Abstract
Noggin is an extracellular cysteine knot protein that plays a crucial role in vertebrate dorsoventral patterning. Noggin binds and inhibits the activity of bone morphogenetic proteins via a conserved N-terminal clip domain. Noncanonical orthologs of Noggin that lack a clip domain (“Noggin-like” proteins) are encoded in many arthropod genomes and are thought to have evolved into receptor tyrosine kinase ligands that promote Torso/receptor tyrosine kinase signaling rather than inhibiting bone morphogenic protein signaling. Here, we examined the molecular function of noggin/noggin-like genes (ApNL1 and ApNL2) from the arthropod pea aphid using the dorso-ventral patterning of Xenopus and the terminal patterning system of Drosophila to identify whether these proteins function as bone morphogenic protein or receptor tyrosine kinase signaling regulators. Our findings reveal that ApNL1 from the pea aphid can regulate both bone morphogenic protein and receptor tyrosine kinase signaling pathways, and unexpectedly, that the clip domain is not essential for its antagonism of bone morphogenic protein signaling. Our findings indicate that ancestral noggin/noggin-like genes were multifunctional regulators of signaling that have specialized to regulate multiple cell signaling pathways during the evolution of animals.
Keywords: Noggins, Noggin-like, bone morphogenic protein, Torso/RTK pathway, dorsal-ventral patterning, terminal patterning, cell signaling evolution
Introduction
In animals, cell-to-cell communication depends on a small number of evolutionarily conserved cell signaling receptor superfamilies, 2 of which are the transforming growth factor-β (TGFβ) and receptor tyrosine kinases (RTK) (Babonis and Martindale 2017). These pathways are unique to metazoans and play multiple roles via regulating hundreds to thousands of genes during embryogenesis, pattern formation, and organogenesis (Hill 2001; Furriols and Casanova 2003; Massague and Gomis 2006; Rokas 2008). For cell-signaling pathways to pattern tissues in development, their activation must be tightly regulated, and this occurs via the functions of a relatively small number of regulatory proteins that act in a spatiotemporal manner during development (Carroll 2001). Despite the deep conservation of these signaling pathways, components are absent in some branches of the animal kingdom or have completely different activities in different animals to generate distinct cellular outcomes (Pires-daSilva and Sommer 2003; Babonis and Martindale 2017).
Noggin, a cysteine knot protein, is an extracellular regulator of the TGFβ/bone morphogenic protein (BMP) cell signaling pathway (Zimmerman et al. 1996; Avsian-Kretchmer and Hsueh 2004). Noggin was first described in the African Clawed Frog Xenopus laevis, where it is secreted from Spemann’s organizer and antagonizes BMP signaling. This activity generates a morphogen gradient along the dorsal-ventral axis, causing prospective ventral mesoderm to become dorsal mesoderm (dorsalization) and prospective epidermis to become neuroectoderm (neutralization; Smith and Harland 1992; Smith et al. 1993; Fang et al. 2000). Noggin may also interact with activin and wnt in head and limb development (Bernatik et al. 2017). In addition to Noggin, many metazoan species possess related “Noggin-like” proteins, which lack a conserved clip domain and have variable sequence length (Molina et al. 2011). Noggin-like functions have not been extensively studied but seem to have different functions than those of Noggins.
While noggin (nog) genes are present in most animal genomes, they were thought to be absent from insects (Skelly et al. 2019). Genome sequencing of hemipteran insects, such as the pea aphid (Acyrthosiphon pisum), first revealed genes similar to nog and noggin-like (nog-like) (Shigenobu et al. 2010). Since then, several nog/nog-like genes have been identified in other arthropod genomes though not in holometabolous insects (Skelly et al. 2019). Phylogenetic analysis indicates that arthropod Noggin/Noggin-like proteins are closely related to 2 well-known signaling proteins in insects, Trunk and Prothoracicotropic hormone (PTTH; Duncan et al. 2013). In Drosophila, Trunk and PTTH proteins regulate embryonic terminal patterning (Casanova 1990) and the timing of developmental transitions (McBrayer et al. 2007), respectively; however, unlike vertebrate Noggins, they act as ligands for RTK signaling. Trunk and PTTH bind to and activate the Torso RTK, triggering signaling via the mitogen-activated protein kinase (MAPK) pathways (Casali and Casanova 2001; Rewitz et al. 2009), rather than interacting with morphogenetic proteins. PTTH is also reported to interact with G-protein coupled (GPCR) receptors (Nagata et al. 2006; Yamanaka et al. 2008), implying it may also signal through G-proteins.
Within the Noggin/Noggin-like/Trunk/PTTH family of proteins (all similar cysteine knot proteins), at least 2 very different signaling activities occur. Canonical Noggins and some Noggin-like proteins function in BMP signaling, while Trunk and PTTH activate RTKs. This implies that this family of proteins has either switched their function entirely from BMP repression to MAPK activation in the insect lineage or that the ancestral Noggin/Noggin-like proteins may be multifunctional and have both MAPK and BMP signaling activity. To distinguish between these possibilities, we focused on understanding the effect on BMP and MAPK signaling of insect nog/nog-like genes. Here, we show the outcome of expressing insect A. pisum nog/nog-like genes during Xenopus dorsoventral (DV) patterning and Drosophila terminal patterning. Our findings suggest that pea aphid Noggin-like 1 can repress BMP signaling in Xenopus DV patterning and activate MAPK pathway in Xenopus animal caps, while pea aphid Noggin-like 2 only activates MAPK pathway in Xenopus animal caps and Drosophila terminal patterning. These findings support a model whereby ancient extracellular regulators could have been a multifunctional protein, which was later co-opted into other developmental processes, losing its primary role.
Materials and methods
Molecular cloning
The clip domain of ApNL1 (encoding the amino acids PVPSNDPGVIDLIEMP) was deleted from ApNL1 to synthesize ApNL1ΔClip and inserted into ApNL2 and Drosophila melanogaster trunk (35–47 and 47–63 aa residues, respectively) to synthesize ApNL2+Clip and DmTrk+Clip. The candidate genes (Supplementary Table 1) were synthesized by GenScript in pBluescript (KS) vector. Genes were subcloned into pCS2 vector for generation of capped polyadenylated mRNA for injection into Xenopus oocytes. Genes were subcloned into pUASP attB vector for the generation of transgenic fly lines.
Xenopus injections
Xenopus embryos were generated and cultured (University of Otago Animal Use Protocol 19/01) as described previously (Beck and Slack 1999). Xenopus embryos were staged according to Nieuwkoop and Faber (1994). mRNA for injection was generated from linearized plasmid using the SP6 mMESSAGE mMACHINE kit (Ambion). Dorsal-anterior index (DAI) scores (Kao and Elinson 1988) were recorded at stage 32. A score of 5 indicated a wild-type phenotype, with lower scores down to 0 indicating the degree of centralization and higher scores up to 10 indicating the degree of dorsalization.
Animal cap assay
The dose for mRNA injection was selected based on the Xenopus functional screening data (Fig. 1, see also Supplementary Fig. 2): XlNog1—5.6 pg, ApNL1—1.5 pg, ApNL2—30 pg, DmTrk—60 pg. The final concentration of human bFGF (Sigma-Aldrich) is 100 ng/ml. Animal caps were dissected as previously described (Ariizumi et al. 2017) and placed at 18°C. N = 15+ caps for control, FGF, and each mRNA injection.
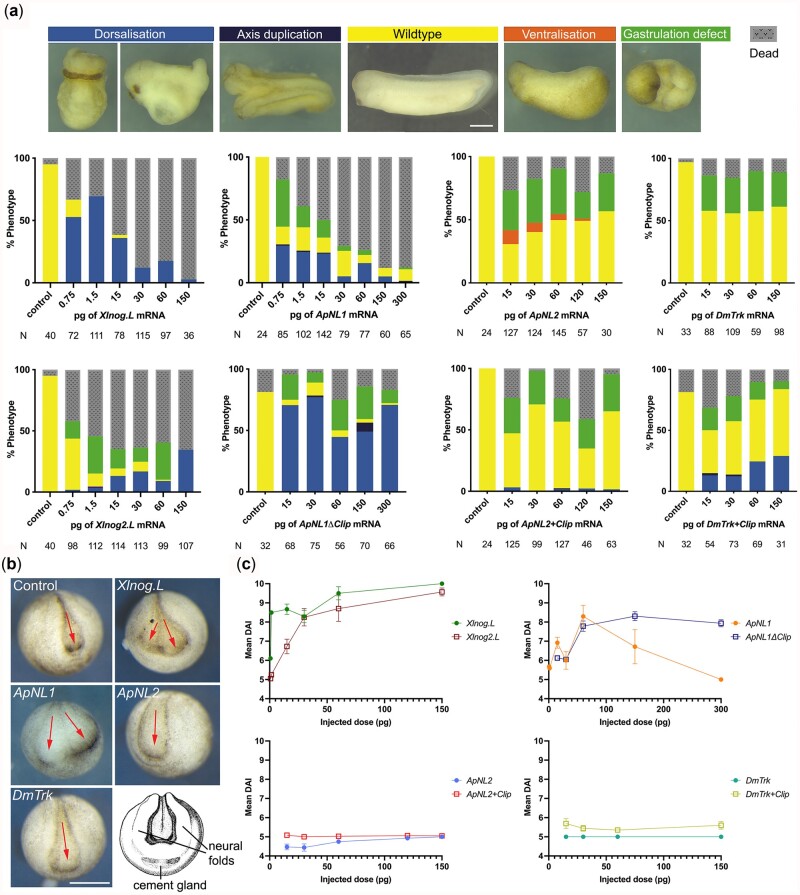
Fig. 1.
ApNL1 induces dorsalization in Xenopus embryos. a) Phenotypes observed at stage 32 Xenopus embryos for microinjections of Xlnog.L, Xlnog2.L, ApNL1, ApNL1ΔClip, ApNL2, ApNL2+Clip, DmTrk, and DmTrk+Clip. Controls were injected with dH2O. N- total Xenopus embryos injected. b) Anterior view of stage 16 Xenopus embryos show presence/absence of axis duplication. c) Mean DAI charts for the microinjections of Xlnog.L, Xlnog2.L, ApNL1, ApNL1ΔClip, ApNL2, ApNL2+Clip, DmTrk, and DmTrk+Clip. Error bars denote SEM. DAI <5—ventralization; DAI 5- wild type: DAI >5—dorsalization. See also Supplementary Fig. 2. Scale bar 1 mm.
Histological analysis
Animal caps were resin-embedded using Technovit 7100 protocols (Kulzer Technik). After 4 days of polymerization, the embedding molds were removed by placing the embedding tray at 60°C for 5 min. Next, animal caps were sectioned at 2 µm using a microtome (Reichert-Jung). The sections were then placed on a water bath at 25°C and transferred to microscope slides, later dried at 60°C for an hour. Microscopic slides were flooded with Polychrome I (Sigma-Aldrich) for 60 s and washed with running water. Then the slides were flooded with Polychrome II (Sigma-Aldrich) for 30 s and later washed with running water, followed by a few dips in 95% ethanol and washing with running water. The slides were then dried in a fume hood. Entellan rapid mounting medium (Merck Millipore) was applied to dried slides, and coverslips were placed on them. The slides were left overnight to dry and imaged using an Olympus BX51 microscope.
RT-PCR
For each sample, 8 animal caps were pooled into a 2 ml micro-centrifuge tube. RNeasy Mini Kit (Qiagen) was used to isolate genomic RNA (gRNA) from the animal caps. Isolated RNAs were then stored at −80°C. The amount of gRNA used in reverse transcription was consistent for each sample (150 ng). The mixture for first cDNA synthesis consisted of 150 ng of template RNA, 2 μl of Random Primers (Invitrogen), 2 μl of 10 × M-MuLV buffer (NEB), 1 μl of M-MuLV Reverse transcriptase (NEB), 1 μl of 10 mM dNTPs (Invitrogen), 0.2 μl of RNase inhibitor (Invitrogen) and the volume was made up to 20 μl with dH2O. The mixture was then placed in a thermocycler (Eppendorf), and the following incubation periods were set up to run: 25°C for 5 min, 42°C for an hour, 65°C for 20 min. The concentrations of cDNA samples were measured using a Nanodrop2000 spectrophotometer and then stored at −20°C. Mesoderm and neural markers (Supplementary Table 2) were selected from the previous studies (Reilly and Melton 1996; Sakata and Maeno 2014).
Drosophila stocks
The following Drosophila stocks were used: Oregon-R (BL5), GAL4::VP16.nos.UTR (BL7253) and trkΔ—a null mutant of trunk (Henstridge et al. 2014). Germline transformants of Drosophila were obtained by microinjection of pUASP plasmid containing the gene of interest into PhiC31 source Drosophila—attP docking site 86F6 (24749) (BestGene Inc). Ectopic expression was driven in the adult ovary by crossing pUASP lines (Supplementary Table 3) with the GAL4::VP16.nos.UTR (BL7253) driver line and trkΔ; GAL4::VP16.nos.UTR driver lines. The flies were maintained, and crosses were set up at 25°C.
In situ hybridization chain reaction and cuticle preparation
Drosophila adults were allowed to lay eggs on apple juice plates containing yeast paste for 4 h before being removed for in situ hybridization chain reaction (HCR) for Dm-tailless (tll) as described previously (Choi et al. 2016). Embryos were imaged using an Olympus FV1000 confocal microscope. Cuticle preparations were prepared by mounting dechorionated Drosophila embryos in Hoyer’s medium using established methods (Henstridge et al. 2014) and visualized using a darkfield modified Olympus BX61 microscope.
Immunostaining and TUNEL assay for Drosophila ovarioles
Flies (2 days old) were left for 24 h in fly food containing Baker’s yeast before dissection. Drosophila ovaries were dissected in 1 × PBS and placed into 400 μl of ice-cold 1 × PBS. The ovaries were fixed with 100 μl of 37% formaldehyde and 500 μl heptane by placing them on a nutator for 20 min. Ovaries were then washed with 1 × PBS for 3 times 5 min each on a rocker. Immunostaining for cleaved Drosophila death caspase 1 (Asp216) (Cell Signaling Technology) was performed using the established protocol (Luo et al. 2013). TUNEL assays with BrdU-Red (Abcam-ab66110) were performed using the manufacturer’s protocol. The ovarioles were separated using tungsten needles and visualized using an Olympus FV3000 confocal microscope.
Results
Acyrthosiphon pisum Noggin-like1 acts as a BMP antagonist in Xenopus embryos
In Xenopus, BMP signaling is blocked locally by Noggin, leading to dorsal development (anterior dorsal structures, i.e. forebrain, cement gland, eye, and neural ectoderm). If noggin is overexpressed in Xenopus embryos, it induces dorsal structures in embryos or induces a secondary axis when ventrally expressed (Fang et al. 2000; Molina et al. 2011). We used this assay to determine if nog/nog-like genes from A. pisum [nog-like 1 (ApNL1) and nog-like 2 (ApNL2), Supplementary Fig. 1] are able to antagonize BMP signaling activity. To do this, we injected mRNA from these genes into Xenopus embryos at the 2-cell stage and quantified dorsalization using the morphological dorsal-anterior index (DAI; Kao and Elinson 1988) at stage 32. Each injected mRNA causes differing degrees and amounts of phenotypes indicating different functions for each protein and different degrees of activity. We began injections with the same amounts of mRNA, but higher doses of some mRNAs were lethal. Micro-injection of ApNL1 dorsalized Xenopus embryos consistent with the dorsalizing effects of micro-injection of X. laevis noggin. L and noggin2.L (Xlnog2.L) but also produced gastrulation defects, not typical of noggin overexpression (Fig. 1a). Injection of either ApNL1 or Xlnog.L mRNA also caused lethality. In contrast, ApNL2 and D. melanogaster trunk (DmTrk) did not dorsalize embryos and instead produced high frequencies of gastrulation defects (>25%), where embryos failed to close their blastopores (Fig. 1a).
Axis duplication, a dorsalization phenotype, can be detected in the early developmental stages of Xenopus (stage 16/mid-neurulation) in the event of dorsalization (Glinka et al. 1997). Axis duplications were frequently observed in Xlnog.L and ApNL1 injected embryos but never in ApNL2 and DmTrk injected embryos (Fig. 1b). Ectopic expression of Xlnog.L, Xlnog2.L, and ApNL1 resulted in mean DAI scores >5 (dorsalized), increasing with dose (Fig. 1c, see also Supplementary Fig. 2) [those embryos that survived a high dose (300 pg) of ApNL1 are wild type, suggesting all properly injected embryos died]. Mean DAI for ApNL2 or DmTrk injected embryos never exceeded 5 (Fig. 1c, see also Supplementary Fig. 2). This evidence suggests ApNL1 encodes a BMP antagonist and is an arthropod noggin, while ApNL2 is not.
The clip domain of ApNL1 is not essential for BMP inhibition
Crystallographic analysis of the interaction between human Noggin (hNoggin) and BMP-7 showed that the clip domain (Gln 28 to Glu 48) of hNoggin mediates its binding to BMP-7, and in vivo studies of substitution mutations at Pro35, Leu46, and Glu48 of hNoggin abolish this interaction (Groppe et al. 2002). To examine whether this clip domain is needed for BMP inhibition by ApNL1, we deleted 15 amino acid residues (PVPSNDPGVIDLIEMP) from ApNL1 (ApNL1ΔClip) and inserted them into ApNL2 (ApNL2+Clip) and DmTrk (DmTrk+Clip) (Supplementary Fig. 1). Intriguingly, ApNL1ΔClip mRNA induced Xenopus embryo dorsalization but was tolerated at higher doses (>30 pg) compared to ApNL1 injections (Fig. 1a), implying that ApNL1ΔClip protein has a weaker BMP inhibiting activity. These data imply that the arthropod Noggin ApNL1 has an inhibitory action on BMP via a molecular mechanism that does not solely require the clip domain. Introducing the clip domain into the D. melanogaster trunk and ApNL2 yielded small but notable numbers of dorsalized embryos (Fig. 1a). As the introduction of a clip domain into trunk confers BMP antagonism, it is clear that the clip domain also contributes to BMP inhibition.
Both ApNL1 and ApNL2 activate RTK/MAPK pathway in Xenopus animal caps
Duncan et al. (2013) suggested that the arthropod-specific genes trunk and PTTH evolved from an ancestral arthropod nog/nog-like gene. If these ancestral arthropod Noggin/Noggin-like proteins were also RTK/MAPK activators, like PTTH and Trunk, this activity might also be conserved in extant Noggin/Noggin-like proteins. To investigate whether arthropod A. pisum Noggin-like proteins can activate the MAPK signaling pathway in Xenopus, animal cap assays (Yamada and Takata 1961; Ariizumi et al. 2009) were performed (Fig. 2, a and b). When cultured in a simple salt solution, the animal cap, an area around the animal pole of mid-blastula stage Xenopus embryos, forms an irregularly shaped epidermis, called the atypical epidermis (Yamada and Takata 1961; Ariizumi et al. 2009). However, activinA (Ariizumi et al. 1999) and Xenopus Noggin (Lamb et al. 1993; Reversade and De Robertis 2005) can induce neural tissues in animal caps by suppressing BMP signaling, while basic fibroblast growth factor (bFGF) mediates mesodermal differentiation in animal caps via the MAPK pathway (LaBonne and Whitman 1994; Umbhauer et al. 1995).
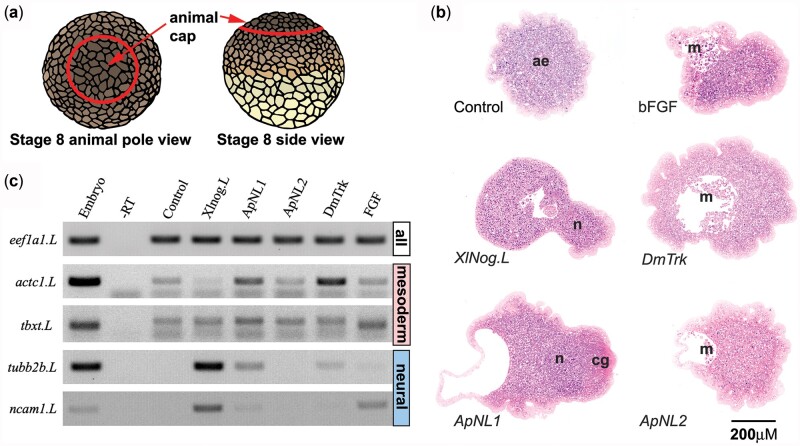
Fig. 2.
ApNL1 and ApNL2 induce mesoderm in Xenopus animal caps. a) Schematic of animal cap explants. b) Representative sections of resin-embedded sections of Polychrome stained animal cap explants. Controls were injected with dH2O. Basic FGF (bFGF) protein was used as a positive control for MAPK activity. ae, atypical epidermis; n, neural; cg, cement gland (neural), m, mesenchyme (ventral mesoderm). Scale bar 200 µm. c) RT-PCR of mesodermal (actc1.L and tbxt. L) and neural (tubb2b.L and ncam1.L) markers. The negative control is genomic RNA from Xenopus embryo without Reverse Transcriptase (RT) enzyme. Ubiquitously expressed elongation factor 1-α (eef1a1.L) is the loading control.
Injections of canonical Noggin Xlnog.L mRNA induced neural fate in animal caps (14/14) as confirmed by RT-PCR for neural markers (tubb2b.L and ncam1.L, Fig. 2, Table 1), consistent with BMP antagonism (Lamb et al. 1993). bFGF (100 ng/ml), an RTK ligand, induced mesenchyme (4/16), part of the ventral mesoderm (Umbhauer et al. 1995), in animal caps (Table 1) and the expression of mesodermal markers [actc1.L, expressed in muscle (ventral mesoderm) (Gotoh et al. 1995) and tbxt. L (brachyury) in pan mesoderm] (Cordenonsi et al. 2007) as confirmed by RT-PCR. Injection of DmTrk also induced mesenchyme (17/20) and expression of the mesoderm marker (actc1.L), indicating that it can activate MAPK signaling activity in Xenopus animal caps, plausibly via Xenopus RTKs (Table 1, Fig. 2c). Microinjection of ApNL1 produced neural tissue (10/21) and cement gland (7/21), consistent with BMP repression, but also produced mesenchyme (9/21, Table 1). RT-PCR confirmed this for neural markers (tubb2b.L and ncam1.L) and mesoderm markers (actc1.L and tbxt. L, Fig. 2). Microinjection of ApNL2 mRNA generated mesenchyme (18/18, Table 1) and expression of tbxt. L (pan mesoderm marker). Our findings imply that when overexpressed, ApNL1 may both repress BMP signaling and activate the MAPK pathway, while ApNL2 and Trunk only activate the MAPK pathway in Xenopus animal caps.
Table 1.
Observed tissue types for each sample.
| Types of cells/tissues observed (number of caps) |
||||||
|---|---|---|---|---|---|---|
| Epidermis | Neural tissue | Cement gland | Mesenchyme | Atypical epidermis | ||
| mRNA injected (number of caps) | Control (9) | — | — | — | — | 9 |
| Xlnog.L (14) | 7 | 14 | — | — | — | |
| ApNL1 (21) | 21 | 10 | 7 | 9 | — | |
| ApNL2 (18) | 18 | — | — | 18 | — | |
| DmTrk (20) | 20 | 6 | 4 | 17 | — | |
| FGF (16) | 16 | — | — | 4 | — | |
ApNL2 activates Torso/MAPK pathway in Drosophila terminal patterning
Next, we wanted to analyze whether arthropod Noggin-like proteins can act as RTK ligands in Drosophila terminal patterning—a robust model system to study the Torso/MAPK pathway (Lu et al. 1993), or can inhibit Decapentaplegic (Dpp), a BMP-2/4 ortholog (Kawabata et al. 1998), in Drosophila DV patterning. We generated nog/nog-like transgenes and performed maternal over-expression using nos-Gal4 driver lines (Wang et al. 1994) and rescue assays using a trunk null (trkΔ) mutant background (Henstridge et al. 2014) during Drosophila oogenesis and embryogenesis.
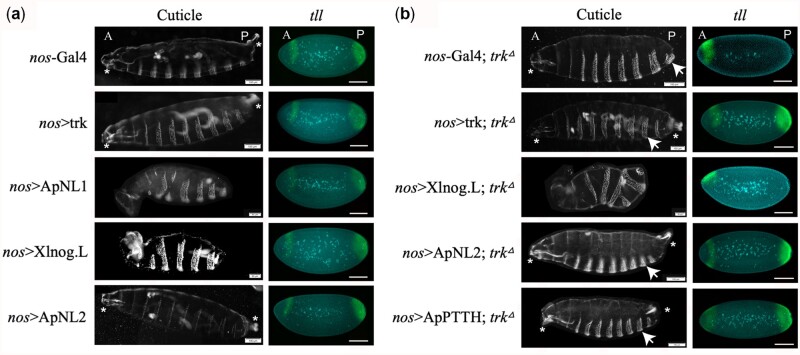
Maternal over-expression of DmTrk during Drosophila embryo patterning did not produce expanded terminal structures in cuticles (240/240), nor cause broadening of the terminal tailless (tll) expression domain (Fig. 3a) as expected since the Torso receptor activation in Drosophila is a rate-limiting step and regulated by Torso-like at the posterior end (Stevens et al. 1990). Maternal over-expression of ApNL1 and Xlnog.L during Drosophila embryo patterning ventralized Drosophila embryos at the expense of anterior and posterior terminal structures (14/16 and 204/209, respectively, Fig. 3a, note loss of the terminal mouth-hooks and filzkörper), an observation consistent with previous studies of Xlnog.L (Holley et al. 1996), implying that they both inhibit Dpp during Drosophila DV patterning. Intriguingly, nos>ApNL1 female flies laid far fewer eggs than females expressing other constructs. In addition, ApNL1 partially suppressed anterior tll expression in stage 4 embryos, implying that ApNL1 may interfere with the activation of Torso/MAPK pathway, perhaps by dimerizing with endogenous trunk and antagonizing its activity. In contrast, over-expression of ApNL2 during Drosophila embryo patterning did not alter either terminal structures of first instar larval cuticles (282/282) or tll expression (Fig. 3a). When we attempted to rescue the terminal phenotypes of trkΔ mutants by over-expressing noggin and noggin-like genes using nos-Gal4 drivers, we again observed strong ventralization by Xlnog.L (71/71) without any rescue of tll expression (Fig. 3b). We did not observe any embryos for the trkΔ rescue assay of ApNL1 as the female flies did not lay eggs. In contrast, both ApNL2 and ApPTTH rescued trkΔ mutant terminal phenotypes (72/72 and 53/53, respectively) and tll expression (Fig. 3b), indicating that these proteins act as Torso ligands.
Fig. 3.
ApNL2 rescues trkΔ mutant phenotypes while Xlnog.L ventralizes Drosophila embryos. a) Maternal expression of genes expressed via the nanos (nos) promoter during Drosophila embryo patterning. Wild-type control (nos-Gal4) first instar larvae cuticle shows normal terminal structures and normal tailless (tll) expression in stage 4 embryos. b) Maternal expression of genes in trunk null mutant flies during Drosophila terminal patterning. The trunk null mutant (nos-Gal4; trkΔ) cuticle lacks abdominal segment after A7 (arrowed) and posterior tll expression in stage 4 embryos. DAPI—staining in centre of embryos, tll—staining at the terminal ends of the embryos. A, anterior; P, posterior. Anterior mouth-hooks and posterior filzkörper are marked by asterisks. Scale bar 100 µm or otherwise stated.
Maternal over-expression of ApNL1 triggers apoptosis during Drosophila oogenesis
When we expressed ApNL1 in Drosophila maternal tissues, nos>ApNL1 female flies laid fewer eggs (n = 16) than females from other crosses and none in our trkΔ rescue assay. Examination of nos>ApNL1 female ovarioles revealed the arrest of stage 9 egg chambers during oogenesis (Fig. 4), a phenotype reminiscent of reduced Dpp signaling (Twombly et al. 1996). Furthermore, immunostaining revealed the presence of the apoptosis marker Dcp-1 (cleaved Drosophila death caspase 1) (85/85, Fig. 4b) and positive TUNEL staining (54/54, Fig. 4c), in stage 9 egg chambers of nos>ApNL1 female flies, indicating that maternal ApNL1 expression induces cell death in late oogenesis. Given that ApNL1 ventralizes Drosophila embryos (Fig. 3a), likely through interfering with Dpp activity, it is plausible that ApNL1 expression also disrupts Dpp signaling during Drosophila oogenesis.
Fig. 4.
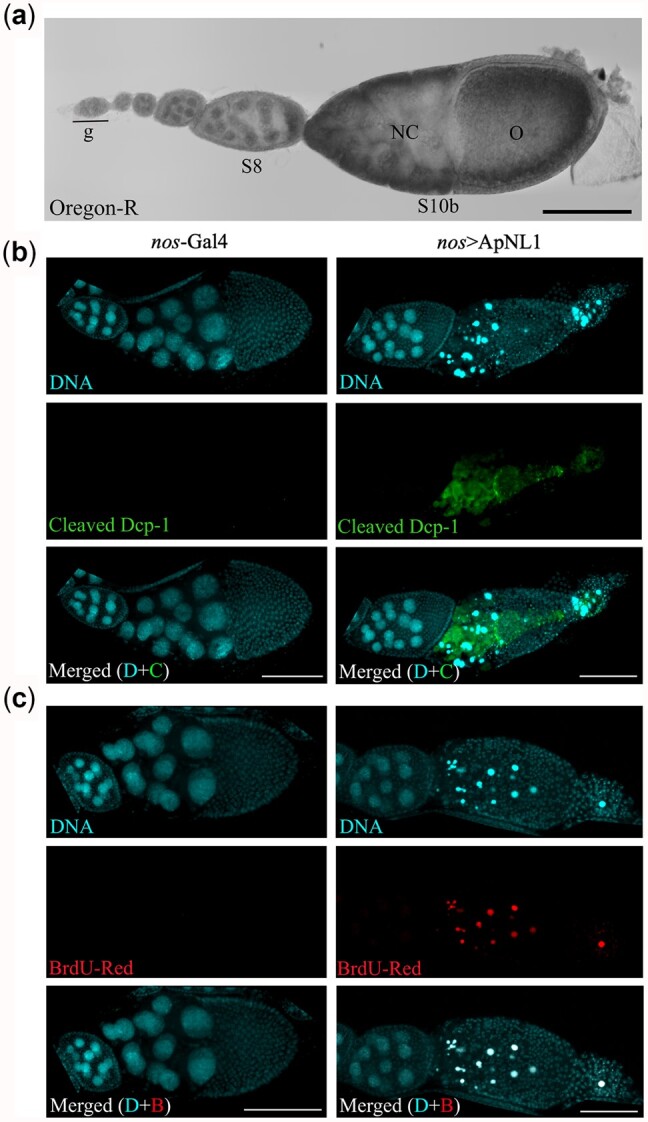
Maternal expression of ApNL1 triggers apoptosis during Drosophila oogenesis. a) A single ovariole of wild-type Oregon-R fly strain. g, germarium; NC, nurse cells; O, oocyte. b) Immunostaining for cleaved Drosophila death caspase 1 (Dcp-1) during late Drosophila oogenesis of nos-Gal4 and nos>ApNL1 female flies. DAPI—cyan, cleaved Dcp-1—green. c) TUNEL assay with BrdU-Red for late stages of Drosophila oogenesis of nos-Gal4 and nos>ApNL1 female flies. Scale bar 100 µm.
Discussion
The relationship between Noggin proteins, which act to repress BMP signaling, and Trunk/PTTH proteins, which activate MAPK, implies that the function of these proteins has switched in their evolution. The evolution of Trunk/PTTH, presumably from a Noggin/Noggin-like ancestor, involved loss of the clip domain, loss of BMP inhibitory activity, and gain of interaction with Torso. In nonholometabolous insect genomes, a number of Noggin and Noggin-like proteins seem to be present, sometimes with Trunk/PTTH proteins as well (Skelly et al. 2019). Here, we show that 2 of these genes in A. pisum have different molecular functions, with ApNL1 acting to inhibit BMP/Dpp signaling and activate MAPK, and ApNL2 acting to activate MAPK/RTK signaling, similar to Trunk and PTTH.
The clip domain, which has been shown to be required for BMP inhibition by hNoggin (Groppe et al. 2002), is not required for inhibition by ApNL1, indicating that some other part of the protein carries out this function. It is possible that BMP inhibition by the clip domain in hNoggin is an evolutionary specialization. The noninhibitory function of the clip domain in ApNL1 may also relate to the very strong BMP inhibition by the protein. In Drosophila over-expression of ApNL1 causes phenotypes in the embryo and ovary, whereas over-expression of Xnog.L does not. It is possible this stronger inhibition of BMP by ApNL1 reflects the fact that it is carried out by a part of the protein other than the clip domain.
That ApNL1, while strongly inhibiting BMP, also has weak MAPK activity in Xenopus implies that this protein may be multifunctional and that the evolutionary history of Noggins and Trunk/PTTH is not a switch from one form of signaling to another, but the specialization of an ancestrally multifunctional protein into either regulating BMP or MAPK signaling. Noggin/Noggin-like proteins have also been shown to have activities on wnt signaling pathways (Eroshkin et al. 2016), and PTTH on GPCRs (Nagata et al. 2006; Yamanaka et al. 2008), implying that Noggin/Noggin-like ancestrally may have been multifunctional extracellular regulators of multiple cell signaling pathways.
Data availability
All data for this study are presented in the manuscript or in the Supplementary data.
Supplemental material is available at GENETICS online.
Supplementary Material
Acknowledgments
The authors thank Nikita Woodhead for Xenopus care, Joanna Ward for technical assistance in molecular cloning, and Matthew Downes for technical support in histological analysis. The authors also thank Shannon Taylor for providing protocols for in situ HCR and Petra Dearden for proofreading. Experimental contributions were as follows: Molecular Cloning (PK, OT, MRL, and EJD); Xenopus injections (PK and OT); animal cap assay, RT-PCR, Drosophila developmental genetics, immunostaining, and TUNEL assay (PK); generation of Drosophila trunk null mutants (GJ). The project was conceived and supervised by EJD, CWB, and PKD. The first draft of the paper was written by PK and edited by CWB, PKD, EJD, TKJ, and OT. CWB, PKD, EJD, TKJ, and PK contributed to the design and interpretation of experiments.
Funding
This research was funded by a University of Otago Research Grant to EJD, CWB, and PKD, and by University of Otago PhD scholarship to PK.
Conflicts of interest
None declared.
Literature cited
- Ariizumi T, Komazaki S, Asashima M.. Activin-treated Urodele animal caps: II. Inductive interactions in newt animal caps after treatment with Activin A. Zool Sci. 1999;16(1):115–124. [Google Scholar]
- Ariizumi T, Michiue T, Asashima M.. In vitro induction of Xenopus embryonic organs using animal cap cells. Cold Spring Harb Protoc. 2017;2017(12):pdb.prot097410. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Takahashi S, Chan TC, Ito Y, Michiue T, AsashimaM.. Isolation and differentiation of Xenopus animal cap cells. Curr Protoc Stem Cell Biol. 2009;Chapter 1:unit 1D.5. [DOI] [PubMed] [Google Scholar]
- Avsian-Kretchmer O, Hsueh AJ.. Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol Endocrinol. 2004;18(1):1–12. [DOI] [PubMed] [Google Scholar]
- Babonis LS, Martindale MQ.. Phylogenetic evidence for the modular evolution of metazoan signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2017;372:20150477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CW, Slack JM.. Gut specific expression using mammalian promoters in transgenic Xenopus laevis. Mech Dev. 1999;88(2):221–227. [DOI] [PubMed] [Google Scholar]
- Bernatik O, Radaszkiewicz T, Behal M, Dave Z, Witte F, Mahl A, Cernohorsky NH, Krejci P, Stricker S, Bryja V, et al. A Novel role for the BMP antagonist noggin in sensitizing cells to non-canonical Wnt-5a/Ror2/Disheveled pathway activation. Front Cell Dev Biol. 2017;5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Chance and necessity: the evolution of morphological complexity and diversity. Nature. 2001;409(6823):1102–1109. [DOI] [PubMed] [Google Scholar]
- Casali A, Casanova J.. The spatial control of Torso RTK activation: a C-terminal fragment of the Trunk protein acts as a signal for Torso receptor in the Drosophila embryo. Development. 2001;128(9):1709–1715. [DOI] [PubMed] [Google Scholar]
- Casanova J. Pattern formation under the control of the terminal system in the Drosophila embryo. Development. 1990;110(2):621–628. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Calvert CR, Husain N, Huss D, Barsi JC, Deverman BE, Hunter RC, Kato M, Lee SM, Abelin ACT, et al. Mapping a multiplexed zoo of mRNA expression. Development. 2016;143(19):3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Montagner M, Adorno M, Zacchigna L, Martello G, Mamidi A, Soligo S, Dupont S, Piccolo S.. Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science. 2007;315(5813):840–843. [DOI] [PubMed] [Google Scholar]
- Duncan EJ, Benton MA, Dearden PK.. Canonical terminal patterning is an evolutionary novelty. Dev Biol. 2013;377(1):245–261. [DOI] [PubMed] [Google Scholar]
- Eroshkin FM, Nesterenko AM, Borodulin AV, Martynova NY, Ermakova GV, Gyoeva FK, Orlov EE, Belogurov AA, Lukyanov KA, Bayramov AV, et al. Noggin4 is a long-range inhibitor of Wnt8 signalling that regulates head development in Xenopus laevis. Sci Rep. 2016;6:23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Marikawa Y, Elinson RP.. Ectopic expression of Xenopus noggin RNA induces complete secondary body axes in embryos of the direct developing frog Eleutherodactylus coqui. Dev Genes Evol. 2000;210(1):21–27. [DOI] [PubMed] [Google Scholar]
- Furriols M, Casanova J.. In and out of Torso RTK signalling. EMBO J. 2003;22(9):1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Onichtchouk D, Blumenstock C, Niehrs C.. Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus. Nature. 1997;389(6650):517–519. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Masuyama N, Suzuki A, Ueno N, Nishida E.. Involvement of the MAP kinase cascade in Xenopus mesoderm induction. EMBO J. 1995;14(11):2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Izpisua Belmonte JC, Choe S, et al. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420(6916):636–642. [DOI] [PubMed] [Google Scholar]
- Henstridge MA, Johnson TK, Warr CG, Whisstock JC.. Trunk cleavage is essential for Drosophila terminal patterning and can occur independently of Torso-like. Nat Communun. 2014;5:3419. [DOI] [PubMed] [Google Scholar]
- Hill CS. TGF-beta signalling pathways in early Xenopus development. Curr Opin Genet Dev. 2001;11(5):533–540. [DOI] [PubMed] [Google Scholar]
- Holley SA, Neul JL, Attisano L, Wrana JL, Sasai Y, O'Connor MB, De Robertis EM, Ferguson EL.. The Xenopus dorsalizing factor noggin ventralizes Drosophila embryos by preventing DPP from activating its receptor. Cell. 1996;86(4):607–617. [DOI] [PubMed] [Google Scholar]
- Kao KR, Elinson RP.. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol. 1988;127(1):64–77. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, Miyazono K.. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9(1):49–61. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Whitman M.. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development. 1994;120(2):463–472. [DOI] [PubMed] [Google Scholar]
- Lamb TM, Knecht AK, Smith WC, Stachel SE, Economides AN, Stahl N, Yancopolous GD, Harland RM.. Neural induction by the secreted polypeptide noggin. Science. 1993;262(5134):713–718. [DOI] [PubMed] [Google Scholar]
- Lu X, Perkins LA, Perrimon N.. The torso pathway in Drosophila: a model system to study receptor tyrosine kinase signal transduction. Dev Suppl. 1993;119(Supplement):47–56. [PubMed] [Google Scholar]
- Luo L, Chai PC, Cai Y.. Immunostaining of germline stem cells and the niche in Drosophila ovaries. In Turksen K, editor. Stem Cell Niche: Methods and Protocols. Totowa (NJ: ): Humana Press; 2013. p. 1–7. [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR.. The logic of TGFbeta signaling. FEBS Lett. 2006;580(12):2811–2820. [DOI] [PubMed] [Google Scholar]
- McBrayer Z, Ono H, Shimell M, Parvy J-P, Beckstead RB, Warren JT, Thummel CS, Dauphin-Villemant C, Gilbert LI, O'Connor MB, et al. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;13(6):857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MD, Neto A, Maeso I, Gómez-Skarmeta JL, Saló E, Cebrià F.. Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr Biol. 2011;21(4):300–305. [DOI] [PubMed] [Google Scholar]
- Nagata S, Namiki T, Ko R, Kataoka H, Suzuki A.. A novel type of receptor cDNA from the prothoracic glands of the silkworm, Bombyx mori. Biosci Biotechnol Biochem. 2006;70(2):554–558. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J.. Normal Table of Xenopus laevis (Daudin): a Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. New York (NY: ): Garland Pub; 1994. [Google Scholar]
- Pires-daSilva A, Sommer RJ.. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4(1):39–49. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Melton DA.. Short-range signaling by candidate morphogens of the TGF beta family and evidence for a relay mechanism of induction. Cell. 1996;86(5):743–754. [DOI] [PubMed] [Google Scholar]
- Reversade B, De Robertis EM.. Regulation of ADMP and BMP2/4/7 at opposite embryonic poles generates a self-regulating morphogenetic field. Cell. 2005;123(6):1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, Gilbert LI, O'Connor MB.. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326(5958):1403–1405. [DOI] [PubMed] [Google Scholar]
- Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. [DOI] [PubMed] [Google Scholar]
- Sakata H, Maeno M.. Nkx2.5 is involved in myeloid cell differentiation at anterior ventral blood islands in the Xenopus embryo. Dev Growth Differ. 2014;56(8):544–554. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Bickel RD, Brisson JA, Butts T, Chang C-C, Christiaens O, Davis GK, Duncan EJ, Ferrier DEK, Iga M, et al. Comprehensive survey of developmental genes in the pea aphid, Acyrthosiphon pisum: frequent lineage-specific duplications and losses of developmental genes. Insect Mol Biol. 2010;19:47–62. [DOI] [PubMed] [Google Scholar]
- Skelly J, Pushparajan C, Duncan EJ, Dearden PK.. Evolution of the Torso activation cassette, a pathway required for terminal patterning and moulting. Insect Mol Biol. 2019;28(3):392–408. [DOI] [PubMed] [Google Scholar]
- Smith WC, Harland RM.. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70(5):829–840. [DOI] [PubMed] [Google Scholar]
- Smith WC, Knecht AK, Wu M, Harland RM.. Secreted noggin protein mimics the Spemann organizer in dorsalizing Xenopus mesoderm. Nature. 1993;361(6412):547–549. [DOI] [PubMed] [Google Scholar]
- Stevens LM, Frohnhöfer HG, Klingler M, Nüsslein-Volhard C.. Localized requirement for torso-like expression in follicle cells for development of terminal anlagen of the Drosophila embryo. Nature. 1990;346(6285):660–663. [DOI] [PubMed] [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM.. The TGF-beta signaling pathway is essential for Drosophila oogenesis. Development. 1996;122(5):1555–1565. [DOI] [PubMed] [Google Scholar]
- Umbhauer M, Marshall CJ, Mason CS, Old RW, Smith JC.. Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature. 1995;376(6535):58–62. [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R.. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199(2):103–115. [DOI] [PubMed] [Google Scholar]
- Yamada T, Takata K.. A technique for testing macromolecular samples in solution for morphogenetic effects on the isolated ectoderm of the amphibian gastrula. Dev Biol. 1961;3:411–423. [DOI] [PubMed] [Google Scholar]
- Yamanaka N, Yamamoto S, Zitnan D, Watanabe K, Kawada T, Satake H, Kaneko Y, Hiruma K, Tanaka Y, Shinoda T, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS One. 2008;3(8):e3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman LB, De Jesús-Escobar JM, Harland RM.. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86(4):599–606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this study are presented in the manuscript or in the Supplementary data.
Supplemental material is available at GENETICS online.