Abstract
Crimean-Congo hemorrhagic fever (CCHF) is one of the utmost broadly distributed tick-borne viruses, with an infection resulting in a fatality rate of up to 30%. During this study period, 25,000 hard adult ticks of Hyalomma species were collected from freshly slaughtered imported camels to determine the presence of Crimean-Congo hemorrhagic fever virus (CCHFV) and genetic lineage of the virus. Ticks were pooled and analyzed for the existence of CCHFV using nested RT- PCR and real-time reverse transcription PCR; the genome was detected in 18 (1.44%) tick pools. Partial genome sequences reveal an adjacent relationship with strains from South Africa to Namibia, Nigeria, Sudan, Senegal, and Mauritania, corresponding to the Africa I and III genotypes. This study indicates the presence of CCHFV in Egypt and illustrates the potential for tick-borne dissemination of the virus. Further studies focused on not only tick samples, but also human samples are epidemiologically valuable to obtain exact data in the region.
Keywords: Crimean-Congo hemorrhagic fever, Hyalomma ticks, Nested RT- PCR, Real-time reverse transcription PCR, Egypt
1. Introduction
Crimean-Congo hemorrhagic fever (CCHF) is one of the tick-borne viral disorders that is broadly distributed in Africa, Europe, and Asia (Okely et al., 2020, Cajimat et al., 2017, Wölfel et al., 2007). It was being detected in the Crimea area of the former Soviet Union in 1944 and became identified as ‘Crimean hemorrhagic fever’. After that, it was identified in the Belgian Congo in 1956 which was called ‘Congo virus’; both names were assembled as CCHF in 1969 (Mazzola and Kelly-Cirino, 2019, Spengler et al., 2019). Since 2000, minor outbreaks of Crimean-Congo hemorrhagic fever virus (CCHFV) have been reported in India, Pakistan, Iran, Turkey, Greece, Spain, Sudan, Uganda, and Georgia. The incidence of CCHFV has been rising since 2008 (Wahid et al., 2019). CCHF has a wide geographical spreading; it transpires in Asia (India, Pakistan, Tajikistan, Kazakhstan, and Russia), the Middle East (Iran and Afghanistan), Europe (Turkey, Bulgaria, Greece, Albania, and Kosovo), and +30 African republics (including Sudan, Mauritania, Kenya, Senegal, and South Africa) (Farhadpour et al., 2016).
CCHFV belongs to the Bunyaviridae family and the genus of Nairovirus (Voorhees et al., 2018, Kazancioğlu et al., 2017). The CCHFV had a spherical shape with a diameter around 80–100 nm, lipid envelope is 5–7 nm, thick and glycoprotein spikes are 8–10 nm long (Wahid et al., 2019). CCHFV is an enveloped virus that acquires a negative-sense RNA genome which contains three segments. The small segment (S, 1.7 kb) translates the nucleocapsid proteins. The medium segment (M, 5.3 kb) encodes M ∼ 1700 amino acids' precursor that promote the glycoproteins' production consisting of mature Gn (37-kDa) and Gc (75-kDa). Meanwhile, the large segment (L, 12.1 kb) encodes the viral polymerase (RNA-dependent RNA-polymerase (450 kDa- L-RdRp) (Papa, 2019, Wahid et al., 2019, Voorhees et al., 2018, Cajimat et al., 2017). CCHFV is marked by excessive genetic variation, based on the series of S segment. CCHFV strains are divided into 7 hereditary lineages (double from Asia, double from Europe, and three from Africa) (Papa, 2019, Tezer and Polat, 2015).
In an enzootic cycle involving humans and vertebrates, CCHFV could be transmitted by ticks. Hyalomma spp. ticks have been reported as the natural vector and reservoir (Shi et al., 2018, Cajimat et al., 2017, Spengler et al., 2016). After being nibbled by infected ticks, several domestic and wild animals, including sheep, goats, cattle, camels, and hares, act as intensifying hosts. But it’s difficult to reveal animal infections as the tick-animal-tick cycle is symptomless (Mazzola and Kelly-Cirino, 2019). Farmers, butchers, housewives, veterinarians, animal dealers, and other people involved in the care of animals exposed to ticks are mostly vulnerable to CCHF. Moreover, health workers are the subsequent most frequently influenced high-risk group, mostly due to its nosocomial nature (Saleem et al., 2020). In the normal transmission pattern of CCHFV to humans, the virus is spread via tick bite, sometimes because of publicity to infected organs or the blood of livestock (Farhadpour et al., 2016).
CCHFV was identified as a vastly pathogenic virus to humans, with a fatality rate of 5–30% is generally notified (Fillâtre et al., 2019, Papa, 2019, Tezer and Polat, 2015). Humans infected with CCHFV may present asymptomatic, mild, or severe disease. Symptoms may include fevers, chills, muscular pain, dizziness, headache, mood changes, pleural effusion, sore throat, neck stiffness, lymphadenopathy, gastrointestinal symptoms, and hemorrhagic diathesis, with multiorgan dysfunction is seen in severe cases (Raabe, 2020, Zohaib et al., 2020, Shi et al., 2018). The existence of CCHFV may be exactly detected in Hyalomma ticks using nucleic acid amplification, viral antigen, and/or nucleic acid amplification testing merged with proteomics (Raabe, 2020). This study was conducted to survey and molecularly characterize CCHFV in ticks assembled from camels, and to investigate the origin of CCHFV in Egypt.
2. Material and methods
2.1. Tick's collections and processing
Hard adult ticks were collected from imported camels to Egypt from December 2018 to April 2021. All ticks were separated from different body parts of camels by fine curved-tip forceps with a great care to avoid the sample's damage. Ticks were placed in a glass jars with porous covers for O2-exchange. Containers were labeled with collection date, transported to the laboratory, and morphologically identified to species level. Subsequently, ticks were divided into pools consisting about 20 hard adult ticks. To eliminate excess particulate contamination from animal hides, each pool was rinsed twice with sterile water, flushed once with EtOH (70%, v/v), and afterward cleansed twice using Minimum Essential Medium (MEM; Sigma-Aldrich, St. Louis, MO) which including antibiotic-antimycotic mixture (GIBCO-BRL; New York, USA).
Tick pools were then crushed in a sterilized mortar and pestle using 90-mesh alundum sand, with 2 mL MEM including 15% fetal bovine serum (Biochrome KG; Berlin, Germany), 2% antibiotic-antimycotic mixture, and 2 % l-glutamine. We clarified the homogenates using centrifugation at low-speed (5000 rpm) and supernatants were collected and kept at −70 °C until the extraction of RNA (Lutomiah et al., 2014, Sang et al., 2006).
2.2. RNA extraction
We extracted viral RNA from a 140 μL aliquot of supernatant using the QIAamp® Viral RNA Mini, Cat No. 52904 (Qiagen, Germany) using the manufacturer’s instruction.
2.3. Nested RT-PCR
Viral RNA was amplified using a nested RT-PCR targeting CCHFV S segment (Voorhees et al., 2018), using Qiagen®OneStep RT-PCR Kit (Qiagen, Germany). The first run was operated using the following primers: R3 (5′-GACAAATTCCCTGCACCA-3′), F2 (5′-TGGACACCTTCACAAACTC-3′), and F2C (5′-TGGATACTTTCACAAACTC-3′). A total of 10 μL of the extracted RNA was inserted to each singular PCR-tube. The reaction was achieved in a 50-μL total volume and the thermal cycling settings were 50 °C for 30 min; 95 °C for 15 min; 5 cycles at 95 °C for 45 s, 37 °C for 45 s, and 72 °C for 45 s; 35 cycles at 95 °C for 45 s, 53 °C for 45 s, 95 °C for 45 s, and a final extension at 72 °C for 5 min. The primers magnify a 536-bp of S-segment by performing 2% agarose gel electrophoresis.
Nested PCR was performed in a 50-μL reaction with 1-μL of first PCR product using the following primers: R2a (5′ GACATCACAATTTCACCAGG-3′), R2b (5′ GACATTACAATTTCGCCAGG-3′), F3 (5′-GAATGTGCATGGGTTAGCTC-3′), and F3C (5′-GAGTGTGCCTGGGTTAGCTC-3′). We used Promega Go Taq G2 master mix (Cat No. M7833, Promega, Germany) with the following cycle conditions: 95 °C for 2 min; 40 cycles of 95 °C for 45 s, 53 °C for 45 s, 72 °C for 45 s; and a final expansion at 72 °C for 5 min. The primers amplify a 260-bp of amplicons by performing 2% agarose gel electrophoresis.
2.4. Real-time reverse transcription PCR
Total RNA extracted for the viral S segment was tested using a real-time reverse transcription PCR (rRT-PCR) assay (Wölfel et al., 2007). This assay was performed in wells triplicate of 48-well plates using the GoTaq Probe 1-Step RT-qPCR System (Cat No. A6120, Promega, Germany). A 5 μL of sample and 25-μL reactions run were used on the Applied Biosystems StepOne™ Real-Time PCR System (Thermo Scientific, Waltham, MA, USA). The reaction was performed at 48 °C for 30 min; 95 °C for 2 min; 40 cycles of 95 °C for 30 s, 60 °C for 60 s, and finally 68 °C for 25 s. The fluorescence data were assembled and measured after each elongation step. The positive and negative control were included in all reactions.
2.5. CCHFV sequencing and analysis
We successfully sequenced 8 samples to determine the CCHFV genotypes. We subjected the amplicons to the modified Sanger sequencing on automated ABI genetic analyzer 3500 using BigDye Terminator Cycle Sequencing Reaction Kit V3.1. We retained the sequences of the CCHFV amplicons used in our study, as well as sequences available in GenBank for phylogenetic analysis. We conducted multiple alignments of sequences with MUSCLE in SeaView - Multiplatform GUI (graphical user interface) for molecular phylogeny (Version 5.0.4 software) (Gouy et al., 2010). Phylogenetic tree of partial small (S) segment sequence (260 nucleotides) of CCHFV was constructed using maximum-likelihood method, Tamura three-parameter model and bootstrap value at 50% cut-off point using Molecular Evolutionary Genetics Analysis (MEGA X version 10.1.8 software) (Kumar et al., 2018). We deposited the nucleotide sequences obtained in this study in GenBank under the accession numbers of MW424419, MW467897, MW467898, MZ330127, MZ342904, MZ322095, MZ326698, and MZ361738.
3. Results
3.1. Tick identification
During this study period, a total of 25,000 adult hard ticks were assembled from imported camels from different African countries (Ethiopia, Somalia, Kenya, and Sudan), and identified under a stereomicroscope. Only one genus was defined as Hyalomma and two species were recorded as the key species infesting camels. Amongst, the most abundant tick species was Hyalomma dromedarii (n = 16,750; 67%), which was followed by Hyalomma rufipes (n = 8250; 33%). All ticks were grouped into 1248 pools, consisting of about 20 adult ticks, by size, species, animal source, and collection date. The numbers and distribution of tick species based on the animal source are tabulated in Table 1.
Table 1.
Distribution of tick species according to the animal sources.
| Animal Source | No. of Ticks |
|
|---|---|---|
| Hyalomma dromedarii (Pools) | Hyalomma rufipes (Pools) | |
| Ethiopia | 3387 (169) | 1426 (71) |
| Somalia | 4141 (207) | 2574 (128) |
| Kenya | 3859 (193) | 1718 (86) |
| Sudan | 5363 (268) | 2532 (126) |
| Total | 16,750 (837) | 8250 (411) |
3.2. Virus detection
All tick pools were imperiled to nested RT-PCR and rRT-PCR to detect CCHF viral genome. The molecular analysis of both RT-PCR products revealed that the CCHFV was detected in 18 tick pools (1.44%), where 2 pools were considered as weak positive by rRT-PCR (Fig. 1 & Fig. 2). The positive control always gave positive amplification signals, but no signal was seen in the negative control. Among the CCHFV-positive pools, 11 pools (61.1%) of H. dromedarii and 7 pools (38.9%) of H. rufipes were infected with the viral genome. The 18 positive pools covered ticks collected from camels imported from Sudan (11 pools), Somalia (5 pools), and Ethiopia (2 pools), as portrayed in Table 2.
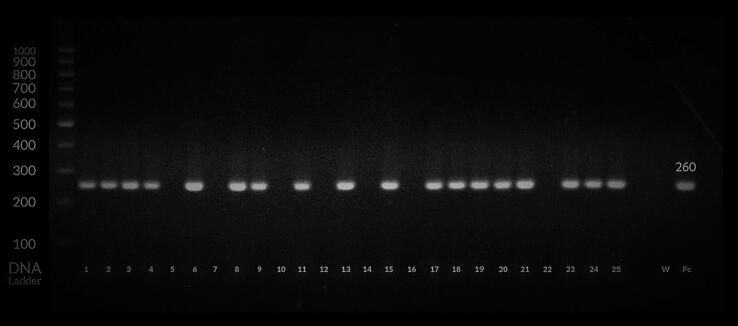
Fig. 1.
Amplification of the S segment of the CCHFV genome using nested RT-PCR representing the DNA bands with 260 base pairs (bp) from infected ticks and positive control (Pc). * W negative control; Pc positive control; samples No. S5, S7, S10, S12, S14, S16 and S22 were negative; S1, S2, S4, S6, S11, S17, S18, S19, S20, S23 and S25 (Hyalomma dromedarii); S3, S8, S9, S13, S15, S21 and S24 (Hyalomma rufipes) were positive.
Fig. 2.
Qualitative real time RT-PCR for CCHFV showing positive and negative controls, and positive samples isolated from infected ticks.
Table 2.
Results of CCHFV detection by nested and real-time RT- PCR in tick pools.
| Animal Source |
No. of CCHFV Positive Pools |
|||
|---|---|---|---|---|
|
Nested RT- PCR |
Real time RT- PCR |
|||
| H. dromedarii | H. rufipes | H. dromedarii | H. rufipes | |
| Ethiopia | 2 | 0 | 3 | 0 |
| Somalia | 3 | 2 | 3 | 2 |
| Kenya | 0 | 0 | 0 | 0 |
| Sudan | 6 | 5 | 7 | 5 |
| Total | 11 | 7 | 13 | 7 |
3.3. Sequencing and phylogenetic analysis
Out of 18 positive pools, eight tick pools were amplified and sequenced the partial S segment. The sequence comparison of eight partial segments with other published CCHFV sequences showed 99.6–95% identity.
Pairwise sequence alignments among the isolates revealed that both the detected gene sequence and genotype were almost identical (Table 3). The partial S segment coding sequences of CCHFV were separated into seven groups and two various genotypes, including Africa I and III were detected after the phylogenetic analysis (Table 4). Nucleotide alignments of the partial genomic sequence of CCHFV exhibited that MW467897 and MW467898 strains clustered together with United Arab Emirate strains (MN516481; 96.9% and 97.3% homology and MN516484; 96.5% and 96.9% homology) and Egyptian strain (JF706233; 97.9% and 98.5% homology). While MZ342904, MW424419, and MZ330127 clustered together with Nigeria strain (KX238958; 99.6%, 97.7, and 97.6% homology) within the Africa III lineage. Other three partial segments (GenBank accession no. MZ326698, MZ361738, and MZ322095) clustered together with strains from Iran, west, and south Africa within the Africa I lineage (Fig. 3).
Table 3.
Genetic distances (%) among the CCHFV genotypes.
| MZ322095 | MZ326698 | MZ361738 | MW424419 | MW467897 | MW467898 | MZ330127 | MZ342904 | |
|---|---|---|---|---|---|---|---|---|
| MZ322095 | – | 95.9% | 100% | 83.5% | 85.1% | 85.5% | 83.5% | 82.7% |
| MZ326698 | 95.9% | – | 96% | 85.1% | 85.4% | 85.5% | 84.9% | 83.7% |
| MZ361738 | 100% | 96% | – | 83.7% | 85.3% | 85.7% | 83.5% | 82.7% |
| MW424419 | 83.5% | 85.1% | 83.7% | – | 90.7% | 90.7% | 100% | 98.4% |
| MW467897 | 85.1% | 85.4% | 85.3% | 90.7% | – | 99.6% | 90.4% | 90.8% |
| MW467898 | 85.5% | 85.5% | 85.7% | 90.7% | 99.6% | – | 90.4% | 90.8% |
| MZ330127 | 83.5% | 84.9% | 83.5% | 100% | 90.4% | 90.4% | – | 98.4% |
| MZ342904 | 82.7% | 83.7% | 82.7% | 98.4% | 90.8% | 90.8% | 98.4% | – |
The percentages of nucleotide identities are presented.
Table 4.
Animal source and tick species of genotyped strains.
| Accession No. | Genotype | Tick Species | Animal Source |
|---|---|---|---|
|
MW424419 MW467897 MW467898 MZ330127 MZ342904 |
Africa III |
Hyalomma dromedarii Hyalomma rufipes Hyalomma dromedarii Hyalomma dromedarii Hyalomma dromedarii |
Sudan Sudan Somalia Sudan Ethiopia |
|
MZ322095 MZ326698 MZ361738 |
Africa I |
Hyalomma rufipes Hyalomma rufipes Hyalomma dromedarii |
Somalia Sudan Sudan |
Fig. 3.
Phylogenetic tree of partial small (S) segment sequence (260 nucleotides) of CCHFV isolated from ticks, Egypt, in year 2019–2021. The maximum-likelihood method was used to construct the tree, MEGA-x software was employed to detect the Tamura three-parameter model and bootstrap value at 50% cut-off point. The isolated virus genome from Egypt ticks clustered in Africa I and Africa III lineage. The nodes’ numbers reflect the values of bootstrap for 1000 replicates. For each isolate, the geographic origin and the GenBank accession number are given.
4. Discussion
CCHF is one of the key geographically prevalent tick-borne viruses, with regions of endemicity includes areas spreading over from Africa to the Middle East, Europe, southern Asia and China (Papa, 2019). The infected hard ticks’ bites or the contact with tissue or blood from viremic domesticated animals can cause the transmission of CCHF to humans. It can also be transmitted from one individual to another by contact with body fluids or blood (Rodriguez et al., 1997). Although the temporariness of animals’ viremia, ticks remain infectious for several years and considered as a reservoir for CCHFV (Bente et al., 2013). Therefore, the detection of CCHFV in vectors provides an opportunity to prevent disease transmission in high-risk abattoir workers.
Although Hyalomma ticks are deemed as the primary reservoir and vector for the CCHFV widespread transmission throughout Asia, Europe, and Africa, CCHFV has also been identified in some other ticks’ genera (Mehravaran et al., 2013, Ergönül, 2006). In this study, the samples of ticks were amassed from newly butchered camels and analyzed for the existence of CCHFV. One genus, namely Hyalomma, and two species were observed. We identified H. dromedarii as the most prevalent species infecting camel ticks (67.28%), followed by H. rufipes (32.72%). This was in accordance with the findings obtained by Bala et al. (2018) in Sudan (Bala et al., 2018), Champour et al. (2016) in Iran (Champour et al., 2016), and Chisholm et al. (2012) in Egypt (Chisholm et al., 2012). They reported that Hyalomma are the key tick species invading camels in these countries. These findings indicated the effect of climate and regional conditions on tick ecology as the relatively arid climate suits Hyalomma ticks (Okely et al., 2021).
The identification of CCHF in ticks and affirmation of their vector ability through venereal and transovarial CCHFV transmission is critical as potential reservoirs of infection (Bente et al., 2013). The technique of utilizing generic PCR assays and subsequent sequencing for the detection of tick-borne viruses was adopted, making this research one of the most comprehensive and huge surveillance efforts conducted in Egypt. Two RT-PCR were employed to detect CCHFV in the tick samples. The CCHFV genome was identified in 18 tick pools (1.44%) that were collected (61.1% of H. dromedarii and 38.9% of H. rufipes). This observation is close to that of Chisholm et al. (2012) in Egypt (Chisholm et al., 2012) who stated that 5 out of 6 positive pools were obtained from H. dromedarii and 4.3% of tested pools were found to be infected by CCHFV. This might indicate that Hyalomma spp. act as the primary vector and reservoir for CCHF in camels and may probably have a superior role in the epidemiology of the virus in Africa. The higher percent of infectivity may be due to these ticks may have fed on various livestock before spreading to unexposed animals, and the sample size is also low relative to our study, while temporal fluctuations between the samples may also account for the discrepancies in infection rate.
Previous serologic studies conducted in Egypt of Morrill et al. (1990) (Morrill et al., 1990) who revealed that antibodies to CCHFV was 14% and that could be attributed to that the samples were collected from camels imported from Sudan and quarantined in southern Egypt. The findings from several reports show that the infectivity rate of CCHF is changeable and is influenced by the geographical diversity and weather, the presence of various species of ticks, and different tick hosts.
Various CCHFV S segment sequences from several areas around the world indicate the genetic diversity among CCHFV strains rely on geographical location and tick species (Whitehouse, 2004, Yashina et al., 2003). From the phylogenetic tree (Fig. 3), CCHFV sequences obtained during the study grouped with clade Africa I and Africa III. CCHFV strains (accession no. MW467897 and MW467898) were closely related to United Arab Emirate strains, that were genetically related to sequences obtained from south and west Africa (Camp et al., 2020), and JF706233 strain from Egypt, which isolated from ticks collected from camels imported from Sudan and Somalia (Chisholm et al., 2012). The CCHFV sequences (accession no. MZ342904, MW424419, and MZ330127) were similar genetically and clustered together with strain from Nigeria (KX238958), suggesting the circulation of the same virus in Africa. Within Africa I lineage, the three partial segments (MZ326698, MZ361738, and MZ322095) clustered together with strains from South Africa, Senegal, and Mauritania. This could indicate that the virus is probably disseminated by migrating birds transmitting infected ticks on their surface or by secondary introductions following the importation of camels.
Overall, this study confirmed the existence of CCHFV in Egypt and illustrate the potential for tick-borne transmission of the virus among camels as well as to human. Since the genetic diversity data of CCHFV in Egypt is scarce, further studies on viruses from tick and human samples are epidemiologically valuable to clearly demonstrate the epidemic situation in the region. Finally, there are no published reports of human infection in Egypt. This could be due to the lack of awareness of physicians and limitation of preventive and control strategies; thus, attention should be paid to this disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge all those who contributed to this study, especially Dr. Hanan Mahmoud for her support in obtaining Genbank accession numbers. We also thank Mr. Nasser Shaban Abosreea for his assistance with the collection of ticks. This work was supported by Researchers Supporting Project Number (RSP-2022R7) King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
References
- Bala, A.E.A., Abakar, A.D., M.S., M., M.A., M., El Tigani, M.A., 2018. Seasonal prevalence and geographical distribution of ticks of camels (Camelus dromedaries) in four states of Great Butana, Sudan. J. Entomol. Zool. Studies, 6(3), 1212-1220.
- Bente D.A., Forrester N.L., Watts D.M., McAuley A.J., Whitehouse C.A., Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100(1):159–189. doi: 10.1016/j.antiviral.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Cajimat M.N.B., Rodriguez S.E., Schuster I.U.E., Swetnam D.M., Ksiazek T.G., Habela M.A., Negredo A.I., Estrada-Peña A., Barrett A.D.T., Bente D.A. Genomic Characterization of Crimean-Congo Hemorrhagic Fever Virus in Hyalomma Tick from Spain, 2014. Vector Borne Zoonotic Dis. 2017;17(10):714–719. doi: 10.1089/vbz.2017.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.V., Kannan D.O., Osman B.M., Shah M.S., Howarth B., Khafaga T., Weidinger P., Karuvantevida N., Kolodziejek J., Mazrooei H., Wolf N., Loney T., Nowotny N. Crimean-Congo Hemorrhagic Fever Virus Endemicity in United Arab Emirates, 2019. Emerg Infect Dis. 2020;26(5):1019–1021. doi: 10.3201/eid2605.191414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champour M., Chinikar S., Mohammadi G., Razmi G., Shah-Hosseini N., Khakifirouz S., Mostafavi E., Jalali T. Molecular epidemiology of Crimean-Congo hemorrhagic fever virus detected from ticks of one humped camels (Camelus dromedarius) population in northeastern Iran. J. Parasit. Dis. 2016;40(1):110–115. doi: 10.1007/s12639-014-0458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm K., Dueger E., Fahmy N.T., Samaha H.A., Zayed A., Abdel-Dayem M., Villinski J.T. Crimean-congo hemorrhagic fever virus in ticks from imported livestock. Egypt. Emerg. Infect. Dis. 2012;18(1):181–182. doi: 10.3201/eid1801.111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergönül O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6(4):203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhadpour F., Telmadarraiy Z., Chinikar S., Akbarzadeh K., Moemenbellah-Fard M.D., Faghihi F., Fakoorziba M.R., Jalali T., Mostafavi E., Shahhosseini N., Mohammadian M. Molecular detection of Crimean-Congo haemorrhagic fever virus in ticks collected from infested livestock populations in a New Endemic Area, South of Iran. Trop. Med. Int. Health. 2016;21(3):340–347. doi: 10.1111/tmi.12667. [DOI] [PubMed] [Google Scholar]
- Fillâtre P., Revest M., Tattevin P. Crimean-Congo hemorrhagic fever: an update. Med. Mal. Infect. 2019;49(8):574–585. doi: 10.1016/j.medmal.2019.09.005. [DOI] [PubMed] [Google Scholar]
- Gouy, M., Guindon, S., Gascuel, O.J.M.b., evolution, 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. 27(2), 221-224. https://doi.org/10.1093/molbev/msp259. [DOI] [PubMed]
- Kazancioğlu S., Akinci E., Bodur H. Crimean-Congo Hemorrhagic Fever. Mediterranean J. Infect. Microbes Antimicrobials. 2017;6:12. doi: 10.4274/mjima.2017.12. [DOI] [Google Scholar]
- Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K., 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol., 35(6), 1547-1549. https://doi.org/10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed]
- Lutomiah J., Musila L., Makio A., Ochieng C., Koka H., Chepkorir E., Mutisya J., Mulwa F., Khamadi S., Miller B.R., Bast J., Schnabel D., Wurapa E.K., Sang R. Ticks and tick-borne viruses from livestock hosts in arid and semiarid regions of the eastern and northeastern parts of Kenya. J. Med. Entomol. 2014;51(1):269–277. doi: 10.1603/ME13039. [DOI] [PubMed] [Google Scholar]
- Mazzola L.T., Kelly-Cirino C. Diagnostic tests for Crimean-Congo haemorrhagic fever: a widespread tickborne disease. BMJ Glob. Health. 2019;4(Suppl. 2):e001114. doi: 10.1136/bmjgh-2018-001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehravaran A., Moradi M., Telmadarraiy Z., Mostafavi E., Moradi A.R., Khakifirouz S., Shah-Hosseini N., Varaie F.S., Jalali T., Hekmat S., Ghiasi S.M., Chinikar S. Molecular detection of Crimean-Congo haemorrhagic fever (CCHF) virus in ticks from southeastern Iran. Ticks Tick Borne Dis. 2013;4(1–2):35–38. doi: 10.1016/j.ttbdis.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Morrill J.C., Soliman A.K., Imam I.Z., Botros B.A., Moussa M.I., Watts D.M. Serological evidence of Crimean-Congo haemorrhagic fever viral infection among camels imported into Egypt. J. Trop. Med. Hyg. 1990;93(3):201–204. [PubMed] [Google Scholar]
- Okely M., Anan R., Gad-Allah S., Samy A.M. Mapping the environmental suitability of etiological agent and tick vectors of Crimean-Congo hemorrhagic fever. Acta Trop. 2020;203:105319. doi: 10.1016/j.actatropica.2019.105319. [DOI] [PubMed] [Google Scholar]
- Okely M., Anan R., Gad‐Allah S., Samy A.M. Hard ticks (Acari: Ixodidae) infesting domestic animals in Egypt: diagnostic characters and a taxonomic key to the collected species. Med. Vet. Entomol. 2021;35(3):333–351. doi: 10.1111/mve.12502. [DOI] [PubMed] [Google Scholar]
- Papa A. Diagnostic approaches for Crimean-Congo hemorrhagic fever virus. Expert Rev. Mol. Diagn. 2019;19(6):531–536. doi: 10.1080/14737159.2019.1615450. [DOI] [PubMed] [Google Scholar]
- Raabe V.N. Diagnostic Testing for Crimean-Congo Hemorrhagic Fever. J. Clin. Microbiol. 2020;58(4):e01580–01519. doi: 10.1128/JCM.01580-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L.L., Maupin G.O., Ksiazek T.G., Rollin P.E., Khan A.S., Schwarz T.F., Lofts R.S., Smith J.F., Noor A.M., Peters C.J., Nichol S.T. Molecular investigation of a multisource outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates. Am. J. Trop. Med. Hyg. 1997;57(5):512–518. doi: 10.4269/ajtmh.1997.57.512. [DOI] [PubMed] [Google Scholar]
- Saleem M., Tanvir M., Akhtar M., Saleem A. Crimean-Congo hemorrhagic fever: etiology, diagnosis, management and potential alternative therapy. Asian Pacific J. Trop. Med. 2020;13(4):143–151. doi: 10.4103/1995-7645.280221. [DOI] [Google Scholar]
- Sang R., Onyango C., Gachoya J., Mabinda E., Konongoi S., Ofula V., Dunster L., Okoth F., Coldren R., Tesh R., da Rossa A.T., Finkbeiner S., Wang D., Crabtree M., Miller B. Tickborne arbovirus surveillance in market livestock, Nairobi Kenya. Emerg. Infect. Dis. 2006;12(7):1074–1080. doi: 10.3201/eid1207.060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Hu Z., Deng F., Shen S. Tick-Borne Viruses. Tick-Borne Viruses. Virol Sin. 2018;33(1):21–43. doi: 10.1007/s12250-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler J.R., Bergeron É., Rollin P.E., Clements A.C.A. Seroepidemiological Studies of Crimean-Congo Hemorrhagic Fever Virus in Domestic and Wild Animals. PLoS Negl. Trop. Dis. 2016;10(1):e0004210. doi: 10.1371/journal.pntd.000421010.1371/journal.pntd.0004210.g00110.1371/journal.pntd.0004210.g00210.1371/journal.pntd.0004210.t00110.1371/journal.pntd.0004210.t00210.1371/journal.pntd.0004210.t003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler J.R., Bergeron É., Spiropoulou C.F. Crimean-Congo hemorrhagic fever and expansion from endemic regions. Curr Opin Virol. 2019;34:70–78. doi: 10.1016/j.coviro.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezer H., Polat M. Diagnosis of Crimean-Congo hemorrhagic fever. Expert Rev. Anti Infect. Ther. 2015;13(5):555–566. doi: 10.1586/14787210.2015.1021782. [DOI] [PubMed] [Google Scholar]
- Voorhees M.A., Padilla S.L., Jamsransuren D., Koehler J.W., Delp K.L., Adiyadorj D., Baasandagwa U., Jigjav B., Olschner S.P., Minogue T.D., Schoepp R.J. Crimean-Congo Hemorrhagic Fever Virus, Mongolia, 2013–2014. Emerg. Infect. Dis. 2018;24(12):2202–2209. doi: 10.3201/eid2412.180175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahid B., Altaf S., Naeem N., Ilyas N., Idrees M. Scoping Review of Crimean-Congo Hemorrhagic Fever (CCHF) Literature and Implications of Future Research. J. Coll. Phys. Surg. Pak. 2019;29(6):563–573. doi: 10.29271/jcpsp.2019.06.563. [DOI] [PubMed] [Google Scholar]
- Whitehouse C.A. Crimean-Congo hemorrhagic fever. Antiviral Res. 2004;64(3):145–160. doi: 10.1016/j.antiviral.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Paweska J.T., Petersen N., Grobbelaar A.A., Leman P.A., Hewson R., Georges-Courbot M.C., Papa A., Günther S., Drosten C. Virus detection and monitoring of viral load in Crimean-Congo hemorrhagic fever virus patients. Emerg. Infect. Dis. 2007;13(7):1097–1100. doi: 10.3201/eid1307.070068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashina L., Petrova I., Seregin S., Vyshemirskii O., Lvov D., Aristova V., Kuhn J., Morzunov S., Gutorov V., Kuzina I., Tyunnikov G., Netesov S., Petrov V. Genetic variability of Crimean-Congo haemorrhagic fever virus in Russia and Central Asia. J. Gen. Virol. 2003;84(Pt 5):1199–1206. doi: 10.1099/vir.0.18805-0. [DOI] [PubMed] [Google Scholar]
- Zohaib A., Saqib M., Athar M.A., Hussain M.H., Sial A.U., Tayyab M.H., Batool M., Sadia H., Taj Z., Tahir U., Jakhrani M.Y., Tayyab J., Kakar M.A., Shahid M.F., Yaqub T., Zhang J., Wu Q., Deng F., Corman V.M., Shen S., Khan I., Shi Z.L. Crimean-Congo Hemorrhagic Fever Virus in Humans and Livestock, Pakistan, 2015–2017. Emerg. Infect. Dis. 2020;26(4):773–777. doi: 10.3201/eid2604.191154. [DOI] [PMC free article] [PubMed] [Google Scholar]