Abstract
The high temperature requirement factor A1 (HTRA1) is a serine protease which modulates an array of signalling pathways driving basal biological processes. HTRA1 plays a significant role in cell proliferation, migration and fate determination, in addition to controlling protein aggregates through refolding, translocation or degradation. The mutation of HTRA1 has been implicated in a plethora of disorders and this has also led to its growing interest as drug therapy target. This review details the involvement of HTRA1 in certain signalling pathways, namely the transforming growth factor beta (TGF-β), canonical Wingless/Integrated (WNT) and NOTCH signalling pathways during organogenesis and various disease pathogenesis such as preeclampsia, age-related macular degeneration (AMD), small vessel disease and cancer. We have also explored possible avenues of exploiting the serine proteases for therapeutic management of these disorders.
Keywords: HTRA1, TGF-β, WNT, NOTCH, Preeclampsia, HTRA inhibitors
1. Introduction
A limited set of cell-signalling pathways account for a wide array of cellular outcomes and behaviours, namely the TGF-β, canonical Wingless/Integrated (WNT) signalling pathway and NOTCH signalling pathway (Guo and Wang, 2009). The versatile nature of biological responses generated via signalling through a handful of extrinsic factors remained a mystery until the discovery of cross-talk between the pathways which lead to higher order networks resulting in the diverse cell fates. These pathways transcriptionally govern cell proliferation, migration, and fate in both embryonic and adult tissues (Attisano and Wrana, 2013). Various studies have implicated HTRAs as regulator of these pathways to govern a myriad of cell responses. In addition to controlling the fate of protein aggregates through refolding, translocation or degradation, these serine proteases also participate in basal biological pathways, modulating cell proliferation, migration and cell fate (Fig. 1 and Fig. 2) (Clausen et al., 2011). Mutations in HTRA have been observed in a wide range of disorders, thus, this is gaining attention as the target molecule for drug therapy in certain diseases.
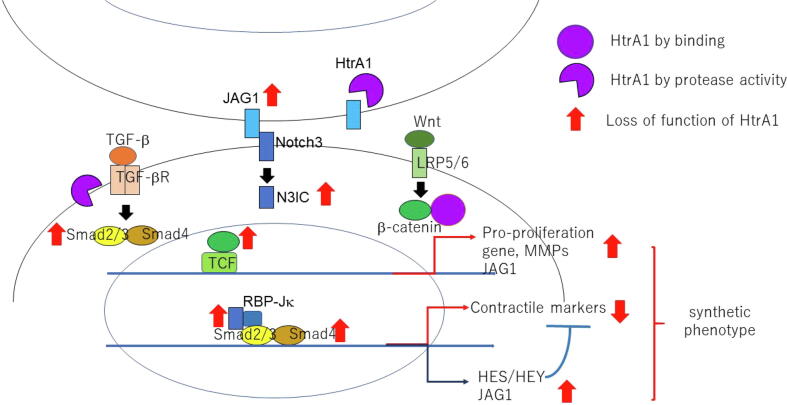
Fig. 1.
Loss of function of HtrA1 induces VSMC synthetic phenotype by activation of TGF-β, Notch3, and Wnt signals.
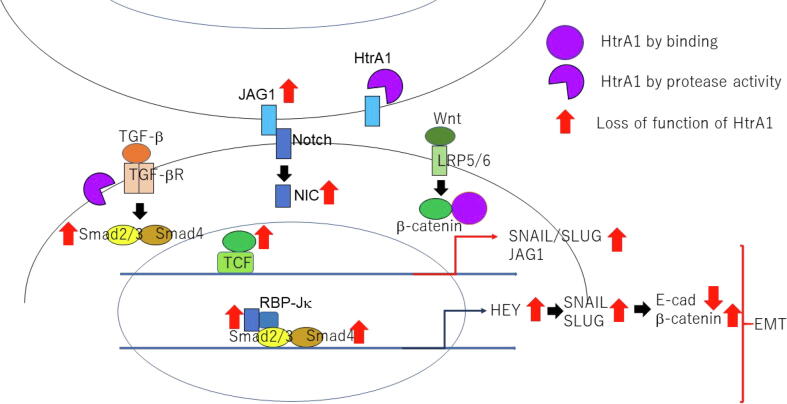
Fig. 2.
Loss of function of HtrA1 induces EMT by activation of TGF-β, Notch and Wnt signals.
The HTRAs are a group of serine proteases involved in protein quality control. This highly conserved protein, expressed in a variety of organisms, ranging from prokaryotes to eukaryotes, is present in humans as four homologues namely HTRA1, HTRA2, HTRA3 and HTRA4 (Oka, 2004). All mammalian HTRA proteins are composed of a protease domain and a PDZ domain (except for short variant of HTRA3(HTRA3S)) in the C-terminal region responsible for proteolytic activity, substrate binding, oligomerization and protein translocation. HTRA2 is unique in its expression in the mitochondria and is mainly involved in protein restructuring and degradation (Moisoi et al., 2009) while the three remaining proteases are similar in domain composition with diverse functionalities. In the N-terminal region, HTRA1, 3 and 4 possess a secretory sequence, IGFBP (Insulin-like growth factor binding protein) domain and a Kazal type protease inhibitor domain. Table 1 depicts structural and functional comparisons of the human HTRAs. A combination of these domains is responsible for the multitude of HTRA functions (Clausen et al., 2011). The mode of HTRA activity remains to be completely elucidated. The following sections elaborate how HTRAs manipulate a host of signalling pathways.
Table 1.
Different structural and functional characteristics of human HTRA proteases.
| Characteristics | HTRA1 | HTRA2 | HTRA3 | HTRA4 | References |
|---|---|---|---|---|---|
| Isoforms | Exists as a single isoform | Two isoforms with the second being proteolytically inactive | Two isoforms HTRA3L and HTRA3S which lacks PDZ domain. | Exists as a single isoform | (Glaza et al., 2015) |
| N-terminal domain | Deletion increases activity 3 folds | Possible roles in directing apoptosis | Deletion increases activity 3 folds | Essential for functional oligomerization. Deletion increases proteolytic activity | (Nguyen, 2017) |
| PDZ domain | Dispensable to activity | PDZ domain restricts entry to active site and so modulates cell apoptotic functions. | Non-essential for protease activity. Needed to form trimeric structure. | Required for conformational changes attributed to activity at temperatures higher than 35 °C | (Bejugam et al., 2013, Runyon et al., 2007) |
| Substrate binding | Resulted in higher order oligomers (from trimeric to 12–24-mers) | Allosteric site binding improves active site catalytic efficiency | Remains a trimer | Substrate binding stabilizes the trimers | (Bejugam et al., 2013, Runyon et al., 2007) |
| Shape | Saucer like form | Trimeric pyramidal shape | Trimer is conical shaped | Trimer | (Glaza et al., 2015, Li et al., 2002) |
| Removing PDZ domain | Remains trimeric in solution | Remains trimeric in solution | Truncated form makes it monomeric in solution | Lowers activity by restricting conformational changes | (Kummari et al., 2019, Runyon et al., 2007) |
| Monomeric form | inactive | inactive | Enzymatically active | Inactive | (Glaza et al., 2015) |
| PD structure | Trimeric structure with a trypsin-like fold | Trypsin like protease domain | Resembles HTRA1 excepting LB loop | Trypsin like protease domain | (Eigenbrot et al., 2012, Walle et al., 2008, Wang et al., 2012) |
2. HTRA1 in different signalling pathways
2.1. Role of HTRA1 in WNT signalling inhibition
The embryonic growth and adult cell homeostasis governed by the WNT signalling pathway is facilitated through the stabilization of the regulatory protein β-catenin. The levels of β-catenin are kept in check by an intracellular destruction complex. The destruction complex composed of Dvl (dishevelled), CK1, Axin, Glycogen synthase kinase 3 (GSK3), anatomous polyposis coli (APC), and βTrcp, is responsible for the phosphorylation, ubiquitination and proteasomal degradation of β-catenin. In the absence of WNT signalling, the β-catenin levels fall through activity of the destruction complex. When the WNT ligand is exosomally transported into the cell, it binds to the Frizzled transmembrane receptor which leads to phosphorylation of the co-receptor low-density lipoprotein receptor related 5 (LRP5). A cascade of events ensues including inactivation of the destruction complex and accumulation of β-catenin. β-catenin is translocated to the nucleus where it complexes with other transcription factors and stimulates WNT-controlled gene expression (Clevers and Nusse, 2012).
A study conducted by Globus et al. indicates that secreted HTRA1 inhibits the WNT signalling pathway by interacting with the β-catenin through its PDZ and Kazal-like domains and lowering downstream gene transcriptions such as Axin2 and CyclinD1. The study also concluded that the catalytic domain of HTRA1 was not involved in WNT signalling inhibition, which was confirmed using both catalytic inactive HTRA1 protein and individual domains transfected into different cell lines (Figs. 1 and 2) (Globus et al., 2017).
Downregulation of HTRA1 has been linked to tumorigenesis, which can be expounded through overexpression of WNT target genes leading to aberrant cellular proliferation (Fig. 1). HTRA1 levels were observed to be reduced in the cancerous cell line compared to the non-cancerous original cell line. Alternatively, cells overexpressing HTRA1 were found to have 60% less proliferative capacity while deliberate depletion of the chaperone protein resulted in 15% increase in the said capacity (Globus et al., 2017).
2.2. Role of HTRA1 in inhibition of TGF- β signalling
TGF-β signalling is elicited by a group of 30 structurally similar cytokines including TGF-βs, bone morphogenic proteins (BMPs), activins and nodals, that bind to the dimeric serine-threonine kinase type 1 and type 2 TGF-β receptors. A third co-receptor, TGF-β type 3, boosts ligand binding to the TGF-β 2 receptor domain. Receptor-ligand binding phosphorylates the type 2 receptor which in turn activates the type 1 receptor serine threonine kinase domain. This type 1 receptor domain recruits SMAD proteins forming a complex which transduces to the nucleus and upregulates gene expression through promoter binding at the TGF-β response elements (Attisano and Labbé, 2004). HTRA1 is postulated to antagonize TGF-β signalling at numerous stages in the pathway through its serine protease domain, beginning at the proteolytic cleavage of the proprotein proTGF-β in the endoplasmic reticulum, to binding to and inactivating various TGF-β receptor-binding ligands such as BMPs, TGF-βs and activins and lastly through cleaving of the TGF-β type 2 and 3 receptors curbing downstream signalling (Figs. 1 and 2) (Graham et al., 2013, Oka, 2004, Shiga et al., 2011).
As the TGF-β signalling pathway is known to regulate various cellular processes in a spatio-temporal manner in both embryos and adult tissues, HTRA1 mediated misregulation leads to various pathologies which can be attributed to the downregulation of HTRA1 precipitating from genetic mutations. HTRA1 mutations have been linked to abnormal osteogenic tissue formation (Filliat et al., 2017) and remodelling as well as a rare disorder known as cerebral autosomal recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CARASIL) (Hara et al., 2009) which is a cerebral small-vessel disorder causing various impairments such as strokes, dementia, mobility issues, alopecia, spondylitis etc (Fukutake and Hirayama, 1995).
2.3. Cross-talk between TGF-β and WNT signalling pathways
Interactions between the signalling pathways occur at various points in the signalling cascade generating diverse molecular responses. Likewise, regulation occurs at multiple sites commencing at the extracellular space in the presence of ligands eventually leading up to the control of downstream signalling by transcription factors in the nucleus. An example of co-operation between the two pathways involves the simultaneous interaction of β-catenin and Smad proteins with TCF, forming the transcriptional regulatory complex responsible for spatio-temporal gene expression of cell fate determinants. Sharing of elements between two distinct pathways is also a well-researched phenomenon demonstrated with axin, which is a component of the destruction complex in WNT signalling, but also associates with Smads to modulate TGF- β signalling (Figs. 1 and 2) (Attisano and Labbé, 2004, Attisano and Wrana, 2013).
2.4. HTRAS in the inhibition of NOTCH signalling pathway
The NOTCH signalling pathway is a linear mechanism which functions in a juxtacrine manner, dependent on neighbouring cell stimulation via specific ligands. The ligands such as JAG1, DLL4 or NOTCH1 interact with the extracellular domain of the membrane-bound NOTCH receptor which undergoes structural alterations on ligand binding and endocytosis, exposing the S2 site for subsequent cleavage (Henrique and Schweisguth, 2019). This releases the Notch intracellular domain (ICD) which proceeds to translocate to the nucleus and associates with specific DNA-binding proteins such as RBP-Jκ to stimulate target gene expressions such as the transcriptional repressors HES and HEY proteins (Figs. 1 and 2) (Klose et al., 2018). In endothelial cells, the Notch ligand JAG1 antagonizes DLL4-mediated Notch signalling and HTRA1 has been found to cleave this JAG1-ICD complex thus activating Notch signalling-directed angiogenesis (Klose et al., 2018). Through the modulation of these signalling mechanisms HTRAs regulate various biological pathways which will be detailed in the subsequent sections.
3. Htra1 in diseases
3.1. Htra1 in placental development and pathology
The placenta performs a host of functions to maintain ideal environment for fetal growth and survival, by taking on the responsibilities of the lungs, gastrointestinal tract, kidneys, liver, endocrine and immune systems. Abnormal placental development not only leads to complications in pregnancy but also predisposes to lifelong illness in both mother and child. To study the different stages of placentation (trophoblast differentiation, feto-maternal vasculature maintenance and utero-placental circulation establishment) various animal models have been investigated, each with their own set of pros and cons (Grigsby, 2016). The expression of HTRA1, associated to both normal placental development as well as the pathogenesis of preeclampsia, has been studied in the placentas of both mouse and humans, demonstrating that HTRA1 is involved in trophoblastic differentiation (Rossant and Cross, 2001, Hasan et al., 2015). As HTRA1 is involved in driving cellular differentiation and vasculature remodelling, aberrant expression can result in defective trophoblastic invasion and subsequent maternal spiral artery remodelling, both of which are involved in the primary stages of preeclamsia.
To analyze the issues of HTRA1 involvement in placental malformation, transgenic HTRA1 knockout mouse were used. HTRA1 null pups were smaller in size than the wild type (Hasan et al., 2015). Embryonic, but not maternal, HTRA1 deficiency was found to be the cause of abnormal placental growth, confirmed by breeding HTRA1-knockout females with wild type males which produced regular placentas during gestation. HTRA1-null placentas showed overall growth retardation. While the placental architecture remained similar histologically, the different regions were much smaller with mislocalization of various trophoblast derived cells, indicative of defective differentiation. The organization of the fetal and placental blood vessels was also more haphazard until the second week. Following E14.5 the phenotypic differences were markedly reduced which can be explained by HTRA3 expression that peaks and remains steady ahead of E14.5 until parturition, and likely to partially compensate for the HTRA1 deficiency (Hasan et al., 2015).
Trophoblast cells which invade the decidua and remodel maternal spiral arteries are affected in HTRA1-null mice during their differentiation and migration phases (Woods et al., 2018). As a result, the maternal arteries are not sufficiently dilated and vessel walls remain thick, reducing potential for blood flow and material exchange within the placenta (Hasan et al., 2015). These aberrant arterial conditions are further exacerbated by lowered matrix metalloproteinase 9 activity that is typically involved in extracellular matrix digestion and cell migration leading to further spiral artery remodelling (Kim et al., 2012).
The differentiation of mouse trophoblast cells is regulated partly by TGF-β signalling which later is modulated by HTRA1 inhibitory activity (Soncin et al., 2015). Absence of HTRA1 early in the pregnancies would thus alter TGF-β mediated cell fate determination accounting for the different cell lineages from trophoblastic progenitors in the HTRA1-null mice and wild type specimens (Hasan et al., 2015). Some studies reported an elevated level of HTRA1 observed in preeclampsia patients which were recorded from sera in later stages of pregnancy and placental blood post parturition (Zong et al., 2013). Further study is required to ascertain whether these elevated levels instigate preeclampsia or are a stress-induced response to it (Dynon et al., 2012, Li et al., 2017, Li et al., 2011, Nie et al., 2006, Wang et al., 2018a).
3.2. HTRA1 in age-related macular degeneration (AMD)
A single nucleotide polymorphism (SNP), rs11200638 in 10q26 genetic locus, was reported to have a significant association with AMD, which was located at the promoter region of HTRA1 gene and suggested that the SNP increased HtrA1 expression in RPE (Chen et al., 2009, Yang et al., 2006). Other two polymorphisms in the 10q26 genetic locus were identified to associate with AMD- a non-synonymous SNP variant (Ala69Ser; rs10490924) in ARMS2 coding region (Rivera et al., 2005), and an insertion/deletion polymorphism (del443ins54) in 3′UTR of ARMS2, which have involved in stabilization of ARMS2 mRNA (Fritsche et al., 2008). Since ARMS2 is located immediately upstream of the HtrA1, the strong linkage disequilibrium existing between these three polymorphisms have made difficult to determine which or both genes have function in AMD susceptibility by genetic linkage analysis. A number of studies in mice overexpressing human or mouse HTRA1 have shown the formation of the Bruch’s membrane and vascular abnormalities and polypoidal lesions in association with AMD (Fu, 2014, Jones et al., 2011, Liu and Hoh, 2015, Nakayama et al., 2014, Vierkotten et al., 2011). ARMS2 is primate specific gene, which makes more difficult the issue to be resolved. However, at least Nakayama et al. (2014) showed that overexpression of wild type ARMS2 or Ala69Ser ARMS2 in mouse had no effect on RPE function. Furthermore, several studies indicated that rs10490924 and del443ins54(in/del) of ARMS2 lie within the regulatory elements of HtrA1 gene and play role in its expression (Iejima et al., 2015, Pan et al., 2021, Yang et al., 2014). Although the possibility that ARMS2 gene function in AMD susceptibility is not completely excluded, it becomes widely accepted that the upregulation of HtrA1 gene by polymorphisms in 10q26 locus is involved in AMD pathogenesis (Liao et al., 2017). In line with this, recently two transcription factors have been identified which bind to the regulatory element in in/del and increase HtrA1 expression level in iPS cells derived from AMD patients (Pan et al., 2021). Variations in the HTRA1 promoter sequence due to SNPs have also been shown to differentiate between PCV and AMD in a study conducted by Ng et al. (2016). They concluded that the SNP rs2672598 is responsible for increased promoter activity while rs11200638 apparently had no impact on transcription factor binding. This former variant also differentially affects transcription in AMD and PCV (Ng et al., 2016).
The expression pattern of HTRA1 in normal eyes was mapped against those suffering from neovascular AMD obtained from post-mortem autopsied tissues. Higher HTRA1 levels were detected in the periphery as compared to the macula in normal eyes. In contrast, a much higher expression of HTRA1 was observed in the macula than periphery in AMD eyes. HTRA1 was detected in the vascular endothelia; anatomically macula has less retinal vessel than periphery in normal eyes. It is possible that enhanced expression of HTRA1 is associated with choroidal neovascularization in macular region or large drusen formation, which are hallmark of AMD. Induced HTRA1 might have proapoptotic activity and protease activity for extracellular matrix protein in macular region.
The initial stages of AMD involve drusen formation and inflammation driven retinal pigment epithelial (RPE) loss affecting central vision. A progressive condition leads to either exudative or non-exudative AMD, both of which have been linked to abnormally elevated levels of HTRA1. Aging and oxidative stress are main causes of AMD. Further it has been shown that oxidative stress induces HtrA1 expression which protects oxidation-induced cell death by promoting senescence of RPE cells and links to AMD (Supanji et al., 2013). Exudative or wet AMD is characterized by choroidal neovascularization (CNV) with the vasculopathy causing serous discharge and haemorrhaging due to abnormal angiogenesis mediated by vascular endothelial growth factors (VEGF) (Ng et al., 2016). Additionally, the elevated HTRA1 levels in drusen, abnormal RPE and CNV lesions have been associated with the increased secretion of VEGF under stress condition and inflammation in human vitreous humors suggesting further that HTRA1 plays a pivotal role in AMD. Dry or non-exudative AMD or geographic atrophy results in complete degeneration of the macular tissue leading to blindness.
There are several possible ways in which HTRA1 may induce AMD. It alternatively mediates the TGF-β pathway, which is responsible for modulation of angiogenesis, through interactions with various TGF-β family receptor binding factors such as TGF β1, TGF β2, activin, BMP4 and growth differentiation factor 5 (GDF5) (Zhang et al., 2012). GDF6 is a member of the TGF-β family responsible for neural and vascular development in the eye and its interplay with HTRA1 is crucial for TGF-β signalling and ocular development (Zhang et al., 2012). A mutation in the GDF6 gene, rs6982567, lowers the level of this mediator while increases the HTRA1 level at the same time, posing a significant risk of AMD (Liu et al., 2018).
Several obstacles lie in studying the pathophysiology of AMD including cell type used, multiple genetic risk factors contributing to the disease, inaccessibility to source tissue and lack of animal models. Two previous studies circumvented some of these issues using wild-type RPE cells and induced pluripotent stem RPE cells derived from an AMD patient who had the high‐risk HTRA1/ARMS2 allele (Yang et al., 2014, Lin et al., 2018). It was found that oxidative stress contributes to the development of AMD (Yang et al., 2014). The other study demonstrated increased expression of HTRA1 and its substrates including EFEMP1 and thrombospondin (TSP1) in cells with high-risk ARMS2/HTRA1 allele (Lin et al., 2018). EFEMP1 is an extracellular matrix protein which forms drusen deposits, and activates the complement pathway instigating an inflammatory response and regulates TGF-β directed angiogenesis. HTRA1 also cleaves TSP1 thus dysregulating its angiogenic inhibitor activity (Lin et al., 2018).
A very recent Phase 1 clinical trial study has confirmed the HTRA1 activity in AMD by determining a new biomarker Dickkopf-related protein 3 (DKK3) (Tom et al., 2020). Another ongoing study funded by the National Health Institute is working on an innovative treatment for AMD with HTRA1 as the therapeutic target. They propose to introduce cloned monoclonal antibodies with high affinity for HTRA1 into RPE cells using gene therapy related tissue-specific delivery techniques. Their work aims to inhibit the proteolytic activity of HTRA1 to prevent CNV leading to wet AMD (HTRA1 as a Therapeutic Target in the Treatment of Wet AMD Shaw, Peter X. University of California, San Diego, La Jolla, CA, United States).
3.3. Htra1 in small vessel disease
A hallmark of vascular disorders is a phenotypic shift in vascular smooth muscle cells (VSMCs). These cells exist on a phenotypic spectrum with two discrete and interchangeable phenotypes on extreme ends, namely the fully differentiated contractile phenotype involved in vessel contractility and the de-differentiated synthetic phenotype responsible for extracellular matrix synthesis and maintenance (Ikawati et al., 2018). These shifts are modulated by a variety of intra and extra cellular stimuli resulting in cell types with distinctive morphological, proliferative, and migratory characteristics and unique marker proteins (Owens et al., 2004). Following this shift to the synthetic phenotype resulting in an initial increase in VSMCs, a rapid loss will ensue, followed by extracellular matrix depletion and fibrotic arteries, all contributing to vascular disorders such CARASIL.
Loss of function study with HTRA1 in mice has demonstrated a link with the modulation of VSMC phenotypes that exhibit stronger migratory and proliferative capacities, higher matrix metalloproteinase activity (especially MMP9) and a propensity towards oxidative stress-induced apoptosis. Characteristic elevated expressions of marker proteins such as vimentin and osteopontin in HTRA1-null mice compared to wild types further solidifies the claim of a synthetic phenotypic switch. However, mice only demonstrate significant changes to aortic vessels in contrast to cerebral blood vessel changes observed in humans leading to CARASIL. These differences can be attributed to anatomical variations between the species in vascular development and metabolism (Ikawati et al., 2018).
HTRA1 mediates both the TGF-β signalling pathway and the NOTCH signalling in the regulation of VSMC differentiation (Fig. 1). The cross-talk occurs between NOTCH and TGF-β in which DNA-binding protein RBP-Jκ associates with phosphor-Smad2/3 and synergistically elevates expression of contractile proteins SM22α and α-SMA (Tang and Liaw, 2010). These signallings direct expression of contractile proteins, also stimulate the expression of transcriptional repressor HEY and HES proteins (Klose et al., 2019). HTRA1 cleaves JAG1 which is a NOTCH receptor ligand and cleaves pro-TGF-β1 or GDF6 of the TGF-β pathway, maintaining a balance between the expression of contractile mediators and the repressor proteins. In the absence of HTRA1, overexpression of HES and HEY repressors are observed which stunt the achievement of the contractile functionality of VSMCs (Klose et al., 2019). The cells remain in the synthetic conformation, prone to oxidative stress induced-degradation leading to small vessel disease complications (Ikawati et al., 2018). It was reported that Wnt/β-catenin signalling induced pro-proliferation genes and MMPs to enhance proliferation and migration of VSMCs (Cheng et al., 2019), which also might involve in the phenotype shift of HTRA1 deficient VSMCs in cooperation with TGF-β and NOTCH signalling (Fig. 1).
Cerebral small vessel diseases (CSVD) such as cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) and the symptomatically similar CARASIL are monogenic disorders. The former is caused by NOTCH3 mutation leading to excessive accumulation of NOTCH3 extracellular domain (NOTCH3-ECD) and vessel wall disruption with complex protein aggregates. HTRA1 has been directly linked to NOTCH3 signalling regulation (Klose et al., 2019). Substantiating the claims of HTRA1 association with the CSVD, proteasome analysis of CADASIL patients exposed colocalization of mutant HTRA1 with NOTCH3-ECD and accumulation of a host of HTRA1 substrates, precipitated by loss of activity (Zellner et al., 2018). Hereditary CARASIL, on the other hand, is direct result of mutations in HTRA1, being mono- or bi allelic in nature. The heterozygous HTRA1 mutations are often asymptomatic but accumulated with NOTCH3 mutation might lead to CADASIL-like symptoms (Verdura et al., 2015).
CADASIL and the closely related CARASIL, characterized by leg muscle spasticity, followed by alopecia, back pain and a plethora of neurological symptoms such as dementia and mood disorders and eventual stroke (Onodera et al., 2019), have been linked directly or indirectly with HTRA1 aberrations (Wu et al., 2018, Zellner et al., 2018, Klose et al., 2019).
3.4. Htra1 and other members in carcinogenesis
The effects of HTRAs including HTRA1 and HTRA3 have been extensively reviewed by Zurawa-Zanicka et al. (Zurawa-Janicka et al., 2017), thus this review mainly focuses on some recent developments in the field. Klose et al. (Klose et al., 2018) recently analyzed the effects of HTRA1 on tumor angiogenesis. Since NOTCH1 signalling is involved in regulating angiogenesis through endothelial cells, this pathway was specifically investigated. The results showed that HTRA1 indeed played a key role in regulating NOTCH1 signalling by cleaving JAG1 which antagonizes the NOTCH ligand DLL4 responsible for anti-angiogenic effects (Klose et al., 2018). JAG1 is an agonist in smooth muscle cell while NOTCH3 signalling appears to be an antagonist in endothelial cell NOTCH1 signalling (Benedito et al., 2009). In the absence of HTRA1 under pathological conditions, haphazard vessel sprouting is observed through uncontrolled VEGF activity leading to poorly vascularized tumor pockets. Thus, HTRA1 depletion might be a viable strategy in impairing tumor angiogenesis (Klose et al., 2018).
WNT signalling plays roles in different stages of cancer pathology including cancerous stem cell proliferation, improving cell viability, promoting tumor angiogenesis and metastasis, suppressing immune response and conferring resistance to chemotherapeutics (Arend et al., 2013). A recent study delineated the mutations in different components of the signalling pathway from β-catenin to members of the destruction complex that precipitate in hyperactivation of β-catenin and result in related gene overexpression, leading to epithelial ovarian cancer (EOC). Aberrant WNT signalling has been linked to all subtypes of EOC (Nguyen et al., 2019). HTRA1 also regulates WNT signalling by interfering with β-catenin directed transcription, and its downregulation has been observed in different subtypes of EOC (Lu et al., 2004). This downregulation can be attributed to epigenetic silencing of HTRA1 in ovarian cancer, which is further confirmed by the discovery of methylated CpG islands in promoter and exon sequences of HTRA1 in non-expressing ovarian cancer cells (Chien et al., 2009). A signalling network among HTRA1, NOTCH, TGF- β, and WNT leading to epithelial mesenchymal transition (EMT) is shown in Fig. 2 during cancer progression. Treatment with the methyltransferase inhibitor 5-aza-cytidine was found to increase HTRA1 expression (Chien et al., 2004) and the higher level of HTRA1 confers better response to chemotherapy in gastric, ovarian (Chien et al., 2009) and breast cancers (Folgueira et al., 2005). Conversely, chemotherapeutics such as paclitaxel and cisplatin modulate HTRA1 upregulation and propagate HTRA1 directed caspase 3/7 mediated cell-death while suppression of HTRA1 by RNA interference triggered chemoresistance (Chien et al., 2006).
HTRA4 possesses similar moieties compared to HTRA1 and HTRA3 (Glaza et al., 2015, Runyon et al., 2007) which stands to reason that it would confer similar tumor suppressor and apoptotic capabilities against cancer cells. Its expression has been shown to negatively correlate with the growth of brain, breast, and prostate cancer (Chien et al., 2009). A recent study analyzed its ability to effect chemotherapeutic drug efficacy in deterring oncogenesis (Wenta et al., 2019). Under stress conditions, HTRA4 directs apoptosis of cancer cells through the proteolytic cleavage of XIAP (X-linked inhibitor of apoptotic proteins), and improves the effectiveness of drugs such as etoposide and cisplatin in driving oncogenic cells towards apoptosis, reducing clonogenic potential and motility of cancer cells, and arresting cell cycle at the G2/M phase, thereby curbs metastatic growth. Both the long and short variants of HTRA3 (HTRA3L and HTRA3S) cleave the anti-apoptotic protein XIAP through proteolytic activity, and the N terminally truncated HtrA4 is more effective in triggering cell death (Wenta et al., 2019). Furthermore, HTRA3 co-localizes with actin, vimentin and β-tubulin which are all propagators of caspase mediated apoptosis (Byun et al., 2001, Wenta et al., 2019). All four HTRAs seem to direct apoptosis and likely to act in conjunction to regulate cancer cell apoptosis depending on various stress conditions.
4. Conclusions
The cross-talk between the TGF-β, WNT and NOTCH signalling pathways govern various cellular mechanisms and imbalances in the intricate mechanisms through ectopic gene expressions that likely to lead to major disease manifestations (Pelullo et al., 2019). Table 2 provides a general outline of the involvement of these pathways in pathogenesis. Sufficient evidence has been detailed to present different forms of interplay in biological pathways such as WNT, TGF-β and NOTCH signalling as mediators of various pathologies, and as such it is evident that manipulating these pathways through HTRA proteases imparts considerable therapeutic value.
Table 2.
HTRA1,3 and 4 mediated pathways involved in various pathologies.
| Biological site | Pathology | Biological Pathway | HTRAs involved | References |
|---|---|---|---|---|
| Placenta | Pre- eclampsia | TGF-β | HTRA1,3 and 4 | (Hasan et al., 2015, Li et al., 2011, Liu et al., 2018, Wang et al., 2018) |
| Eyes | Age related macular disorder (AMD) | TGF-β | HTRA1 | (Friedrich et al., 2015) |
| Cerebral blood vessel | Small vessel disease (CARASIL, CADASIL) | TGF-β, NOTCH3 | HTRA1 | (Beaufort et al., 2014, Klose et al., 2019, Tikka et al., 2014) |
| Multiple sites | Cancer | WNT, NOTCH1 | HTRA1, 3 and 4 | (Cheng et al., 2019, Globus et al., 2017, Klose et al., 2018) |
| Neurons | Alzheimer’s disease | TGF-β | HTRA1 | (Grau et al., 2005, Launay et al., 2008) |
| Bones, cartilage | Osteoarthritis | TGF- β | HTRA1, 3 | (Graham et al., 2013, Larkin et al., 2013) |
Elevated level of HTRA1 is correlated to preeclampsic and AMD pathophysiologies that are propagated through TGF-β signalling-directed cell fate determination and choroidal neovascularization respectively. As such, the application of HTRA1 inhibitors could form the possible basis of a therapeutic regimen. To design HTRA1 inhibitors, a previous study proposed the exploitation of allosteric sites on these chymotrypsin-like serine proteases alongside exploring the active site (Zurawa-Janicka et al., 2010). Since the active sites of HTRAs have been well conserved, allosteric sites can serve as a more viable focus for developing target specific inhibitors (Zurawa-Janicka et al., 2010). The non-ribosomal peptide inhibitors mimic substrate like binding to the serine protease active sites without undergoing proteolysis, and Ahp-cyclodepsipeptide scaffold has been suggested for the synthesis of tailored human HTRA protease inhibitors (Köcher et al., 2017). The tailored inhibitors are likely to have improved target specificity, circumventing the issue of unintended physiological side effects. A previous study through in-silico screening of allosteric sites between the proteolytic and PDZ domains of Helicobater pylori HTRA proposed a site-specific bioactive ligand (Perna et al., 2014). Similar studies can be conducted on human HTRA1-3 to derive relevant allosteric inhibitors.
Carcinogenesis has been negatively correlated with HTRAs, demonstrating the beneficial role they play in deterring cancer metastasis via anoikis, caspase-dependant and independent apoptosis as well as TGF-β signalling inhibition which is imperative in oncogenesis (Chien et al., 2009). HTRA1-4 proteolytically degrade anti-apoptotic protein XIAP, and together with other anti-cancer drugs, such as cisplatin, improve response to chemoresistance.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
Authors would like to thank Razmin Bari for English editing and proof-reading.
Authors’ contributions
All authors have contributed substantially to conception, design, acquisition of data, analysis and interpretation. All authors were involved in writing and critical revision of the manuscript. All authors have read and approved the current version of manuscript for publication.
Funding
None.
Availability of data and materials
Not applicable.
Footnotes
Peer review under responsibility of King Saud University.
References
- Arend R.C., Londoño-Joshi A.I., Straughn J.M., Buchsbaum D.J. The Wnt/β-catenin pathway in ovarian cancer: A review. Gynecol. Oncol. 2013;131(3):772–779. doi: 10.1016/j.ygyno.2013.09.034. [DOI] [PubMed] [Google Scholar]
- Attisano L., Labbé E. TGFβ and Wnt pathway cross-talk. Cancer Metastasis Rev. 2004;23:53–61. doi: 10.1023/a:1025811012690. [DOI] [PubMed] [Google Scholar]
- Attisano L., Wrana J.L. Signal integration in TGF-β, WNT, and Hippo pathways. F1000Prime Rep. 2013;5 doi: 10.12703/P10.12703/P5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaufort N., Scharrer E., Kremmer E., Lux V., Ehrmann M., Huber R., Houlden H., Werring D., Haffner C., Dichgans M. Cerebral small vessel disease-related protease HtrA1 processes latent TGF-β binding protein 1 and facilitates TGF-β signaling. Proc. Natl. Acad. Sci. 2014;111(46):16496–16501. doi: 10.1073/pnas.1418087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejugam P.R., Kuppili R.R., Singh N., Gadewal N., Chaganti L.K., Sastry G.M., Bose K., Srinivasula S.M. Allosteric regulation of serine protease HtrA2 through novel non-canonical substrate binding pocket. PLoS One. 2013;8(2):e55416. doi: 10.1371/journal.pone.005541610.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito R., Roca C., Sörensen I., Adams S., Gossler A., Fruttiger M., Adams R.H. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Byun Y., Chen F., Chang R., Trivedi M., Green K.J., Cryns V.L. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001;8(5):443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- Chen W., Xu W., Tao Q., Liu J., Li X., Gan X., Hu H., Lu Y. Meta-analysis of the association of the HTRA1 polymorphisms with the risk of age-related macular degeneration. Exp. Eye Res. 2009;89(3):292–300. doi: 10.1016/j.exer.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Cheng H., Zhu H., Cao M., Lu C., Bao S., Pan Y. HtrA1 suppresses the growth of pancreatic cancer cells by modulating Notch-1 expression. Brazilian J. Med. Biol. Res. 2019;52 doi: 10.1590/1414-431X20187718. e7718–e7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J., Aletti G., Baldi A., Catalano V., Muretto P., Keeney G.L., Kalli K.R., Staub J., Ehrmann M., Cliby W.A. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J. Clin. Invest. 2006;116:1994–2004. doi: 10.1172/JCI27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J., Campioni M., Shridhar V., Baldi A. HtrA serine proteases as potential therapeutic targets in cancer. Curr. Cancer Drug Targets. 2009;9:451–468. doi: 10.2174/156800909788486704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J., Staub J., Hu S.-I., Erickson-Johnson M.R., Couch F.J., Smith D.I., Crowl R.M., Kaufmann S.H., Shridhar V. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene. 2004;23(8):1636–1644. doi: 10.1038/sj.onc.1207271. [DOI] [PubMed] [Google Scholar]
- Clausen T., Kaiser M., Huber R., Ehrmann M. HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. Mol. cell Biol. 2011;12(3):152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- Clevers, H., Nusse, R., 2012. Review Wnt/b -Catenin Signaling and Disease. 10.1016/j.cell.2012.05.012. [DOI] [PubMed]
- Dynon K., Heng S., Puryer M., Li Y., Walton K., Endo Y., Nie G., Wang H. HtrA3 as an early marker for preeclampsia: specific monoclonal antibodies and sensitive high-throughput assays for serum screening. PLoS One. 2012;7(9):e45956. doi: 10.1371/journal.pone.004595610.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrot C., Ultsch M., Lipari M.T., Moran P., Lin S.J., Ganesan R., Quan C., Tom J., Sandoval W., van Lookeren Campagne M. Structural and functional analysis of HtrA1 and its subdomains. Structure. 2012;20:1040–1050. doi: 10.1016/j.str.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Filliat G., Mirsaidi A., Tiaden A.N., Kuhn G.A., Weber F.E., Oka C., Richards P.J., Papaccio G. Role of HTRA1 in bone formation and regeneration: In vitro and in vivo evaluation. PLoS One. 2017;12(7):e0181600. doi: 10.1371/journal.pone.0181600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folgueira M.A.A.K., Carraro D.M., Brentani H., da Costa Patrão D.F., Barbosa E.M., Netto M.M., Caldeira J.R.F., Katayama M.L.H., Soares F.A., Oliveira C.T., Reis L.F.L., Kaiano J.H.L., Camargo L.P., Vêncio R.Z.N., Snitcovsky I.M.L., Makdissi F.B.A., da Silva e Silva P.J., Góes J.C.G.S., Brentani M.M. Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin. Cancer Res. 2005;11(20):7434–7443. doi: 10.1158/1078-0432.CCR-04-0548. [DOI] [PubMed] [Google Scholar]
- Friedrich U., Datta S., Schubert T., Plössl K., Schneider M., Grassmann F., Fuchshofer R., Tiefenbach K.-J., Längst G., Weber B.H.F. Synonymous variants in HTRA1 implicated in AMD susceptibility impair its capacity to regulate TGF-β signaling. Hum. Mol. Genet. 2015;24(22):6361–6373. doi: 10.1093/hmg/ddv346. [DOI] [PubMed] [Google Scholar]
- Fritsche L.G., Loenhardt T., Janssen A., Fisher S.A., Rivera A., Keilhauer C.N., Weber B.H.F. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat. Genet. 2008;40(7):892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- Fu Y. A functional approach to examine the role of HTRA1 versus ARMS2 in AMD. Invest. Ophthalmol. Vis. Sci. 2014;55(10):6524. doi: 10.1167/iovs.14-15670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutake T., Hirayama K. Familial young-adult-onset arteriosclerotic leukoencephalopathy with alopecia and lumbago without arterial hypertension. Eur. Neurol. 1995;35(2):69–79. doi: 10.1159/000117096. [DOI] [PubMed] [Google Scholar]
- Glaza P., Osipiuk J., Wenta T., Zurawa-Janicka D., Jarzab M., Lesner A., Banecki B., Skorko-Glonek J., Joachimiak A., Lipinska B., van Raaij M.J. Structural and functional analysis of human HtrA3 protease and its subdomains. PLoS One. 2015;10(6):e0131142. doi: 10.1371/journal.pone.0131142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globus O., Evron T., Caspi M., Siman-Tov R., Rosin-Arbesfeld R. High-temperature requirement A1 (Htra1)-a novel regulator of canonical Wnt signaling. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-18203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.R., Chamberland A., Lin Q., Li X.J., Dai D., Zeng W., Ryan M.S., Rivera-Bermúdez M.A., Flannery C.R., Yang Z. Serine protease HTRA1 antagonizes transforming growth factor-β signaling by cleaving its receptors and loss of HTRA1 in vivo enhances bone formation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau S., Baldi A., Bussani R., Tian X., Stefanescu R., Przybylski M., Richards P., Jones S.A., Shridhar V., Clausen T. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc. Natl. Acad. Sci. 2005;102:6021–6026. doi: 10.1073/pnas.0501823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigsby, P.L., 2016. Animal models to study placental development and function throughout normal and dysfunctional human pregnancy, B: Seminars in reproductive medicine. NIH Public Access, c 11. [DOI] [PMC free article] [PubMed]
- Guo X., Wang X.-F. Signaling cross-talk between TGF-β/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Shiga A., Fukutake T., Nozaki H., Miyashita A., Yokoseki A., Kawata H., Koyama A., Arima K., Takahashi T. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N. Engl. J. Med. 2009;360:1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- Hasan M.Z., Ikawati M., Tocharus J., Kawaichi M., Oka C. Abnormal development of placenta in HtrA1-deficient mice. Dev. Biol. 2015;397:89–102. doi: 10.1016/j.ydbio.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Henrique D., Schweisguth F. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development. 2019;146 doi: 10.1242/dev.172148. [DOI] [PubMed] [Google Scholar]
- Iejima D., Itabashi T., Kawamura Y., Noda T., Yuasa S., Fukuda K., Oka C., Iwata T. HTRA1 (high temperature requirement A serine peptidase 1) gene is transcriptionally regulated by insertion/deletion nucleotides located at the 3′ end of the ARMS2 (age-related maculopathy susceptibility 2) gene in patients with age-related macular degener. J. Biol. Chem. 2015;290:2784–2797. doi: 10.1074/jbc.M114.593384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawati M., Kawaichi M., Oka C. Loss of HtrA1 serine protease induces synthetic modulation of aortic vascular smooth muscle cells. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A., Kumar S., Zhang N., Tong Z., Yang J.-H., Watt C., Anderson J., Fillerup H., McCloskey M., Luo L. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc. Natl. Acad. Sci. 2011;108:14578–14583. doi: 10.1073/pnas.1102853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.H., Kwon H.-J., Kim D.-S. Matrix metalloproteinase 9 (MMP-9)-dependent processing of βig-h3 protein regulates cell migration, invasion, and adhesion. J. Biol. Chem. 2012;287:38957–38969. doi: 10.1074/jbc.M112.357863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R., Adam M.G., Weis E.-M., Moll I., Wüstehube-Lausch J., Tetzlaff F., Oka C., Ehrmann M., Fischer A. Inactivation of the serine protease HTRA1 inhibits tumor growth by deregulating angiogenesis. Oncogene. 2018;37:4260–4272. doi: 10.1038/s41388-018-0258-4. [DOI] [PubMed] [Google Scholar]
- Klose R., Prinz A., Tetzlaff F., Weis E.-M., Moll I., Rodriguez-Vita J., Oka C., Korff T., Fischer A. Loss of the serine protease HTRA1 impairs smooth muscle cells maturation. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-54807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köcher S., Rey J., Bongard J., Tiaden A.N., Meltzer M., Richards P.J., Ehrmann M., Kaiser M. Tailored Ahp-cyclodepsipeptides as Potent Non-covalent Serine Protease Inhibitors. Angew. Chemie Int. Ed. 2017;56:8555–8558. doi: 10.1002/anie.201701771. [DOI] [PubMed] [Google Scholar]
- Kummari R., Dutta S., Chaganti L.K., Bose K. Discerning the mechanism of action of HtrA4: a serine protease implicated in the cell death pathway. Biochem. J. 2019;476:1445–1463. doi: 10.1042/BCJ20190224. [DOI] [PubMed] [Google Scholar]
- Larkin D.J., Kartchner J.Z., Doxey A.S., Hollis W.R., Rees J.L., Wilhelm S.K., Draper C.S., Peterson D.M., Jackson G.G., Ingersoll C., Haynie S.S. Inflammatory markers associated with osteoarthritis after destabilization surgery in young mice with and without Receptor for Advanced Glycation End-products (RAGE) Frontiers in physiology. 2013;4:121. doi: 10.3389/fphys.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay S., Maubert E., Lebeurrier N., Tennstaedt A., Campioni M., Docagne F., Gabriel C., Dauphinot L., Potier M.C., Ehrmann M. HtrA1-dependent proteolysis of TGF-β controls both neuronal maturation and developmental survival. Cell Death Differ. 2008;15:1408–1416. doi: 10.1038/cdd.2008.82. [DOI] [PubMed] [Google Scholar]
- Li W., Srinivasula S.M., Chai J., Li P., Wu J.-W., Zhang Z., Alnemri E.S., Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 2002;9:436–441. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- Li Y., Puryer M., Lin E., Hale K., Salamonsen L.A., Manuelpillai U., Tong S., Chan W., Wallace E.M., Nie G. Placental HtrA3 is regulated by oxygen tension and serum levels are altered during early pregnancy in women destined to develop preeclampsia. J. Clin. Endocrinol. Metab. 2011;96:403–411. doi: 10.1210/jc.2010-1405. [DOI] [PubMed] [Google Scholar]
- Li Y., Salamonsen L.A., Hyett J., da Silva Costa F., Nie G., Costa S. Maternal HtrA3 optimizes placental development to influence offspring birth weight and subsequent white fat gain in adulthood. Sci. Rep. 2017;7:1–14. doi: 10.1038/s41598-017-04867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S.-M., Zheng W., Zhu J., Lewis C.A., Delgado O., Crowley M.A., Buchanan N.M., Jaffee B.D., Dryja T.P. Specific correlation between the major chromosome 10q26 haplotype conferring risk for age-related macular degeneration and the expression of HTRA1. Mol. Vis. 2017;23:318. [PMC free article] [PubMed] [Google Scholar]
- Lin, M.K., Yang, J., Hsu, C.W., Alexander, A.G., Lewis, G.B., Ryan, M.B., Jesse, C., Mahajan, V.B., Tsang, S.H., 2018. Processes extracellular matrix proteins EFEMP1 and TSP1. 10.1111/acel.12710. [DOI] [PMC free article] [PubMed]
- Liu C., Xing F., He Y., Zong S., Luo C., Li C., Duan T., Wang K., Zhou Q. Elevated HTRA1 and HTRA4 in severe preeclampsia and their roles in trophoblast functions. Mol. Med. Rep. 2018;18:2937–2944. doi: 10.3892/mmr.2018.9289. [DOI] [PubMed] [Google Scholar]
- Liu J., Hoh J. Postnatal overexpression of the human ARMS2 gene does not induce abnormalities in retina and choroid in transgenic mouse models. Invest. Ophthalmol. Vis. Sci. 2015;56:1387–1388. doi: 10.1167/iovs.14-15914. [DOI] [PubMed] [Google Scholar]
- Lu K.H., Patterson A.P., Wang L., Marquez R.T., Atkinson E.N., Baggerly K.A., Ramoth L.R., Rosen D.G., Liu J., Hellstrom I. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin. cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A., Plun-Favreau H., Edwards R.E., Teismann P., Esposti M.D. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009;16:449–464. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- Nakayama M., Iejima D., Akahori M., Kamei J., Goto A., Iwata T. Overexpression of HtrA1 and exposure to mainstream cigarette smoke leads to choroidal neovascularization and subretinal deposits in aged mice. Invest. Ophthalmol. Vis. Sci. 2014;55:6514–6523. doi: 10.1167/iovs.14-14453. [DOI] [PubMed] [Google Scholar]
- Ng T.K., Liang X.Y., Lai T.Y.Y., Ma L., Tam P.O.S., Wang J.X., Chen L.J., Chen H., Pang C.P. HTRA1 promoter variant differentiates polypoidal choroidal vasculopathy from exudative age-related macular degeneration. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep28639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, C., 2017. Characterizing the Function of the N-Terminal Domain of Omi/.
- Nguyen V.H.L., Hough R., Bernaudo S., Peng C. Wnt/β-catenin signalling in ovarian cancer: Insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019;12:1–17. doi: 10.1186/s13048-019-0596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie G., Li Y., Hale K., Okada H., Manuelpillai U., Wallace E.M., Salamonsen L.A. Serine peptidase HTRA3 is closely associated with human placental development and is elevated in pregnancy serum. Biol. Reprod. 2006;74:366–374. doi: 10.1095/biolreprod.105.047324. [DOI] [PubMed] [Google Scholar]
- Oka C. HtrA1 serine protease inhibits signaling mediated by Tgf family proteins. Development. 2004;131:1041–1053. doi: 10.1242/dev.00999. [DOI] [PubMed] [Google Scholar]
- Onodera, O., Nozaki, H., Fukutake, T., 2019. HTRA1 disorder.
- Owens G.K., Kumar M.S., Wamhoff B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Pan Y., Iejima D., Nakayama M., Suga A., Noda T., Kaur I., Das T., Chakrabarti S., Guymer R.H., DeAngelis M.M. Binding of Gtf2i-β/δ transcription factors to the ARMS2 gene leads to increased circulating HTRA1 in AMD patients and in vitro. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelullo M., Zema S., Nardozza F., Checquolo S., Screpanti I., Bellavia D. Wnt, Notch, and TGF-β pathways impinge on hedgehog signaling complexity: an open window on cancer. Front. Genet. 2019;10:711. doi: 10.3389/fgene.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna A.M., Reisen F., Schmidt T.P., Geppert T., Pillong M., Weisel M., Hoy B., Simister P.C., Feller S.M., Wessler S. Inhibiting Helicobacter pylori HtrA protease by addressing a computationally predicted allosteric ligand binding site. Chem. Sci. 2014;5:3583–3590. doi: 10.1039/C4SC01443J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera A., Fisher S.A., Fritsche L.G., Keilhauer C.N., Lichtner P., Meitinger T., Weber B.H.F. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- Rossant J., Cross J.C. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Runyon S.T., Zhang Y., Appleton B.A., Sazinsky S.L., Wu P., Pan B., Wiesmann C., Skelton N.J., Sidhu S.S. Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3. Protein Sci. 2007;16:2454–2471. doi: 10.1110/ps.073049407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga A., Nozaki H., Yokoseki A., Nihonmatsu M., Kawata H., Kato T., Koyama A., Arima K., Ikeda M., Katada S. Cerebral small-vessel disease protein HTRA1 controls the amount of TGF-β1 via cleavage of proTGF-β1. Hum. Mol. Genet. 2011;20:1800–1810. doi: 10.1093/hmg/ddr063. [DOI] [PubMed] [Google Scholar]
- Soncin F., Natale D., Parast M.M. Signaling pathways in mouse and human trophoblast differentiation: a comparative review. Cell. Mol. Life Sci. 2015;72:1291–1302. doi: 10.1007/s00018-014-1794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supanji, Shimomachi M., Hasan M.Z., Kawaichi M., Oka C. HtrA1 is induced by oxidative stress and enhances cell senescence through p38 MAPK pathway. Experimental eye research. 2013;112:79–92. doi: 10.1016/j.exer.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Tang Y., Liaw L. Notch and transforming growth factor-beta signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. FASEB J. 2010;24:822–829. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikka S., Baumann M., Siitonen M., Pasanen P., Pöyhönen M., Myllykangas L., Viitanen M., Fukutake T., Cognat E., Joutel A. Cadasil and Carasil. Brain Pathol. 2014;24:525–544. doi: 10.1111/bpa.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom I., Pham V.C., Katschke K.J., Li W., Liang W.-C., Gutierrez J., Young A.A., Figueroa I., Eshghi S.T., Lee C.V. Development of a therapeutic anti-HtrA1 antibody and the identification of DKK3 as a pharmacodynamic biomarker in geographic atrophy. Proc. Natl. Acad. Sci. 2020;117:9952–9963. doi: 10.1073/pnas.1917608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdura E., Herve D., Scharrer E., Amador M.D.M., Guyant-Marechal L., Philippi A., Corlobe A., Bergametti F., Gazal S., Prieto-Morin C., Hervé D., Scharrer E., Amador M.D.M., Guyant-Maréchal L., Philippi A., Corlobé A., Bergametti F., Gazal S., Prieto-Morin C., Beaufort N., Le Bail B., Viakhireva I., Dichgans M., Chabriat H., Haffner C., Tournier-Lasserve E. Heterozygous HTRA1 mutations are associated with autosomal dominant cerebral small vessel disease. Brain. 2015;138:2347–2358. doi: 10.1093/brain/awv155. [DOI] [PubMed] [Google Scholar]
- Vierkotten S., Muether P.S., Fauser S. Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch’s membrane via cleavage of extracellular matrix components. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walle L. Vande, Lamkanfi M., Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- Wang L.-J., Cheong M.-L., Lee Y.-S., Lee M.-T., Chen H. High-temperature requirement protein A4 (HtrA4) suppresses the fusogenic activity of syncytin-1 and promotes trophoblast invasion. Mol. Cell. Biol. 2012;32:3707–3717. doi: 10.1128/MCB.00223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Li, Y., Hyett, J., Costa, S., Nie, G., 2018. HtrA3 Isoform – Specific ELISAs for Early Detection of Preeclampsia 27–31. 10.1177/1087057116682425. [DOI] [PubMed]
- Wang Y., Li Y., Hyett J., da Silva Costa F., Nie G. HtrA3 Isoform-Specific ELISAs for Early Detection of Preeclampsia. SLAS Discov. Adv. Life Sci. R&D. 2018;23:1092–1099. doi: 10.1177/1087057116682425. [DOI] [PubMed] [Google Scholar]
- Wenta T., Rychlowski M., Jurewicz E., Jarzab M., Zurawa-Janicka D., Filipek A., Lipinska B. The HtrA3 protease promotes drug-induced death of lung cancer cells by cleavage of the X-linked inhibitor of apoptosis protein (XIAP) FEBS J. 2019;286:4579–4596. doi: 10.1111/febs.14977. [DOI] [PubMed] [Google Scholar]
- Woods L., Perez-Garcia V., Hemberger M. Regulation of placental development and its impact on fetal growth—new insights from mouse models. Front. Endocrinol. 2018;Lausanne). 9:570. doi: 10.3389/fendo.2018.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li C., Mao J., Li L., Liu Y., Hou Y. Heterozygous HTRA1 missense mutation in CADASIL-like family disease. Brazilian J. Med. Biol. Res. 2018;51 doi: 10.1590/1414-431X20176632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li Y., Chan L., Tsai Y.-T., Wu W.-H., Nguyen H.V., Hsu C.-W., Li X., Brown L.M., Egli D. Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Hum. Mol. Genet. 2014;23:3445–3455. doi: 10.1093/hmg/ddu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Camp N.J., Sun H., Tong Z., Gibbs D., Cameron D.J., Chen H., Zhao Y., Pearson E., Li X. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science (80-.) 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Zellner A., Scharrer E., Arzberger T., Oka C., Domenga-Denier V., Joutel A., Lichtenthaler S.F., Müller S.A., Dichgans M., Haffner C. CADASIL brain vessels show a HTRA1 loss-of-function profile. Acta Neuropathol. 2018;136:111–125. doi: 10.1007/s00401-018-1853-8. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lim S.L., Du H., Zhang M., Kozak I., Hannum G., Wang X., Ouyang H., Hughes G., Zhao L. High temperature requirement factor A1 (HTRA1) gene regulates angiogenesis through transforming growth factor-β family member growth differentiation factor 6. J. Biol. Chem. 2012;287:1520–1526. doi: 10.1074/jbc.M111.275990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong L., Wang L., Huang P., Shao W., Song Y., Gou W. High temperature requirement A1 in placental tissues and serum from pre-eclamptic pregnancies with or without fetal growth restriction. Arch. Med. Sci. AMS. 2013;9:690. doi: 10.5114/aoms.2013.34989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawa-Janicka D., Skorko-Glonek J., Lipinska B. HtrA proteins as targets in therapy of cancer and other diseases. Expert Opin. Ther. Targets. 2010;14:665–679. doi: 10.1517/14728222.2010.487867. [DOI] [PubMed] [Google Scholar]
- Zurawa-Janicka D., Wenta T., Jarzab M., Skorko-Glonek J., Glaza P., Gieldon A., Ciarkowski J., Lipinska B. Structural insights into the activation mechanisms of human HtrA serine proteases. Arch. Biochem. Biophys. 2017;621:6–23. doi: 10.1016/j.abb.2017.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.