-S2c plasmid was digested using restriction enzyme BbsI and run on a 0.8% agarose gel. The sample was cut out and purified. The digested plasmid was ligated with the annealed oligonucleotide to generate the designer construct. Correct insertion of target sequences into the vectors was confirmed by Sanger sequencing using the following primer: GAGGGCCTATTTCCCATGATT.
Ligand control of designer all-in-one plasmid with MBL-binding units in human cells
HeLa cells were seeded in a six-well plate 24 h prior to transfection. Unless stated otherwise, 4 × 105 HeLa cells were transfected with 2.5 μg of each complete plasmid using 5 μl lipofectamine 3000, following manufacturer's instructions. Each complete plasmid was transfected as biological triplicates in three separate transfections. After 4 h of transfection, the medium of the ‘control groups’ and the ‘experimental groups’ was replaced by the same medium, but in the ‘experimental groups’ it contained different concentrations of NCD. Then, the groups of cells were incubated for another 24 h. The cells were then lysed and genomic DNA was extracted and PCR-amplified around the target locus. Finally, the T7E1 assay was performed for measuring endogenous gene editing. Data were normalized on samples without NCD treatment.
Statistical analysis
Statistical significance was determined with an unpaired, two-tailed Student's t-test with Origin software (OriginLab Corporation, Northampton, MA). P values <0.05 were considered statistically significant.
RESULTS
Rational gRNA engineering for switching CRISPR/Cas9
The goal of this study is to develop a small molecule strategy to control CRISPR/Cas9. The methods toward modulating the RNA structure in response to external stimuli have drawn our interests. However, the rational design of small molecules targeting the higher order structures in gRNA is not well developed (40,41). We envision that rational engineering of the sequence of gRNAs can be utilized. We recognized that some privileged RNA motif-small molecule pairs were defined (42–44). It is noteworthy that some small-molecule ligands can recognize the mismatched base pairs and further lock DNA and RNA into specific structures (45,46). Previous studies demonstrated that some small-molecule ligands have a remarkable ability to bind with specific triads in the duplex form (5′-HGG-3′/5′-HGG-3′ or 5′-GGH-3′/5′-GGH-3′; H = A, C or T) (29,37). In these triads, two NCD molecules bound to four guanines by disrupting hydrogen bonding of two H-G base pairs and a G-G mismatch. We envision that this property can be utilized for modulating the structure of gRNAs and their interaction with Cas9.
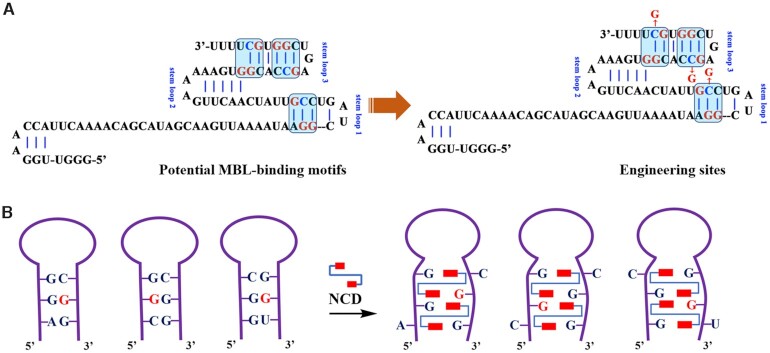
The most commonly used Cas9 nuclease is the one originating from S. pyogenes (1,47). The gRNA for this enzyme is made up of two parts: CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA). While crRNAs and tracrRNAs exist as two separate RNA molecules in nature, they can be combined as a single RNA molecule (sgRNA) (1,2). In this respect, we tend to explore ligand-induced inactivation of tracrRNA and sgRNA. Since MBL binding is favored at the specific triads, it is important to identify potential MBL-binding motifs in gRNAs. However, there is no such typical motif in gRNA scaffolds. A potential alternative way to construct RNA binders is through rational design using information about the RNA motifs ligands prefer to bind. We therefore conducted sequence and structure analysis of gRNA scaffolds to identify potential units with close resemblance with typical MBL-binding motifs. We have identified a total of three units with potential for sequence engineering (Figures 1A and 2A). Engineering of these units holds the promise of reducing much disturbance to the original structure. According to the design of this strategy, the workflow would proceed in two steps. For the first step, specific sequence alterations are designed to introduce MBL-binding motifs into gRNA scaffolds (sgRNA and tracrRNA). At this point, the sequence engineering is desired not affect their structure and activity dramatically. Secondly, MBLs are used to introduce structural constraints into gRNAs (Figure 2B).
Figure 2.
Rational gRNA engineering for switching CRISPR/Cas9. (A) Sequence and structure analysis of gRNA scaffold (tracrRNA demonstration). We have identified a total of three motifs with close resemblance with the units possessing affinity for MBL binding. (B) MBLs are used to introduce structural constraints into gRNAs through hydrogen bonding and stacking. Sequence modification sites are indicated in red. Red rectangles: 2-amino-1,8-naphthyridine.
Selective binding of MBL to designer model RNAs
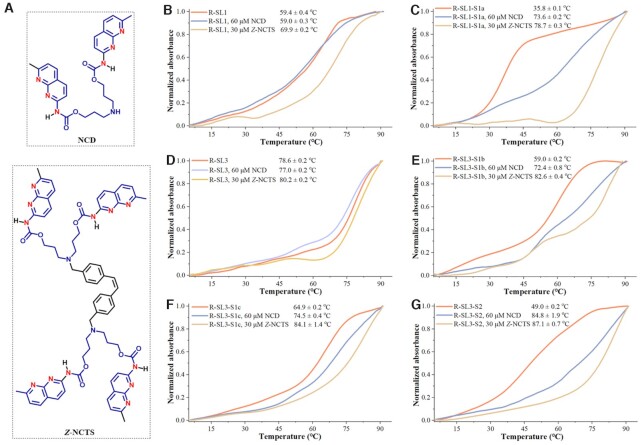
The above structural analysis of gRNA scaffold demonstrates several candidate units for engineering (r(AGG)/r(CCG) in stem–loop 1, r(CCG)/r(CGG) and r(GGC)/r(GCU) in stem–loop 3). Considering the highly polymorphic nature of the gRNA sequence, two model sequences containing these units were examined first (R-SL1, R-SL3 in Supplementary Table S1). We generated a series of variants (R-SL1-S1a, R-SL3-S1b, R-SL3-S1c and R-SL3-S2 in Supplementary Table S1). From the structural point of view, the R-SL1-S1a was stem–loop 1-engineered sequence, and the other sequences (R-SL3-S1b, R-SL3-S1c and R-SL3-S2) were stem–loop 3-engineered sequences. The R-SL1-S1a, R-SL3-S1b, R-SL3-S1c have only one potential MBL-binding sites, whereas R-SL3-S2 were designed to form a MBL-dependent structure containing two of these sites existing in proximity. To address the MBL-binding mechanism, here we focus on two well-known MBLs, i.e. NCD and Z-NCTS (structures in Figure 3A). The synthesis of NCD and Z-NCTS is performed according to previous literatures (Scheme 1 and Scheme 2 in Supporting information).
Figure 3.
Ligand-induced changes of melting temperature of wild-type and designer model RNAs Experiments were performed as described in the Experimental Section. All measurements were performed in three biological replicates. The average of three data sets of denaturation profiles was used for the plots. (A) The structural demonstration of NCD and Z-NCTS. (B–G) Thermal melting profiles of R-SL1 (B), R-SL1-S1a (C), R-SL3 (D), R-SL3-S1b (E), R-SL3-S1c (F), R-SL3-S2 (G) in the absence or presence of each MBL. The melting curve was measured by observing the absorbance of each model RNA at 260 nm in a Tris–HCl buffer (10 mM, pH 7.4) containing 50 mM NaCl. All normalized melting curves were temperature-overlaid for illustration purposes.
Circular Dichroism (CD) study is used to quickly investigate the binding ability of each MBL to these designer model sequences (35). To this aim, thermal melting temperatures (Tm) of each model RNA in the absence and presence of each MBL were examined for comparison (29). We first evaluated the binding of each MBL to the model sequences originated from stem–loop 1. The original sequence (R-SL1) produced a stem–loop containing an AGG/CCG sequence with a Tm of 59.4°C in 50 mM NaCl buffer. Further results demonstrate that a decrease in Tm (∼0.4°C) was detectable after incubation of this wild-type RNA with 60 μM NCD (Figure 3B). In contrast, an increase in the Tm (ΔTm, 10.5°C) was observed for the same sequence in the presence of a lower concentration of Z-NCTS (30 μM). This phenomenon may be due to the unintended binding of Z-NCTS to guanine-rich sites of R-SL1. The binding of each MBL was next investigated for R-SL1-S1a where one cytosine of the AGG/CCG sequence was substituted with a guanine. Thermal melting profiles of R-SL1-S1a showed the presence of stable secondary structure only in the presence of MBLs (Figure 3C). These results indicate that a single base alteration in this model sequence renders a binding pocket for each MBL.
We next evaluated the binding of each MBL to model sequences originated from stem–loop 3. At the employed conditions (10 mM Tris–HCl at pH 7.4, 50 mM NaCl), the fully matched R-SL3 had a melting temperature of 78.6°C (Figure 3D), whereas the mismatch-containing sequences R-SL3-S1b, R-SL3-S1c and R-SL3-S2 denaturated at significantly lower temperatures (59.0, 64.9 and 49.0°C in Figure 3E–G). In the presence of Z-NCTS (30 μM) or NCD (60 μM), small changes in Tm was observed for R-SL3 (ΔTm = –1.6, 1.6°C, respectively), which may be attributed to a certain extent of non-specific binding (Figure 3D). Notably, the structure of mismatch-containing variants were preferentially stabilized with the tested MBLs. The MBL-induced stabilization effect was more pronounced for multi-nucleotide variants (R-SL3-S2) than it was for single-site variants (R-SL3-S1b and R-SL3-S1c). The largest ΔTm of 38.1°C was observed for the Z-NCTS-treated R-SL3-S2, which exhibited the highest Tm of 87.1°C among the ‘stem–loop 3’ variants (Figure 3G). Under the reaction conditions of these experiments, the MALDI-TOF Mass measurements also indicate that the binding of NCD is specific to the designer RNAs with MBL-binding units (Supplementary Figures S1 and S2). On the basis of these results, both NCD and Z-NCTS strongly stabilize the structure of mismatch-containing variants, with NCD showing a better selectivity.
Ligand control of designer sgRNAs for switching CRISPR/Cas9
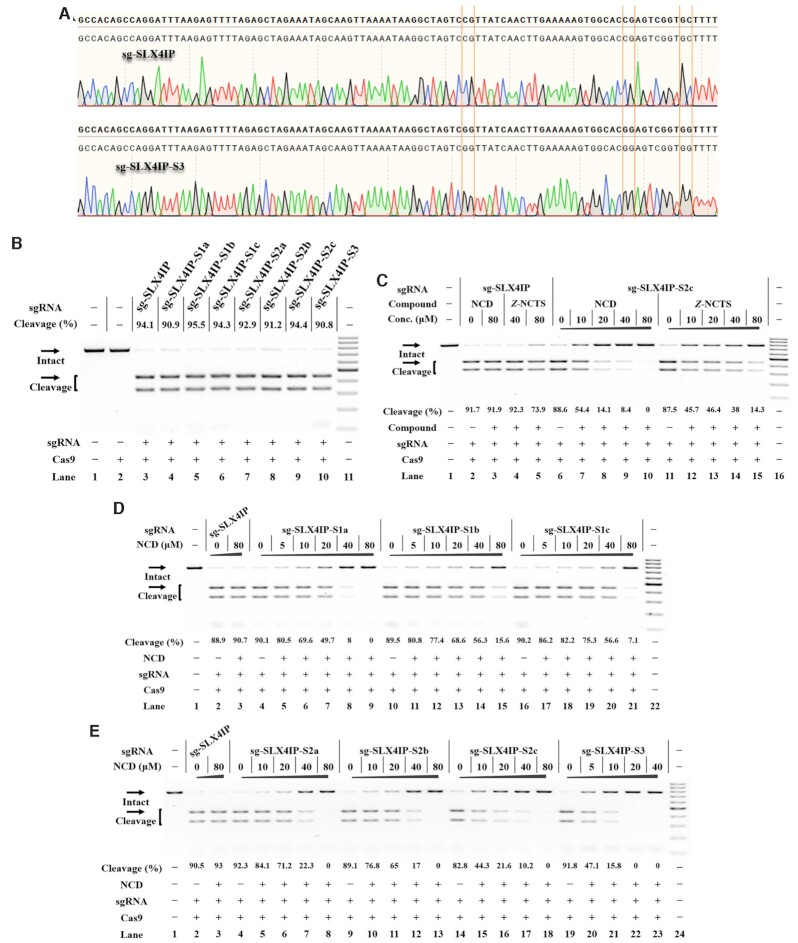
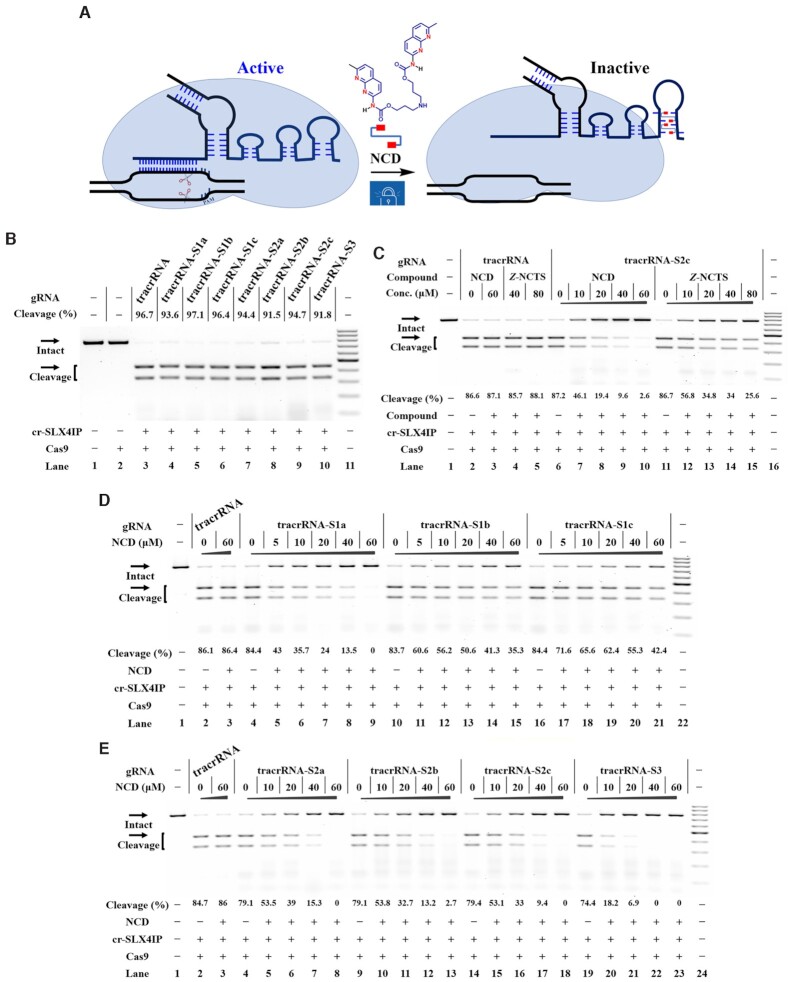
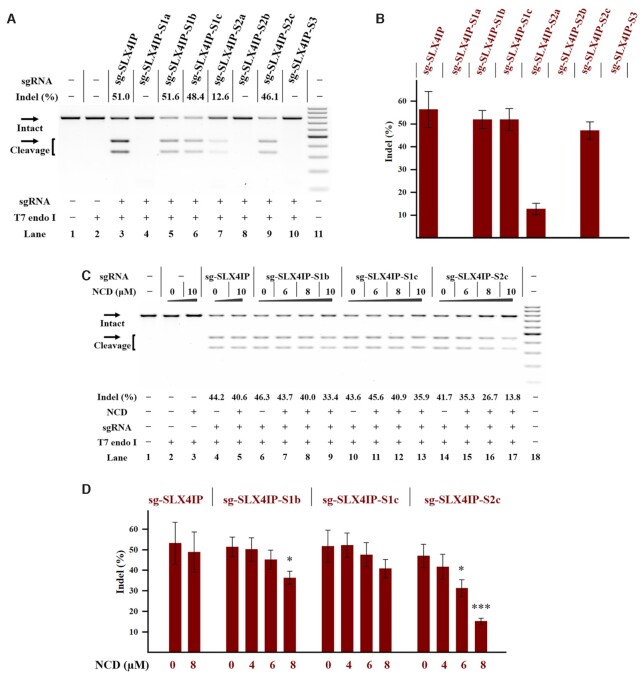
The above studies demonstrate that each MBL exhibits strong interactions with designer model RNAs. We were encouraged to investigate whether such a chemistry-based approach can be used to control CRISPR/Cas9. Currently, the sgRNA represents the most popular format for CRISPR/Cas9 (1,48). To determine the universal effectiveness of our strategy, we selected a total of 4 sgRNAs with different spacer sequences as model systems (sg-SLX4IP, sg-HPRT1, sg-HBEGF, sg-GFP in Supplementary Table S1) (15,49–51). As a first example, the target SLX4IP DNA was amplified by PCR (polymerase chain reaction) from human genomic DNA (the SLX4 interacting protein, SLX4IP target in Supplementary Figure S3A). The sg-SLX4IP was designed to target the selected positions within the SLX4IP DNAs (50). We systematically designed and generated a series of designer sgRNAs (sg-SLX4IP-S1a, sg-SLX4IP-S1b, sg-SLX4IP-S1c, sg-SLX4IP-S2a, sg-SLX4IP-S2b, sg-SLX4IP-S2c and sg-SLX4IP-S3). The sg-SLX4IP-S1a, sg-SLX4IP-S1b and sg-SLX4IP-S1c had only one potential MBL-binding site, whereas sg-SLX4IP-S2a, sg-SLX4IP-S2b and sg-SLX4IP-S2c were designed to form a MBL-dependent structure containing two of these sites. Moreover, the sg-SLX4IP-S3 was designed to have three of these sites (Figure 1A). Sanger sequencing was performed to confirm the sequence of each designer sgRNA. In this study, each in vitro-transcribed (IVT) sgRNA was reverse transcribed, PCR amplified and then cloned into a T-vector for sequencing analysis. The results confirmed the correct engineering of sgRNAs (Figure 4A, Supplementary Figure S4).
Figure 4.
Ligand control of designer sgRNAs for switching CRISPR/Cas9 Reactions were performed as described in the Experimental Section. Uncleaved SLX4IP DNA (773 bp) cut to shorter cleavage fragments (441 bp and 332 bp) are demonstrated. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Sanger sequencing analysis of selected sgRNAs. The sites for sequence modification are indicated. (B) The tolerance of Cas9 to each designer sgRNA. Lane 1: target control; lane 2: Cas9-only control; lane 3 contains original sg-SLX4IP; lanes 4–10 contain designer sgRNAs harboring different MBL-binding units; lane 11: DNA marker (GeneRuler 100-bp DNA Ladder). (C) Responsiveness of designer sgRNAs to different MBLs. Lane 1: no Cas9 control; lanes 2–5 contain original sg-SLX4IP; lanes 6–10, 11–15 contain sg-SLX4IP-S2c; lane 16: DNA marker. (D) The NCD-dependent inhibition of CRISPR/Cas9 with single-site variants. Lane 1: no Cas9 control; lanes 2–3 contain original sg-SLX4IP; lanes 4–9 contain sg-SLX4IP-S1a; lanes 10–15 contain sg-SLX4IP-S1b; lanes 16–21 contain sg-SLX4IP-S1c; lane 22: DNA marker. (E) The NCD-dependent inhibition of CRISPR/Cas9 with multi-nucleotide variants. Lane 1: no Cas9 control; lanes 2–3 contain original sg-SLX4IP; lanes 4–8 contain sg-SLX4IP-S2a; lanes 9–13 contain sg-SLX4IP-S2b; lanes 14–18 contain sg-SLX4IP-S2c; lanes 19–23 contain sg-SLX4IP-S3; lane 24: DNA marker.
Before we investigate the effect of MBL on functional control of sgRNA, it is important to determine if Cas9 is tolerant of the desired sequence modifications in sgRNAs. We therefore conducted a parallel analysis to determine the function of these variants of sg-SLX4IP to support the Cas9-mediated DNA cleavage. When designer sgRNAs with single-site modification were used (sg-SLX4IP-S1a, sg-SLX4IP-S1b, sg-SLX4IP-S1c), the Cas9 cleavage was performed with an efficiency comparable to that observed for original sgRNA (Figure 4B). Besides single-site variants, Cas9 enzymes are demonstrated to tolerate more divergent sequences, such as sg-SLX4IP-S2a, sg-SLX4IP-S2b, sg-SLX4IP-S2c and even sg-SLX4IP-S3. Based on these results, all tested nucleotides (C41, C66 and C76) in the scaffold of sgRNAs targeting SLX4IP are highly tolerant to modification.
More studies were performed to determine if these criteria are common for sgRNAs with different target sequences (Supplementary Figure S3B–D). Different variants of another three sgRNAs were used (sequencing results in Supplementary Figures S5–S7), and their cleavage activities were studied. We found that there were major performance differences among the variants of sgRNAs with different target sequences. Like sg-SLX4IP, all variants of sg-HPRT1 and sg-GFP can be efficiently used to support Cas9-mediated DNA cleavage (Supplementary Figures S8A, B). However, for sg-HBEGF, when nucleotide C41 was mutated to G at stem–loop 1, the cleavage efficacy was decreased significantly relative to that of the original sg-HBEGF (sg-HBEGF-S1a, sg-HBEGF-S2a, sg-HBEGF-S2b and sg-HBEGF-S3 in Supplementary Table S1, results in Supplementary Figure S8C). In contrast, all ‘stem–loop 3’ variants of sg-HBEGF have activities comparable to that of the original sgRNA (sg-HBEGF-S1b, sg-HBEGF-S1c and sg-HBEGF-S2c). These data suggest that Cas9 is more tolerant to sequence variation in the stem–loop 3 compared to the stem–loop 1. We suggest the reason could be that the stability of stem–loop 3 with six base pairs is higher than that of the stem–loop 1 with only four base pairs. Hence, the stem–loop 3 of sgRNA scaffold can accomodate more sequence variations without affecting its original structure.
We next investigate the responsiveness of each designer sgRNA to two compounds of particular interest (NCD and Z-NCTS) for their potential to bind with MBL-binding units. In this study, designer sgRNA/Cas9 complexes were exposed to different concentrations of NCD or Z-NCTS. We demonstrated that NCD and Z-NCTS could each efficiently inhibit the function of designer sgRNAs to support Cas9-mediated DNA cleavage (Figure 4C, Supplementary Figure S9). It is interesting that NCD appears slightly more potent, inhibiting all designer sgRNAs at the concentration of 80 μM. These conclusions held true for three additional studies using another three sgRNAs with different target sequences, suggesting a general principle (Supplementary Figures S10-S12). The activity of original sgRNA was not significantly affected by addition of NCD, while Z-NCTS becomes nonspecific at concentrations higher than 40 μM. These results are basically consistent with the above observation that NCD generally exhibits a weaker interaction with wild-type model RNAs compared to Z-NCTS (Figure 3B). Hence, NCD is more specific at inhibiting CRISPR/Cas9 with designer sgRNAs than Z-NCTS does.
We next compared the responsiveness of each single-site variant to NCD at different concentrations. Notably, the ‘stem–loop 1’ variant of sgRNA was preferentially targeted with NCD. Taken the guide sequence of SLX4IP as an example, an almost total inhibition of DNA cleavage was observed for sg-SLX4IP-S1a when the ligand concentration was increased to 40 μM. However, the inhibitory effect of NCD on two ‘stem–loop 3’ variants was less pronounced than it was for sg-SLX4IP-S1a (Figure 4D). This result is basically consistent with the greater stabilization of the MBL-binding structure in the ‘stem–loop 1’ variant versus each ‘stem–loop 3’ variant (Figure 3C, E, F). We further studied the responsiveness of each multi-nucleotide variant to NCD. The inhibitory effect of NCD on Cas9-mediated DNA cleavage was more pronounced for multi-nucleotide variants than it was for single-site variants (Figure 4D, E). This is consistent with the above melting study that the MBL-induced stabilization effect was more pronounced for multi-nucleotide variants than it was for single-site variants (Figure 3E–G). Additional results demonstrate a general principle that is more widely applicable (Supplementary Figures S13–S15). Hence, the interaction between ligand and the MBL-binding unit of designer sgRNAs provides a handle for controlling CRISPR/Cas9.
Ligand control of designer tracrRNAs for switching CRISPR/Cas9
In the above studies, Cas9 was guided with the form of sgRNA. Unlike with the sgRNA, the tracrRNA does not need to be changed for every unique target sequence (52). It is useful to have the form of 2-part gRNA (crRNA:tracrRNA) as a result of its facile modularity. We next determined the efficieny of our strategy to control the function of designer tracrRNAs (Figure 5A). In this study, IVT tracrRNAs and chemically synthesized crRNAs were used to guide DNA cleavage. In parallel to designer sgRNAs, a panel of designer tracrRNAs were created by introducing single and multiple variations on the stem–loop 1 and/or stem–loop 3 (tracrRNA-S1a-c, tracrRNA-S2a-c, and tracrRNA-S3 in Supplementary Table S1). From the structural point of view, the tracrRNA2a, tracrRNA2b and tracrRNA3 contain multiple MBL-binding units existing in proximity at both stem–loop 1 and stem–loop 3. Successful construction of each designer tracrRNA was confirmed by Sanger sequencing (Supplementary Figure S16).
Figure 5.
Ligand control of designer tracrRNAs for switching CRISPR/Cas9 Reactions were performed as described in the Experimental Section. All samples were tested in three biological replicates. Image of representative data was shown here. (A) Illustration of MBL-responsive CRISPR/Cas9 with designer tracrRNAs. Red rectangles: 2-amino-1,8-naphthyridine. (B) The tolerance of Cas9 to each designer tracrRNA. Lane 1: target control; lane 2: Cas9-only control; lane 3 contains cr-SLX4IP and original tracrRNA; lanes 4–10 contain cr-SLX4IP and designer tracrRNAs harboring different MBL-binding units; lane 11: DNA marker (GeneRuler 100-bp DNA Ladder). (C) Dose-dependent response of the indicated tracrRNAs to each MBL. Lane 1: no Cas9 control; lanes 2–5 contain cr-SLX4IP and original tracrRNA; lanes 6–10, 11–15 contain cr-SLX4IP and tracrRNA-S2c; lane 16: DNA marker. (D) Effects of NCD on the function of tracrRNA and its single-site variants. Lane 1: no Cas9 control; lanes 2–3 contain cr-SLX4IP and original tracrRNA; lanes 4–9 contain cr-SLX4IP and tracrRNA-S1a; lanes 10–15 contain cr-SLX4IP and tracrRNA-S1b; lanes 16–21 contain cr-SLX4IP and tracrRNA-S1c; lane 22: DNA marker. (E) Effects of NCD on the function of tracrRNA and its multi-nucleotide variants. Lane 1: no Cas9 control; lanes 2–3 contain cr-SLX4IP and original tracrRNA; lanes 4–8 contain cr-SLX4IP and tracrRNA-S2a; lanes 9–13 contain cr-SLX4IP and tracrRNA-S2b; lanes 14–18 contain cr-SLX4IP and tracrRNA-S2c; lanes 19–23 contain cr-SLX4IP and tracrRNA-S3; lane 24: DNA marker.
As the starting point, it is desirable to modify the tracrRNA sequence without introduce significant perturbation to its original function. The in vitro DNA cleavage reactions were performed to determine the tolerance of Cas9 to each designer tracrRNA. The results demonstrate that CRISPR/Cas9 tolerates each designer tracrRNA very well. In parallel with the sgRNA studies, ‘stem–loop 1’ variants of tracrRNA induced variable cleavage activity for different target sites. All designer tracrRNAs function well to support Cas9-mediated DNA cleavage when they were used together with the cr-SLX4IP, cr-HPRT1 or cr-GFP (Figure 5B, Supplementary Figures S17A, S17B). When we performed in vitro DNA cleavage using cr-HBEGF, only ‘stem–loop 3’ variants of tracrRNA (tracrRNA-S1b, tracrRNA-S1c and tracrRNA-S2c) showed a considerable activity comparable to that of original tracrRNA (Supplementary Figure S17C). All ‘stem–loop 1’ variants of tracrRNA were functionally impaired when paired with cr-HBEGF. These combined data support the conclusion that sequence modifications are better tolerated in stem–loop 3 compared to stem–loop 1.
We next investigate the propensity of MBLs to switch off the function of designer tracrRNAs. In this study, different concentrations of each MBL were added to the incubation mixture containing Cas9:crRNA:tracrRNA prior to the addition of target DNAs. Being consistent with the above sgRNA studies, both NCD and Z-NCTS showed a considerable activity against CRISPR/Cas9 with designer tracrRNAs, with NCD appearing to be the more potent (Figure 5C, Supplementary Figure S18). We also observed a non-specific action of Z-NCTS, which may be attributed to the non-specific binding of this molecule to structural units in tracrRNAs. Similar results were also seen with another three crRNAs with different target sites (cr-HBEGF, cr-GFP, cr-SLX4IP in Supplementary Table S1, results in Supplementary Figures S19–S21). Likewise, NCD exhibited a pronounced inhibition of the single-site variants but with a better performance for the multiple-site variants (Figure 5D, E). This pattern was further characterised for each designer tracrRNA with another three crRNAs (Supplementary Figures S22–S24). The dose response behavior for each designer tracrRNA exposed to NCD was very similar to that of designer sgRNA with the same MBL-binding unit, indicating the impotant role of ligand–RNA interactions for controlling CRISPR/Cas9.
Ligand control of designer sgRNAs in Cas9-expressing cells
Encouraged by the above studies, we proceeded to test the effects of our strategy by ligand control of designer sgRNAs in human cells. The HeLa-OC cell line, which constitutively expresses Cas9, was used as a model to demonstrate proof of principle of our approach (51). We selected a number of endogenous genes, including HPRT1 gene, HBEGF gene and SLX4IP gene. Because of the better performance of NCD in the above studies, this compound was used in the following cellular studies. In parallel to the above studies, the same set of sgRNAs was applied to HeLa-OC cells to support the Cas9-mediated gene editing. A complete media change was performed at 4 h post-transfection and cells were allowed to culture for an additional 24 h. After harvesting the genomic DNA of these cells, a T7 endonuclease I assay (T7E1) was performed to analyze the amplified genomic target regions. When we performed in vitro DNA cleavage using sgRNAs with the SLX4IP target site, all designer sgRNAs were functional. However, the gene editing induced by the same set of designer sgRNAs revealed a dramatic difference in efficiency (Figure 6A, B). We demonstrate that only ‘stem–loop 3’ variants of sgRNA (sg-SLX4IP-S1b, sg-SLX4IP-S1c and sg-SLX4IP-S2c) showed a considerable activity for supporting the Cas9-mediated gene editing. Remarkably, all constructs with modification in stem–loop 1 displayed drastically diminished editing efficiency, as seen by inefficient gene editing at 24 h after sgRNA transfection (sg-SLX4IP-S1a, sg-SLX4IP-S2a, sg-SLX4IP-S2b and sg-SLX4IP-S3). These conclusions held true for another set of sgRNAs with a different target site, suggesting a general principle (Supplementary Figure S25). These results together indicate that the nucleotides in stem–loop 1 are less tolerant to variation than the ones in stem–loop 3.
Figure 6.
Ligand control of designer sgRNAs in a stable Cas9-expressing cell line Cellular studies were performed using HeLa-OC cells as described in the Experimental Section. The treatment for each sample is indicated by the signs at the bottom of each lane. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Editing of SLX4IP gene in HeLa-OC cells using the indicated sgRNAs. Lane 1: target control; lane 2: no sgRNA control; lane 3 contains original sg-SLX4IP; lanes 4–10 contain designer sgRNAs harboring different MBL-binding units; lane 11: DNA marker (GeneRuler 100-bp DNA Ladder). (B) The effect of sequence modification on the function of sgRNAs in cells. (C) Ligand control of designer sgRNAs in HeLa-OC cells. Hela-OC cells were exposed to the NCD ligand for 24 h before being harvested for DNA cleaving activity assessments. Lane 1: target control; lanes 2–3: no sgRNA control; lanes 4–5 contain original sg-SLX4IP; lanes 6–9 contain sg-SLX4IP-S1b; lanes 10–13 contain sg-SLX4IP-S1c; lanes 14–17 contain sg-SLX4IP-S2c; lane 18: DNA marker. (D) Bar graph shows the effect of NCD on the function of sgRNAs in HeLa-OC cells. In each group, the indel formation of NCD-treated cells were compared to that of mock-treated cells. P values less than 0.05 are given one asterisk, and P values <0.001 are given three asterisks. For (A) and (C), uncleaved SLX4IP DNA (773 bp) cut to shorter cleavage fragments (441 bp and 332 bp) are demonstrated. For (B) and (D), data represent the mean of three replicates and were shown as mean ± SEM.
We next investigated the effect of ligand binding to control the function of ‘stem–loop 3’ variants of sg-SLX4IP in cells. A MTT cell viability assay was used to find the suitably used concentrations of NCD. The results demonstrate that HeLa-OC cells can tolerate well up to 16 μM NCD after 24 h of incubation (Supplementary Figure S26). HeLa-OC cells were transfected with each sgRNA and then treated with different concentrations of NCD. Upon 24 h post-treatment, the cells were harvested to analyze the gene editing efficiencies. The relative indel formation of each sgRNA was normalized to untreated cells. Among our tested designer sgRNAs, sg-SLX4IP-S2c stood out as the best construct, which displayed the highest degree of response upon NCD treatment (Figure 6C, D). We also found that the sg-HPRT1-S2c construct was responsive to NCD in HeLa-OC cells (Supplementary Figure S27). In parallel experiments, the wild-type sgRNA showed less change in the indel formation by the addition of NCD. We further performed a time-dependent experiment to characterize in cellular potency of NCD. The results demonstrated that NCD can be removed from cells after 12 h of incubation (Supplementary Figure S28). These studies provided evidence that ligand control of designer sgRNAs could be employed in cells for controlling gene editing.
Ligand control of designer genetically encoded sgRNAs
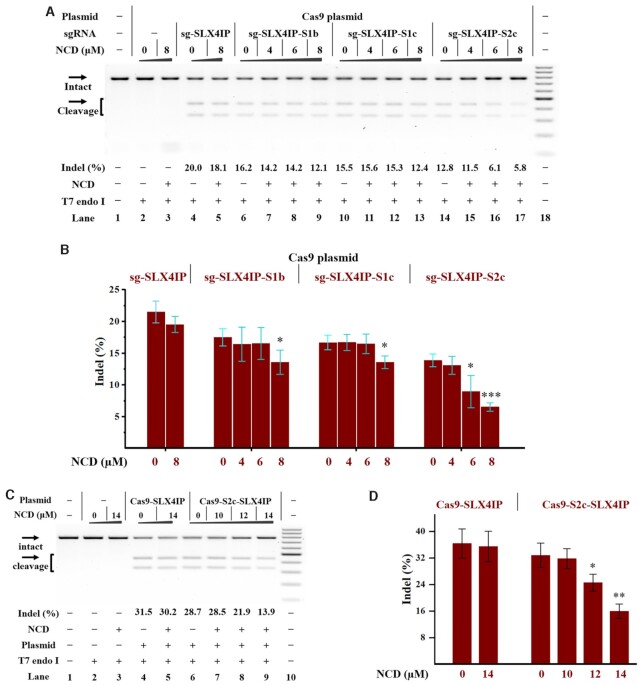
In the above studies, the Cas9 was directly integrated into the genome of model cells (HeLa-OC); however, this is not appropriate for a general genome-editing protocol. A more generalizable approach is to use a plasmid-based system, thus avoiding dependence on specific cells. We first attempted a hybrid system using a Cas9-only plasmid (PX165) and IVT sgRNAs (5,53). To investigate whether genomic loci can be targeted using this hybrid system, the SLX4IP gene was studied in HeLa cells. We therefore delivered the SLX4IP targeting sgRNAs 4 h after the PX165 transfection. The amount of indel formation was determined 24 h after transfection. Being consistent with the above HeLa-OC studies, each ‘stem–loop 3’ variant of sgRNA (sg-SLX4IP-S1b, sg-SLX4IP-S1c or sg-SLX4IP-S2c) in combination with PX165 showed considerable intracellular activities for editing the SLX4IP gene (Supplementary Figure S29). Hence, this hybrid system is a viable strategy to edit target gene in human cells. We further investigated the effect of NCD on the function of hybrid systems. The PX165 plasmid and IVT sgRNAs were delivered into cells as above before increasing amounts of NCD were added. We observe a decrease in the indel formation upon NCD treatment (Figure 7A, B). More studies were performed to demonstrate the generality of our approach (Supplementary Figure S30). We further performed Cas9 ribonucleoprotein (RNP) study of using Cas9 protein along with different IVT sgRNAs. The results also showed a trend to reduction after NCD treatment (Supplementary Figure S31).
Figure 7.
Ligand control of plasmid-based gene editing in human cells Cellular studies were performed as described in the Experimental Section. The plasmids and/or sgRNAs were delivered into HeLa cells before the treatment with NCD. Hela cells were exposed to the NCD ligand for 24 h before being harvested for DNA cleaving activity assessments. All samples were tested in three biological replicates. Image of representative data is shown here. (A) Ligand control of the hybrid system with IVT sgRNAs and Cas9-only plasmids. Lane 1: target control; lanes 2–3: no sgRNA control; lanes 4–5 contain PX165 and sg-SLX4IP; lanes 6–9 contain PX165 and sg-SLX4IP-S1b; lanes 10–13 contain PX165 and sg-SLX4IP-S1c; lanes 14–17 contain PX165 and sg-SLX4IP-S2c; lane 18: DNA marker (GeneRuler 100-bp DNA Ladder). (B) Bar graph shows the effect of NCD on the function of the hybrid system (IVT sgRNAs and PX165). (C) Ligand control of all-in-one plasmids with MBL-binding units. Lane 1: target control; lanes 2–3: no plasmid control; lanes 4–5 contain PX459-SLX4IP; lanes 6–9 contain PX459-S2c-SLX4IP; lane 10: DNA marker. (D) Bar graph shows the effect of NCD on the function of all-in-one plasmids. For (A) and (C), uncleaved SLX4IP DNA (773 bp) cut to shorter cleavage fragments (441 bp and 332 bp) are demonstrated. For (B) and (D), the data are presented as the means ± SEM from three independent experiments. In each group, the indel formation of NCD-treated cells were compared to that of mock-treated cells. P values less than 0.05 are given one asterisk, P values <0.01 are given two asterisks, and P values <0.001 are given three asterisks.
It will be more useful to provide a genetically encoded system toward the control of gene editing in human cells. The most straightforward approach is to use an all-in-one plasmid encoding Cas9 and sgRNA from the same vector, thus avoiding multiple transfections of different components (5,54). For our purpose, the first step will be the engineering of a widely used all-in-one plasmid (PX459). Hence, in-fusion cloning technology was adopted to engineer the sgRNA expression cassette. Because of the better performance of the ‘stem–loop 3’ multi-nucleotide variant of sgRNA, this sgRNA-expression cassette was used for our studies. Sanger sequencing was performed to confirm the correct engineering of PX459. The correctly constructed designer plasmid is named as PX459-S2c.
It is necessary to validate the activity of designer plasmids for gene editing in human cells. We therefore designed different pairs of oligonucleotide corresponding to SLX4IP and HBEGF genes, respectively (Oligo-SLX4IP-F and Oligo-SLX4IP-R; Oligo-HBEGF-F and Oligo-HBEGF-R in Supplementary Table S1). The cloning site of each plasmid (PX459 or PX459-S2c) was digested with BbsI for accepting two annealed oligonucleotides containing the spacer of the gRNA. We next delivered the complete plasmids into HeLa cells to perform gene editing. Upon 24 h post-transfection, the cells were harvested to analyze the indel formation by using T7E1 assay. We demonstrate that the transfection of designer complete plasmid had comparable gene editing efficacy to that of original PX459 (lanes 4, 6 in Figure 7C).
We next investigated the effect of NCD after the transfection of each complete plasmid targeting the SLX4IP gene. We demonstrate that NCD effectively inhibits the indel formation in HeLa cells receiving the designer plasmid bearing the MBL-binding units, whereas its effect on HeLa cells receiving original plasmid is much less pronounced (Figure 7C, D). These results were replicated using designer complete plasmid targeting the HBEGF gene (PX459-S2c-HBEGF in Supplementary Figure S32).
DISCUSSION
RNA structures and their interactions with small molecules are generally less well elucidated than their protein counterparts (44). Currently, the chemistry-based approaches have been hindered by difficulties in the design of small-molecule ligands that binds to RNA molecules (27,55–57). The present study marked the first step toward the generation of designer gRNAs by using a rational design of small molecules that bind to the predetermined RNA sequences. Using existing structural data and the principles of RNA folding and small molecule interaction (25,35), we develop an original and useful strategy to reprogram gRNAs that are capable of higher order structures in response to small synthetic molecules. The introduction of MBL-binding units into gRNAs offers a great potential for small-molecule interventions. In the current study, we have surveyed the tolerated sequence diversity of different MBL-binding motifs to assess their utility as a scaffold for gRNA engineering. We demonstrate that a ligand–RNA pair is an essential element in the creation of our designer gRNAs. Although the ‘stem–loop 1’ variants of sgRNA induced variable cleavage activity for different target sequences, all ‘stem–loop 3’ variants of sgRNA are well tolerated for CRISPR/Cas9 across different target sequences. Both NCD and Z-NCTS, are basically derivatives of the same naphthyridine-based molecules but still show such significant differences in their specificity. The comparison of the structures of these two molecules demonstrates that Z-NCTS has linker moieties with restricted conformational flexibility. The (Z)-stilbene linker in Z-NCTS may cause unintended interactions leading to reduced specificity.
The present study seeks to develop a general method to identify lead ligands that bind to highly structured RNA targets and modulate their activities. Ligand control of designer gRNAs will likely be a promising alternative to conventional anti-CRISPR approaches (22). The engineering of gRNA sequence could also become a complementary approach to directed evolution of Cas9 proteins in an attempt to strengthen CRISPR/Cas9 (9,12). The understanding from the current study will hopefully help guide the development of future generations of designer gRNAs with desired properties for small molecule intervention. Challenges also remain to improve the potency and selectivity of currently used compounds. NCD was known to bind to guanine residues present both in DNA and RNA biomolecules. These possible binding events at genome wide scale will leave behind some toxic side effects. This study takes NCD as a starting compound and further structural optimization is required to propose new molecules with more desirable properties.
In the current study, a genetically encodable MBL-operated module is introduced to put chemical brakes on genome editing. Our system enables chemogenetic control of gene editing by using small molecules. These small molecules are not designed in a target agnostic fashion but rather focus on modulating specific targets with a known structure. We have established the control of three different CRISPR/Cas9 systems in human cells to perform conditional gene editing: (i) the plasmid-free system, (ii) the plasmid-based system, and (iii) the hybrid system (a Cas9-only plasmid and IVT sgRNAs). For the plasmid-based strategy, the endogenous U6 small nuclear RNA promoter in human cells was used to express designer sgRNAs. For the hybrid system, a combination of Cas9-only plasmids and IVT sgRNAs is employed. Desirable advantages of the current strategy therefore include (i) the easy accessibility of small molecules, (ii) a single-component system without the introduction of extra RNA units, (iii) excellent compatibility with other tools. These desirable features may pave the way for further application of the designer IVT sgRNAs and designer all-in-one plasmids in chemistry and biology. It is likely that this approach can be applied to other RNA-based systems.
CONCLUSIONS
Overall, the present study describes an efficient strategy to reprogram gRNA and CRISPR/Cas9 in response to small molecules. This study will extend our strategic choices for the modulation of structured RNA functions using small molecules.
Supplementary Material
ACKNOWLEDGEMENTS
The authors also thank Prof. Wensheng Wei (Peking University), Prof. Ping Yin (Huazhong Agricultural University) and Prof. Shaoru Wang (Wuhan University) for their valuable help.
Author contributions: T.T. conceived the original idea, designed the studies and led the project. X.Y.L. and Q.Q.Q. performed all biological studies. W.X. synthesized NCD and Z-NCTS. Y.T.Z., H.M.J. and S.Y.C. performed agarose gel electrophoresis. X.M.S. performed statistical analysis. J.A. and H.Y. performed RNP study. X.Z. provided consistent support during the running of this project. T.T. wrote the manuscript. All the authors provided feedback on the study and on the manuscript. The authors declare no competing financial interests.
Contributor Information
Xingyu Liu, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Wei Xiong, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Qianqian Qi, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Yutong Zhang, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Huimin Ji, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Shuangyu Cui, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Jing An, Frontier Science Center for Immunology and Metabolism, Medical Research Institute, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, Hubei, China.
Xiaoming Sun, Department of Human Anatomy, School of Basic Medical Sciences, Hubei University of Medicine, Shiyan 442000, Hubei, China.
Hao Yin, Frontier Science Center for Immunology and Metabolism, Medical Research Institute, Zhongnan Hospital of Wuhan University, Wuhan University, Wuhan 430071, Hubei, China.
Tian Tian, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
Xiang Zhou, Key Laboratory of Biomedical Polymers of Ministry of Education, College of Chemistry and Molecular Sciences, Hubei Province Key Laboratory of Allergy and Immunology, Wuhan University, Wuhan 430072, Hubei, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [22177089, 91853119, 21721005, 91753201, 21877086, 22177088]; Hubei Natural Science Foundation for Distinguished Young Scholars [2019CFA064]; National Major Scientific and Technological Special Project for ‘Significant New Drugs Development’ [2017ZX09303013]; Fundamental Research Funds for the Central Universities [2042019kf0189]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Niu Y., Shen B., Cui Y., Chen Y., Wang J., Wang L., Kang Y., Zhao X., Si W., Li W.et al.. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014; 156:836–843. [DOI] [PubMed] [Google Scholar]
- 7. Peng R., Lin G., Li J.. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016; 283:1218–1231. [DOI] [PubMed] [Google Scholar]
- 8. Wu X., Kriz A.J., Sharp P.A.. Target specificity of the CRISPR-Cas9 system. Quant Biol. 2014; 2:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N., Zeina C.M., Gao X., Rees H.A., Lin Z.et al.. Evolved cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018; 556:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S., Bae T., Hwang J., Kim J.S.. Rescue of high-specificity cas9 variants using sgRNAs with matched 5′ nucleotides. Genome Biol. 2017; 18:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R.et al.. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo J., Liu Q., Morihiro K., Deiters A.. Small-molecule control of protein function through staudinger reduction. Nat. Chem. 2016; 8:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hemphill J., Borchardt E.K., Brown K., Asokan A., Deiters A.. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015; 137:5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jain P.K., Ramanan V., Schepers A.G., Dalvie N.S., Panda A., Fleming H.E., Bhatia S.N.. Development of light-activated CRISPR using guide RNAs with photocleavable protectors. Angew. Chem. Int. Ed. Engl. 2016; 55:12440–12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang S.R., Wu L.Y., Huang H.Y., Xiong W., Liu J., Wei L., Yin P., Tian T., Zhou X.. Conditional control of RNA-guided nucleic acid cleavage and gene editing. Nat. Commun. 2020; 11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Habibian M., McKinlay C., Blake T.R., Kietrys A.M., Waymouth R.M., Wender P.A., Kool E.T.. Reversible RNA acylation for control of CRISPR-Cas9 gene editing. Chem. Sci. 2019; 11:1011–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aschenbrenner S., Kallenberger S.M., Hoffmann M.D., Huck A., Eils R., Niopek D. Coupling cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci. Adv. 2020; 6:eaay0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B.et al.. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell. 2019; 177:1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L.et al.. Naturally occurring off-switches for CRISPR-Cas9. Cell. 2016; 167:1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bubeck F., Hoffmann M.D., Harteveld Z., Aschenbrenner S., Bietz A., Waldhauer M.C., Borner K., Fakhiri J., Schmelas C., Dietz L.et al.. Engineered anti-CRISPR proteins for optogenetic control of CRISPR-Cas9. Nat. Methods. 2018; 15:924–927. [DOI] [PubMed] [Google Scholar]
- 22. Marino N.D., Pinilla-Redondo R., Csorgo B., Bondy-Denomy J.. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods. 2020; 17:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watters K.E., Shivram H., Fellmann C., Lew R.J., McMahon B., Doudna J.A.. Potent CRISPR-Cas9 inhibitors from staphylococcus genomes. Proc. Natl. Acad. Sci. USA. 2020; 117:6531–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rusk N. CRISPR inhibitors. Nat. Methods. 2019; 16:577. [DOI] [PubMed] [Google Scholar]
- 25. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu L., Li X., Ma J., Li Z., You L., Wang J., Wang M., Zhang X., Wang Y.. The molecular architecture for RNA-Guided RNA cleavage by cas13a. Cell. 2017; 170:714–726. [DOI] [PubMed] [Google Scholar]
- 27. Suresh B.M., Li W., Zhang P., Wang K.W., Yildirim I., Parker C.G., Disney M.D.. A general fragment-based approach to identify and optimize bioactive ligands targeting RNA. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:33197–33203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haniff H.S., Tong Y., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O’Leary C.A., Benhamou R.I., Moss W.N.et al.. Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 2020; 6:1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng T., Dohno C., Nakatani K.. Mismatch-binding ligands function as a molecular glue for DNA. Angew. Chem. Int. Ed. Engl. 2006; 45:5623–5626. [DOI] [PubMed] [Google Scholar]
- 30. Dohno C., Uno S.N., Nakatani K.. Photoswitchable molecular glue for DNA. J. Am. Chem. Soc. 2007; 129:11898–11899. [DOI] [PubMed] [Google Scholar]
- 31. Hong C., Otabe T., Matsumoto S., Dohno C., Murata A., Hagihara M., Nakatani K.. Formation of a ligand-assisted complex of two RNA hairpin loops. Chemistry. 2014; 20:5282–5287. [DOI] [PubMed] [Google Scholar]
- 32. Ernst R.J., Song H., Barton J.K.. DNA mismatch binding and antiproliferative activity of rhodium metalloinsertors. J. Am. Chem. Soc. 2009; 131:2359–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang C., Pu F., Lin Y., Ren J., Dohn C., Nakatani K., Qu X.. Molecular-glue-triggered DNA assembly to form a robust and photoresponsive nano-network. Chemistry. 2011; 17:8189–8194. [DOI] [PubMed] [Google Scholar]
- 34. Hong C., Hagihara M., Nakatani K.. Ligand-assisted complex formation of two DNA hairpin loops. Angew. Chem. Int. Ed. Engl. 2011; 50:4390–4393. [DOI] [PubMed] [Google Scholar]
- 35. Nakatani K., Hagihara S., Goto Y., Kobori A., Hagihara M., Hayashi G., Kyo M., Nomura M., Mishima M., Kojima C.. Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats. Nat. Chem. Biol. 2005; 1:39–43. [DOI] [PubMed] [Google Scholar]
- 36. Yamada T., Miki S., Ul’Husna A., Michikawa A., Nakatani K.. Synthesis of naphthyridine carbamate dimer (NCD) derivatives modified with alkanethiol and binding properties of G-G mismatch DNA. Org. Lett. 2017; 19:4163–4166. [DOI] [PubMed] [Google Scholar]
- 37. Peng T., Nakatani K.. Binding of naphthyridine carbamate dimer to the (CGG)n repeat results in the disruption of the G-C base pairing. Angew. Chem. Int. Ed. Engl. 2005; 44:7280–7283. [DOI] [PubMed] [Google Scholar]
- 38. Dohno C., Kohyama I., Kimura M., Hagihara M., Nakatani K.. A synthetic riboswitch that operates using a rationally designed ligand–RNA pair. Angew. Chem. Int. Ed. Engl. 2013; 52:9976–9979. [DOI] [PubMed] [Google Scholar]
- 39. Shah S., Rangarajan S., Friedman S.H.. Light-activated RNA interference. Angew. Chem. Int. Ed. Engl. 2005; 44:1328–1332. [DOI] [PubMed] [Google Scholar]
- 40. Costales M.G., Childs-Disney J.L., Haniff H.S., Disney M.D.. How we think about targeting RNA with small molecules. J. Med. Chem. 2020; 63:8880–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warner K.D., Hajdin C.E., Weeks K.M.. Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 2018; 17:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stelzer A.C., Frank A.T., Kratz J.D., Swanson M.D., Gonzalez-Hernandez M.J., Lee J., Andricioaei I., Markovitz D.M., Al-Hashimi H.M.. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 2011; 7:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Disney M.D. Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc. 2019; 141:6776–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ursu A., Childs-Disney J.L., Andrews R.J., O’Leary C.A., Meyer S.M., Angelbello A.J., Moss W.N., Disney M.D.. Design of small molecules targeting RNA structure from sequence. Chem. Soc. Rev. 2020; 49:7252–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dohno C., Kimura M., Nakatani K.. Restoration of ribozyme tertiary contact and function by using a molecular glue for RNA. Angew. Chem. Int. Ed. Engl. 2018; 57:506–510. [DOI] [PubMed] [Google Scholar]
- 46. Mukherjee S., Dohno C., Asano K., Nakatani K.. Cyclic mismatch binding ligand CMBL4 binds to the 5′-T-3′/5′-GG-3′ site by inducing the flipping out of thymine base. Nucleic Acids Res. 2016; 44:7090–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A.. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015; 519:199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen B., Zhang J., Wu H., Wang J., Ma K., Li Z., Zhang X., Zhang P., Huang X.. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013; 23:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gasperini M., Findlay G.M., McKenna A., Milbank J.H., Lee C., Zhang M.D., Cusanovich D.A., Shendure J.. CRISPR/Cas9-Mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 2017; 101:192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Panier S., Maric M., Hewitt G., Mason-Osann E., Gali H., Dai A., Labadorf A., Guervilly J.H., Ruis P., Segura-Bayona S.et al.. SLX4IP antagonizes promiscuous BLM activity during ALT maintenance. Mol. Cell. 2019; 76:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., Wei W.. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014; 509:487–491. [DOI] [PubMed] [Google Scholar]
- 52. Karvelis T., Gasiunas G., Miksys A., Barrangou R., Horvath P., Siksnys V.. crRNA and tracrRNA guide Cas9-mediated DNA interference in streptococcus thermophilus. RNA Biol. 2013; 10:841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang H.X., Song Z., Lao Y.H., Xu X., Gong J., Cheng D., Chakraborty S., Park J.S., Li M., Huang D.et al.. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu C., Zhang L., Liu H., Cheng K.. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release. 2017; 266:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kadina A., Kietrys A.M., Kool E.T.. RNA cloaking by reversible acylation. Angew. Chem. Int. Ed. Engl. 2018; 57:3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Velema W.A., Kietrys A.M., Kool E.T.. RNA control by photoreversible acylation. J. Am. Chem. Soc. 2018; 140:3491–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meyer S.M., Williams C.C., Akahori Y., Tanaka T., Aikawa H., Tong Y., Childs-Disney J.L., Disney M.D.. Small molecule recognition of disease-relevant RNA structures. Chem. Soc. Rev. 2020; 49:7167–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.