Abstract
Background
Pseudomonas aeruginosa has the ability to exhibit resistance to a broad range of antibiotics, highlighting the importance of identifying alternative or adjunctive treatment options, such as phages.
Patients and methods
We report the case of a 25-year-old male who experienced an accidental electrocution resulting in exposed calvarium in the left parieto-temporal region, complicated by a difficult-to-treat P. aeruginosa (DTR-P. aeruginosa) infection. Cefiderocol was the sole antibiotic with consistent activity against six bacterial isolates obtained from the infected region over a 38 day period.
Results
WGS analysis identified a blaGES-1 gene as well as the MDR efflux pumps MexD and MexX in all six of the patient’s ST235 DTR-P. aeruginosa isolates, when compared with the reference genome P. aeruginosa PA01 and a P. aeruginosa ST235 isolate from an unrelated patient. After debridement of infected scalp and bone, the patient received approximately 6 weeks of cefiderocol in conjunction with IV phage Pa14NPøPASA16. Some improvement was observed after the initiation of cefiderocol; however, sustained local site improvement and haemodynamic stability were not achieved until phage was administered. No medication-related toxicities were observed. The patient remains infection free more than 12 months after completion of therapy.
Conclusions
This report adds to the growing literature that phage therapy may be a safe and effective approach to augment antibiotic therapy for patients infected with drug-resistant pathogens. Furthermore, it highlights the importance of the GES β-lactamase family in contributing to inactivation of a broad range of β-lactam antibiotics in P. aeruginosa, including ceftolozane/tazobactam, ceftazidime/avibactam and imipenem/relebactam.
Introduction
Pseudomonas aeruginosa is a ubiquitous organism that remains an important nosocomial pathogen.1 It has an impressive capacity to exhibit resistance to all available antibiotics through the development of mutations in or induction of chromosomal genes, in conjunction with the emergence and dissemination of transferable resistance determinants.1 Four β-lactam agents (i.e. ceftolozane/tazobactam, ceftazidime/avibactam, imipenem/cilastatin/relebactam, and cefiderocol) with activity against P. aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa) have received approval by the US FDA.2 Unfortunately, acquisition of resistance to all of these agents by P. aeruginosa has been encountered, leading to a renewed interest in phage therapy. Lytic phages have been used therapeutically for the last century for their ability to infect and lyse bacteria for the treatment of bacterial infections.3 We report a case of a patient with DTR-P. aeruginosa cranial osteomyelitis that was successfully treated with cefiderocol in conjunction with phage therapy.
Methods and results
Clinical case
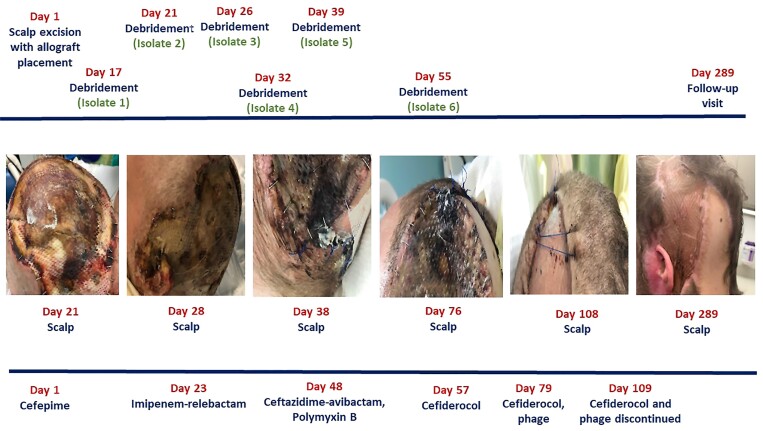
A 25-year-old male with no significant past medical history presented to medical care after an accidental electrocution (Figure 1). He experienced 22% total body surface area third- and fourth-degree burns. Burn wounds of the skull were complicated by a soft tissue defect resulting in exposed calvarium in the left parieto-temporal region. The patient underwent excision of the scalp wound with surgical debridement of the skull, allograft placement and partial closure of the defect with a musculocutaneous flap. On Day 16, the patient developed a temperature to 39.8°C and haemodynamic instability. A fluid collection was identified under his flap. A WBC count and C-reactive protein (CRP) measurement obtained on Day 16 were 42 600 cells/mL and 12.8 mg/dL, respectively. The collection was drained on Day 17 and DTR-P. aeruginosa was recovered. Additional debridement occurred on Day 21 with recovery of DTR-P. aeruginosa once again. The patient was transitioned to imipenem/cilastatin/relebactam on Day 23 (Table 1). He underwent additional skull debridement on Day 26 and Day 32. Cultures from both procedures recovered DTR-P. aeruginosa isolates exhibiting non-susceptibility to imipenem/relebactam, prompting a transition to ceftazidime/avibactam and polymyxin B on Day 48.
Figure 1.
General overview of clinical course. Cultures without recovery of P. aeruginosa not included. Only antibiotic agents with Gram-negative coverage included for simplicity.
Table 1.
AST results using BMD for six P. aeruginosa isolates recovered from the same patient over a 38 day period
| Isolate number | Day of culture | MIC (mg/L) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | ATM | CAZ | CZA | CIP | CST | C/T | FDC | FEP | GEN | IPM | IMR | MEM | PLZ | TOB | TZP | ||
| Isolate 1 | Day 17 | >32 | 16 | >16 | >32 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 2 | >8 | >4 | >8 | 32 |
| Isolate 2 | Day 21 | >32 | >16 | >16 | >32 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 2 | >8 | >4 | >8 | 32 |
| Isolate 3 | Day 26 | >32 | >16 | >16 | 32 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 2 | >8 | >4 | >8 | 32 |
| Isolate 4 | Day 32 | >32 | >16 | >16 | 32 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 4 | >8 | >4 | >8 | >64 |
| Isolate 5 | Day 39 | >32 | >16 | >16 | 32 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 4 | >8 | >4 | >8 | >64 |
| Isolate 6 | Day 55 | >32 | >16 | >16 | 4 | >2 | ≤1 | >8 | 0.5 | >16 | >8 | 16 | 4 | >8 | >4 | >8 | >64 |
The reduction in MIC of CZA for isolate 6 was confirmed by repeat testing.
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CZA, ceftazidime/avibactam; CIP, ciprofloxacin; CST, colistin; C/T, ceftolozane/tazobactam; FDC, cefiderocol; FEP, cefepime; GEN, gentamicin; IPM, imipenem; IMR, imipenem/relebactam; MEM, meropenem; PLZ, plazomicin; TOB, tobramycin; TZP, piperacillin/tazobactam.
Although cefiderocol was the only remaining β-lactam agent exhibiting consistent activity against all six isolates (Table 1), in an effort to preserve its activity, a decision was made to withhold its administration until surgical interventions were exhausted. After the patient underwent full thickness craniectomy on Day 55, he was transitioned to cefiderocol on Day 57 and phage therapy was requested. Between Days 57 and 78 the affected region of the skull showed some improvement; however, the patient continued to have episodes of temperature and haemodynamic stability.
Three of the six P. aeruginosa isolates were shipped to Adaptive Phage Therapeutics (APT) to identify suitable phage matches. The isolates were susceptible to a lytic P. aeruginosa phage, Pa14NPøPASA16, originally identified in Israel. A mono-phage preparation with Pa14NPøPASA16, manufactured under cGMP conditions (Supplementary Methods, available as Supplementary data at JAC-AMR Online), was initiated on Day 79 at a concentration of 1.7 × 1011 PFU diluted in 25 mL of normal saline (NS) and administered IV every 12 h as a 30 min infusion. NS bags containing the phage were thoroughly mixed before each infusion. Tubing was flushed with at least 25 mL of NS after each infusion. Phage infusions were spaced at least 3 h apart from the cefiderocol, to theoretically enable phage to proliferate in sufficient quantities of host before lysis.
Within 24 h of the first dose of phage therapy the CRP increased from 4.6 mg/dL to 6.4 mg/dL and the WBC count increased from 6580 to 14 800 cells/mL. Within 48 h of administration of the first dose of phage therapy, both inflammatory markers normalized. The patient demonstrated local wound improvement with no further episodes of temperature or haemodynamic instability. No additional DTR-P. aeruginosa isolates were recovered. During receipt of phage therapy, weekly inflammatory markers, electrolytes, liver function tests and complete blood counts were obtained. No abnormal laboratory values or findings on clinical exam suggestive of medication toxicities were observed. Antibiotic and phage therapy were discontinued on Day 109 and the patient was discharged from the hospital shortly after. The patient’s left scalp region showed significant improvement at follow-up visits 6 months and 12 months after the discontinuation of therapy (Figure 1). The patient remains infection free more than 12 months after completion of anti-infective therapy.
Antimicrobial susceptibility testing (AST)
Bacterial genus and species were identified using MALDI TOF (Bruker Daltonics Inc.). Initial AST results were determined using the BD Phoenix Automated System (BD Diagnostics). After resistance to traditional β-lactam agents was identified, AST for novel β-lactams was determined using lyophilized Sensititre broth microdilution (BMD) GN7F and MDRGN2F BMD panels (Thermo Fisher Scientific). CLSI interpretive criteria were applied to determine P. aeruginosa susceptibility.4 Isolates were stored at −80°C in Microbank tubes (Pro-Lab Diagnostics) until further testing was performed.
Phage susceptibility testing
APT evaluated their existing phage library to identify a phage with activity against three clinical isolates provided by the patient. The library consists of a collection of well characterized phages isolated from environmental samples and identified as pure, free of deleterious genes or elements of lysogeny, and capable of lysing pathogenic bacteria. APT performed phage susceptibility testing by evaluating the bacteria’s ability to establish a logarithmic growth phase in the presence of phage compared with an uninfected bacterial control (Supplementary Methods). Pa14NPøPASA16 inhibited growth of the patient’s isolates for at least 20 h longer compared with a control tested in the absence of the phage. The prolonged duration of growth inhibition suggests that Pa14NPøPASA16 had independent activity against the patient’s P. aeruginosa isolates.
WGS
WGS was conducted to understand the mechanisms contributing to the DTR-P. aeruginosa phenotype. Frozen isolates were subcultured twice to tryptic soy agar with 5% sheep blood. Genomic DNA was extracted from the six isolates using the DNeasy PowerSoil Pro Kit (QIAGEN Inc.). WGS was conducted using Illumina MiSeq short-read sequencing (Illumina) and bioinformatics analysis was performed by Ares Genetics, both as previously described.5 All six P. aeruginosa isolates belonged to multilocus ST235.
Multiple sequence alignment of the six isolates were compared to both the reference genome P. aeruginosa PA01 as well as to a clinical isolate of P. aeruginosa ST235 from an unrelated patient. The ‘reference’ clinical P. aeruginosa ST235 isolate was obtained from an 81-year-old male with a complicated urinary tract infection at The Johns Hopkins Hospital in December 2016. AST of the reference ST235 isolate revealed resistance to all traditional β-lactam agents. The reference ST235 isolate exhibited susceptibility to all the novel β-lactams: cefiderocol (2 mg/L), ceftazidime/avibactam (4 mg/L), ceftolozane/tazobactam (1/4 mg/L) and imipenem/relebactam (2/4 mg/L).
Compared with the PA01 isolate, the patient’s isolates contained a blaOXA-488 (member of the blaOXA-50 family), blaPDC-35 and blaGES-1. Several protein mutations were identified in AmpR, AmpD, AmpC and OprD. MexD and MexX efflux pumps were identified in the patient’s isolate; no full-length close homologies of these proteins were identified in the reference PA01 isolate. Compared with the ‘reference’ ST235 isolate, the only additional β-lactamase gene identified was blaGES-1. No differences in AmpR, AmpD, AmpC or OprD sequences were identified. No close homologues of MexD or MexX identified in the clinical isolates were identified in the reference ST235 isolate.
Discussion
We report the case of an unfortunate man who was previously healthy and sustained an electrocution-associated exit wound resulting in extensive injury to his left parieto-temporal skull and scalp. His course was complicated by the development of a skull osteomyelitis due to DTR-P. aeruginosa. WGS revealed elevated MICs of all clinically available β-lactam lactams, except cefiderocol, which were likely due to the presence of blaGES-1, a β-lactamase gene, in combination with the presence of the MDR efflux pumps MexD and MexX,6 as this combination of resistance markers was present in the patient’s isolates but not in the PA01 reference genome or the reference clinical ST235 isolate.
The blaGES family are Class A β-lactamase genes generally encoded on class 1 integrons—increasing their potential for widespread dissemination, especially when found in high-risk clones such as ST235.7,8 Although GES enzymes have traditionally been considered ESBLs, point mutations can expand their activity to include carbapenem substrate utilization (e.g. GES-5, GES-6).9 Several reports have demonstrated that P. aeruginosa producing GES enzymes are capable of exhibiting resistance to ceftolozane/tazobactam,9–11 ceftazidime/avibactam10,11 and imipenem/relebactam,12–15 similar to the current case. The MexD and MexX MDR efflux pumps identified in our patient’s isolates likely provided a furthering of β-lactam resistance, as they have been previously associated with the extrusion of β-lactam antibiotics.6 Although less common in the USA, in several regions of the world, GES enzymes are becoming an important contributor to DTR-P. aeruginosa phenotypes.7,8,16,17 Our patient had no history of international travel and was otherwise healthy prior to his current hospitalization suggesting likely nosocomial acquisition.
Despite the heavy burden of disease with a DTR-P. aeruginosa isolate, our patient achieved clinical cure of his infection, and in the absence of any treatment-related adverse events. The lack of toxicities in this case is consistent with the consistent safety record of therapeutic phages as reported in the literature.3 Marked improvement in the appearance of the wound and normalization of vital signs occurred only after the initiation of phage therapy and was less apparent during the interim period when cefiderocol therapy was administered alone. Nevertheless, it is unknown whether improvement would have eventually occurred without the addition of phage therapy. The vast majority of clinical experiences generated over the past two decades have evaluated the role of phages in conjunction with, rather than in place of, antibiotic therapy providing limited insight into the incremental benefit of phages in contributing to clinical cure.3 We postulate, however, that there may be a synergistic effect of the phage-antibiotic combination. Bacteria can readily downregulate receptors used by phages to penetrate bacteria and lead to phage resistance, while simultaneously reducing bacterial virulence, underscoring an advantage in using both treatments concurrently.18,19
As an example, to ward off the activity of ΦFG02 and ΦCO01 phages, which use the Acinetobacter baumannii capsule as a receptor, A. baumannii isolates developed loss-of-function mutations in genes responsible for capsular biosynthesis—disrupting phage entry while increasing the vulnerability of A. baumannii mutants to human complement.18 This is further exemplified by the clinical case of a 76-year-old patient with a P. aeruginosa aortic graft infection successfully treated with a phage-antibiotic combination that exploited a similar evolutionary trade-off. The patient was treated with phage OMKO1, which uses the MexAB/XY-OprM MDR efflux systems of P. aeruginosa as bacterial receptors.19,20 As P. aeruginosa isolates reduced expression of these efflux systems to inhibit OMKO1 binding, they also inadvertently reinstated the activity of antibiotics that were previously removed by efflux pumps.19 Knock-out experiments to identify the receptor for Pa14NPøPASA16 used in the current case were not undertaken and its receptor remains unknown.
In summary, the above case supports a growing body of literature that phage therapy may be a safe and effective approach that may have additive value when combined with antibiotic therapy for patients infected with challenging infections—including those caused by DTR bacterial isolates. Clinical trials are currently underway to define the specific subpopulations and infection types that may benefit the most from potential antibiotic-phage synergy.
Supplementary Material
Acknowledgements
We would like to express our gratitude to Ronan Hazan, PhD and his colleagues from Hebrew University in Jerusalem, Israel who originally isolated the phage used in this clinical case.
Funding
This work was supported by a National Institutes of Health R21-AI153580 (P.D.T.) and an American Lung Association Research Grant (P.D.T.). This work was also supported by the National Institute of Health T32 AI007291 (J.C.).
Transparency declarations
P.J.S. reports grants and personal fees from Accelerate Diagnostics, OpGen Inc. and BD Diagnostics; grants from bioMérieux Inc., Affinity Biosensors and Hardy Diagnostics; and personal fees from Roche Diagnostics, Shionogi Inc. and GeneCapture, outside the submitted work. S.B. is an employee of Ares Genetics. J.F., M.L. and R.J.H. are employees of Adaptive Phage Therapeutics (APT). APT provided the phage used in this clinical case. G.A.S. reports a business relationship with APT. All other authors have nothing to disclose.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary data
Supplementary Methods are available as Supplementary data at JAC-AMR Online.
References
- 1. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22: 582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tamma PD, Aitken SL, Bonomo RAet al. Infectious Diseases Society of America Guidance on the treatment of extended-spectrum β-lactamase producing Enterobacterales (ESBL-E), carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin Infect Dis 2021; 72: e169–83. [DOI] [PubMed] [Google Scholar]
- 3. Tamma PD, Suh GA. Phage are all the rage: bacteriophage in clinical practice. J Pediatric Infect Dis Soc 2021; 10: 749–753. [DOI] [PubMed] [Google Scholar]
- 4. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Thirty-Second Edition: M100. 2022. [Google Scholar]
- 5. Tamma PD, Beisken S, Bergman Yet al. Modifiable risk factors for the emergence of ceftolozane-tazobactam resistance. Clin Infect Dis 2021; 73: e4599–606. [DOI] [PubMed] [Google Scholar]
- 6. Morita Y, Tomida J, Kawamura Y. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol 2012; 3: 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Viedma E, Juan C, Acosta Jet al. Nosocomial spread of colistin-only-sensitive sequence type 235 Pseudomonas aeruginosa isolates producing the extended-spectrum β-lactamases GES-1 and GES-5 in Spain. Antimicrob Agents Chemother 2009; 53: 4930–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hishinuma T, Tada T, Kuwahara-Arai Ket al. Spread of GES-5 carbapenemase-producing Pseudomonas aeruginosa clinical isolates in Japan due to clonal expansion of ST235. PLoS One 2018; 13: e0207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poirel L, De La Rosa JM O, Kieffer Net al. Acquisition of extended-spectrum β-lactamase GES-6 leading to resistance to ceftolozane-tazobactam combination in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2019; 63: e01809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ortiz de la Rosa JM, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother 2019; 74: 1934–9. [DOI] [PubMed] [Google Scholar]
- 11. Khan A, Tran TT, Rios Ret al. Extensively drug-resistant Pseudomonas aeruginosa ST309 harboring tandem Guiana extended spectrum β-lactamase enzymes: a newly emerging threat in the United States. Open Forum Infect Dis 2019; 6: ofz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young K, Painter RE, Raghoobar SLet al. In vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol 2019; 19: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlowsky JA, Lob SH, Kazmierczak KMet al. In vitro activity of imipenem/relebactam against Gram-negative ESKAPE pathogens isolated in 17 European countries: 2015 SMART surveillance programme. J Antimicrob Chemother 2018; 73: 1872–9. [DOI] [PubMed] [Google Scholar]
- 14. Karlowsky JA, Lob SH, Kazmierczak KMet al. In vitro activity of imipenem/relebactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples: SMART Surveillance United States 2015-2017. J Glob Antimicrob Resist 2020; 21: 223–8. [DOI] [PubMed] [Google Scholar]
- 15. Fraile-Ribot PA, Zamorano L, Orellana Ret al. Activity of imipenem-relebactam against a large collection of Pseudomonas aeruginosa clinical isolates and isogenic β-lactam-resistant mutants. Antimicrob Agents Chemother 2020; 64: e02165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garza-Ramos U, Barrios H, Reyna-Flores Fet al. Widespread of ESBL- and carbapenemase GES-type genes on carbapenem-resistant Pseudomonas aeruginosa clinical isolates: a multicenter study in Mexican hospitals. Diagn Microbiol Infect Dis 2015; 8: 135–7. [DOI] [PubMed] [Google Scholar]
- 17. Laudy AE, Rog P, Smolinska-Krol Ket al. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS One 2017; 12: e0180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordillo Altamirano F, Forsyth JH, Patwa Ret al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat Microbiol 2021; 6: 157–61. [DOI] [PubMed] [Google Scholar]
- 19. Chan BK, Sistrom M, Wertz JEet al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 2016; 6: 26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan BK, Turner PE, Kim Set al. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018; 2018: 60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.