Abstract
Escherichia coli is an important pathogen that shows increasing antimicrobial resistance in isolates from both animals and humans. Our laboratory recently described Salmonella isolates from food animals and humans that expressed an identical plasmid-mediated, AmpC-like β-lactamase, CMY-2. In the present study, 59 of 377 E. coli isolates from cattle and swine (15.6%) and 6 of 1,017 (0.6%) isolates of human E. coli from the same geographic region were resistant to both cephamycins and extended-spectrum cephalosporins. An ampC gene could be amplified with CMY-2 primers in 94.8% of animal and 33% of human isolates. Molecular epidemiological studies of chromosomal DNA revealed little clonal relatedness among the animal and human E. coli isolates harboring the CMY-2 gene. The ampC genes from 10 animal and human E. coli isolates were sequenced, and all carried an identical CMY-2 gene. Additionally, all were able to transfer a plasmid containing the CMY-2 gene to a laboratory strain of E. coli. CMY-2 plasmids demonstrated two different plasmid patterns that each showed strong similarities to previously described Salmonella CMY-2 plasmids. Additionally, Southern blot analyses using a CMY-2 probe demonstrated conserved fragments among many of the CMY-2 plasmids identified in Salmonella and E. coli isolates from food animals and humans. These data demonstrate that common plasmids have been transferred between animal-associated Salmonella and E. coli, and identical CMY-2 genes carried by similar plasmids have been identified in humans, suggesting that the CMY-2 plasmid has undergone transfer between different bacterial species and may have been transmitted between food animals and humans.
The Escherichia genus comprises a large group of organisms, many of which reside as normal commensals in the intestinal tracts of animals and humans while others serve as important intestinal and extraintestinal pathogens. Pathogenic human and animal Escherichia coli resistant to many classes of antimicrobial agents have been reported worldwide (6, 32). These multiresistant pathogens present an important challenge to achieving effective therapy. Antimicrobial resistance in commensal strains of E. coli, however, may also play an important role in the ecology of resistance and clinical infectious diseases. Transmission of resistance genes from normally nonpathogenic species to more virulent organisms within the animal or human intestinal tract may be an important mechanism for acquiring clinically significant antimicrobial-resistant organisms. E. coli may serve as an important reservoir for these transmissible resistances, since it is clear that this organism has developed a number of elaborate mechanisms for acquiring and disseminating plasmids, transposons, phage, and other genetic determinants (21).
Fecal-oral and food-borne transmission of E. coli are well documented (19). Epidemiologic studies have frequently traced E. coli outbreaks to meat products, particularly those of bovine origin or food products contaminated with manure (19). Transmission has been best documented when the strain involved expresses an unusual serotype, virulence factor, or antibiotic resistance gene.
Our laboratory recently identified Salmonella isolates from food animals of bovine and porcine origin that express resistance to the cephamycins and extended-spectrum cephalosporins. These isolates carried a plasmid-mediated ampC, CMY-2 (31). An identical CMY-2 gene has also been identified in Salmonella isolated from humans, with one case demonstrating epidemiologic associations between a human isolate and a chromosomally related bovine isolate that expressed an identical plasmid-mediated CMY-2 gene (9, 10, 31). In our present study, we have identified clinically significant isolates of E. coli from bovine, porcine, and human sources that express an identical CMY-2 β-lactamase. Plasmid analysis and molecular hybridization techniques demonstrate significant homology between the E. coli and Salmonella plasmids from food animals and humans. This new antimicrobial resistance gene may provide a unique tool for analyzing food-borne transmission of a highly antimicrobial-resistant organism, transfer of resistance between enteric organisms, and possibly the transfer of resistance determinants from commensals to more virulent organisms.
MATERIALS AND METHODS
Organisms.
Since 1998, the University of Iowa has been conducting an antibiotic resistance surveillance project, Emerging Infections and the Epidemiology of Iowa Organisms (EIEIO), aimed at defining antimicrobial resistance patterns in the state of Iowa. The College of Veterinary Medicine at Iowa State University has served as a collaborator on surveillance of isolates from farm animals. From November 1998 to December 1999, 377 clinical bovine and porcine E. coli isolates were analyzed at the Iowa State University Veterinary Diagnostic Microbiology Laboratory and were referred to the Medical Microbiology Division of the Department of Pathology at the University of Iowa College of Medicine for further characterization. From November 1998 to March 2000, 1,017 random clinical human E. coli isolates were referred from 15 medical centers throughout Iowa. Isolates were stored at −70°C on porous beads (ProLab, Austin, Tex.) until further use. Human and food animal Salmonella isolates were described previously (31).
Antimicrobial susceptibility testing.
MICs of selected antimicrobial agents were determined by broth microdilution. Custom-designed microdilution trays containing dilutions of selected antimicrobial agents in cation-adjusted Mueller-Hinton broth (TREK Diagnostic Systems Inc., Westlake, Ohio) were inoculated and analyzed using guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (12, 20). Streptomycin, chloramphenicol, and sulfisoxazole susceptibility tests were performed by disk diffusion as described by the NCCLS (12, 20).
Isoelectric focus analysis.
Crude β-lactamase extracts were prepared by freeze-thaw lysis of bacterial cultures grown exponentially in tryptic soy broth as previously described (4). Analytical isoelectric focusing was performed using a Multiphore II electrophoresis system with commercially prepared ampholine-polyacrylamide plates, pI 3.5 to 9.5 (Amersham Pharmacia Biotech, Piscataway, N.J.). β-Lactamase activity was detected with 0.5 mg of nitrocefin (BD, Franklin Lakes, N.J.)/ml. TEM-1, TEM-4, SHV-1, SHV-3, and SHV-5 β-lactamases expressed in E. coli C600 were used as isoelectric focus standards. These enzymes are known to migrate with isoelectric points of 5.4, 5.9, 7.6, 7.0, and 8.2, respectively (13). Known pI values of each standard were plotted against the distance from the cathode, and a regression analysis was performed (Microsoft Excel 98). Unknown β-lactamase pIs were calculated using the regression curve generated from each gel.
Pulsed-field gel electrophoresis (PFGE).
Genomic DNA was isolated and digested with XbaI (New England Biolabs, Beverly, Mass.) as previously described (22). Electrophoresis was performed on the contour-clamped homogeneous electric field-DRII (Bio-Rad Laboratories, Richmond, Calif.) with the following conditions: 0.5× TBE (Tris-borate-EDTA), 1% agarose, 13°C, 6 V/cm, 23 h with switch times ranging from 5 to 30 s. The running buffer contained 0.5× TBE and 0.38% thiourea. A λ ladder that contains concatemers of the 48.5-kb phage DNA was used as a molecular size standard (FMC BioProduct, Rockland, Maine). Gels were stained with ethidium bromide and photographed using a Bio-Rad Gel Doc 1000 system. Strains which contained restriction fragment patterns that differed by more than three bands were considered unique (1).
Molecular techniques.
Plasmid DNA was isolated using a protocol described for isolation of large bacterial artificial chromosome plasmid DNA (25) or the NucleoBond BAC Maxi kit (Clontech). Plasmid DNA preparations were digested with PlasmidSafe DNase according to the manufacturer's recommendations (Epicentre Technologies, Madison, Wis.). Transformation of plasmid DNA was performed using standard electroporation techniques with DH10B electrocompetent E. coli (Gibco BRL Life Technologies, Grand Island, N.Y.). Transformants were selected on Luria-Bertani agar containing 32 μg of cefoxitin (Sigma Aldrich Chemicals, St. Louis, Mo.)/ml. DNA restriction fragment length polymorphisms (RFLPs) were analyzed using agarose gel electrophoresis of plasmid DNA cleaved with various restriction endonucleases (New England Biolabs, Beverly, Mass.).
PCR analysis was performed on total DNA as prepared using the cetyltrimethylammonium bromide protocol described previously (2) or plasmid DNA treated with PlasmidSafe DNase. Amplification was performed with consensus primers ampC1 and ampC2, which have been shown to recognize bla BIL-1, bla LAT-1, bla LAT-2, bla CMY-2, and the ampC gene of Citrobacter freundii OS60 (14) or TEM-1 forward and reverse primers (31) (see Table 1). PCR fragments were isolated using Qiaquick PCR cleanup columns (Qiagen, Valencia, Calif.). DNA sequence analysis was performed using Big Dye terminator cycle sequencing chemistry with AmpliTaq polymerase, FS enzyme (PE Applied Biosystems, Foster City, Calif.). The reactions were performed and analyzed with an Applied Biosystems Model 373A stretch fluorescent automated sequencer at the University of Iowa DNA Core Facility.
TABLE 1.
Oligonucleotide primers used for PCR amplifications
| Gene | Primera | Sequence | Product size (gene size) | Reference |
|---|---|---|---|---|
| C. freundii ampC family | ampC1 | 5′-ATGATGAAAAAATCGTTATGC-3′ | 1,143 bp | 14 |
| ampC2 | 5′-TTGCAGCTTTTCAAGAATGCGC-3′ | (Full length) | ||
| TEM-1 | TEM-1 forward | 5′-CCCGAATTCGGAAGAGTATGAGTATTC-3′ | 878 bp | 31 |
| TEM-1 reverse | 5′-CCCGGATCCCAGTTACCAATGCTTAATC-3′ | (Full length) | ||
| Inserted gene cassettes for type I integrons | 5′ CS | 5′-GGCATCCAAGCAGCAAG-3′ | Variable | 16 |
| 3′ CS | 5′-AAGCAGACTTGACCTGA-3′ |
TEM-1 forward encodes an EcoRI restriction site, shown underlined. TEM-1 reverse encodes a BamHI restriction site, shown underlined. CS, conserved sequence.
Southern blot analysis was performed as previously described (26). The 1,143-bp CMY-2 PCR product was cloned into PCR2.1 (Invitrogen). The CMY-2 gene was then isolated and radiolabeled using the random priming method (Roche Molecular Biochemicals). Digested DNA was transferred to Nytran (Schleicher & Schuell) and probed with the α-32P-labeled CMY-2 probe. Blots were washed with a solution containing 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate at 65°C.
Type 1 integron gene cassettes were detected using PCR with primers previously described to amplify gene cassettes inserted between the 5′ and 3′ conserved sequences present in these integrons (16) (see Table 1). Gene cassette products were analyzed by DNA sequence analysis.
RESULTS
Antimicrobial susceptibility analysis of bovine, porcine, and human E. coli isolates.
Of 377 E. coli isolates of bovine or porcine origin tested, the extended-spectrum cephalosporin and cefoxitin MICs for 59 (15.6%) isolates were elevated (MIC of ceftriaxone, ceftazidime, or aztreonam, ≥8 μg/ml; MIC of cefoxitin, ≥8 μg/ml), but cefepime MICs were low (≤4 μg/ml), consistent with an AmpC phenotype (17). The addition of clavulanic acid at 2 μg/ml had no effect on ampicillin or ticarcillin MICs. Among these 59 isolates, 100% were also resistant to sulfisoxazole and tetracycline, 98% were resistant to streptomycin, 92% were resistant to chloramphenicol, 70% were resistant to gentamicin, 63% were resistant to tobramycin, 19% were resistant to nalidixic acid, and 15.2% were resistant to ciprofloxacin (Table 2).
TABLE 2.
Animal and human E. coli isolates from Iowa expressing an AmpC phenotypea
| Isolateb | Coresistance (%)c
|
IEF results (%) ford:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chloramphenicol | Tetracycline | Sulfisoxazole | Streptomycin | Gentamicin | Tobramycin | Ciprofloxacin | pI 5.4 | pI ∼8.9 | CMY-2e | |
| Animal (n = 59) | 54 (91.5) | 59 (100) | 59 (100) | 58 (98.3) | 41 (69.5) | 37 (62.7) | 9 (15.2) | 20 (33.9) | 59 (100) | 55 (94.8) |
| Human (n = 6) | 4 (66.7) | 4 (66.7) | 4 (66.7) | 4 (66.7) | 3 (50) | 2 (33.3) | 2 (33.3) | 2 (33.3) | 6 (100) | 2 (33.3) |
Ceftriaxone, ceftazidime, and aztreonam MICs were ≥8 μg/ml, the cefoxitin MIC was ≥8 μg/ml, and the cefepime MIC was ≤4 μg/ml.
Fifty nine (15.6%) of the 377 animal isolates tested expressed the AmpC phenotype. Of those, 42 (22.2%) were bovine isolates (n = 189) and 17 (9.0%) were porcine isolates (n = 188). Six (0.6%) of the 1,017 human isolates tested expressed the AmpC phenotype.
Coresistances associated with the organisms expressing an AmpC phenotype.
Percentage of isolates with an AmpC phenotype with a band that migrates at pI 5.4 or pI ∼8.9 on isoelectric focus (IEF) analysis.
Percentage of isolates with an AmpC phenotype that amplify a CMY-2-like gene using PCR.
These 59 cephalosporin-resistant E. coli isolates were obtained from bovine and porcine sources located throughout the state of Iowa. The majority of isolates (83%) were obtained from intestinal biopsy samples, though eight isolates (13.5%) came from feces and two isolates (3.4%) were derived from milk. None of the isolates belonged to the 0157:H7 serotype.
Over 1,000 E. coli isolates from human sources from Iowa were also characterized. Six (0.6%) were resistant to the cephamycins, monobactams, and extended-spectrum cephalosporins. Coresistance rates were lower than those seen with the animal isolates (Table 2). There was no geographic clustering of the six cephalosporin-resistant human E. coli isolates. Five of these E. coli isolates were recovered from urine and one was recovered from blood.
The cephalosporins tested for the animal and human isolates included cephalothin, cefoxitin, ceftriaxone, ceftazidime, and cefepime. Though the ceftriaxone MICs for most AmpC-producing isolates were elevated, the range included MICs of 1, 2, and 4 μg/ml, values that would be considered susceptible by NCCLS criteria. The ceftazidime MICs for all isolates were ≥8 μg/ml, and the cefoxitin MICs were ≥32 μg/ml. Ceftiofur was tested against only 10 AmpC isolates, but the MICs of ceftiofur ranged from 2 to >8 μg/ml. As has been shown for organisms expressing a chromosomal AmpC, all isolates remained susceptible by in vitro criteria to cefepime, with a range of cefepime MICs of ≤0.12 to 4 μg/ml (17, 23).
Molecular epidemiology.
To determine whether a particular clone was responsible for statewide dissemination of cephalosporin-resistant E. coli, all 65 animal and human isolates were analyzed by PFGE of XbaI-digested chromosomal DNA. Only two groups of isolates demonstrated similar though nonidentical PFGE patterns. Three isolates were recovered from bovine specimens obtained from three towns in Iowa that are located within 60 miles of each other. Two other isolates, one human and one bovine, shared similar PFGE patterns, despite the fact that the isolates were obtained from hosts located in geographically unrelated locations in Iowa. The remaining human and animal isolates demonstrated unique PFGE patterns. These data indicated that the dissemination of cephalosporin-resistant E. coli throughout Iowa could not be explained by the clonal spread of a particular strain.
Molecular analysis.
The cephalosporin resistance pattern of these E. coli isolates was similar to the resistance pattern of bovine, porcine, and human Salmonella isolates previously identified in our laboratory. These Salmonella isolates were shown to express a plasmid-encoded AmpC β-lactamase, CMY-2 (31). Crude protein extracts from the cephalosporin-resistant E. coli isolates were characterized by isoelectric focus analysis. All isolates expressed a β-lactamase that migrated at pI ∼8.9 to 9.0; this band comigrated with the β-lactamase expressed by the cephalosporin-resistant Salmonella isolates (Table 2). Additionally, a third of the animal and human isolates expressed a pI 5.4 β-lactamase that comigrated with the TEM-1 β-lactamase standard.
Given the similarities between the Salmonella and E. coli isolates, PCR was performed using consensus primers (ampC1 and ampC2) known to amplify the citrobacter family of ampC genes, which includes CMY-2. PCR was performed on total bacterial DNA from all 65 animal and human E. coli isolates. A 1,143-bp band that comigrated with a fragment amplified from C. freundii and a CMY-2-expressing Salmonella isolate was detected in 95% of animal and 33% of human isolates (Table 2). The ampC1 and ampC2 primers did not amplify products from antibiotic-susceptible E. coli isolates.
From the isolates that amplified a citrobacter-like ampC product, eight animal and both human E. coli isolates were chosen for further analysis. To increase the likelihood of analyzing a diverse population of organisms, the animal isolates selected expressed various antimicrobial coresistance patterns. Bacterial transformation studies were performed to determine whether cephalosporin resistance could be transferred to a laboratory strain of E. coli, DH10B. All 10 cephalosporin-resistant E. coli isolates studied could transfer cephalosporin resistance to a laboratory strain of E. coli. Two animal-derived transformants contained a 10-kb plasmid, while the others carried a large plasmid that migrated at 80 to 90 kb. Streptomycin and chloramphenicol resistances cosegregated with the cephalosporin resistance in all transformants (Table 3); tetracycline resistance cosegregated in 90% of transformants, sulfamethoxazole in 80% of transformants, gentamicin in 50% of transformants, and trimethoprim-sulfamethoxazole in 40% of transformants. Ciprofloxacin resistance was not transferred to E. coli DH10B.
TABLE 3.
MICs for E. coli isolates
| E. coli isolate | MIC (μg/ml)a
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amp | Pip | Pip/tazo | CTAZ | CTX | CFTI | CFOX | CPIM | Tet | T/S | Gent | Cipro | Strepc | Chlorc | Sulfc | |
| 9 | >16 | 32 | 16 | >16 | 32 | >8 | >32 | 1 | >8 | >1 | 1 | 0.03 | 6 | 6 | 6 |
| DH10B 9b | >16 | 64 | 16 | >16 | >32 | NTd | >32 | 1 | ≤4 | ≤0.5 | 0.5 | ≤0.015 | 6 | 6 | 28 |
| 164 | >16 | >128 | >64 | >16 | >32 | >8 | >32 | 2 | >8 | >1 | 8 | 0.03 | 6 | 6 | 6 |
| DH10B 164 | >16 | >128 | >64 | >16 | >32 | NT | >32 | 2 | >8 | >1 | 0.25 | ≤0.015 | 6 | 6 | 6 |
| 279 | >16 | >128 | 16 | 16 | 8 | 2 | >32 | 0.25 | >8 | >1 | >16 | >2 | 6 | 6 | 6 |
| DH10B 279 | >16 | >128 | 16 | >16 | 16 | NT | >32 | 0.5 | >8 | ≤0.5 | 16 | ≤0.015 | 6 | 6 | 6 |
| 292 | >16 | >128 | 16 | >16 | 32 | >8 | >32 | 0.5 | >8 | >1 | >16 | >2 | 6 | 6 | 6 |
| DH10B 292 | >16 | 128 | 16 | >16 | >32 | NT | >32 | 1 | >8 | ≤0.5 | >16 | ≤0.015 | 6 | 6 | 34 |
| 321 | >16 | >128 | 8 | >16 | 32 | 8 | >32 | 0.5 | >8 | >1 | 16 | ≤0.015 | 6 | 6 | 6 |
| DH10B 321 | >16 | >128 | 16 | >16 | >32 | NT | >32 | 1 | >8 | >1 | 16 | ≤0.015 | 6 | 14 | 6 |
| 665 | >16 | >128 | 16 | 16 | 16 | >8 | >32 | 1 | >8 | >1 | 8 | >2 | 6 | 6 | 6 |
| DH10B 665 | >16 | >128 | 32 | >16 | 32 | NT | >32 | 2 | >8 | >1 | 16 | ≤0.015 | 6 | 6 | 6 |
| 725 | >16 | >128 | 16 | >16 | 32 | >8 | >32 | 4 | >8 | >1 | 1 | >2 | 6 | 6 | 6 |
| DH10B 725 | >16 | >128 | 32 | >16 | >32 | NT | >32 | 4 | >8 | ≤0.5 | 1 | ≤0.015 | 6 | 6 | 6 |
| 771 | >16 | >128 | 8 | 8 | 16 | >8 | >32 | 1 | >8 | >1 | 8 | ≤0.015 | 6 | 6 | 6 |
| DH10B 771 | >16 | >128 | 16 | 16 | 32 | NT | >32 | 1 | >8 | ≤0.5 | 8 | ≤0.015 | 6 | 6 | 6 |
| Human 109 | >16 | >128 | 8 | >16 | 32 | >8 | >32 | 1 | >8 | >1 | >16 | >2 | 6 | 11.6 | 6 |
| DH10B H109 | >16 | >128 | 8 | >16 | 32 | NT | >32 | 0.25 | >8 | ≤0.5 | 0.5 | ≤0.015 | 6 | 6 | 6 |
| Human 826 | >16 | 128 | 8 | 16 | 8 | 8 | >32 | 0.5 | >8 | >1 | >16 | >2 | 6 | 6 | 6 |
| DH10B H826 | >16 | 128 | 8 | >16 | 16 | NT | >32 | 0.5 | >8 | >1 | 0.25 | ≤0.015 | 6 | 6 | 6 |
| DH10B | 4 | 4 | 2 | 0.5 | ≤0.25 | NT | 8 | ≤0.12 | ≤4 | ≤0.5 | 0.5 | ≤0.015 | 6 | 22 | 22 |
Amp, ampicillin; Pip, piperacillin; Pip/tazo, Piperacillin combined with a fixed concentration of tazobactam (4 μg/ml); CTAZ, ceftazidime; CTX, ceftriaxone; CFTI, ceftiofur; CFOX, cefoxitin; CPIM, cefepime; Tet, tetracycline; T/S, trimethoprim-sulfamethoxazole; Gent, gentamicin; Strep, streptomycin; Chlor, chloramphenicol; Sulf, sulfisoxazole; Cipro, ciprofloxacin.
E. coli DH10B transformants carrying the CMY-2 plasmid from the corresponding isolate.
Antimicrobial susceptibility determined by disk methodology.
NT, not tested.
The transformants each expressed a β-lactamase with a pI of ∼8.9, and PCR with the ampC1 and ampC2 primers amplified a 1,143-bp fragment from all transformants. Each PCR fragment was isolated, and the entire nucleotide sequence of the ampC-like gene was determined from both strands. All isolates carried CMY-2, with complete homology between all cephalosporin-resistant animal and human E. coli and Salmonella CMY-2 genes examined (31).
Plasmid analysis.
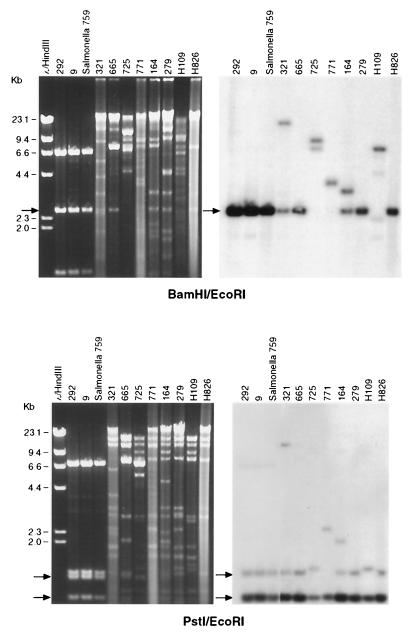
Restriction fragment analysis of the cephalosporin-resistant E. coli transformants demonstrated two isolates, isolates 9 and 292, with an identical 10-kb plasmid (Fig. 1). These two isolates were obtained from cattle located on farms that showed no geographic relationship to each other. The remaining isolates contained 80- to 90-kb plasmids that shared selected common fragments. However, these larger plasmids did not show absolute identity to each other. During our previous studies analyzing cephalosporin-resistant Salmonella, a single bovine isolate from Missouri was found to carry a 10-kb plasmid that shares an identical restriction digest pattern with the two bovine E. coli isolates from Iowa.
FIG. 1.
The left panels represent restriction endonuclease-treated plasmid DNA isolated from the E. coli DH10B transformants. Plasmids were digested with BamHI and EcoRI (top panel) or PstI and EcoRI (lower panel). After gel electrophoresis, DNA was transferred to Nytran and a Southern blot was performed using a random primed α-32P-labeled CMY-2 probe (right panels). A single Salmonella isolate (759) was included for comparison; H109 and H826 are plasmids derived from human E. coli carrying CMY-2. The first three lanes from the PstI/EcoRI Southern blot represent a 2-h autoradiogram, while the remaining lanes were exposed on film for 6 h.
To further analyze the CMY-2 plasmids, Southern blot analysis was performed using a radiolabeled CMY-2 probe (Fig. 1). Seven of nine E. coli CMY-2 plasmids analyzed revealed a 2.4-kb band in BamHI/EcoRI-digested DNA that hybridized to the CMY-2 probe. Additionally, the single bovine Salmonella isolate from Missouri showed a hybridization profile identical to that of the two bovine E. coli plasmids that shared a restriction pattern. Two isolates, bovine 725 and human 109, demonstrated a 6.6-kb fragment that hybridized to the CMY-2 probe. When plasmid DNAs were digested with PstI/EcoRI, all isolates demonstrated at least two bands, all contained a ∼760-bp fragment, and most demonstrated a 1,100-bp fragment. PstI is known to cleave the CMY-2 gene 414 nucleotides from the start codon, thus explaining the presence of two bands. Isolates 725 and H109 demonstrated a slightly larger second band, which migrated at approximately 1,200 bp.
Isolates 771, 164, and 321 each demonstrated extra fragments that hybridized to the CMY-2 probe, though the size of each fragment differed in each isolate. These additional fragments could represent a second β-lactamase that has homology to the CMY-2 gene or partial or complete duplications of CMY-2. There is no significant DNA homology between CMY-2 and TEM-1, and the presence of a pI 5.4 enzyme, consistent with expression of TEM-1, did not correlate with the presence of these additional bands. Therefore, the presence of a TEM-1-like β-lactamase does not explain the additional fragments.
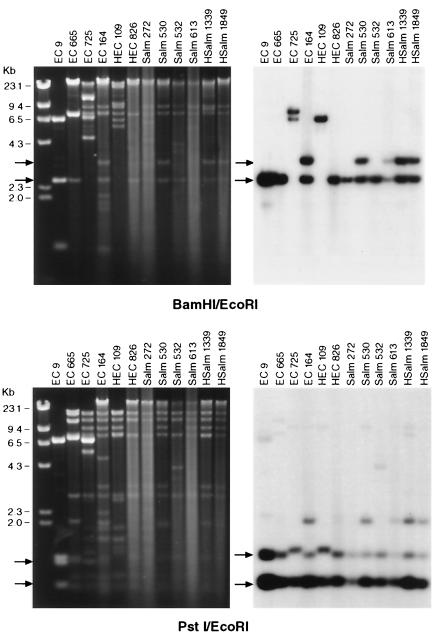
These results suggest that the molecular sequences surrounding the CMY-2 gene are closely related in CMY-2 plasmids from animal and human E. coli. This similar molecular environment is seen in both the 10-kb and 80- to 90-kb plasmids. To further analyze the association between the E. coli and Salmonella plasmids, six CMY-2 plasmids described previously (31) were digested with BamHI/EcoRI and PstI/EcoRI and compared to the E. coli plasmids (Fig. 2). The RFLP patterns between the E. coli and Salmonella CMY-2 plasmids showed many similar fragments, and Southern blot analysis demonstrated identical hybridization profiles between various human and animal isolates from both genera of bacteria.
FIG. 2.
The left panels represent restriction endonuclease-treated plasmid DNA isolated from E. coli (EC) DH10B transformants. Plasmids were digested with BamHI and EcoRI (top panel) or PstI and EcoRI (lower panel). After gel electrophoresis, DNA was transferred to Nytran and a Southern blot was performed using a random primed α-32P-labeled CMY-2 probe (right panels). HSalm, human Salmonella.
Integron analysis.
Type 1 integrons have been shown to carry antimicrobial resistance gene cassettes in many Salmonella DT104 isolates (7, 27). To evaluate whether the CMY-2 gene was located within a type 1 integron, primers homologous to the 5′ conserved and 3′ conserved sequences which flank the insertion site for gene cassettes were used to amplify DNA from the original E. coli isolates or transformed E. coli DH10B carrying the CMY-2 plasmid (16). Six of the eight animal E. coli isolates contained DNA inserted within the type 1 integron. DNA sequence analysis demonstrated that three contained genes with strong homology to the dhfrI genes and three contained a gene that showed homology to the aadAI aminoglycoside modifying enzymes. Only three transformants amplified DNA fragments using the type 1 integron primers, and these three carried genes homologous to the original parent strain, two with dhfr-like genes (11) and one with an aadA-like gene (28). CMY-2 was not located within the type I integrons. Thus, type 1 integrons are common in animal E. coli, and in some cases integrons have become inserted on CMY-2 plasmids. However, this mechanism for mobilizing resistance genes does not explain the transfer of the CMY-2 gene.
DISCUSSION
Expanded-spectrum cephalosporins are important therapeutic agents in veterinary and human medicine and are often used as first-line agents for invasive gram-negative infections. Ceftiofur, an expanded-spectrum cephalosporin, was approved in 1991 for therapeutic veterinary use in cattle in the United States and in 1995 for use in swine. The prevalence of cephalosporin resistance in pathogenic veterinary strains of E. coli described in this study suggests that this agent may be rapidly losing efficacy in food animals. Additionally, high rates of coresistance to other classes of antimicrobial agents were noted. Over 15% of the cephalosporin-resistant strains were resistant to ciprofloxacin (MIC of >2 μg/ml), with seven of these isolates also demonstrating resistance to gentamicin, tobramycin, streptomycin, tetracycline, trimethoprim-sulfamethoxazole, and chloramphenicol. These isolates are now resistant to most, if not all, classes of therapeutic agents available in veterinary medicine.
Plasmid-mediated AmpC-type β-lactamases have been identified in Klebsiella pneumoniae, E. coli, Proteus mirabilis, Enterobacter aerogenes, and Salmonella human clinical isolates from the United States, Europe, and other parts of the world (3, 6, 30). The CMY-2 gene belongs to a small family of plasmid-mediated AmpC-like enzymes (LAT-1, LAT-2, BIL-1, CMY-2 and -2b, CMY-3, CMY-4, CMY-5) that share homology with the chromosomal ampC from C. freundii (3, 14, 30, 33). CMY-2 has now been identified in E. coli and Salmonella from food animal and human isolates in the United States as well as Klebsiella and Salmonella human isolates from Greece and Algeria (3, 10, 14, 31). The E. coli isolates examined in this study were obtained in Iowa. Though not included in this study, two cephalosporin-resistant animal E. coli isolates from Missouri and North Carolina have also been identified, and a single cephalosporin-resistant Salmonella isolate from Missouri was shown in this study to carry an identical 10-kb plasmid, as seen in several Iowa E. coli isolates. Bovine isolates of E. coli with resistance to the expanded-spectrum cephalosporins have also been identified in North Dakota (5). Additionally, the National Antimicrobial Resistance Monitoring System has identified 15 cephalosporin-resistant human isolates from eight different states. All isolates expressed the CMY-2 cephalosporinase (9). These data suggest that CMY-2 is more widespread than initially recognized and that a pool of this resistance gene resides in bacteria endogenous to food animals.
Ninety-five percent of extended-spectrum cephalosporin-cephamycin-resistant animal E. coli isolates in this study expressed a CMY-like enzyme. The few isolates that expressed a highly basic β-lactamase but showed no evidence of having a CMY-like gene have likely undergone upregulation of the native E. coli ampC. Further studies are under way to examine this possibility. Cephalosporin resistance in human E. coli is less predictably associated with expression of CMY-2 (O. Gudlaugsson and P. L. Winokur, unpublished data).
In this study, two different plasmid backgrounds that carried the CMY-2 gene were identified. Both plasmid types were identified in E. coli and Salmonella isolates. These data suggest that the CMY-2 plasmids have been transmitted between different genera of bacteria, as shown with other resistance plasmids (24). The Southern blot hybridization studies demonstrated significant similarities between the 10- and 90-kb plasmids, suggesting that the molecular environment surrounding the CMY-2 gene was similar within the two plasmid types. Transfer of the CMY-2 gene by a common transposon or other genetic transfer mechanism could explain the close homology observed in the microenvironments surrounding CMY-2. Though some differences were seen in the plasmid restriction fragment analyses and the antibiotic coresistances, these may reflect rapid evolution of the plasmids as they are exposed to different environmental stresses. Plasmids have been shown to evolve rapidly, sometimes over a period as short as several months (15, 29).
Nearly 16% of E. coli isolates from clinically ill animals and 5.1% of Salmonella isolates carried CMY-2 (31). The rates in human isolates were much lower: 0.6% for Salmonella and 0.2% for E. coli. The absolute homology between the CMY-2 genes identified in animal and human isolates, the strong similarities between the plasmids, the higher rates of carriage in food animals, and the fact that over 95% of cases of Salmonellosis are thought to occur through food-borne transfer (18) all argue that this resistance may be transmitted to humans through food sources or animal contact. The two human E. coli isolates in this study were obtained from patients with community-acquired infections. One patient developed a bacteremia following an outpatient transrectal prostate biopsy, and the second was isolated from the urine of a 17-year-old woman suffering from an uncomplicated urinary tract infection. Though these isolates could represent pathogenic strains acquired from a food-borne transmission, it is also possible that these strains acquired the resistance plasmid from other nonpathogenic enteric organisms.
National surveillance studies for food-borne pathogens have concentrated on antimicrobial resistance in Salmonella, Campylobacter jejuni, and E. coli 0157:H7 (8). The results of the present study suggest that analysis of other species or serotypes might provide additional insight into antibiotic resistance in food sources.
The findings of our study have important ramifications for clinical veterinary and human medical practice. CMY-2 resistance plasmids appear to move readily between different organisms. There is also a strong possibility that CMY-2 cephalosporin-resistant organisms have been transmitted from food animals to humans. The recent detection of cephalosporin-resistant E. coli and Salmonella in retail meat products provides strong support for food-borne transmission (S. Zhao, D. G. White, R. D. Walker, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. Maurer, and R. Holland, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. Z-52, 2001). However, many questions remain unanswered. More detailed analyses of the use of ceftiofur on farms and the use of other antimicrobials whose resistance genes are coexpressed on the CMY-2 plasmids are needed. Additionally, an analysis of CMY-2 in wildlife, soil, and groundwater samples could provide information regarding environmental contamination.
ACKNOWLEDGMENTS
We thank the Bacteriology Section of the Iowa State University Veterinary Diagnostics Laboratory for providing the animal E. coli isolates. We thank S. Coffman, P. Rhomberg, H. Huynh, K. Heilmann, Josi Taylor, and Jessi Richey for technical assistance.
P.L.W. was supported, in part, by a Merit Review grant from the Department of Veterans Affairs. E.K.U., L.J.H., and the Iowa State Veterinary Diagnostic Laboratory were supported, in part, by the Iowa Healthy Livestock Advisory Council Supplemental Appropriations Grants Program.
REFERENCES
- 1.Arbeit R A. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: American Society for Microbiology; 1995. pp. 190–208. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Stzuhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons Inc.; 1993. [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford P A, Cherubin C E, Idemyor V, Rasmussen B A, Bush K. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing beta-lactamases in a single isolate. Antimicrob Agents Chemother. 1994;38:761–766. doi: 10.1128/aac.38.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford P A, Petersen P J, Fingerman I M, White D G. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J Antimicrob Chemother. 1999;44:607–610. doi: 10.1093/jac/44.5.607. [DOI] [PubMed] [Google Scholar]
- 6.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. National antimicrobial resistance monitoring system: enteric bacteria 1999 annual report. Atlanta, Ga: Centers for Disease Control and Prevention; 1999. [Google Scholar]
- 9.Dunne E F, Fey P D, Kludt P, Reporter R, Mostashari F, Shillam P, Wicklund J, Miller C, Holland B, Stamey K, Barrett T J, Rasheed J K, Tenover F C, Ribot E M, Angulo F J. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA. 2000;284:3151–3156. doi: 10.1001/jama.284.24.3151. [DOI] [PubMed] [Google Scholar]
- 10.Fey P D, Safranek T J, Rupp M E, Dunne E F, Ribot E, Iwen P C, Bradford P A, Angulo F J, Hinrichs S H. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med. 2000;342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 11.Huovinen P, Sundstrom L, Swedberg G, Skold O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isenberg H D. Clinical microbiology procedures handbook. Washington, D.C.: American Society for Microbiology; 1992. [Google Scholar]
- 13.Jacoby G A, Carreras I. Activities of beta-lactam antibiotics against Escherichia coli strains producing extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1990;34:858–862. doi: 10.1128/aac.34.5.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koeck J L, Arlet G, Philippon A, Basmaciogullari S, Thien H V, Buisson Y, Cavallo J D. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol Lett. 1997;152:255–260. doi: 10.1111/j.1574-6968.1997.tb10436.x. [DOI] [PubMed] [Google Scholar]
- 15.Kron M A, Shlaes D M, Currie-McCumber C, Medeiros A A. Molecular epidemiology of OHIO-1 beta-lactamase. Antimicrob Agents Chemother. 1987;31:2007–2009. doi: 10.1128/aac.31.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque C, Piche L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore D M. Beta-lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. . (Erratum, 11:403.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing ninth informational supplement. 19 number 1. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 21.Neidhardt F C. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 22.Pfaller M A, Hollis R J, Sader H S. Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis. In: Isenberg H D, editor. Clinical microbiology procedures handbook, supplement 1. Washington, D.C.: American Society for Microbiology; 1994. pp. 10.5c.1–10.5c.12. [Google Scholar]
- 23.Pfaller M A, Jones R N, Marshall S A, Coffman S L, Hollis R J, Edmond M B, Wenzel R P. Inducible Amp C beta-lactamase producing gram-negative bacilli from blood stream infections: frequency, antimicrobial susceptibility, and molecular epidemiology in a national surveillance program (SCOPE) Diagn Microbiol Infect Dis. 1997;28:211–219. doi: 10.1016/s0732-8893(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 24.Rice L B, Willey S H, Papanicolaou G A, Medeiros A A, Eliopoulos G M, Moellering R C, Jr, Jacoby G A. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemother. 1990;34:2193–2199. doi: 10.1128/aac.34.11.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rondon M R, Raffel S J, Goodman R M, Handelsman J. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc Natl Acad Sci USA. 1999;96:6451–6455. doi: 10.1073/pnas.96.11.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 28.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thal L A, Chow J W, Evans Patterson J, Peri M B, Donabedian S, Clewell D B, Zervos M J. Molecular characterization of highly gentamicin-resistant Enterococcus faecalis isolates lacking high-level streptomycin resistance. Antimicrob Agents Chemother. 1993;37:134–137. doi: 10.1128/aac.37.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdet C, Arlet G, Ben Redjeb S, Ben Hassen A, Lagrange P H, Philippon A. Characterisation of CMY-4, an AmpC-type plasmid-mediated beta-lactamase in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol Lett. 1998;169:235–240. doi: 10.1111/j.1574-6968.1998.tb13323.x. [DOI] [PubMed] [Google Scholar]
- 31.Winokur P L, Brueggemann A, DeSalvo D L, Hoffmann L, Apley M D, Uhlenhopp E K, Pfaller M A, Doern G V. Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob Agents Chemother. 2000;44:2777–2783. doi: 10.1128/aac.44.10.2777-2783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winokur P L, Canton R, Casellas J-M, Legakis N. Variations in the prevalence of strains expressing an extended spectrum b-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific. Clin Infect Dis. 2001;2(Suppl.):S94–S103. doi: 10.1086/320182. [DOI] [PubMed] [Google Scholar]
- 33.Wu S W, Dornbusch K, Kronvall G, Norgren M. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC beta-lactamase. Antimicrob Agents Chemother. 1999;43:1350–1357. doi: 10.1128/aac.43.6.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]