Abstract
A gold nanoparticle strip was developed for rapidly evaluating FMDV type O antibody level and simultaneously discriminating FMDV vaccinated animals from infected animals. The strip was established depending on the colloidal gold nanoparticle labeling technique. Staphylococcal protein A colloidal gold nanoparticles were used as a probe. The epitope antigens of FMDV structural proteins and nonstructural proteins were dispensed on a nitrocellulose membrane as two test lines, respectively, and goat anti-pig antibody IgG was used as a control line. The assay was evaluated with FMDV immunized, infected sera and positive sera for another virus. The results showed the specificities of the T1 and T2 lines were 95.17% and 100% respectively. The sensitivity was in accordance with commercial ELISA kits. The coincidence rate of the new strip with 3ABC Mab-bELISA and LPB-ELISA was 95.5% and 93.13%, respectively. In summary, this experimental strip could provide a simple, inexpensive and rapid approach for onsite detection of FMDV type O antibody level and discrimination of FMDV vaccinated from infected animals without any expensive instrument.
A gold nanoparticle strip was developed for rapidly evaluating FMDV type O antibody level and simultaneously discriminating FMDV vaccinated animals from infected animals.
1. Introduction
Foot-and-mouth disease (FMD) is induced by a highly contagious virus named FMD virus (FMDV),1 which is mostly pandemic and severely impacts the swine industry and food security.2 The multiple modes of transmission, fast replication rate and rapid viral excretion have rendered FMD an acute and highly contagious disease of cloven-hoofed animals.3 Countries in Southeast and East Asia show varying FMD endemic levels, among which, Cambodia, Thailand, Laos, China and Vietnam show relatively high FMD incidence.4
FMDV is a single-stranded, positive-sense RNA virus. The FMDV genome contains a single open reading frame (ORF) that encodes structural proteins (SPs) and non-structural proteins (NSPs).2,5 The SPs include VP1, VP2, VP3, VP4, and VP1 is one of the most important protective antigens which contain critical neutralizing antigen epitopes.6 Whereas the NSPs of FMDV are relatively conserved.7–9
In developing countries, the eradication and control strategy of FMD mainly relies on vaccine immunization. Therefore, for FMDV infected and immunized animals, there are a series of questions that need to be investigated thoroughly. Firstly, how to rapidly evaluate the maternal antibody and vaccine-induced antibody level, which is the basis of setting a proper immunizing program. Secondly, how to accurately differentiate FMDV infected from immunized animals is an important challenge. Normally, the serological methods for FMDV antibody level evaluation are all based on SPs and whole virus, including virus neutralization test,10 liquid phase blocking enzyme linked immunosorbent assay (LPB-ELISA),11 solid-phase competition ELISA (SPCE),12 while ELISAs based on NSPs are normally used to discriminate infected animals from vaccinated animals.13–16 Recently, the immunochromatographic strip was be successfully used in various fields due to its specificity, sensitivity, rapidity, low cost and adaptability for field detection and high sample throughput.17,18 FMDV strips for antibody detection and vaccinated-infected animal discrimination have been successfully developed based on recombinant VP1 protein, and antigen epitope of NSPs by our team, previously.19–22 To make a more convenient detection strip, a gold nanoparticle assay for simultaneous discriminating infected from immunized animal and evaluating immunized antibody level of FMDV was developed using neutralize epitopes of VP1 and B cell epitopes of NSPs in this study.
2. Experimental
2.1. Sera origins
A total of 379 swine sera were used to evaluate the performance of the gold nanoparticle strip. The sera came from different source as follows: (I) FMDV vaccinated swine sera were collected from different day after immunization (n = 145). (II) FMDV (A/GDMM/CHA/2013) infected swine sera were collected between 0–63 days post-infection (DPI) (n = 58). (III) Field sera from pigs infected with FMDV O/MYA/98 strain (n = 8). (IV) Positive sera against PRRSV, PRV, CSFV, PCV2 and JEV (n = 20). (V) Unvaccinated healthy pigs sera were collected from a FMDV-free region (n = 36). (VI) FMDV (type O, type Asia1 and type A) standard immunized sera (n = 3) and infected sera (n = 3) came from the laboratory of Lanzhou veterinary research institution in China (LVRI). (VII) Field sera (n = 118) came from 4 different intensive swine farms.
2.2. Antigens
In China, FMDV type O vaccine strain mainly include GX/09-7, ZK/93, TAW/97, MYA/98, PanAsia/TZ/2011, OS/99. According to these vaccine strain, four peptides were designed on the basis of the epitope140–160 amino acids of VP1 (Table 1). Five peptides (Table 1) on the basis of NSPs (2B, 3B, 3C) were synthesized according to previous reported.19 All peptides were synthesized by Gil Biochemical Inc. (Shanghai, China), then conjugated to BSA (Sigma, St. Louis, MO) using a hetero-bifunctional cross-linker Sulfo-SMCC (Pierce, Rockford, IL). Then, the four conjugates of SPs peptides and BSA (BSA-SPs) were diluted to 0.5 mg ml−1 in PBS respectively and mixing with the same volume. The BSA-NSPs antigen were given similar treatment.
Synthetic peptides sequence of foot-and-mouth disease virus serotype O.
| Location | Peptide ID | Amino acid sequence |
|---|---|---|
| SPs | SP 1 | CSTNNVRGDLQVLAQKAERALP |
| SP 2 | CRVSNVRGDLQVLAQKAERALP | |
| SP 3 | CSTNNVRGDLQVLAQKAERTLP | |
| SP 4 | CSLTNVRGDLQVLAQKAARPLP | |
| NSPs | NSP1 | CRSTPEDLERAEKQ |
| NSP2 | CLEKQRDLNDPSKYKEAKE | |
| NSP3 | CELHEKVSSHPIFKQ | |
| NSP4 | CGPYTGPLERQKPLK | |
| NSP5 | CGPYAGPMERQKPLK |
2.3. Gold nanoparticle preparing
Colloidal gold nanoparticle (GNP) was prepared according to the reported method.23
2.4. Preparation of colloidal gold labeled staphylococcal protein A (SPA)
In this study, SPA (Sigma, St Louis, MO, USA) was used to label nanogold due to it can combine mammal IgG Fc. Briefly, 0.5 mg SPA was dissolved into 0.5 ml normal saline, then added into 50 ml colloidal gold solution (pH 9.0) drop by drop, standing 30 min at RT. After the addition of 0.5 ml of 3% casein solution, the mixture was incubated for another 10 min, then centrifuged (12 000 × g) at 4 °C for 30 min. The precipitate was resuspended with 5 ml 1% BSA buffer and stored at 4 °C for use.
2.5. Immobilization of test lines and control line reagents
BSA-NSPs, BSA-SPs and goat anti-pig antibody IgG were dispensed with 1 μl cm−1 by a XYZ3000 Dispenser (Bio-Dot, Richmond, CA, USA) on nitrocellulose (NC) membrane (Millipore, Bedford, MA, USA) as the test line 1 (T1), test line 2 (T2) and control line (C), respectively. T1 line can react to FMDV infected antibody only, and T2 can combine with FMDV type O immunized or infected antibody specifically. The three lines were situated at the center of the membrane with 0.3 cm apart. After drying for 30 min at 70 °C, the membrane was stored at room temperature.
2.6. Preparation of the gold nanoparticle strip
The gold nanoparticle strip was prepared using three pads (sample pads, 300 × 15 mm; conjugate pads, 300 × 8 mm; absorbent pads, 300 × 18 mm) and a NC membrane, all which were assembled on a semirigid polyethylene sheet. Briefly, sample pads were soaked with sample pads treatment buffer (20 mM Tris, 2.0% (w/v) BSA, 2.0% (w/v) sucrose, pH 9.0), and were dried at 40 °C for 30 min. The conjugate pads were coated with a mixture of gold–SPA conjugates using a XYZ3000 Dispenser at jetting rate of 6 μl cm−1 and dried at 40 °C for 30 min. Then the blotted membrane, conjugate pad, sample pad, and absorbent pad were assembled on the semirigid polyethylene sheet sequentially with a 1–2 mm overlap (Fig. 1A). The assembled card was cut to 2.85 mm width strips.
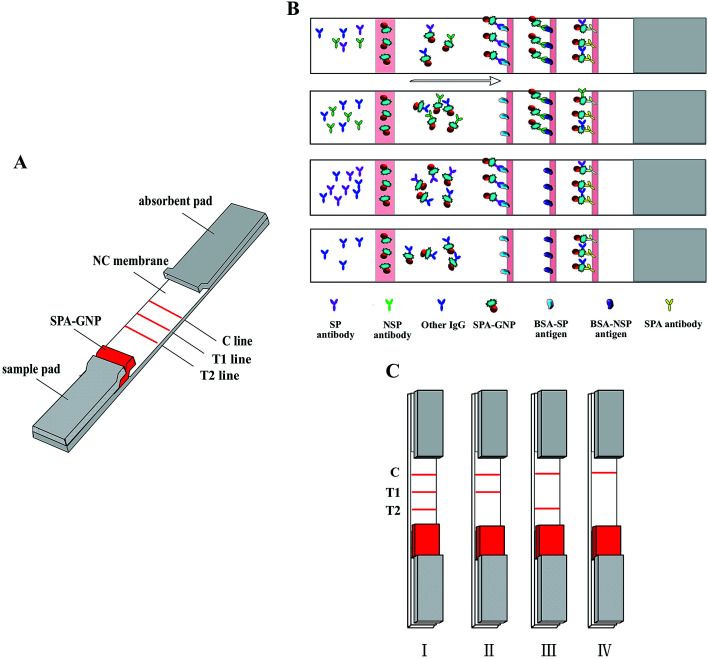
Fig. 1. (A) Schematic illustration of the gold nanoparticle strip. T1 line: BSA-NSPs antigen. T2 line: BSA-SPs antigen. (B) Principle diagram. (C) Test results of the test strip. I: FMDV type O infected serum. II: FMDV infected but not type O. III: FMDV type O immunized serum. IV: FMDV negative serum, neither infected nor immunized with type O vaccine or immunized antibody was not enough to resist FMDV virus.
2.7. Sensitivity of the test strip
The sensitivity of the T1 line was evaluated with sera group II and III. The sensitivity of the T2 line was evaluated with sera group VII. The test procedures of the strip was as follows: serum was 1 : 200 diluted with normal saline (NS), then 100 μl diluted sample was added into the sample well, the results was observed in 5–10 min.
The detection limit of the strip was evaluated with 2 fold serially diluted FMDV type O vaccinated serum (LPB ELISA titer ≥ 1 : 128), type O, A, Asia1 infected sera respectively.
2.8. Specificity of the test strip
The specificity of the test strip T1 line was analyzed with sera group I and V, FMDV type O, type Asia1 and type A infected sera were used as positive control. Furthermore, the specificity of the T2 line was estimated with sera group IV and VI, FMDV type O immunized sera, FMDV A, O, Asia1 infected sera and FMDV negative sera were used as control.
2.9. Synchronously comparison study of strip and ELISA
To estimate the diagnose accordance rate of the test strip with commercial ELISA kits, all 379 sera (sera group V–VII) were also parallelly screened by the experimental strip, LVRI® FMDV LPB ELISA kit and LVRI® 3ABC Mab blocking ELISA kit (3ABC Mab-bELISA).
2.10. Ethics statement
The animal experiments were carried out according to the Animal Experiment Committee of Henan Academy of Agricultural Sciences (Approval number SYXK 2014-0007). All animals received humane care in compliance with good animal practice according to the animal ethics procedures and guidelines of China.
3. Results
3.1. Strip mechanism and results judgment
The mechanism of the gold nanoparticle strip based on the antibody–antigen reaction which form into a sandwich format (GNP–SPA–antibody–antigen). Briefly, diluted serum sample migrated to the gold labeled SPA, all IgGs in serum are combined by SPA gold probe whenever the sera is FMDV positive or negative. GNP–SPA–IgG migrate to the NC membrane, the SPs antigen on T2 line will combine with FMDV SP antibody, and NSPs antigen on T1 line will specifically capture FMDV NSPs antibody, control line can specifically combine SPA (Fig. 1B).
As shown in Fig. 1C, when T1, T2 and C lines all turn red in 5–10 min, which represent the tested pig is infected with type O FMDV (Fig. 1C-I). If only C and T1 lines give visible red, which means the tested pig is infected by FMDV but not type O FMDV (Fig. 1C-II). When only C and T2 lines give visible red, which means the tested pig is immunized with type O FMDV vaccine and the antibody level was enough resistant to type O FMDV (Fig. 1C-III). If only C line turn red, the pig is nether infected FMDV nor immunized with type O FMDV (Fig. 1C-IV).
3.2. Optimization of the gold nanoparticle strip system
To increase the accuracy of the gold nanoparticle strip, many factors should be optimized such as the type of the NC membrane, pH of conjugate and sample pads and different blocking buffers. In this strip system, 135s membrane was selected. The optimal sample pad buffer was 20 mM TBS (pH 9.0), containing 2.0% BSA and 2.0% sucrose. Then, 20 mmol l−1 sodium borate (pH 9.0) containing 10% BSA was chosen as the optimal conjugate pad solution.
3.3. Sensitivity of the gold nanoparticle strip
The sensitivity of the T1 line can be seen in Table 2. There are four of the 11 FMDV infected swine sera showed positive at 11 DPI, and all samples gave positive results at 14 DPI, until to 63 DPI. All of the 8 field FMDV infected swine sera (DPI was unknown) gave positive results. This sera were also detected by a commercial ELISA kit (LVRI® 3ABC Mab-bELISA) simultaneously, and results were compared with strip detected results one by one, they were approximate.
The sensitivity of T1 line of the test strip.
| Serotype | Days post-infection (DPI) | Total | T1 line of the test strip | LVRI® 3ABC Mab-bELISA kit | ||
|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |||
| A | 1 | 2 | 0 | 2 | 0 | 2 |
| 11 | 11 | 4 | 7 | 4 | 7 | |
| 13 | 2 | 1 | 1 | 1 | 1 | |
| 14 | 4 | 4 | 0 | 4 | 4 | |
| 18 | 2 | 2 | 0 | 2 | 0 | |
| 21 | 7 | 7 | 0 | 7 | 0 | |
| 23 | 2 | 2 | 0 | 2 | 0 | |
| 28 | 9 | 9 | 0 | 7 | 2 | |
| 30 | 2 | 2 | 0 | 2 | 0 | |
| 35 | 7 | 7 | 0 | 7 | 0 | |
| 37 | 1 | 1 | 0 | 1 | 0 | |
| 42 | 4 | 4 | 0 | 4 | 0 | |
| 63 | 5 | 5 | 0 | 5 | 0 | |
| O | Unknown | 8 | 8 | 0 | 8 | 0 |
The sensitivity of the T2 line can be seen in Table 3. There have 11 serum sample gave different results and 107 serum sample give the same results between the test strip and commercial LPB ELISA, so the sensitivity of the strip was 90.68% (107/118).
The sensitivity of T2 line of the gold nanoparticle strip.
LPB ELISA titer > 26.
LPB ELISA titer ≤ 26.
The detection limit of the gold nanoparticle strip was evaluated as described above. The test lines of the strip were scanned with a membrane strip reader (Bio-Dot, Richmond, CA, USA), the result showed that the G/D × A (area) and G/peak of the relative optical density (ROD) was decreasing as the dilution of samples increased (Fig. 2). When G/D × A lower than 15, the color of test lines almost could not be seen by eyes. Fig. 2 illustrated that the detection limit of T1 line for FMDV O, A, Asia 1 infected serum and type O immunized serum were 1 : 6400, 1 : 12 800, 1 : 800, 1 : 6400, respectively. The detection limit of T2 line for FMDV O infected serum and immunized serum were 1 : 6400 and 1 : 12 800, severally.
Fig. 2. The detection limit of the gold nanoparticle strip. (A) FMDV type O infected serum, (B) FMDV type A infected serum, (C) FMDV type Asia1 infected serum, (D) FMDV type O immunized serum.
3.4. Specificity of the strip
The specificity of T1 line was shown in Table 4. There are only 7 false positive results in 145 FMDV type O immunized pig sera, and all 36 unvaccinated healthy pig sera gave negative results. The 3 FMDV infected sera gave clear positive results. The specificity of T1 line was 95.17% (138/145), which illustrated that the T1 line of the experimental strip can be applied to differentiation FMDV vaccinated animal from infected animal.
The specificity of T1 line of the test strip.
| Serum sample | Serum source | Immune state | T1 line of the test strip | |
|---|---|---|---|---|
| Positive | Negative | |||
| S1–S20 | Pregnant sows | Immunized 4 times with type O FMDV vaccine | 4 | 17 |
| S21–S45 | Nursing sows | Immunized 4 times with type O FMDV vaccine | 3 | 22 |
| S46–S65 | 20 day-old piglets | Unvaccinated | 0 | 20 |
| S66–S85 | 40 day-old piglets | Unvaccinated | 0 | 20 |
| S86–S105 | 60 day-old pigs | Immunized 1 time with type O FMDV vaccine | 0 | 20 |
| S106–S125 | 80 day-old pigs | Immunized 2 time with type O FMDV vaccine | 0 | 20 |
| S126–S145 | 100 day-old pigs | Immunized 2 time with type O FMDV vaccine | 0 | 20 |
| P1–P36 | Fattening pigs | Unvaccinated healthy pig | 0 | 36 |
| Positive control | FMDV infected pig | 3 | 0 | |
| Total | 9 | 148 | ||
As shown in Fig. 3, the test results of type Asia1 and A FMDV immunized sera, PRRSV, PRV, CSFV, PCV2, JEV positive sera and FMDV negative control sera displayed only one red band at the C line. The T2 line results of the test strip were summarized in Table 5. All FMDV type O positive sera (included immunized and infected swine) showed positive results, FMDV type Asia1 and type A immunized and infected positive sera were negative tested by the strip. The results proved that the T2 line of the strip were specific for detecting type O FMDV antibody with the specificity at 100%.
Fig. 3. The specificity of the gold nanoparticle strip. Note: 1: FMDV O infected positive serum, 2: FMDV A infected positive serum, 3: FMDV Asia1 infected positive serum, 4: FMDV O immunized positive serum, 5: FMDV A immunized positive serum, 6: FMDV Asia1 immunized positive serum, 7: PRRSV positive serum, 8: PRV positive serum, 9: CSFV positive serum, 10: PCV2 positive serum, 11: JEV positive serum, 12: FMDV negative serum.
The specificity of T2 line of the gold nanoparticle strip.
| Serum source | Total | T2 line | T1 line | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| PRRSV positive | 4 | 0 | 4 | 0 | 4 |
| PRV positive | 4 | 0 | 4 | 0 | 4 |
| CSFV positive | 4 | 0 | 4 | 0 | 4 |
| PCV2 positive | 4 | 0 | 4 | 0 | 4 |
| JEV positive | 4 | 0 | 4 | 0 | 4 |
| FMDV O immunized positive | 1 | 1 | 0 | 0 | 1 |
| FMDV A immunized positive | 1 | 0 | 1 | 0 | 1 |
| FMDV Asia1 immunized positive | 1 | 0 | 1 | 0 | 1 |
| FMDV O infected positive | 1 | 1 | 0 | 1 | 0 |
| FMDV A infected positive | 1 | 0 | 1 | 1 | 0 |
| FMDV Asia1 infected positive | 1 | 0 | 1 | 1 | 0 |
| FMDV negative | 5 | 0 | 5 | 0 | 5 |
3.5. Comparative studies between the test strip and ELISAs
The comparative experiment between the strip and two commercial ELISA kits can be seen in Table 6, the results indicated that the coincidence rate of the test strip with the 3ABC Mab-bELISA and LPB ELISA was 95.5% (362/379) and 93.13% (353/379), respectively. This results indicated that the strip was suitable for evaluating FMDV serotype O immunized antibody level and differentiating FMDV vaccinated animal from infected animal simultaneously.
The comparison experiment results of the strip and two commercial ELISA kitsa.
| T1 line of strip | T2 line of strip | ||||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| 3ABC Mab-bELISA | Positive | 65 | 10 | — | — |
| Negative | 7 | 277 | — | — | |
| LVRI® LPB ELISA | Positive | — | — | 154 | 16 |
| Negative | — | — | 10 | 199 | |
All 379 swine serum samples were detected with the strip and two commercial ELISA kit methods synchronously. The coincidence rate were calculated. 3ABC Mab-bELISA positive represented the percentage inhibition (PI) > 50%. LVRI® LPB ELISA positive represented the serum titer > 26.
4. Discussion
Recent years, a series of new label material, such as up-converting phosphors nanoparticles,24 colloidal selenium,25 colloidal carbon,26 lanthanide fluorescent nanoparticles,27 quantum dots28 were successfully applied to lateral flow immunoassay. All these methods are built on the combination of markers and antigens or antibodies. Usually, the purpose of using label materials is to enhance the sensitivity of lateral flow immunoassay, but these materials are expensive and difficult to prepare, than the conventional colloidal gold and the methods based on which need instruments to analyze the results. In current antibody detection system, colloidal gold was used, because of it was stable, sensitive and specific. And the tested results of this strip can be determined by naked eyes in 5–10 min.
For an antibody test strip, the traditional principle is select two antigens as colloidal gold labeled antigen protein and test line antigen protein. In this study, the purpose is to detect two antibodies on one strip, so the colloidal gold labeled proteins need combine the two antibodies; over the course of the study, we have tried to label two different antigens by gold, but the result was not specific; and anti-pig IgG antibody was also used as colloidal gold labeled antigen but the poor sensitivity limited its use in follow test. So, SPA was selected to prepare the nanogold labeled particle duo to SPA can combine the Fc fragment of any mammal IgG.
Over the course of the study, the T1 and T2 lines capture reagents of the test strip production were preliminarily characterized with ELISA technique. The test results of T2 line (SPs antigen) were same with ELISA. Interestingly, the location of NSPs antigen on the NC membrane was very important, it would give nonspecific results when reverse the position of T1 and T2 lines, so they must be placed the correct location as shown in Fig. 1A.
The definition of the optimal concentration of the antigens on T1 and T2 line was the one that give the best specificity and sensitivity. During antigens concentration optimization, tested accuracy of the strip was evaluated by use different peptide as antigens. The results shown that the four BSA-SPs mixed equally on T1 was best, and the optimal concentration was 0.5 mg ml−1. In addition, among the NSPs, the NSP1 and NSP2 peptides were not suitable for the strip system although they are specific in ELISA system. Therefore, BSA-NSP1, BSA-NSP3 and BSA-NSP5 were mixed with the same volume as detecting antigen for NSP antibody, the optimal concentration was 0.2 mg ml−1 (data was not shown).
5. Conclusion
A gold nanoparticle strip has been developed in this study based on the epitope antigens and SPA gold labeled nanoparticle, which offers simultaneous evaluation of immunized antibody level and differentiation of FMDV vaccinated from infected animal. NSPs and SPs epitope antigens were immobilized on the NC membrane as T1 and T2 test lines, and gold labeled SPA was used as detector enabling the simultaneous detection of two kind of antibodies on one single test strip in 10 min without any expensive instrument. The experimental strip system showed good accuracy, reproductivity and good coincidence with commercial ELISAs. In summery, this developed strip could provide a simple, inexpensive and rapid approach for the onsite detection of FMDV type O immunized antibody level and for discrimination of FMDV vaccinated from infected animal without any expensive instrument.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Key Research and Development Program of China (no. 2016YFD0501503). We also thank PHD Pan Li (Lanzhou Veterinary Research Institution in China) for serum samples supply.
References
- Alexandersen S. Zhang Z. Donaldson A. I. Garland A. J. M. J. Comp. Pathol. 2003;129:1–36. doi: 10.1016/S0021-9975(03)00041-0. [DOI] [PubMed] [Google Scholar]
- Grubman D. O. M. J. M. J. J. Virol. 1985;56:120–126. doi: 10.1128/jvi.56.1.120-126.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert R. R. Wimmer E. J. Virol. 1984;50:957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumption K. Rweyemamu M. Wint W. Transboundary Emerging Dis. 2008;55:5–13. doi: 10.1111/j.1865-1682.2007.01017.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sainz I. Medina G. N. Ramirez-Medina E. Koster M. J. Grubman M. J. de Los S. T. Virology. 2017;502:123–132. doi: 10.1016/j.virol.2016.12.021. [DOI] [PubMed] [Google Scholar]
- Wang J. H. Liang C. J. Shieh J. J. Jong M. H. Lin Y. L. Sieber M. Vaccine. 2003;21:3721–3729. doi: 10.1016/S0264-410X(03)00363-3. [DOI] [PubMed] [Google Scholar]
- Clavijo A. Wright P. Kitching P. Vet. J. 2004;167:9–22. doi: 10.1016/S1090-0233(03)00087-X. [DOI] [PubMed] [Google Scholar]
- Mason P. W. Grubman M. J. Baxt B. Virus Res. 2003;91:9–32. doi: 10.1016/S0168-1702(02)00257-5. [DOI] [PubMed] [Google Scholar]
- Domingo E. B. C. E. E. Comp. Immunol. Microbiol. Infect. Dis. 2002;25:297–308. doi: 10.1016/S0147-9571(02)00027-9. [DOI] [PubMed] [Google Scholar]
- Wekesa S. N. Namatovu A. Sangula A. K. Dhikusooka M. T. Muwanika V. B. Tjørnehøj K. Trop. Anim. Health Prod. 2014;46:575–581. doi: 10.1007/s11250-013-0530-0. [DOI] [PubMed] [Google Scholar]
- Sharma G. K. Mahajan S. Matura R. Subramaniam S. Mohapatra J. K. Pattnaik B. Biologicals. 2015;43:158–164. doi: 10.1016/j.biologicals.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Chénard G. Miedema K. Moonen P. Schrijver R. S. Dekker A. J. Virol. Methods. 2003;107:89–98. doi: 10.1016/S0166-0934(02)00196-9. [DOI] [PubMed] [Google Scholar]
- Mahajan S. Mohapatra J. K. Pandey L. K. Sharma G. K. Pattnaik B. J. Virol. Methods. 2013;193:405–414. doi: 10.1016/j.jviromet.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Lu Z. Zhang X. Fu Y. Cao Y. Tian M. Sun P. Li D. Liu Z. Xie Q. J. Virol. Methods. 2010;170:128–133. doi: 10.1016/j.jviromet.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Bergmann I. E. Malirat V. Neitzert E. Beck E. Panizzutti N. Sánchez C. Falczuk A. Arch. Virol. 2000;145:473–489. doi: 10.1007/s007050050040. [DOI] [PubMed] [Google Scholar]
- Sørensen K. J. Madsen K. G. Madsen E. S. Salt J. S. Nqindi J. Mackay D. K. Arch. Virol. 1998;143:1461–1476. doi: 10.1007/s007050050390. [DOI] [PubMed] [Google Scholar]
- Zhang G. Wang X. Zhi A. Bao Y. Yang Y. Qu M. Luo J. Li Q. Guo J. Wang Z. Food Addit. Contam., Part A: Chem., Anal., Control, Exposure Risk Assess. 2008;25:413–423. doi: 10.1080/02652030701561452. [DOI] [PubMed] [Google Scholar]
- Zhang G. P. Guo J. Q. Wang X. N. Yang J. X. Yang Y. Y. Li Q. M. Li X. W. Deng R. G. Xiao Z. J. Yang J. F. Vet. Parasitol. 2006;137:286–293. doi: 10.1016/j.vetpar.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Yang S. Z. Yang J. F. Zhang G. P. Qiao S. L. Wang X. N. Zhao D. Li X. W. Deng R. G. Zhi A. M. You L. M. J. Vet. Diagn. Invest. 2010;22:412–415. doi: 10.1177/104063871002200311. [DOI] [PubMed] [Google Scholar]
- Fu Y. Li P. Cao Y. Wang N. Sun P. Shi Q. Ji X. Bao H. Li D. Chen Y. Bai X. Ma X. Zhang J. Lu Z. Liu Z. PLoS One. 2017;12:e170560. doi: 10.1371/journal.pone.0170560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. P. Liu Y. Luo X. B. Isobe M. Yuan G. L. Liu Y. Q. Hua Y. Song X. Y. Yang J. W. Li X. Rev. Sci. Instrum. 2014;85:53502. doi: 10.1063/1.4872385. [DOI] [PubMed] [Google Scholar]
- Yang S. Yang J. Zhang G. Wang X. Qiao S. Zhao D. Zhi Y. Li X. Xing G. Luo J. J. Virol. Methods. 2010;165:139–144. doi: 10.1016/j.jviromet.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Frens G. Controlled Nucleation for the Regulation of the Particle Size in Monodisperse Gold Suspensions. Nat. Phys. Sci. 1973:20–22. doi: 10.1038/physci241020a0. [DOI] [Google Scholar]
- Song C. Zhi A. Liu Q. Yang J. Jia G. Shervin J. Tang L. Hu X. Deng R. Xu C. Biosens. Bioelectron. 2013;50:62. doi: 10.1016/j.bios.2013.06.022. [DOI] [PubMed] [Google Scholar]
- Wang Z. Zhi D. Zhao Y. Zhang H. Wang X. Ru Y. Li H. Int. J. Nanomed. 2014;9:1699. doi: 10.2147/IJN.S58942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera P. Posthuma-Trumpie G. A. Tuil M. V. Wal F. J. V. D. Boer A. D. Moers A. P. H. A. Amerongen A. V. Anal. Bioanal. Chem. 2011;399:831. doi: 10.1007/s00216-010-4334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Zou M. Chen Y. Li J. Wang Y. Qi X. Xue Q. Biosens. Bioelectron. 2014;51:29–35. doi: 10.1016/j.bios.2013.06.065. [DOI] [PubMed] [Google Scholar]
- Mohamadi E. Moghaddasi M. Farahbakhsh A. Kazemi A. J. Photochem. Photobiol., B. 2017;174:291–297. doi: 10.1016/j.jphotobiol.2017.08.005. [DOI] [PubMed] [Google Scholar]