Abstract
Recently, the SARS-CoV-2 Omicron has spread very quickly worldwide. Several studies have indicated that the Omicron variant causes a substantial evasion of the humoral immune response and the majority of existing SARS-CoV-2 neutralizing antibodies. Here we address this challenge by applying a spherical cocktail neutralizing aptamer-gold nanoparticle (SNAP) to block the interaction of Omicron receptor binding domain (RBD) and host Angiotensin-Converting Enzyme 2 (ACE2). With the synergetic blocking strategy based on multivalent multisite aptamer binding and steric hindrance by the size-matched gold scaffold, the SNAP conjugate tightly binds to Omicron RBD with a dissociation constant of 13.6 pM, almost completely blocking the infection of Omicron pseudovirus with a half-maximal inhibitory concentration of 35.9 pM. Overall, the SNAP strategy not only fills the gap of the humoral immune evasion caused by clustered mutations on Omicron, but also provides a clue for the development of new broad neutralizing reagents against future variants.
Keywords: SARS-CoV-2 Omicron, Aptamer, Neutralization, Spherical nucleic acids
Graphical Abstract
Introduction
The accumulated mutations of SARS-CoV-2 result in modulating the binding affinity to host receptors and immune escape. At the end of November 2021, the SARS-CoV-2 variant B.1.1.529 was first detected and designated as Omicron, which was immediately declared as a variant of concern by the World Health Organization [1], [2], [3], [4]. Omicron appears to be very quickly spreading and rising in frequency worldwide [5], [6], [7]. An unprecedented number of mutations have been found in the Omicron virus, including 37 residue substitutions of the spike [8]. Fifteen of the mutations are clustered in the receptor binding domain (RBD), which is involved in interacting with the host cell receptor (Angiotensin-Converting Enzyme 2, ACE2) [9], [10], and the main target of neutralizing therapy and neutralizing antibodies after vaccination or infection [11], [12], [13], [14]. Therefore, these mutations suggest that Omicron may escape the existing neutralizing antibody-based therapy and vaccine-elicited antibodies.
Several studies have indicated that the neutralizing activity of the majority of existing SARS-CoV-2 neutralizing antibodies has been abolished, and only a few still function at reduced efficacy against the Omicron variant [15], [16], [17]. In addition, preliminary reports showed that the neutralizing activities of plasma against Omicron are significantly lower than that against Delta in individuals who completed the mRNA vaccination (BioNTech BNT162b2) [18], [19], [20]. Together, these indicate that the Omicron variant causes a substantial, albeit incomplete evasion, of the humoral immune system. Therefore, it is urgent to discover other therapies that can effectively resist Omicron mutation escape.
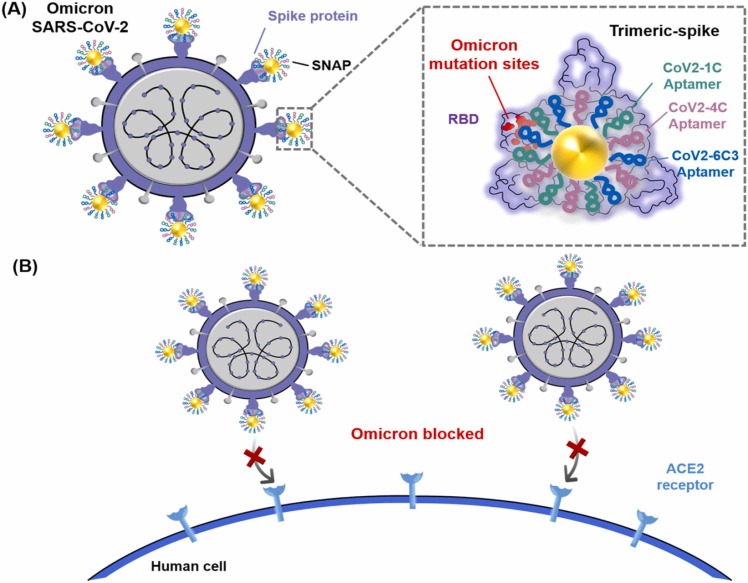
Recently, our group has developed a spherical cocktail neutralizing aptamer-gold nanoparticle (SNAP) to block the interaction between SARS-CoV-2 RBD and host ACE2 [21]. As a functional nucleic acid, the neutralizing aptamer can be programmatically engineered and assembled more easily than a neutralizing antibody. Combining multivalent hetero-neutralizing aptamers and the steric hindrance effect of the gold nanoparticle, SNAP exhibits excellent neutralization against wild and three mutant SARS-CoV-2 pseudoviruses. To establish an effective neutralizing treatment against Omicron, here we investigate how Omicron affects the binding performance of the selected neutralizing aptamers and the neutralization capability of SNAP ( Fig. 1).
Fig. 1.
(A) Schematic diagram of the interaction of Omicron SARS-CoV-2 and SNAP. The cocktail aptamers (CoV2-1C, CoV2-4C and CoV2-6C3) of SNAP tolerate the mutations of RBD on Omicron SARS-CoV-2-spike trimer. Trimeric-spike protein and SNAP are shown in top view, and the mutation sites of Omicron are marked in red. (B) Simplified mechanism of SNAP neutralization: Infection by Omicron SARS-CoV-2 virus is blocked via the binding of SNAP to the spike trimer protein at the binding surface of the ACE2 receptor.
Material and methods
All experimental materials and methods are included in the Supporting Information.
Results and discussion
To avoid mutation-induced neutralization escape of Omicron variant ( Fig. 2A), three neutralizing aptamers (CoV2‐1C, CoV2‐4C and CoV2-6C3) with nonoverlapping epitopes on SARS-CoV-2 RBD were chosen for SNAP assembly. We first investigated the binding performance of the three aptamers against Omicron RBD. As shown in the flow cytometry results, the three aptamers still maintained some binding capacity to Omicron RBD-beads (Fig. 2B). Among the three aptamers, the fluorescence intensities of aptamers CoV2‐4C and CoV2-6C3 against Omicron RBD-beads were 5.0 and 2.7 times that of the random sequence, respectively, while the fluorescence value of aptamer CoV2‐1C against Omicron RBD-beads was only 1.5 times that of the random sequence. These results suggest that these aptamers tolerate the multiple synergetic mutations on Omicron RBD, especially aptamer CoV2‐4C and CoV2-6C3. Moreover, the results of cross-competition experiments between any two of these three aptamers suggested that these three aptamers are likely to have less than 30% interference with each other and likely interact with non-overlapping epitopes on Omicron RBD (Fig. 2C).
Fig. 2.
(A) Mapping Omicron RBD mutations on the surface of SARS-CoV-2 spike trimer (PDB: 6VSB). Detailed mutant residues in Omicron variants are denoted on RBD protomer in red spheres. (B) Flow cytometric analysis of CoV2-1C, CoV2-4C and CoV2-6C3 binding to Omicron RBD. (C) Epitope binning of any two of CoV2-1C, CoV2-4C, and CoV2-6C3 aptamers for Omicron RBD binding using cross-competition experiments. The abscissa sequences indicate the target aptamers with fluorescence, and the ordinate sequences indicate the competitive aptamers without fluorescence. (D-F) Amino acid sites for CoV2-6C3, CoV2-4C and CoV2-1C binding are marked in green spheres, and the label for the amino acid sites of Omicron mutation is red.
To further understand the interaction between the selected aptamers and Omicron RBD, we compared the simulated aptamer binding RBD amino acids with 15 mutations on Omicron RBD. The simulated results indicated that the interaction between of CoV2-6C3 and RBD involves two consecutive binding fragments, and that neither of the amino acids are involved in interaction-contained mutation sites. Specifically, aptamer CoV2-6C3 was predicted to bind to one shoulder of RBD, extensively overlapping with the binding interface of ACE2, including Phe486 and Gln474 (Fig. 2D). Fortunately, Phe486 is critically involved in spike-ACE2 binding, and therefore it does not generally escape mutationally [8], [9]. The epitopes of aptamer CoV2‐4C are suggested to overlap extensively with the middle segment of the ACE2-RBD “bridge” interaction [22]. This segment may be insensitive to the Lys417 single site change on Omicron RBD due to the formation of hydrogen bonds with the other two conserved binding sites (Gln409 and Try421) (Fig. 2E). The simulated binding epitopes of CoV2‐1C aptamer partly overlap or occur near to mutations on Omicron RBD, causing certain reeducation of the aptamer’s binding (Fig. 2F). Therefore, in order to avoid the escape induced by the accumulated mutations of Omicron and new mutant strains in the future, we still chose these three neutralizing aptamers for the SNAP assembly cocktail.
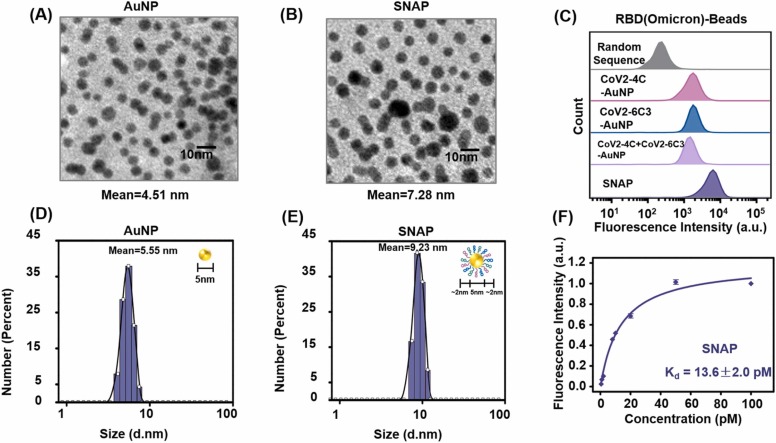
To provide multivalent binding and neutralization, we applied 5-nm gold nanoparticles (AuNPs), which have sizes similar to that of the trimeric-RBD of SARS-CoV-2, as the scaffold for neutralizing aptamer cocktail assembly to form SNAP conjugates. The SNAP conjugates can be easily obtained by the freeze-thaw procedure of AuNPs and SH-labeled cocktail aptamers [23]. As shown by transmission electron microscopy (TEM) images ( Fig. 3A, B) and dynamic light scattering (DLS) (Fig. 3C, D), the SNAP conjugates have slightly larger average diameters than bare AuNPs, indicating the successful aptamer modification of SNAPs. With nearly 50 aptamers loaded on each 5 nm AuNP (Figs. S1 and S2), there is sufficient affinity to form SNAP-Omicron RBD interactions for improved neutralization by the multivalent effect. As a result of simultaneous binding against distinct epitopes on Omicron RBD based on homo- and hetero-neutralizing aptamers, the amount of SNAP binding to Omicron RBD beads is about 3.5–4.7 times greater than that of one or two sequentially modified gold nanoparticles (Fig. 3E). Taking advantage of the synergistic effect, the Kd value of SNAP conjugate against Omicron RBD-bead is 13.6 pM (Fig. 3F), about 4 or 5 orders of magnitude better than its monomeric aptamer or a neutralizing antibody (Figs. S3 and S4), and about 650-fold higher than the reported binding affinity of Omicron RBD and ACE2 [24]. These results indicate the feasibility of improving the binding performance using SNAP as the recognition element for Omicron neutralization compared to the single aptamer.
Fig. 3.
(A, B) Transmission electron microscopy images of AuNPs and SNAPs. (C, D) Hydrodynamic size distribution histograms of AuNPs and SNAPs using dynamic light scattering. (E) Flow cytometry analysis of CoV2-4C-modified AuNP, CoV2-6C3-modified AuNP, and a mixture of CoV2-4C- and CoV2-6C3- modified AuNP and SNAP against Omicron RBD-beads. Inserts: cartoon illustration with the theoretical diameters. (F) Binding curve of SNAP against Omicron RBD-beads, where the SNAP concentration equals the corresponding concentration of AuNP. Data are reported as the mean ± SEM of three independent replicates.
To evaluate the neutralization effect of SNAP against Omicron, we used a pseudotyped Omicron virus, which contains the genome consisting of the Omicron spike protein and GFP reporter. The results indicated that SNAP can protect more than 98% of the ACE2-transfected cells from pseudotyped Omicron infection, displaying much higher potency than CoV2‐4C (53.22%), CoV2-6C3 (50.74%) and the respective aptamer cocktail (65.53%) ( Fig. 4A). Moreover, the inhibitory ability of SNAP is also greater than that of a commercial neutralizing antibody (32.65%) at the same concentration. In addition, the binding of SNAP to Omicron SARS-CoV-2 spike pseudotyped virus was verified by transmission electron microscopy (TEM) (Fig. S5) and SNAP showed no obvious cell cytotoxicity on ACE2-transfected 293 T cells (Fig. S6), which is similar to the results in the reported literature [21], [25], [26].
Fig. 4.
(A) Images of monomeric aptamer, aptamer-modified AuNP, SNAP, and the antibody inhibition of Omicron SARS-CoV-2 spike pseudotyped virus infection of ACE2-expressing 293 T cells. Monomeric aptamer: CoV2-4C, CoV2-6C3 and aptamer cocktail (the mixture of CoV2-1C, CoV2-4C and CoV2-6C3); aptamer-modified AuNP: CoV2-4C-AuNP and CoV2-6C3-AuNP. (B) The neutralization of Omicron SARS-CoV-2 pseudovirus by SNAP assessed through IC50. Assays are repeated as three independent replicates, and data are presented as the mean ± SEM.
Similar to our previous results, the inhibition effect was greatly improved after the aptamer was modified on gold nanoparticles, due to the synergistic benefits of multiple valent binding from the cocktail aptamer and the steric hindrance effect from the nano-scaffold [21]. Although CoV2‐4C‐AuNP and CoV2-6C3-AuNP achieved nearly 90% cell protection, combination with our previous investigation [21], SNAP conjugates always have the best neutralization potency against wild type and a series of SARS-CoV-2 variants, including Omicron. In addition, SNAP conjugates exhibit excellent neutralizing activity against pseudotyped Omicron virus with a half-maximal inhibitory concentration (IC50) of 35.9 pM (Fig. 4B). Different from neutralizing antibodies, which is usually based on one-site blocking, SNAP tolerates clustered mutations and completely blocks infection by Omicron pseudovirus, due to multi-site recognition and steric hindrance. Overall, the SNAP strategy circumvents the humoral immune evasion caused by Omicron, and provides a new direction for neutralization against SARS-CoV-2 variants and other emerging viruses.
Conclusions
Omicron is reported to escape the majority of SARS-CoV-2 neutralizing antibodies identified from convalescents and vaccines. To address this challenge, we investigated the application of the previously developed SNAP strategy, which displays broad neutralizing activity against several mutants, to suppress Omicron escape. As shown by our results, SNAP tightly binds to Omicron RBD and almost completely blocks infection by pseudotyped Omicron virus. The radically different result compared to neutralizing antibodies is mainly due to several outstanding characteristics of the SNAP strategy. First, different aptamers with distinct epitopes are more insensitive to the accumulated mutations of SARS-CoV-2. Second, the size-matched nano-scaffold not only ensures multi-homologous and multi-heterogeneous recognition of cocktail aptamers, but also provides suitable steric hindrance to inhibit the interaction of RBD and ACE2. Moreover, the combination of SNAP and the photothermal effect of gold nanoparticles has the potential to kill viruses [27], [28], [29]. And compared with neutralizing antibodies, neutralizing aptamers are small in size, easy to modify and assemble, and easier to combine with nanotechnology. In addition, considering an AuNP based drug in the clinical phase I experiment and no obvious cell cytotoxicity of SNAP [21], [25], [26], [30], it is expected that the clinical advance of this method could be accelerated after the subsequent evaluation the antiviral efficiency of SNAP against authentic SARS-CoV-2 Omicron variants in vitro and in vivo. Overall, the SNAP strategy provides a direction for the development of new nano-neutralizing reagents against current or emerging SARS-CoV-2 variants.
CRediT authorship contribution statement
Miao Sun: Investigation, Methodology, Validation, Formal analysis, Writing – original draft. Zijing Wu: Investigation, Validation, Formal analysis. Jialu Zhang: Resources, Validation. Mingying Chen: Investigation, Visualization. Yao Lu: Formal analysis. Chaoyong Yang: Conceptualization, Supervision, Writing – review & editing, Funding acquisition. Yanling Song: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Chaoyong Yang reports financial support was provided by the National Natural Science Foundation of China.
Acknowledgments
We thank the National Natural Science Foundation of China (Grants 22022409, 21735004, and 21874089), the Program for Changjiang Scholars and Innovative Research Teams in University (Grant IRT13036), National Science Fund for Fostering Talents in Basic Science (NFFTBS, J1310024), and XMU Training Program of Innovation and Entrepreneurship for Undergraduates for their financial support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nantod.2022.101499.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal A., Stella A.O., Walker G., Akerman A., Milogiannakis V., Brilot F., Amatayakul-Chantler S., Roth N., Coppola G., Schofield P., Jackson J., Henry J.Y., Mazigi O., Langley D., Lu Y., Forster C., McAllery S., Mathivanan V., Fichter C., Hoppe A.C., Munier M.L., Jack H.-M., Cromer D., Darley D., Matthews G., Christ D., Khoury D., Davenport M., Rawlinson W., Kelleher A.D., Turville S. SARS-CoV-2 Omicron: evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. medRxiv. 2021 doi: 10.1101/2021.12.14.21267772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He X., Hong W., Pan X., Lu G., Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., Choga W.T., Colquhoun R., Davids M., Deforche K., Doolabh D., Du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N.-Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S.L., Kraemer M.U.G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R.J., Lockman S., Lucaci A.G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M.B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M.S., Mphoyakgosi T., Msomi N., Mwangi P.N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J.E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., Van Wyk S., Weaver S., Wibmer C.K., Wilkinson E., Wolter N., Zarebski A.E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O.G., Bhiman J., Glass A., Martin D.P., Rambaut A., Gaseitsiwe S., Von Gottberg A., De Oliveira T. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulliam J.R.C., Van Schalkwyk C., Govender N., Von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. medRxiv. 2021 doi: 10.1101/2021.11.11.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grabowski F., Kochańczyk M., Lipniacki T. The spread of SARS-CoV-2 variant Omicron with the doubling time of 2.0–3.3 days can be explained by immune evasion. medRxiv. 2021 doi: 10.1101/2021.12.08.21267494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y., Yisimayi A., Bai Y., Huang W., Li X., Zhang Z., Yuan T., An R., Wang J., Xiao T., Du S., Ma W., Song L., Li Y., Li X., Song W., Wu J., Liu S., Li X., Zhang Y., Su B., Guo X., Wei Y., Gao C., Zhang N., Zhang Y., Dou Y., Xu X., Shi R., Lu B., Jin R., Ma Y., Qin C., Wang Y., Feng Y., Xiao J., Xie X.S. Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res. 2021;31:732–741. doi: 10.1038/s41422-021-00514-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu H.J., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y.X., Teng Y., Zhao Z.P., Cui Y.J., Li Y.C., Li X.F., Li J.F., Zhang N.N., Yang X.L., Chen S.L., Guo Y., Zhao G.Y., Wang X.L., Luo D.Y., Wang H., Yang X., Li Y., Han G.C., He Y.X., Zhou X.J., Geng S.S., Sheng X.L., Jiang S.B., Sun S.H., Qin C.F., Zhou Y.S. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., Acton O.J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J.C., Mele F., Jovic S., Rodriguez B.F., Gupta S.V., Jin F., Piumatti G., Lo Presti G., Pellanda A.F., Biggiogero M., Tarkowski M., Pizzuto M.S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H.W., Lanzavecchia A., Corti D., Veesler D. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., Wang J., Wang Y., Niu X., Yang S., Liang H., Sun H., Li T., Yu Y., Cui Q., Liu S., Yang X., Du S., Zhang Z., Hao X., Shao F., Jin R., Wang X., Xiao J., Wang Y., Xie X.S. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., Vanblargan L.A., De Marco A., Zepeda S.K., Iulio J.D., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Veesler D., Snell G., Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv. 2021 doi: 10.1101/2021.12.12.472269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.-H., Porrot F., Staropoli I., Lemoine F., Péré H., Veyer D., Puech J., Rodary J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina -Bokalanga T., Martí-Carreras J., Cuypers L., Sève A., Hocqueloux L., Prazuck T., Rey F., Simon-Loriere E., Bruel T., Mouquet H., André E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 18.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D., Karim F., Bernstein M., Lustig G., Archary D., Smith M., Ganga Y., Jule Z., Reedoy K., Hwa S.-H., Giandhari J., Blackburn J.M., Gosnell B.I., Karim S.S.A., Hanekom W., Von Gottberg A., Bhiman J., Lessells R.J., Moosa M.-Y.S., Davenport M.P., De Oliveira T., Moore P.L., Sigal A. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 19.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., Metzler M., Kohmer N., Hoehl S., Helfritz F.A., Wolf T., Goetsch U., Ciesek S. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021 doi: 10.1101/2021.12.07.21267432. [DOI] [Google Scholar]
- 20.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., Simons D., Blomquist P.B., Zaidi A., Nash S., Aziz N.I.B.A., Thelwall S., Dabrera G., Myers R., Amirthalingam G., Gharbia S., Barrett J.C., Elson R., Ladhani S.N., Ferguson N., Zambon M., Campbell C.N., Brown K., Hopkins S., Chand M., Ramsay M., Bernal J.L. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. 2021 doi: 10.1101/2021.12.14.21267615. [DOI] [Google Scholar]
- 21.Sun M., Liu S., Song T., Chen F., Zhang J., Huang J.-A., Wan S., Lu Y., Chen H., Tan W., Song Y., Yang C. Spherical neutralizing aptamer inhibits SARS-CoV-2 infection and suppresses mutational escape. J. Am. Chem. Soc. 2021;143:21541–21548. doi: 10.1021/jacs.1c08226. [DOI] [PubMed] [Google Scholar]
- 22.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 23.Liu B., Liu J. Freezing directed construction of bio/nano interfaces: reagentless conjugation, denser spherical nucleic acids, and better nanoflares. J. Am. Chem. Soc. 2017;139:9471–9474. doi: 10.1021/jacs.7b04885. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X.T., Wu S.J., Wu B.L., Yang Q.R., Chen A.C., Li Y.Z., Zhang Y.W., Pan T., Zhang H., He X. SARS-CoV-2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct. Target. Ther. 2021;6:430. doi: 10.1038/s41392-021-00852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S., Chen C., Xue C., Chang D., Xu H., Salena B.J., Li Y., Wu Z.S. Ribbon of DNA lattice on gold nanoparticles for selective drug delivery to cancer cells. Angew. Chem. Int. Ed. 2020;59:14584–14592. doi: 10.1002/anie.202005624. [DOI] [PubMed] [Google Scholar]
- 26.Yu S., Zhou Y., Sun Y., Wu S., Xu T., Chang Y.C., Bi S., Jiang L.P., Zhu J.J. Endogenous mRNA triggered DNA-Au nanomachine for in situ imaging and targeted multimodal synergistic cancer therapy. Angew. Chem. Int. Ed. 2021;60:5948–5958. doi: 10.1002/anie.202012801. [DOI] [PubMed] [Google Scholar]
- 27.Ren M., Zhou J., Song Z., Mei H., Zhou M., Fu Z.F., Han H., Zhao L. Aptamer and RVG functionalized gold nanorods for targeted photothermal therapy of neurotropic virus infection in the mouse brain. Chem. Eng. J. 2021;411 [Google Scholar]
- 28.Kuo W.S., Chang C.N., Chang Y.T., Yang M.H., Chien Y.H., Chen S.J., Yeh C.S. Gold nanorods in photodynamic therapy, as hyperthermia agents, and in near-infrared optical imaging. Angew. Chem. 2010;122:2771–2775. doi: 10.1002/anie.200906927. [DOI] [PubMed] [Google Scholar]
- 29.Park S., Kim H., Lim S.C., Lim K., Lee E.S., Oh K.T., Choi H.G., Youn Y.S. Gold nanocluster-loaded hybrid albumin nanoparticles with fluorescence-based optical visualization and photothermal conversion for tumor detection/ablation. J. Control. Release. 2019;304:7–18. doi: 10.1016/j.jconrel.2019.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Alkilany A.M., Lohse S.E., Murphy C.J. The gold standard: gold nanoparticle libraries to understand the nano-bio interface. Acc. Chem. Res. 2013;46:650–661. doi: 10.1021/ar300015b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material