Abstract
The increasing prevalence of antibiotic resistance among bacterial pathogens prompted a microbiological study of fluoroquinolone structure-activity relationships with resistant mutants. Bacteriostatic and bactericidal activities for 12 fluoroquinolones were examined with a gyrase mutant of Mycobacterium smegmatis and a gyrase-topoisomerase IV double mutant of Staphylococcus aureus. For both organisms C-8 halogen and C-8 methoxy groups enhanced activity. The MIC at which 99% of the isolates tested were inhibited (MIC99) was reduced three- to fivefold for the M. smegmatis mutant and seven- to eightfold for the S. aureus mutant by C-8 bromine, chlorine, and methoxy groups. With both organisms a smaller reduction in the MIC99 (two- to threefold) was associated with a C-8 fluorine moiety. In most comparisons with M. smegmatis the response to a C-8 substituent was similar (within twofold) for wild-type and mutant cells. In contrast, mutant S. aureus was affected more than the wild type by the addition of a C-8 substituent. C-8 halogen and methoxy groups also improved the ability to kill the two mutants and the respective wild-type cells when measured with various fluoroquinolone concentrations during an incubation period equivalent to four to five doubling times. Collectively these data help define a group of fluoroquinolones that can serve (i) as a base for structure refinement and (ii) as test compounds for slowing the development of fluoroquinolone resistance during infection of vertebrate hosts.
Antimicrobial resistance is now well documented for many pathogens, and studies with a variety of bacteria indicate that resistance can develop within just a few years (1, 3, 13, 19, 25). As an antimicrobial agent becomes ineffective, it tends to be replaced by another agent; then, resistance develops to the second agent. For example, in the mid-1990s many isolates of Streptococcus pneumoniae exhibited resistance to penicillin (1). The fluoroquinolones ciprofloxacin and levofloxacin were substituted for penicillin, and now fluoroquinolone resistance is appearing (6). Preserving existing agents may require interruption of the process by which resistance develops to one agent after another.
One approach for slowing the development of resistance is to identify and use derivatives that exhibit preferential activity against resistant mutants (31). Such a feature has been seen with fluoroquinolones, initially with derivatives containing C-8 chlorine and C-8 methoxy moieties (17, 20). These observations suggested a reason that sparfloxacin, the first C-8-substituted fluoroquinolone in clinical use, is more active against resistant Mycobacterium tuberculosis than the C-8 hydrogen compound ciprofloxacin (8, 27). Subsequent comparisons between C-8 methoxy and C-8 hydrogen derivatives strengthened the observation that activity against resistant mutants can be improved preferentially (8, 9, 31).
In the present work we examined a set of fluoroquinolones for activity against two resistant mutants to rank the C-8 moieties for activity enhancement. Mycobacterium smegmatis was chosen as a representative of organisms thought to have only gyrase as the fluoroquinolone target (7), while Staphylococcus aureus represented those having both gyrase and topoisomerase IV as targets (11, 12). In most cases the C-8-substituted fluoroquinolones blocked mutant growth and killed mutants better than comparable C-8 hydrogen compounds. Enhancement of activity was usually greater with a C-8 methoxy group and lower with a C-8 fluorine moiety. Structural similarities, plus enhanced activity against resistant mutants, characterize C-8 halogen and methoxy compounds as members of a distinct group of fluoroquinolones.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. smegmatis mc2155 and its gyrA quinolone-resistant mutant KD2003 have been described previously (32). Strain KD2003, which contains an Asp-95-to-Gly substitution in GyrA, was selected as a first-step mutant on agar containing 13 times the MIC at which 99% of isolates tested are inhibited (MIC99) of PD160788, a C-8 hydrogen fluoroquinolone (32). M. smegmatis was grown as described previously (18). Incubation was at 37°C. The doubling time in 7H9 medium was 3.5 to 4 h for both strains.
S. aureus strain ISP794 (4) and its gyrA parC (grlA)-resistant mutant EN1252 were obtained from David Hooper (Massachusetts General Hospital, Boston). The construction of EN1252, which contains a Ser-84-to-Leu substitution in GyrA and a Ser-80-to-Phe substitution in ParC (Gr1A), was described in reference (4). S. aureus strains were cultured with vigorous shaking at 37°C in CY medium (24). The doubling time in CY medium was 25 min.
Fluoroquinolones.
The fluoroquinolones used (and their sources) were as follows: ciprofloxacin (Miles Laboratories, Kankakee, Ill.; currently available from Bayer AG, West Haven, Conn.); moxifloxacin and Bay y 3114 (Bayer AG) and sparfloxacin and investigational compounds (Parke-Davis Pharmaceutical Co., Ann Arbor, Mich.). Fluoroquinolone solutions were prepared as described in reference (26). Agar plates containing fluoroquinolone were prepared by adding concentrated solutions to molten agar.
Measurement of bacteriostatic and bactericidal activity.
To measure MIC99s, stationary phase cells, prepared by overnight growth, were diluted using liquid medium, and 10-μl aliquots of each dilution were spotted on quinolone-containing agar plates. Colonies on each plate were counted after incubation at 37°C for 1 day (S. aureus) or 2 to 3 days (M. smegmatis). MIC99 was defined as the fluoroquinolone concentration required to inhibit colony formation by 99%. Preliminary determinations using twofold dilutions of fluoroquinolone provided an approximate value of MIC99. This measurement was followed by a second measurement, plus a replicate, that utilized linear drug concentration increments that increased by 10 to 20% at each step. Numbers of colonies recovered were plotted against drug concentration to determine the MIC99 by interpolation.
To measure bactericidal activity, small aliquots of frozen cultures were introduced into liquid medium to give a cell density of 107 CFU/ml. Cells were then grown to exponential phase (108 CFU/ml) by shaking at 37°C. Cultures were distributed into tubes containing liquid medium and various concentrations of fluoroquinolone (see Fig. 2 through 5). Incubation with shaking was continued for either 2 h (S. aureus) or 18 h (M. smegmatis). Serial dilutions, which eliminated drug carryover, were prepared, and aliquots from the dilutions were then spotted on drug-free agar plates. Plates were incubated at 37°C for 1 day (S. aureus) or 3 days (M. smegmatis), and the colonies were counted. Bactericidal activity was expressed as percent survival relative to the CFU per milliliter at the time of fluoroquinolone addition. The 90% lethal dose (LD90) was the fluoroquinolone concentration at which survival was 10% that observed with an untreated control plated at the time of fluoroquinolone addition.
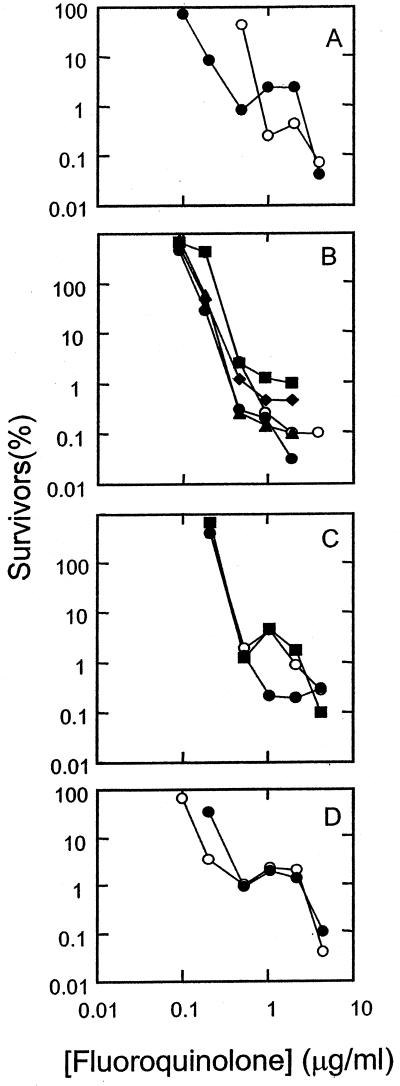
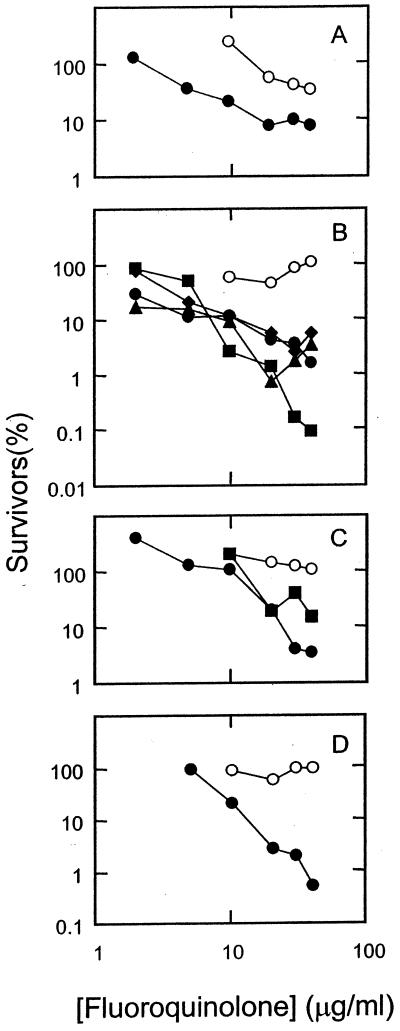
FIG. 2.
Bactericidal activities of fluoroquinolones against a wild-type strain of M. smegmatis. M. smegmatis strain mc2155 was grown to mid-log phase and then treated with the indicated concentration of fluoroquinolone for 18 h. Dilutions were prepared, and aliquots were applied to drug-free agar plates. After incubation for 2 to 3 days, colonies were counted, and the fraction of surviving cells was calculated. (A) Ciprofloxacin (C-8-H, open circles) and PD129603 (C-8-C1, filled circles). (B) PD138032 (C-8-H, open circles), PD125275 (C-8-F, filled diamonds), PD 163753 (C-8-Br, filled squares), PD138124 (C-8-Cl, filled triangles), and PD135432 (C-8-methoxy, filled circles). (C) PD158804 (C-8-H, open circles), sparfloxacin (C-8-F, filled circles), and PD125232 (C-8-F, filled squares). (D) Bay y-3114 (C-8-H, open circles) and moxifloxacin (C-8-methoxy, closed circles). Similar results were obtained in a replicate experiment.
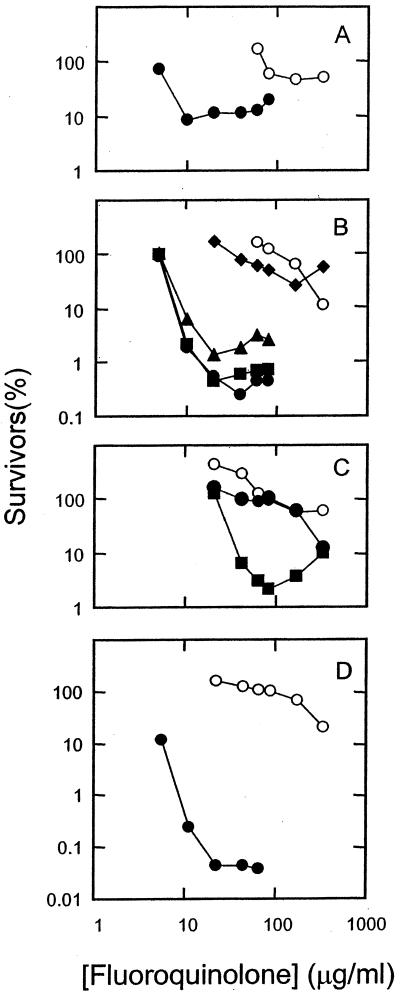
FIG. 5.
Bactericidal activities of fluoroquinolones against a parC gyrA resistance mutant of S. aureus. Strain EN1252 was grown to mid-log phase and then treated with the indicated concentration of fluoroquinolone for 2 h. Dilutions were prepared and aliquots were applied to drug-free agar plates. After incubation for 1 day, colonies were counted and the fraction of surviving cells was calculated. Symbols and panels are the same as those in Fig. 3. Similar results were obtained in a replicate experiment.
Selection of third-step mutants of S. aureus.
S. aureus strain EN1252 was grown to stationary phase by overnight incubation in CY medium, the cells were harvested by centrifugation (4,000 × g for 10 min), and they were then resuspended in CY medium at a dilution of 1:10 relative to the culture prior to centrifugation. Fluoroquinolones (see Table 3) were added at lethal concentrations (fourfold the MIC99 for each fluoroquinolone; see reference 5) to cell suspensions to enrich third-step mutant fractions of the populations. Incubation was for 5 h, after which cells were again concentrated by centrifugation. Cells (5 × 107 CFU/ml) were then applied to agar plates containing the same fluoroquinolone used for enrichment of mutants at concentrations of two and threefold the MIC99. Colonies were recovered after incubation at 37°C for 2 to 3 days. Cells from colonies were spread on drug-free GL agar to obtain single colonies after incubation for 1 day at 37°C; subsequent testing on quinolone-containing GL agar confirmed that the putative third-step mutants were capable of growth on the selective fluoroquinolone at the selection concentration.
TABLE 3.
Effect of C-8 substituent on identity of third-step resistance allele in S. aureus
| Straina | Fluoroquinolone for selectionb | C-8 substituent | C-7 ring alkyl | Variants recoveredc
|
|
|---|---|---|---|---|---|
| GyrA | ParC | ||||
| KD2062 | PD138032 | H | 3′ C-methyl | S85P | |
| KD2063 | PD138032 | H | 3′ C-methyl | S85P | — |
| KD2064 | PD138032 | H | 3′ C-methyl | S85P | — |
| KD2065 | PD163753 | Br | 3′ C-methyl | S85P | — |
| KD2066 | PD163753 | Br | 3′ C-methyl | S85P | — |
| KD2067 | PD163753 | Br | 3′ C-methyl | —d | —d |
| KD2068 | PD138124 | Cl | 3′ C-methyl | S85P | — |
| KD2069 | PD138124 | Cl | 3′ C-methyl | —d | —d |
| KD2070 | PD138124 | Cl | 3′ C-methyl | E84K | |
| KD2071 | PD135432 | OMe | 3′ C-methyl | S85P | — |
| KD2072 | PD135432 | OMe | 3′ C-methyl | S85P | — |
| KD2073 | Ciprofloxacin | H | None | S85P | — |
| KD2074 | Ciprofloxacin | H | None | S85P | — |
| KD2075 | Ciprofloxacin | H | None | —d | —d |
| KD2076 | Sparfloxacin | F | 3′,5′ dimethyl | S85P | — |
| KD2077 | Sparfloxacin | F | 3′,5′ dimethyl | S85P | — |
| KD2078 | Sparfloxacin | F | 3′,5′ dimethyl | S85P | — |
Parental strain for selection of mutants was S. aureus EN1252, a GyrA (S84L)-ParC (S80F) double mutant. Strain numbers listed are for third-step mutants.
Mutants were selected on GL agar plates containing two to three -times the MIC99 (listed in Table 1) for each compound.
GyrA (DNA gyrase) and ParC (DNA topoisomerase IV; also called GrlA in S. aureus) variants. Variants are indicated with the wild-type amino acid preceeding the amino acid number, which is followed by the variant amino acid. Amino acid abbreviations: A, alanine; E, glutamic acid; F, phenylalanine, K, lysine; P, proline; S, serine; V, valine. —, QRDR had no changes relative to the parental strain.
QRDR of gyrB and parE (grlB) had no changes relative to the parental strain.
DNA sequence determination.
The nucleotide sequences of the quinolone-resistance-determining region (QRDR) of gyrA, parC, gyrB, and parE were determined after amplification of the respective DNA fragments from S. aureus chromosomal DNA templates using PCR. Cells in 1 ml of late-log-phase culture were harvested by centrifugation and resuspended in 100 μl of SST buffer (50 mM NaCl, 20% sucrose [wt/vol], 50 mM Tris-HCl [pH 7.6]). Lysostaphin (AMBI Inc., Purchase, N.Y.) and RNase A (Sigma Chemical Corp., St. Louis, Mo.) were added to final concentrations of 100 and 200 μg/ml, respectively. After incubation at 37°C for 10 min followed by 3 min on ice, samples were mixed with 300 μl of general lysis buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.9], 1 mM EDTA [pH 8.0]) plus 20 μl of 20% Triton X-100. Samples were then boiled for 10 min. Cell debris was removed by centrifugation, and 8 μl of DNA-containing supernatant fluid was used for each 100-μl PCR.
PCRs involved incubations at 95°C for 2 min followed by 30 cycles at 93°C for 1 min 20 s, 49°C for 1 min, and 72°C for 1 min. After each cycle the 72°C elongation phase was extended by 1 s. The PCR products were purified with a Qiagen PCR purification kit, and nucleotide sequences were determined in the forward direction with an automated DNA sequencer. Primers SA-parC.seq (5′ ACG TCG TAT TTT ATA TGC AA-3′), SA-gyrAfwd (5′-AGA TTA TGC GAT GAG TGT TAT CGT TGC-3′), SA parE.seq (5′-AAA AAG CGA TTA AAG CAC AAC AAG CAA-3′), and SA gyrB.seq (5′-GTC GCA CGT ACA GTG GTT GAA AAA GG-3′) were used for sequencing after PCR amplification of DNA fragments using primers SA-parCfwd (5′ TGA TGA GGA GGA AAT CTA GTG-3′), SA-parCrev (5′ GGA AAT CTT GAT GGC AAT AC-3′), SA-gyrAfwd (defined above), SA-gyrArev (5′-TAG TCA TAC GCG CTT CAG TAT AAC GCA-3′), SA-parEfwd (5′-CAA ACG AAA TCT AAA TTG GGT ACT TCT-3′), SA-parErev (5′-TCT TCG TCT GTC CAA GCG TAT T-3′), SA-gyrBfwd (5-TGG TAC GCA TGA AGA CGG ATT C-3′), and SA-gyrBrev (5′-GAC CTT TGT ATA GCG CAA TAG ACC ATT-3′).
RESULTS
Effect of C-8 substituents on bacteriostatic activity.
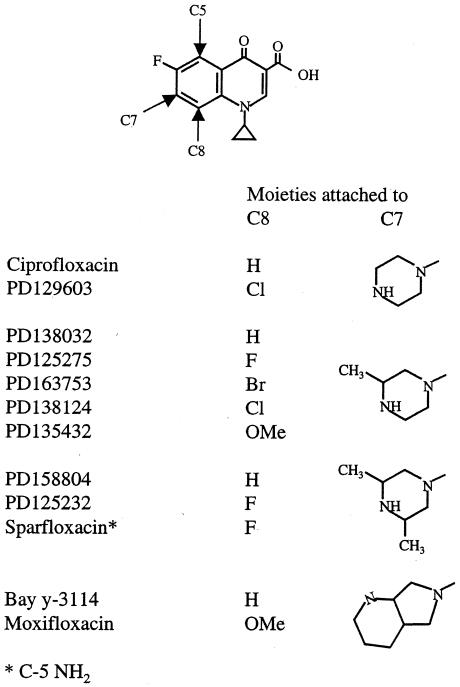
We have argued that inhibition of growth by quinolones occurs at lower drug concentrations than cell death when the latter is measured using short incubation times (5, 31). To better distinguish between the two processes and to attain greater accuracy, we measured bacteriostatic activity by determining the MIC99 rather than the more standard inhibition of 99.99 to 99.999% (23). MIC99s for wild-type M. smegmatis are listed in Table 1, where the fluoroquinolones are grouped according to their C-7 ring structure (Fig. 1 shows structures of compounds). With M. smegmatis the C-8 halogen and methoxy groups improved the ability to block growth by about threefold when the compounds had small C-7 rings. The larger C-7 ring systems present on PD158804 and Bay y-3114 rendered these C-8 hydrogen compounds more active than their C-8-modified derivatives. With the gyrase mutant, however, all of the C-8-modified compounds were more active than their C-8 hydrogen derivatives (Table 1). Thus, a C-8-halogen or methoxy group increases bacteriostatic activity against a resistant variant of M. smegmatis in which glycine has been substituted for aspartic acid at GyrA position 95.
TABLE 1.
Effect of C-8 substituents on inibition of growth by fluoroquinolones
| Fluoroquinolonea | C-7 ring alkyld |
M. smegmatisb
|
S. aureusc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
|
gyrA+
|
gyrA
|
parC+gyrA+
|
parC gyrA
|
||||||
| MIC99 (μg/ml)e | MIC(C-8-H)f MIC(C-8-X) | MIC99 (μg/ml) | MIC(C-8-H) MIC(C-8-X) | MIC99 μg/ml) | MIC(C-8-H) MIC(C-8-X) | MIC99 (μg/ml) | MIC(C-8-H) MIC(C-8-X) | ||
| Ciprofloxacin (H) | None | 0.25 | 5.4 | 0.10 | 23 | ||||
| PD129603 (Cl) | None | 0.08 | 3.1 | 1.0 | 5.4 | 0.065 | 1.6 | 2.7 | 8.5 |
| PD138032 (H) | Me | 0.10g | 2.8 | 0.12 | 21 | ||||
| PD125275 (F) | Me | 0.036g | 2.8 | 1.2 | 2.3 | 0.1 | 1.2 | 7.3 | 2.9 |
| PD163753 (Br) | Me | 0.032g | 3.1 | 0.96 | 2.9 | 0.066 | 1.8 | 2.9 | 7.2 |
| PD138124 (Cl) | Me | 0.029g | 3.4 | 0.66 | 4.2 | 0.056 | 2.1 | 2.4 | 8.7 |
| PD135432 (OMe) | Me | 0.030g | 3.3 | 0.49 | 5.7 | 0.05 | 2.4 | 2.5 | 8.4 |
| PD158804 (H) | di-Me | 0.043 | 2.9 | 0.12 | 29 | ||||
| PD125232 (F) | di-Me | 0.049 | 0.88 | 1.2 | 2.5 | 0.15 | 0.81 | 14 | 2.1 |
| Bay y 3114 (H) | NA | 0.06 | 1.2 | 0.022 | 14 | ||||
| Moxifloxacin (OMe) | NA | 0.08 | 0.75 | 0.8 | 1.5 | 0.035 | 0.63 | 1.7 | 8.5 |
| Sparfloxacinh (F) | di-Me | 0.07 | 1.3 | 0.038 | 9 | ||||
Parenthetical information indicates moiety at the C-8 position. PD135432 is identical to gatifloxacin. OMe, methoxy.
M. smegmatis strains were mc2 155 (gyrA+) and KD2003 (gyrA).
S. aureus strains were ISP794 (gyrA+ parC+) and EN1252 (gyrA parC); in S. aureus parC was originally termed grlA.
Alkyls attached to C-7 ring carbon. NA, not applicable because the C-7 ring systems are not comparable to other members of the set (see Fig. 1).
Identical results for each MIC99 were obtained for two independent determinations.
MIC99 for the C-8-H fluoroquinolone of each series having a given C-7 ring structure, divided by MIC99 for indicated compounds having C-8 substitutents, where X = F, Br, Cl, or OMe (methoxy) groups.
Data taken from reference (26).
Identical to PD125232 but with additional C-5 NH2.
FIG. 1.
Quinolone structures. OMe, methoxyl moiety.
We next examined S. aureus as an example of the more complex situation in which two mutant quinolone targets are present. In this mutant, position 84 is Leu in GyrA (gyrase) and position 80 is Phe in ParC (also known as GrlA, a subunit of topoisomerase IV; we use the ParC designation below to make S. aureus nomenclature consistent with that of other bacteria). The general pattern of susceptibility was similar to that observed with M. smegmatis (Table 1), supporting the conclusions drawn above. However, the enhancing effect of the moieties attached to the C-8 position was less with wild-type S. aureus than with wild-type M. smegmatis and more with mutant S. aureus than with mutant M. smegmatis (Table 1). These differences between the two organisms were expected because wild-type M. smegmatis, like many other mycobacterial species, behaves as though it is a moderately resistant mutant (14, 15). With mutants of both species enhancement was usually least for a C-8 fluorine group and most for the C-8 chlorine and C-8 methoxy substituents.
An appropriate C-8 hydrogen compound was unavailable for directly evaluating the effect of the C-8 fluorine in sparfloxacin (sparfloxacin contains a C-5 amino group absent from the other compounds tested). However, the closely related C-8 fluorine derivative, PD125232, was more active against mutants than its C-8-hydrogen cognate PD158804 (Table 1). Thus, the C-8 fluorine of sparfloxacin probably contributes to activity against mutants.
Effect of C-8 substituents on bactericidal activity.
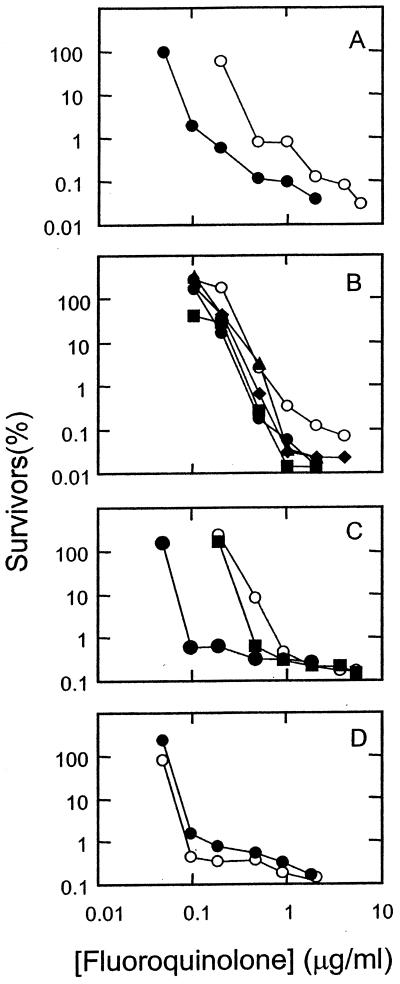
Lethal activity, when measured with short incubation times, probably arises from the formation of quinolone-topoisomerase-DNA complexes followed by release of double-strand DNA breaks from the complexes (5). To assess the action of C-8 substituents on the combined effect of these two events, cultures of M. smegmatis and S. aureus were incubated with each of the 12 fluoroquinolones. Then, the fraction of surviving cells was determined (see Materials and Methods). Plots of survival at various quinolone concentrations show that few of the C-8-substituted compounds were more lethal than their C-8 hydrogen derivatives against wild-type M. smegmatis (Fig. 2) and S. aureus (Fig. 3). The effect of the C-8 moieties on lethal action was more pronounced with the resistant GyrA variant of M. smegmatis (Fig. 4) and with the GyrA ParC variant of S. aureus (Fig. 5) than with wild-type strains, as shown by LD90 determinations (Table 2). Against the mutants the C-8 bromine and methoxy groups conferred the most activity; in some cases a C-8-fluorine moiety improved activity, while in others it did not.
FIG. 3.
Bactericidal activities of fluoroquinolones against a wild-type strain of S. aureus. Strain ISP794 was grown to mid-log phase and then treated with the indicated concentration of fluoroquinolone for 2 h. Dilutions were prepared and aliquots were applied to drug-free agar plates. After incubation for 1 day, colonies were counted and the fraction of surviving cells was calculated. (A) Ciprofloxacin (C-8-H, open circles) and PD129603 (C-8-Cl, filled circles). (B) PD138032 (C-8-H, open circles), PD125275 (C-8-F, filled diamonds), PD 163753 (C-8-Br, filled squares), PD138124 (C-8-C1, filled triangles), and PD135432 (C-8-OMe, filled circles). (C) PD158804 (C-8-H, open circles), sparfloxacin (C-8-F, filled circles), and PD125232 (C-8-F, filled squares). (D) Bay y-3114 (C-8-H, open circles) and moxifloxacin (C-8-methoxy, closed circles). Similar results were obtained in a replicate experiment.
FIG. 4.
Bactericidal activities of fluoroquinolones against a gyrA resistance mutant of M. smegmatis. M. smegmatis strain KD2003 was grown to mid-log phase and then treated with the indicated concentration of fluoroquinolone for 18 h. Dilutions were prepared, and aliquots were applied to drug-free agar plates. After incubation for 2 to 3 days, colonies were counted, and the fraction of surviving cells was calculated. Panels and symbols are the same as those in Fig. 2. Similar results were obtained in a replicate experiment.
TABLE 2.
Effect of C-8 substituents on lethal action of fluoroquinolones
| Fluoroquinolonea | C-7 ring alkylb | LD90 (μg/ml)c
|
|||
|---|---|---|---|---|---|
|
M. smegmatis
|
S. aureus
|
||||
| gyrA+ | gyrA | parC+ gyrA+ | parC gyrA | ||
| Ciprofloxacin (H) | 0.6 | >120 | 0.28 | >320 | |
| PD129603 (Cl) | 0.19 | 11 | 0.07 | 9.4 | |
| PD1380232 (H) | Me | 0.38 | >120 | 0.34 | 320 |
| PD125275 (F) | Me | 0.29 | 12 | 0.27 | >320 |
| PD163753 (Br) | Me | 0.38 | 7.2 | 0.23 | 7.5 |
| PD138124 (Cl) | Me | 0.25 | 9.0 | 0.32 | 9.0 |
| PD135432 (OMe) | Me | 0.22 | 8.0 | 0.21 | 7.3 |
| PD158804 (H) | di-Me | 0.37 | >120 | 0.47 | >320 |
| PD125232 (F) | di-Me | 0.36 | 100 | 0.31 | 35 |
| Bay y 3114 (H) | NA | 0.15 | 120 | 0.07 | >320 |
| Moxifloxacin (OMe) | NA | 0.28 | 12 | 0.08 | 5.2 |
Parenthetical information indicates moiety at the C-8 position. PD135432 is identical to gatifloxacin. OMe, methoxy.
Alkyls attached to C-7 ring. NA, not applicable because the C-7 ring systems are not comparable with other members of the set (see Fig. 1).
Concentration required to reduce survival by 90% as determined from Fig. 2 to 5. Similar results were obtained in two independent experiments. The S. smegmatis strains were mc2155 (gyrA+) and KD2003 (gyrA); the S. aureus strains were ISP794 (parC+ gyrA+) and EN1252 (parC gyrA). In S. aureus parC was originally designated grlA.
Determination of fluoroquinolone target in S. aureus.
Since S. aureus has two topoisomerase targets for the fluoroquinolones, we did not know a priori whether the data described above for the mutant reflected activity against resistant gyrase or resistant topoisomerase IV. Since the more susceptible target is expected to be under more selective pressure than the less susceptible one, the identity of a third-step mutation should reveal the principle target in the double mutant. We obtained third-step (triple) mutants from the double mutant by selection with six of the fluoroquinolones and then determined the nucleotide sequences of the QRDR (28) of gyrA and parC. Most of the compounds selected GyrA variants in which serine-85 was changed to proline (Table 3). We were able to use levofloxacin, a fluoroquinolone whose structure is distantly related to that of compounds in the present set, to rule out the possibility that third-step ParC variants were inviable in the genetic background tested: of three independent mutants selected by levofloxacin, two were variants of ParC; one lacked a mutation in the QRDR of either gyrA or parC (data not shown). Taken together the data indicate that resistant gyrase was the target in the S. aureus mutant for the compounds listed in Table 3.
Several additional features of Table 3 require comment. First, the propensity for selection of the Ser-85-to-Pro variant, rather than a more common position 88 variant, may be related to the preexisting amino acid change at position 84. A comparable change occurred when the reverse experiment was performed with Escherichia coli: Ser-83 (equivalent to position 84 in S. aureus)-to-Leu variants were selected when the parental strain contained a Ser-84-to-Pro change (data not shown). Rules governing selection of multiple changes in gyrase are under investigation. Second, the C-8 chlorine derivative PD138124 selected both GyrA and ParC variants. With this compound some cells in the population appear to have mutant gyrase as the primary target while other cells have mutant topoisomerase IV as the main target. Apparently a change in either protein lowers susceptibility. Third, the QRDRs for three mutants lacked nucleotide sequence changes in either gyrA or parC. For these cases additional work is required to determine whether resistance is due to nontarget mutations, as reported previously for second-step mutants (11), or to amino acid changes in GyrA that are outside the QRDR (16). Changes in the GyrB or ParE proteins are not likely explanations because nucleotide sequence analysis indicated that the QRDRs of these two proteins were identical in the three mutants to those of the parental strain (data not shown).
DISCUSSION
The experiments described above indicate that C-8 fluorine, chlorine, bromine, and methoxy moieties increase the activity of fluoroquinolones against fluoroquinolone-resistant mutants of M. smegmatis and S. aureus. The potency measures used in the present work (MIC99 and LD90) differ from those used by clinical laboratories (MIC and minimum bactericidal concentration). However, the rank order of the compounds is likely to be similar; thus, we expect the activity increase associated with C-8 halogen or methoxy groups to be observed when NCCLS assays are employed.
Measurement of MIC99 with S. aureus (Table 1) showed that C-8 substituents enhance the ability to inhibit growth of mutants more than growth of wild-type cells. This phenomenon, which has also been observed for a C-8 methoxy compound with E. coli (31), was not as obvious with M. smegmatis (Table 1). Since wild-type gyrase of M. smegmatis has an amino acid sequence and behavior similar to first-step mutants of other bacteria (14, 15), the similarity between wild-type and mutant M. smegmatis is consistent with a preferential enhancement of activity with mutants.
Biochemical experiments with an engineered, quinolone-resistant gyrase of E. coli support the assumption that the differences among the compounds reflect effects on topoisomerases. Purified, resistant enzyme showed greater quinolone susceptibility, relative to wild-type enzyme, according to the following order (from greatest to least activity): C-8 bromine and C-8 chlorine, C-8 fluorine and C-8 ethoxy, and C-8 hydrogen (2). Our intracellular data (Table 1) showed a similar series, although we examined C-8 methoxy rather than C-8 ethoxy effects. The methoxy group was generally among the most active. Preferential improvement of activity against resistant mutants can be explained if the recognition helix of the GyrA protein (22) serves as an important portion of the quinolone binding site (21). Resistance mutations that lower susceptibility the most tend to reduce the electron-rich character of the recognition helix. The C-8 halogen and C-8 methoxy fluoroquinolone substituents may partially restore that microenvironment, thereby improving drug binding. Although it is likely that resistant gyrase was the primary target for M. smegmatis (the related M. tuberculosis lacks genes encoding topoisomerase IV [7]) and in most cases for S. aureus (Table 3), the principle sketched above could also apply to topoisomerase IV (16).
Enhanced lethal activity, which was most noticeable with the mutants (Fig. 2 through 5; Table 2), is probably due in part to increased ability to form ternary complexes (lower MIC99s; Table 1). We have argued previously that C-8 moieties confer an additional activity against mutants that is likely to involve the release of breaks from the complexes (8, 31). Determination of the LD90, which reflects both events, shows that C-8 halogen and methoxy groups increase lethal activity, although with some compounds the C-8 fluorine has little effect (Table 2).
We conclude that the presence of a C-8 fluorine, chlorine, bromine, or methoxy moiety helps define a set of fluoroquinolones that is distinguished by increased activity against resistant gyrase mutants. In addition to findings with M. smegmatis and S. aureus, described above and in the references (17, 26, 30), selected representatives of the C-8-substituted group have shown increased activity with mutants of E. coli (21, 31), Pseudomonas aeruginosa (20), Mycobacterium bovis BCG (8, 29), and M. tuberculosis (8, 29). Thus, it is likely that the properties of the group apply to many bacterial species. Since some members of the group, such as sparfloxacin, gatifloxacin, and moxifloxacin, are being used clinically, it may be possible to test the idea (10) that these compounds will cause resistance to develop more slowly than a C-8 hydrogen fluoroquinolone such as ciprofloxacin.
ACKNOWLEDGMENTS
We thank Jerome Schentag for suggesting a study of quinolone relationships and the following for critical comments on the manuscript: Marila Gennaro, Samuel Kayman, and Anthony Maxwell. We also thank Anthony Maxwell for communicating unpublished observations.
This work was supported by grants from the NIH (AI35257), Mylan Pharmaceutical Co., and Bayer AG.
REFERENCES
- 1.Baquero F. Trends in antibiotic resistance of respiratory pathogens: an analysis and commentary on a collaborative surveillance study. J Antimicrob Chemother. 1996;38(Suppl. A):117–132. doi: 10.1093/jac/38.suppl_a.117. [DOI] [PubMed] [Google Scholar]
- 2.Barnard F, Maxwell A. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser83 and Asp87. Antimicrob Agents Chemother. 2001;45:1994–2000. doi: 10.1128/AAC.45.7.1994-2000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennish M, Salam M A, Hossain M A, Myaux J, Khan E H, Chakraborty J, Henry F, Ronsmans C. Antimicrobial resistance of Shigella isolates in Bangladesh, 1983–1990: increasing frequency of strains multiply resistant to ampicillin, trimethoprim-sulfamethoxazole, and nalidixic acid. Clin Infect Dis. 1992;14:1055–1060. doi: 10.1093/clinids/14.5.1055. [DOI] [PubMed] [Google Scholar]
- 4.Bisognano C, Vaudaux P, Lew D, Ng E, Hooper D. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother. 1997;41:906–913. doi: 10.1128/aac.41.5.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C-R, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 6.Chen D K, McGeer A, DeAzavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S, Eiglmeier K, Gas S, Barry C E, Tekaia F, Babcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on bacterial growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica K. A strategy for fighting antibiotic resistance. ASM News. 2001;67:27–33. [Google Scholar]
- 11.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 13.Gorwitz R, Nakashima A, Moran J, Knapp J. Sentinel Surveillance for antimicrobial resistance in Neisseria gonorrhoeae—United States, 1988–1991. Morb Mortal Wkly Rep. 1993;42:29–39. [PubMed] [Google Scholar]
- 14.Guillemin I, Jarlier V, Cambau E. Correlation between quinolone sensitivity patterns and sequences in the A and B subunits of DNA gyrase in mycobacteria. Antimicrob Agents Chemother. 1998;42:2084–2088. doi: 10.1128/aac.42.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemin I, Sougakoff W, Cambau E, Revel-Viravau V, Moreau N, Jarlier V. Purification and inhibition by quinolones of DNA gyrases from Mycobacterium avium, Mycobacterium smegmatis and Mycobacterium fortuitum bv. peregrinum. Microbiology. 1999;145:2527–2532. doi: 10.1099/00221287-145-9-2527. [DOI] [PubMed] [Google Scholar]
- 16.Ince D, Hooper D. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob Agents Chemother. 2000;44:3344–3350. doi: 10.1128/aac.44.12.3344-3350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T, Matsumoto M, Nishino T. Improved bactericidal activity of Q-35 against quinolone-resistant staphylococci. Antimicrob Agents Chemother. 1995;39:1522–1525. doi: 10.1128/aac.39.7.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs W R, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems in mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 19.Johnson A P. Antibiotic resistance among clinically important Gram-positive bacteria in the UK. J Hosp Infect. 1998;40:17–26. doi: 10.1016/s0195-6701(98)90020-2. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura A, Hoshino K, Kimura Y, Hayakawa I, Sato K. Contribution of the C-8 substituent of DU-6859a, a new potent fluoroquinolone, to its activity against DNA gyrase mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1467–1471. doi: 10.1128/aac.39.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu T, Zhao X, Drlica K. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activity by the C-8-methoxy group. Antimicrob Agents Chemother. 1999;43:2969–2974. doi: 10.1128/aac.43.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morais-Cabral J H, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved Standard M7–A4, section 5.3.4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.Novick R P, Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972;68:285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph K, Parkinson A, Reasonover A, Bulkow L, Parks D, Butler J. Serotype distribution and antimicrobial resistance patterns of invasive isolates of Streptococcus pneumoniae: Alaska, 1991–1998. J Infect Dis. 2000;182:490–496. doi: 10.1086/315716. [DOI] [PubMed] [Google Scholar]
- 26.Sindelar G, Zhao X, Liew A, Dong Y, Zhou J, Domagala J, Drlica K. Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrob Agents Chemother. 2000;44:3337–3343. doi: 10.1128/aac.44.12.3337-3343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu C, Kreiswirth B N, Sreevatsan S, Musser J M, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174:1127–1130. doi: 10.1093/infdis/174.5.1127. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B-Y, Pine R, Domagala J, Drlica K. Fluoroquinolone action against clinical isolates of Mycobacterium tuberculosis: effects of a C-8 methoxyl group on survival in liquid media and in human macrophages. Antimicrob Agents Chemother. 1999;43:661–666. doi: 10.1128/aac.43.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Wang J-Y, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42:956–958. doi: 10.1128/aac.42.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J-F, Dong Y, Zhao X, Lee S, Amin A, Ramaswamy S, Domagala J, Musser J M, Drlica K. Selection of antibiotic resistance: allelic diversity among fluoroquinolone-resistant mutations. J Infect Dis. 2000;182:517–525. doi: 10.1086/315708. [DOI] [PubMed] [Google Scholar]