Abstract
Infection or vaccination leads to the development of germinal centers (GC) where B cells evolve high affinity antigen receptors, eventually producing antibody-forming plasma cells or memory B cells. Here we follow the migratory pathways of B cells emerging from germinal centers (BEM) and find that many BEM cells migrate into the lymph node subcapsular sinus (SCS) guided by sphingosine-1-phosphate (S1P). From the SCS, BEM cells may exit the lymph node to enter distant tissues, while some BEM cells interact with and take up antigen from SCS macrophages, followed by CCL21-guided return towards the GC. Disruption of local CCL21 gradients inhibits the recycling of BEM cells and results in less efficient adaption to antigenic variation. Our findings thus suggest that the recycling of antigen variant-specific BEM cells and transport of antigen back to GC may support affinity maturation to antigenic drift.
Subject terms: Germinal centres, Immunological memory, Lymph node, Imaging the immune system
Activated B cell enter germinal centers (GC) to become plasma cells and memory B cells. Here the authors show that some memory B cells recycle to GC via CCL-21 mediated chemotaxis to deliver antigens from the lymph node subcapsular sinus (SCS) to potentially contribute to affinity maturation and antigenic drift.
Introduction
The hallmark of adaptive immunity is memory, which is mediated by the expansion and long-term survival of antigen-specific lymphocytes, affinity maturation of B lymphocytes, and the long-term production of neutralizing antibody. Affinity maturation of B cells occurs via molecular evolution in germinal centers (GCs)1. This involves cycles of B cell proliferation and the mutation of B cell receptor genes, followed by natural selection of B cells expressing the highest affinity B cell receptors. The outputs of the GC reaction are high-affinity antibody-producing plasma cells and memory B cells, both providing long-term immunity2–5.
Plasma cells can be very long-lived6, as are memory B cells7,8. Interestingly, the affinity-dependent selection of memory B cells in the GC is less stringent than that seen for plasma cells, resulting in a highly variable pool of antigen-specific cells9–11. As long-term immunity can be provided by long-lived plasma cells, the advantage of a low-quality B cell output from the GC is not immediately obvious12. However, their high variability may provide a pool of cells with the potential to protect against pathogen variants. Memory B cells can sense specific antigen, rapidly enter secondary responses, immediately present antigen to memory T cells13,14, and generate new plasma cells within days15–17.
Lymph nodes are important sites for the initiation of the adaptive immune response. They represent a platform where immunological information is sequestered and exchanged. Resident cells, including B cells, occupy distinct anatomical niches, and their movement between different areas of the lymph node is required for the progression of a GC reaction18. One important structure in this regard is the subcapsular sinus (SCS), the primary area into which tissue-derived lymph fluid drains, bringing antigens and pathogens. The SCS houses a subset of CD169+ macrophages that are specialized for antigen acquisition and pathogen defense19 and shuttle antigen to naïve and memory B cells16,20–22.
Here we follow migration of GC-derived memory-like B cells and show that they enter the SCS. From here they may move on to other lymphoid tissues or interact with SCS macrophages and recycle back to the GC. We show that the migration to the SCS and back is organized S1P and CCL21, respectively, and present data supporting the hypothesis that memory B cell recycling may be a mechanism to survey for and adapt to antigen variants.
Results
Appearance of memory-like B cells entering the SCS guided by S1PR
In order to track the migration of antigen-specific B cells and plasma cells as they emerged from primary GCs in draining lymph nodes (drLN) following immunization, we adoptively transferred 4-hydroxynitrohpenyl (NP)-specific B cells from B1-8i mice23, which express eGFP under the control of the Prdm1 promoter24 (labeling plasmablasts and plasma cells with eGFP) and Cdt1-mKO2 hybrid protein25 (labeling cells in G0/G1 phase of cell cycle with mKO2), and immunized with NP coupled to the carrier protein chicken gamma globulin (CGG). As we previously described4, plasmablasts emerged from the interface between the GC dark zone and T cell areas (Fig. 1a). Large numbers of antigen-specific B cells were located in the outer follicle surrounding GCs, typically close to the LN SCS (Fig. 1a). Cdt1-mKO2 labeling of these B cells suggests that they were recently activated B cells that emerged from adjacent GCs (BEM). This is reminiscent of historical observations describing the accumulation of marginal zone-like memory B cells under the SCS26,27, and recent descriptions of switched memory B cells in follicles around GCs and under the SCS16,28.
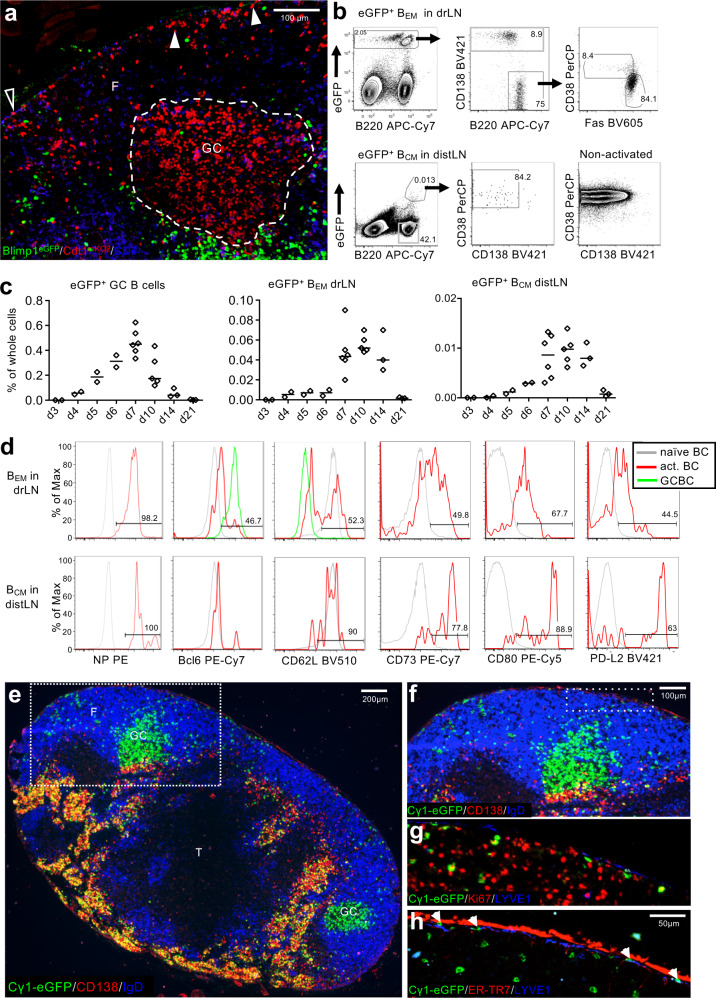
Fig. 1. Appearance of antigen-activated memory-like B cells in drLN and distLN.
a Location of B1-8i/k−/−/Blimp1GFP/Cdt1mKO2 B cells in drLN 6 d after immunization. Antigen-specific B cells in the G0/G1 phase of cell cycle (red) inside the GC (dashed line) and in the follicle (F) close to the SCS (arrow heads). Interfollicular region (open triangle). Blimp-1+ PCs are eGFP (green). Hoechst33342-labeled naïve T cells (blue). Scale bar: 100 μm. Representative image of 3 lymph nodes. b Gating of eGFP+ BEM in drLN and BCM and distLN 8 d after immunization of recipients of NP-specific Cγ1Cre QM mTmG B cells. c Kinetic of eGFP+ B cell appearance in drLN and distLN. Data merged from two independent experiments (n = 2–3). Each datapoint represents one animal. d Memory B cell typical markers on BEM and BCM in drLN and distLN. e drLN of recipient of QM Cγ1Cre mTmG cells 8 d after immunization. T zone (T). f Enlargement of box in e showing the eGFP+ BEM close to the subcapsular sinus, g Ki-67 expression in BEM, h same area showing BEM location below the LYVE1+ ER-TR7-ve SCS floor endothelium and inside the SCS (arrowheads). Image is a representative of three lymph nodes.
In order to identify GC-derived BEM in drLNs and their migration to non-reactive distant lymphoid tissues (distLNs), we used the well-established Cγ1-Cre reporter strain, which induces constitutive expression of GFP in B cells after T cell-dependent activation, which includes GC-lineage B cells29. We crossed these with mice expressing B cell receptors specific for the hapten 4-hydroxynitrophenyl (NP)30,31 and a Cre-inducible eGFP reporter (QM Cγ1Cre mTmG mice)29,32. We immunized wild type (WT) host mice that had received a small number of antigen-specific B cells from QM Cγ1Cre mTmG. We observed that GC B cells (eGFP+NP+CD38lowFas+) were detectable from 4 d after immunization with maximum numbers seen at 6–10 d (Fig. 1b–d). Within a day of GCs reaching maximum size, there was the emergence of a population of B cells that were eGFP+, NP-binding, CD38high, Fasint, CD138-, Bcl6low (Fig. 1b) in the drLN (Fig. 1c, d). CD38 is expressed when GC B cells acquire a memory-like phenotype5. These cells also started to express other markers associated with memory B cells such as CD73, CD80, and PD-L23. At the same time, antigen-activated B cells were observed in distant lymphoid tissues (Fig. 1b–d). These eGFP+ circulating memory B cells (BCM) were confirmed to be antigen-specific, expressed CD62L at similar levels to naïve B cells, and high levels of CD73, CD80, and PDL2 (Fig. 1d). This suggests that the presence of BEM close to the SCS at the peak of the GC response is related to emigration of antigen-activated B cells from the drLN through the SCS, generating systemic cellular B cell immunity.
Further immunohistological examination of drLNs around the peak of BEM migration (Fig. 1e–h) showed that the eGFP+ BEM in B cell follicles surrounding the GC were still in cell cycle (Fig. 1g). Staining with Lyve-1 and ER-TR7, to identify the SCS floor and ceiling respectively, showed that indeed some BEM had moved into the SCS (Fig. 1h). These data suggest that BEM move from the GC into the SCS, from where they may join the efferent lymph flow33, leaving the drLN to disseminate via blood into distant lymphoid tissues.
Intravital imaging of drLN of Cγ1Cre mTmG mice confirmed that a large number of eGFP+ BEM had actively migrated between the GC and the SCS (Fig. 2a, Suppl. movie 1). BEM entered the SCS lumen (Fig. 2b), where some moved along the SCS (Fig. 2c) presumably migrating towards efferent lymphatics. Surprisingly, some BEM, after a short pause around macrophages in the SCS, recycled into the LN follicles through the SCS floor and migrated back towards the GC (Fig. 2b, d, Suppl. Fig. 1, Suppl. movie 2).
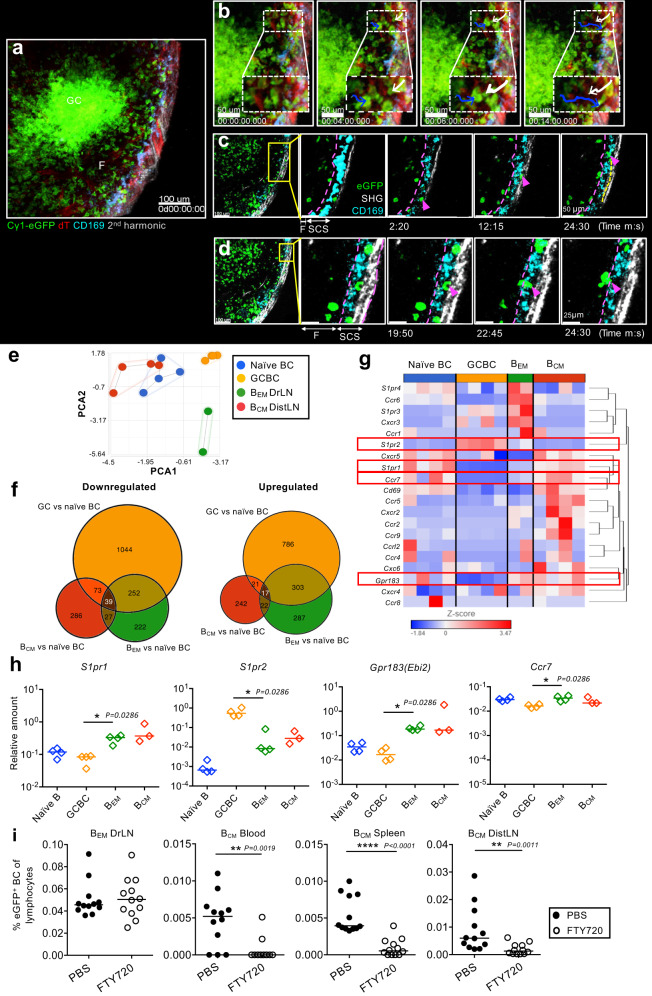
Fig. 2. BEM movement in the drLN.
a Intravital observation of popliteal lymph node from Cγ1Cre mTmG mice 8 d after NP-CGG foot immunization (see suppl. movie 1). b Still images showing eGFP+ BEM entering the SCS (blue arrow) and reentering the lymph node follicle from the SCS (white arrow). c Images showing an eGFP+ B cells migrating along the SCS. d Images showing a BEM reentering the lymph node follicle. Representative of 7 Cγ1Cre mTmG mice (n = 7). RNA-Seq data from B cell populations sorted from drLN 8 d after immunization of recipients of Cγ1-Cre QM mTmG B cells. e Principal component analysis of global gene expression in sorted populations, f Numbers of differentially expressed genes. g Heat map of selected chemotactic receptors among mRNA. Genes were hierarchically clustered by Euclidean distance measure. Data are from two independent experiments with four samples (each sample from pooled popliteal lymph nodes from three mice). h S1pr1, S1pr2, Ebi2, and Ccr7 mRNA expression analyzed by qRT-PCR. Each diamond represents pooled lymph nodes from four mice. All values are relative to B2m mRNA. Two-tailed Mann-Whitney test, *: p = 0.0286. Data representative of three independent experiments (total n = 12). i BEM in drLN and BCM in blood, spleen, and distLN 8 d after immunization. Mice received FTY720 over 2 d before collection. Each dot represents one mouse, data merged from three independent experiments (total n = 12). Two-tailed Mann-Whitney test, **: p < 0.01, ****: p < 0.0001.
To examine the factors that regulate the migration of BEM from GCs, we performed RNA-Seq analysis of FACS sorted eGFP+ B220+ CD38+ CD95int/+ CD138– BEM in LNs comparing them to eGFP+ B220+ CD38– CD95+ CD138– GC B cells, and eGFP+ B220+ CD38+ CD138– BCM and eGFP– dTomato– B220+ CD38+ CD138– naïve B cells from distLNs. Principle component analysis of all genes expressed by these four subsets of cells confirmed a close relationship of eGFP+ BEM with GC B cells, whereas eGFP+ BCM in distLN are much closer to naïve B cells (Fig. 2e). This was also evident in the number of individual genes differentially expressed, with a greater number of genes differentially expressed regarding the transition from naïve to GC B cells, and a larger overlap in genes co-expressed in GC B cells and BEM (Fig. 2f). Analysis of migratory receptors during the transition from GC B cells into BEM by qRT-PCR revealed a loss of expression chemokine receptors known to be associated with B cells location in the GC (Cxcr5, Cxcr4, S1pr2)34, and increased expression of the receptors S1pr1, S1pr3, S1pr4, Ebi2, Cxcr3, Ccr6, and Ccr7 (Fig. 2g, h, Suppl. Fig. 3). CCR6, EBI2, and CXCR3 are known to be expressed on memory B cells10,35. Blockade or deletion of these receptors, however, did not lead to a noticeable change in the appearance of BCM in distLNs (Suppl. Fig. 4a–c). S1P receptors, particularly S1PR1 and S1PR2, are known to direct the location of B cells in the follicle center and their emigration into lymph vessels34,36. In vivo S1PR blockade using FTY720 led to a dramatic reduction of BCM in blood and distLNs (Fig. 2i, Suppl. Fig. 4d), while there was no noticeable effect on the numbers of other lymphocytes in distant lymphoid tissues (Suppl. Fig. 4d). This suggests that S1PR guides memory B cell migration into the SCS and to lymphatic vessels.
CCR7 dependent recycling of BEM
The intravital imaging we performed (Fig. 2a–d) showed that many BEM after entering the SCS returned to the follicles. Dendritic cells (DC), arriving in the SCS from afferent lymph, migrate into the lymph node guided by local CCL21 gradients that are sensed by CCR7 on DC37. As B cells upregulate CCR7 during the transition from GC B cell to BEM (Figs. 2H, 3A), we hypothesized that a local CCL21 gradient might have a similar role for BEM return into the drLN, as it has for DCs. While CCL21 has not been observed in the subcapsular sinus region surrounding follicles of nonimmunized lymph nodes37,38, Ccl19 and Ccl21 expression have been shown in immunized lymph nodes in marginal reticular cells (MRC) located under the SCS39. In order to test whether CCL21 is expressed in this area, we screened for tdTomato in drLN of Ccl21tdTom mice 8 d after foot immunization. This showed substantial Ccl21 gene expression in stroma located close to the SCS of reactive follicles and in the SCS floor (Fig. 3b, Suppl. Fig. 5a). Immunohistology for CCL21 protein produced a similar pattern, with weak staining for CCL21 around the SCS floor close to follicles containing GCs (Suppl. Fig. 5b).
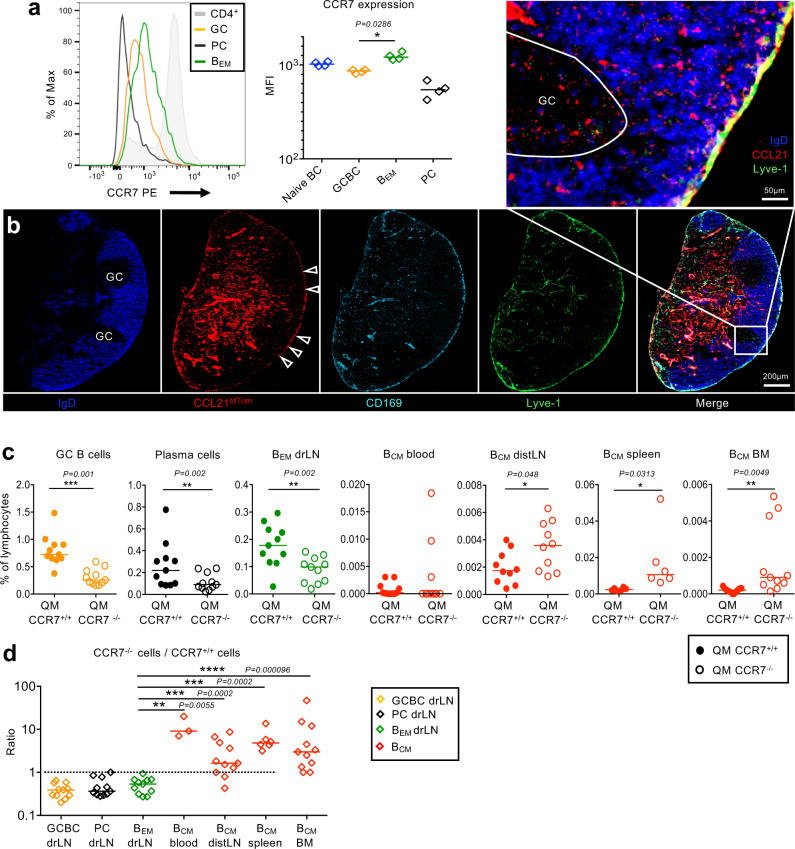
Fig. 3. CCR7-dependent migration of memory B cells.
a CCR7 expression on different lymphocyte subsets measured by flow cytometry. Each symbol represents one animal. Two-tailed Mann-Whitney test, *p = 0.0286. b drLN from a heterozygous Ccl21tdTom/wt reporter mouse 8 d after foot immunization. The SCS ceiling endothelium is missing. Images show dTomato expression in red, and IgD (blue), CD169 (turquoise) and Lyve-1 (green)-specific antibody staining. Arrow heads: Ccl21 expression in SCS. Enlarged box in shows Ccl21a expression under and in the SCS floor endothelium. Image is a representative from 4 lymph nodes. c GC B cells, plasma cells (PC), BEM from drLN 8 d after foot immunization of recipients of a mix of QM mT CCR7+/+ and QM eYFP CCR7−/− B cells. BCM in blood, distLN, spleen, and bone marrow (BM). Two- tailed Wilcoxon matched-pairs test. d Ratio of CCR7−/− to CCR7+/+ QM B cells in different tissues. Two-tailed unpaired t test. Each symbol represents one animal. Data are from two independent experiments with 5–6 mice each.
In order to test how CCR7 ligands affect BEM migration, QM CCR7+/+ mT+ and QM CCR7−/− eYFP+ B cells were co-transferred into WT mice and their migration was assessed after immunization. CCR7 is required for the initial activation of naïve B cells, enabling B cell migration into T cell zones40,41. Therefore, Ccr7-deficient B cells were underrepresented in activated B cell populations (antigen-specific GC B cells, plasma cells, and BEM) in the drLN (Fig. 3c). Despite this, there was an increase in Ccr7−/− BCM in blood, distLNs, spleen, and bone marrow (Fig. 3c, d). This is compatible with a role for CCL21 in orchestrating BEM re-entry into the follicle from the SCS. Without cues sensed by CCR7, BEM is unable to move back from the SCS into the LN parenchyma and therefore appears in larger numbers in blood and distant lymphoid tissues. While CCR7 is an important factor directing the entry of lymphocytes from blood into distant lymphoid tissues, CXCR4 and CXCR5 can provide similar functions42. Both are expressed at elevated levels in BCM (Fig. 1, Suppl. Fig. 3).
The non-signaling atypical chemokine receptor 4 (ACKR4) is expressed in the SCS ceiling endothelium and shares the ligands CCL19 and CCL21 with CCR7. To test whether CCR7-mediated retention of BEM in the drLN is dependent on an ACKR4-generated chemokine gradient, we co-transferred QM mT+ Ackr4+/+ and QM eYFP+ Ackr4−/− B cells into Ackr4+/+ or Ackr4−/− hosts and immunized with NP-CGG. While ACKR4-deficiency on B cells had no significant effect on the size of the GC compartment nor affected BEM numbers in the drLN, ACKR4-deficiency of the LN environment led to decreased numbers of antigen-specific BEM being retained in the drLN and higher numbers appearing in the blood (Fig. 4a). This suggests that chemotactic cues generated in the SCS environment organize BEM reentry into the drLN. In the absence of these, BEM leave the SCS in larger numbers to appear as BCM in the efferent lymph and distLNs.
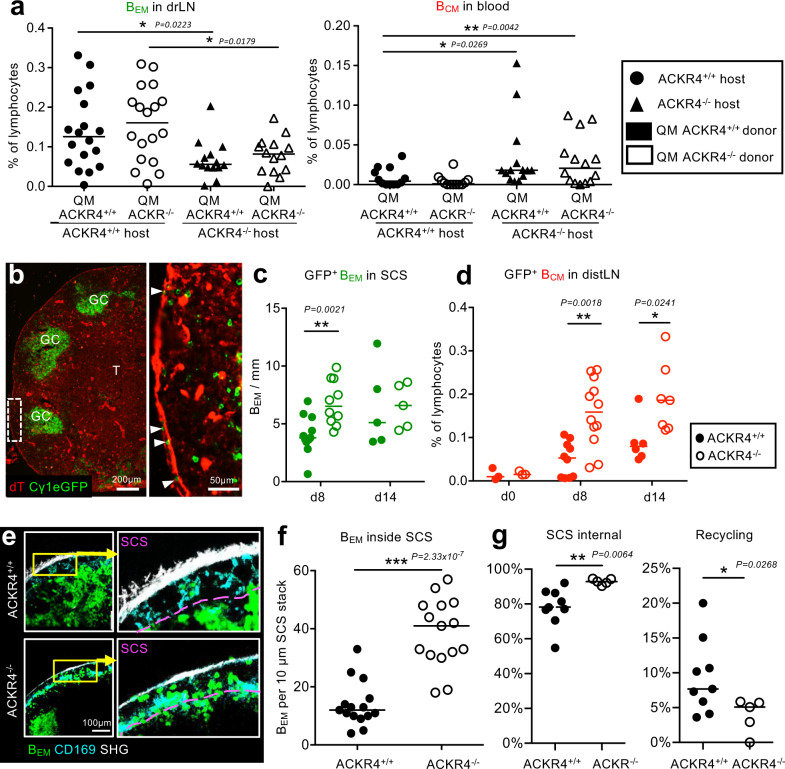
Fig. 4. Local ACKR4-generated chemokine gradients at the SCS direct migration of memory B cells.
a Frequency of QM ACKR4+/+ and QM ACKR4−/− BEM in drLN or BCM in blood of ACKR4+/+ and ACKR4−/− hosts. Data merged from three independent experiments with 4–5 mice each. Two-tailed Mann-Whitney test. b drLN from an Cγ1-Cre mTmG mouse 8 d after immunization. Arrows: eGFP+ BEM inside the SCS. Representative image of nine lymph nodes. c eGFP+ BEM in the SCS of drLN in Cγ1Cre mTmG ACKR4+/+ or ACKR4−/− mice at d8 and d14 after foot immunization. d BCM in distLN in ACKR4+/+ or ACKR4−/− mice after foot immunization. Data in c and d are merged from two independent experiments with 3–5 mice each. Two-tailed Mann-Whitney test. e Intravital observation of BEM entry into SCS in Cγ1Cre mTmG ACKR4+/+ or ACRK4−/− drLN. f Quantitation of intravital eGFP+ BEM in SCS of ACKR4+/+ or ACKR4−/− drLN. Each dot represents cell counts per field of view of a 10 μm Z stack. Pooled data from imaging of three ACKR4+/+ and two ACKR4−/− popliteal LNs. Two-tailed unpaired t test, ***P = 2.33 × 10−7. g Automated B cell tracking showing fraction of BEM tracks that are inside the SCS or recycling from the SCS into the lymph node parenchyma. Images from five ACKR4+/+ and 2 ACKR4−/− popliteal LNs were analyzed. Each symbol corresponds to the average fraction of tracks from one video, total n of tracks = 2478. One-tailed unpaired t test, *p = 0.0268; **p = 0.0064.
To further test this, we followed the migration of BEM in the SCS of drLN by fluorescence microscopy in drLNs of Cγ1Cre mTmG mice that were Ackr4+/+ or Ackr4−/−. This revealed a significantly increased retention of BEM in the SCS of ACKR4−/− drLN 8 d post immunization when BEM recycle (Fig. 4b, c). There was also an increased number of BCM arriving in ACKR4−/− distLNs, and this difference persisted until d 14 (Fig. 4d). Intravital two-photon microscopy confirmed the increased numbers of BEM in the SCS of drLNs of ACKR4-deficient mice (Fig. 4e, f, Suppl. Fig. 6, Suppl. movie 3), and a significantly reduced number of tracks of BEM recycling into the drLN when ACKR4 was absent (Fig. 4g, Suppl. movie 4).
BEM recycling supports adaption to antigenic drift
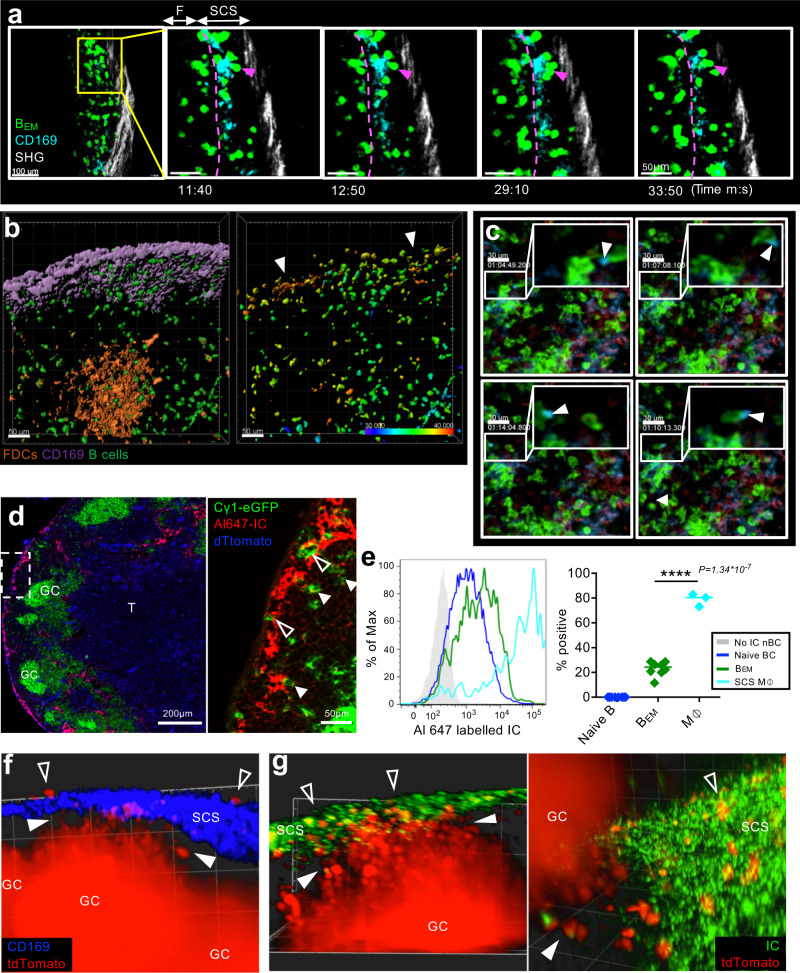
We next considered the functional significance of BEM LN re-entry from the SCS into the B cell follicle. Of note, we observed that BEM appeared to undergo prolonged interaction with CD169-positive SCS macrophages (Fig. 5a, Suppl. Fig. 7, Suppl. movie 5) before reentering the LN parenchyma. Intravital imaging of cytoplasmic calcium showed an increase in calcium specifically in B cells contacting SCS macrophages (Fig. 5b, Suppl. Fig. 8, Suppl. movie 6), suggesting an antigen-specific interaction between BEM and antigen-carrying SCS macrophages. SCS macrophages are known to transfer antigens to naïve B cells20,21,43. A recent study showed similar interactions of antigen-specific memory B cells during secondary responses16. For some BEM, we observed that they acquired CD169-labeled material from SCS macrophages (Fig. 5c, Suppl. movie 7), suggesting that BEM can acquire and transport antigen from SCS macrophages into the GC. To test this, mice were immunized with rabbit-IgG and eight days later injected with AlexaFluor647-labeled mouse anti-rabbit immune complex (IC) into the foot. Within 10 min, IC was seen associated with SCS macrophages of drLNs. IC was also present inside intranodal lymphatics and entering the lymph node parenchyma. BEM within the SCS were in intimate contact with IC-carrying cells, whereas inside the LN parenchyma, those BEM that were close to the SCS carried speckles of IC (Fig. 5d). Flow cytometry confirmed that 20–30% of BEM carried increased amounts of IC within minutes of IC injection (Fig. 5e, Suppl. Fig. 9)44.
Fig. 5. BEM interaction with antigen on SCS macrophages.
a Intravital observation of interactions of eGFP+ BEM (green) and SCS macrophages (CD169 turquois). b Intravital Ca2+ levels in BEM. Left: Surface rendering of CD169-stained macrophages (purple) and GC (orange, CD21/35Atto590), B1-8hi TN-XXL+ B cells (green). Right: FRET intensity in BEM of same frame with color-coded mean FRET intensity. Arrowheads: clusters of FRET-positive BEM in contact with SCS macrophages. Animation in movie 6. Scale bar 50 µm. c Intravital observation (clockwise from top left) of BEM (green) in SCS containing CD169+ material (blue, arrowhead). d drLN from an Cγ1Cre mTmG mouse 10 min after foot injection with Alexa647 labeled immune complex (IC). Open Arrow heads: eGFP+ BEM in SCS. Closed arrow heads: Alexa647-IC colocalizing with eGFP+ BEM in follicle. Representative image of three lymph nodes. e Left: Alexa647-IC on CD11b+ CD169+ SCS macrophage (green), eGFP+ BEM (red), naïve B cells (blue), or naïve B cells without Alexa647 injection (gray). Right: Percent Alexa647-IC positive BEM or macrophages compared to naïve B cells. Overton subtraction from two independent experiments with three mice each. SCS M⏀: CD169+ subcapsular sinus macrophages. Two-tailed unpaired Student’s t test, ****p = 1.34 × 10−7. f Light-sheet microscopy of live explanted drLN, showing S1PR2tdTom BEM (red) in located between GC and SCS (closed arrowheads). SCS macrophages (blue). Some BEM are seen in SCS above the SCS layer (open arrowheads). See also movie 8. g Same as (f) showing Alexa488 labeled IC (green) in the SCS and S1pr2 expression dependent tdTomato positive BEM (red) between GC and SCS (closed arrowheads) or in contact with IC in SCS (open arrowheads). Representative image of four lymph nodes. See also Suppl. movies 9, 10.
While the majority of B cells with a history of Cγ1Cre expression is GC-derived, Ig class switch recombination is induced before GCs develop15,45 and some memory B cells can develop before GCs appear3. To confirm that BEM seen recycling are GC derived, we made use of mice with tamoxifen-inducible Cre expressed under the S1pr2 promotor9 crossed onto Ai14 mice with Cre-inducible tdTomato expression (S1pr2CreERT2 Ai14). These mice were foot immunized and treated with tamoxifen 2 d before drLNs were removed at day 8. Light sheet microscopy of live drLNs showed large numbers of tdTomato+ BEM in follicles between GC and SCS (Fig. 5f, Suppl. movie 8). When these mice had been injected with immune complex 10 min before drLNs were taken, immune complexes could be seen in close association with BEM (Fig. 5g, Suppl. movies 9, 10). Taken together, these data suggest that BEM can be activated by specific antigen in the SCS, and then may transport this back towards the GC.
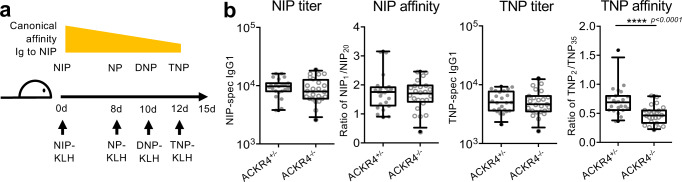
GCs typically contain large amounts of antigen deposited on follicular dendritic cells. Therefore, additional antigen deposition by BEM seems unnecessary, unless the antigen is changing during the course of an infection. BEM, are a GC output with highly variable affinity and specificity for antigen, and would therefore include cells that may interact with antigenic variants. To test the hypothesis that BEM recycling has a role in adaption to antigenic drift, we used variants of the hapten NP and measured the adaption of affinity maturation to these variants. The B cell response of C57BL/6 mice is dominated by a canonical IgH VDJ BCR combination that has natural affinity to 4-hydroxy-iodo-phenyl (NIP), and reduced affinity to the variants NP, dinitrophenyl (DNP) and trinitrophenyl (TNP) (Fig. 6a). ACKR4+/− mice were immunized with NIP-KLH. After the onset of BEM recycling, we rechallenged in the same foot with NP, DNP, followed by TNP-KLH. Three days after the last injection we observed a shift in antibody affinity towards TNP (Suppl. Fig. 10). In order to test whether this was dependent on BEM recycling, the experiment was repeated in Ackr4−/− mice, where BEM cannot undergo recycling. This showed that without BEM recycling, the drift toward the new antigenic variant was significantly reduced (Fig. 6b). This occurred despite ACKR4 deficiency itself not affecting GC sizes, antibody generation, or affinity maturation to immunization when there was no antigenic drift (Suppl. Fig. 11).
Fig. 6. BEM interaction with antigen in SCS affects response to antigenic variants.
a Design of antigenic drift experiment using variants of the NIP hapten. b NIP-specific and TNP-specific IgG1 antibody titer and affinity in ACKR4+/− and ACKR4−/− mice. Merged data from 4 independent experiments with five mice each (total n = 20). Boxes show medians and 50% range, whiskers: 5–95%. Symbols represent individual mice. Two-tailed Mann-Whitney test, ****p < 0.0001.
Discussion
We set out to detect the migratory pathways taken by memory B cells generated from GCs. Lymphocytes leaving lymph nodes are thought to move into intranodal lymphatics before leaving via the medulla and efferent lymph. It was therefore surprising to see memory B cells taking the journey in the opposite direction toward the SCS. Along the SCS they are still able to travel to the hilum and join efferent lymph46, but this is not the short route. The experiments presented here suggest that S1PR directs the migration of memory B cells into the periphery, leading to their appearance in other lymphoid organs, while CCL21 gradients, organized by ACKR4 expressed in the SCS ceiling, direct recycling of BEM cells back into the lymph node. Another ligand of ACKR4, CCL19, may also be involved, but current detection methods are not sensitive enough to confirm this47,48. ACKR4 is also expressed by splenic perimarginal sinus cells, where it may have a similar function49. We showed that BEM further express Ccr6, Ebi2 and Cxcr3 mRNA. While inhibition of these receptors did not affect the appearance of memory B cells in distant lymph nodes, this does not mean that they are not playing a role in directing migration of BEM to non-lymphoid tissues or within the reactive LN. CCR6/CCL20 interactions are involved in the migration of follicular GC-derived B cells35 and CCL20 expressed by subcapsular lymphatics can organize the accumulation of innate-like lymphocytes near the SCS36. EBI2 is known to organize the movement of activated B cells to the periphery of the lymph node,50,51 and its ligands are produced by MRC52.
The B cell subsets analyzed here are defined by their anatomical location and differential gene expression. For the purpose of this study, they were labeled BEM and BCM, although, being freshly generated from GCs they are not memory cells in the classical sense. The two subsets behave differently, with one cell type still in active interaction with antigen and able to re-enter affinity maturation, whereas BCM leave the cell cycle and are probably differentiating towards long-term survival and surveillance for antigen. We have not tested whether epigenetic changes have fixed the differential gene expression in these subsets, so do not know whether the cells represent different lineages evolving or mere differentiation stages that are still plastic and under the influence of the more inflammatory or quiescent microenvironments in different lymph nodes.
SCS macrophages are well known to accumulate and present antigen entering the lymph node via afferent lymph53. Naïve B cells scan these macrophages and acquire immune complex via complement receptor cells44. Specific antigen-BCR interaction leads to efficient activation of naïve B cells and their migration to the T zone–follicle border, where they can interact with Tfh cells15,20,21,43. Others have shown that after secondary immunization, memory B cells can interact with SCS macrophages presenting recall antigen, which leads to interaction with local Tfh cells and rapid plasma cell generation16,28. In the current study, we tested the migration and output of memory B cells during the early peak of the GC reaction. While some memory B cells leave the lymph node and appear in other organs, others interact with SCS macrophages. At this stage, we did not notice substantial plasma cell generation, but found B cells that recycled towards the GC.
Memory B cells are not selected for high affinity and have highly variable BCRs9–11. As recycling BEM showed signs of BCR signaling and were seen to transport antigen, we assumed that memory B cell recycling may represent a secondary selection step after BEM test their mutated BCR for the first time on antigen arriving outside the GC. Memory B cells expressing BCRs that are able to interact with antigen in the SCS would be allowed to recycle to the GC together with antigen, where they could undergo further affinity maturation. Many pathogens mutate, leading to antigenic drift. For RNA viruses this happens over different seasons or within individual hosts, such as escape mutants developing during HIV infection. For HIV mutation rates have been reported to be as high as 4 bases per kbp per infected cell54. Other RNA viruses mutate slower, with mutation rates for SARS-CoV estimated at around 1 bp per genome per week55. The antigenic drift experiment suggests that memory B cell recycling is a mechanism for adaptation to antigenic drift inside the host.
Ackr4-deficiency had a noticeable impact on the adaption to antigenic drift, most likely caused by an inhibition of memory B cell recycling. Dendritic cells entering the LN through the SCS are dependent on the same ACKR4-induced chemokine gradients, however, dendritic cells are more important during the initial of antibody responses when T cells need to be primed37.
A memory B cell pool with highly variable BCRs is likely to contain BEM variants capable of interacting with antigen variants. BEM recycling would therefore not only transport antigen into the GC but, more importantly, would return the genomic information for BCR variants that may enhance affinity maturation towards the antigen variant. This could contribute to the high number of BCR mutations observed in HIV infection56, which seem to be caused by repeated B cell re-entry into the GC. While this is not able to stop the emergence of HIV variants, for other viruses this process may be an efficient mechanism of keeping mutations at bay.
Methods
Mice and immunization
All procedures on animals were approved by the University of Birmingham Ethics Committee AWERB (Animal Welfare Ethical Review Board) and done under UK Home Office licenses PPL 70/8011 and PP8702596. Host C57BL/6J mice (wild type, WT) were purchased from Harlan laboratories (stock number: 632c57bl/6J). WT or heterozygous gene-deficient control animals were bred in the same colony as gene-deficient animals. All mice were housed in the Biomedical Services Unit (University of Birmingham) under specific pathogen-free conditions/SPF. At the end experiments mice were humanely killed under anesthesia by cervical dislocation or exsanguination. Mice were mixed sexes and used at 8–12 weeks age. Ackr4tm1.1Rjbn (Ackr4−/−) mice57 were a gift from R. Nibbs (University of Glasgow). Cγ1Cre mTmG mice were generated by crossing Ighg1tm1(cre)Cgn 29 (gift from Stefano Casola, Istituto FIRC di Oncologia Molecolare, Milan) with Gt(ROSA)26Sortm4(ACTB-tdTomato, -EGFP)Luo mice32 (Jackson Laboratory, Stock No. 007914). For all adoptive transfer experiments, variants of QM mice were used, which were homozygous for a NP-specific Ig heavy chain variable region from Igh-Jtm1(VDJ-17.2.25)Wabl and Ig kappa light chain deficient (Igktm1Dhu)30. Some QM mice contained a constitutively expressed enhanced yellow fluorescent protein (eYFP) derived from Gt(ROSA)26Sortm1.1(EYFP)Cos,58 (QM eYFP mice), or were crossed onto Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo (QM mTmG mice). QM CCR7−/− mice are QM crossed with CCR7tm1Rfor mice59. Ccl21tdTom knock-in mice38,60 were a kind gift from G Anderson (University of Birmingham). Ccl21tdTom heterozygous mice were used to detect Ccl21a gene expression. S1pr2CreERT2 Ai14 mice were generated by crossing S1pr2CreERT2 9 onto Gt(ROSA)26Sortm14(CAG-tdTomato)Hze mice61 (Jackson Laboratory, Stock No. 007908) to get S1pr2CreERT2 Ai14 mice.
NP (4-hydroxy-3-nitrophenyl acetyl) was conjugated to CGG (Chicken γ- globulin) at a ratio of NP18-CGG. Mice were immunized into the plantar surface of their rear feet with 20 μg alum precipitated NP18-CGG plus 105 chemically inactivated Bordetella pertussis (B.p.) (LEE laboratories, BC, USA)62. Popliteal lymph nodes draining the injection site (drLN) and distant axillary and brachial lymph nodes (distLN) were analyzed.
S1PR inhibition: Wt mice received adoptive transfer of 2 × 105 NP+ B220+ cells from QM Cγ1Cre mTmG. One day later they were foot immunized with NP-CGG. FTY720 (Caymanchem, USA) was given at 1 mg/kg body weight i.p. 6 and 7 d later. Tissues were analyzed 8 d after immunization.
EBI2 and CXCR3 inhibition: Wt mice were immunized as above, and NIBR189 (Ebi2 antagonist) or NBI7433 (CXCR3 antagonist) (both from Tocris, UK) were injected i.p. at 60 μg per mouse 5 h before being killed at 8 d after immunization.
For immune complex injections, mice were primed with 20 μg rabbit IgG (PRABP01, BioRad UK) alum precipitated with 105 Bordetella pertussis (B.p) in the foot. Eight days later 4 μg of Alexafluor647 (or Alexafluor488) labeled immune complex (IC) was made with 1:1 ratio of Alexafluor647 (or Alexafluor488) conjugated mouse anti-rabbit IgG and rabbit IgG (Jackson Immunoresearch). This was mixed for 30 min before injecting into the foot. Tissues were usually taken 30 min after injection.
For antigenic drift experiments ACKR4−/− mice and littermate ACKR4+/− control mice were primed with 10 μg of NIP-KLH in alum precipitated with 105 B.p. in the rear feet. 8 d later, mice were boosted with 1 μg of soluble NP-KLH, DNP-KLH, and TNP-KLH on the same feet every 2 days (NP conjugates from Biosearch Technologies, USA).
To label early GC derived memory B cells, 4 mg tamoxifen (SIGMA) was given once by gavage to S1pr2CreERT2 Ai14 mice 6 d after antigen injection into the foot. Popliteal lymph nodes were collected 2 d later.
Immunohistology
Lymph node sections were prepared and stained as described previously31,63. Frozen sections were cut at 6 μm, and then fixed in acetone. Lymph nodes from Ccl21tdTom heterozygous mouse were fixed in 1% paraformaldehyde overnight followed by 30% sucrose before frozen. Sections were stained with CD138, IgD, CD3, biotinylated peanut agglutinin (PNA), Ki-67 and LYVE-1, Fibroblasts Specific monoclonal antibody (ER-TR7), CD169, and CCL21. NP-rabbit was house made to detect NP antigen-specific B cells. Secondary antibodies were Cy3-conjugated donkey anti-rat, donkey anti-rabbit, or donkey anti-goat and Alexafluor405 (or Alexafluor488) conjugated streptavidin. Slides were mounted in ProLong Gold antifade reagent (Invitrogen, UK) and left to dry in a dark chamber for 24 h. Images were taken on a Leica DM6000 fluorescence-microscope (Leica Germany), or Zeiss Axio ScanZ1, and Zeiss LSM880 with Airyscan Fast. Image data were processed using Fiji 2.3.064, ZEN black (ver. 8.1) or Zen Blue (ver. 1.1.2.0) (Carl Zeiss Germany). All antibodies are listed in Supplementary Tables 1 and 2.
Flow cytometry and adoptive transfer
Cells from spleens and lymph nodes were prepared as described31. Red blood cells were lysed by ACK lysing buffer (Gibco). Cell suspensions were blocked by CD16/32 diluted in FACS buffer (PBS supplemented with 0.5% BSA plus 2 mM EDTA), followed with staining cocktail. NP was conjugated in house with PE to detect antigen-specific B cells31. For detecting transcription factor BCL6, cell suspensions were fixed with BD Cytofix fixation buffer (BD Biosciences) and permeabilized with BD Phosflow Perm buffer III (BD Biosciences), before intracellular staining with Bcl6 PE-Cy7 (BD Biosciences). Samples were analyzed using BD LSRFortessa Analyzer (BD Biosciences, USA) with the software BD FACSDiva (BD Biosciences). Data were analyzed offline with FlowJo (ver. 9 and 10) (FlowJo LLC, USA). All antibodies are listed in Supplementary Tables 1 and 2. All samples were analyzed after gating out dead cells by using live/dead fixable Near-IR dead cell stain kit (Cat: L10119, Invitrogen) or propidium iodide solution (Cat: 431301, BioLegend).
For adoptive transfer experiment, 2 × 105 NP+ B220+ cells from spleens of fluorescent protein labeled QM background mice were transferred into C57BL6/J hosts 1 d before immunization with NP-CGG in alum on rear feet. In co-transfer experiments, a mix of 1 × 105 of NP+B220+ B cells of each genotype respectively were injected i.v. Gating scheme see Suppl. Fig. 13.
Cell sorting for qRT-PCR, RNA-seq library preparation, and data analysis
DrLN and distLN in mice 8 d after foot immunization with NP-CGG in alum and B.p were stained as described above. Naive B cells, GC B cells, plasma cells, BEM cells from drLN and BCM from distLN were sorted using a high-speed cell sorter (MoFlo, Beckman-Coulter). Gating scheme see Suppl. Fig. 12.
For real-time PCR, RNA was purified by using the RNeasy Mini kit (QIAGEN), cDNA preparation was as described as before 31. Real-time PCR from cDNA (qRT-PCR) was done in multiplex with β2-microglobulin and gene expression related to β2-microglobulin gene expression levels. Primers and probes are listed in Supplementary Table 3.
For RNA-seq, cells were directly sorted into 500 μl of Trizol. The total RNA was purified using the RNeasy Plus Micro kit (QIAGEN) according to the manufacturer’s instructions. Un-stranded, non-rRNA, non-polyA+ selected libraries were prepared using the SMARTer Ultra Low Input RNA kit for Sequencing v3 (Clontech Laboratories). The libraries were sequenced on the Illumina HiSeq 2000 platform (Illumina, Crick advanced sequencing) as 75 bp paired-end runs. The datasets generated during the current study are available in the Gene Expression Omnibus (GEO) Database under accession code GSE188687, [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188687].
The sequencing data were analyzed using Partek Flow software, version 8.0.19 (Partek Inc., St. Louis, MO, USA). Paired sequencing data was imported and then aligned to mouse genome GRCm38 (mm10). t-SNE analysis was performed on normalized RNA counts to generate a 2D plot by dimensional reduction. Gene-specific analysis (GSA) tool was used to identify differentially expressed genes against naïve B cells subset as a control. GSA used the lognormal and negative binomial response distribution under the multi-model approach and the lowest maximum coverage of 1.0 was used as the low-value filter. Venn diagrams were produced from differential expression of genes with a log fold change >1 or <−1 and P value < 0.05 using BioVenn65. The heatmap was produced from GSA data as described earlier from a predetermined list of genes of interest using the Hierarchical Clustering tool; genes were clustered based on their average Euclidean distance from one another.
Intravital multiphoton microscopy
Intravital microscopy of popliteal lymph nodes of Cγ1Cre mTmG mice was performed 8 days after subcutaneous immunization with NP-CGG in alum ppt in the hind hock. Subcapsular sinus macrophages were labeled with CD169-A647 antibody (BioLegend) injected subcutaneously in the hind hock before imaging. The popliteal LN was surgically exposed under a dissecting microscope, and imaged with a Chameleon Vision-S tuneable Ti:Sapphire multiphoton laser and Leica SP8 microscope, with the mouse under inhalational anesthesia and the imaging box kept at 36 °C throughout. Images were acquired using a 25× objective, with one Z stack every 30–40 s, and processed using either Bitplane Imaris 8.2 or Fiji ImageJ 2.3.066.
Automated cell tracking
Tracking of BEM was performed in 3D using the Fiji 2.3.0 plugin TrackMate67. Individual cells were detected using the local maxima of a Laplacian of Gaussian filtered (radius 10 µm) image volume with a quality threshold of 15. A linear assignment problem tacking algorithm was used with maximum frame-to-frame linking distance, maximum tack gap close distance, and maximum track gap closing interval set to 20 µm, 20 µm and two frames, respectively68. All tracks less than 5 frames were discarded. A customized Fiji 2.3.0 groovy script was written to automate the tracking and export statistics, available at https://github.com/JeremyPike/SCS-cell-tracking69.
Subcapsular sinus segmentation
The SCS was segmented using a random forest-based pixel classifier implemented and trained using ilastik 1.3.3post370. Four classes were used representing the inner SCS wall, the outer SCS wall, the internal SCS region, and background (no signal). All default ilastik features were used for training which include intensity, edge and texture-based measures over a range of scales. Before training, and prediction, movies were down-sampled by a factor of four in xy. Three time-points (selected from either start, middle, or end of movies) were annotated for training. Automated production of training data and prediction was performed using customized ImageJ macros available at https://github.com/JeremyPike/SCS-cell-tracking69.
Quantification of interaction between cells and SCS
All tracking results and SCS segmentations were cropped to 13 z-slices and 32 time-points (for consistency across movies). The ilastik probability maps for individual timepoints were temporally smoothed using a rolling average of three frames. This reduces noise but still allows for movement and drift of the SCS over time. Each voxel was then assigned the class with the maximal probability. For each class (apart from background) all but the largest connected component were discarded. To produce a full SCS segmentation the inner-wall, internal and outer-wall classes were combined. Any holes in the SCS segmentation were filled. The cell tracks were then analyzed with respect to the SCS segmentation, counting occurrences of tracks which entered, left and stayed within the SCS. Entering tracks were defined as starting at least 5 µm from the inner SCS surface and ending within the SCS. Conversely leaving tracks start within the SCS and end at least 5 µm from SCS. Internal tracks start and end within the SCS. This post-processing was performed using Matlab R2020b and a customized script69.
Calcium imaging by intravital multiphoton microscopy
C57BL6/J mice received B cells from B18hi mice (carrying the Vh186.2 heavy chain with high affinity to NP) that contained genetically encoded Ca2+ indicator TN-XXL71 under the control of the CD19 promoter72. These were immunized with 10 µg NP-CGG emulsified in complete Freund’s adjuvant into the right foot. The popliteal LN was analyzed on day 7. One day prior to imaging, a mixture of 10 µg anti-CD21/35 Fab (clone 7G6) -Atto590 (produced at the DRFZ) for staining follicular dendritic cells and 10 µg CD169-efluor660 (eBioscience) to label SCS macrophages were injected into the footpad.
Intravital two-photon microscopy was performed as described before73, using a TrimScope II from Lavision Biotec, at an excitation of 850 nm (TiSa) and 1100 nm (OPO). The detection of the fluorescence signals was accomplished with photomultiplier tubes in the ranges of (466 ± 20) nm, (525 ± 25) nm, and (655 ± 20) nm.
TN-XXL is a genetically encoded calcium indicator that consists of a chicken troponin C domain connecting the fluorescent proteins eCFP and Citrine (Suppl. Fig. S8A). These act as a Förster resonance energy transfer (FRET) pair with ECFP as the donor and Citrine as the acceptor fluorophore. Troponin C contains four binding lobes for Ca2+ ions. If Ca2+ is present or cytosolic concentrations are elevated this leads to a conformation change of the linker peptide that causes donor and acceptor to come into sufficient proximity for FRET emission. When quenched ECFP is excited with one photon at 475 nm, or two photons at 850 nm, citrine will emit fluorescence at 530 nm. If no calcium is present, emission in the blue range of the donor group will be more prominent.
Measurements from six immunized mice were analyzed with image analysis software Imaris 9.2 (Bitplane AG). Raw data were pre-processed using a linear unmixing algorithm74 to minimize interference of red fluorescence from antibody staining into the green channel of the citrine fluorescence. Relative FRET ratio was calculated by dividing green fluorescence gain by the sum of blue and green fluorescence, and corrected for instrument-specific values and spectral overlap. A colocalization channel was used to measure contact intensity between B cells (citrine-positive, masked on eCFP to exclude OPO influence) and CD169-efluor660 signal. Using the histogram of the colocalization intensity mean of the B cell surfaces, we identified distinguishable populations of B cells (Suppl. Fig. 8F) with either no contact (–) or tight contact (+). All B cells with colocalization intensity of 0 AU were assigned to the (−) group. To choose a threshold value of colocalization intensity for B cells to be assigned to the (+) group, we biexponentially fitted the decay of the histogram and determined the point in which cell numbers intersect the plateau of y = 9509 to be 717 AU. Non-contacting B cells and B cells with colocalization intensities >717 AU were filtered and corresponding FRET intensities of all cells at all time points exported for plotting.
Light-sheet microscopy
A plane illumination microscope (Zeiss LightSheet Z1) was used to detect memory B cells in popliteal lymph nodes of S1pr2CreERT2 Ai14 mice 30 min after injecting AlexFluo488 labeled immune complex at 8 d after immunization with Rabbit IgG on the plantar surface of rear feet. Subcapsular sinus macrophages were labeled with CD169-A647 antibody (BioLegend) injected subcutaneously into the plantar surface of rear foot before imaging. Images were acquired using a 20× Plan-Apochromat objective. Z planes were scaled to 4 µm. The laser wavelengths used were 638 nm, 488 nm, and 561 nm. Images were processed using Vision4D (Arivis).
Statistical analysis
All analysis was performed using GraphPad Prism 6 software. To calculate significance two-tailed Mann-Whitney nonparametric test was used. In the experiments where two parameters from the same individual mouse are compared, Wilcoxon matched-pairs signed-rank test (paired nonparametric test) was used to calculate significance. Statistics throughout were performed by comparing data obtained from all independent experiments. P values < 0.05 were considered significant (*). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All data are available from the authors upon request.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional Supplementary File
Acknowledgements
Y.Z., C.J.M.B., J.C.Y.P., and K.M.T. were funded by the BBSRC (BB/S003800/1, BB/M025292/1) and MRC (MR/P001874/1). L.G.I. and G.B. were supported by EU Marie Curie Initial Training Network DECIDE. V.L.J.T. was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001194), the UK Medical Research Council (FC001194), and the Wellcome Trust (FC001194). For the purpose of Open Access, the author has applied a CC-BY public copyright licence to any Author Accepted Manuscript version arising from this submission. MRC is supported by a Medical Research Council New Investigator Research Grant (MR/N024907/1), a Chan-Zuckerberg Initiative Human Cell Atlas Technology Development Grant, a Versus Arthritis Cure Challenge Research Grant (21777), and an NIHR Research Professorship (RP-2017-08-ST2-002). A.E.H. was supported by Deutsche Forschungsgemeinschaft (DFG) TRR130, TP17 and C01, and DFG HA5354/10-1. The authors would like to acknowledge the Flow Cytometry Platform, University of Birmingham for support of flow cytometry experiments.
Source data
Author contributions
Conceptualization: Y.Z., A.R., A.E.H., M.R.C., K.M.T., Methodology: Y.Z., L.G.I., C.U., M.R.C., L.S.C.K., Formal analysis: L.S.C.K., C.J.B., Investigation: Y.Z., L.G.I., C.U., L.S.L., T.W.D., J.R.F., J.M.W., C.J.B., J.Y.P., L.Z., L.S.C.K., R.M.A., Automated tracking and segmentation analysis: J.A.P., Writing: Y.Z., L.G.I., C.U., M.R.C., K.M.T., Provision of gene manipulated animals: Y.T., I.O., T.K., Writing - review and editing: G.B., T.K., V.L.T., A.E.H., M.R.C., K.M.T., Visualization: C.U., L.S.L., T.W.D., J.R.F., Supervision: Y.Z., V.L.T., A.E.H., M.R.C., K.M.T., Funding acquisition: Y.Z., L.S.L., G.B., V.L.T., A.E.H., M.R.C., K.M.T.
Peer review
Peer review information
Nature Communications thanks the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
Source data are provided as a Source Data file. The gene expression data have been deposited in the gene expression omnibus (GEO) repository under the accession code GSE188687.
Code availability
Customized Fiji code and Matlab script for postprocessing of intravital tracking data is available at https://github.com/JeremyPike/SCS-cell-tracking (DOI is 10.5281/zenodo.6351432)69.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yang Zhang, Laura Garcia-Ibanez.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-29978-y.
References
- 1.Zhang Y, Garcia-Ibanez L, Toellner K-M. Regulation of germinal centre B cell differentiation. Immunol. Rev. 2016;270:8–19. doi: 10.1111/imr.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida T, et al. Memory B and memory plasma cells. Immunol. Rev. 2010;237:117–139. doi: 10.1111/j.1600-065X.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- 3.Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. Plasma cell output from germinal centers is regulated by signals from Tfh and stromal cells. J. Exp. Med. 2018;215:1227–1243. doi: 10.1084/jem.20160832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laidlaw BJ, et al. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med. 2017;214:639–649. doi: 10.1084/jem.20161461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landsverk OJ, et al. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 2017;214:309–317. doi: 10.1084/jem.20161590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gitlin AD, et al. Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity. 2016;44:769–781. doi: 10.1016/j.immuni.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pape KA, et al. Cells with high-avidity germline-encoded antigen receptors produce persistent IgM(+) and transient IgG(+) memory B cells. Immunity. 2018;48:1135–1143 e1134. doi: 10.1016/j.immuni.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinnakasu R, et al. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 2016;17:861–869. doi: 10.1038/ni.3460. [DOI] [PubMed] [Google Scholar]
- 10.Suan D, et al. CCR6 defines memory B cell precursors in mouse and human germinal centers, revealing light-zone location and predominant low antigen affinity. Immunity. 2017;47:1142–1153 e1144. doi: 10.1016/j.immuni.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 11.Viant C, et al. Antibody affinity shapes the choice between memory and germinal center B cell fates. Cell. 2020;183:1298–1311 e1211. doi: 10.1016/j.cell.2020.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinkernagel RM. What if protective immunity is antigen-driven and not due to so-called “memory” B and T cells? Immunol. Rev. 2018;283:238–246. doi: 10.1111/imr.12648. [DOI] [PubMed] [Google Scholar]
- 13.Ise W, et al. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proc. Natl Acad. Sci. USA. 2014;111:11792–11797. doi: 10.1073/pnas.1404671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelcic I, et al. Memory B cells activate brain-homing, autoreactive CD4(+) T cells in multiple sclerosis. Cell. 2018;175:85–100 e123. doi: 10.1016/j.cell.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toellner K-M, Gulbranson-Judge A, Taylor DR, Sze DM-Y, MacLennan ICM. Immunoglobulin switch transcript production in vivo related to the site and time of antigen-specific B cell activation. J. Exp. Med. 1996;183:2303–2312. doi: 10.1084/jem.183.5.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran I, et al. Memory B cells are reactivated in subcapsular proliferative foci of lymph nodes. Nat. Commun. 2018;9:3372. doi: 10.1038/s41467-018-05772-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mesin L, et al. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell. 2020;180:92–106 e111. doi: 10.1016/j.cell.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi T, et al. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37:535–548. doi: 10.1016/j.immuni.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moseman EA, et al. B cell maintenance of subcapsular sinus macrophages protects against a fatal viral infection independent of adaptive immunity. Immunity. 2012;36:415–426. doi: 10.1016/j.immuni.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27:160–171. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Junt T, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–114. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 22.Arnon TI, et al. GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science. 2011;333:1898–1903. doi: 10.1126/science.1208248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonoda E, et al. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/S1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 24.Kallies A, et al. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 2004;200:967–977. doi: 10.1084/jem.20040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Stein H, et al. Immunohistologic analysis of the organization of normal lymphoid tissue and non-Hodgkin’s lymphomas. J. Histochem. Cytochem. 1980;28:746–760. doi: 10.1177/28.8.7003001. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y-J, Oldfield, S, MacLennan, ICM. Thymus-independent type 2 responses in lymph nodes. In: Advances in Experimental Medicine and Biology (eds Fossum S, Rolstad B). In Proc. of the Ninth International Conference on Lymphatic Tissues and Germinal Centres in Immune Reactions, August 10–14 1987 (Oslo, Norway edn. Plenum Press, 1988).
- 28.Aiba Y, et al. Preferential localization of IgG memory B cells adjacent to contracted germinal centers. Proc. Natl Acad. Sci. USA. 2010;107:12192–12197. doi: 10.1073/pnas.1005443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casola S, et al. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl Acad. Sci. USA. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cascalho M, Ma A, Lee S, Masat L, Wabl M. A quasi-monoclonal mouse. Science. 1996;272:1649–1652. doi: 10.1126/science.272.5268.1649. [DOI] [PubMed] [Google Scholar]
- 31.Marshall JL, et al. Early B blasts acquire a capacity for Ig class switch recombination that is lost as they become plasmablasts. Eur. J. Immunol. 2011;41:3506–3512. doi: 10.1002/eji.201141762. [DOI] [PubMed] [Google Scholar]
- 32.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 33.Girard JP, Moussion C, Forster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat. Rev. Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 34.Green JA, Cyster JG. S1PR2 links germinal center confinement and growth regulation. Immunol. Rev. 2012;247:36–51. doi: 10.1111/j.1600-065X.2012.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoler-Barak L, et al. B cell dissemination patterns during the germinal center reaction revealed by whole-organ imaging. J. Exp. Med. 2019;216:2515–2530. doi: 10.1084/jem.20190789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Migratory and adhesive cues controlling innate-like lymphocyte surveillance of the pathogen-exposed surface of the lymph node. Elife. 2016;5:e18156. doi: 10.7554/eLife.18156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulvmar MH, et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 2014;15:623–630. doi: 10.1038/ni.2889. [DOI] [PubMed] [Google Scholar]
- 38.Kozai M, et al. Essential role of CCL21 in establishment of central self-tolerance in T cells. J. Exp. Med. 2017;214:1925–1935. doi: 10.1084/jem.20161864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang HY, et al. Identification of a new subset of lymph node stromal cells involved in regulating plasma cell homeostasis. Proc. Natl Acad. Sci. USA. 2018;115:E6826–E6835. doi: 10.1073/pnas.1712628115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reif K, et al. Balanced responsiveness to chemoattractants from adjacent zones determines B-cell position. Nature. 2002;416:94–99. doi: 10.1038/416094a. [DOI] [PubMed] [Google Scholar]
- 41.Okada T, et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3:e150. doi: 10.1371/journal.pbio.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okada T, et al. Chemokine requirements for B cell entry to lymph nodes and Peyer’s patches. J. Exp. Med. 2002;196:65–75. doi: 10.1084/jem.20020201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 44.Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 2009;10:786–793. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roco JA, et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350 e337. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jalkanen S, Salmi M. Lymphatic endothelial cells of the lymph node. Nat. Rev. Immunol. 2020;20:566–578. doi: 10.1038/s41577-020-0281-x. [DOI] [PubMed] [Google Scholar]
- 47.Ngo VN, Tang HL, Cyster JG. Epstein-Barr virus-induced molecule 1 ligand chemokine is expressed by dendritic cells in lymphoid tissues and strongly attracts naive T cells and activated B cells. J. Exp. Med. 1998;188:181–191. doi: 10.1084/jem.188.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cremasco V, et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat. Immunol. 2014;15:973–981. doi: 10.1038/ni.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werth K, et al. Expression of ACKR4 demarcates the “peri-marginal sinus,” a specialized vascular compartment of the splenic red pulp. Cell Rep. 2021;36:109346. doi: 10.1016/j.celrep.2021.109346. [DOI] [PubMed] [Google Scholar]
- 50.Gatto D, Paus D, Basten A, Mackay CR, Brink R. Guidance of B cells by the orphan G protein-coupled receptor EBI2 shapes humoral immune responses. Immunity. 2009;31:259–269. doi: 10.1016/j.immuni.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Kelly LM, Pereira JP, Yi T, Xu Y, Cyster JG. EBI2 guides serial movements of activated B cells and ligand activity is detectable in lymphoid and nonlymphoid tissues. J. Immunol. 2011;187:3026–3032. doi: 10.4049/jimmunol.1101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodda LB, et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity. 2018;48:1014–1028 e1016. doi: 10.1016/j.immuni.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batista FD, Harwood NE. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 2009;9:15–27. doi: 10.1038/nri2454. [DOI] [PubMed] [Google Scholar]
- 54.Cuevas JM, Geller R, Garijo R, Lopez-Aldeguer J, Sanjuan R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vega VB, et al. Mutational dynamics of the SARS coronavirus in cell culture and human populations isolated in 2003. BMC Infect. Dis. 2004;4:32. doi: 10.1186/1471-2334-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoofs T, et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity. 2019;50:1513–1529 e1519. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comerford I, et al. The atypical chemokine receptor CCX-CKR scavenges homeostatic chemokines in circulation and tissues and suppresses Th17 responses. Blood. 2010;116:4130–4140. doi: 10.1182/blood-2010-01-264390. [DOI] [PubMed] [Google Scholar]
- 58.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forster R, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/S0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 60.Riken Institute Accession Number CDB1030K, http://www.clst.riken.jp/arg/mutant%20mice%20list.html).
- 61.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Garcia-Ibanez L, Brown G, Toellner KM. Detecting gene expression in lymphoid microenvironments by laser microdissection and quantitative RT-PCR. Methods Mol. Biol. 2017;1623:21–36. doi: 10.1007/978-1-4939-7095-7_2. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 2013;210:457–464. doi: 10.1084/jem.20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clatworthy MR, et al. Immune complexes stimulate CCR7-dependent dendritic cell migration to lymph nodes. Nat. Med. 2014;20:1458–1463. doi: 10.1038/nm.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tinevez JY, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 68.Jaqaman K, et al. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, Y, et al. Recycling of memory B cell between germinal center and lymph node subcapsular sinus supports affinity maturation to antigenic drift. github.com/JeremyPike/SCS-cell-tracking10.5281/zenodo.6351432, (2022). [DOI] [PMC free article] [PubMed]
- 70.Berg S, et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 71.Mank M, et al. A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods. 2008;5:805–811. doi: 10.1038/nmeth.1243. [DOI] [PubMed] [Google Scholar]
- 72.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hauser AE, et al. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 2007;26:655–667. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 74.Rakhymzhan A, et al. Synergistic strategy for multicolor two-photon microscopy: application to the analysis of germinal center reactions in vivo. Sci. Rep. 2017;7:7101. doi: 10.1038/s41598-017-07165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional Supplementary File
Data Availability Statement
Source data are provided as a Source Data file. The gene expression data have been deposited in the gene expression omnibus (GEO) repository under the accession code GSE188687.
Customized Fiji code and Matlab script for postprocessing of intravital tracking data is available at https://github.com/JeremyPike/SCS-cell-tracking (DOI is 10.5281/zenodo.6351432)69.