Abstract
The dysregulation of transcription factors is widely associated with tumorigenesis. As the most well-defined transcription factor in multiple types of cancer, c-Myc can transform cells by transactivating various downstream genes. Given that there is no effective way to directly inhibit c-Myc, c-Myc targeting strategies hold great potential for cancer therapy. In this study, we found that WSB1, which has a highly positive correlation with c-Myc in 10 cancer cell lines and clinical samples, is a direct target gene of c-Myc, and can positively regulate c-Myc expression, which forms a feedforward circuit promoting cancer development. RNA sequencing results from Bel-7402 cells confirmed that WSB1 promoted c-Myc expression through the β-catenin pathway. Mechanistically, WSB1 affected β-catenin destruction complex-PPP2CA assembly and E3 ubiquitin ligase adaptor β-TRCP recruitment, which inhibited the ubiquitination of β-catenin and transactivated c-Myc. Of interest, the effect of WSB1 on c-Myc was independent of its E3 ligase activity. Moreover, overexpressing WSB1 in the Bel-7402 xenograft model could further strengthen the tumor-driven effect of c-Myc overexpression. Thus, our findings revealed a novel mechanism involved in tumorigenesis in which the WSB1/c-Myc feedforward circuit played an essential role, highlighting a potential c-Myc intervention strategy in cancer treatment.
KEY WORDS: Transcription factors, c-Myc, WSB1, Ubiquitination-proteasome pathway, β-Catenin destruction complex, Feedback loop, Tumorigenesis, Cancer treatment
Abbreviations: ATM, serine-protein kinase ATM; CHIP, chromatin immunoprecipitation; CK1, casein kinase 1; c-Myc, proto-oncogene c-Myc; EBP2, probable rRNA-processing protein EBP2; eIF4F, eukaryotic translation initiation factor 4F; ESC complex, elongin B/C-cullin 2/5-SOCS box containing ubiquitin ligase protein complex; GSK3β, glycogen synthase kinase 3β; HCC, hepatocellular carcinoma; HIF1-α, hypoxia induced factor 1-alpha; IHC, immunohistochemistry; PLK1, serine/threonine-protein kinase PLK1; PP2A, serine/threonine protein phosphatase 2A; PROTAC, proteolysis targeting chimaera; RhoGDI2, Rho GDP dissociation inhibitor 2; TFs, transcription factors; WSB1, WD repeat and SOCS box containing 1
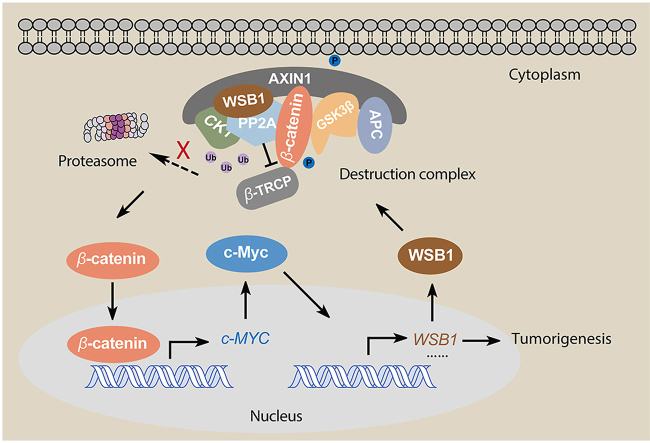
Graphical abstract
WSB1 promoted the expression of c-Myc through WNT/β-catenin pathway, which enhanced the cancer promoting effect induced by c-Myc in a E3 ligase activity independent manner.
1. Introduction
Transcription factors (TFs) recognize DNA in a sequence-specific manner and are responsible for genome transcription control1,2. Given that cancers arise from the abnormal activation of oncogenes or the inactivation of anti-oncogenes, it is not surprising that mutations and dysregulation of TFs are associated with tumorigenesis3. Moreover, the dysregulated activity of TFs could in turn make cancer cells become highly demanding for certain regulators of gene expression, which represents a common tumor-associated phenomenon called “transcriptional addiction”4. To date, hundreds of tumor-associated TFs have been identified. Proto-oncogene c-Myc (c-Myc) is one of the most well-documented tumor-associated TFs in multiple types of cancer and directly transforms cells by transactivating downstream genes involved in cell division, cell cycle progression, angiogenesis, cell senescence and tumor progression5, 6, 7.
Although c-Myc has been widely accepted as a promising target for cancer therapy, lacking of a specific ligand binding domain limits the development of targeting strategies8. Moreover, as c-Myc is located in the nucleus, monoclonal antibody drugs with macromolecules are not able to target it effectively9,10. Thus, there is still no effective way to directly target c-Myc. Recently, many studies have shown that the transactivation of c-Myc is regulated by upstream cytokine signaling, transcription factors and related binding proteins11, among which the WNT/β-catenin pathway is the most classical signaling12. Upon WNT signaling activation, the ubiquitination of β-catenin is inhibited. As a result, β-catenin is able to accumulate and translocate into the nucleus to activate the transcription of c-Myc13,14. However, the molecular mechanism of the upstream regulation of c-Myc is not fully understood, which hinders the theoretical development of c-Myc targeting strategies.
WD repeat and SOCS box containing 1 (WSB1), a member of the elongin B/C-cullin 2/5-SOCS box containing ubiquitin ligase protein complex (ESC complex), functions as an adaptor for recognizing substrates for ubiquitination and has been considered as the target of multiple onco-genes including c-Myc15, 16, 17. Increasing evidence suggests that WSB1 is correlated with tumor progression, metastasis and drug resistance18, 19, 20. Moreover, a recent study discovered that WSB1 promoted the degradation of the DNA damage repair associated protein serine-protein kinase ATM (ATM) and participated in cell senescence induced by GTPase RAS16. In our previous study, we found that WSB1 was the target gene of hypoxia induced factor 1-alpha (HIF1-α) and could promote the degradation of its substrate Rho GDP dissociation inhibitor 2 (RhoGDI2), leading to metastasis of osteosarcoma21. Thus, further exploration of WSB1 and its substrate as well as the regulatory mechanism will not only clarify the important role of WSB1 in tumor progression but also provide new targets for cancer treatment.
As far as we know, the relationship of WSB1 and c-Myc has been predicted and preliminary confirmed by different research teams in the past few years16. Based on the literature search and experimental data, we analyzed the promoter sequence of WSB1 and found that there was exactly a putative c-Myc binding site. Recently, multiple studies have proposed that c-Myc can activate several downstream target proteins, such as probable rRNA-processing protein EBP2 (EBP2), eukaryotic translation initiation factor 4F (eIF4F), and serine/threonine-protein kinase PLK1 (PLK1), to form feedback loops in tumor progression22, 23, 24, which greatly aroused our interest to find out whether WSB1 could regulate c-Myc in turn? In this study, we report that WSB1 is a direct target gene of c-Myc. Moreover, WSB1 could promote the expression of c-Myc through the WNT/β-catenin pathway, which enhances the cancer-promoting effect induced by c-Myc. Thus, our findings reveal a novel mechanism involved in tumorigenesis in which the WSB1/c-Myc feedforward circuit plays an essential role, highlighting that targeting WSB1 might be a potential method to regulate c-Myc in cancer treatment.
2. Materials and methods
2.1. Reagents and antibodies
Proteasome inhibitor MG132, β-catenin inhibitor ICG-001 and XAV939 were purchased from MedChemExpress. Phosphatase inhibitor cocktail I (#C002) was purchased from Target Molecule Corp. Leupeptin, Na3VO4, pyruvic acid sodium salt (PN) and WSB1 antibody (#HPA003293) was purchased from Sigma–Aldrich. Non-essential amino acid and l-glutamine was purchased from Invitrogen (Thermo Fisher Scientific) and polyethylenimine 40000 (PEI-40000) was purchased from Polysciences. TRIzol, T4 ligase and KOD PCR kit were purchased from Takara Bio. SYBR Green supermixes for real-time PCR was purchased from BIO-RAD. The BCA protein quantitative kit was purchased from Shanghai Yisheng Biotechnology Co., Ltd. And c-Myc (#db1667), HA (#db2603), GAPDH (#db106), ubiquitin (#db935), GSK3β (#db2953) and CK1 (#db3190) antibodies were purchased from Diagnostic Biosystems. c-Myc (#sc-40) antibody for immunohistochemistry was purchased from Santa Cruz Biotechnology. Phosphatase PP1/PP2A inhibitor calyculin A (#9902), as well as p-β-catenin (#9561S), β-catenin (#9562S), AXIN1 (#2087S), β-TRCP (#4394S) and phospho-(Ser/Thr) Phe (#9631) antibodies were purchased from Cell Signaling Technology. FLAG antibody was purchased from GenScript. PPP2CA (I3482-I-AP), lamin B (66095-I-Ig) antibodies were purchased from Proteintech Group. WNT 3A conditional medium was generated according to previous report25.
2.2. Cell culture
Human embryonic kidney cell line HEK 293FT which was purchased from Invitrogen (Thermo Fisher Scientific), human liver hepatocellular carcinoma cell line HepG2, human ovarian cancer cell line A2780 and human colon cancer cell line HCT116 which were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences, were cultured in Dulbecco's modified Eagle's medium (Gibco, Thermo Fisher Scientific). Human osteosarcoma cell line KHOS/NP was kindly provided by Dr. Lingtao Wu in University of Southern California, and was cultured in Dulbecco's modified Eagle's medium (Gibco, Thermo Fisher Scientific). Human hepatocellular carcinoma cell line Bel-7402 as well as HuH-7, human ovarian cancer cell line OVCAR-8, human osteosarcoma cell line U2OS, human colon cancer cell line SW620 and human lung cancer cell line NCI-H460 which were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences, were cultured in RPMI-1640 (Gibco, Thermo Fisher Scientific). Human non-small cell lung cancer cell line A549 which was purchased from the Shanghai Cell Bank of Chinese Academy of Sciences, was cultured in Ham's F12 nutrient medium (Gibco, Thermo Fisher Scientific). All cells were cultured at 37 °C with 5% CO2 atmosphere, and all mediums were supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific) as well as 100 IU/mL penicillin and 100 μg/mL streptomycin. All cell lines used were authenticated by STR profiling. The cell lines were monitored for mycoplasma contamination every six months.
2.3. Plasmid construction
The pCDH, pGL4.14, Renilla, pMXs-Hu-N-Myc and pMXs-Hu-L-Myc plasmids were obtained from Addgene. The pCCL-WSB1 and pCCL-c-Myc plasmids were purchased from Genscript, and were re-constructed to pCDH vector with N-terminal FLAG tag. pGL4.14-WSB1 promoter and pGL4.14-c-Myc promoter plasmids were cloning from the genome promoter and re-constructed into pGL4.14 vector. The pGM-TCF/LEF1-luciferase plasmid was purchased from Shanghai Yisheng Biotechnology Co., Ltd. The wild type AXIN1 and its deletion mutants D1–D6 plasmids were constructed into pCDH vector with N-terminal HA tag. CK1, GSK3β, AXIN1, β-TRCP as well as β-catenin plasmids were constructed into pCDNA 3.0 vector with N-terminal HA tag. PPP2CA plasmid was reconstructed into pCDH vector with N-terminal FLAG tag. The pCMV-R8.91 and pMD2-VSVG plasmids were kindly provided by Dr. Lingtao Wu in University of Southern California (USA). The lentivirus vector pLKO.1 was purchased from Thermo-Open-Biosystem. The shRNAs of WSB1 and β-catenin were synthesized by Boshang Biotechnology and constructed into pLKO.1 vector, sequences were as follows (5′–3′):
shWSB1-1#: CCGGGAGTTTCTCTCGTATCGTATTCTCGAGAATACGATACGAGAGAAACTCTTTTTG;
shWSB1-2#: CCGGGATCGTGAGATTACGTACTATCTCGAGATAGTACGTAATCTCACGATCTTTTTG;
shβ-catenin-1#: CCGGCAGATGGTGTCTGCTATTGTACTCGAGTACAATAGCAGACACCATCTGTTTTTG;
shβ-catenin-2#: CCGGGCTTGGAATGAGACTGCTGATCTCGAGATCAGCAGTCTCATTCCAAGCTTTTTG.
2.4. Lentivirus transduction
Lentivirus was produced by transfecting 293FT cells with pCMV-R8.91 (packaging vector), pMD2-VSVG (envelope vector) and shRNA plasmids or pCDH plasmids by PEI-40000 with the ratio of 5:1:5 in opti-MEM (Gibco, Thermo Fisher Scientific). Virions were collected after 48 h after transfection. Bel-7402, 293FT and H460 cells were transduced with lentivirus companied by polybrene (6 mg/mL) at the MOI of 5–10 to obtain stable cell strains.
2.5. Cell lysate preparation and immunoprecipitation
Cells were collected with pre-cold PBS and lysed with 1% NP-40 buffer (25 mmol/L Tris, 150 mmol/L NaCl, 10% glycerol and 1% Nonidet P-40, pH = 8.0) with 0.1 mmol/L PMSF, 0.1 mmol/L Na3VO4 and 5 μg/mL leupeptin. After centrifugation at 14,000×g, 4 °C for 30 min, the protein quantification was operated by Bicinchoninic acid kit from Yeasen Biotech. Flag affinity beads (Genscript) or HA magnetic beads (Bimake) were mixed with cell lysate at 4 °C overnight, and washed for 4 times with washing buffer (25 mmol/L Tris, 300 mmol/L NaCl, 10% glycerol and 0.2% Nonidet P-40, pH = 8.0). Cell lysates or the affinity beads were heated at 95 °C for 15 min with loading buffer (25 mmol/L Tris–HCl, 2% SDS, 1 mg/mL bromophenol blue, 50% glycerol and 5% β-mercaptoethanol, pH = 6.8).
2.6. Nuclear and cytoplasmic separation
Cells were collected and gently resuspended with cytoplasmic lysis solution lysis buffer B (100 mmol/L Tris–HCl, 140 mmol/L NaCl, 1.5 mmol/L MgCl2·6H2O, pH = 8.4), and then incubated on ice for 5 min before centrifuging at 1000×g at 4 °C for 3 min. The supernatant was the cytoplasm part while the precipitate were the nucleus components. The prepared detergent (sodium deoxych 3 mg/mL with Tween 40) was added to lysis buffer B according to the ratio of 1:10, and then the mixture was vortexed 2–3 times to wash the nuclei sufficiently. The tube was incubated on ice for 5 min before centrifuging at 1000×g at 4 °C for 3 min. Then the supernatant was discarded, and washing was repeated for 5–10 times. An appropriate amount of 1% NP40 lysate buffer was added to the nucleus and put it in liquid nitrogen for 1 min, then took it out and thawed on ice. Repeated the freezing and thawing process twice, and then vortex vigorously before centrifuging at 16,363×g together with the cytoplasmic components at 4 °C for 30 min.
2.7. GST pull down assay
GST and GST-WSB1 expressed in E. coli BL21 were purified according to previous report26 and the purified protein was used freshly. The whole cell lysate of 293FT and GST or GST-WSB1 protein was incubated in 1% NP-40 buffer (25 mmol/L Tris, 150 mmol/L NaCl, 10% glycerol and 1% Nonidet P-40, pH = 8.0) with protease inhibitor cocktails at 4 °C for 4 h, and the glutathione-agarose beads were subsequently added to incubate for another 2 h. The beads were washed for 4 times with washing buffer (25 mmol/L Tris, 300 mmol/L NaCl, 10% glycerol and 0.2% Nonidet P-40, pH = 8.0) and the proteins were resolved by heating at 95 °C with loading buffer (25 mmol/L Tris–HCl, 2% SDS, 1 mg/mL bromophenol blue, 50% glycerol and 5% β-mercaptoethanol, pH = 6.8) for Western blotting.
2.8. Western blotting
Proteins were separated by 8% or 10% Tris-glycine-SDS PAGE gels for 1 h and transferred on PVDF membrane (0.45 μm, Millipore). Then blocked the membrane with 5% skimmed milk in TBS-T buffer (20 mmol/L Tris–HCl, 150 mmol/L NaCl, 0.1% Tween 20, pH = 7.6) for 1 h. The antibodies were diluted with TBS-T buffer (FLAG, HA and GAPDH are 1:5000, others are 1:1000) and the membrane was put into the antibody buffer at room temperature for 1 h or 4 °C overnight. The membrane was washed with TBS-T for 3 times and the membrane incubated with secondary antibody 1:5000 diluted with 5% skimmed milk in TBS-T for 1 h at room temperature. ECL reagent (NEL103E001EA, PerkinElmer) was applied to catch the chemiluminescence signal using Amersham Imager 600 (GE Healthcare).
2.9. Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation assay was performed using a CHIP assay kit from Millpore. Briefly, 293FT cells transfected with pCDH-c-Myc plasmid were collected and subsequently cross-linked with formaldehyde. Chromatin was transferred on the ice with sonic treatment to obtain about 300 bp fragments. Anti-IgG or anti-c-Myc antibody were incubated with the chromatin fragments at 4 °C overnight, and the immunoprecipitated fragments were detected by PCR amplification using P1, P2 and P3 primers of WSB1 promoter region.
P1 forward: TCAAGACCAGCCTGGCCAATATCGTG;
P1 reverse: GCTCTGTCTCCCAGAGCAGAGTG;
P2 forward: GATGAAGTAAACATGCCCTACAGTG;
P2 reverse: TTCACCCTGTTGGCCAGGCTAGT;
P3 forward: GCAATGTTTAGGGTCCACACGAG;
P3 reverse: ATCCCACTGTCCTGGCCAAGATG.
2.10. RNA extraction, reverse transcriptional PCR and quantitative real-time PCR
RNA was extracted with TRIzol reagent (#9109, Takara) from cultured cells according to the manufacturer's instructions. cDNA was acquired by Transcript one-step gDNA removal and cDNA synthesis supermix (#AT311-03, Tansgene Biotech). The quantitative real-time RT-PCR analysis was performed by iTaqTM Universal SYBR Green Supremix (#172-5124, BIO-RAD). The reaction mixtures containing SYBR Green were composed following the manufacturer's protocol and then CT values were obtained using a qPCR platform (QuantStudio 6 Flex Real-Time PCR System, ThermoFisher Scientific). Real-time PCR data was monitored using QuantStudio 6 Design and Analysis Software Version 2.3. Relatively expression levels were normalized to the internal control β-actin. Primers used in quantitative real-time PCR were as follows:
WSB1 forward: ACTGTGGAGATATAGTCTGGAGTCT;
WSB1 reverse: GTAGCAAGAAGTAGCTGATCTTGTC;
CTNNB1 forward: GTTCAGTTGCTTGTTCGTGC;
CTNNB1 reverse: GTTGTGAACATCCCGAGCTAG;
c-MYC forward: GGACCCGCTTCTCTGAAAG;
c-MYC reverse: GTCGAGGTCATAGTTCCTGTTG;
MYCN forward: GCGACCACAAGGCCCTCAGTACCTC;
MYCN reverse: AATGTGGTGACAGCCTTGGTGTTGG;
MYCL forward: CTGGAGAGAGCTGTGAGCGACCGG;
MYCL reverse: GAGCAGGCCTGGGTCTTGGGTTCG;
β-Actin forward: TCACCCACACTGTGCCCATCTACGA;
β-Actin reverse: CAGCGGAACCGCTCATTGCCAATGG.
2.11. Dual-luciferase reporter assay
Cells were seeded in 96-well plate with density of 80% and transfected with pCDH-c-Myc and pGL4.14-WSB1 promoter-luciferase plasmids using jetPRIME (Polyplus Transfection), the ratio of which was 1:2. After transfection on 293FT cells for 36 h, the supernatant medium was discarded and the cells were lysed using 1 × PLB buffer in the luciferase reporter kit (Promega) for 15 min. An aliquot of 20 μL cell lysis was transferred to white plate and added by 50 μL LAR II per well. Then the Firefly Fluc values were detected in the Microplate reader. Stop & Glo reagent was used to stop the Firefly reaction and start Renilla luciferase reaction at the same time, the value of which was used as an internal control.
pCDH-WSB1 and pGL4.14-c-Myc promoter-luciferase plasmids as well as pCDH-WSB1 and pGL4.14-TCF/LEF1-luciferase plasmids were transfected on H460 and tested with the luciferase reporter kit (Promega) in the same way. Stop & Glo reagent was used to stop the Firefly reaction and start Renilla luciferase reaction at the same time, the value of which was used as an internal control.
2.12. Cell proliferation assay
Bel-7402 stably overexpressing WSB1 or WSB1 with c-Myc and H460 cells knocking down WSB1 were seeded in 6-well plate with density of 10% respectively. After 2 weeks, cells were stained by the sulforhodamine B and photographed by E-Gel Imager (Bio-Rad). The cloning numbers in each group were counted to calculate the inhibition rate.
2.13. Sphere formation assay
The medium of sphere assay was DMEM/F-12 accompanied with penicillin/streptomycin antibiotic, basic fibroblast growth factor (bFGF, 10 ng/mL), human recombinant epidermal growth factor (EGF, 10 ng/mL) and N-2 supplements (1 ×). Bel-7402 cells and HuH-7 cells were collected and washed with DMEM/F-12 medium to remove the serum, and the cells were counted to reseed with the density of 3000 cells per 2 mL medium per well of the ultra-low attachment 6-well plate. The plate was incubated at 37 °C with 5% CO2 for about 5–8 days when the cells formed and reached the size of 100 μm. Photographs were taken by upright microscope (Olympus).
2.14. Immunofluorescence assay
Cells were seeded in confocal plates and fixed with 4% paraformaldehyde for about 15 min. The fixed cells were blocked with 1% BSA for 30 min at room temperature and incubated with β-catenin antibody (dilution at 1: 100) at 4 °C overnight. Anti-rabbit secondary antibody was performed at room temperature for 1 h and the nuclear was stained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) for 1–4 min. Photographs were taken by the confocal laser scanning microscope (Leica).
2.15. Xenograft tumor model
Four to six weeks old female M-NSG mice (Shanghai Model Organisms Center, Shanghai, China) were used for experiments. The mice were randomly divided into 4 groups. Bel-7402 cells after lentivirus infection were subcutaneous injected into the right side of the mice with the density of 5 × 106 and the weights, tumor numbers as well as the tumor size recorded until the mice were sacrificed. Tumor size (mm3) was measured with vernier calipers and calculated with Eq. (1):
| (1) |
in which W is the width and L is the length. The animal experiment was approved by the Institutional Animal Care and Use Committee in Zhejiang University, and was carried out following the institutional guidelines. The IACUC number of the animal experiment was IACUC-s21-027.
2.16. Immunohistochemistry (IHC)
HCC tissue array (BC03119b) was purchased and processed from Alena Biotechnology (Xi'an, China) and photographs were taken by upright microscope (Olympus). And the informed written consent from all participants or next of kin was obtained prior to the research. The nude mice were sacrificed at the end of the xenograft experiment after the tumor dissected. The tissues were fixed with 4% paraformaldehyde. Paraffin sections were prepared and the antigen retrieval was applied with EDTA solution (50 mmol/L Tris, 10 mmol/L EDTA, pH = 9.0) after dewaxing treatment. The tissues were incubated with WSB1 antibody (1:200) and c-Myc antibody (1: 200) at 4 °C overnight. DAB chromogenic kit was used to react with the second antibody. Photographs were taken by upright microscope (Olympus). The evaluation standard of staining intensity is as follows: 0 point for no staining, 1 point for yellow, 2 points for brown and yellow, 3 points for yellowish brown. The score criteria for the proportion of positive cells are as follows: 0 point for less than 10%; 1 point for 10%–40%; 2 points for 40%–70%; 3 points for ≥70%. The sum of the two scores is 2 points for 0–70%; 3 points for ≥70%. Adding the two scores of 0–2 is divided into weak expression; 3 to 6 is divided into strong expression.
2.17. RNA-seq
Bel-7402 cells stably overexpressing pCDH, pCDH-WSB1 and pCDH-ΔSOCS by lentivirus transfection were harvested and washed for 3 times with pre-cold PBS. Total RNA of the cells was extracted by TRIzol reagent and sent to Bohao Biotechnology (Shanghai, China) for RNA expression profiling. RNA-Seq data were uploaded to GEO database with the GEO accession number GSE182635, which has been publicly available on Oct 01, 2021.
2.18. Statistical analysis
Data are presented as mean ± standard deviation (SD) and unpaired two-tailed Student's t-test was applied for statistical analysis. The group differences are considered significantly with P values less than 0.05 (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
3. Results
3.1. WSB1 is a target gene of c-Myc and can in turn regulate c-Myc expression
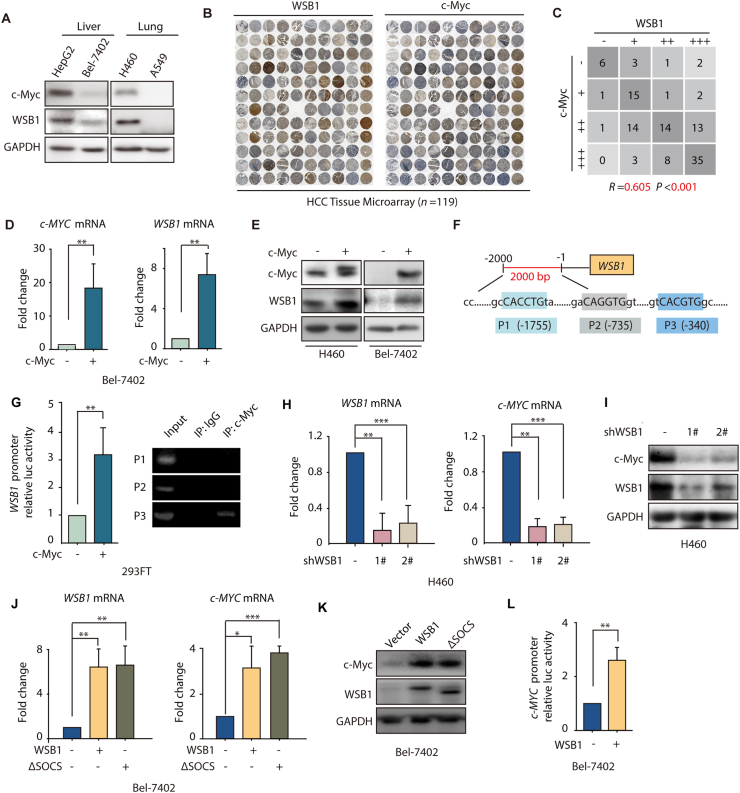
In previous reports, we and other groups proved that WSB1 is the target gene of HIF1-α by its direct binding to −339 bp region of the gene21. Given that the proto-oncogene c-Myc binding E boxes (CACGTG) match the hypoxia response element consensus sequence (A/GCGT), which results in a similar pattern of gene regulation between HIFs and c-Myc, and that the WSB1 promoter also contains putative c-Myc binding E boxes in its −340 bp region, we reasonably speculated that WSB1 might be a target gene of c-Myc. To confirm this prediction, we first examined the protein levels of WSB1 and c-Myc in 10 cancer cell lines and found that there was a highly positive correlation between the expression of WSB1 and c-Myc in these cell lines from various tissues (Fig. 1A, Supporting Information Fig. S1A and S1B). Moreover, we applied IHC analysis to a tissue microarray with 119 hepatocellular carcinoma (HCC) clinical samples, and the results showed that there was a statistically positive correlation between WSB1 and c-Myc protein expression levels in HCC tissues (Fig. 1B and C).
Figure 1.
WSB1 is a target gene of c-Myc and could in turn regulate c-Myc expression. (A) Western blotting of WSB1 and c-Myc expression in different tumor cell lines. (B) IHC staining of WSB1 and c-Myc in hepatocellular carcinoma tissues. (C) Statistical analysis of IHC results of WSB1 and c-Myc in hepatocellular carcinoma tissues (n = 119). (D) Relative mRNA levels of c-MYC and WSB1 in Bel-7402 cells transfected with pCDH-c-Myc plasmid for 48 h. (E) Western blotting of WSB1 and c-Myc in H460 and Bel-7402 cells transfected with pCDH-c-Myc plasmid for 48 h. (F) Schematic representation of the human WSB1 promoter sequence spanning 2.0 kb upstream of the transcriptional start site. P1 (−1755), P2 (–735) and P3 (–340) are predicted potential c-Myc binding sites. (G) 293FT cells were co-transfected with vector or pCDH-c-Myc and pGL4.14-WSB1 promotor for 48 h. Luciferase activity was detected by M5 and normalized by Renilla activity. 293FT cells were transfected with pCDH-c-Myc plasmid for 48 h, anti-IgG and anti-c-Myc antibodies were used in the chromatin ChIP assay. (H) Relative mRNA levels of c-MYC and WSB1 in H460 cells infected with 2 specific lentivirus WSB1 shRNAs for 72 h. (I) Western blotting of WSB1 and c-Myc in H460 cells infected with lentivirus WSB1 shRNA for 72 h. (J) Relative mRNA level of WSB1 and c-MYC in Bel-7402 cells infected with lentivirus pCDH or pCDH-WSB1 or pCDH-ΔSOCS for 72 h. (K) Western blotting of WSB1 and c-Myc in Bel-7402 cells infected with pCDH or pCDH-WSB1 or pCDH-ΔSOCS lentivirus for 72 h. (L) Bel-7402 cells were co-transfected with vector or pCDH-WSB1 and pGL4.14-c-Myc promotor luciferase for 48 h. Luciferase activity was normalized by cell numbers and shown as fold change. Data are represented as mean ± SD, n = 3; statistical significance was determined by Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (versus vector group).
Next, we investigated whether WSB1 was transcriptionally regulated by c-Myc. We overexpressed c-Myc in Bel-7402 cells with low WSB1 expression under basal conditions and subsequently examined both the mRNA and protein levels of WSB1. As shown in Fig. 1D and E, both the mRNA and protein levels were significantly increased upon c-Myc overexpression in Bel-7402 cells. An increase in WSB1 protein expression was also observed in H460 cells with high WSB1 expression under basal conditions (Fig. 1E). Additionally, WSB1 promoter luciferase reporter assay and chromatin ChIP were performed in 293FT cells to confirm whether WSB1 was the direct target gene of c-Myc (Fig. 1F). As shown in Fig. 1G, c-Myc overexpression significantly increased WSB1 promoter-based luciferase activity, and the ChIP-PCR results further suggested that c-Myc was able to transcriptionally activate WSB1 expression by directly binding to the −340 bp region of the WSB1 promoter.
We also tested the effect of other Myc family proteins, N-Myc and L-Myc, on WSB1 expression in Bel-7402 cells. The results showed that N-Myc had no effect on WSB1 expression, while L-Myc could promote the mRNA levels of WSB1 (Fig. S1C–S1F). As there is a lack of evidence to show the role of L-Myc in tumorigenesis27, we mainly focused on c-Myc in our study. Collectively, our data indicated that WSB1 is a target gene of c-Myc.
Encouraged by the fact that c-Myc can transactivate several downstream genes, such as EBP2, eIF4F, and PLK1, to form feedback loops in tumor progression22, 23, 24, we tested whether WSB1 was able to conversely regulate c-Myc. Notably, decreased c-Myc mRNA and protein levels were detected in H460 cells after WSB1 was knocked down by two specific shRNAs (Fig. 1H and I). Consistent with these findings, overexpressing wild-type WSB1 in Bel-7402 cells increased both the mRNA and protein levels of c-Myc (Fig. 1J and K). Moreover, c-MYC promoter luciferase reporter assay also showed that WSB1 could transactivate c-Myc (Fig. 1L). Interestingly, when we overexpressed the C-terminal deletion mutant of WSB1 (named ΔSOCS), whose SOCS-box domain was missing and lacked E3 ligase activity, both the mRNA and protein levels of c-Myc were also increased, and comparable effects were observed compared to those of wild-type WSB1 in Bel-7402 cells (Fig. 1J and K), suggesting that the regulatory effect of WSB1 on c-Myc was independent of its E3 ligase activity. Taken together, all the data mentioned above suggest that WSB1 was a target gene of c-Myc and could in turn regulate c-Myc expression, thus forming a feedforward circuit.
3.2. WSB1 enhances c-Myc expression through the WNT/β-catenin pathway
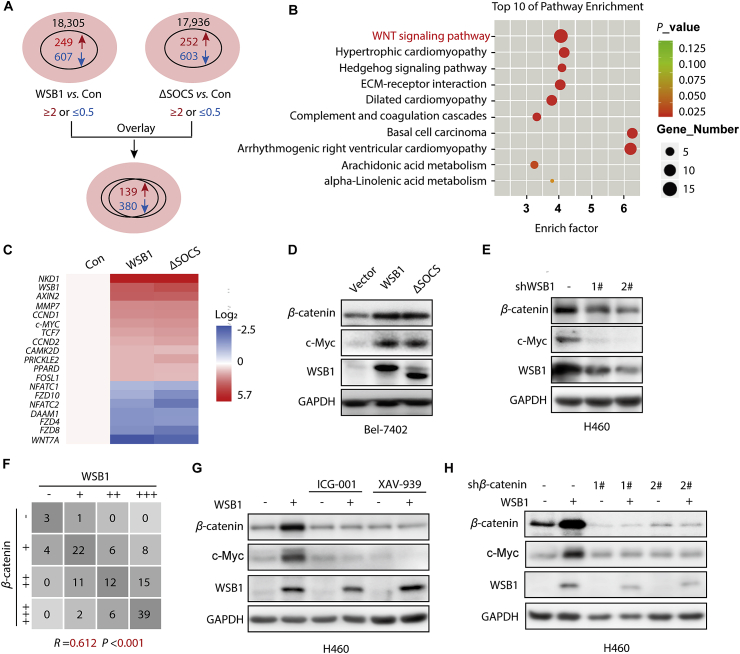
Since WSB1 is not a transcription factor, we aimed to decipher the mechanism of how WSB1 promoted the transcriptional level of c-Myc. Thus, RNA-seq was performed to characterize the potential factor in this regulatory process. We stably overexpressed wild-type WSB1 as well as ΔSOCS WSB1 in Bel-7402 cells, and introduced the vector control by lentivirus infection, then the cells were collected to profile gene expression. RNA-seq results show that 249 genes were upregulated, and 607 genes were downregulated with 2-fold changes out of 18,305 genes in WSB1-overexpressing group compared to the vector control. Moreover, 252 genes were upregulated and 603 genes were downregulated with 2-fold changes out of 17,936 genes in the ΔSOCS-overexpressing group compared to the vector control (Fig. 2A and Supporting Information Table S1). Among the genes with altered expression, 519 genes (139 upregulated genes and 380 downregulated genes) were identified from the two data sets (Fig. 2A).
Figure 2.
WSB1 enhances c-Myc expression through WNT/β-catenin pathway. (A) Bel-7402 cells were transduced with pCDH vector or pCDH-WSB1 or pCDH-ΔSOCS for 72 h. The total RNA was isolated and subsequently performed the RNA-seq. Differential genes (with at least 2-fold change) of WSB1 vs. Control and ΔSOCS vs. Control were overlaid. (B) Differential genes of WSB1 vs. Control and ΔSOCS vs. Control were analyzed by KEGG pathway enrichment analysis and the top 10 pathways were displayed as scatter diagram. (C) Differential genes in WNT/β-catenin signaling pathway are shown as heatmap. (D) Western blotting of β-catenin, WSB1 and c-Myc in Bel-7402 cells infected with lentivirus pCDH-WSB1 for 72 h. (E) Western blotting of β-catenin, WSB1 and c-Myc in H460 cells infected with lentivirus WSB1 shRNAs for 72 h. (F) Statistical analysis of IHC results of WSB1 and β-catenin in hepatocellular carcinoma tissue (n = 119). (G) H460 cells infected with lentivirus vector or lentivirus pCDH WSB1 were treated with β-catenin inhibitors ICG-001 (10 μmol/L) and XAV-939 (10 μmol/L) for 24 h. The protein expression of β-catenin, WSB1 and c-Myc were evaluated by Western blotting. (H) H460 cells overexpressing WSB1 or control cells were infected with lentivirus vector or lentivirus β-catenin shRNA for 72 h. The protein expression of β-catenin, WSB1 and c-Myc were evaluated by Western blotting.
Given that the effect of WSB1 on c-Myc expression was independent of its E3 ligase activity (Fig. 1J and K), we reasoned that both the wild-type and ΔSOCS WSB1 would activate the factor that regulates c-Myc expression. Thus, KEGG pathway analysis of these 519 overlapping genes was performed, and the results indicate that among the top 10 pathways, arrhythmogenic right ventricular cardiomyopathy, basal cell carcinoma and the WNT signaling pathway were the top 3 candidates (Fig. 2B). Considering that the WNT/β-catenin signaling pathway is a classical upstream pathway of c-Myc transcription, we performed further analysis of WNT-regulated gene enrichment. Consistent with the results above, 16 WNT/β-catenin downstream genes were enriched, including c-MYC, that were obviously regulated by WSB1 in our RNA-seq results, indicating the regulation of WSB1 by the WNT/β-catenin signaling pathway (Fig. 2C).
To confirm this finding, we overexpressed WSB1 and ΔSOCS in Bel-7402 cells and found that WSB1 increased the protein levels of both β-catenin and c-Myc (Fig. 2D). When we knocked down WSB1 in H460 cells with 2 specific shRNAs, β-catenin and c-Myc levels were downregulated as expected (Fig. 2E). Moreover, immunohistochemistry analysis of HCC clinical samples also suggested a highly positive correlation between WSB1 and β-catenin expression (Fig. 2F and Supporting Information Fig. S2). To prove that WSB1 regulated c-Myc expression through β-catenin, we treated WSB1-overexpressing cells with two pharmacological β-catenin inhibitors, ICG-001 and XAV-939, and subsequently measured β-catenin and c-Myc expression. As shown in Fig. 2G, the β-catenin inhibitors blocked the upregulation of both β-catenin and c-Myc expression induced by WSB1. Similar results were also observed when β-catenin was knocked down by lentivirus infection (Fig. 2H). Taken together, our data indicate that β-catenin was the key factor participating in WSB1-mediated promotion of c-Myc expression.
3.3. WSB1 promotes β-catenin nuclear translocation and inhibits its ubiquitination
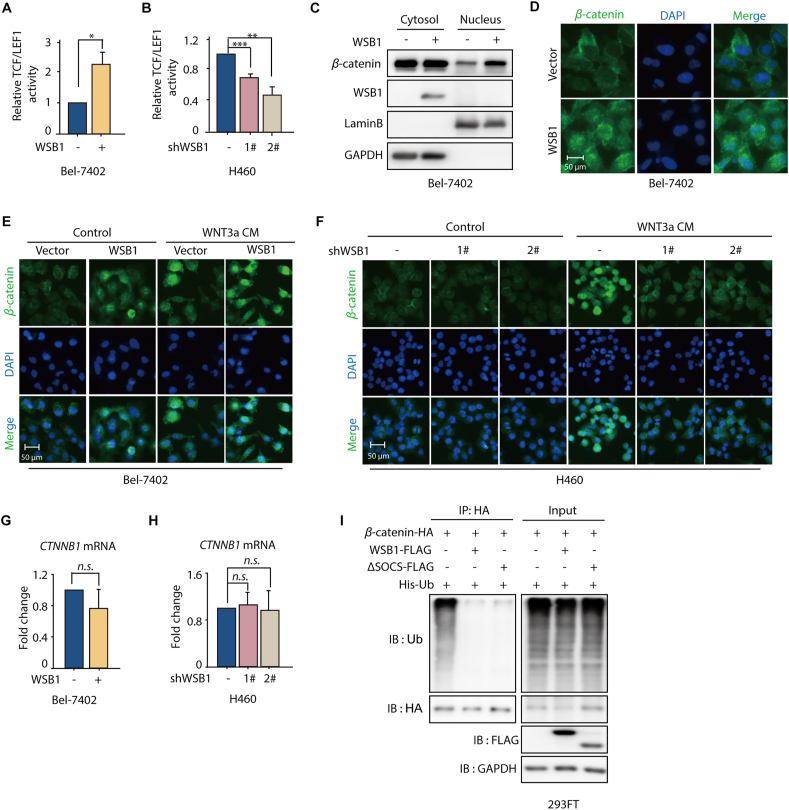
In our previous data, we found that WSB1 increased the expression of β-catenin (Fig. 2D). In line with this, manipulating the WSB1 expression level could also affect the transcriptional activity of β-catenin in the nuclear transcription factor TCF/LEF luciferase reporter assay (Fig. 3A and B). Under normal conditions without WNT induction, β-catenin is captured by the destruction complex, subsequently losing its ability to enter the nucleus and then recognized by its E3 ligase adaptor β-TRCP followed by ubiquitination-mediated proteasomal degradation. Upon WNT stimulation, β-catenin translocates into the nucleus, then interacts with TCF/LEF and transactivates downstream genes28,29. Based on these findings, we explored the effect of WSB1 on the cellular location and ubiquitination of β-catenin.
Figure 3.
WSB1 promotes β-catenin nucleus translocation and inhibits its ubiquitination. (A) Bel-7402 cells were co-transfected with vector or pCDH-WSB1 and pGL4.14-TCF/LEF1-luciferase for 48 h. Luciferase activity was normalized by cell numbers and shown as fold change. ∗P < 0.05 (versus vector group). (B) H460 cells were transduced with lentivirus WSB1 shRNA followed by transfection with TCF/LEF1-luciferase for 48 h. Luciferase activity was normalized by cell numbers and shown as fold change. ∗∗P < 0.01, ∗∗∗P < 0.001 (versus control group). (C) Bel-7402 cells were infected with lentivirus pCDH Vector or lentivirus pCDH-WSB1 for 72 h and collected for cell fraction (nuclei and cytoplasm) separation. Cellular localization of β-catenin was detected by Western blotting in cell fraction. GAPDH was used as loading control for cytoplasm fraction and lamin B was for nucleus fraction. (D) Immunofluorescence staining of β-catenin in Bel-7402 cells infected with lentivirus pCDH vector or lentivirus pCDH-WSB1. (E) Bel-7402 cells infected with lentivirus pCDH vector or lentivirus pCDH-WSB1 were treated with WNT3a condition medium (WNT3a CM) for 16 h, and stained with β-catenin antibody by immunofluorescence. (F) H460 cells infected with lentivirus vector or lentivirus WSB1 shRNAs were treated with WNT3a condition medium (WNT3a CM) for 16 h, and stained with β-catenin antibody by immunofluorescence. (G) Relative mRNA level of WSB1 and CTNNB1 in Bel-7402 cells infected with lentivirus pCDH-WSB1 or 72 h. (H) Relative mRNA level of WSB1 and CTNNB1 in H460 cells infected with lentivirus WSB1 shRNA for 72 h. (I) 293FT cells were infected with lentivirus pCDH vector or lentivirus pCDH-WSB1 or lentivirus pCDH-ΔSOCS for 72 h. Then the cells were plated and transfected with pCDNA-3.0-β-catenin-HA and His-Ub for 48 h and treated with MG132 (10 μmol/L) for 8 h before collecting cells. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-ubiquitination and anti-HA antibody. Data are represented as mean ± SD, n = 3; statistical significance was determined by Student's t-test. n.s., no significant difference (P > 0.05).
First, we determined the effect of WSB1 on the nuclear translocation of β-catenin by performing a nucleocytoplasmic separation assay. As shown in Fig. 3C, WSB1 overexpression increased nuclear β-catenin expression, while cytoplasmic β-catenin was not significantly changed, suggesting a promoting effect of WSB1 on β-catenin nuclear translocation. Similarly, an immunofluorescence assay in Bel-7402 cells also proved that WSB1 overexpression could promote β-catenin entry into the nucleus compared to control group (Fig. 3D). Given that WNT signaling could stabilize β-catenin and promote its nuclear translocation, we next tested the effect of WSB1 on β-catenin nuclear translocation under WNT stimulation conditions. As expected, Bel-7402 cells treated with WNT3a-conditioned medium (WNT3a-CM) markedly induced β-catenin nuclear translocation (Fig. 3E). Interestingly, WSB1 overexpression further promoted the nuclear translocation of β-catenin in coordination with WNT3a (Fig. 3E). Moreover, knocking down WSB1 in H460 cells abolished the promoting effect of WNT3a-conditioned medium (Fig. 3F). And related quantitative data of the nuclear to cytoplasmic fluorescence ratio was shown in Supporting Information Fig. S3A and S3B. Therefore, these data clearly suggest that WSB1 promoted β-catenin nuclear translocation under both basal and WNT stimulation conditions.
To further address the role of WSB1 on β-catenin ubiquitination status, RT-PCR was performed to investigate whether WSB1 influenced the mRNA level of CTNNB1 (the gene encoding β-catenin). As shown in Fig. 3G and H, manipulating the expression of WSB1 in Bel-7402 and H460 cells had no influence on CTNNB1. Together with the fact that WSB1 increased the expression of β-catenin (Fig. 2D), these results further indicate that WSB1 regulated β-catenin at the posttranscriptional level. Subsequently, we investigated whether WSB1 participated in the ubiquitination process of β-catenin. As illustrated in Fig. 3I, wild-type WSB1, as well as ΔSOCS WSB1, inhibited the ubiquitination of β-catenin. Thus, our data further suggest that WSB1 significantly inhibited the ubiquitination of β-catenin, which resulted in β-catenin protein stabilization and nuclear translocation. As the phosphorylation and degradation of β-catenin has been a well-established mechanism, we performed phosphorylation assay in Bel-7402 as well as β-catenin activation cell line HCT116. And results show that WSB1 promoted the nucleus translocation of β-catenin through its phosphorylation and ubiquitination in Bel-7402 cells (Fig. S3C). However, in the cells that β-catenin has already been constitutively activated such as HCT116, WSB1 overexpression did not further affect its stability anymore (Fig. S3D). Nucleocytoplasmic separation assay and immunofluorescence assay were subsequently performed to validate that WSB1 had no effect on the translocation of β-catenin in HCT116 cells (Fig. S3E and S3F).
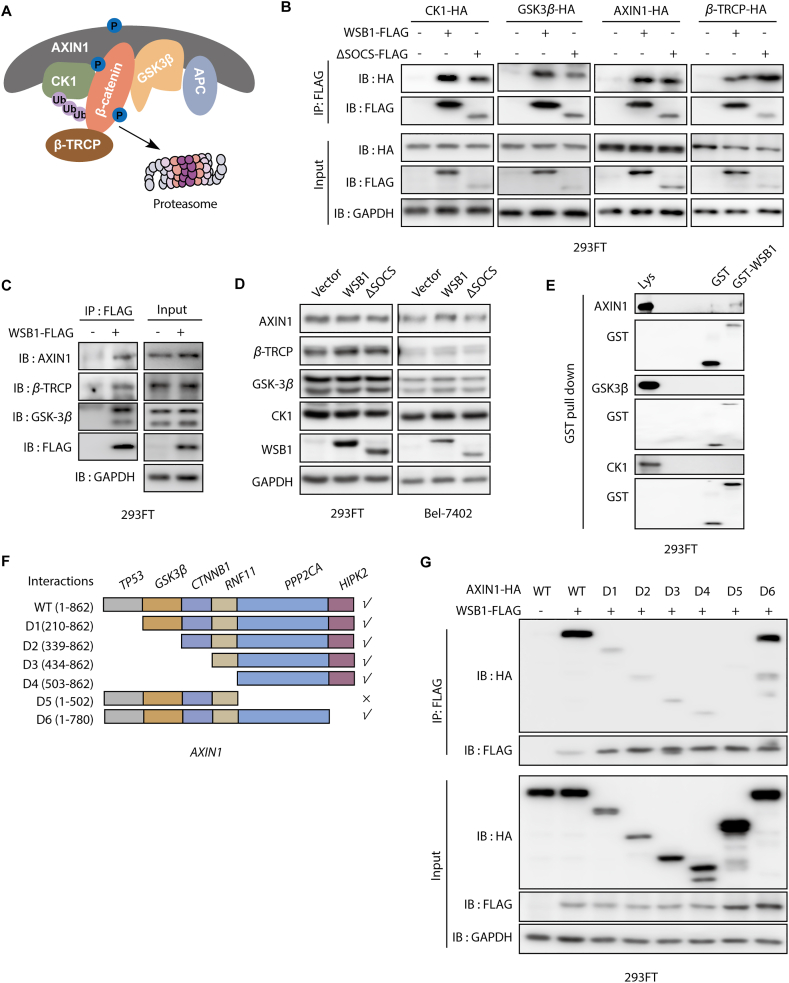
3.4. WSB1 interacts with the destruction complex through the scaffold protein AXIN1
It is known that in the absence of WNT, β-catenin is captured by the destruction complex composed of AXIN1, APC, casein kinase 1 (CK1) and glycogen synthase kinase 3β (GSK3β), subsequently losing the ability to enter the nucleus and being recognized by its E3 ligase adaptor β-TRCP14 (Fig. 4A). Given that (i) WSB1 decreased the ubiquitination of β-catenin (Fig. 3I) and (ii) co-immunoprecipitation experiments indicated that both wild-type and ΔSOCS WSB1 could interact with the four major exogenous and endogenous components (AXIN1, CK1, GSK3β, and β-TRCP) of the β-catenin destruction complex (Fig. 4B and C), we wanted to investigate how WSB1 modulated the β-catenin destruction complex. Thus, we first checked whether WSB1 has an effect on the protein expression of the major components of the destruction complex. As shown in Fig. 4D, overexpressing wild-type WSB1 and ΔSOCS did not inhibit AXIN1, CK1, GSK3β, or β-TRCP expression in either 293FT or Bel-7402 cells, which roughly ruled out the possibility of WSB1-driven inhibition of destruction complex components.
Figure 4.
WSB1 interacts with the destruction complex through the scaffold protein AXIN1. (A) Schematic representation of the β-catenin destruction complex. (B) 293FT cells were transfected with β-catenin destruction complex component plasmids (pCDNA-3.0-CK1-HA or pCDNA-3.0-GSK3β-HA or pCDNA-3.0-AXIN1-HA or pCDNA-3.0-β-TRCP-HA) or co-transfected with pCDH-WSB1-FLAG or pCDH-ΔSOCS-FLAG for 48 h. Then the cells were lysed and for immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-HA antibody. (C) 293FT cells were transfected with pCDH vector or pCDH-WSB1-FLAG for 48 h. Then the cells were lysed and for immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-AXIN1 or anti-β-TRCP or anti-GSK3β or anti-FLAG antibody. (D) Western blotting of AXIN1, β-TRCP, GSK3β and CK1 in 293FT cells and Bel-7402 cells infected with lentivirus pCDH vector or lentivirus pCDH-WSB1 or lentivirus pCDH-ΔSOCS for 72 h. (E) 293FT cells were transfected with pCDNA-3.0-AXIN1-HA or pCDNA-3.0-CK1-HA or pCDNA-3.0-GSK3β-HA for 48 h. The cells were lysed by 4% SDS lysis buffer and then diluted in PBS buffer to make the SDS concentration was 0.1%. GST protein or WSB1-GST fusion protein (5 μg) were incubated with 10 μL GST-seflinose™ resin for 2 h at 4 °C followed by incubating with the lysates overnight at 4 °C and immunoblotting with anti-HA antibody and anti-GST antibody. (F) Schematic representation of AXIN1 domains and a series of AXIN1-HA deletion mutations. (G) 293FT cells were transfected with pCDH-WSB1-FLAG and a series of AXIN1-HA deletion mutations for 48 h. Then the cells were lysed and for immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-HA antibody.
Interestingly, when we incubated the recombinant GST-tagged WSB1 with cell lysate to identify the potential direct interaction proteins, the results indicated that AXIN1 could directly interact with WSB1, while the other complex components could not (Fig. 4E). Given that AXIN1 is the scaffold protein in the destruction complex, acting as the recruiter of the other proteins30,31, we further hypothesized that AXIN1 played a critical role in the interaction between destruction complex components and WSB1. To further determine the regions mediating the interaction between WSB1 and AXIN1, we constructed a series of truncated AXIN1 isoforms and tested their interactions with WSB1 (Fig. 4F). As shown in Fig. 4G, we found that the D5 region truncation of AXIN1 (Δ503–862) abolished the interaction of WSB1 and AXIN1. Collectively, these data imply that WSB1 directly interacted with the destruction complex through the scaffold protein AXIN1, and the 503–780 region of AXIN1 was responsible for this interaction.
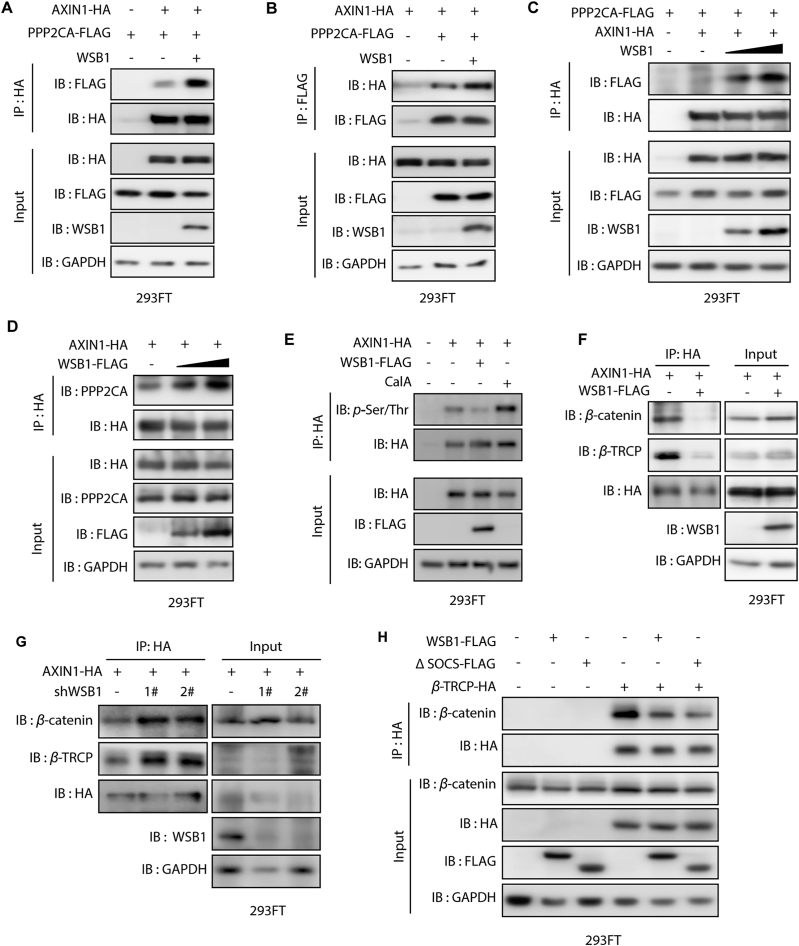
3.5. WSB1 affects destruction complex-PPP2CA assembly and E3 ubiquitin ligase adaptor β-TRCP recruitment
Previous reports have suggested that the 503–582 region of AXIN1 is the binding site of PPP2CA, which is the catalytic subunit of the serine/threonine protein phosphatase 2A (PP2A)32. As we have already proved that WSB1 had a direct interaction with AXIN1, we further investigated whether WSB1 could directly bind with PPP2CA as well to regulate the β-catenin complex. Intracellular exogenous overexpression and GST-pull down assay were conducted and the results of which suggest that WSB1 could form a complex with PPP2CA through AXIN1; however, WSB1 had no direct interaction with PPP2CA (Supporting Information Fig. S4A and S4B). As PP2A can dephosphorylate the β-catenin destruction complex and affect the stability of β-catenin as well as its downstream genes33,34, we explored whether the interaction of AXIN1 and WSB1 regulated the recruitment of PPP2CA and its function. Co-immunoprecipitation assays were carried out in 293FT cells overexpressing WSB1, AXIN1 and PPP2CA, and the results indicate that WSB1 overexpression promoted the interaction between AXIN1 and PPP2CA (Fig. 5A and B). Importantly, the promoting effect of WSB1 on AXIN1–PPP2CA complex formation was dose-dependent (Fig. 5C and D).
Figure 5.
WSB1 affects the destruction complex-PPP2CA assembly and the E3 ubiquitin ligase adaptor β-TRCP recruitment. (A) 293FT cells were transfected with pCDNA3.0-PPP2CA-FLAG, or co-transfected with pCDNA3.0-PPP2CA-FLAG and pCDNA3.0-AXIN1-HA or co-transfected with pCDNA3.0-PPP2CA-FLAG, pCDNA3.0-AXIN1-HA and pCDH-WSB1-FLAG for 48 h. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-FLAG antibody. (B) 293FT cells were transfected with pCDNA3.0-AXIN1-HA, or co-transfected with pCDNA3.0-AXIN1-HA and pCDNA3.0-PPP2CA-FLAG or co-transfected with pCDNA3.0-PPP2CA-FLAG, pCDNA3.0-AXIN1-HA and pCDH-WSB1 for 48 h. Then the cells were lysed and for immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-HA antibody. (C) 293FT cells were transfected with pCDNA3.0-PPP2CA-FLAG, or co-transfected with pCDNA3.0-PPP2CA-FLAG and pCDNA3.0-AXIN1-HA, or co-transfected with pCDNA3.0-PPP2CA-FLAG, pCDNA3.0-AXIN1-HA and different doses of pCDH-WSB1 (1 or 2 μg) for 48 h. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-FLAG antibody. (D) 293FT cells were transfected with pCDNA3.0-AXIN1-HA, or co-transfected with pCDNA3.0-AXIN1-HA and different doses of pCDH-WSB1-FLAG (1 or 2 μg) for 48 h. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-PPP2CA antibody. (E) 293FT cells were transfected with pCDNA3.0-AXIN1-HA, or co-transfected with pCDNA3.0-AXIN1-HA and pCDH-WSB1-FLAG for 48 h and treated with phosphatase PP1/PP2A inhibitor calyculin A (2 μmol/L) for 1 h before collecting cells. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-p-Ser/Thr antibody. (F) 293FT cells were transfected with pCDNA3.0-AXIN1-HA, or co-transfected with pCDNA3.0-AXIN1-HA and pCDH-WSB1-FLAG for 48 h. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-β-catenin, anti-β-TRCP, anti-WSB1 and anti HA antibody. (G) 293FT cells were infected with lentivirus shRNA vector or lentivirus WSB1 shRNA for 72 h. Then the cells were plated and transfected with pCDNA-3.0-AXIN1-HA for 48 h. Finally, the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-β-catenin, anti-β-TRCP, anti-WSB1 and anti HA antibody. (H) 293FT cells were infected with lentivirus pCDH vector or lentivirus pCDH-WSB1 or lentivirus pCDH-ΔSOCS for 72 h. Then the cells were plated and transfected with pCDHA3.0 vector or pCDNA-3.0-β-TRCP-HA for 48 h. Then the cells were lysed and for immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-β-catenin and anti-HA antibody.
Although the interaction between PPP2CA and AXIN1 is required for the proper function of the β-catenin destruction complex, recent evidence further suggests that the dynamic interaction of AXIN1 and PPP2CA, rather than a steady-state AXIN1–PPP2CA complex formation, is pivotal in maintaining the β-catenin destruction complex function28. Thus, we wondered whether the interaction between AXIN1 and PPP2CA promoted by WSB1 would regulate the assembly and function of the β-catenin destruction complex. The results show that the promoting effect of WSB1 overexpression on AXIN1 and PPP2CA interaction significantly reduced the serine/threonine phosphorylation level of AXIN1 (Fig. 5E), which might inhibit the systematical recruitment and formation of the destruction complex. Next, to validate our hypothesis, we overexpressed WSB1 in 293FT cells and found that WSB1 significantly inhibited the interaction between β-catenin and AXIN1 as well as β-TRCP and AXIN1 (Fig. 5F), while knocking down WSB1 increased the interactions (Fig. 5G), which was consistent with the results of WSB1 overexpression. To further clarify the function of WSB1 in β-catenin stability regulation, we investigated whether WSB1 affected the interaction between β-catenin and its E3 ligase adaptor β-TRCP. As shown in Fig. 5H, both wild-type and ΔSOCS WSB1 inhibited the interaction between β-catenin and β-TRCP. Taken together, our data show that WSB1 promoted the interaction of AXIN1 and the Ser/Thr phosphatase subunit PPP2CA, which significantly reduced the serine/threonine phosphorylation level of AXIN1, and subsequently inhibited β-catenin destruction complex assembly and E3 ubiquitin ligase β-TRCP recruitment and therefore promoted the stability of β-catenin.
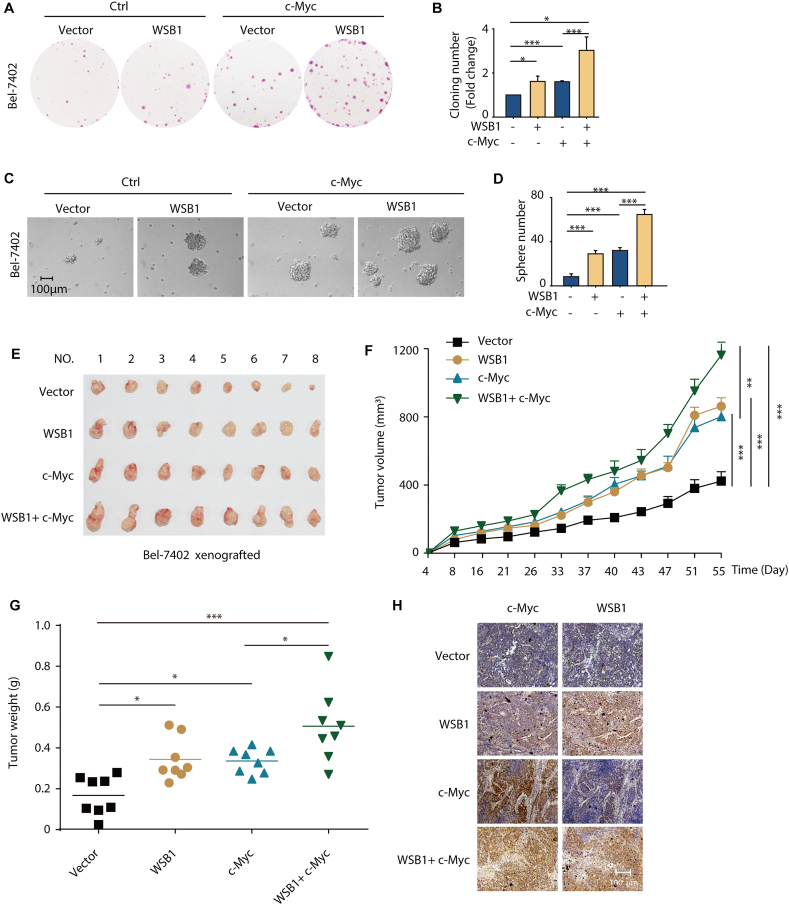
3.6. The WSB1/c-Myc feedforward circuit participates in the development of cancer
Next, we explored the physiological relevance of the WSB1/c-Myc feedforward circuit. For the reason that c-Myc and the WNT/β-catenin pathway both play important roles in tumor initiation and tumor stemness regulation, we conducted colony formation and sphere formation assays to study whether the WSB1/c-Myc feedforward circuit regulated cell proliferation and stemness. The results indicate that overexpressing WSB1 by lentivirus infection of Bel-7402 cells increased the colony numbers, and when co-overexpressed with c-Myc, WSB1 further promoted colony formation compared to WSB1 overexpression alone (Fig. 6A and B). To further validate this result, we knocked down WSB1 in H460 cells under both basal conditions and c-Myc overexpression conditions. As expected, the overexpression of c-Myc significantly increased colony formation in H460 cells (Supporting Information Fig. S5A and S5B). Of note, WSB1 knockdown significantly inhibited the increase in colony formation induced by c-Myc (Fig. S5A and S5B). These results suggest that the WSB1/c-Myc feedforward circuit promoted cancer cell proliferation.
Figure 6.
WSB1/c-Myc feedforward circuit participated in the development of cancer. (A) Cloning formation assay was performed in Bel-7402 cells transduced with lentivirus pCDH or pCDH-WSB1 or pCDH-c-Myc or both. Cells were stained with the sulforhodamine B and photographed by E-Gel Imager. (B) Cloning numbers were counted and shown as fold change. ∗P < 0.05, ∗∗∗P < 0.001 (versus vector group). (C) Sphere formation assay was performed in Bel-7402 cells transduced with lentivirus pCDH or pCDH-WSB1 or pCDH-c-Myc or both. Representative photographs were taken by microscope. (D) Statistical analysis of cell colonies. ∗∗∗P < 0.001 (versus vector group). (E) Photographs of tumors from different groups. In the end of the experiment, the M-NSG mice were sacrificed and tumors were dissected, and the tumor formation rate was 8/8 in each group. (F) Analysis of tumor volumes in different groups. ∗∗P < 0.01, ∗∗∗P < 0.001 (versus vector group or c-Myc singly overexpressed group). (G) Analysis of tumor weight in different groups. ∗P < 0.05; ∗∗∗P < 0.001 (versus vector group or c-Myc singly overexpressed group). (H) IHC staining of WSB1 and c-Myc in the tumor tissue. Data are represented as mean ± SD, n = 3 (B, D) or means ± SEM, n = 8 (F, G); statistical significance was determined by Student's t-test.
To examine the effect of WSB1 on cell stemness, sphere formation assay was performed in Bel-7402 and HuH7 cells. As shown in Fig. S5C–S5F, both wild-type and ΔSOCS WSB1 overexpression significantly increased the cell sphere numbers, suggesting that WSB1 played a critical role in cell stemness. And when c-Myc was also overexpressed, WSB1 further promoted sphere formation (Fig. 6C and D).
Encouraged by the in-cell results, we wondered whether this effect could be recapitulated in vivo. Thus, we performed the Bel-7402 cells xenograft tumor assay in M-NSG mice, 32 mice were divided randomly to 4 groups and different lentivirus-infected cells (Vector, WSB1, c-Myc and WSB1+c-Myc) were subcutaneously injected into the right side of the mice. The tumor growth and the mice weight were monitored since Day 4 after transplantation. Upon the end of the experiment, the mice were sacrificed and tumors were dissected. As shown in Fig. 6E and F, in line with the in-cell results, overexpression of WSB1 or c-Myc could significantly promoted the tumor growth, and when co-overexpressed with WSB1, c-Myc further promoted the tumor growth compared with c-Myc overexpression alone, which strongly support our in-cell results that the WSB1/c-Myc feedforward circuit promoted cancer progression. The statistics results of tumor weight also convinced this conclusion (Fig. 6G). The immunohistochemistry assay was performed subsequently to validation the expression of Myc and WSB1 levels in tumors (Fig. 6H). All of these data prove that the WSB1/c-Myc feedforward circuit existed in vivo and participated in the development of cancer.
4. Discussion
c-Myc is documented to be related not only with tumor initiation but also the malignant progression, such as tumor resistance and recurrence7. Apart from its nuclear location, the amplification or transcriptional dysregulation of c-Myc is accompanied by an anabolic transcriptional response that promotes the metabolic adaptation of cancer cells, thus limiting the development of cancer therapy strategies directly targeting c-Myc35,36. Studies have implied that the regulation of c-Myc is mainly characterized by transcription and protein stability. The half-life of c-Myc is approximately 20–30 min, indicating that the protein level of c-Myc changes dynamically in cellular activity, which means that it is difficult to target37,38. In contrast, as the transcription of c-Myc is regulated by multiple signaling pathways, including the classic WNT/β-catenin pathway14,39, the upstream regulation of c-Myc through β-catenin would provide a critical theoretical basis for c-Myc targeting strategies. In this regard, our results suggest that the WSB1/c-Myc feedforward circuit plays an important role in tumorigenesis, and WSB1 inhibition can effectively block the tumor-promoting effect of c-Myc.
Composed of seven WD40 repeat domains and a SOCS box responsible for elongin B/C binding and substate recognition, WSB1 has been reported to regulate the degradation of multiple substrates, all of which are based on its E3 enzyme activity. However, in our study, we found that WSB1 could enhance the expression of c-Myc even when lacking the SOCS box, which means that the promoting effect of WSB1 is independent of its E3 ligase activity. As an adaptor of the E3 ligase, WSB1 has other biological functions in addition to mediating substrate recognition of the ubiquitination process. From this perspective, our findings propose a new role for WSB1 in cancer development for the first time, which enriches the theoretical knowledge of WSB1 and the ubiquitin proteasome system. Although the WSB1 inhibitor could be ideally designed based on the interaction between WSB1 and its substrates, our results further suggest that there are other functional domains regulating tumor progression. Thus, it might be worth further developing a proteolysis targeting chimaera (PROTAC) molecule to degrade WSB1 protein, rather than a protein–protein binding inhibitor, to fully inhibit its tumor promoting effect.
Many studies have reported that c-Myc can form feedback loops in tumor regulation. For example, c-Myc can directly transactivate EBP2, while EBP2 affects FBW7, the E3 ligase of c-Myc, to inhibit c-Myc degradation, which ultimately promotes cancer progression40,41. Moreover, c-Myc can form a feedback loop with its downstream gene eIF4F to promote cell proliferation induced by c-Myc23. In our study, we found that WSB1 forms a feedforward circuit with c-Myc through the WNT/β-catenin pathway to significantly promote cancer development, and at the same time, c-Myc can transactivate WSB1 to form a circuit loop. Thus, the WSB1/c-Myc feedforward circuit, which can also be considered as a β-catenin/c-Myc loop, provides new insight into the relationship between β-catenin and c-Myc. Moreover, β-catenin and c-Myc have been reported to be associated with cancer stemness in several studies42,43, which endows the β-catenin/c-Myc loop with stemness-promoting ability compared with other reported feedback loops.
However, there are some limitations to our mechanistic study. As the precise regulatory role and the direct substrate protein of PPP2CA in this feedback loop are still unclear and remain to be further explored in future studies. Though WSB1 interacts with AXIN1 through the same domain as PPP2CA, our data interestingly suggest that WSB1 overexpression doesn't compete with PPP2CA to bind AXIN1. Instead, WSB1 enhances the interaction between AXIN1 and PPP2CA. Though the mechanism is still unknown, the interaction of WSB1 might allosterically regulate the AXIN1–PPP2CA interaction. In addition, numerous studies have pointed out that the phosphorylation and dephosphorylation of the destruction complex, especially of AXIN1, precisely control β-catenin stability and WNT pathway activation44. The phosphatases PP2A and PP1 are both reported to be correlated with the complex; of note, PP2A can interact with both AXIN1 and APC, while PP1 only binds with AXIN132,44,45. These facts indicate that it is necessary to explore whether the promotional effects of WSB1 on PP2A are specific to the WSB1/c-Myc loop; in other words, determine whether other phosphatases also participate in the regulation of WSB1. Moreover, as PP1 is reported to dephosphorylate AXIN1 and induce a conformational switch44, it is interesting to determine whether different structural characteristics and assembly status of AXIN1 play a critical role in WSB1 regulation. More importantly, a recent study proved that Myc regulated the immune response through PD-L16, which greatly aroused our interest to explore whether WSB1 could affect PD-L1 and immune therapy through the WSB1/c-Myc feedforward loop.
5. Conclusions
In summary, our results prove that WSB1 is a direct target gene of c-Myc and can form a feedforward circuit with c-Myc through β-catenin, leading to cancer initiation and development, which enriches the c-Myc and WSB1 crosstalk network theory. Considered that there is still no effective way to directly target c-Myc, our research pointed out that WSB1 inhibitors might be an alternative choice to achieve indirectly regulation of c-Myc. Thus, WSB1 inhibitor development could be considered as a promising therapeutic strategy in c-Myc driven tumors. From the other aspect, this finding not only provides new targeting strategies for c-Myc in cancer therapy, but also reminds us that WSB1 could be employed as a marker of poor prognostic of cancer as well. What's more, the discovery of WSB1/β-catenin/c-Myc axis clarifies the critical role of WSB1 in the WNT/β-catenin signaling pathway, which also provides new possibilities for indirectly intervening in the β-catenin pathway in cancer treatment.
Acknowledgments
This work was supported by grants from Zhejiang Provincial Natural Science Foundation (No. Y18H310001 to Ji Cao, China), the National Natural Science Foundation of China (No. 81872885 to Ji Cao; No.81625024 to Bo Yang), and the Talent Project of Zhejiang Association for Science and Technology (No.2018YCGC002 to Ji Cao, China).
Author contributions
Ji Cao, Yanling Gong, Xiaomeng Gao, and Bo Yang designed the research; Ji Cao, Xiaomeng Gao, and Bo Yang wrote the manuscript; Yanling Gong, Xiaomeng Gao, Jieqiong You, Meng Yuan, Liang Fang, and Haiying Zhu performed the biochemical and cellular studies; Yanling Gong, Xiaomeng Gao, and Jieqiong You conducted the animal studies; Ji Cao, Yanling Gong, Xiaomeng Gao analyzed the results; Ji Cao, Hong Zhu, Meidan Ying, Qiaojun He and Bo Yang directed the study.
Conflicts of interest
The authors declare no competing financial interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.10.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Maston G.A., Evans S.K., Green M.R. Transcriptional regulatory elements in the human genome. Annu Rev Genom Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Nunez V., Rodríguez R.E. In: The neuroscience of cocaine. Preedy V.R., editor. Academic Press; San Diego: 2017. Chapter 12-cocaine and transcription factors; pp. 107–124. [Google Scholar]
- 3.Darnell J.E. Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 4.Bradner J.E., Hnisz D., Young R.A. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang C.V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey S.C., Tong L., Li Y., Do R., Walz S., Fitzgerald K.N., et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz W., Leon J., Eilers M. Contributions of myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 8.Bidwell G.L., 3rd, Davis A.N., Raucher D. Targeting a c-Myc inhibitory polypeptide to specific intracellular compartments using cell penetrating peptides. J Control Release. 2009;135:2–10. doi: 10.1016/j.jconrel.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean M., Kent R.B., Sonenshein G.E. Transcriptional activation of immunoglobulin alpha heavy-chain genes by translocation of the c-myc oncogene. Nature. 1983;305:443–446. doi: 10.1038/305443a0. [DOI] [PubMed] [Google Scholar]
- 11.Levens D. You don't muck with MYC. Genes Cancer. 2010;1:547–554. doi: 10.1177/1947601910377492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacDonald B.T., Tamai K., He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S., Li Y., Wu Y., Shi K., Bing L., Hao J. Wnt/β-catenin signaling pathway upregulates c-Myc expression to promote cell proliferation of P19 teratocarcinoma cells. Anat Rec. 2012;295:2104–2113. doi: 10.1002/ar.22592. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes D.R., Chinnaiyan A.M. Integrative analysis of the cancer transcriptome. Nat Genet. 2005;37 Suppl:S31–S37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.J., Lee S.B., Yi S.Y., Han S.A., Kim S.H., Lee J.M., et al. WSB1 overcomes oncogene-induced senescence by targeting ATM for degradation. Cell Res. 2017;27:274–293. doi: 10.1038/cr.2016.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benita Y., Kikuchi H., Smith A.D., Zhang M.Q., Chung D.C., Xavier R.J. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haque M., Kendal J.K., MacIsaac R.M., Demetrick D.J. WSB1: from homeostasis to hypoxia. J Biomed Sci. 2016;23:61. doi: 10.1186/s12929-016-0270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poujade F.A., Mannion A., Brittain N., Theodosi A., Beeby E., Leszczynska K.B., et al. WSB-1 regulates the metastatic potential of hormone receptor negative breast cancer. Br J Cancer. 2018;118:1229–1237. doi: 10.1038/s41416-018-0056-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi D.W., Seo Y.M., Kim E.A., Sung K.S., Ahn J.W., Park S.J., et al. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem. 2008;283:4682–4689. doi: 10.1074/jbc.M708873200. [DOI] [PubMed] [Google Scholar]
- 21.Cao J., Wang Y., Dong R., Lin G., Zhang N., Wang J., et al. Hypoxia-induced WSB1 promotes the metastatic potential of osteosarcoma cells. Cancer Res. 2015;75:4839–4851. doi: 10.1158/0008-5472.CAN-15-0711. [DOI] [PubMed] [Google Scholar]
- 22.Liao P., Wang W., Shen M., Pan W., Zhang K., Wang R., et al. A positive feedback loop between EBP2 and c-Myc regulates rDNA transcription, cell proliferation, and tumorigenesis. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2013.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin C.J., Cencic R., Mills J.R., Robert F., Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res. 2008;68:5326–5334. doi: 10.1158/0008-5472.CAN-07-5876. [DOI] [PubMed] [Google Scholar]
- 24.Palomero T., Lim W.K., Odom D.T., Sulis M.L., Real P.J., Margolin A., et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X., Rubin J.S., Kimmel A.R. Rapid, Wnt-induced changes in GSK3β associations that regulate β-catenin stabilization are mediated by Gα proteins. Curr Biol. 2005;15:1989–1997. doi: 10.1016/j.cub.2005.10.050. [DOI] [PubMed] [Google Scholar]
- 26.Dentice M., Bandyopadhyay A., Gereben B., Callebaut I., Christoffolete M.A., Kim B.W., et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7:698–705. doi: 10.1038/ncb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nau M.M., Brooks B.J., Battey J., Sausville E., Gazdar A.F., Kirsch I.R., et al. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 28.van Kappel E.C., Maurice M.M. Molecular regulation and pharmacological targeting of the β-catenin destruction complex. Br J Pharmacol. 2017;174:4575–4588. doi: 10.1111/bph.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harb J., Lin P.J., Hao J. Recent development of Wnt signaling pathway inhibitors for cancer therapeutics. Curr Oncol Rep. 2019;21:12. doi: 10.1007/s11912-019-0763-9. [DOI] [PubMed] [Google Scholar]
- 30.Abitbol S., Dahmani R., Coulouarn C., Ragazzon B., Mlecnik B., Senni N., et al. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of β-catenin activation. J Hepatol. 2018;68:1203–1213. doi: 10.1016/j.jhep.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Zucman-Rossi J., Benhamouche S., Godard C., Boyault S., Grimber G., Balabaud C., et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 32.Hsu W., Zeng L., Costantini F. Identification of a domain of Axin that binds to the serine/threonine protein phosphatase 2A and a self-binding domain. J Biol Chem. 1999;274:3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J.J., Williams C.S. Protein phosphatase 2A in the regulation of Wnt signaling, stem cells, and cancer. Genes. 2018;9:121. doi: 10.3390/genes9030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratcliffe M.J., Itoh K., Sokol S.Y. A positive role for the PP2A catalytic subunit in Wnt signal transduction. J Biol Chem. 2000;275:35680–35683. doi: 10.1074/jbc.C000639200. [DOI] [PubMed] [Google Scholar]
- 35.Posternak V., Cole M.D. Strategically targeting MYC in cancer. F1000Res. 2016;5 doi: 10.12688/f1000research.7879.1. F1000 Faculty Rev-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topper M.J., Vaz M., Chiappinelli K.B., DeStefano Shields C.E., Niknafs N., Yen R.C., et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell. 2017;171:1284–1300.e21. doi: 10.1016/j.cell.2017.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin C.P., Liu C.R., Lee C.N., Chan T.S., Liu H.E. Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J Hepatol. 2010;2:16–20. doi: 10.4254/wjh.v2.i1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas L.R., Tansey W.P. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 39.Sears R.C. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 40.Weber A., Kristiansen I., Johannsen M., Oelrich B., Scholmann K., Gunia S., et al. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008;8:369. doi: 10.1186/1471-2407-8-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malz M., Bovet M., Samarin J., Rabenhorst U., Sticht C., Bissinger M., et al. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60:1241–1250. doi: 10.1002/hep.27218. [DOI] [PubMed] [Google Scholar]
- 42.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cliff T.S., Wu T., Boward B.R., Yin A., Yin H., Glushka J.N., et al. MYC controls human pluripotent stem cell fate decisions through regulation of metabolic flux. Cell Stem Cell. 2017;21:502–516.e9. doi: 10.1016/j.stem.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.E., Huang H., Zhao M., Zhang X., Zhang A., Semonov M.V., et al. Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science. 2013;340:867–870. doi: 10.1126/science.1232389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto H., Kishida S., Kishida M., Ikeda S., Takada S., Kikuchi A. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3beta regulates its stability. J Biol Chem. 1999;274:10681–10684. doi: 10.1074/jbc.274.16.10681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.