Abstract
The DNA damage response is a highly orchestrated process. The involvement of the DNA damage response factors in DNA damage response depends on their biochemical reactions with each other and with chromatin. Using online live-cell imaging combined with heavy ion microbeam irradiation, we studied the response of the scaffold protein X-ray repair cross-complementary protein 1 (XRCC1) at the localized DNA damage in charged particle irradiated HT1080 cells expressing XRCC1-tagged RFP. The results showed that XRCC1 was recruited to the DNA damage with ultrafast kinetics in a poly ADP-ribose polymerase-dependent manner. The consecutive reaction model well explained the response of XRCC1 at ion hits, and we found that the XRCC1 recruitment was faster and dissociation was slower in the G2 phase than those in the G1 phase. The fractionated irradiation of the same cells resulted in accelerated dissociation at the previous damage sites, and the dissociated XRCC1 immediately recycled with a higher recruitment efficiency. Our data revealed XRCC1’s new rescue mechanism and its high turnover in DNA damage response, which benefits our understanding of the biochemical mechanism in DNA damage response.
Significance
DNA damage response and repair play a deterministic role in cell proliferation, carcinogenesis, aging, and genetic diseases. This work analyzed the rapid kinetics of the scaffold protein XRCC1 in human cells quantitatively using online live-cell imaging and heavy ion microbeam irradiation. The experimental data revealed that XRCC1 response to the DNA damage had ultrafast kinetics of the consecutive reaction, and the kinetics of recruitment and dissociation are dependent on the cell cycle. Through fractionated irradiation of the same cells, we believe that XRCC1’s high turnover and novel rescue mechanism in DNA damage response are discovered for the first time.
Introduction
DNA is the hereditary material in humans and almost all other organisms, also vulnerable to attacks. Human cells are constantly repairing a large number of DNA damages, which are caused by endogenous factors (including spontaneous decay, replication errors, and cell metabolism) or exogenous factors (such as ionizing radiation, chemicals, anticancer drugs, and environmental stress) (1). Compared with other factors, charged particle radiation with high linear energy transfer (LET: the amount of energy transferred to the medium from the ionizing particle per unit length along ion trajectory (2)), such as radiations in heavy ion therapy and space radiation, deposits localized ionizing damage at the nanoscale surroundings along the ion trajectory with an identical physical dose for each ion hit (3), inducing dominant DNA break directly. These charged particles are highly effective in causing clustered DNA damage due to this unique way of energy deposition, which includes strand breaks, base damage, apurinic/apyrimidine (AP) site, etc., in one or two DNA helical turns (4). In particular, the probability and complexity of clustered DNA damage increase with LET (5). The repair of these damages is slower and inefficient, which is more likely to cause cell death and mutations like deletions, translocations, and chromosomal aberrations in surviving cells (6).
DNA damage response (DDR) factors accumulate to the DNA damage sites, and the scaffold proteins are considered to play a central role in the coordination of DDR factors recruitment/dissociation (7). These scaffold proteins have no enzyme activity in themselves, but they can build a three-dimensional loading platform around the broken DNA strand to stabilize the DNA break and its surrounding environment (8). Then DDR proteins are recruited to DNA damage sites to form local enrichment, allowing more effective repair (9). XRCC1 (x-ray cross complementary factor 1) is a vital scaffold protein in both base excision repair and single-strand break (SSB) repair pathway, providing binding sites for a variety of enzyme active components, such as PARP1/2, DNA polymerase β, DNA ligase IIIα, and so on (10). It is also reported that the XRCC1 repair complex is involved in the activation of microhomology-mediated end joining in irradiated human cells (11).
The structure and function of XRCC1 have been extensively studied over the past decades. The spatiotemporal kinetics of XRCC1 in response to base excision repair and strand break repair is also revealed as a transient process within a time-span of minutes using live-cell imaging (LCI) of beamline microscopies (12, 13, 14) and laser micro-irradiation (15, 16, 17). The dynamics of the DDR and strand break repair process reflect the biochemical reaction process around the DNA damage. At the same time, the reaction rate constant is an important parameter to describe the reaction process, which indicates the relationship between the concentration of reactants (DNA damages and repair factors) and the rate of a chemical reaction (such as phosphorylation, ubiquitinoylation, PARylation, polymerization, etc.) (18). The fast recruitment and dissociation of XRCC1 are based on the biochemical reaction of XRCC1 molecules involved in the DNA repair cascade, and the recruitment and dissociation rate constant represent the XRCC1 interaction at DNA damage. However, the previous LCI works mainly focus on the dynamics of XRCC1 response; how the factors of DDR process influencing XRCC1’s rate constant in vivo using systematic and quantitative analysis have not been reported until now.
In this paper, an LCI system based on the Lanzhou Interdisciplinary Heavy Ion Microbeam (LIHIM) facility was used to investigate the early recruitment and dissociation of XRCC1 at DNA damage induced by high-LET charged particle irradiation in HT1080 cells (expressing XRCC1-RFP). The LIHIM microbeam is established on the TR0 terminal of Lanzhou Heavy Ion Research Facility (HIRF) in the Institute of Modern Physics (IMP, Chinese Academy of Sciences). The microbeam can focus a 80.5 MeV/u 12C6+ beam into microns in the air, and many interdisciplinary experiments such as irradiation of living cells (19) and mice (20), single ion hit of polymers (21), and single event effect study of microelectronics (22) have been successfully carried out. The micro-irradiation and LCI results showed that XRCC1 was recruited to the DNA damage within 1 s, and its recruitment was mainly a PARP-dependent manner. Fractionated radiation induced a time-dependent stimulated XRCC1 recruitment. The G2 phase cells displayed faster recruitment and slower dissociation due to the higher local damage density, but the reaction constants are independent of the number of ion hits. This study demonstrates the fast response, high turnover, and rescue effect of XRCC1 in handling DNA damage induced by high-LET radiation. Our results deepen the understanding of the DDR from the biochemical reaction fundamentals of XRCC1 dynamics in vivo.
Materials and methods
Materials
Materials included Dulbecco’s Modified Eagle’s Medium (DMEM, high glucose) (Gibco, Product Number: 11,965,092); fetal calf serum (FBS) (Sigma-Aldrich, Product Number: F8687); penicillin and streptomycin (Sigma-Aldrich, Product Number: V900929); trypsin-EDTA (0.25%) (Gibco, Product Number: 25,200,056); LCI solution (ThermoFisher, Product Number: A14291DJ); phosphate buffer saline (pH = 7.4, Sigma-Aldrich, Product Number: P5496); thymidine (Sigma-Aldrich, Product Number: T9250); olaparib (Selleck, Product Number: S1060); cell adherent agent (Applygen, Product Number: C1010); and ddH2O (Milli-Q).
Cell culture
The human fibrosarcoma cell line HT1080 expressing XRCC1 tagged RFP was a gift from Dr. Gen Yang (Peking University). Cells were cultured in flasks in DMEM (high glucose) supplemented with 10% heat-inactivated fetal calf serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C and 5% CO2. Cells were seeded on the 170-μm glass bottom (for 2150 MeV Kr irradiation) or 8-μm polypropylene membrane (for 473 MeV Kr and 358 MeV Ni irradiation) of the customized cell plate about 24 h before the irradiation.
Double thymidine (TdR) block was used to arrest cells at the G1/S boundary. Briefly, cells were cultured in high-glucose DMEM complete medium containing 2 mM thymidine for 18 h, and then the cells were cultured in complete medium for 9 h. After another culture in medium with 2 mM thymidine for 18 h, the cells were arrested in the G1/S phase. Afterward, the G1/S cells were incubated in a complete medium for 8 h to prepare the G2 phase cell population.
Olaparib, a selective inhibitor of PARP1/2, was used to stop PARP from working properly. In the experiment, 10 μM olaparib was added to the culture medium 30 min before cell irradiation and LCI.
Irradiation and LCI
Charged particle radiation was carried out at the LIHIM microbeam facility. The LCI system at the microbeam was used for the long-term observation of cells before and after irradiation. The HT1080 cells were irradiated in an LCI solution with 473 MeV and 2150 MeV Krypton ions, 358 MeV Nickel ions. The LET is about 5273 keV/μm, 3637 keV/μm, 3585 keV/μm (at cell entrance) respectively, as calculated using SRIM code with an additional concern for the dish bottom thickness (23).
An inverted fluorescence microscope Olympus IX81 with a Hamamatsu ORCA-FLASH4.0 COMS camera (Model 11,440-22CU) was used to acquire the time-lapse images. The image acquisition was started a few seconds before the irradiation with Micro-manager, the open-source software for microscope control, time-lapse, and multichannel imaging (24). The objective used was a 40× objective (Olympus LUCPLFLN, NA 0.60, WD 3.4 mm, FN 22).
Images and kinetic analysis
ImageJ was employed to analyze the time-lapse images (25). The regions of foci and cell nucleus area were selected as the region of interest (ROI) in each image, and the mean intensity of the ROIs was measured. To correct the photo-bleaching effects, the intensity Ifoci was normalized to the intensity Inucl of the whole cell nucleus for each image. The relative fluorescence intensity of the DNA damage region (foci) is defined as Irel = Ifoci/Inucl, which is used to represent the time-dependent recruitment and dissociation of the XRCC1.
The obtained relative XRCC1 fluorescence showed a dual-exponential curve (Fig. 1 B), which represents a typical consecutive reaction model of biochemical reaction between nuclear XRCC1 and DNA damage. The reaction rate constant is a proportionality constant that indicates the relationship between the molar concentration of reactants and the rate of a chemical reaction. According to the Arrhenius equation, the reaction rate constant depends on the effective reaction collision frequency, the activation energy, and the reaction temperature (26). The dual-exponential function used to fit the XRCC1 fluorescence is defined as follows:
| (1) |
| (2) |
in which I0 is the fluorescence proportional to the XRCC1 reactant concentration (denoted as [X]) at ion hit before the irradiation; the second part in Eq. 2 is the parameter proportional to the first-order reaction product (DNA damage-bound XRCC1), and I1 is also proportional to [X], and the DNA damage reactant concentration (denoted as [D]); k1 represents the recruitment reaction rate constant of XRCC1 molecule accumulating to the damage sites, and k2 is the dissociation reaction rate constant of the bound XRCC1 molecules released at the damage sites; T0 represents the starting time when XRCC1 begins to accumulate at the ion hits. In this paper, T0 is also the starting time of the irradiation because we found that the XRCC1 foci formed immediately after irradiation. A simplified approach has been described in our previous work (19,27).
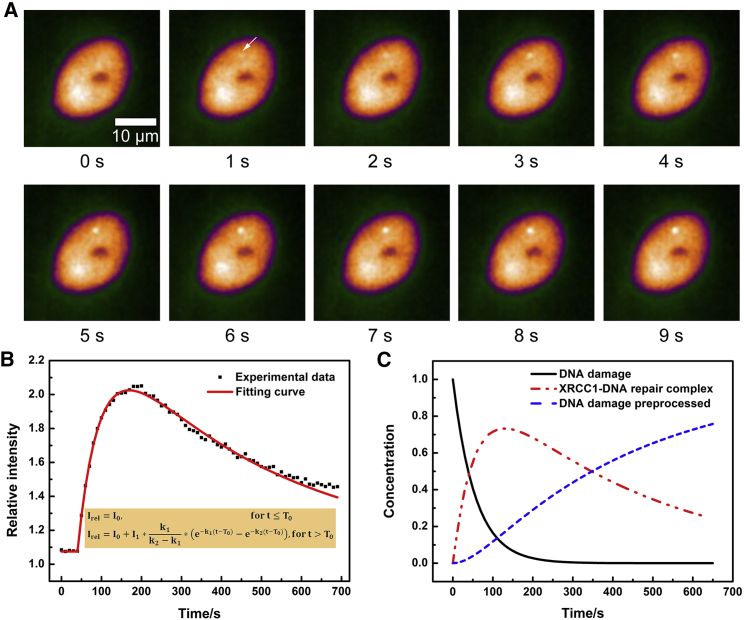
Figure 1.
The early kinetics analysis of XRCC1 based on live-cell imaging. (A) The time-lapse images of HT1080 cells (expressing XRCC1-RFP) irradiated with 2150 MeV Kr ions at 0 s. XRCC1 was recruited to the damage site immediately after irradiation. (B) The typical dual-exponential fitting of the XRCC1 fluorescence at ion hit using the consecutive reaction model. (C) Kinetics simulation of reactant (DNA damage), transient intermediate (the XRCC1-DNA repair complex), and the final product (DNA damage preprocessed) in the consecutive reaction model. The initial concentration of DNA damages (I1) is normalized to 1 at 0 s, and the first and second reaction constant are taken from the experimental data in (B); namely, k1 is 1.8 × 10−2 s−1, and k2 is 2.4 × 10−3 s−1. To see this figure in color, go online.
All data were presented as the mean ± standard error (SEM). The Student’s t-test was used to compare if the means of two sets of data were significantly different from each other. The criterion for statistical significance was taken at P < 0.05.
Results
The quantitative analysis of the ultrafast XRCC1 kinetics
Using LCI of 1-s interval, we first examined the instantaneous behavior of the XRCC1 response to irradiation of 2150 MeV Kr ions. As shown in the time-lapse images of Fig. 1 A, it can be discerned that XRCC1 was recruited to the DNA damage site within 1 s (irradiation performed at 0 s). The XRCC1 accumulation and foci formation took place with no noticeable delay. A typical relative fluorescence intensity curve of the ion hit ROI in longer LCI measurements is shown in Fig. 1 B, which demonstrates the fast kinetics of XRCC1 molecules and its well-fitting of the dual-exponential model (R2 = 0.99, the exponential parameters are described subsequently). XRCC1 molecules accumulated at the damage sites quickly and reached the peak at about 2 min post-IR. The accumulated XRCC1 dissociated rapidly, and the intensity of bound XRCC1 decreased more than 50% from its maximum within 10 min after the irradiation. These data show the ultrafast response and kinetics of XRCC1 to the DNA damage induced by heavy ions, and such transient kinetics cannot be observed with traditional biochemical approaches using cell fixation.
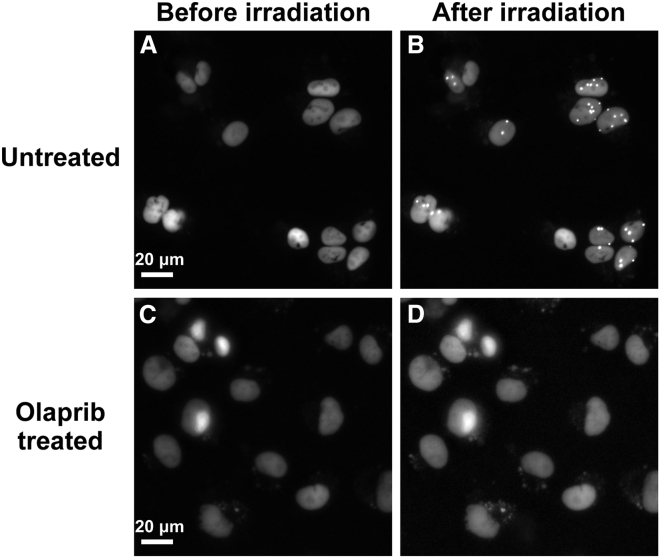
However, when cells were treated with olaparib, a selective inhibitor of PARP1/2, no accumulation of XRCC1 was found at ion hit sites within the 600-s observation, which is in distinct contrast with nontreated control shown in Fig. 2. These data demonstrated that disabling the PARP1/2 impeded the recruitment of XRCC1 after charged particle radiation, which was in good agreement with previous reports (28,29).
Figure 2.
The PARP-dependent recruitment of XRCC1 at the ion hits in HT1080 cells (expressing XRCC1-RFP). The fluorescence images were taken before and after irradiation for untreated cells (A and B) and cells treated with PARP 1/2 inhibitor of 10 μM olaparib (C and D). The cells were irradiated with 473 MeV Kr ions, and no XRCC1 foci were found when cells were treated with olaparib within 600 s observation.
Given that the early kinetics of XRCC1 at the DNA damage induced by ion hit follows dual-exponential fitting, we proposed a consecutive reaction model to study the XRCC1 response quantitatively (30). As shown in Eq. 3, after DNA damage induction by ion irradiation, the reactant (DNA damage) and nuclear XRCC1 form a transient intermediate (the XRCC1-DNA repair complex) in the first step reaction with a reaction rate constant k1. Then the intermediate disassembles to the final product (DNA damage preprocessed, which represents the DNA damage has no more recruitment of XRCC1 and is ready for other DDR steps) and XRCC1 in the second step with a reaction rate constant k2.
| (3) |
It is assumed that abundant nuclear XRCC1 involved the reaction as we observed little change of nuclear XRCC1 concentration at a dose of several ion hits per cell. Using parameters obtained from the dual-exponential fitting, we simulated the kinetics and evolution of the reactant (DNA damage), transient intermediate (the XRCC1-DNA repair complex), and the final product (DNA damage preprocessed) (Fig. 1 C). These data show that the intact DNA damages recruited the scaffold protein XRCC1 to stabilize the DNA break in high-speed kinetics, namely the damage decreases by 51% in 40 s and by 99% in 260 s. On the other hand, the XRCC1-preprocessed DNA damage increased gradually, and about 73% of the damaged breaks are preprocessed with XRCC1 in 10 min, and 95% in 25 min (data not shown). The change of the intermediate product (repair complex) follows the dual-exponential curve of the XRCC1 fluorescence recruited at the ion hits. These data demonstrate that the consecutive reaction model explains the kinetics of XRCC1 response to DNA damage after ion irradiation very well, and the obtained reaction rate constants k1 and k2 are used to study the XRCC1 response quantitatively thereinafter.
The high turnover and rescue effect of XRCC1 in fractionated irradiation
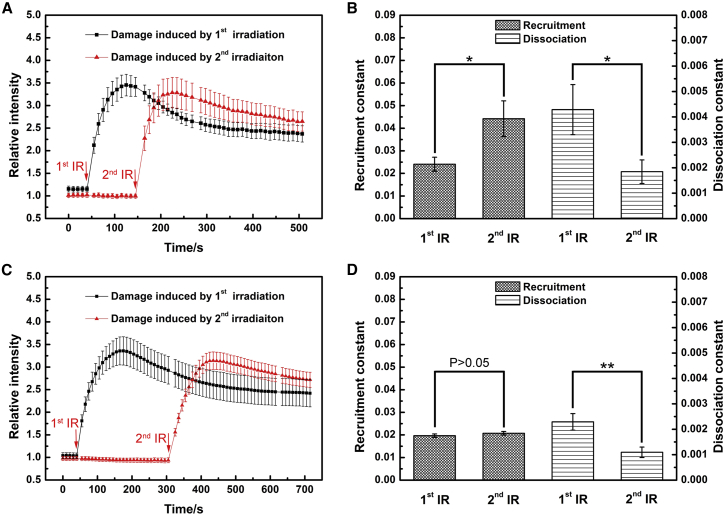
To study the XRCC1 response at the different stages after stimulation of ion irradiation, the same individual cells were exposed to fractionated irradiation of 473 MeV Kr ions with an interval of 100 s first. The 100-s interval was chosen so the XRCC1 molecules bound at the DNA damages of the first ion irradiation reached the maximum when the second ion irradiation was performed (a typical LCI movie is given as Video S1). No significant difference was observed from the intensity maximum and the curve shape between the XRCC1 responses after these two stimulations, as shown in Fig. 3 A. Surprisingly, the calculated recruitment rate constant k1 and dissociation rate constant k2 of the second stimulation were changed obviously from the first stimulation, as shown in Fig. 3 B. The recruitment constant k1 of the first irradiation (2.41 ± 0.31 × 10−2 s−1) is close to that of cells irradiated only once (2.47 ± 0.29 × 10−2 s−1, as shown in Fig. S1), which is about half of that in the second irradiation (4.42 ± 0.79 × 10−2 s−1); but the dissociation k2 of the first irradiation (4.29 ± 0.98 × 10−3 s−1) is about 2.3 times faster than that of the second stimulation (1.84 ± 0.46 × 10−3 s−1). These data suggest that the XRCC1 recruitment becomes faster in the second irradiation, and the dissociation of the first irradiation is stimulated by the second irradiation.
Figure 3.
The kinetics of XRCC1 protein in HT1080 (XRCC1-RFP) cells exposed to fractionated irradiation of 473 MeV Kr ions. The data represent the measured mean value and the standard error of eight cells (A and B, 100-s interval) and 10 cells (C and D, 260-s interval). In the 100-s interval experiment, ion irradiated is 3.5 ± 1.1 (first irradiation) and 2.4 ± 0.6 (second irradiation) per cell. In the 260-s interval experiment, ion irradiated is 2.8 ± 0.5 (first irradiation) and 2.3 ± 0.5 (second irradiation) per cell. ∗P < 0.05, ∗∗P < 0.01. To see this figure in color, go online.
Interestingly, when we extended the interval between the two irradiations (from 100 s to 260 s), it turned out that the difference of recruitment between the twice irradiation became almost unnoticeable (1.97 ± 0.76 × 10−2 s−1 vs. 2.07 ± 0.78 × 10−2 s−1). The dissociation constants of both irradiations decreased (2.29 ± 0.33 × 10−3 s−1 vs. 1.10 ± 0.21 × 10−3 s−1) as shown in Fig. 3 D. The dissociation constant of the second irradiation was nearly identical to that of cells irradiated only once (1.28 ± 0.17 × 10−3 s−1, as shown in Fig. S1), demonstrating that the fractionated irradiation with longer interval has little effect on its dissociation.
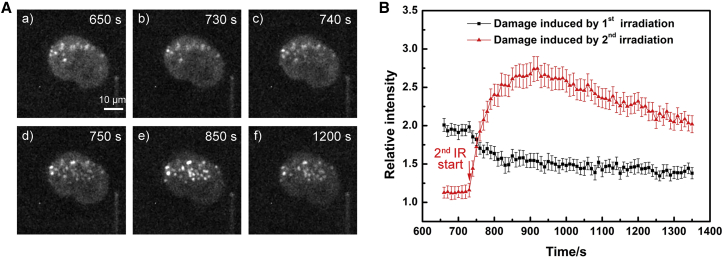
Then we exposed the same cells to fractionated irradiation with a large number of ion hits to examine the dose influence. As shown in Fig. 4, a sharp decrease of XRCC1 fluorescence in the first ion irradiation damage sites occurred immediately after the second ion irradiation, demonstrating the accelerated XRCC1 dissociation from old damage initiated by new damage. Taking the accelerated dissociation from old damage and quicker recruitment of XRCC1 to new damage together, the XRCC1 demonstrates its recyclable capability in DDR and its rescue mechanism to process newly generated damage with high priority, whereas its role at the old repair factory has not been completed (discussed hereafter).
Figure 4.
The accelerated XRCC1 dissociation at old damage was initiated by the second irradiation. The HT1080 cell was exposed to irradiation of 358 MeV Ni ions twice, with an interval of 12 min, 15 and 18 ions respectively. (A) The fluorescence images of the irradiated cell; the bright spots represent the fluorescence of recruited XRCC1 at individual ion hits. (B) The XRCC1 kinetics at the first and second ion irradiation damage. The data represent the mean fluorescence value and its standard error at these two groups of ion hits. To see this figure in color, go online.
XRCC1 recruitment is dependent on cell phase
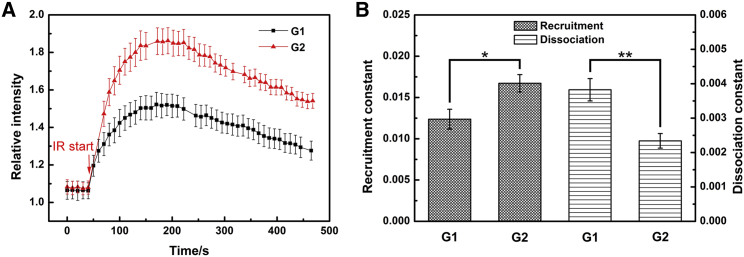
The amount and organization of nuclear DNA are variant and metabolic through the cell cycle, impacting the DNA damages response and repair processes (31,32). To determine the influence of the cell cycle on the early XRCC1 response in the DDR process, HT1080 cells in the G1 phase and G2 phase were irradiated with 2150 MeV ions, and the XRCC1 kinetics was investigated. As shown in Fig. 5 A, the XRCC1 kinetics in both groups followed the dual-exponential curve, but the XRCC1 maximum recruited to the damage sites in G2 phase cells (90% increment) was almost twice that in G1 phase cells (50% increment). The recruitment in G2 phase cells was also faster in G1 phase cells (1.67 ± 0.11 × 10−2 s−1 vs. 1.24 ± 0.12 × 10−2 s−1), but the dissociation was slower (2.34 ± 0.21 × 10−3 s−1 vs. 3.83 ± 0.32 × 10−3 s−1), and both their recruitment and dissociation rate constants are significantly different in the t-test, as shown in Fig. 5 B. The obtained normalized parameter I1 in Eq. 2, which is proportional to the DNA damage concentration in the consecutive reaction model, was significantly higher in G2 cells (1.1) than that in G1 cells (0.7). These data show that the density of DNA damage per ion hit in G2 cells is higher than that in G1 cells, and the XRCC1 kinetics is dependent on the cell cycle.
Figure 5.
The XRCC1 kinetics in HT1080 cells of G1 and G2 cell cycles irradiated with 2150 MeV Kr ions. (A) The relative fluorescence intensity curves of XRCC1 before and after irradiation. (B) The recruitment and dissociation constant of XRCC1 in G1 and G2 cells after irradiation. The data represent the mean value and its standard error of 10 G1 cells and 10 G2 cells; ion irradiated in both samples is 3.6 ± 0.7 ion per cell. ∗P < 0.05, ∗∗P < 0.01. To see this figure in color, go online.
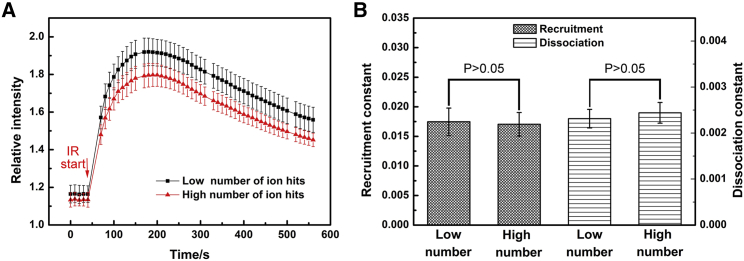
Finally, we examined the effect of the number of irradiated ions on the kinetics of XRCC1. The HT1080 cells of G2 phase were irradiated with 2150 MeV Kr ions with a different number of ions, and the cell synchronization could help to exclude the influence of the cell cycle. The dose of a single Kr ion on a cell is about 1.44 Gy (nuclear area assumed as 400 μm2). When the irradiated ions were of low numbers (1–2 ions for each cell), the relative fluorescence intensity of damage sites was higher than that caused by irradiation with multiple ions, as shown in Fig. 6 A, demonstrating more XRCC1 molecules were recruited there. However, the recruitment and the dissociation rate constants of XRCC1 were not significantly different (P > 0.05), as shown in Fig. 6 B.
Figure 6.
The XRCC1 kinetics in HT1080 cells of the G2 phase. A) The relative fluorescence intensity curves of XRCC1 before and after irradiation. B) The recruitment and dissociation constant of XRCC1 in cells irradiated with a different number of ions. Cells were irradiated with 2150 MeV Kr ions; the low number of ion hits is 1.8 ± 0.2 ion per cell (2.52 Gy), and the high number of ion hits is 5.3 ± 1.8 ion per cell (7.56 Gy). The data represent the mean value and its standard error of eight cells respectively. There is no significant difference in the recruitment and dissociation reaction constant between the two groups (P > 0.05). To see this figure in color, go online.
Discussion
The early spatiotemporal dynamics analysis based on LCI has greatly improved the knowledge on the hierarchical organization and process of the DDR. Upon DNA strand damage, XRCC1 provides an important scaffolding platform to recruit, activate, or regulate the strand break repair factors. Thanks to the fast LCI at the heavy ion microbeam facility, the XRCC1 response kinetics to the localized DNA damage at each ion hit was measured. Since most biological processes, including DDR, result from the biochemical reaction inside the cellular organelle, we introduced the consecutive reaction model and the reaction rate constants to describe the dynamics of XRCC1 in the DDR process after ion irradiation. The model well explained the behavior of XRCC1 at the damage site of ion hit, and the reaction rate constant k1 and k2 of the two-step reaction elucidated the recruitment and dissociation of XRCC1 in different conditions quantitatively. Compared with previous reports focused on the observations of XRCC1 change or its recruitment half time (12,33), this work deepened the understanding of the DDR from the biochemical reaction fundamentals of XRCC1 dynamics in vivo.
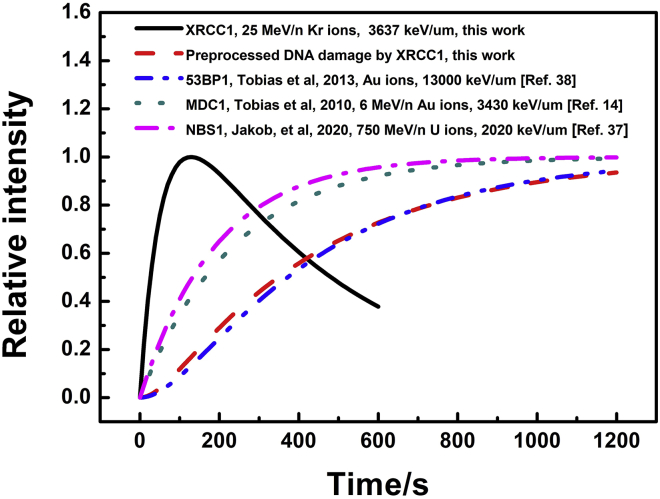
We found that the response of XRCC1 is extremely fast (obvious increase within 1 s and accumulation maximum at ∼120 s) after high-LET irradiation through Figs. 1 and 3, 4, 5, and 6. The inhibition of PARP prevented the recruitment of XRCC1 at the ion hit (Fig. 2), which verified that recruitment of XRCC1 is mainly PARP dependent in response to DNA strand breaks after ion irradiation, and no apparent oxidative base damage appeared at the ion hit region, which can induce PARP-independent XRCC1 recruitment (34). Upon DNA strand break, the nuclear PARP recognizes and binds to nicked DNA, then PARP is rapidly activated by the formation of chains of poly-ADP-ribose (PAR) onto itself (29). At the same time, XRCC1 is recruited to the PARylated PARP1 by its PAR-binding BRCT domain and helps DNA break repair (10). The participation of PARP1 possesses a transient kinetics as the PARylation of PARP1 reduces its binding affinity with DNA subsequently and induces the PARP1 dissociation from bound DNA (35). Our observation on XRCC1 kinetics is similar to the quick and transient process of upstream binding/activation of PARP at DNA strand breaks (36). Although it is well known that XRCC1 is mainly involved in SSB repair and its role in DNA double-strand break (DSB) repair is still not fully understood, we found that the kinetics of XRCC1-processed DNA damage (the final product of the consecutive reaction) is comparable to the kinetics of the DSB repair factors MDC1 (14), NBS1 (37), and 53BP1 (38). The coincidental match of the XRCC1-preprocessed DNA damage and 53BP1 kinetics, as shown in Fig. 7, indicates that the PARP-XRCC1 might be more involved in the repair of the DSB at the damage sites induced by charged particle radiation (29).
Figure 7.
Kinetics of typical proteins involved in SSB and DSB repair after heavy ion irradiation. XRCC1 is an SSB repair protein, and 53BP1, MDC1, and NBS1 are involved in the repair of DSB. To see this figure in color, go online.
Interestingly, exposure to an extremely high dose of fractionated irradiation induced visible accelerated dissociation from the first damage initiated by the second irradiation (Fig. 4), and this accelerated dissociation is verified by the higher dissociation k2 of the first damage in the short-interval fractionated irradiation. At the same time, the XRCC1 recruitment k1 at the second damages is faster than (about two-fold) that at the first damages in the same cells of the short-interval fractionated irradiation (Fig. 3 B). The significant increase of k1 indicates a new form of XRCC1 molecule is participating in DDR response, and this promotion effect is very transient and varnished in the extended interval experiment (Fig. 3 D). Considering all the aforementioned results, it is reasonable to conclude that the freshly dissociated XRCC1 molecules are immediately reusable and participate in the DDR response with a higher recruitment rate constant. Some dissociated XRCC1 molecules may possess a particular state with lower recruitment energy (e.g., partly ADP-ribosylated/phosphorylated) within seconds after dissociation. From another point of view, the accelerated dissociation of XRCC1 from the old damage where XRCC1 is still interacting with the repair factory and its re-interaction at new damage with a prepared form (with higher recruitment k2) reveal the rescue capability of XRCC1 in the DDR response. The high turnover of XRCC1 ensures its dynamic and recyclable functioning on demand and might be responsible for the high efficiency.
Although the XRCC1 kinetic is independent of ion hits and remains similar in the long-interval fractionated irradiation (Figs. 5 and 3), we found that the XRCC1 kinetics is different in synchronized G1-phase and G2-phase cells; namely, the XRCC1 molecules are recruited faster and dissociated slower in G2 phase cells than G1 cells (Fig. 5). Consistent with the fact that the DNA content in G2 is duplex, almost doubled DNA damage concentration in G2 cells was obtained compared with G1 cells using the identical ion irradiation. Probably the demonstrated higher affinity of DNA damage and XRCC1 in G2 cells (higher k1 and low k2) results from the high DNA damage density at the ion hits of G2 cells, as the diffusion of XRCC1 molecule is limited in a complicated nuclear environment in vivo instead of the ideal reaction condition in vitro. The competition of the limited XRCC1 molecule at higher DNA damage density can subsequently result in faster recruitment and slower dissociation. However, other factors might also play a role behind the XRCC1 kinetic dependence on the cell cycle, such as the evolution of chromatin structure and nuclear factors that can facilitate XRCC1 response.
Conclusion
The effective repair of DNA damages requires the coordination of multiple repair factors. Using the online LCI at the heavy ion microbeam facility, we found that the response of XRCC1 is extremely fast at the localized DNA damage of heavy ion hits. The consecutive reaction model well explained the kinetics of XRCC1, and with the quantitative analysis of the recruitment and dissociation rate constant, we found the dependence of XRCC1 response on the cell cycle. At higher DNA damage concentration, the XRCC1 reaction model is limited in a complicated nuclear environment. Especially, using fractionated irradiation of the same cells, we found that the dissociated XRCC1 is immediately recycled, and even with higher efficiency within a short time, indicating its rescue capability in the DDR response. In summary, our work discovered XRCC1’s new rescue mechanism in DDR and its high flexibility with the quantitative analysis of the XRCC1 kinetics.
Author contributions
G.D. and W.L. designed the experiments, R.W., W.L., and C.S. completed the experiments and analyzed the data, J.G., J.Z., G.M., H.M., and L.Z. provided technical support, and G.D. and W.L. interpreted the data and wrote the manuscript.
Acknowledgments
This work is supported by the National Key R&D Program of China (2021YFA1601400), China; by the National Natural Science Foundation of China (11975283 and U1632271), China; and by Heavy Ion Research Facility in Lanzhou (HIRFL). The authors acknowledge Professor Gen Yang (Peking University) for providing cells in the experiments, and they thank colleagues in the accelerator department for delivering the beam.
Editor: Gijs Wuite.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.03.011.
Supporting material
material
References
- 1.Giglia-Mari G., Zotter A., Vermeulen W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zirkle R.E., Tobias C.A. Effects of ploidy and linear energy transfer on radiobiological survival curves. Arch. Biochem. Biophys. 1953;47:282–306. doi: 10.1016/0003-9861(53)90467-6. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Zhao J., et al. Duan H. Sub-5 nm lithography with single GeV heavy ions using inorganic resist. Nano Lett. 2021;21:2390–2396. doi: 10.1021/acs.nanolett.0c04304. [DOI] [PubMed] [Google Scholar]
- 4.Mavragani I.V., Nikitaki Z., et al. Georgakilas A.G. Ionizing radiation and complex DNA damage: from prediction to detection challenges and biological significance. Cancers. 2019;11:29. doi: 10.3390/cancers11111789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill M.A. Radiation track structure: how the spatial distribution of energy deposition drives biological response. Clin. Oncol. 2020;32:75–83. doi: 10.1016/j.clon.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Sage E., Shikazono N. Radiation-induced clustered DNA lesions: repair and mutagenesis. Free Radic. Biol. Med. 2017;107:125–135. doi: 10.1016/j.freeradbiomed.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochs F., Karemore G., et al. Lukas C. Stabilization of chromatin topology safeguards genome integrity. Nature. 2019;574:571–574. doi: 10.1038/s41586-019-1659-4. [DOI] [PubMed] [Google Scholar]
- 9.London R.E. XRCC1-strategies for coordinating and assembling a versatile DNA damage response. DNA Repair. 2020;93:5. doi: 10.1016/j.dnarep.2020.102917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldecott K.W. XRCC1 protein; form and function. DNA Repair. 2019;81:102664. doi: 10.1016/j.dnarep.2019.102664. [DOI] [PubMed] [Google Scholar]
- 11.Dutta A., Eckelmann B., et al. Mitra S. Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Res. 2017;45:2585–2599. doi: 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bourret S., Vianna F., et al. Barberet P. Fluorescence time-lapse imaging of single cells targeted with a focused scanning charged-particle microbeam. Nucl. Instrum. Methods Phys. Res. B. 2014;325:27–34. [Google Scholar]
- 13.Jakob B., Splinter J., et al. Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobias F., Durante M., et al. Jakob B. Spatiotemporal analysis of DNA repair using charged particle radiation. Mutat. Res. 2010;704:54–60. doi: 10.1016/j.mrrev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Mortusewicz O., Ame J.C., et al. Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mok M.C.Y., Campalans A., et al. Junop M.S. Identification of an XRCC1 DNA binding activity essential for retention at sites of DNA damage. Sci. Rep. 2019;9:13. doi: 10.1038/s41598-019-39543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menoni H., Wienholz F., et al. Vermeulen W. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018;46:7747–7756. doi: 10.1093/nar/gky579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neves S.R., Iyengar R. Models of spatially restricted biochemical reaction systems. J. Biol. Chem. 2009;284:5445–5449. doi: 10.1074/jbc.R800058200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo N., Du G.H., et al. Wei J.Z. Live cell imaging combined with high-energy single-ion microbeam. Rev. Sci. Instrum. 2016;87:5. doi: 10.1063/1.4943257. [DOI] [PubMed] [Google Scholar]
- 20.Du G., Guo J., et al. Li H. The first interdisciplinary experiments at the IMP high energy microbeam. Nucl. Instrum. Methods Phys. Res. B. 2015;348:18–22. [Google Scholar]
- 21.Li Y.N., Du G.H., et al. Liu W.J. Electrical field regulation of ion transport in polyethylene terephthalate nanochannels. ACS Appl. Mater. Interfaces. 2019;11:38055–38060. doi: 10.1021/acsami.9b13088. [DOI] [PubMed] [Google Scholar]
- 22.Guo J.L., Du G.H., et al. Ma S.Y. Development of single-event-effects analysis system at the IMP microbeam facility. Nucl. Instrum. Methods Phys. Res. B. 2017;404:250–253. [Google Scholar]
- 23.Ziegler J.F. In: Ion Implantation Science and Technology. Ziegler J.F., editor. Academic Press; 1984. The stopping and range of ions in solids; pp. 51–108. [Google Scholar]
- 24.Edelstein A., Amodaj N., et al. Stuurman N. Computer control of microscopes using Manager. Curr. Protoc. Mol. Biol. 2010;14 doi: 10.1002/0471142727.mb1420s92. Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Northrup S.H., Hynes J.T. Reaction-rate constants and rate kernels. Chem. Phys. Lett. 1978;54:248–252. [Google Scholar]
- 27.Liu W.J., Du G.H., et al. Li X.Y. Influence of the environment and phototoxicity of the live cell imaging system at IMP microbeam facility. Nucl. Instrum. Methods Phys. Res. B. 2017;404:125–130. [Google Scholar]
- 28.Masson M., Niedergang C., et al. de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaudhuri A.R., Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ball D.W. Kinetics of consecutive reactions: first reaction, first-order; second reaction, zeroth-order. J. Chem. Educ. 1998;75:917–919. [Google Scholar]
- 31.Blackford A.N., Stucki M. How cells respond to DNA breaks in mitosis. Trends Biochem. Sci. 2020;45:321–331. doi: 10.1016/j.tibs.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Ambrosio S., Di Palo G., et al. Majello B. Cell cycle-dependent resolution of DNA double-strand breaks. Oncotarget. 2016;7:4949–4960. doi: 10.18632/oncotarget.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aleksandrov R., Dotchev A., et al. Stoynov S.S. Protein dynamics in complex DNA lesions. Mol. Cell. 2018;69:1046–1061.e5. doi: 10.1016/j.molcel.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 34.Campalans A., Kortulewski T., et al. Radicella J.P. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013;41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleschke J.M., Kleczkowska H.E., et al. Althaus F.R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 36.Pascal J.M. The comings and goings of PARP-1 in response to DNA damage. DNA Repair. 2018;71:177–182. doi: 10.1016/j.dnarep.2018.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakob B., Dubiak-Szepietowska M., et al. Taucher-Scholz G. Differential repair protein recruitment at sites of clustered and isolated DNA double-strand breaks produced by high-energy heavy ions. Sci. Rep. 2020;10:12. doi: 10.1038/s41598-020-58084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobias F., Lob D., et al. Jakob B. Spatiotemporal dynamics of early DNA damage response proteins on complex DNA lesions. PLoS One. 2013;8:14. doi: 10.1371/journal.pone.0057953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
material