Abstract
Biogenic synthesis using medicinal plants has less harmful effects as compared to chemically synthesized nanoparticles. Here, for the first time, we successfully demonstrated the eco-friendly synthesis of zinc oxide nanoparticles (ZnO NPs) using an aqueous extract of Papaver somniferum L. The phyto-mediated ZnO NPs were characterized using UV-visible spectroscopy, XRD (X-ray diffraction), FT-IR (Fourier transform infrared spectroscopy), SEM (scanning electron microscopy) and TEM (transmission electron microscopy). They were also evaluated for anti-diabetic activity, biocompatibility with RBCs and bactericidal biological applications. The UV spectrum showed a strong surface plasmon peak for ZnO NPs at 360 nm. The optical band gap was observed to be 2.93 eV using UV spectroscopy data. The crystalline nature and the crystal size (48 nm) of the prepared ZnO NPs were confirmed by XRD. FT-IR analysis confirmed the formation of functional bio-molecules linked with ZnO NPs. SEM and TEM images revealed irregular and spherical morphology. The ZnO NPs demonstrated moderate enzyme inhibition (30.8%) at a concentration of 200 mg ml−1. No potential damage was caused by ZnO NPs to red blood cells, if used in low doses. P. somniferum aqueous extract has the potency to combat drug-resistant bacteria but comparatively, ZnO NPs synthesized from the same plant were found to be more effective against resistant pathogenic strains. It is concluded from the above study that phyto-fabricated ZnO NPs have strong potential as theranostic agents and can be adopted in drug delivery systems.
Introduction
Recently, the bridge between nanoscience and biology has extended their applications and increased research attention in this area. The biogenic synthesis of Ag and Au nanoparticles (NPs) was of primary focus but currently the synthesis of oxide nanomaterials using green routes has furthered applications in various fields.1–5 Among them, zinc oxide (ZnO) nanoparticles are frequently exploited owing to their tunable nature and multifarious applications.6 Their applications are well known in every area, ranging from electronic devices to cosmetics. The wide direct bandgap and high excitation energy of 3.37 eV and 60 meV, respectively, mean that ZnO NPs are considered ideal material for many devices, such as semiconductor diodes, transistors, and UV photodetectors.7,8 Moreover, ZnO also plays a key role in biomedical applications, such as in bioimaging and drug delivery. It is already being applied in cosmetics, particularly in lotions, ointments, and sunscreen.9 Thanks to their overall bio-safe nature, different morphological shapes and sizes, ZnO NPs can be characterized as a prime candidate for biomedical applications.
Physical and chemical routes are mostly used for the synthesis of ZnO NPs. Physical methods that have been used for the synthesis include chemical laser deposition, thermal evaporation, molecular beam epitaxy, and pulsed laser deposition. The chemical procedures used for preparing nanosized ZnO NPs include solvothermal, sol–gel, sonochemical, electrodeposition, hydrothermal and spray pyrolysis.10–14 However, synthesis by physical and chemical means has disadvantages. Physical methods require large amounts of energy and are time-consuming while chemical methods produce noxious chemicals, which are unfriendly for the environment.3 To reduce the risk of these problems, greener routes have been demonstrated successfully for the synthesis of ZnO NPs; consequently, a bio-nano combination can be employed. Among all biological resources, plants provide an ideal platform for the biogenic synthesis of NPs because of their easy availability and they are a rich source of metabolites.15 Data on metallic nanoparticles are already available but there is limited data on metal oxide NPs and their applications.16
In view of the limited data availability, in this work, we successfully demonstrated the green synthesis of ZnO NPs using P. somniferum for the first time. P. somniferum is a rich source of useful metabolites, which can act as effective oxidizing/reducing and capping agents. The medicinal uses of P. somniferum (poppy plant) are well documented, and it is mostly used in sedative analgesics or for treatment of coughs, diarrhea and some tumors.17,18
Material and methods
Plant extract preparation
The P. somniferum part (pod) was harvested from the KP province of Pakistan. With the help of an electric grinder, a fine powder was obtained for extract preparation. The obtained powder (30 g) was mixed with 300 ml of deionized water. The mixture was heated at 150 °C for 20 min using a hot plate stirrer. The mixture was incubated overnight for maximum extraction, which was followed by filtration. The resulting product was cooled and stored at 4 °C in a refrigerator for further analysis. The overall scheme of materials and methods is indicated in Fig. 1.
Fig. 1. Biogenic synthesis of ZnO-NPs using P. somniferum.
Preparation of ZnO NPs
For the preparation of zinc oxide nanoparticles, 6.0 g of the zinc nitrate [Zn (NO3)·6H2O] precursor salt was added to 100 ml of P. somniferum plant extract, followed by heating at 100 °C for 2 h.4 The obtained white precipitate was washed three times with deionized water and centrifuged at 1000 rpm for 10 min. The supernatant was then discarded and the product was kept in an oven at 100 °C. The dried sample was then annealed at a high temperature (500 °C) for 2 h to obtain highly crystalline ZnO NPs.
Characterization of ZnO NPs
The obtained Zn-NPs were first characterized by UV-vis absorption spectroscopy (Halo DB-20 UV-vis double beam). The reaction mixtures were scanned in the wavelength (λ) range between 200 and 800 nm. The crystalline nature of the ZnO NPs synthesized using P. somniferum was studied by XRD (Model-D8 Advance, Germany). To determine the functional groups of the ZnO NPs, FT-IR analysis was conducted (Spectrum One, PerkinElmer, Waltham, MA, USA). SEM Microscopy was used to study the surface morphology of the ZnO NPs. The morphological investigation and particle size distribution analysis were carried out using TEM microscopy. The particle size was calculated using ImageJ software after digitizing the various TEM images.
Alpha-amylase enzyme inhibition assay
The alpha-amylase in vitro inhibition assay was carried out according to the previous investigation reported in ref. 19. A 96-well plate was used to perform this activity. To the reaction mixture (15 ml phosphate-buffered saline (PBS)/25 ml α-amylase enzyme) the test samples (10 ml) and starch solution (40 ml) were added in a stepwise manner and incubated at 50 °C for 30 min. After that, 20 μl of 1 M HCl was added, which was followed by the addition of 90 μl of iodine solution. A mixture of buffer solution, starch, and dH2O was used as a blank while dH2O with acarbose was assigned as a positive control. The results were determined using the microplate reader. The following formula was used to calculate the percent enzyme inhibition for alpha-amylase:% Enzyme inhibition = [{ODS − ODN} ÷ {ODB − ODN}] × 100ODS, ODN and ODB correspond to the optical densities of the sample, negative control, and blank, respectively.
Hemolytic activity (biocompatibility with human RBCs)
To check the biocompatibility of P. somniferum-mediated ZnO NPs, fresh RBCs were collected from a healthy individual. An EDTA tube was used and 3 ml of fresh blood was added for isolation of erythrocytes. The blood was then centrifuged for 5 min at 14 000 rpm. The resulting pellet was washed three times with PBS. 9.8 ml of PBS was added to 200 μl of the pellet. The prepared suspension of erythrocytes was added to test samples of different concentrations. It was then incubated at 37 °C for 1 h, followed by centrifugation for 10 min at 2500 rpm. The supernatant was collected and distributed in a 96-well plate and release of hemoglobin was observed at 540 nm. DMSO and Triton X-100 were used as negative and positive controls, respectively. The following equation was used to calculate the percent hemolysis:%Hemolysis = [{(sample absorbance) − (negative control absorbance)} ÷ {(positive control absorbance) − (negative control absorbance)}] × 100
Antibacterial susceptibility test
Antibacterial susceptibility tests were carried out against four drug-resistant bacterial strains using the agar disc diffusion method. The four pathogenic drug-resistant bacteria used in the experiment were Bacillus subtilis, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Staphylococcus epidermidis. The bacterial broth was consistently distributed on a nutrient agar plate at a density of 1 × 108 CFU ml−1. First of all, the bacterial broth was refreshed using a shaking incubator for 24 h at 37 °C, 200 rpm. Ampicillin, as a standard antibiotic (10 μg ml−1), and zinc nitrate salt (2 mM) were also added to the test as negative and positive controls, respectively. Previously, deionized water was used to dissolve samples. The sterile disc made from filter paper was loaded with 10 mg ml−1 of sample, followed by incubation at 37 °C for 24 h. Afterwards, the zone of inhibition was measured and the assay was performed in triplicate.
Result and discussions
UV-visible spectroscopic analysis of ZnO NPs
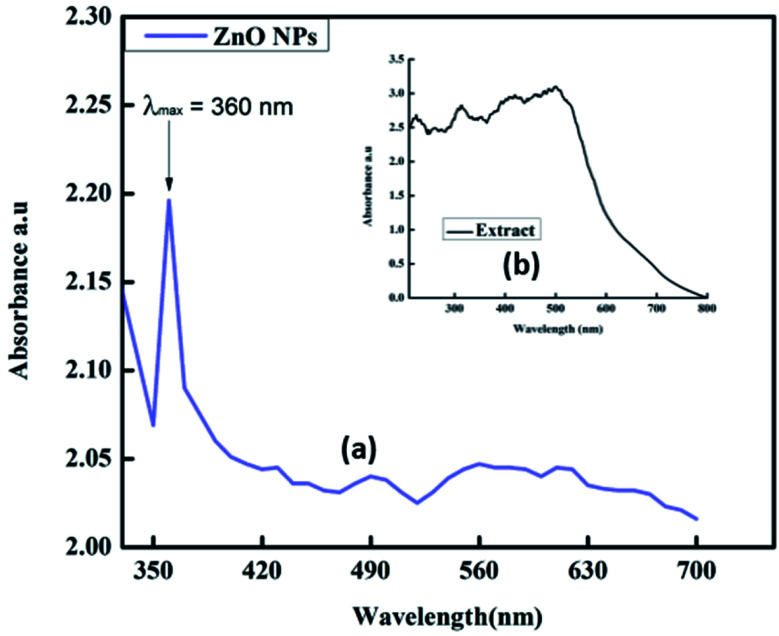
UV-visible spectroscopic analysis was conducted to confirm the biogenic synthesis of ZnO NPs. For this typical analysis, the sample was dissolved in deionized water. The UV-visible wavelength range was between 200 and 800 nm. The sharp peak obtained at 360 nm confirmed the presence of ZnO NPs in the mixture (Fig. 2a).20 The broad absorption band that ranges towards longer wavelength might be owing to the movement of the electronic cloud on the overall skeleton of the ZnO NPs. UV-vis analysis was also performed for the plant extract and the results showed many peaks at different wavelengths ranging from 200 to 520 nm (Fig. 2b). The plant extract was found to contain starch, proteins and antioxidant molecules. The presence of these molecules in the plant extract was responsible for the reduction of Zn metal salt to form ZnO NPs.
Fig. 2. UV-visible spectra of (a) synthesized ZnO NPs and (b) plant extract.
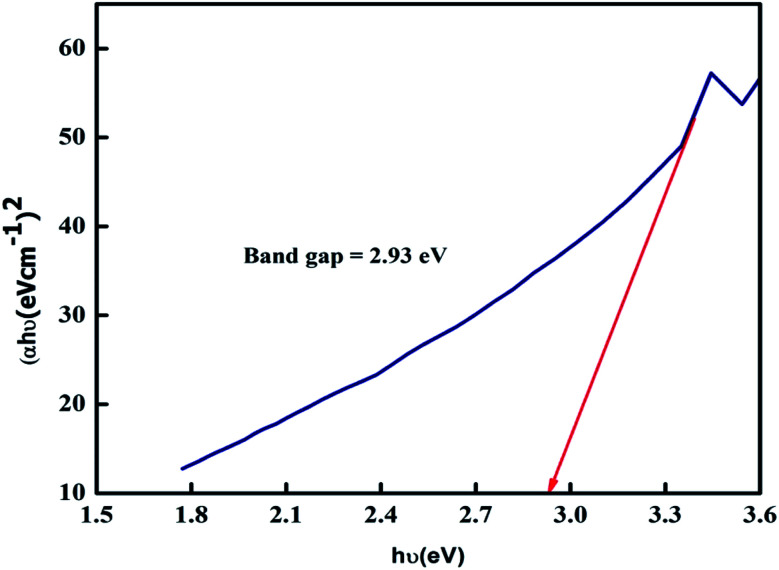
The energy gap (bandgap) was determined from the UV-visible spectroscopy data. By using the following eqn (1), the optical band gap of the ZnO NPs was found to be 2.93 eV, as shown in Fig. 3.
| αhυ = A(hυ − Eg)n | 1 |
where α is the absorption coefficient, hυ represents the energy of the photon, A is the proportionality constant and varies with the material, and n represents the index.21
Fig. 3. The bandgap of ZnO NPs.
X-ray diffraction analysis
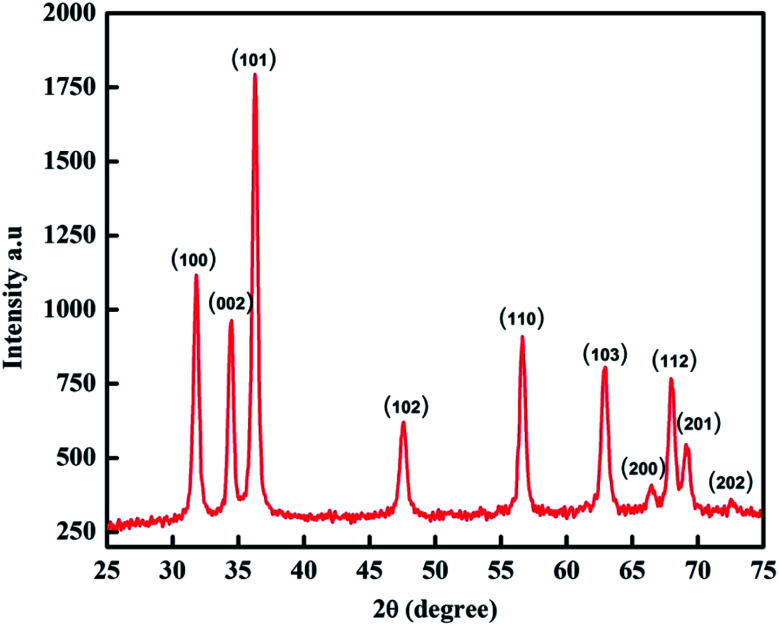
XRD analysis is used to provide insight into the crystalline structure and crystallite size of particles. The XRD pattern for the biogenic synthesized ZnO NPs is shown in Fig. 4. The XRD peaks at 2θ = 31.8355°, 34.5207°, 36.2871°, 47.5808°, 56.6487°, 62.8917°, 66.4115°, 67.9331°, 69.1842° and 77.0106° correspond to the (100), (002), (101), (102), (110), (103), (200), (112), (201) and (202) crystal planes and hexagonal crystal geometry according to JCPSD card no. 01-007-2551. The average particle size of the NPs was determined by using the Scherrer equation (2):
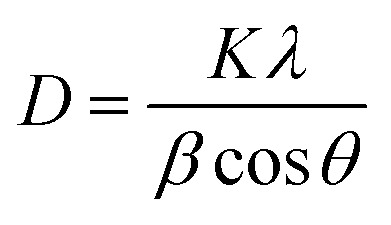 |
2 |
where D is the crystallite size of the particle, K represents the Scherrer constant, which is equal to 0.9, λ is the wavelength of light used for diffraction (λ = 1.54 Å), β is the FWHM (full width at half maximum) of the diffraction peak and θ is the angle of reflection.22 The average size of the ZnO NPs calculated from the XRD pattern was 48 nm.
Fig. 4. XRD pattern of ZnO NPs.
FT-IR analysis
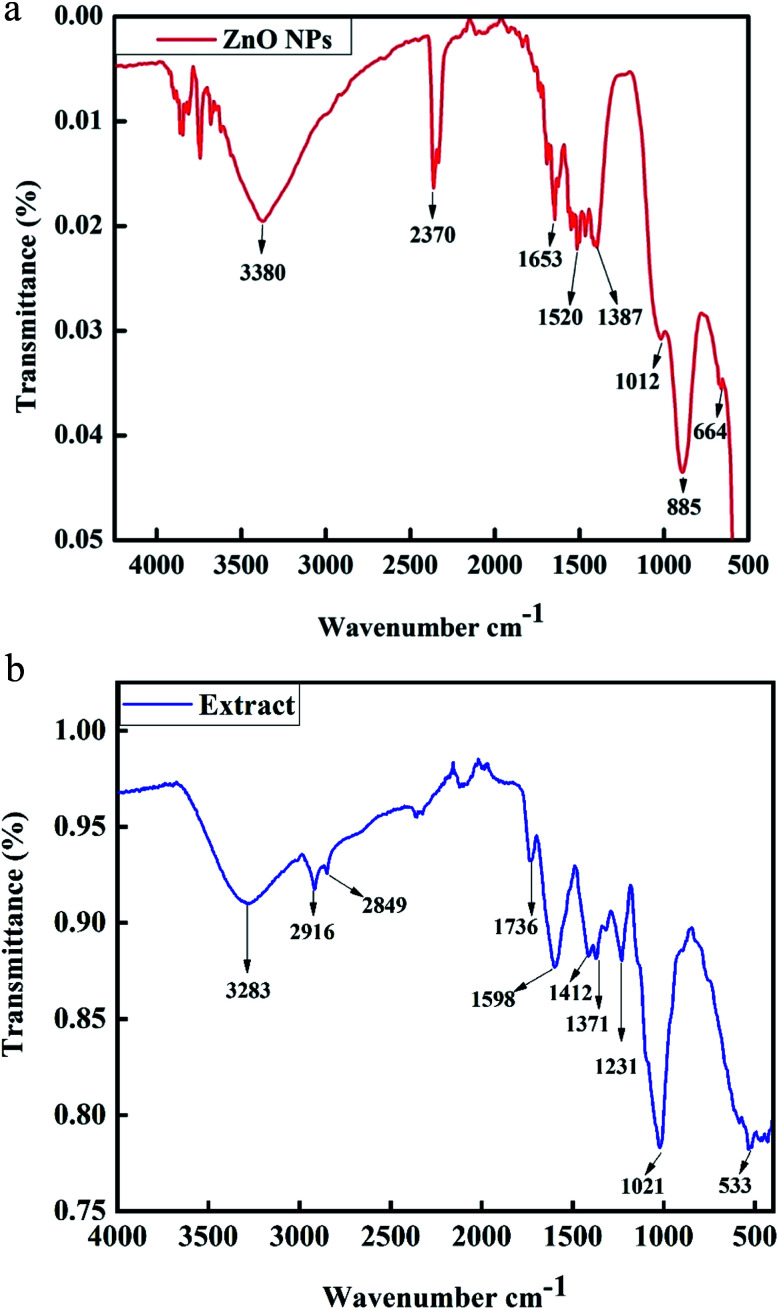
Fourier transform infrared spectroscopy is a characterization technique used for the detection of functional groups in compounds. The FT-IR spectrum of phyto-fabricated ZnO NPs in the wavenumber range from 500 to 4500 cm−1 is shown in Fig. 5a. The broad peak at 3380 cm−1 corresponds to the O–H stretching vibration of the amide group. The peaks at 2370 cm−1 are the H–O–H vibration of a cluster of water molecules of crystallization. The peaks at 1652 cm−1, 1520 cm−1, 1387 cm−1, 1012 cm−1, and 885 cm−1 correspond to the C C stretching of an alkane group, C C stretching in an aromatic ring and stretching in polyphenol (C O), C–H bending vibration of an alkane group, stretching of C–N, and bending vibration of C–H, respectively.20 The ignorable small peak at 664 cm−1 reveals the presence of C-alkyl chloride and ZnO hexagonal phase. FT-IR analysis of the plant extract was also carried out, as shown in Fig. 5b. The band at 3283 cm−1 shows symmetric O–H stretching.23 The peak at 2916 cm−1 corresponds to cholesterol, phospholipids and creatine. The band at 2849 cm−1 in the plant extract confirmed C–H stretching.24 The peak at 1736 cm−1 was attributed to C O stretching, which corresponds to lipids.25 The peak at 1598 cm−1 was attributed to C N and NH2 adenine, the peak at 1412 cm−1 represents stretching C–N, deformation N–H, and deformation C–H, the peak at 1371 cm−1 was assigned to deformation of N–H and C–H, and the peak at 1231 cm−1 was attributed to the overlapping of the protein amide III and the nucleic acid phosphate vibration.23 The peak at 1021 cm−1 corresponds to glycogen while that at 533 cm−1 corresponds to sulfur compounds.26
Fig. 5. (a) FT-IR spectrum of biosynthesized ZnO NPs. (b) FT-IR spectrum of P. somniferum plant extract.
SEM analysis of ZnO NPs
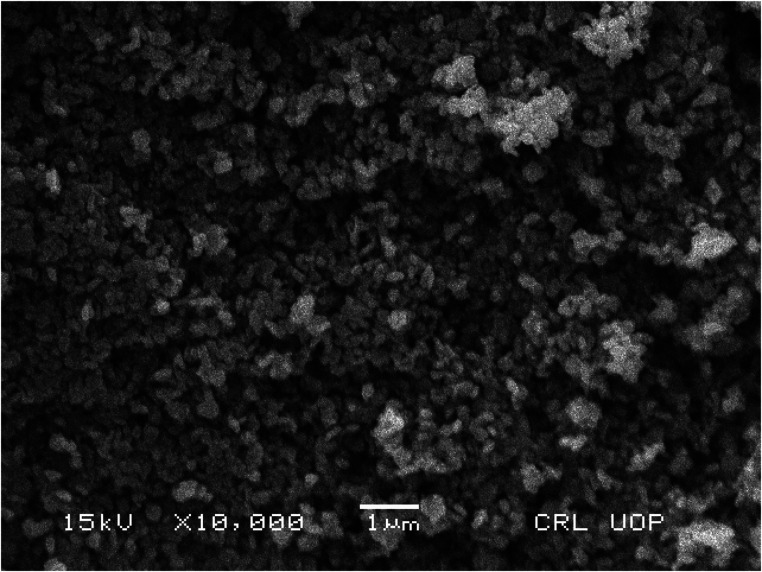
For further characterization, the ZnO NPs were analyzed by SEM to observe their morphology and structure. The current study revealed that the ZnO NPs are spherical in nature and are agglomerates of nanocrystallites, as shown in Fig. 6. A similar result for SEM analysis was reported by ref. 27.
Fig. 6. SEM analysis of ZnO NPs.
Transmission electron microscopy (TEM)
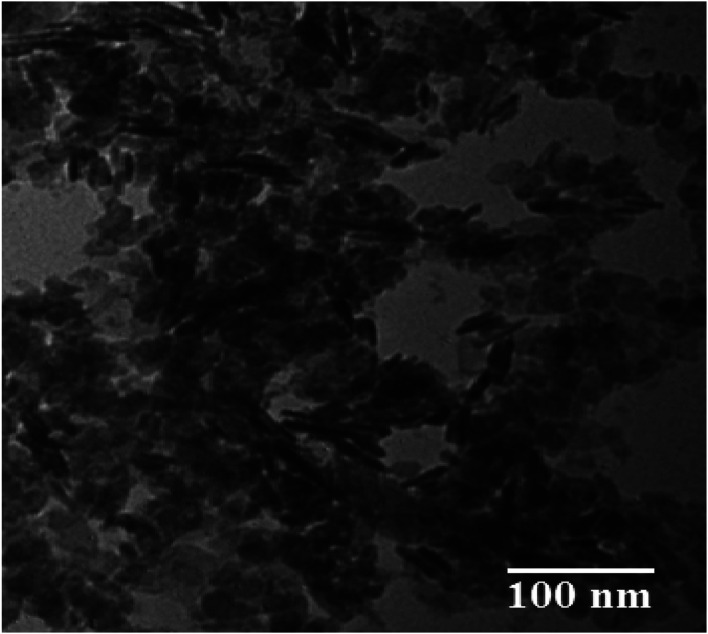
The morphology and particle size distribution of the ZnO NPs was also studied through TEM, as indicated in Fig. 7. From the TEM results, the shape of the particles can be deduced as being irregular and spherical. After digitization of the various TEM images, the particle distribution was calculated and the results were found to be in agreement with those of the XRD study.
Fig. 7. TEM image of ZnO NPs.
Anti-diabetic activity (α-amylase) of plant-mediated ZnO NPs
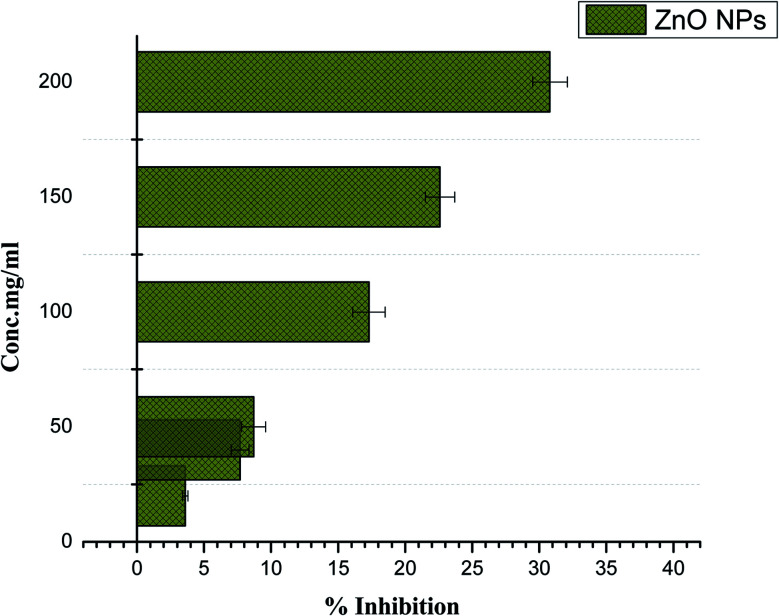
The breakdown of carbohydrates into glucose is catalyzed by α-amylase and therefore it has been related with postprandial glucose excursions in patients suffering from diabetes. Thus, α-amylase enzyme inhibitors signify a vital area in diabetes research. The α-amylase enzyme effectiveness was checked for phytosynthesized ZnO NPs and moderate enzyme inhibition was reported (30.8%) at the higher concentration of 200 mg ml−1, as indicated in Fig. 8. No inhibition was observed at 5 mg ml−1, which is the lowest concentration. Further research is recommended to determine the potency of phyto-fabricated ZnO NPs against diabetes.
Fig. 8. Alpha-amylase enzyme inhibition assay of Papaver somniferum-mediated ZnO NPs.
Live subject statement
This study was performed in compliance with the laws of QAU bioethical committee for health care. It was approved by the ethical committee of Quaid-i-Azam University. Informed consent was obtained from the human subjects prior to obtaining blood.
Biocompatibility of ZnO NPs with human RBCs
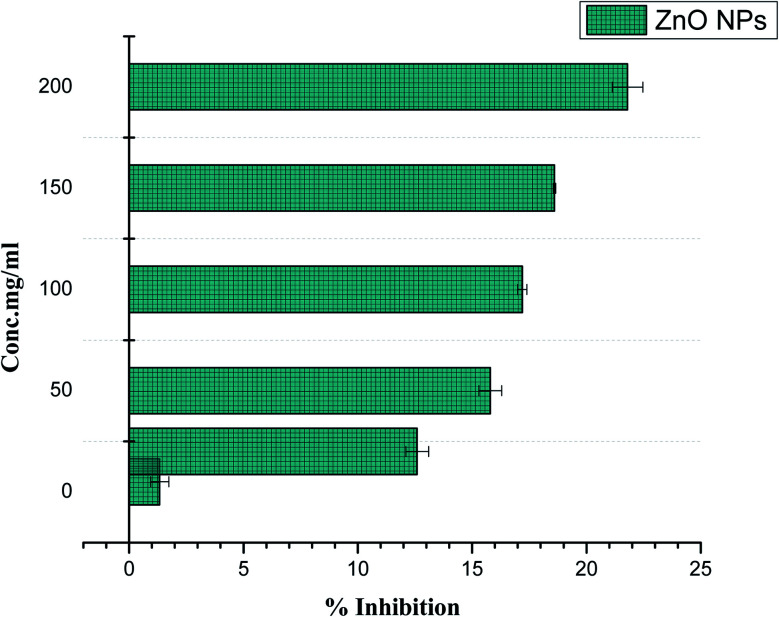
A sample causes rupturing of RBCs if the sample is found to be hemolytic. In order to observe the bio-safe nature of ZnO NPs with human RBCs, hemolytic activity was assessed at different concentrations ranging from 1 μg ml−1 to 200 μg ml−1. Cytotoxicity of ZnO NPs was observed (21.8%) at the higher concentration of 200 μg ml−1, while no hemolysis was detected at doses of <5 μg ml−1. Significantly lowering the concentration decreases the hemolysis activity. The results summarized in Fig. 9 show the less cytotoxic effect of ZnO NPs to freshly isolated RBCs. Our results have strong agreement with previous data reported by Khalil et al.28
Fig. 9. Biocompatibility potential of ZnO NPs with human RBCs.
Bactericidal activity of ZnO NPs
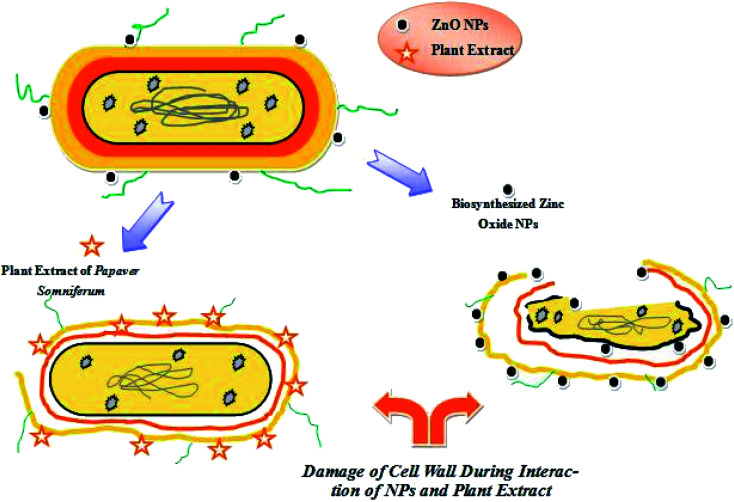
Nowadays, bacteria are showing increased resistance to drugs in the clinical field according to the literature. For this purpose, the scientific community is generating new alternative sources of antibiotics to minimize the risk of bacterial infections. The essential alkaloids present in P. somniferum exhibit a bactericidal effect against several human pathogenic bacteria. Kumaravel and Alagusundaram reported that phytochemicals present in P. somniferum have a natural inhibitory mechanism by which they either inhibit or completely kill the pathogens.29 ZnO NPs prepared using P. somniferum were found to be more potent against Gram-positive bacteria as compared to Gram-negative. Our results shown in Table 1 were in accordance with the results of.30 The permeability of ZnO NPs and their ability to bind with the cell membrane generate reactive oxygen species (ROS). Generation of ROS enhances the efficiency against drug-resistant bacterial strains.31 It was assumed that the cell wall of Gram-positive bacteria possesses specific components that help ZnO NPs to be embedded within the cellular wall. As a result, ZnO NPs break down the cellular and internal components of Gram-positive bacteria. A small amount of ZnO NPs bonds with the surface of Gram-negative bacteria and causes partial leakage of the internal contents. Our results revealed that the generation of ROS is considered as a primary bridge for cytotoxicity to the cell. Species of negatively charged ions do not have the potential to enter into the cell, thus they are converted into hydrogen peroxide and then easily penetrate inside the cells of bacteria. ZnO NPs bind to the cell wall and easily rapture the wall and interfere with the cellular machinery, which produces oxidative stress and genotoxicity and hence causes damage to the bacteria, as shown in Fig. 10.32
Antibacterial activity of biosynthesized ZnO NPs against two Gram-positive and two Gram-negative bacteria.
| Zone of inhibition (mm) | ||||
|---|---|---|---|---|
| Tested bacterial strains | Ampicillin antibiotic (10 μg ml−1) (mean ± SD) | Zinc nitrate salt (2 mM) (mean ± SD) | Plant extract (10 mg ml−1) (mean ± SD) | ZnO NPs (10 mg ml−1) (mean ± SD) |
| Gram-positive | ||||
| Staphylococcus epidermidis | 21 ± 1.08 | — | 9 ± 1.02 | 15 ± 0.78 |
| Bacillus subtilis | 16 ± 0.82 | — | 10 ± 1.01 | 13 ± 0.70 |
| Gram-negative | ||||
| Klebsiella pneumoniae | 23 ± 2.28 | — | 9 ± 1.11 | 12 ± 0.99 |
| Pseudomonas aeruginosa | 20 ± 0.87 | — | 8 ± 0.87 | 12 ± 0.85 |
Fig. 10. The proposed antibacterial mechanism presented by ZnO NPs and plant extract.
Conclusions
Our results show that P. somniferum possesses essential active bio-compounds, which are responsible for combatting different diseases, such as diabetes and infectious diseases. An optical study was done using UV-vis spectroscopy. The FT-IR results revealed that phytochemicals present in P. somniferum are responsible for capping of ZnO NPs. The spherical morphology of the ZnO NPs was confirmed by SEM imaging and the hexagonal crystal system was confirmed by XRD. The anti-diabetic activity was determined using an alpha-amylase inhibition assay, and a moderate percentage enzyme inhibition was observed at the higher concentration. At low concentration, ZnO NPs were found to be more compatible with human cells and tissues. The biocompatibility of ZnO NPs depends upon their morphology, pH, size and surface chemistry. The ZnO NPs were found to be more effective against Gram-positive drug-resistant bacteria as compared to Gram-negative bacteria strains. Based on their incredible potency, ZnO NPs prepared from P. somniferum could play a vital role in the field of nanomedicine. They could be adapted to be used in the treatment of diabetes, and also for infectious diseases. Thus, further research must be carried out to expose the toxic nature of ZnO NPs.
Conflicts of interest
No potential conflict of interest was reported by the authors.
Supplementary Material
Acknowledgments
The authors are thankful to the Department of Chemistry, Quaid-i-Azam University, Islamabad, National Center for Physics, Islamabad and Institute of Space and Technology, Islamabad for providing us the research facilities. We are also thankful to the ethical committee for approving the study.
References
- Thema F. Beukes P. Gurib-Fakim A. Maaza M. J. Alloys Compd. 2015;646:1043–1048. doi: 10.1016/j.jallcom.2015.05.279. [DOI] [Google Scholar]
- Thovhogi N. Diallo A. Gurib-Fakim A. Maaza M. J. Alloys Compd. 2015;647:392–396. doi: 10.1016/j.jallcom.2015.06.076. [DOI] [Google Scholar]
- Diallo A. Ngom B. Park E. Maaza M. J. Alloys Compd. 2015;646:425–430. doi: 10.1016/j.jallcom.2015.05.242. [DOI] [Google Scholar]
- Thema F. Manikandan E. Dhlamini M. Maaza M. Mater. Lett. 2015;161:124–127. doi: 10.1016/j.matlet.2015.08.052. [DOI] [Google Scholar]
- Sone B. Manikandan E. Gurib-Fakim A. Maaza M. J. Alloys Compd. 2015;650:357–362. doi: 10.1016/j.jallcom.2015.07.272. [DOI] [Google Scholar]
- Singh J. Kumar S. Alok A. Upadhyay S. K. Rawat M. Tsang D. C. Bolan N. Kim K.-H. J. Cleaner Prod. 2019;214:1061–1070. doi: 10.1016/j.jclepro.2019.01.018. [DOI] [Google Scholar]
- Kaminsky F. Earth-Sci. Rev. 2012;110:127–147. doi: 10.1016/j.earscirev.2011.10.005. [DOI] [Google Scholar]
- Kalusniak S. Sadofev S. Puls J. Henneberger F. Laser Photonics Rev. 2009;3:233–242. doi: 10.1002/lpor.200810040. [DOI] [Google Scholar]
- Mirzaei H. Darroudi M. Ceram. Int. 2017;43:907–914. doi: 10.1016/j.ceramint.2016.10.051. [DOI] [Google Scholar]
- Fan F. Feng Y. Tang P. Li D. Mater. Lett. 2015;158:290–294. doi: 10.1016/j.matlet.2015.05.109. [DOI] [Google Scholar]
- Suntako R. Mater. Lett. 2015;158:399–402. doi: 10.1016/j.matlet.2015.06.061. [DOI] [Google Scholar]
- Mani G. K. Rayappan J. B. B. Mater. Lett. 2015;158:373–376. doi: 10.1016/j.matlet.2015.05.006. [DOI] [Google Scholar]
- Sonker R. K. Sabhajeet S. Singh S. Yadav B. Mater. Lett. 2015;152:189–191. doi: 10.1016/j.matlet.2015.03.112. [DOI] [Google Scholar]
- Wang M.-H. Ma X.-Y. Zhou F. Mater. Lett. 2015;142:64–66. doi: 10.1016/j.matlet.2014.11.126. [DOI] [Google Scholar]
- Ovais M. Khalil A. T. Raza A. Khan M. A. Ahmad I. Islam N. U. Saravanan M. Ubaid M. F. Ali M. Shinwari Z. K. Nanomedicine. 2016;12:3157–3177. doi: 10.2217/nnm-2016-0279. [DOI] [PubMed] [Google Scholar]
- Singh J. Dutta T. Kim K.-H. Rawat M. Samddar P. Kumar P. J. Nanobiotechnol. 2018;16:84. doi: 10.1186/s12951-018-0408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wali M. Sajjad A. Sumaira S. Nano Sci. Nano Technol. 2017;11:118. [Google Scholar]
- Muhammad W. Khan M. A. Nazir M. Siddiquah A. Mushtaq S. Hashmi S. S. Abbasi B. H. Mater. Sci. Eng. C. 2019:109740. doi: 10.1016/j.msec.2019.109740. [DOI] [PubMed] [Google Scholar]
- Ali A. Ambreen S. Javed R. Tabassum S. ul Haq I. Zia M. Mater. Sci. Eng. C. 2017;74:137–145. doi: 10.1016/j.msec.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Santhoshkumar J. Kumar S. V. Rajeshkumar S. Resour.-Effic. Technol. 2017;3:459–465. [Google Scholar]
- Bhattacharya P. Dhibar S. Hatui G. Mandal A. Das T. Das C. K. RSC Adv. 2014;4:17039–17053. doi: 10.1039/C4RA00448E. [DOI] [Google Scholar]
- Jacob R. Isac J. Int. J. Chem. Stud. 2015;2:12–21. [Google Scholar]
- Dovbeshko G. I. Gridina N. Y. Kruglova E. B. Pashchuk O. P. Talanta. 2000;53:233–246. doi: 10.1016/S0039-9140(00)00462-8. [DOI] [PubMed] [Google Scholar]
- Huleihel M. Salman A. Erukhimovitch V. Ramesh J. Hammody Z. Mordechai S. J. Biochem. Biophys. Methods. 2002;50:111–121. doi: 10.1016/S0165-022X(01)00177-4. [DOI] [PubMed] [Google Scholar]
- Fabian H. Jackson M. Murphy L. Watson P. H. Fichtner I. Mantsch H. H. Biospectroscopy. 1995;1:37–45. doi: 10.1002/bspy.350010106. [DOI] [Google Scholar]
- Wood B. R. Quinn M. A. Tait B. Ashdown M. Hislop T. Romeo M. McNaughton D. Biospectroscopy. 1998;4:75–91. doi: 10.1002/(SICI)1520-6343(1998)4:2<75::AID-BSPY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Vidya C. Hiremath S. Chandraprabha M. Antonyraj M. L. Gopal I. V. Jain A. Bansal K. Int. J. Curr. Eng. Technol. 2013;1:118–120. [Google Scholar]
- Khalil A. T. Ovais M. Ullah I. Ali M. Shinwari Z. K. Khamlich S. Maaza M. Nanomedicine. 2017;12:1767–1789. doi: 10.2217/nnm-2017-0124. [DOI] [PubMed] [Google Scholar]
- Kumaravel S. Alagusundaram K. J. Pure Appl. Microbiol. 2014;8:1–6. [Google Scholar]
- Premanathan M. Karthikeyan K. Jeyasubramanian K. Manivannan G. Nanomedicine. 2011;7:184–192. doi: 10.1016/j.nano.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Abinaya C. Mayandi J. Osborne J. Frost M. Ekstrum C. Pearce J. M. Mater. Res. Express. 2017;4:075401. doi: 10.1088/2053-1591/aa796d. [DOI] [Google Scholar]
- Sirelkhatim A. Mahmud S. Seeni A. Kaus N. Ann L. Bakhori S. Hasan H. Mohamad D. Nano-Micro Lett. 2015;7:219. doi: 10.1007/s40820-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]