Abstract
The antifungal effect of GM 237354, a sordarin derivative, was studied in an in vitro pharmacokinetic (PK)-pharmacodynamic dynamic system (bioreactor) which reproduces PK profiles observed in a previously described model of drug efficacy against murine systemic candidiasis. Immunocompetent mice infected intravenously with 105 CFU of Candida albicans were treated with GM 237354 at 2.5, 10, and 40 mg/kg of body weight every 8 h subcutaneously for 7 days. Free concentrations in serum were calculated by multiplying total concentrations measured in vivo by 0.05, the free fraction determined in vitro by equilibrium dialysis. In the bioreactor the inoculum was ≈106 CFU/ml; and a one-compartment PK model was used to reproduce the PK profiles of free and total GM 237354 in serum obtained in mice, and clearance of C. albicans was measured over 48 h. A good correlation was observed when the in vivo fungal kidney burden and the area under the survival time curve were compared with the in vitro broth “burden,” although only when free in vivo levels in serum were reproduced in vitro. GM 237354 displayed a 3-log decrease effect both in vivo and in vitro. The very few reports available on in vitro-in vivo correlations have been obtained with antibiotics. The good in vitro-in vivo correlation obtained with an antifungal agent shows that the in vitro dynamic system could constitute a powerful investigational tool prior to assessment of the efficacy of an anti-infective agent in animals and humans.
Pharmacokinetic (PK)-pharmacodynamic (PD) studies assess relationships between plasma drug concentrations, the in vitro susceptibilities of target microorganisms, and efficacy. The aim of these studies is to identify the PK-PD parameter that can best predict the treatment outcome based on in vitro antimicrobial activity (MIC), efficacy, and PK properties in animals and humans. The correlation between the best PK-PD parameter and efficacy in animals for key pathogens provides, together with PKs in humans, the base for optimization of dosing regimens for early clinical trials (15, 29).
There has been growing interest in dynamic PK-PD in vitro experimental models that simulate in vivo animal and human PK profiles (29). These models offer the possibility of evaluating the intrinsic activity of an antibiotic, antiviral, or antifungal agent against a given microorganism without the added in vivo PK variability and without the influence of host immune response. Numerous repeated-dosing regimens for key microorganism strains and for selected animal species and humans can thus be evaluated more easily, completely, and precisely in vitro than they can in vivo. These models can also be used to study the emergence of resistance during drug exposure (29).
Although such in vitro PK-PD models have been extensively used for antibacterial, antiviral, and antifungal compounds (18, 19, 22, 28, 35, 44), few reports show cross-validation with in vivo results (6; C. R. Bonapace, L. V. Friedrich, R. L. White, and J. A. Bosso, 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 998, 1999). Such a validation is especially important when one considers that a main advantage of these dynamic in vitro models is the possibility of easily simulating the anti-infective activity of any PK profile and hence any dosing regimen in animals and humans. However, crucial parameters such as protein binding of drugs must be taken into account to determine the in vitro-in vivo activity relationships.
Protein binding per se is not a direct determinant of efficacy, but it affects the PK characteristics of a compound. Only unbound drug is able to diffuse across physiological membranes and hence to reach the physiological spaces where therapeutic action is required. The level of free drug in plasma sets the gradient for tissue diffusion (27, 34). For tetracyclines there is a good relationship between protein binding, renal clearance, and half-life. However, this does not apply to penicillins and cephalosporins, which also undergo tubular secretion (27). In vitro, protein binding must be taken into account when studying the effects of an antibiotic, as medium constituents could bind to the compound, thus affecting its in vitro activity (48). Reviews that describe methods for quantification of protein binding in vitro and correlation of their results with in vivo results are available (34, 36). However, most consider only the molecular aspects of binding, without taking into account its microbiological implications either in vitro or in vivo (36, 39). Plasma binding had been shown to influence the in vivo activity of antibiotic (16, 20, 26, 27, 34, 40, 45, 48, 49), antimalarial (25), and antiviral (3, 4, 5) agents.
Sordarin derivatives are a new class of antifungal compounds (14) with demonstrated in vitro activity against a broad spectrum of fungal microorganisms with clinical effects (24). In addition, sordarin derivatives have been shown to have in vivo activity against systemic and local experimental infections involving Candida albicans and Pneumocystis carinii (1, 2, 32). They also showed efficacy against murine histoplasmosis (23) and coccidioidomycosis (10). Studies aimed at the identification of the PK-PD parameter that best predicts in vivo efficacy were carried out for GM 237354, a representative of the sordarins. For this compound, the area under the concentration-time curve (AUC) was the PK-PD parameter predictive of efficacy, with a very good agreement between several efficacy end points, such as survival and kidney burden (1).
The objective of the work described here was to establish the correlation between the clearance of C. albicans determined by using an in vitro dynamic system (bioreactor) which reproduces different in vivo PK profiles for GM 237354 (representing various dosing regimens) and the results of studies of therapeutic efficacy (fungal kidney burden) in a model of systemic candidiasis in mice.
(Part of this work was presented at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., 26 to 29 September 1999 [Aviles, P., C. Falcoz, M. J. Guillén, R. San Román, and D. Gargallo, 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2000, 2000].)
MATERIALS AND METHODS
Antifungal agent.
GM 237354 (as the potassium salt) was synthesized at GlaxoWellcome S.A. (Tres Cantos, Madrid, Spain). For the determination of in vitro activity with the bioreactor, a stock solution of 5,000 μg/ml was prepared in sterile deionized water and diluted appropriately in broth medium. For in vivo tests, the compound was dissolved extemporaneously in sterile deionized water to reach the desired concentrations. Doses are expressed as milligrams of base per kilogram of body weight.
C. albicans isolates.
C. albicans 4711E and C. albicans 2005 (from the culture collection of GlaxoWellcome, Greenford, United Kingdom) were used throughout the study. The GM 237354 MIC was determined to be 0.001 μg/ml (24). Strains were maintained in Sabouraud dextrose agar (SDA; Difco, Detroit, Mich.) with 15% glycerol at −70°C until required. For inoculum preparation, yeasts were cultured on SDA (Difco) plates. The resulting growth was collected from the plates in sterile 0.9% NaCl and adjusted to the appropriate concentration (colony counts were verified on SDA plates).
Determination of serum protein binding.
Protein binding was determined by equilibrium dialysis as described previously (1). Briefly, [3H]GM 237354 was incubated at six concentrations ranging from 1 to 80 μg/ml in 1 ml of blank mouse serum at 37°C for 2 h. Dialysis was performed in chambers (Cellu · Sep; Spectrum) which were filled with red blood cell buffer (RBCB; 150 mM sodium chloride, 0.8 mM magnesium chloride, 0.2 mM calcium chloride in deionized water) and with serum spiked with the compound on each side of the membrane, respectively. Dialysis chambers were separated with visking membranes previously soaked with RBCB. The chambers were then placed in a rotator (Dianorm, Munich, Germany), and dialysis was carried out at 16 rpm at 37°C for 24 h. Following incubation, the aliquots in both compartments were counted and the free fraction was calculated.
Mice.
Male CD-1 mice (weight, 25 to 30 g; Charles River, France Inc., Lyon, France) were used after an acclimatization period (at least 7 days). Mice had free access to food and water throughout the experimental period. The research complied with Spanish national legislation and with company policy on the care and use of animals and related codes of practice.
Systemic infection.
The systemic infection model and other experimental conditions have been described previously (1). Briefly, immunocompetent male CD-1 mice were challenged intravenously with 200 μl of the appropriate inoculum of C. albicans 4711E (ca. 105 CFU) in the lateral tail vein. Infected animals were left untreated, and the rest were randomly assigned to treatment groups of 13 animals each. GM 237354 was administered subcutaneously (s.c.) at 2.5, 10, and 40 mg/kg every 8, 12, or 24 h. Therapy was initiated 1 h after inoculation and was continued for 7 days. Deaths were recorded daily up to 28 days postinoculation. The kidney burden was measured only for the 8-h dosing interval, and consequently, only the data collected for that dosing interval were used in the present analysis. Twelve hours after the end of treatment, three randomly selected animals were killed. The kidneys were removed, weighed, and homogenized with 5 ml of cold sterile saline in a blender (Stomacher 400; Seward Medical, London, United Kingdom). Samples from each specimen were diluted and spread onto SDA plates. After 24 h of incubation at 35°C, the log10 CFU (log CFU) per gram of kidney was calculated.
Pharmacokinetics in mice and dosing regimens simulated.
The drug was administered s.c. to noninfected mice at 40 mg/kg in 500 μl of vehicle, as described previously (1). At different times (0, 0.25, 0.5, 0.75, 1.5, 2, 2.5, and 3 h) three animals were killed and blood samples were collected by cardiac puncture. The blood samples were then centrifuged, and the serum was stored at −20°C until analysis. Serum samples were analyzed for compound concentrations by a reversed-phase high-performance liquid chromatography procedure (1), and the mean of four values obtained for each sampling time was used for PK analysis. PK parameters were derived from the total concentrations in serum measured after the administration of 40 mg/kg s.c. on the basis of a one-compartment open PK model with first-order absorption-elimination kinetics. Then, the PK profiles were simulated for doses of 2.5, 10, and 40 mg/kg administered every 8 h for 2 consecutive days by using WinNonlin PK software (Scientific Consulting, Inc., Apex, N.C.). The dose proportionality of the PKs was previously verified for doses between 5 and 50 mg/kg (1). These simulations corresponded to total concentrations in serum. In addition, PK profiles for the free compound in serum were calculated by applying the formula PKf = PKt · fu, where PKt is the total in vivo concentration simulated with WinNonlin software, fu is the unbound fraction (5 or 0.05%, as determined in vitro by equilibrium dialysis [1]), and PKf is the free, unbound calculated in vivo concentration. This was applied for all three doses used in vivo (2.5, 10, and 40 mg/kg). No accumulation was assumed between doses, which is reasonable considering the half-life of the compound. The predicted profiles of the total and free concentrations in serum were then directly reproduced in the in vitro dynamic PK experiments as free concentrations in vitro (no proteins are present in the broth).
In vitro PK-PD dynamic system. (i) Bioreactor.
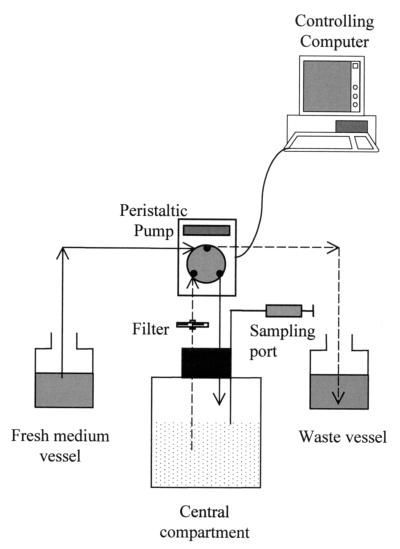
A previously described in vitro model was used in this study (22), with some modifications. The model (Fig. 1) consisted of three connected vessels. The first vessel contained fresh, antifungal agent-free culture medium. The second vessel (also called the central compartment) was filled with the same medium containing only a fungal culture (control growth experiments) or fungal culture and antifungal agent at changing concentrations (killing experiments). The third vessel was a waste reservoir. Peristaltic pumps (Minipuls 3; Gilson, Middleton, Wis.) continuously supplied and removed culture broth from the central compartment at flow rates adjusted to mimic the target PK profile within the central compartment. Calibrated (inner diameter, 0.5 mm) polyvinyl chloride tubes (Gilson) were connected to the peristaltic pumps. The system was controlled with software running on a personal computer (740 ProTech System controller software; Gilson).
FIG. 1.
Schematic representation of the system used for in vitro PK simulation. Continuous and broken lines represent central compartment inflow and outflow, respectively. The filter is used to prevent microorganism loss from the central vessel.
(ii) Simulation of PK profiles in bioreactor.
The system was filled with the basal protein-free medium RPMI 1640 (GIBCO BRL, Life Technologies, Renfrewshire, United Kingdom). The medium contained l-glutamine (Merck, Darmstadt, Germany), was buffered to pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS; Sigma Chemical Co., St. Louis, Mo.), and was supplemented with 2% sorbitol (Sigma-Aldrich S.A., Madrid, Spain) (RPMI–2% sorbitol) to minimize filamentous forms during C. albicans growth (J. L. Rodriguez-Tudela, personal communication). The bioreactor was placed in a shaking water bath adjusted to 30°C. Fifty milliliters of RPMI–2% sorbitol was added to the central compartment, and this volume was kept constant throughout the experiment. In order to closely reproduce PK profiles within the central compartment, flow rates (and, as a consequence, the rotations per minute in the peristaltic pumps) were scheduled to change in the course of the experiment. The central compartment was inoculated with a C. albicans 4711E suspension to yield a final concentration of about 106 CFU/ml. Adherence of the organisms to the surfaces of the bioreactor was avoided by using glass vessels and sufficient shaking. After inoculation, GM 237354 was perfused into the central compartment at predetermined rates for in vitro PK simulations. The solubility of GM 237354 (as the potassium salt) in broth was >5 mg/ml, well above the highest concentration used in the bioreactor. The robustness of the system for the reproduction of PK profiles was first checked in a preliminary experiment. Then, a total of six PK profiles were reproduced in the in vitro model by using (i) concentrations equal to the total levels in serum simulated in vivo after dosing with 2.5, 10, and 40 mg/kg every 8 h for 2 consecutive days or (ii) concentrations equal to the corresponding in vivo free levels in serum (see “Pharmacokinetics in mice and dosing regimens simulated” above).
(iii) Determination of in vitro antifungal activity following dosing with GM 237354 with different PK profiles.
Throughout each experiment, samples (0.1 ml each) were taken from the central compartment for microbiological assessment at times 0, 8, 16, 24, 32, 40, and 48 h. The samples were centrifuged and washed twice with cold saline as a standard method to avoid drug carryover (46). Samples were cultured in duplicate on SAD plates by using an autoplate (Spiral Biotech, Aplicaciones Analíticas, Barcelona, Spain) and incubated for 24 h at 35°C. Then, the colonies in each plate were counted and the numbers of CFU per milliliter were calculated. All the experiments were performed in triplicate.
(iv) Determination of GM 237354 concentrations in medium.
Broth samples (0.2 ml) were taken from the central compartment to measure GM 237354 concentrations. Concentrations were measured by the agar diffusion bioassay method. C. albicans 2005 was used as the indicator microorganism. The medium for the bioassay was prepared by supplementing yeast nitrogen base agar (YNB; Difco) with 10% d-glucose (Sigma-Aldrich S.A.) and 6% sodium citrate (Merck). Then, C. albicans 2005 was added to melted medium stabilized at 45°C to yield a final concentration of 5 × 105 CFU/ml. Supplemented YNB (100 ml) with microorganisms was poured into square plastic Nunc (Nalge Nunc International, Roskilde, Denmark) bioassay plates (245 by 245 mm). The agar was allowed to settle to room temperature for 1 h, and 5-mm-diameter wells were cut with a 36-well template. The wells were loaded with 20 μl of fluid. Standard curves were generated with RPMI–2% sorbitol, with the concentrations used being 0.625, 1.25, 2.5, 5, and 10 μg/ml (accuracy, >95%; precision, <12%). Each standard sample was assayed in triplicate, while unknown samples were loaded in duplicate. The plates were incubated overnight at 35°C, and the inhibition zone was measured with a digital caliper (Mitutoyo Ltd., Warwick, United Kingdom). When a sample was outside of the calibration range, it was diluted with RPMI–2% sorbitol prior to assay.
In vitro-in vivo correlations.
The effect of GM 237354 was measured as the level of reduction in viable counts of C. albicans at the end of the treatment period. The observed effects (EOBSs) in vivo, as fungal counts measured previously in the kidney (1), and in vitro during the PK-PD experiments were expressed as arithmetic means ± standard deviations (SDs) of log CFU per gram of kidney and per milliliter of broth, respectively. In addition, net effects (ENETs) were calculated both in vitro and in vivo. In vitro, ENET was defined as the difference in log CFU per milliliter of broth at 48 h of therapy between the system exposed to changing GM 237354 concentrations and the untreated control system (inoculated with C. albicans 4711E but without GM 237354). In vivo, ENET was expressed as the difference in log CFU per gram of kidney at 7 days of therapy between infected, treated and infected, untreated animals. In addition, the observed area under the killing time curve (AUKTCOBS) was calculated by using the mean observed in vitro data and the linear trapezoidal rule, as implemented in the noncompartmental PK option of the WinNonlin software.
In vitro-in vivo correlations were studied by a simple linear regression between the effects observed in vivo and the effects obtained in vitro by reproducing total or free in vivo serum GM 237354 concentrations.
RESULTS
Serum protein binding.
The binding of GM 237354 to mouse serum was a reversible process in equilibrium. The proportion bound was 95% and proved constant over the concentration range of 1 to 80 μg/ml.
Systemic infection (1).
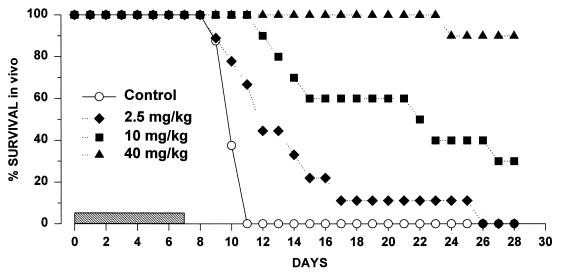
Considering the data generated for the 8-h dosing interval, all untreated animals died within between 8 and 11 days postinfection. GM 237354 produced a statistically significant increase in survival compared with no treatment, even at the lower dose (2.5 mg/kg). Survival was dose dependent. Table 1 presents the area under the survival-time curve (AUSTC) up to 28 days postinfection, the mean survival day, the percentage of survivors at 28 days postinfection, and the kidney burden at 7 days postinfection, at the end of the 7 days of therapy; very close agreement was observed between the different end points. The variability in kidney burden at the end of therapy was very low (the coefficient of variation [CV] ranged from 3 to 10%) except at the high dose (CV = 70%). Figure 2 in turn shows the survival-time curves.
TABLE 1.
Summary of in vivo efficacy data throughout the 28-day treatment period after GM 237354 dosing every 8 h for 7 consecutive days in mice inoculated systemically with C. albicans
| Dose (mg/kg every 8 h) | AUSTC over 28 days (% · day)
|
Mean survival daya | % Survivors | Kidney burden (log CFU)/g)
|
||
|---|---|---|---|---|---|---|
| AUSTCOBSb | AUSTCNETb | EOBSa | ENET | |||
| Untreated | 975 | 0 | 10.25 ± 0.25 | 0 | 6 ± 0.2 | 0 |
| 2.5 | 1,349 | 374 | 15 ± 2.2c | 0 | 5.8 ± 0.3 | −0.2 |
| 10 | 2,065 | 1,090 | 21 ± 2.04c | 30 | 4.2 ± 0.4 | −1.8 |
| 40 | 2,755 | 1,780 | 25.8 ± 0.25c | 90 | 2.3 ± 1.6 | −3.7 |
Values are expressed as means ± SDs.
The AUSTCOBS and AUSTCNET data were taken from reference 1 (in reference 1, AUSTCNET was quoted as ES (effect on survival) and AUSTCOBS was quoted as AUSTC). AUSTCOBS is the AUSTC for the measured, observed, effect. AUSTCNET is calculated as the difference in AUSTCOBS values between untreated and treated animals.
P < 0.05 versus no treatment by Kaplan-Meier survival analysis.
FIG. 2.
Cumulative survival of C. albicans 4711E-infected mice treated with GM 237354 every 8 h for 7 consecutive days. The shaded bar indicates the treatment period.
PKs and in vitro PK simulations. (i) In vitro preliminary PKs.
A preliminary experiment with the dose of 50 mg/kg demonstrated that the in vitro PK-PD system accurately reproduced in vivo GM 237354 PK profiles. In fact, over three in vitro experiments, the maximum concentration in serum (Cmax) was 32.6 ± 4.8 μg/ml, the AUC from time zero to infinity (AUC0–∞) was 52.6 ± 5.8 μg · h/ml, and the terminal half-life (t1/2) was 0.90 ± 0.09 h, with all values expressed as mean ± SD. The values observed in vivo, determined by using the mean levels in serum obtained at the same dose, were 23 μg/ml for Cmax, 46.0 μg · h/ml for AUC0–∞, and 0.85 h for t1/2 (32).
(ii) In vivo and in vitro PK.
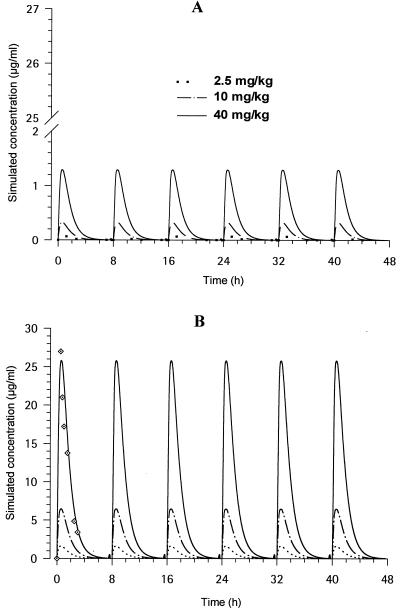
The PK of GM 237354 in mice that received a single s.c. dose of 40 mg/kg was best described by a one-compartment model with a t1/2 of 0.5 h. A Cmax value of 25.72 μg/ml was reached at 0.64 h postdosing. AUC0–∞ was calculated as 44.7 μg · h/ml (1). The CV of the estimated PK parameters was low (range, 2 to 28%). Figure 3 shows the in vivo PK profiles reproduced in vitro with doses of 2.5, 10, and 40 mg/kg by using the PK parameters derived from data observed in vivo with a dose of 40 mg/kg in terms of the free (Fig. 3A) and the total (Fig. 3B) simulated concentrations in serum. These profiles were all reproduced in the broth of the central compartment of the in vitro system. The GM 237354 concentrations measured in vitro were close to the simulated, target in vivo values for the 40-mg/kg regimen, as shown in Fig. 3B. After application of a one-compartment model to these in vitro observed data, the calculated t1/2 was 0.8 h, with the Cmax (33.9 μg/ml) being slightly higher than that obtained after dosing in animals. AUC0–∞ was calculated as 48.4 μg · h/ml, which was close to the value observed in vivo.
FIG. 3.
In vitro PK profiles simulated in the bioreactor with concentrations in broth (free concentrations) equal to free (A) and total (B) GM 237354 in vivo concentrations in serum following administration of 2.5, 10, and 40 mg/kg every 8 h. ·⃟, observed values in the central compartment of the bioreactor when simulating 40 mg/kg every 8 h.
In vitro PK-PD model.
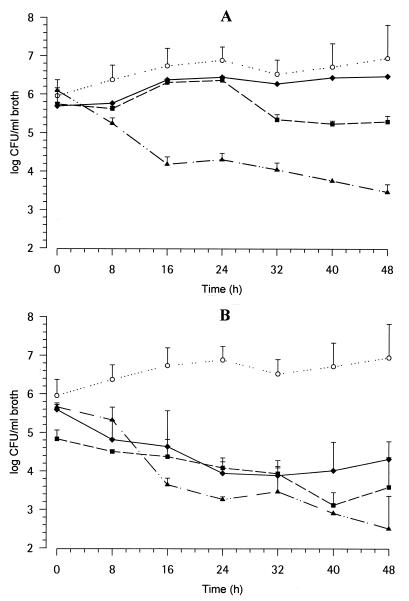
The time course of cells (log CFU per milliliter) of C. albicans 4711E exposed to the PK profiles simulated with 2.5, 10, and 40 mg of GM 237354 per kg every 8 h for 2 days are shown in Fig. 4. Free (Fig. 4A) and total (Fig. 4B) concentrations in serum are mimicked. C. albicans 4711E counts slowly increased over 2 days in untreated cultures (initial and final inocula, 5.7 ± 0.4 and 6.9 ± 0.9 log CFU/ml, respectively). The PK profiles simulated with most doses showed activities against C. albicans viability, as displayed in Table 2. The variability in the burden in broth at the end of the 48-h period was very low (CVs, 13% for the control and 2 and 6% for free and total concentrations, respectively). The profiles for mimicked free GM 237354 PKs displayed dose-dependent antifungal activity, starting earlier with the high dose, although the differentiation between doses was not immediate (Fig. 4A). At higher concentrations (mimicking total compound concentrations; Fig. 4B) the effect was higher, and as a result the differences among doses were less pronounced. The median MIC of GM 237354 was very low, 0.001 μg/ml; however, the time at which the concentration was greater than the MIC (T > MIC) had previously been shown not to predict efficacy in vivo (1).
FIG. 4.
In vitro time-kill plots of C. albicans 4711E exposed to changing GM 237354 (free) concentrations in broth equal to in vivo PK profiles as free concentrations (A) or total concentrations (B). Symbols: ○, untreated cultures; ⧫, 2.5 mg/kg every 8 h; ▪, 10 mg/kg every 8 h; ▴, 40 mg/kg every 8 h.
TABLE 2.
Number of CFU of C. albicans 4711E in vitro for different simulated PK profiles (concentrations in broth equal to in vivo total or free concentrations in serum) of GM 237354 after 48 h of incubation
| Dose (mg/kg every 8 h) | AUKTCOBS over 48 h (log CFU/ml) · h
|
Broth burden (log CFU/ml) at end of 48 h of therapy
|
||||
|---|---|---|---|---|---|---|
|
EOBSa
|
ENET
|
|||||
| Total concn | Free concn | Total concn | Free concn | Total concn | Free concn | |
| Untreated | 317 | 317 | 6.9 ± 0.9 | 6.9 ± 0.9 | 0 | 0 |
| 2.5 | 210 | 299 | 4.3 ± 0.5 | 6.5 ± 0.1 | −2.6 | −0.4 |
| 10 | 194 | 275 | 3.6 ± 0.8 | 5.3 ± 0.2 | −3.3 | −1.6 |
| 40 | 181 | 210 | 2.5 ± 0.8 | 3.5 ± 0.2 | −4.4 | −3.4 |
Values are expressed as means ± SDs.
In vivo-in vitro correlation.
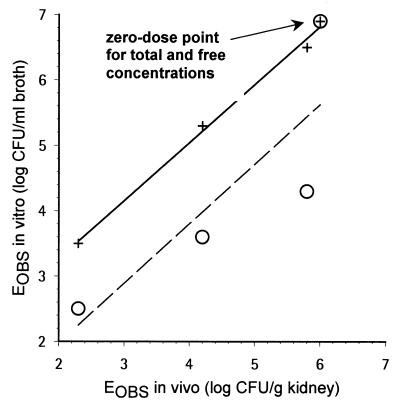
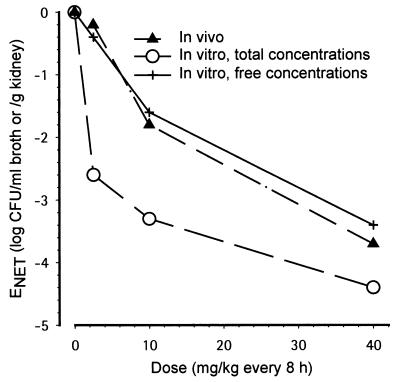
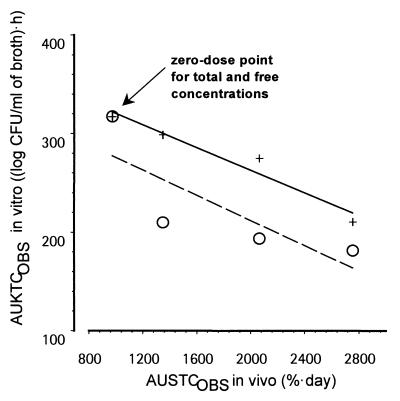
As described before (1) and as shown in Table 1, there was a good agreement between the various in vivo efficacy end points such as AUSTC, mean survival day, and kidney burden. For the in vitro-in vivo correlation, kidney burden could thus be selected as an in vivo end point directly comparable to the broth burden. However, a good correlation between the in vivo (log CFU per gram of kidney) and in vitro (log CFU per milliliter of broth) burden was observed only when using free concentrations and not when using total concentrations. Figure 5 displays EOBS in vitro (EOBS, IN VITRO) as a function of EOBS in vivo (EOBS, IN VIVO) (kidney burden) measured at the end of therapy. There was a clear lack of correlation when total concentrations were used: (i) at 2.5 mg/kg, ENET was −2.4 in vitro, whereas it was −0.2 in vivo; (ii) at 10 mg/kg, ENET was −3.3 in vitro, whereas it was −1.8 in vivo; and (iii) at 40 mg/kg, ENET was −4.4 in vitro, whereas it was −3.7 in vivo (Table 2). When ENET in vivo and ENET in vitro were compared (Fig. 6) as a function of dose, a similar dose-response curve was observed in vivo and in vitro only when free concentrations were used. In addition, the relationship was more gradual compared with that obtained when total concentrations were used in vitro (Fig. 6). Integrating efficacy over the whole experimental periods, Fig. 7 displays AUKTCOBS in vitro (AUKTCOBS, IN VITRO) as a function of AUSTC observed in vivo (AUSTCOBS, IN VIVO) (survival). A very good correlation was also observed only when free concentrations in serum were reproduced in vitro.
FIG. 5.
Relationship between in vivo and in vitro EOBSs at the end of therapy (log CFU per milliliter of broth versus log CFU per gram of kidney). Symbols: ○, total in vivo serum GM 237354 concentrations reproduced in vitro; +, free in vivo serum GM 237354 concentrations reproduced in vitro. Linear regressions were EOBS, IN VITRO = 1.5 + 0.88 · EOBS, IN VIVO for free concentrations (R2 = 0.995) and EOBS, IN VITRO = 0.16 + 0.91 · EOBS, IN VIVO for total concentrations (R2 = 0.700).
FIG. 6.
Dose-response curve for the in vivo and in vitro ENETs for fungal counts at the end of therapy. Symbols: ▴, in vivo systemic infection; ○, in vitro, total GM 237354 concentrations; +, in vitro, free GM 237354 concentrations.
FIG. 7.
Relationship between in vivo and in vitro EOBSs over the whole experimental period as summarized by AUKTCOBS, IN VITRO for broth burden versus AUSTCOBS, IN VIVO for survival. Symbols: ○, total in vivo serum GM 237354 concentrations reproduced in vitro; +, free in vivo serum GM 237354 concentrations reproduced in vitro. Linear regressions were AUKTCOBS, IN VITRO = 378 − 0.058 · AUSTCOBS, IN VIVO for free concentrations simulated in vitro (R2 = 0.946) and AUKTCOBS, IN VITRO = 340 to 0.064 · AUSTCOBS, IN VIVO for total concentrations (R2 = 0.655) simulated in vitro.
DISCUSSION
For antibiotics, prediction of whether a therapeutic dosing regimen is likely to be effective in humans is usually determined by combining PK profiles for human plasma with the PK-PD correlation determined from an in vivo thigh infection model with immunosuppressed mice (Craig's model [17]). Determination of the relationship between intrinsic in vitro activity (MIC), PK characteristics, and in vivo efficacy provides information on the best “PK-PD predictor of efficacy” (15, 47). T > MIC, peak concentration/MIC, and/or AUC/MIC are the parameters most frequently used to determine the efficacies of several antibiotics (11, 17, 38, 41, 42).
Although during the last decade the MIC concept has been revised (9, 12, 18, 30, 33), it remains the most important parameter for defining intrinsic antimicrobial activity and predicting antibiotic potency against a microorganism. Despite the advantages that MICs offer, especially in the clinical setting, one drawback is the lack of information that they provide concerning antimicrobial activity over time. Several methods yield information not only on killing behavior over time (21, 43) (such as killing rates) but also on the duration of the inhibitory activity of the compound, such as the postantibiotic effect. These parameters improve knowledge of the PDs of an anti-infective agent for a set of constant concentrations; however, they do not take into account the PK profiles obtained after dosing in either animals or humans. In vitro PK dynamic models do allow the determination of killing rates by use of PK profiles. The latter could be the best approach for describing the time course of the anti-infective activity while incorporating the link between PKs and PDs (11). They allow the simulation of more dosing regimens and inoculum sizes for different species (including humans) and different microorganisms than is feasible in vivo. Consequently, they can lead to a reduction in the number of in vivo experiments required.
The present study evaluated the correlation between an in vitro PK dynamic model and efficacy in an animal model for an antifungal agent representative of the sordarin class: GM 237354. The same C. albicans strain was used in vitro and in vivo. The influence of protein binding was also assessed.
The in vivo results used to assess the correlation with in vitro results were taken from another study (1); however, they were comparable to those obtained in the same model of systemic candidiasis in mice (32) and are in line with those of other efficacy models (31).
The fungal count at a single time point, namely, the end of the treatment period, was selected in vitro and in vivo as the simplest measure of efficacy. Other measures have been described, although no consensus seems to exist (29). In vivo, the fungal count was measured in the kidney tissue. A good agreement between various in vivo efficacy end points has previously been shown for GM 237354 (1).
Over the 2.5- to 40-mg/kg dose range, a good in vitro-in vivo correlation was obtained between the kidney burden in vivo and the broth “burden” in vitro when PK profiles of the free concentration in serum in vivo were reproduced in vitro. In vivo, following administration of a 40-mg/kg dose every 8 h, the kidney burden at the end of the 7-day treatment period was 2.3 log CFU/g, whereas it was 6.0 log CFU/g in untreated animals. This corresponds to a 3.7-log drop. In vitro, the broth contained 3.5 log CFU/ml at the end of the 2-day GM 237354 exposure period, whereas it contained 6.9 log CFU/ml when the antifungal agent was not added. This corresponds to a 3.4-log drop.
The correlation was observed even though the effect was measured at different times. The durations of the in vitro and in vivo treatments were 2 and 7 days, respectively. This time period was probably sufficient to achieve a maximal effect in both systems. In vitro, the fungal counts stabilized after about 40 h (Fig. 4). In vivo, it remains to be explored if a treatment duration shorter than 7 days would be sufficient to obtain stabilized kidney counts.
In vitro, the numbers of CFU were measured both at time zero and over the whole experiment. In untreated cultures, counts slowly increased over 2 days from 5.7 to 6.9 log CFU/ml. In vivo, in infected, untreated animals, the kidney burden could be determined only at the end of the 7-day treatment period (as 6.0 log CFU/g). The fungal burden in kidney tissue at the start of antifungal therapy is unknown, and it would be interesting to determine the time curve for the kidney burden and compare it to that in vitro. At the end of therapy, the kidney burden in vivo was close to that in the in vitro situation (6.9 log CFU/ml); however, there was a ≈1-log CFU difference between the two systems over the dose range used in the study. This may result from the inoculum size in vitro, which was based a priori on what is commonly accepted for MIC determinations. It remains to be explored whether an even closer in vitro-in vivo agreement could be achieved by selecting, in a preliminary experiment involving untreated systems, an in vitro inoculum that would provide a CFU count close to that observed in vivo at the end of treatment for both systems.
The previous correlation was shown by measurement of the effects at the end of therapy both in vitro (48 h) and in vivo (7 days). By summarizing effect profiles by calculating the areas under the effect-time curves, namely, AUKTC for killing time curves in vitro (0 to 48 h) and AUSTC for survival time curves in vivo (0 to 28 days, with the end of therapy at day 7), a good in vitro-in vivo correlation was also obtained (when the free concentrations are considered).
A good correlation was obtained between the data obtained from the in vitro system and the immunocompetent animal infection model, even though the in vitro system was devoid of immune function.
The good correlation obtained would allow in vitro a complete assessment of the PK-PD behavior of this class of compounds and also investigation of whether or not similar critical values for AUC/MIC would be applicable to several strains for which MICs vary and several sordarin derivatives.
The few reports available on in vitro-in vivo correlations have been obtained with antibiotics. For Staphylococcus aureus, Staphylococcus epidermidis, and Escherichia coli, a good correlation was observed for the reduction in the number of bacteria between the in vitro PK dynamic system and the in vivo tissue cage model in guinea pigs (6). The effect of ticarcillin against Pseudomonas aeruginosa was evaluated in an in vitro dynamic model, and the results were compared with results previously obtained with a murine thigh infection model with dose escalation and dose fractionation. There was a very similar relationship in vitro and in vivo between T > MIC and the change in log CFU (Bonapace et al., 39th ICAAC).
It remains to be assessed whether an in vitro-in vivo correlation can also be observed in the case of cellular PK limitations and intracellular pathogens or when the immune reaction plays an important role in the in vivo response.
The in vitro-in vivo agreement obtained in the present study indirectly suggests that free serum GM 237354 concentrations were an adequate surrogate for free levels at the site of infection. Kidney is a soft tissue in rapid equilibrium with serum, and efficacy was directly linked to the levels in serum. GM 237354 is excreted by the biliary route, with negligible amounts excreted in the urine. Consequently, the concentration of intact compound in urine cannot be related to efficacy. Use of the profiles of the concentration in serum in the in vitro dynamic model is a valid approximation when there is no significant distribution delay between serum and the infection location (i.e., extracellular fluid [ECF]). On the contrary, an alternative approach would be to directly measure PK profiles as the free concentration in ECF at the site of infection by microdialysis and then reproduce these profiles in the bioreactor (13).
It is generally accepted that only unbound drug is pharmacologically relevant, since only free drug can cross membranes and thus access ECF, where soft tissue infections are usually located. Drug uptake by microorganisms likewise depends on free drug levels, except maybe in situations of pinocytosis. In principle, only free drug is able to reach a molecular target and then produce a therapeutic effect, even in the case of diffusion limitations via an active transport mechanism in or out of the cell. These principles have been demonstrated with antibacterial agents (16, 20, 26, 27, 34, 40, 45, 48, 49), as well as with other anti-infective compounds such as antiviral (3, 4, 5) and antimalarial (25) drugs. For example, six penicillins had MICs (determined by using small steps rather than twofold dilutions) very similar to the concentrations in broth and the free concentrations in serum over a 22 to 95% binding range; the level of binding could even be predicted directly from the MICs determined in broth and in serum present at 100% (26, 27). For seven penicillins, in vivo activity increased in direct correlation to the amount of the unbound fraction present (50% effective dose range, 0.74 to 49.7 mg/kg; binding range, 36 to 98%) for the same MIC and similar PK characteristics (34). In vitro PK-PD dynamic systems have already been used to show the influence of protein binding for antibiotic and antiviral agents. These studies have been carried out with broth supplemented with either serum (16, 40) or purified proteins (4, 37). However, the addition of serum to the broth introduces several additional variables that can negatively affect the result, such as compound stability in the presence of serum components or the impairment of microorganism growth. The addition of purified proteins at physiological levels for relevant species (mice, humans) offers an alternative, allowing assessment of the influence of infection when the concentration of binding proteins changes during the infectious process (4). Another method, adopted in the present study, is to simply correct the in vivo PK profiles for total concentration in serum observed in a species for binding by using the free fraction determined in vitro for this species. The in vitro PK-PD dynamic system can then be perfused with drug at these calculated free concentrations.
The experiments described herein could be part of an overall strategy initiated early during drug development and based on the following steps: (i) determination of the compound's PK behavior in the animal efficacy model; (ii) identification, in the in vitro dynamic system, of the PK-PD parameter best predictive of an effect against a key microorganism by using dose escalation and dose fractionation (a PK-PD model may also be established to describe the killing curves for different dosing regimens); (iii) determination of the validity of the PK-PD relationship established in vitro in the animal efficacy model (for screening purposes, the most frequently used animal models are the murine systemic infection model for antifungal agents and the thigh infection model in mice for antibiotics; the experimental design can be optimized on the basis of information collected previously, thus reducing the number of experiments required); (iv) use of the in vitro dynamic system to compare different microorganisms and strains likely to be encountered in the clinical setting, as well as to assess the compound's potential to build resistance (and dosing regimens that may overcome resistance); (v) determination of the compound's PK behavior in humans, initially by allometric scaling and then directly during the first trials with humans (7, 8); (vi) simulation via mathematical modeling of dosing regimens likely to be efficacious in humans to guide the design of early clinical trials; and (vii) performance of confirmatory simulations in the in vitro system (or eventually in an animal efficacy model) by using the human PK profiles for the dosing regimens planned for the trial. The ultimate aim of this strategy should be to derive sufficient information on the pharmacological behaviors of new anti-infective entities before entering the clinical trial phase.
In conclusion, the present study shows a good correlation between an in vivo murine model of systemic candidiasis and an in vitro dynamic system that reproduces in vivo PK profiles. The study was performed with GM 237354, a sordarin derivative, by using various dosing regimens and the same C. albicans strain in vitro and in vivo. GM 237354 showed a reduction in CFU greater than 3 logs both in vitro and in vivo. As suggested, the in vitro dynamic system could be a powerful investigational tool prior to assessment of the efficacy of any anti-infective agent first in animals and then in humans.
ACKNOWLEDGMENTS
We thank Carla D'Angeli and Lisa Squassante for assistance with the statistical analysis and Peter Mutch for performing metabolic studies.
P.A. and C.F. contributed equally to this work.
REFERENCES
- 1.Aviles P, Falcoz C, San Román R, Gargallo-Viola D. Pharmacokinetics-pharmacodynamics of a sordarin derivative (GM 237354) in a murine model of lethal candidiasis. Antimicrob Agents Chemother. 2000;44:2333–2340. doi: 10.1128/aac.44.9.2333-2340.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviles P, Aliouat E M, Martinez A, Dei-Cas E, Herreros E, Dujardin L, Gargallo-Viola D. In vitro pharmacodynamic parameters of sordarin derivatives in comparison with those of marketed compounds against Pneumocystis carinii isolated from rats. Antimicrob Agents Chemother. 2000;44:1284–1290. doi: 10.1128/aac.44.5.1284-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilello J A, Drusano G L. Relevance of plasma protein binding to antiviral activity and clinical efficacy of inhibitors of human immunodeficiency virus protease. J Infect Dis. 1996;173:1524–1526. doi: 10.1093/infdis/173.6.1524. [DOI] [PubMed] [Google Scholar]
- 4.Bilello J A, Bilello P A, Kort J J, Dudley M N, Leonard J, Drusano G L. Efficacy of constant infusion of A-77003: an inhibitor of the human immunodeficiency virus type 1 (HIV-1) protease, in limiting acute HIV-1 infection in vitro. Antimicrob Agents Chemother. 1995;39:2523–2527. doi: 10.1128/aac.39.11.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilello J A, Bilello P A, Stellrecht K, Leonard J, Norbeck D W, Kempf D J, Robins T, Drusano G L. Human serum α1 acid glycoprotein reduces uptake, intracellular concentration, and antiviral activity of A-80987, an inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1996;40:1491–1497. doi: 10.1128/aac.40.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser J, Vergeres P, Widmer A F, Zimmerli W. In vivo verification of in vitro model of antibiotic treatment of device-related infection. Antimicrob Agents Chemother. 1995;39:1134–1139. doi: 10.1128/aac.39.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boxenbaum H. Interspecies pharmacokinetic scaling and the evolutionary-comparative paradigm. Drug Metab Rev. 1984;15:1071–1121. doi: 10.3109/03602538409033558. [DOI] [PubMed] [Google Scholar]
- 8.Boxenbaum H, DiLea C. First-time in human dose selection: allometric thoughts and perspectives. J Clin Pharmacol. 1995;35:957–966. doi: 10.1002/j.1552-4604.1995.tb04011.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown D F J. The comparative methods of antimicrobial susceptibility testing—time for a change? J Antimicrob Chemother. 1990;25:307–312. doi: 10.1093/jac/25.3.307. [DOI] [PubMed] [Google Scholar]
- 10.Clemons K V, Stevens D A. Efficacies of sordarin derivatives GM193663, GM211676, and GM237354 in a murine model of systemic coccidioidomycosis. Antimicrob Agents Chemother. 2000;44:1874–1877. doi: 10.1128/aac.44.7.1874-1877.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig W A. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis. 1997;26:1–12. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 12.Davies B I. The importance of the geometric mean MIC. J Antimicrob Chemother. 1990;25:471–472. doi: 10.1093/jac/25.3.471. [DOI] [PubMed] [Google Scholar]
- 13.Delacher S, Derendorf H, Hollenstein U, Brunner M, Joukhadar C, Hofmann S, Georgopoulos A, Eichler H G, Muller M. A combined in vivo pharmacokinetic-in vitro pharmacodynamic approach to simulate target site pharmacodynamics of antibiotics in humans. J Antimicrob Chemother. 2000;46:733–739. doi: 10.1093/jac/46.5.733. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez J M, Kelly V A, Kinsman O S, Marriott M S, Gomez de las Heras F, Martin J J. Sordarins: a new class of antifungals with selective inhibition of the protein synthesis elongation cycle in yeasts. Antimicrob Agents Chemother. 1998;42:2274–2278. doi: 10.1128/aac.42.9.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drusano G L. Role of pharmacokinetics in the outcome of infections. Antimicrob Agents Chemother. 1988;32:289–297. doi: 10.1128/aac.32.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dudley M N, Blaser J, Gilbert D, Zinner S H. Significance of “extravascular” protein binding for antimicrobial pharmacodynamics in an in vitro capillary model of infection. Antimicrob Agents Chemother. 1990;34:98–101. doi: 10.1128/aac.34.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fantin B, Leggett J E, Ebert S, Craig W A. Correlation between in vitro and in vivo activity of antimicrobial agents against gram-negative bacilli in a murine infection model. Antimicrob Agents Chemother. 1991;35:1413–1422. doi: 10.1128/aac.35.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firsov A A, Savarino D, Ruble M, Gilbert D, Manzano B, Medeiros A A, Zinner S H. Predictors of effect of ampicillin-sulbactam against TEM-1 β-lactamase-producing Escherichia coli on an in vitro dynamic model: enzyme activity versus MIC. Antimicrob Agents Chemother. 1996;40:734–738. doi: 10.1128/aac.40.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Firsov A A, Vostrov S N, Shevchenko A A, Cornaglia G. Parameters of bacterial killing and regrowth kinetics and antimicrobial effect examined in terms of area under the concentration-time curve relationships: action of ciprofloxacin against Escherichia coli in an in vitro dynamic model. Antimicrob Agents Chemother. 1997;41:1281–1287. doi: 10.1128/aac.41.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrison M W, Vance K, Larson T A, Toscano J P, Rotschafer J C. Assessment of effects of protein binding on daptomycin and vancomycin killing of Staphylococcus aureus by using an in vitro pharmacodynamic model. Antimicrob Agents Chemother. 1990;34:1925–1931. doi: 10.1128/aac.34.10.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein E J C, Citron D M, Cherubin C E. Comparison of the inoculum effect of cefoxitin and other cephalosporins and of β-lactamase inhibitors and their penicillin-derived components on the Bacteroides fragilis group. Antimicrob Agents Chemother. 1991;35:1868–1874. doi: 10.1128/aac.35.9.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grasso S, Meinardi G, de Carneri I, Tamassia V. New in vitro model to study the effect of antibiotic concentration and rate of elimination on antibacterial activity. Antimicrob Agents Chemother. 1978;13:570–576. doi: 10.1128/aac.13.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graybill J R, Najvar L K, Fothergill A, Bocanegra R, Gomez de las Heras F. Activities of sordarins in murine histoplasmosis. Antimicrob Agents Chemother. 1999;43:1716–1718. doi: 10.1128/aac.43.7.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herreros E, Martinez C M, Almela M J, Marriott M S, Gomez de las Heras F, Gargallo-Viola D. Sordarins: in vitro activities of new antifungal derivatives against pathogenic yeasts, Pneumocystis carinii, and filamentous fungi. Antimicrob Agents Chemother. 1998;42:2863–2869. doi: 10.1128/aac.42.11.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humberstone A J, Cowman A F, Horton J, Charman W N. Effect of altered serum lipid concentration on the IC50 of halofantrine against Plasmodium falciparum. J Pharm Sci. 1998;87:256–258. doi: 10.1021/js970279q. [DOI] [PubMed] [Google Scholar]
- 26.Kunin C M. I. The importance of serum protein binding in determining antimicrobial activity and concentration in serum. Clin Pharmacol Ther. 1965;7:168–179. doi: 10.1002/cpt196672166. [DOI] [PubMed] [Google Scholar]
- 27.Kunin C M, Craig W A, Kornguth M, Monson R. Influencing of binding on the pharmacological activity of antibiotics. Ann N Y Acad Sci. 1973;226:214–224. doi: 10.1111/j.1749-6632.1973.tb20483.x. [DOI] [PubMed] [Google Scholar]
- 28.Lewis R E, Lund B C, Klepser M E, Ernst E J, Pfaller M A. Assessment of antifungal activities of fluconazole and amphotericin B administered alone and in combination against Candida albicans by using a dynamic in vitro mycotic infection model. Antimicrob Agents Chemother. 1998;42:1382–1386. doi: 10.1128/aac.42.6.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacGowan A, Rogers C, Bowler K. The use of in vitro pharmacodynamic models of infection to optimize fluoroquinolone dosing regimens. J Antimicrob Chemother. 2000;46:163–170. doi: 10.1093/jac/46.2.163. [DOI] [PubMed] [Google Scholar]
- 30.Malouin F, Chamberland S, Brochu N, Parr T R. Influence of growth media on Escherichia coli cell composition and ceftazidime susceptibility. Antimicrob Agents Chemother. 1991;35:477–483. doi: 10.1128/aac.35.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez A, Regadera J, Jimenez E, Santos I, Gargallo-Viola D. Antifungal efficacy of GM237354, a sordarin derivative, in experimental oral candidiasis in immunosuppressed rats. Antimicrob Agents Chemother. 2001;45:1008–1013. doi: 10.1128/AAC.45.4.1008-1013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez A, Aviles P, Jimenez E, Caballero J, Gargallo-Viola D. Activities of sordarins in experimental models of candidiasis, aspergillosis, and pneumocystosis. Antimicrob Agents Chemother. 2000;44:3389–3394. doi: 10.1128/aac.44.12.3389-3394.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattie H, Zang L-C, van Strijen E, Sekh B R, Douwes-Idema A E A. Pharmacokinetic and pharmacodynamic models of the antistaphylococcal effects of meropenem and cloxacillin in vitro and in experimental infection. Antimicrob Agents Chemother. 1997;41:2083–2088. doi: 10.1128/aac.41.10.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrikin D J, Briant J, Rolinson G N. Effect of protein binding on antibiotic activity in vivo. J Antimicrob Chemother. 1983;11:233–238. doi: 10.1093/jac/11.3.233. [DOI] [PubMed] [Google Scholar]
- 35.Mouton J W, Hollander J G d. Killing of Pseudomonas aeruginosa during continuous and intermittent infusion of ceftazidime in an in vitro pharmacokinetic model. Antimicrob Agents Chemother. 1994;38:931–936. doi: 10.1128/aac.38.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pacifici G M, Vianni A. Methods of determining plasma and tissue binding of drugs. Clin Pharmacokinet. 1992;23:449–468. doi: 10.2165/00003088-199223060-00005. [DOI] [PubMed] [Google Scholar]
- 37.Palmer S M, Kang S L, Cappellety D M, Rybak M J. Bactericidal killing activities of cefepime, ceftazidime, cefotaxime, and ceftriaxone against Staphylococcus aureus and β-lactamase-producing strains of Enterobacter aerogenes and Klebsiella pneumoniae in an in vitro infection model. Antimicrob Agents Chemother. 1995;39:1764–1771. doi: 10.1128/aac.39.8.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peck C C, Barr W H, Benet L Z, Collins J, Desjardins R E, Furst D E, Harter J G, Levy G, Ludden T, Rodman J H, Sanathanan L, Schentag J J, Shah V P, Sheiner L B, Skelly J P, Stanski D R, Temple R J, Viswanathan C T, Weissinger J, Yacobi A. Opportunities for integration of pharmacokinetics, pharmacodynamics, and toxicokinetics in rational drug development. J Pharm Sci. 1992;81:605–610. doi: 10.1002/jps.2600810630. [DOI] [PubMed] [Google Scholar]
- 39.Rowland M. Protein binding and drug clearance. Clin Pharmacokinet. 1984;9:10–17. doi: 10.2165/00003088-198400091-00002. [DOI] [PubMed] [Google Scholar]
- 40.Scaglione F, Demartini G, Arcidiacono M M, Dugnani S, Fraschini F. Influence of protein binding on the pharmacodynamics of ceftazidime or ceftriaxone in an in vitro infection model. J Chemother. 1998;10:29–34. doi: 10.1179/joc.1998.10.1.29. [DOI] [PubMed] [Google Scholar]
- 41.Soriano F. Optimal dosage of beta-lactam antibiotics: time above the MIC and inoculum effect. J Antimicrob Chemother. 1992;30:566–569. doi: 10.1093/jac/30.5.567. [DOI] [PubMed] [Google Scholar]
- 42.Soriano F, Ponte C, Santamaria M, Jimenez-Arriero M. Relevance of the inoculum effect of antibiotics in the outcome of experimental infections caused by Escherichia coli. J Antimicrob Chemother. 1990;25:621–627. doi: 10.1093/jac/25.4.621. [DOI] [PubMed] [Google Scholar]
- 43.Soriano F, Edwards R, Greenwood D. Effect of inoculum size on bacteriolytic activity of cefminox and four other beta-lactam antibiotics against Escherichia coli. Antimicrob Agents Chemother. 1992;36:223–226. doi: 10.1128/aac.36.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein D S, Drusano G L. Mathematical modeling of the interrelationship of CD4 lymphocyte count and viral load changes induced by the protease inhibitor indinavir. Antimicrob Agents Chemother. 1998;42:358–361. doi: 10.1128/aac.42.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tawara S, Matsumoto S, Kamimura T, Goto S. Effect of protein binding in serum on therapeutic efficacy of cephem antibiotics. Antimicrob Agents Chemother. 1992;36:17–24. doi: 10.1128/aac.36.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Ogtrop M L, Mattie H, Guiot H F L, van Strijen E, Hazekamp-van Dokkum A M, van Furth R. Comparative study of the effects of four cephalosporins against Escherichia coli in vitro and in vivo. Antimicrob Agents Chemother. 1990;34:1932–1937. doi: 10.1128/aac.34.10.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogelman B, Gudmundsson S, Leggett J E, Turnidge J, Ebert S, Craig W A. Correlation of antimicrobial pharmacokinetic parameters with therapeutic efficacy in an animal model. J Infect Dis. 1988;158:831–847. doi: 10.1093/infdis/158.4.831. [DOI] [PubMed] [Google Scholar]
- 48.Wise R. The relevance of pharmacokinetics to in-vitro models: protein binding—does it matter? J Antimicrob Chemother. 1985;15(Suppl. A):77–83. doi: 10.1093/jac/15.suppl_a.77. [DOI] [PubMed] [Google Scholar]
- 49.Wise R. The clinical relevance of protein binding and tissue concentrations in antimicrobial therapy. Clin Pharmacokinet. 1986;11:471–482. doi: 10.2165/00003088-198611060-00004. [DOI] [PubMed] [Google Scholar]