Abstract
The widespread use of ganciclovir (GCV) to treat cytomegalovirus (CMV) infections in immunosuppressed patients has led to the development of drug resistance. Phenotypic assays for CMV drug resistance are presently too time-consuming to be therapeutically useful. To support the development of genotypic assays for GCV resistance, the complete sequences of the UL97 phosphotransferase genes in 28 phenotypically GCV-sensitive CMV clinical isolates were determined. The gene was found to be highly conserved, with nucleotide sequence identity among strains ranging from 98.6 to 100% and amino acid sequence identity of >99%. Primers for a genotypic assay were designed to amplify codons 400 to 707, because all known UL97 mutations conferring drug resistance occur at three sites within this region. This part of the UL97 gene was amplified from over 50 clinical isolates, and two sequencing reactions for the coding strand were successfully used to identify GCV resistance mutations. This genotypic assay can be performed in 48 h using genomic DNA extracted from cell monolayers at very low levels of virus infectivity, thus rapidly providing therapeutically useful results.
Ganciclovir (GCV), the most widely used antiviral drug for treatment of systemic cytomegalovirus (CMV) disease, has proven effective in significantly reducing the CMV viral load in transplant recipients and patients with AIDS. A consequence of this antiviral drug therapy is the development of GCV-resistant CMV strains, which have been well documented in human immunodeficiency virus-infected patients (3, 20, 21, 24, 36). More recently resistance has been increasingly detected in isolates from transplant recipients following extended prophylactic or preemptive therapy (1, 2, 9, 16, 17, 19, 23, 25).
The UL97 phosphotransferase (kinase), a virus-encoded product, activates GCV by monophosphorylation (32). Subsequent di- and triphosphorylation are carried out by cellular enzymes. A second viral product, the DNA polymerase, incorporates GCV triphosphate into the viral genome, leading to marked attenuation of viral DNA replication (12, 13, 15). Although mutations conferring GCV resistance have been mapped to both genes, mutations in the UL97 gene appear to be detected during antiviral therapy much more frequently than DNA polymerase gene mutations (7, 9, 17, 31). The occurrence of these resistance mutations has prompted efforts to develop genotypic methods for rapid antiviral susceptibility testing.
The standard phenotypic method for detection of drug resistance has been the plaque reduction assay, which requires lengthy viral propagation to obtain sufficient infectivity for performance of the assay (22). The slow growth of cell-associated clinical isolates limits the value of this assay for therapeutic decisions. In addition, the assay is inherently subjective in the differentiation of true drug-resistant plaques from foci of drug-sensitive infectivity that slowly resolve in the presence of drug. Additional phenotypic assays which are more objective and which require less time to determine the effect of antiviral drugs on CMV replication have been developed. These include detection of the CMV immediate-early antigen by flow cytometry (28) and detection of viral DNA by DNA hybridization (14). However, these assays still require several weeks of cell culture to produce the initial CMV inoculum for the assay.
Genotypic methods based on identification of mutations in the viral genome lack the subjectivity of the plaque reduction assay and have the potential to detect drug resistance in a therapeutically relevant time frame (4, 8). The CMV UL97 gene is an important viral target for genotypic assays, because all documented mutations conferring GCV resistance have been found at one of three sites within the coding region for the C-terminal half of the phosphotransferase (2, 7, 9, 11, 19, 26, 29, 38). At two of the corresponding sites in the UL97 gene there are point mutations in single codons (460 and 520), and at the third there are point mutations or short deletions within the codon range 590 to 607. The limitation of resistance mutations to only these three sites enhances the feasibility of UL97 genotypic GCV susceptibility analysis.
The genotypic assay that we describe in this report is based on direct automated DNA sequencing of PCR products amplified from CMV genomic DNA. This effort represents the combined work of a group of four laboratories, which have collaboratively studied CMV clinical isolates from widely separated geographic regions. Others have used direct sequencing, both manual and automated, to identify resistance mutations (2, 8, 9, 16, 19, 29, 36, 37). Our approach has been to define the baseline variability that occurs in the absence of drug resistance by sequencing the complete UL97 gene from a large number of clinical isolates that are phenotypically sensitive to GCV. These sequences from GCV-sensitive isolates combined with the discrete sites of GCV resistance mutations that have been previously identified (2, 9, 11, 19, 26, 29, 38) provide the basis for the assay. Our results show that amplification of the portion of the UL97 gene encoding the C-terminal half of the enzyme followed by two sequencing reactions can provide rapid identification of all presently known sites of GCV resistance attributed to this gene.
MATERIALS AND METHODS
Clinical isolates.
Over 50 clinical CMV isolates were obtained through the clinical virology laboratories at the participating institutions and were tested for GCV susceptibility by the plaque reduction assay (see below). A set of 28 phenotypically GCV-sensitive isolates were randomly selected for sequencing the complete UL97 coding sequence as described below. Twenty of these isolates were from transplant recipients, and 8 were from human immunodeficiency virus-infected patients. The isolates were routinely received as infected human foreskin fibroblast (HFF) monolayers in tube cultures. Infected monolayers were trypsinized and passed to fresh uninfected monolayers in T-25 tissue culture flasks. The level of infectivity was increased by repeated rounds of trypsinization and redistribution of infected monolayers as well as passage to new monolayers. The total number of passages required to reach a level of 60 to 80% infectivity for use in the plaque reduction assay was usually five to seven, and all strains remained cell associated. Some of the tube cultures were used directly for DNA extraction to decrease the time required to perform the genotypic assay (see below).
Plaque reduction assay.
Infected cells generated by passage of clinical strains as described above were used as the inoculum for the assay. HFF monolayers in 24-well plates were inoculated with the number of trypsinized, infected cells calculated to produce 60 to 80 plaques per well (22). The medium in the wells contained GCV concentrations of 3, 6, 12, 24, and 48 μM. Each concentration was represented in quadruplicate. One set of four wells served as the virus control without drug. The plates were read at a minimum of 7 days postinoculation. The 50% inhibitory concentration (IC50) was the concentration of drug producing a 50% reduction in the number of plaques relative to the control wells. GCV IC50 values ≤6 μM indicate sensitivity, and those >6 μM indicate resistance (22).
DNA extraction and PCR amplification.
CMV genomic DNA was extracted either directly from infected HFF tube monolayers or from infected HFF monolayers in T-25 flasks passed from the original tube monolayers. Visible cytopathic effect was the minimum requirement for extraction. Infected monolayers were trypsinized and centrifuged. DNA was extracted from the cell pellets using a lysis buffer containing 10 mM Tris-HCl, pH 8.0, 50 mM KCl, 2.5 mM MgCl2 0.9% Nonidet P-40, and 100 μg of proteinase K/ml. Alternatively the extraction was carried out using the QIAamp DNA Mini Kit (Qiagen, Inc., Valencia, Calif.).
The extracted viral genomic DNA was used as the template for amplification of either the complete UL97 coding region or codons 400 to 707, which contain the known drug resistance mutations. The GenAmp XL kit (PE Applied Biosystems, Foster City, Calif.) was used for these amplifications, using the following conditions: 30 cycles of 94°C for 1 min and 60°C for 10 min. The complete gene was amplified with primers CPL97-F and CPL97-R, shown in Table 1, yielding a product 2,254 bp in length. Codons 400 to 707 were amplified with primers HLF97-F and HLF97-R (Table 1), which produces a 1,038-bp product.
TABLE 1.
Primer sequences for PCR amplication and sequencing of UL97 gene GCV resistance region
| Primer function | Primer sequence |
|---|---|
| PCR amplification of complete UL97 gene coding sequence | 5′-GGAAGACTGTCGCCACTATGTCC-3′ (CPL97-F) |
| 5′-CTCCTCATCGTCGTCGTAGTCC-3′ (CPL97-R) | |
| PCR amplification of codons 400 to 707 | 5′-CTGCTGCACAACGTCACGGTACATC-3′ (HLF97-F) |
| 5′-CTCCTCATCGTCGTCGTAGTCC-3′ (HLF97-R) | |
| Sequencing of coding strand for resistance mutations | 5′-ATCGACAGCTACCGACGTGCC-3′ (460/520) |
| 5′-GTCGGAGCTGTCGGCGCTGGG-3′ (590–607) | |
| Sequencing of coding strand for resistance mutations (alternative primers) | 5′-GTGGAAGCTGGCGTGCAT-3′ (460/520) |
| 5′-GGATTACAACGAGCGCTG-3′ (590–607) | |
| Sequencing of both strands for resistance mutations (alternative primers) | 5′-GTGGAAGCTGGCGTGCAT-3′ (460/520)a |
| 5′-CGACACGAGGACATCTTG-3′ (590–607)b |
Same as top sequence of alternative primer for sequencing of coding strand for resistance, mutations.
Noncoding sequence.
DNA sequencing.
The PCR products were purified by Microcon centrifugal filters (Millipore Corp., Bedford, Mass.) or enzymatic digestion with exonuclease I and shrimp alkaline phosphatase (U.S. Biochemical Corporation, Cleveland, Ohio) using the protocol recommended by the manufacturer. The purified templates were sequenced using the ABI Prism BigDye Terminator Cycle Sequencing kit (PE Applied Biosystems) and analyzed on an ABI 377 automated DNA sequencer.
For rapid sequencing analysis, two primers for the coding strand were used to cover the region containing the drug resistance mutations (Table 1). Alternative primers for the coding strand are also shown in Table 1, as well as a pair of primers that cover the coding and noncoding strands in the same region. All primer pairs produce the required sequences. DNA sequences were analyzed using Align Plus, version 4.0 (Scientific and Educational Software, Durham, N.C.).
Nucleotide sequence accession numbers.
The sequences of the GCV-sensitive strains have been submitted to GenBank and have been given accession no. AF34548 through AF34573.
RESULTS
Phenotypic antiviral drug susceptibility.
The collaborating laboratories routinely performed plaque assays for GCV susceptibility on CMV isolates. The assay was used to select a set of 28 CMV isolates from 20 transplant recipients and 8 patients with AIDS, which were determined to be GCV sensitive, since their IC50 values were below 6 μM. Phenotypically resistant isolates (IC50 > 6 μM) were also identified by the plaque assay. The usual time required to complete the assay was 1 to 2 months from detection of virus in cell culture. Some isolates replicated more slowly, which increased the period required for propagation. The time to completion of the assay in these cases exceeded 2 months.
DNA sequences of UL97 genes from GCV-sensitive clinical isolates.
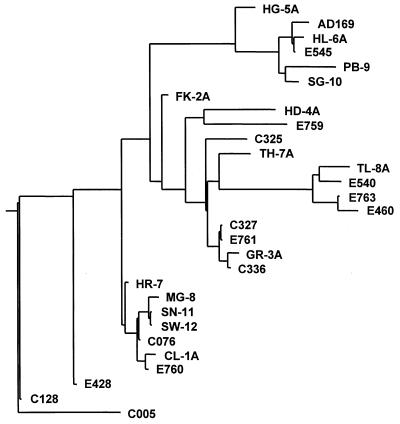
The UL97 gene coding sequence of reference strain AD169 (EMBL accession no. X17403) is 2,124 bases in length. Comparison of the 28 complete UL97 gene DNA sequences from GCV-sensitive strains indicates that the gene is highly conserved among these strains. Compared to the AD169 reference sequence there are 88 variant nucleotide positions, 39 of which are present in more than one strain. Figure 1 shows the dendrogram generated from the DNA sequences by Align Plus, which uses the neighbor-joining algorithm to construct the distance-based tree. Strains PB-9 and C005 are the most divergent, with 30 base changes relative to each other. Pairwise alignments of strains that are tightly grouped on the dendrogram (e.g., MG-8, SN-11, SW-12, C076, CL-1A, and E760) differ by single nucleotides. Two pairs of strains have identical DNA sequences: SN-11 and SW-12 and C327 and E761. Nucleotide changes are distributed across the entire coding region rather than occurring in clusters or patterns. The sequence identity among all strains ranges from 98.6 to 100%, with an average of 99.2%. This is slightly higher than the average 98.8% nucleotide identity previously determined for the DNA polymerase gene (10).
FIG. 1.
Phylogenetic relationship of UL97 gene DNA sequences from 28 GCV-sensitive isolates. The dendrogram was generated by the Align Plus program, version 4.0, using the neighbor-joining algorithm. Strains PB-9 and C005 differ by the largest number of nucleotides, a total of 30.
Amino acid substitutions in GCV-sensitive clinical isolates.
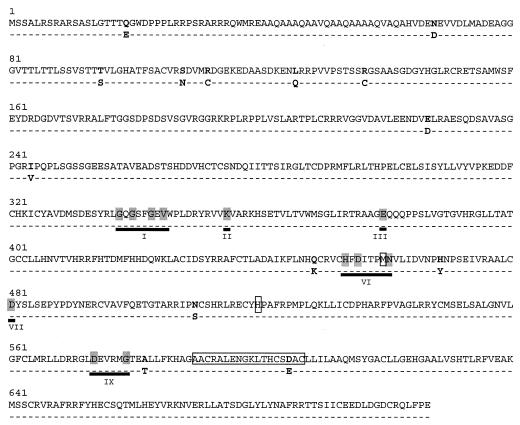
The amino acid sequences encoded by the UL97 genes from the 28 GCV-sensitive isolates are even more highly conserved than the DNA sequences, indicating that the majority of nucleotide changes are synonymous. The variant residues and the resulting amino acid substitutions are shown in Fig. 2. With AD169 as the reference there are 14 amino acid residues that differ in at least one isolate, with only 6 of these residues occurring in more than one isolate. The maximum variation observed in pairwise alignments was between TH-7A and C128, which differ by six amino acids. Thus, the percent sequence identity at the amino acid level ranges from 99.2 to 100%, which is also slightly higher than that of the DNA polymerase (10). The same amino acid is replaced at any given site in all isolates that are altered at that site, and more than one-half of the amino acid substitutions are nonconservative.
FIG. 2.
Sites of amino acid substitutions (in boldface) identified in GCV-sensitive CMV isolates using the AD169 sequence as the reference. Numbers are amino acid residues based on the coding sequence of AD169. Dashes indicate sequence identity. Variant residues are shown below the AD169 sequence. Conserved protein kinase regions (roman numerals) defined by Hanks et al. (18) that are found in UL97 are underlined, with conserved residues shaded. Sites of drug resistance mutations are boxed.
The UL97 gene encodes a predicted protein kinase that fortuitously phosphorylates GCV (33). Proposed functional domains for protein kinases designated I through XI are shared by protein kinases derived from widely diverse species (6, 18). The degree of amino acid conservation within these domains is significantly lower than that observed for functional regions of DNA polymerases (5, 34). There are amino acid residues, however, that are invariant among the members of this group of kinases. The conserved amino acids in UL97 and the corresponding domains, shown in Fig. 2, are as follows: GXGXXGXV (codons 337 to 345 for region I), lysine 354 (region II), glutamate 379 (region III), HXDXXXXN (codons 454 to 461 for region VI), aspartate 481 (region VII), and DXXXXG (codons 574 to 579 for region IX). The amino acid substitutions in the drug-sensitive strains do not occur at any of the conserved amino acid residues, nor does any resistance mutation occur at any of the invariant kinase residues. Although the methionine 460 codon, which is a site for drug resistance mutations, lies within region VI of the human CMV and human herpesvirus 6 homologues, this methionine residue is not present in other protein kinases (6, 18).
It should be noted that in these GCV-sensitive isolates no substitutions occurred at amino acid residues directly linked to drug resistance. The amino acid change D605E observed in baseline isolate C325 has not been linked to phenotypic GCV resistance even though its codon is within the range 590 to 607, where many GCV resistance mutations are clustered.
Genotyping of clinical isolates.
Primers HLF97-F and HLF97-R were successfully used to amplify UL97 codons 400 to 707 in over 50 clinical isolates obtained by each of the participating laboratories. The forward primer (HLF97-F) covers a portion of the coding sequence that has no base substitutions in any drug-sensitive or drug-resistant strain. The reverse primer (HLF97-R) is derived from sequences downstream of the 3′ end of the UL97 gene. There were no viral DNA templates that did not yield the appropriately sized PCR-amplified product (1,038 bp) using this primer set.
Resistance mutations were identified by two forward (coding strand) sequencing reactions using PCR templates from codons 400 to 707. Table 2 lists the amino acid substitutions that were identified in phenotypically GCV-resistant isolates by the genotypic assay. Although no problems were encountered in performing the sequencing reactions with the first set of sequencing primers listed in Table 1, the possibility exists that there are clinical isolates containing base changes at these primer binding sites which will inhibit the sequencing reactions. The alternative primer sets shown in Table 1 have also provided reliable sequencing data from the region containing the UL97 resistance mutations.
TABLE 2.
Mutations identified in GCV-resistant isolates
| UL97 gene codon | Amino acid of wild type | Mutant amino acid detected with genotypic assay (no. of isolates) |
|---|---|---|
| 460 | M | I (1) |
| V (3) | ||
| 520 | H | Q (5) |
| 592 | C | G (7) |
| 594 | A | V (4) |
| P (1)a | ||
| 595 | L | S (14) |
| W (1) | ||
| 603 | C | W (2) |
| 607 | C | Y (1) |
Not confirmed by marker transfer.
DISCUSSION
The demand for antiviral susceptibility testing has risen in response to the increased awareness that drug resistance can result from the widespread use of antiviral therapy. GCV has been the drug of choice for treatment of systemic CMV disease. However, long-term therapy and suboptimal drug concentrations increase the risk of development of GCV resistance. The UL97 gene is by far the most frequent site of mutations conferring GCV resistance, and extensive work by a number of groups (2, 7, 9, 17, 29, 31, 38) has failed to identify any nucleotide changes associated with GCV resistance outside codons 460, 520, and 590 to 607 (Fig. 2). Not all of the mutations at these sites have been confirmed by marker transfer. For some of the unconfirmed mutations, a phenotypically sensitive isolate has been obtained prior to the isolation of a resistant isolate from the same patient. Sensitive and resistant isolates from the same patient, which differ only in a specific point mutation or deletion in association with GCV therapy, provide strong support for the role of that mutation in GCV resistance.
A genotypic assay designed to detect several of the UL97 mutations has been used by a number of investigators (2, 8, 30). The assay is based on detection of loss or gain of restriction enzyme cleavage sites directly resulting from the presence of UL97 GCV resistance mutations. There are several advantages to this type of assay. The assay is rapid because it can be performed directly from the initial laboratory tube culture, it relies on PCR amplification of short sequences surrounding each potential mutation site, and it requires relatively simple equipment to perform. However, potential problems may be encountered if products are incompletely digested, new restriction sites are created by nucleotide changes unrelated to resistance, or resistance results from a DNA polymerase mutation. In addition, previously unmapped resistance mutations will not be detected.
We believe that automated sequencing represents the state-of-the-art approach to genotypic detection of drug resistance. The results of the sequencing of the UL97 gene from GCV-sensitive isolates demonstrate that the UL97 gene is highly conserved among clinical isolates, with an average 99% sequence identity. These baseline sequences along with the well-defined sites for drug resistance mutations provide the foundation for a rapid genotypic drug resistance assay based on direct sequencing of PCR-amplified products containing these sites.
The assay as outlined in this report has a minimal requirement of visible cytopathic effect in the original tube culture obtained from the clinical laboratory, which usually requires 7 to 14 days postinoculation. Amplification and sequencing can be completed within approximately 48 h. This approach drastically shortens the time to detection of drug resistance mutations compared to the plaque reduction assay, which requires at least 2 weeks of propagation to acquire enough virus. In addition a minimum of a week is required before the assay can be read. The potential exists to further shorten the time required for the genotypic assay by extracting viral DNA directly from patient samples. Portions of the UL97 and polymerase coding sequences can be amplified from DNA extracted from blood cells or plasma (35, 37, 38; N. S. Lurain, unpublished data) and directly sequenced to detect resistance mutations. We are currently focusing on developing this modification of the assay.
The primers listed in Table 1 for amplification and sequencing of the viral templates have been used successfully in the assays that we have performed. Some of the isolates contain single base changes within the sequencing primer binding sites, but there was no apparent effect on the ability of these primers to produce the predicted sequences. It is possible that future isolates may contain base changes that will affect either amplification or sequencing reactions. We have successfully tested several alternative primers (Table 1) for both types of reactions; thus there is considerable flexibility in primer selection, although problems in primer design can arise from the high G+C content of the UL97 coding region.
By monitoring sequential isolates from the same patient over the course of long-term antiviral therapy, the appearance of known resistance mutations can be observed (2, 17, 30, 31). In many cases double peaks representing mixed sensitive and resistant viral populations become apparent on sequencing chromatograms as resistance develops. Another advantage of closely monitoring patients on GCV therapy is that new mutations potentially conferring resistance can be detected when sequences of drug-sensitive and drug-resistant strains from the same patient are compared.
Although this assay was developed specifically for GCV resistance resulting from UL97 mutations, the same approach can be used for genotyping drug-resistant DNA polymerase mutants. We have previously reported PCR conditions for amplification of the complete polymerase gene coding region. PCR products were sequenced to determine the variability of the DNA polymerase gene in a large group of drug-sensitive CMV isolates (10). Detection of drug resistance mutations in the polymerase gene, however, is more difficult than detection of such mutations in the UL97 gene, because the polymerase gene is larger and the codons known to confer resistance are widely dispersed across the coding sequence (11, 12, 27, 31). Therefore, a genotypic assay for the polymerase gene will require a minimum of six sequencing reactions to cover the presently known sites of drug resistance mutations in the coding strand. Further sequencing and marker transfer experiments will also be required to determine whether there are as yet unidentified polymerase drug resistance mutations in phenotypically resistant CMV isolates.
Phenotypic assays are still important to corroborate genotypic results, because the genotypic approach assumes that the same drug resistance mutation in all genetic backgrounds will confer resistance. Phenotypic assays are also still required to identify targets of new drugs. While phenotypic assays have the advantage of relating drug resistance directly to the biological functions of the virus, the time required to perform the assays is too long to provide useful therapeutic information. The genotypic assay that we have developed for UL97 mutants is very rapid and covers all of the known drug resistance mutations. This method can be readily expanded and adapted to include the complete UL97 gene, the polymerase gene, and other viral targets to identify mutations conferring resistance to new antiviral drugs.
ACKNOWLEDGMENTS
This work was supported in part by subcontracts to N.S.L., A.W., C.S.C., and S.C. from Social and Scientific Systems for NIH/NIAID Cooperative Agreement AI38858, a research grant from the American Lung Association of Metropolitan Chicago to N.S.L., NIH grant AI39938 to S.C., and NIH grant AI41690 to C.S.C. Sequences obtained at the University of Colorado Health Sciences Center were provided by the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility supported by NIH/NCI Cancer Core Support Grant CA46934.
We thank Emmanuel Robert, Kathi Kapell, Anne Senters, Galen Cai, and Shaobing Li for excellent technical support.
REFERENCES
- 1.Alain S, Honderlick P, Grenet D, Stern M, Vadam C, Sanson-Le Pors M J, Mazeron M C. Failure of ganciclovir treatment associated with selection of a ganciclovir-resistant cytomegalovirus strain in a lung transplant recipient. Transplantation. 1997;63:1533–1536. doi: 10.1097/00007890-199705270-00031. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti F, Simoncini L, Sarasini A, Zavattoni M, Grossi P, Revello M G, Gerna G. Ganciclovir resistance as a result of oral ganciclovir in a heart transplant recipient with multiple human cytomegalovirus strains in blood. Transplantation. 1998;66:324–329. doi: 10.1097/00007890-199808150-00008. [DOI] [PubMed] [Google Scholar]
- 3.Bowen E F, Emery V C, Wilson P, Johnson M A, Davey C C, Sabin C A, Farmer D, Griffiths P D. Cytomegalovirus polymerase chain reaction viraemia in patients receiving ganciclovir maintenance therapy for retinitis. AIDS. 1998;12:605–611. doi: 10.1097/00002030-199806000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Bowen E F, Johnson M A, Griffiths P D, Emery V C. Development of a point mutation assay for the detection of human cytomegalovirus UL97 mutations associated with ganciclovir resistance. J Virol Methods. 1997;68:225–234. doi: 10.1016/s0166-0934(97)00131-6. [DOI] [PubMed] [Google Scholar]
- 5.Braithwaite D K, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee M S, Lawrence G L, Barrell B G. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J Gen Virol. 1989;70:1151–1160. doi: 10.1099/0022-1317-70-5-1151. [DOI] [PubMed] [Google Scholar]
- 7.Chou S. Antiviral drug resistance in human cytomegalovirus. Transplant Infect Dis. 1999;1:105–114. doi: 10.1034/j.1399-3062.1999.010204.x. [DOI] [PubMed] [Google Scholar]
- 8.Chou S, Erice A, Jordan M C, Vercellotti G M, Michels K R, Talarico C L, Stanat S C, Biron K K. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J Infect Dis. 1995;171:576–583. doi: 10.1093/infdis/171.3.576. [DOI] [PubMed] [Google Scholar]
- 9.Chou S, Guentzel S, Michels K R, Miner R C, Drew W L. Frequency of UL97 phosphotransferase mutations related to ganciclovir resistance in clinical cytomegalovirus isolates. J Infect Dis. 1995;172:239–242. doi: 10.1093/infdis/172.1.239. [DOI] [PubMed] [Google Scholar]
- 10.Chou S, Lurain N S, Weinberg A, Cai G Y, Sharma P L, Crumpacker C S. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Adult AIDS Clinical Trials Group CMV Laboratories. Antimicrob Agents Chemother. 1999;43:1500–1502. doi: 10.1128/aac.43.6.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou S, Marousek G, Guentzel S, Follansbee S E, Poscher M E, Lalezari J P, Miner R C, Drew W L. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J Infect Dis. 1997;176:786–789. doi: 10.1086/517302. [DOI] [PubMed] [Google Scholar]
- 12.Cihlar T, Fuller M D, Cherrington J M. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J Virol. 1998;72:5927–5936. doi: 10.1128/jvi.72.7.5927-5936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crumpacker C S. Ganciclovir. N Engl J Med. 1996;335:721–729. doi: 10.1056/NEJM199609053351007. [DOI] [PubMed] [Google Scholar]
- 14.Dankner W M, Scholl D, Stanat S C, Martin M, Sonke R L, Spector S A. Rapid antiviral DNA-DNA hybridization assay for human cytomegalovirus. J Virol Methods. 1990;28:293–298. doi: 10.1016/0166-0934(90)90122-v. [DOI] [PubMed] [Google Scholar]
- 15.Erice A. Resistance of human cytomegalovirus to antiviral drugs. Clin Microbiol Rev. 1999;12:286–297. doi: 10.1128/cmr.12.2.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erice A, Borrell N, Li W, Miller W J, Balfour H H., Jr Ganciclovir susceptibilities and analysis of UL97 region in cytomegalovirus (CMV) isolates from bone marrow recipients with CMV disease after antiviral prophylaxis. J Infect Dis. 1998;178:531–534. doi: 10.1086/517467. [DOI] [PubMed] [Google Scholar]
- 17.Erice A, Gil-Roda C, Perez J L, Balfour H H, Jr, Sannerud K J, Hanson M N, Boivin G, Chou S. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J Infect Dis. 1997;175:1087–1092. doi: 10.1086/516446. [DOI] [PubMed] [Google Scholar]
- 18.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 19.Hanson M N, Preheim L C, Chou S, Talarico C L, Biron K K, Erice A. Novel mutation in the UL97 gene of a clinical cytomegalovirus strain conferring resistance to ganciclovir. Antimicrob Agents Chemother. 1995;39:1204–1205. doi: 10.1128/aac.39.5.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jabs D A, Enger C, Dunn J P, Forman M. Cytomegalovirus retinitis and viral resistance: ganciclovir resistance. CMV Retinitis and Viral Resistance Study Group. J Infect Dis. 1998;177:770–773. doi: 10.1086/514249. [DOI] [PubMed] [Google Scholar]
- 21.Jabs D A, Martin B K, Forman M S, Dunn J P, Davis J L, Weinberg D V, Biron K K, Baldanti F. Mutations conferring ganciclovir resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J Infect Dis. 2001;183:333–337. doi: 10.1086/317931. [DOI] [PubMed] [Google Scholar]
- 22.Landry M L, Stanat S, Biron K, Brambilla D, Britt W, Jokela J, Chou S, Drew W L, Erice A, Gilliam B, Lurain N, Manischewitz J, Miner R, Nokta M, Reichelderfer P, Spector S, Weinberg A, Yen-Lieberman B, Crumpacker C. A standardized plaque reduction assay for determination of drug susceptibilities of cytomegalovirus clinical isolates. Antimicrob Agents Chemother. 2000;44:688–692. doi: 10.1128/aac.44.3.688-692.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limaye A P, Corey L, Koelle D M, Davis C L, Boeckh M. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet. 2000;356:645–649. doi: 10.1016/S0140-6736(00)02607-6. [DOI] [PubMed] [Google Scholar]
- 24.Liu W, Kuppermann B D, Martin D F, Wolitz R A, Margolis T P. Mutations in the cytomegalovirus UL97 gene associated with ganciclovir-resistant retinitis. J Infect Dis. 1998;177:1176–1181. doi: 10.1086/515293. [DOI] [PubMed] [Google Scholar]
- 25.Lurain N S, Ammons H C, Kapell K S, Yeldandi V V, Garrity E R, O'Keefe J P. Molecular analysis of human cytomegalovirus strains from two lung transplant recipients with the same donor. Transplantation. 1996;62:497–502. doi: 10.1097/00007890-199608270-00012. [DOI] [PubMed] [Google Scholar]
- 26.Lurain N S, Spafford L E, Thompson K D. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganeiclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lurain N S, Thompson K D, Holmes E W, Read G S. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J Virol. 1992;66:7146–7152. doi: 10.1128/jvi.66.12.7146-7152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McSharry J M, Lurain N S, Drusano G L, Landay A, Manischewitz J, Nokta M, O'Gorman M, Shapiro H M, Weinberg A, Reichelderfer P, Crumpacker C. Flow cytometric determination of ganciclovir susceptibilities of human cytomegalovirus clinical isolates. J Clin Microbiol. 1998;36:958–964. doi: 10.1128/jcm.36.4.958-964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez J C, Sia I G, Tau K R, Espy M J, Smith T F, Chou S, Paya C V. Novel mutation in the CMV UL97 gene associated with resistance to ganciclovir therapy. Transplantation. 1999;67:755–757. doi: 10.1097/00007890-199903150-00020. [DOI] [PubMed] [Google Scholar]
- 30.Prix L, Hamprecht K, Holzhuter B, Handgretinger R, Klingebiel T, Jahn G. Comprehensive restriction analysis of the UL97 region allows early detection of ganciclovir-resistant human cytomegalovirus in an immunocompromised child. J Infect Dis. 1999;180:491–495. doi: 10.1086/314877. [DOI] [PubMed] [Google Scholar]
- 31.Smith I L, Cherrington J M, Jiles R E, Fuller M D, Freeman W R, Spector S A. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J Infect Dis. 1997;176:69–77. doi: 10.1086/514041. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan V, Biron K K, Talarico C, Stanat S C, Davis M, Pozzi L M, Coen D M. A point mutation in the human cytomegalovirus DNA polymerase gene confers resistance to ganciclovir and phosphonylmethoxyalkyl derivatives. Antimicrob Agents Chemother. 1993;37:19–25. doi: 10.1128/aac.37.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan V, Talarico C L, Stanat S C, Davis M, Coen D M, Biron K K. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature. 1992;359:85. doi: 10.1038/359085a0. [DOI] [PubMed] [Google Scholar]
- 34.Teo I A, Griffin B E, Jones M D. Characterization of the DNA polymerase gene of human herpesvirus 6. J Virol. 1991;65:4670–4680. doi: 10.1128/jvi.65.9.4670-4680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinberg A, Hodges T N, Li S, Cai G, Zamora M R. Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol. 2000;38:768–772. doi: 10.1128/jcm.38.2.768-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolf D G, Lee D J, Spector S A. Detection of human cytomegalovirus mutations associated with ganciclovir resistance in cerebrospinal fluid of AIDS patients with central nervous system disease. Antimicrob Agents Chemother. 1995;39:2552–2554. doi: 10.1128/aac.39.11.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf D G, Smith I L, Lee D J, Freeman W R, Flores-Aguilar M, Spector S A. Mutations in human cytomegalovirus UL97 gene confer clinical resistance to ganciclovir and can be detected directly in patient plasma. J Clin Investig. 1995;95:257–263. doi: 10.1172/JCI117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf D G, Yaniv I, Ashkenazi S, Honigman A. Emergence of multiple human cytomegalovirus ganciclovir-resistant mutants with deletions and substitutions within the UL97 gene in a patient with severe combined immunodeficiency. Antimicrob Agents Chemother. 2001;45:593–595. doi: 10.1128/AAC.45.2.593-595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]