Abstract
Disease-associated variants (DAVs) are commonly considered either through a genomic lens that describes variant function at the DNA level, or at the protein function level if the variant is translated. Although the genomic and proteomic effects of variation are well-characterized, genetic variants disrupting post-transcriptional regulation is another mechanism of disease that remains understudied. Specific RNA sequence motifs mediate post-transcriptional regulation both in the nucleus and cytoplasm of eukaryotic cells, often by binding to RNA binding proteins or other RNAs. However, many DAVs map far from these motifs, which suggests deeper layers of post-transcriptional mechanistic control. Here, we consider a transcriptomic framework to outline the importance of post-transcriptional regulation as a mechanism of disease-causing single-nucleotide variation in the human genome. We first describe the composition of the human transcriptome and the importance of abundant yet overlooked components such as introns and Untranslated Regions (UTRs) of messenger RNAs (mRNAs). We present an analysis of Human Gene Mutation Database variants mapping to mRNAs and examine the distribution of causative disease-associated variation across the transcriptome. Although our analysis confirms the importance of post-transcriptional regulatory motifs, a majority of DAVs do not directly map to known regulatory motifs. Therefore, we review evidence that regions outside these well-characterized motifs can regulate function by RNA structure-mediated mechanisms in all four elements of an mRNA: exons, introns, 5’ and 3’ UTRs. To this end, we review published examples of riboSNitches, which are single-nucleotide variants that result in a change in RNA structure that is causative of the disease phenotype. In this review we present the current state of knowledge of how DAVs act at the transcriptome level, both through altering post-transcriptional regulatory motifs and by the effects of RNA structure.
Keywords: riboSNitch, Single Nucleotide Variant, messenger RNA, structure, rare variant
Introduction
Disease-associated human genetic variation has been identified by numerous genome-wide association studies (Visscher et al. 2017). Typically, these variants are identified and characterized through a genomic perspective at the DNA level. When identifying disease-associated variation, investigators focus most often on exonic variants that change the protein sequence and thereby impair protein function. However, the majority of disease-associated variation occurs in noncoding and intergenic regions of genes (Telenti et al. 2016). These mutations do not change the protein sequence and therefore it is often difficult to determine if they are causative of the disease phenotype. As an intermediate messenger, RNA has a key regulatory function in determining protein expression by a variety of mechanisms, including alternative splicing, RNA stability, and RNA localization (Solem et al. 2015). In this review, we focus on the RNA-based molecular mechanisms of causative disease-associated variation within the transcriptome.
When considering the transcriptome, one should consider the entire lifespan of RNA transcripts, from nascent transcription to translation. RNA is first transcribed from DNA in the nucleus as precursor messenger RNA, or pre-mRNA. Pre-mRNA contains numerous components encoded in the gene, including the coding exons, intervening introns, and both 5’ and 3’ untranslated regions (UTRs) (Fig 1a). Of these components, typically only the exons will be translated into protein, although the other components still provide important regulatory functions. For example, a 5’ cap and a 3’ polyadenylated tail are added co-transcriptionally to the ends of the transcript; these modifications are required for mature messenger RNA (mRNA) stability in the cytoplasm (Wilusz et al. 2001). Furthermore, the 5’ cap recruits the ribosome for cap-dependent translation, which is the main means of translation (Leppek et al. 2018). The 5’ and 3’ UTR both contain key post-transcriptional regulatory motifs important for translational control, despite only rarely being translated themselves (Leppek et al. 2018). The 5’ UTR begins at the first nucleotide of the transcript and ends at the start codon of the first exon, whereas the 3’ UTR begins immediately following the stop codon in the final exon and continues through to the end of the transcript (Leppek et al. 2018; Steri et al. 2018). It is important to note that in most genes, the start and stop codon do not align with exon/exon boundaries, and as such the identification of the open reading frame in a messenger RNA requires a start and stop codon in frame in the mature mRNA sequence. Because the noncoding components of RNA serve as regulatory sites, variation in these parts of the transcriptome is likely relevant for disease states, even though noncoding sequences are not translated into the final protein sequence.
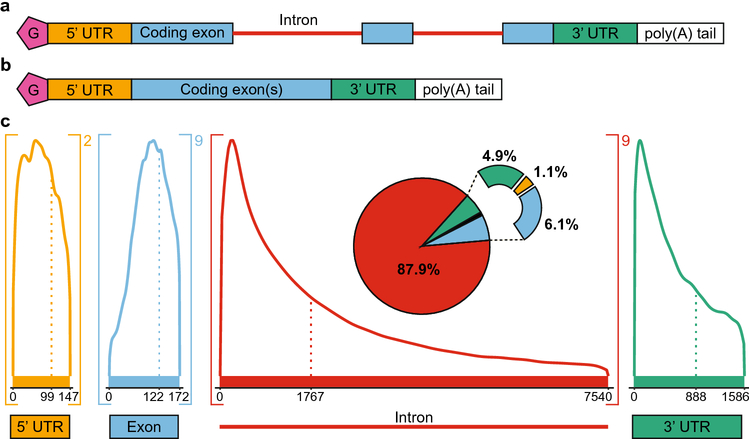
Fig 1.
RNA composition and length distributions. a. Precursor mRNA (pre-mRNA) is transcribed from DNA. Pre-mRNA typically contains a 5’ cap (pink, G), a 5’ untranslated region (UTR, orange), coding exons (blue), intervening introns (red), a 3’ untranslated region (green), and a polyadenylated tail (poly(A) tail, white). b. RNA splicing creates mature mRNA by removing introns. c. Length distributions of the components of the transcriptome. The dotted line represents the median length of each component, whereas the maximum value shown on the graph denotes the average length. Values greater than the average are not shown due to the extremely long outliers that skew the distribution. The number outside brackets indicates the average number of times a component occurs in a transcript. The pie chart breaks down the relative amount of sequence space of each component, accounting for length.
Prior to its export to the cytoplasm where translation occurs, a fully mature mRNA must undergo splicing to remove introns (Fig 1b). Introns are noncoding sequences that interrupt exons and must be removed prior to translation (Scotti and Swanson 2016). The spliceosome, a large macromolecular ribonucleoprotein complex, removes introns based on splice site sequence recognition at locations where intron meets exon (Lee and Rio 2015). In the absence of correct splicing, introns are retained and the transcript undergoes nonsense-mediated decay, which degrades the RNA and prevents its translation (Kurosaki et al. 2019). However, incorrect splicing can cause disease, and many disease-associated single-nucleotide variants (SNVs) have been implicated in disrupting splicing (Scotti and Swanson 2016). Splicing is controlled by an interplay between the spliceosome and RNA sequence elements in the transcript. Variation in sequence and even RNA structure around a splice site can lead to improper splicing and disease phenotypes (Xu et al. 2021). Thus, despite the lack of direct coding functionality, introns are still important for proper gene expression, and variants that alter splicing can lead to disease.
Although introns are removed prior to mRNA translation, alternative splicing of introns has an important function in determining what proteins are produced. Alternative splicing produces different isoforms of a protein by selective retention or skipping of particular exons based on splice sites that vary somewhat in sequence, which greatly diversifies the coding potential of the human genome. (Lee and Rio 2015). Interestingly, the only pre-mRNA component that does not undergo alternative splicing is the 3’ UTR because of how exon junction complexes are recognized in relation to termination codons during nonsense-mediated decay (Kurosaki et al. 2019). As mentioned above, nonsense-mediated decay protects the cell from incorrectly spliced transcripts, and is an important component of quality control for a healthy cell. However, alternative splicing, in addition to diversifying protein-coding capacity, creates more opportunities for splicing to go awry. Indeed, some transcripts exhibit a mixed ratio of splice isoforms that can be shifted to a single isoform by SNVs, which results in disease (Niblock and Gallo 2012; Scotti and Swanson 2016).
To understand disease-associated variation, it is important to consider the composition of RNA transcripts. Because of alternative splicing, the average human pre-mRNA transcript contains two 5’ UTR segments, 9 exons, 9 introns, and a single 3’ UTR (Fig 1c). Introns take up an incredible amount of sequence space in the human transcriptome because of both their length and abundance. The median length of an intron is 1767 nucleotides, and the average length is 7540 nucleotides (Fig 1c). Such a dramatically greater average length compared to the median length indicates that some extremely long introns skew the distribution. In contrast, the median length of transcribed exons is 122 nucleotides, and the average length is 172 nucleotides (Fig 1c). By length, most pre-mRNA transcripts are composed of 87.9% intron RNA, with smaller proportions being exon (6.1%), 3’ UTR (4.9%), and 5’ UTR (1.1%) sequences (Fig 1c). Therefore, although the general tendency of investigators is to focus on exonic coding sequences, a large amount of the genome is transcribed that is noncoding but contains regulatory potential.
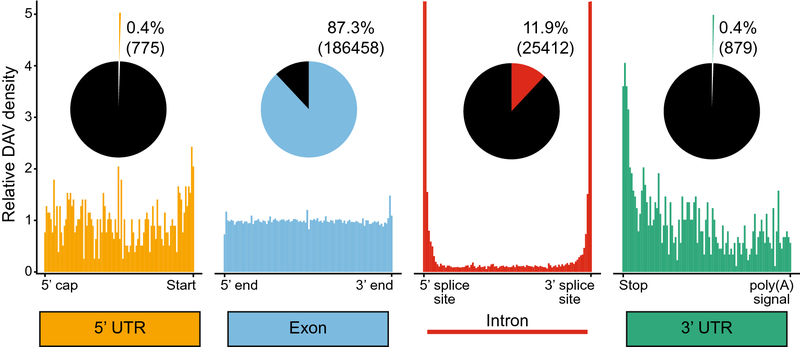
Most SNVs have little to no observable phenotype, however some variants in both coding and noncoding regions of transcripts do cause disease (Fig 2). In this review, we analyze causative disease-associated variants (DAVs) using data from the Human Gene Mutation Database (HGMD) (Stenson et al. 2003, 2020). From these data, we observe that although most DAVs are found in exons (87.3%), thousands of DAVs are in noncoding components of mRNA (Fig 2 and Table 1). Nearly all exonic DAVs (98.5%) are nonsynonymous mutations, which change the final protein sequence and demonstrates the importance of protein coding functionality for human health. However, the remaining 2,708 (1.5%) exonic DAVs are synonymous, which cause disease without altering the protein sequence and suggest a noncoding regulatory disruption. In addition, HGMD is curated from literature focusing on mutations of clinical significance, which most often are identified by a gene-targeted or whole exome sequencing approach, leading to an overrepresentation of exonic sequences and therefore exonic DAVs (Meienberg et al. 2016). Another way to examine DAV abundance is to account for the total length of each component in the genome (Table 1). DAV density is by far the highest among nonsynonymous exonic DAVs, with approximately 5 DAVs per every thousand nucleotides (5.16 × 10−3), whereas synonymous DAVs are present at a much lower frequency (7.61 × 10−5). DAVs in introns and the 3’ UTR occur at similar frequencies to synonymous DAVs (1.86 × 10−5 and 2.13 × 10−5 respectively), however DAVs in the 5’ UTR are present at slightly higher frequencies (1.09 × 10−4).
Fig 2.
Causative disease-associated variant (DAV) distributions from mutations in the Human Gene Mutation Database (HGMD). The relative abundance of DAVs is shown positionally relative to the length of each component, with the ends labeled. The fractional abundance of DAVs for each component is shown in a pie chart, with the total number of mutants for each in parentheses. Note the intron DAV density graph is cut off at 5 for visualization purposes; intron DAV density is nearly 40 near the splice sites (Fig 6a).
Table 1:
DAV density data by component
| DAVs | Total Length | DAVs/nucleotide | |
|---|---|---|---|
| 5’ UTR | 775 | 7117500 | 1.09 × 10−4 |
| Exon (total) | 186458 | 35588738 | 5.24 × 10−3 |
| Exon (nonsynonymous) | 183750 | 35588738 | 5.16 × 10−3 |
| Exon (synonymous) | 2708 | 35588738 | 7.61 × 10−5 |
| Intron | 25412 | 1368110257 | 1.86 × 10−5 |
| 3’ UTR | 879 | 41236033 | 2.13 × 10−5 |
We observe more interesting trends when comparing the positional distribution of DAVs across the length of each component. Exonic DAVs are uniformly distributed across the length of exons, whereas other noncoding sequence elements are enriched for DAVs around post-transcriptional regulatory sequence motifs (Fig 2). For example, DAVs in intronic sequences are dramatically more abundant at each end of the intron, which correspond to splice sites (Fig 2). Similar trends are observed in the 3’ UTR near the stop codon and the 5’ UTR near the Kozak sequence, the site for translation initiation (Kozak 1987) (Fig 2). Mutations near these key regulatory regions are likely to contribute disproportionately to disease states by disrupting key RNA sequence information; however, there are still a majority of disease variants that map far from these key regions that cause disease through RNA-mediated mechanisms.
One disease relevant cis-regulatory mechanism is RNA-binding protein (RBP) binding, which can be specified based on sequence or RNA structure (Rouault 2006; Glisovic et al. 2008; Solem et al. 2015). RBPs regulate gene expression post-transcriptionally through a wide variety of mechanisms, including modulating RNA stability, splicing, and even translation (Glisovic et al. 2008). RNA structure can mediate RBP binding, such that even variants outside the binding site that disrupt structure can disrupt RBP binding (Rouault 2006; Solem et al. 2015). The software RADAR incorporates several levels of data, including eCLIP, Bind-n-Seq, and RNA-Seq experiments after knockdown to identify the impact of variants in RBP binding sites (Zhang et al. 2020). Based on population level polymorphism data, RBP binding site sequences and structures are often conserved, as demonstrated by an enrichment of rare variants (Zhang et al. 2020). Enrichment for rare variants is indicative of purifying selection on these regions as that implies common variants are selected against and the variation is disruptive, potentially causing disease (Khurana et al. 2013). In coding regions, 88.4% of RBP binding sites are enriched for rare variants, whereas in noncoding regions 93.8% of binding sites are enriched for rare variants, suggesting that these RBP binding sites are functionally important (Zhang et al. 2020). Additionally, regions with multiple RBP binding sites are more enriched for rare variants, indicating that RBP hubs are under greater selective pressure (Zhang et al. 2020). Therefore, RBP binding sites are an important aspect of DAV function in both coding and noncoding regions of the genome, and a prime example of how DAVs can act at the transcriptome level.
Another example where noncoding DAVs can be causal is through microRNA (miRNA) mediated regulation. miRNAs are short (~22 nucleotide) RNAs that regulate gene expression by guiding RNA silencing. miRNAs target miRNA binding sites typically located in the 3’ UTR of a transcript (Gebert and MacRae 2019). miRNA binding guides the miRNA-induced silencing complex to translationally repress or degrade the target RNA transcript (Gebert and MacRae 2019). The sequence dependence of miRNA binding provides the opportunity for DAVs to disrupt function, as highlighted in several databases that describe the intersection of SNVs and miRNAs (Fehlmann et al. 2019; Liu et al. 2021). When examining if DAVs are enriched in miRNA binding sites as catalogued in the TargetScanHuman database (Agarwal et al. 2015), we observed 46 DAVs in miRNA binding sites in the 3’ UTR, compared to 879 DAVs across the entire 3’ UTR (Table 2). Comparing the ratio of DAVs in the miRNA binding sites to the total DAVs in the 3’ UTR (0.052) against the corresponding ratio of common SNPs (0.0096) revealed a significant 5.4-fold enrichment for DAVs (chi-squared test, p < 2.2 × 10−16). Therefore, both RBP and miRNA binding sites are enriched for DAVs, suggesting that disease-associated variation in these sites is an important component of disease etiology in humans.
Table 2:
3’ UTR miRNA DAV enrichment analysis
| 3’ UTR miRNA binding sites | 3’ UTR total | |
|---|---|---|
| Number of DAVs | 46 | 879 |
| Number of Common SNPs | 4232 | 441142 |
Although both RBP and miRNA binding sites are key motif elements of post-transcriptional regulation, RNA structure throughout the entire transcriptome is also essential for many cellular processes (Wan et al. 2011, 2014). However, SNVs throughout the transcriptome can alter these structures, disrupting functions such as RBP binding or splicing, and resulting in disease (i.e., DAVs) (Wan et al. 2014). One recent analysis demonstrated how SNVs predicted to change structure are less abundant in the population proportional to the degree of structural change (Gaither et al. 2021). The riboSNitch is an RNA structure-based mechanism of disease-associated genetic variation. In a riboSNitch, a single-nucleotide change shifts the RNA structural ensemble; this shift affects gene function and results in a disease phenotype (Halvorsen et al. 2010). RiboSNitches have been identified by analyzing RNA folding with various computational methods, such as SNPfold (Halvorsen et al. 2010), RNAsnp (Sabarinathan et al. 2013), remuRNA (Salari et al. 2013), and recently Riprap (Lin et al. 2020). It is also essential to verify putative riboSNitches with structural probing approaches, such as parallel analysis of RNA structure (PARS) (Kertesz et al. 2010; Wan et al. 2014) and selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) (Wilkinson et al. 2006; Siegfried et al. 2014). Recently, Lin et al. published a database of riboSNitches (RiboSNitchDB) (Lin et al. 2020) that reanalyzed a previously analyzed dataset (Corley et al. 2015). RiboSnitchDB lists 1058 putative riboSNitches and further describes their degree of validation. Of these putative riboSNitches, 63 are considered “validated” by allele specific mapping between parents and children, whereas 11 are considered “probed”, which is the highest level of certainty whereby the structures have been experimentally characterized by chemical probing (Lin et al. 2020). Therefore, if one has a SNV that is suspected to be a riboSNitch, the first step is to predict if there are structural differences between the wild type and the variant computationally using one of the tools described above, such as Riprap (Lin et al. 2020). To experimentally validate a riboSNitch, structural differences are most often validated experimentally using chemical probing, for example with SHAPE-MaP (Wilkinson et al. 2006; Siegfried et al. 2014). However, identifying the exact regulatory function of a riboSNitch requires customized experimental design based on the hypothesized mechanism and the gene being regulated or the disease in question. Unfortunately, there is no single high-throughput experiment that can be used to functionally validate a riboSNitch and as such their ultimate validation still requires significant experimental work. Here, we describe patterns of causative disease-associated variation from SNVs in the HGMD (Stenson et al. 2003, 2020) for each component of an RNA transcript. In addition to describing positional hotspots for disease associated variation, we highlight specific examples of disease-causing riboSNitches for each component of an mRNA and the molecular mechanism of the associated disease phenotype.
Disease-associated variation in the 5’ UTR and the FTL riboSNitch
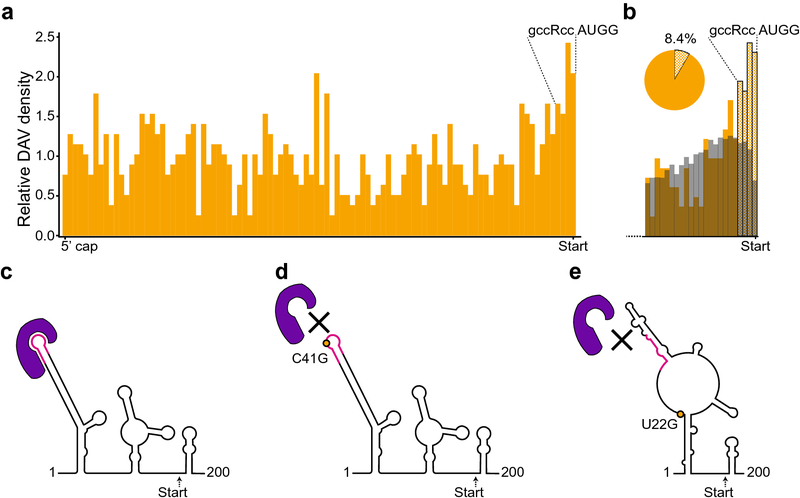
The 5’ UTR has an important function in translation initiation and translational control; thus, DAVs in this region likely disrupt translation (Leppek et al. 2018). Consistent with this expectation, we observed an increase in DAVs near the start codon (Fig 3a). In particular, the 3’ end of the 5’ UTR contains part of the Kozak consensus sequence that guides the translation machinery to the AUG start codon, the site of translation initiation (Kozak 1987). Therefore, the Kozak sequence is a post-transcriptional regulatory motif in which we observed a higher density of DAVs (Fig 3a and b). To more carefully examine this enrichment, we compared the DAV distribution around the 3’ end of the 5’ UTR against the background level of the common single-nucleotide polymorphism (SNP) frequency determined by a minor allele frequency (MAF) greater than or equal to 0.01 (Fig 3b). With this comparison, DAVs were clearly enriched above the background common SNP frequency at the 3’ end near the Kozak sequence (Fig 3b). Furthermore, there was a clear drop-off of common SNPs at the last nucleotide of the 5’ UTR, which suggested that this specific region is functionally important and is maintained by selective pressures (Fig 3b). Despite the importance of the Kozak sequence as a post-transcriptional regulatory motif, only 8.4% of DAVs occur within the 3’ end of the UTR that approximately corresponds to the Kozak sequence (Fig 3b). Therefore, the majority of DAVs map outside the Kozak region of the 5’ UTR, which suggests that other sequence- or structure-based regulatory elements are important.
Fig 3.
Disease-associated variants in the 5’ UTR. a. DAV distribution across 5’ UTRs. The Kozak sequence is labeled above the start codon. b. The 3’ end of the 5’ UTR, particularly the Kozak sequence (labeled above the graph) is enriched for DAVs (orange) when compared with common SNPs (gray), defined as a minor allele frequency (MAF) of >= 0.01. Checkered bars indicate potential post-transcriptional regulatory motifs at the 3’ end of the 5’ UTR. The checkered portion of the pie chart shows the proportion of 5’ UTR transcripts that contain a mutation approximately within the Kozak sequence. c. The ferritin light chain (FTL) 5’ UTR forms an RNA secondary structure that enables the binding of the IRE-binding protein (IREBP, purple), which recognizes both RNA structure and a sequence-specific binding site (pink). The secondary structure of the 5’ UTR is shown, from the first nucleotide of the 5’ UTR to just past the start codon (178 nucleotides, “Start”). d. The mutation changing C to a G at position 41 (C41G, orange) disrupts IREBP (purple) binding by changing the sequence of the binding site (pink). e. A single nucleotide change from U to G at position 22 (orange dot, rs886037623) changes the secondary structure of the RNA and the IREBP binding site (purple), disrupting protein binding without changing the sequence of the binding site, thereby behaving as a riboSNitch.
In addition to the Kozak sequence, the 5’ UTR of genes often contains structured regions important for translational control that could harbor DAVs. Translation initiation begins in the 5’ UTR and typically involves scanning along the UTR by the ribosome. RNA secondary structure in the 5’ UTR can serve as a physical barrier to translation that must be unwound for the ribosome to proceed (Kozak 1986; Leppek et al. 2018). However, RNA structures in the 5’UTR can also recruit important translation factors and even the ribosome itself (Leppek et al. 2018). For example, some 5’ UTRs contain an internal ribosome entry site, which is a highly structured region of RNA that recruits the ribosome and allows for cap-independent translation (Macejak and Sarnow 1991; Jackson 2013; Leppek et al. 2018). Furthermore, numerous RNA binding proteins (RBPs) often bind specific structures in 5’ UTRs, and binding can occlude the ribosome, thereby, blocking translation (Rouault 2006; Leppek et al. 2018). With all these sites of regulation and activity, the 5’ UTR is a broad platform for translational regulation by both sequence elements and RNA structure that are also relevant to disease phenotypes.
Although mutations in the Kozak sequence are an obvious source of DAVs, other variants across the 5’ UTR can cause disease by more subtle mechanisms. One example is the riboSNitch in the 5’ UTR of the ferritin light chain (FTL) gene, which was one of the first riboSNitches to be identified and structurally characterized (Halvorsen et al. 2010; Martin et al. 2012). FTL, also known as IRE1, is important for regulating iron homeostasis because it codes for the light chain of the iron-binding protein ferritin. Ferritin captures excess iron and overexpression of FTL leads to iron deficiency (Rouault 2006; Solem et al. 2015). Normally, RNA structures called iron-responsive elements (IREs) in the 5’ UTR of the FTL gene are bound by IRE-binding proteins (IREBP) (Fig 3c) that downregulate translation of FTL (Rouault 2006). However, mutations in the IRE binding site that prevent IREBP binding (Fig 3d) lead to unregulated overexpression of FTL and hyperferritinemia cataract syndrome (Allerson et al. 1999; Rouault 2006). Remarkably, another variant in the 5’ UTR of FTL (rs886037623) can disrupt binding of the IREBPs without changing the sequence of the binding site (Figure 3e). Instead, this variant changes the structure of the RNA and sequesters the binding site such that the IREBP cannot reach it (Martin et al. 2012). This mutation is considered a riboSNitch because the SNP leads to a change in the RNA structure that disrupts IREBP binding, resulting in overexpression of the FTL protein and hyperferritinemia cataract syndrome.
Although the FTL riboSNitch was one of the first discovered and characterized, numerous other putative riboSNitches in 5’ UTR regions have been identified and associated with diseases including hypertension, B-thalassemia, chronic obstructive pulmonary disease, and retinoblastoma (Halvorsen et al. 2010; Kutchko et al. 2015). By structural probing, investigators have experimentally verified some of these riboSNitches, such as the retinoblastoma riboSNitch (Kutchko et al. 2015). With numerous examples of riboSNitches, it is clear that DAVs outside known post-transcriptional regulatory motifs deserve consideration because RNA structure can provide both a molecular mechanism of the disease and an avenue for therapeutics.
Disease-associated variation in the 3’ UTR and the FKB5 riboSNitch
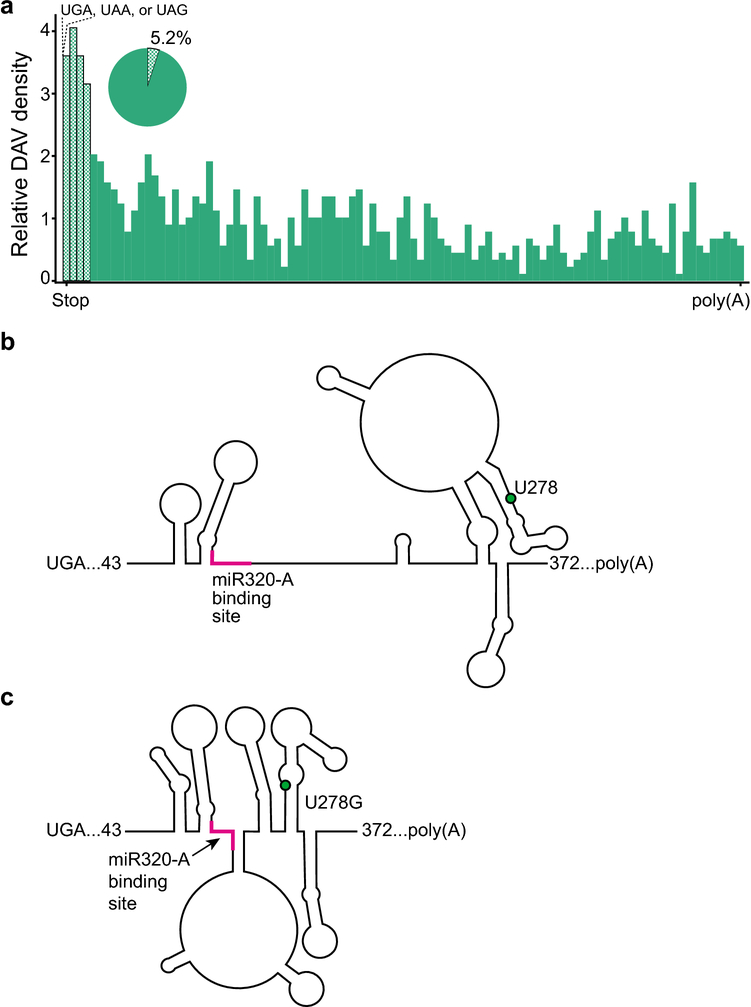
Similar to the 5’ UTR, we observed fewer DAVs in the 3’ UTR than in exons or introns (Fig 2). However, we observed a higher density of DAVs near the stop codon at the 5’ end of the 3’ UTR (Fig 4a), which is another post-transcriptional regulatory motif important for proper translational control via termination. In particular, 5.2% of DAV transcripts have a mutation within the ribosome footprint of the stop codon (15 nucleotides, based on a 30-nucleotide ribosome footprint) (Lareau et al. 2014). However, most DAVs (94.8%) occur outside the potential post-transcriptional regulatory region of the ribosome footprint around the stop codon; thus, other mechanisms such as RBPs or microRNA (miRNA) binding likely are at play. The 3’ UTR serves as a common site for microRNA binding (Gebert and MacRae 2019), and disruption of either RNA sequence or structure of a microRNA binding site provides a prime environment for causing disease. Mechanistically, miRNAs base pair with the 3’ UTRs of target mRNAs, which leads to RNA decay and the repression of translation (Gebert and MacRae 2019). Therefore, mutations in the miRNA binding site can disrupt miRNA regulation of mRNA stability and impair translational control. In addition to binding site mutations, changes in RNA structure can inhibit the availability of the miRNA binding site by intramolecular base pairing that repositions an available binding site into an inaccessible stem-loop.
Fig 4.
Disease associated variants in the 3’ UTR. a. DAV distribution across 3’ UTRs. The checkered bars indicate potential post-transcriptional regulatory motifs approximately around the ribosomal footprint at the stop codon. Stop codons are labeled above the graph. The pie chart shows proportion of 3’ UTRs that have a DAV within the ribosomal footprint (~15 nucleotides). b. The RNA secondary structure of a section of the FKBP5 3’UTR. The 3’UTR contains the binding site for the miRNA miR320-A (pink). c. The minor allelic SNP rs3800373 changes U to G at position 278 (green dot) causing a structural change that makes the miRNA binding site (pink) less accessible because a greater proportion of the binding site is base paired in a novel stem-loop.
Although the 3’ UTR has been described to be resistant to riboSNitches due to its high functional importance for RBP and miRNA binding (Wan et al. 2014), other analyses describe the 3’ UTR as a potential mechanistic source of disease-causing riboSNitches (Solem et al. 2015). The riboSNitch in the 3’ UTR of the FKB5 gene is an example of a disease-associated riboSNitch that disrupts miRNA binding (Linnstaedt et al. 2018). FKB5 is an important regulator of the stress response and improper regulation of FKB5 leads to increased vulnerability to post-traumatic chronic musculoskeletal pain (Linnstaedt et al. 2018). Normally, miRNA-320a binds the 3’ UTR of FKB5 mRNA (Fig 4b), but the presence of the rs3800373 variant changes the structure of the 3’ UTR and partially sequesters the binding site in a stem-loop (Fig 4c). Notably, the rs3800373 variant is more than 100 nucleotides from the miRNA-320a binding site, and rs3800373 does not change the sequence of the binding site. The structural change caused by this variant, which was validated by SHAPE probing, is a novel stem-loop that includes three nucleotides of the miRNA-320a binding site (Fig 4c). In the rs3800373 variant, more than half of the miRNA-320a binding site is engaged in intramolecular base pairing, dramatically reducing its accessibility for miRNA binding (Fig 4c). The disruption in miRNA binding leads to increased translation of FKBP5, which, in turn, causes glucocorticoid resistance and an increased vulnerability to post traumatic pain (Linnstaedt et al. 2018). Furthermore, rs3800373 has a global minor allele-frequency greater than 0.28, demonstrating that riboSNitch mechanisms are causative in common alleles as well. The FKBP5 riboSNitch is a clear example of how a single nucleotide change in the 3’ UTR can cause a disease phenotype by a change in RNA structure instead of a change in the sequence of a post-transcriptional regulatory element.
Disease-associated variation in exons and the PNPO riboSNitch
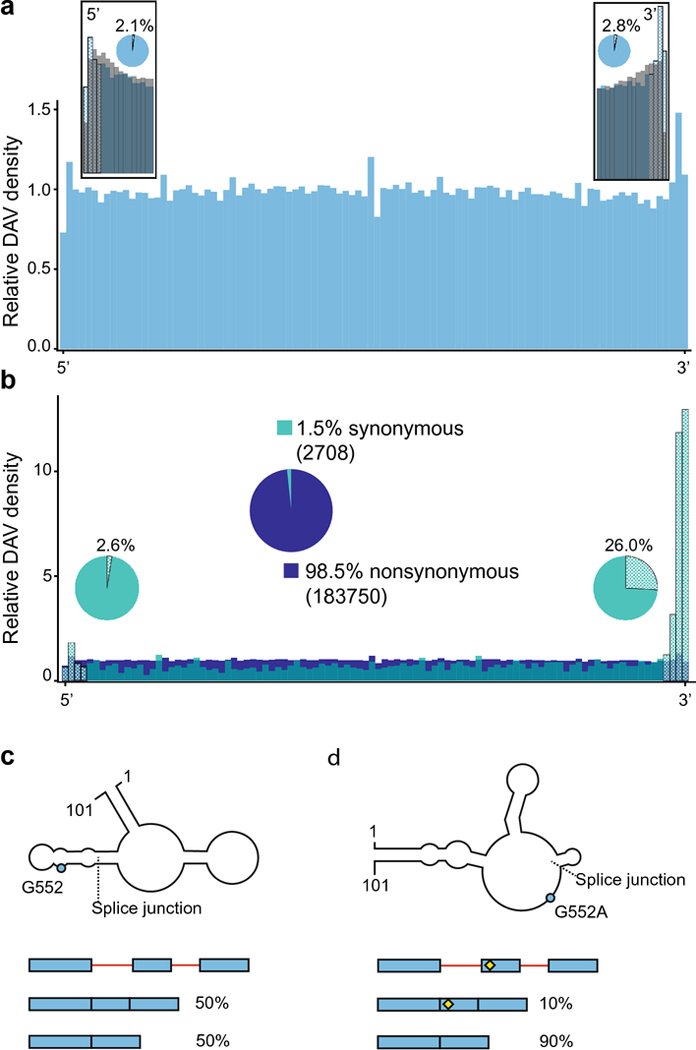
Almost all DAVs occur in exons (87.3%) (Fig 5a), which is consistent with the requirements of protein-coding functionality. It is also likely a reflection of the sampling bias in the HGMD data, due to the historical use of exonic sequencing in clinical genomics settings. Exonic DAVs demonstrate a relatively even distribution across exons, with slight enrichment around the 5’ and 3’ ends of exons that correspond to the post-transcriptional regulatory motifs of the 5’ and 3’ splice sites (Fig 5a). In contrast, the common SNPs (MAF >= 0.01) demonstrate a dramatic drop off at the very ends of the exons, which suggests that these regions are conserved and testifies to their functional importance (Fig 5a). Furthermore, only 4.9% of DAVs are found near the 5’ and 3 ends of the exons that correspond to the splice sites (three nucleotides from each end), which suggests that, although these regions are important, disease-associated variation is widely distributed throughout the entire exon sequence space.
Fig 5.
Disease associated variants in exons. a. Exonic DAV distribution (blue), with inset boxes showing the relative abundance of DAVs (blue) against common SNPs with a MAF >= 0.01 (gray) at the 5’ and 3’ ends of exons. The checkered bars indicate potential post-transcriptional regulatory motifs around the splice sites. The pie charts show the proportion of transcripts with a DAV at the splice sites, within 3 nucleotides of each end. b. Exonic DAVs divided into synonymous (blue-green) and nonsynonymous (dark blue). The large, centered pie chart shows the relative abundance of synonymous versus nonsynonymous DAVs in exons, with the absolute number of mutants in parentheses. Nonsynonymous DAVs are found at a uniform distribution across the length of the exon, whereas the synonymous DAVs are enriched at the 3’ end of exons. The smaller pie charts near each end of show the proportion of transcripts with a synonymous DAV in a post-transcriptional regulatory motif compared to all synonymous DAV-containing transcripts. c. The RNA secondary structure of a 101-nucleotide section of the mature PNPO transcript is shown, with the splice junction between exons 4 and 5 marked. The schematic below the structure illustrates how the PNPO gene exhibits a mixture of alternative splicing events showing both exons (blue) and introns (red), where splice isoforms are approximately equally distributed in the wild-type background. d. The mutation of G552A (rs4378657), just 6 nucleotides from the splice junction at the 5’ end of exon 5, creates a riboSNitch in PNPO (blue dot) by disrupting the binding of the TARDBP RNA binding protein, which results in exon skipping. The SNP is shown in the splicing schematic with a yellow diamond.
For disease-associated variation within exons, it is important to consider whether a variant is nonsynonymous or synonymous. Nonsynonymous mutations change the amino acid sequence of the protein product, whereas synonymous mutations change the DNA and RNA sequence, but they do not change the sequence of the final protein product. It should also be noted that nonsynonymous DAVs can still have an RNA-based mechanism in addition to changing the protein sequence. For example, a nonsynonymous mutation in an exon can change splicing by disrupting exonic splicing enhancers or exonic splicing silencers (Woolfe et al. 2010). Still, when we consider how variation in the transcriptome affects disease, the distinction between nonsynonymous and synonymous DAVs becomes particularly valuable and synonymous DAVs deserve particular attention. Most DAVs (98.5%) are nonsynonymous, with an even distribution as expected from the exon DAV distribution (Fig 5b). However, there are 2708 synonymous DAVs, which are more prevalent around the ends of exons, particularly the 3’ end (Fig 5b). In fact, 26% of synonymous mutations are in the very 3’ end of the exon (~3 nucleotides) (Fig 5b). Despite the high concentration of synonymous DAVs at these post-transcriptional regulatory motifs, 71.4% of synonymous DAVs are outside these regions, a condition that leaves many potential RNA-based mechanisms to be explained.
Recently, a synonymous riboSNitch (rs4378657) was discovered in the pyridoxamine 5’-phosphate oxidase (PNPO) gene, which is associated with epilepsy (Mills et al. 2014; Sun et al. 2021). Although the rs4378657 variant of PNPO is not part of the splice site sequence, it is only 6 nucleotides from a splice junction and was computationally predicted to form a different RNA structure as a putative riboSNitch (Sun et al. 2021). To examine this putative riboSNitch, Sun et al. compared a cell line that contained a single copy of the minor allele against a cell line that had only the major allele (Sun et al. 2021). By analyzing ic-SHAPE RNA-seq libraries from both cell lines, it was determined that the two alleles form different RNA structures (Fig 5c and d) (Sun et al. 2021). The variant structure moves the SNP from a bulge in a stem-loop (Figure 5c) to an accessible region (Fig 5d) (Sun et al. 2021). Furthermore, by combining computational prediction with experimental CLIP-seq data from the two cell lines, Sun et al. demonstrated that the TARDBP RNA-binding protein had a higher affinity for the variant (Sun et al. 2021). TARDBP binding affects alternative splicing, and this riboSNitch favored the skipped exon isoform of PNPO (Fig 5c and d, bottom), consistent with the change in TARDBP-binding affinity (Sun et al. 2021). The riboSNitch-TARDBP-mediated exon skipping was further confirmed by examining splicing after knockdown of the TARDBP protein (Sun et al. 2021). Therefore, the rs4378657 variant changes RNA structure and demonstrates a higher affinity for the TARDBP RNA-binding protein, which pushes alternative splicing to favor an increase in exon skipping (Fig. 5c, d). In summary, this example shows how a synonymous riboSNitch leads to higher rates of epilepsy by a change in RNA structure that affects RBP binding and, thereby, changes the splice isoform ratio.
Disease-associated variation in introns and a MAPT riboSNitch
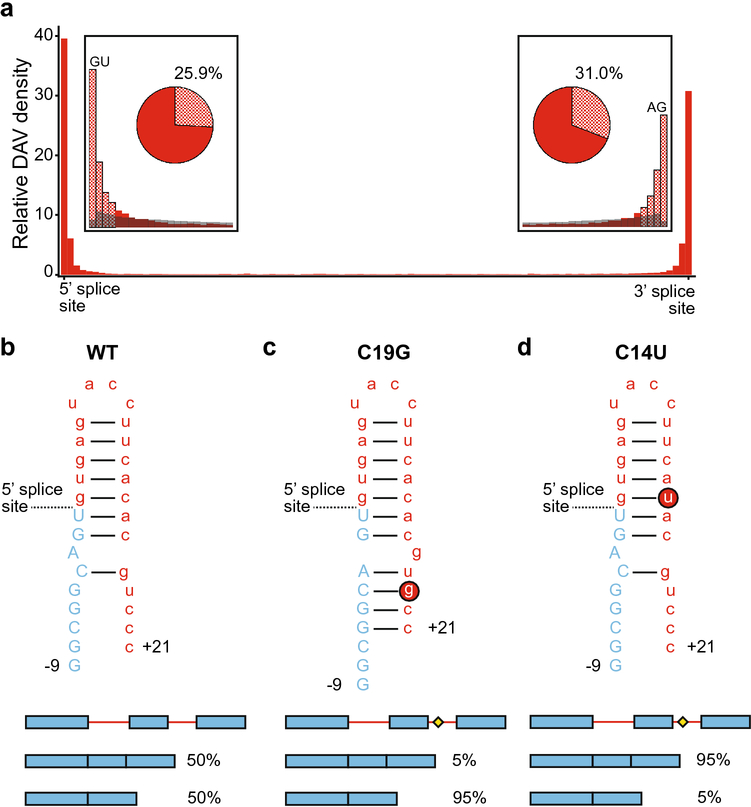
Despite their removal in the final transcript, introns have a key regulatory function in gene expression. Eukaryotes use introns in alternative splicing to diversify their gene products by generating multiple protein isoforms from a single gene (Xu et al. 2021). Splicing is dependent on the ability of the spliceosome to find authentic splice sites, and the existence of multiple potential splice sites enables alternative splicing. However, mutations at these locations can confound this process and are likely to generate DAVs (Scotti and Swanson 2016). Accordingly, DAVs in introns are found at high densities around both the 5’ and 3’ splice sites (Fig 6a). Particularly, for correct splicing, the 5’ splice site requires an invariable GU, and the 3’ splice site requires an invariable AG (Fig 6a inset). Beyond these invariable sequence elements, the 5’ splice site exhibits conserved sequence for 6 nucleotides and the 3’ splice site for approximately 35 nucleotides up to the conserved branchpoint, which is typically an adenosine. These conserved regions are clearly important, and 25.9% of intronic DAVs occur within 6 nucleotides of the 5’ splice site and 31% occur within 35 nucleotides of the 3’ splice site (Fig 6a inset). Interestingly, common SNPs are depleted at these regions (Fig 6a inset), consistent with these regions being highly conserved and functionally important for correct splicing. However, it should be noted that 43.1% of DAVs occur outside of these post-transcriptional regulatory regions. Furthermore, as noted previously, the DAVs reported here are based on data in the HGMD, and the HGMD has a bias towards sequences of exons and exon-intron junctions. Thus, the internal regions of introns have been neglected in sequencing experiments, and DAVs farther from the exon-intron junctions may be underrepresented. Regardless, the maintenance of splice sites is important for proper cellular function and intronic DAVs can occur by disrupting splicing.
Fig 6.
Disease associated variants in introns. a. Intronic DAV distribution is shown in red, where intronic DAVs are dramatically more abundant around splice sites. Note the differing scale from Fig 2. The insets compare the DAV frequency (red) and the common SNP frequency (gray) at the very 5’ and 3’ ends of introns. The checkered bars indicate the approximate location of post-transcriptional regulatory motifs. The pie charts show the proportion of transcripts containing an intronic DAV in a post-transcriptional regulatory motif (i.e. within 6 nucleotides of the 5’ splice site, or 35 nucleotides of the 3’ splice site). b. The MAPT transcript RNA forms a hairpin structure between exon 10 (blue, uppercase) and the following intron (red, lowercase), shown here as nucleotides and numerically labeled from the splice site, where the upstream exon nucleotides are labeled with a minus (−9) and the downstream intron nucleotides are labeled with a plus (+21). MAPT RNA splices as a roughly even mixture of splice isoforms skipping or retaining exon 10, shown as the schematic below with exons (blue) and introns (red), where accumulation of the exon-inclusion isoform is associated with disease phenotypes. c. The rs63750162 mutation of C to G at +19 (C19G, red circled G) creates a riboSNitch shifting the splice isoforms to favor exon skipping. The splicing diagram shows the DAV SNP as a yellow diamond. d. The rs63750972 mutation of C to U at +1 (C14U, red circled U) minimally changes the RNA structure but weakens the hairpin strength, shifting the isoform ratio to favor exon inclusion.
The effect of DAVs on splicing agree with previous reports and speculation that riboSNitches can act through splicing (Wan et al. 2014). One well-characterized group of intronic DAVs occurs in the MAPT gene; these DAVs have many neurodegenerative disease implications, particularly frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) tauopathies (Niblock and Gallo 2012). The MAPT gene exhibits different splice isoforms that can be grouped into 3R and 4R isoforms depending on the variable splicing of exon 10 (Niblock and Gallo 2012). In healthy tissue, MAPT is spliced into a roughly equal mixture of 3R and 4R isoforms of the Tau protein. However, SNVs in the intron distal to exon 10 can produce isoform ratios that are nearly exclusively 3R or 4R and result in disease (Niblock and Gallo 2012). Exclusion of exon 10 yields the 3R Tau protein, which accumulates into Pick bodies associated with a tauopathy known as Pick’s disease (de Silva et al. 2006). In contrast, the inclusion of exon 10 produces the 4R Tau isoform, which is associated with progressive supranuclear palsy and corticobasal degeneration (Ingelsson et al. 2007). Therefore, the delicate balance of splice isoforms can be disturbed by SNVs.
Some of the MAPT DAVs have been biochemically characterized, and molecular mechanisms that affect splicing have been identified (Tan et al. 2019). The wild-type MAPT transcript forms an RNA hairpin (Fig 6b) between exon 10 and the following intron, which must be unfolded for splicing to occur correctly, and disruptions to this hairpin lead to disruptions in the 3R:4R splice isoform ratio (Grover et al. 1999; Varani et al. 1999; Buratti and Baralle 2004; Donahue et al. 2006; Tan et al. 2019). For example, changing C to G 19 nucleotides after the splice junction (C19G, rs63750162) introduces a new base pairing interaction and results in a longer stem-loop with 3 additional base pairs (Fig 6c), strengthening the RNA hairpin and requiring more energy to unfold (Tan et al. 2019). The C19G variant shifts the splice isoform ratio to almost exclusively 3R. In contrast, a mutation 14 nucleotides after the splice junction (C14U, rs63750972) destabilizes the stem-loop by introducing a G-U wobble base pair that makes the hairpin easier to unfold and shifts the ratio to almost exclusively 4R Tau (Fig 6d) (Tan et al. 2019). As described previously, changing the 3R to 4R ratio to a single isoform results in various tauopathies; thus, these variants act as DAVs by disrupting the splice isoform ratio. These two variants are not within the splice site itself; however through the structure of RNA, mutations at a distance interact with the splice site and regulate splicing and cause disease, providing yet another example whereby RNA structure can dictate disease phenotypes.
Conclusions
Human genetic variation can cause disease by a wide range of mechanisms. Here, we described distributions of causative disease-associated variation throughout the components of the transcriptome and acknowledged the importance of post-transcriptional regulatory motifs. Furthermore, we highlighted examples of DAVs that are outside these regulatory motifs and cause disease by a change in RNA structure. Particularly, we illustrated examples of riboSNitches that affect each component of an mRNA, whereby a single nucleotide variant changes RNA structure and leads to disease. We also described mechanisms for how changes in RNA structure can affect gene function, such as changing RBP binding affinity, miRNA binding affinity, and splice site accessibility. Overall, understanding human variation and how riboSNitches cause disease is a powerful prospective for the future of personalized medicine and RNA-targeting therapeutics.
Methods:
Data collection
NCBI RefSeq-Curated hg38 bed files for coding exon, intron, 5’ UTR and 3’ UTR coordinates were downloaded from the UCSC table browser (Karolchik 2004). Hereby, component refers to either an exon, intron, 5’ UTR or 3’ UTR. Disease mutations were obtained from the Human Genetic Mutations Database (HGMD) (Stenson et al. 2003, 2020) version 2021.1. Common SNPs were downloaded from dbSNP Build150 (Smigielski 2000). Hg19 genome coordinates of miRNA binding sites were downloaded from TargetScanHuman Release 7.2 (Agarwal et al. 2015) from the file Genome coordinates of Predicted Conserved Targets (default predictions). The UCSC genome browser tool liftOver was used to convert hg19 coordinates to hg38.
Length distribution analysis
Noncoding transcripts were filtered out using “NR” field in the RefSeqID column. Only transcripts with unique start and end coordinates were kept, and duplicate transcripts were removed. The length for each component was calculated by taking the difference between start and end coordinates. Components included in plots had a length less than the mean length of all components to discard long tails generated from outlier lengths. Histograms of lengths were quantified with Matplotlib (Hunter 2007) python package and plotted using ggplot2 (Wickham 2016) R package.
Mutation and SNP distribution analysis
Only single nucleotide disease mutations were considered in the analysis. Insertions and deletions were removed. Mutations were then separated into coding (n=186,487) and noncoding (n=29,380) by using the presence of the “PROT” field to determine whether they affected the protein sequence. Coding mutations were divided into synonymous (n=2708) and nonsynonymous (n=183,779) using information within the “PROT” field that indicates the change in amino acid. Bedtools intersect (Quinlan and Hall 2010) was used to link coding mutations to their associated exonic regions and noncoding mutations to intronic, 5’ UTR and 3’ UTR regions. If a mutation intersected multiple transcripts, one transcript was randomly chosen to associate with the mutation. The distance from the mutation to the 5’ start coordinate of its associated component was calculated and normalized to the length of the component. Histograms of normalized distances were plotted using ggplot2 (Wickham 2016) R package. A similar analysis was performed for common SNPs (n=39,097,002). However, there was no information about a SNP’s effect on protein, hence, they were not separated into coding or noncoding SNPs. Only SNPs found within 50 nucleotides of the 5’ start coordinate of an intersected component were plotted.
To determine the proportion of transcripts with a DAV in a previously described post-transcriptional regulatory motif (e.g. Kozak sequence or splice sites), the position of the motif was calculated with respect to the normalized transcript length. A normalized cutoff was calculated by dividing the length of the motif (i.e., 6 nucleotides of Kozak sequence, 3 nucleotides of splice sites in exons, 6 nucleotides of 5’ intron splice site, 35 nucleotides of 3’ intron splice site, and 15 nucleotides of the 3’ UTR as half the ribosomal footprint) by the average length of the component. Any DAV between the cutoff and the appropriate end of the sequence was considered to be within the post-transcriptional regulatory motif.
Enrichment of variants in miRNA binding sites was analyzed by determining the single nucleotide 3’ UTR DAVs found in miRNA binding sites using bedtools intersect. This was repeated for common SNPs. A chi-squared test in R was used to statistically test the difference in the proportion of 3’ UTR variants found within versus outside miRNA binding sites between DAVs and common SNPs.
Acknowledgments
Funding: This work was supported by US National Institutes of Health R35 GM140844, R01 GM101237 and R01 HL111527 to AL
Footnotes
Conflicts of interest: None declared
Declarations:
Availability of data and material: Code is available upon request.
References:
- Agarwal V, Bell GW, Nam J-W, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. eLife 4:e05005. 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerson CR, Cazzola M, Rouault TA (1999) Clinical Severity and Thermodynamic Effects of Iron-responsive Element Mutations in Hereditary Hyperferritinemia-Cataract Syndrome. Journal of Biological Chemistry 274:26439–26447. 10.1074/jbc.274.37.26439 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2004) Influence of RNA Secondary Structure on the Pre-mRNA Splicing Process. Mol Cell Biol 24:10505–10514. 10.1128/MCB.24.24.10505-10514.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M, Solem A, Qu K, et al. (2015) Detecting riboSNitches with RNA folding algorithms: a genome-wide benchmark. Nucleic Acids Research 43:1859–1868. 10.1093/nar/gkv010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Silva R, Lashley T, Strand C, et al. (2006) An immunohistochemical study of cases of sporadic and inherited frontotemporal lobar degeneration using 3R- and 4R-specific tau monoclonal antibodies. Acta Neuropathol 111:329–340. 10.1007/s00401-006-0048-x [DOI] [PubMed] [Google Scholar]
- Donahue CP, Muratore C, Wu JY, et al. (2006) Stabilization of the Tau Exon 10 Stem Loop Alters Pre-mRNA Splicing. Journal of Biological Chemistry 281:23302–23306. 10.1074/jbc.C600143200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlmann T, Sahay S, Keller A, Backes C (2019) A review of databases predicting the effects of SNPs in miRNA genes or miRNA-binding sites. Briefings in Bioinformatics 20:1011–1020. 10.1093/bib/bbx155 [DOI] [PubMed] [Google Scholar]
- Gaither JBS, Lammi GE, Li JL, et al. (2021) Synonymous variants that disrupt messenger RNA structure are significantly constrained in the human population. GigaScience 10:giab023. 10.1093/gigascience/giab023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert LFR, MacRae IJ (2019) Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 20:21–37. 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Letters 582:1977–1986. 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A, Houlden H, Baker M, et al. (1999) 5′ Splice Site Mutations in tau Associated with the Inherited Dementia FTDP-17 Affect a Stem-Loop Structure That Regulates Alternative Splicing of Exon 10. Journal of Biological Chemistry 274:15134–15143. 10.1074/jbc.274.21.15134 [DOI] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A (2010) Disease-Associated Mutations That Alter the RNA Structural Ensemble. PLoS Genet 6:e1001074. 10.1371/journal.pgen.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007) Matplotlib: A 2D Graphics Environment. Comput Sci Eng 9:90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Ingelsson M, Ramasamy K, Russ C, et al. (2007) Increase in the relative expression of tau with four microtubule binding repeat regions in frontotemporal lobar degeneration and progressive supranuclear palsy brains. Acta Neuropathol 114:471–479. 10.1007/s00401-007-0280-z [DOI] [PubMed] [Google Scholar]
- Jackson RJ (2013) The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 5:a011569. 10.1101/cshperspect.a011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D (2004) The UCSC Table Browser data retrieval tool. Nucleic Acids Research 32:493D–496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, et al. (2010) Genome-wide measurement of RNA secondary structure in yeast. Nature 467:103–107. 10.1038/nature09322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Colonna V, et al. (2013) Integrative Annotation of Variants from 1092 Humans: Application to Cancer Genomics. Science 342:1235587. 10.1126/science.1235587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucl Acids Res 15:8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. PNAS 83:2850–2854. 10.1073/pnas.83.9.2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, Maquat LE (2019) Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20:406–420. 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchko KM, Sanders W, Ziehr B, et al. (2015) Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. RNA 21:1274–1285. 10.1261/rna.049221.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO (2014) Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3:e01257. 10.7554/eLife.01257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC (2015) Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 84:291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19:158–174. 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Chen Y, Zhang Y, Ouyang Z (2020) Identification and analysis of RNA structural disruptions induced by single nucleotide variants using Riprap and RiboSNitchDB. NAR Genomics and Bioinformatics 2:lqaa057. 10.1093/nargab/lqaa057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Riker KD, Rueckeis CA, et al. (2018) A Functional riboSNitch in the 3′ Untranslated Region of FKBP5 Alters MicroRNA-320a Binding Efficiency and Mediates Vulnerability to Chronic Post-Traumatic Pain. J Neurosci 38:8407–8420. 10.1523/JNEUROSCI.3458-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-J, Fu X, Xia M, et al. (2021) miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Research 49:D1276–D1281. 10.1093/nar/gkaa783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak D, Sarnow P (1991) Internal initiation of translation mediated by the 5’ leader of a cellular mRNA. Nature 353:90–94. 10.1038/353090a0 [DOI] [PubMed] [Google Scholar]
- Martin JS, Halvorsen M, Davis-Neulander L, et al. (2012) Structural effects of linkage disequilibrium on the transcriptome. RNA 18:77–87. 10.1261/rna.029900.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meienberg J, Bruggmann R, Oexle K, Matyas G (2016) Clinical sequencing: is WGS the better WES? Hum Genet 135:359–362. 10.1007/s00439-015-1631-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills PB, Camuzeaux SSM, Footitt EJ, et al. (2014) Epilepsy due to PNPO mutations: genotype, environment and treatment affect presentation and outcome. Brain 137:1350–1360. 10.1093/brain/awu051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblock M, Gallo J-M (2012) Tau alternative splicing in familial and sporadic tauopathies. Biochemical Society Transactions 40:677–680. 10.1042/BST20120091 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA (2006) The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol 2:406–414. 10.1038/nchembio807 [DOI] [PubMed] [Google Scholar]
- Sabarinathan R, Tafer H, Seemann SE, et al. (2013) RNA snp: Efficient Detection of Local RNA Secondary Structure Changes Induced by SNP s. Human Mutation 34:546–556. 10.1002/humu.22273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari R, Kimchi-Sarfaty C, Gottesman MM, Przytycka TM (2013) Sensitive measurement of single-nucleotide polymorphism-induced changes of RNA conformation: application to disease studies. Nucleic Acids Research 41:44–53. 10.1093/nar/gks1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS (2016) RNA mis-splicing in disease. Nat Rev Genet 17:19–32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, et al. (2014) RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. 10.1038/nmeth.3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigielski EM (2000) dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Research 28:352–355. 10.1093/nar/28.1.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem AC, Halvorsen M, Ramos SBV, Laederach A (2015) The potential of the riboSNitch in personalized medicine: Potential of the riboSNitch. WIREs RNA 6:517–532. 10.1002/wrna.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, et al. (2003) Human Gene Mutation Database (HGMD ®): 2003 update: HGMD 2003 UPDATE. Hum Mutat 21:577–581. 10.1002/humu.10212 [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, et al. (2020) The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting. Hum Genet 139:1197–1207. 10.1007/s00439-020-02199-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steri M, Idda ML, Whalen MB, Orrù V (2018) Genetic variants in mRNA untranslated regions. WIREs RNA 9:e1474. 10.1002/wrna.1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xu K, Huang W, et al. (2021) Predicting dynamic cellular protein–RNA interactions by deep learning using in vivo RNA structures. Cell Res 31:495–516. 10.1038/s41422-021-00476-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Yang L, Ong AAL, et al. (2019) A Disease-Causing Intronic Point Mutation C19G Alters Tau Exon 10 Splicing via RNA Secondary Structure Rearrangement. Biochemistry 58:1565–1578. 10.1021/acs.biochem.9b00001 [DOI] [PubMed] [Google Scholar]
- Telenti A, Pierce LCT, Biggs WH, et al. (2016) Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA 113:11901–11906. 10.1073/pnas.1613365113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Hasegawa M, Spillantini MG, et al. (1999) Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proceedings of the National Academy of Sciences 96:8229–8234. 10.1073/pnas.96.14.8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, et al. (2017) 10 Years of GWAS Discovery: Biology, Function, and Translation. The American Journal of Human Genetics 101:5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Kertesz M, Spitale RC, et al. (2011) Understanding the transcriptome through RNA structure. Nat Rev Genet 12:641–655. 10.1038/nrg3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, et al. (2014) Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505:706–709. 10.1038/nature12946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM (2006) Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW (2001) The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2:237–246. 10.1038/35067025 [DOI] [PubMed] [Google Scholar]
- Woolfe A, Mullikin JC, Elnitski L (2010) Genomic features defining exonic variants that modulate splicing. Genome Biol 11:R20. 10.1186/gb-2010-11-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Meng Y, Jin Y (2021) RNA structures in alternative splicing and back- splicing. WIREs RNA 12:. 10.1002/wrna.1626 [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu J, Lee D, et al. (2020) RADAR: annotation and prioritization of variants in the post-transcriptional regulome of RNA-binding proteins. Genome Biol 21:151. 10.1186/s13059-020-01979-4 [DOI] [PMC free article] [PubMed] [Google Scholar]