Summary
Background
Autopsies are an important tool in medicine, dissecting disease pathophysiology and causes of death. In COVID-19, autopsies revealed e.g., the effects on pulmonary (micro)vasculature or the nervous system, systemic viral spread, or the interplay with the immune system. To facilitate multicentre autopsy-based studies and provide a central hub supporting autopsy centres, researchers, and data analyses and reporting, in April 2020 the German COVID-19 Autopsy Registry (DeRegCOVID) was launched.
Methods
The electronic registry uses a web-based electronic case report form. Participation is voluntary and biomaterial remains at the respective site (decentralized biobanking). As of October 2021, the registry included N=1129 autopsy cases, with 69271 single data points including information on 18674 available biospecimens gathered from 29 German sites.
Findings
In the N=1095 eligible records, the male-to-female ratio was 1·8:1, with peaks at 65-69 and 80-84 years in males and >85 years in females. The analysis of the chain of events directly leading to death revealed COVID-19 as the underlying cause of death in 86% of the autopsy cases, whereas in 14% COVID-19 was a concomitant disease. The most common immediate cause of death was diffuse alveolar damage, followed by multi-organ failure. The registry supports several scientific projects, public outreach and provides reports to the federal health authorities, leading to legislative adaptation of the German Infection Protection Act, facilitating the performance of autopsies during pandemics.
Interpretation
A national autopsy registry can provide multicentre quantitative information on COVID-19 deaths on a national level, supporting medical research, political decision-making and public discussion.
Funding
German Federal Ministries of Education and Research and Health.
Hintergrund: Obduktionen sind ein wichtiges Instrument in der Medizin, um die Pathophysiologie von Krankheiten und Todesursachen zu untersuchen. Im Rahmen von COVID-19 wurden durch Obduktionen z.B. die Auswirkungen auf die pulmonale Mikrovaskulatur, das Nervensystem, die systemische Virusausbreitung, und das Zusammenspiel mit dem Immunsystem untersucht. Um multizentrische, auf Obduktionen basierende Studien zu erleichtern und eine zentrale Anlaufstelle zu schaffen, die Obduktionszentren, Forscher sowie Datenanalysen und -berichte unterstützt, wurde im April 2020 das deutsche COVID-19-Autopsieregister (DeRegCOVID) ins Leben gerufen.
Methoden: Das elektronische Register verwendet ein webbasiertes elektronisches Fallberichtsformular. Die Teilnahme ist freiwillig und das Biomaterial verbleibt am jeweiligen Standort (dezentrales Biobanking). Im Oktober 2021 umfasste das Register N=1129 Obduktionsfälle mit 69271 einzelnen Datenpunkten, die Informationen über 18674 verfügbare Bioproben enthielten, die von 29 deutschen Standorten gesammelt wurden.
Ergebnisse: In den N=1095 ausgewerteten Datensätzen betrug das Verhältnis von Männern zu Frauen 1,8:1 mit Spitzenwerten bei 65-69 und 80-84 Jahren bei Männern und >85 Jahren bei Frauen. Die Analyse der Sequenz der unmittelbar zum Tod führenden Ereignisse ergab, dass in 86 % der Obduktionsfälle COVID-19 die zugrunde liegende Todesursache war, während in 14 % der Fälle COVID-19 eine Begleiterkrankung war. Die häufigste unmittelbare Todesursache war der diffuse Alveolarschaden, gefolgt von Multiorganversagen. Das Register unterstützt mehrere wissenschaftliche Projekte, die Öffentlichkeitsarbeit und liefert Berichte an die Bundesgesundheitsbehörden, was zu einer Anpassung des deutschen Infektionsschutzgesetzes führte und die Durchführung von Obduktionen in Pandemien erleichtert.
Interpretation: Ein nationales Obduktionsregister kann multizentrische quantitative Informationen über COVID-19-Todesfälle auf nationaler Ebene liefern und damit die medizinische Forschung, die politische Entscheidungsfindung und die öffentliche Diskussion unterstützen.
Finanzierung: Bundesministerien für Bildung und Forschung und für Gesundheit.
Keywords: Autopsy, Registry, COVID-19, SARS-CoV-2, Cause of Death
Research in context.
Evidence before this study
Autopsies are a valuable tool for understanding novel diseases, such as COVID-19; however, most COVID-19 autopsy studies are conducted at a single center, with a small to medium cohort size, and no central (national) registries for COVID-19 autopsies existed. We reviewed the literature available on PubMed using the search terms "COVID-19" AND "cause of death" AND “autopsy” (through January 11, 2022). All autopsy studies were included; verbal autopsy studies, case reports and review articles were excluded. Review articles and meta-analyses on autopsy studies were searched separately using the terms “COVID-19” AND “autopsy” (through January 11, 2022). Included articles were published in English, Russian or Hungarian language.
Added value of this study
This first report of the German Registry of COVID-19 Autopsies (DeRegCOVID) represents the largest, multicentric, national autopsy study to date, with N=1129 COVID-19 autopsy cases from N=29 autopsy centers. We analyzed N=1095 eligible cases with positive clinical or post-mortem SARS-CoV-2 test regarding autopsy rate per calendar week, patient sex and age, disease duration, SARS-CoV-2 RNA detection at autopsy, and cause of death. Beyond the central data analyses, we also present our experience on how such a central autopsy registry can facilitate research, for example, by providing information on available decentrally archived biomaterial, and how it can deliver data for policy making and public outreach. We also discuss the limitations of the study, framing further development of the registry.
Implications of all the available evidence
Our study demonstrates the potential benefit of the autopsy registry in supporting medical research and serving as a central information hub for various actors in the health sector.
Alt-text: Unlabelled box
Introduction
Autopsies are an important tool in medicine, offering insights into disease pathophysiology and causes of death. In COVID-19, autopsies revealed the crucial role of pulmonary (micro)vascular thromboembolism and remodelling, systemic viral spread, and the interplay between viral effects and the immune system.1,2
A number of COVID-19 autopsy studies have been published. However, most of these studies were conducted at single centres and thus had variable, but mostly small to medium cohort sizes (median cohort size N=14), including several studies from Germany. To our knowledge, the only multicentre studies published to date are two survey studies, one from the USA and Brazil, and one from eight European countries and Russia, including N=135 and N=313 COVID-19 autopsies from pathology or forensic medicine, respectively.3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14
There were no national registries that would collect multicentre autopsy data of COVID-19 or even other diseases. Therefore, in April 2020, we launched the German COVID-19 Autopsy Registry, Germany's unified national response to the pandemic (http://www.deregcovid.ukaachen.de/).15,16 We defined the primary objectives of the registry as follows:
-
1.
to collect data from as many COVID-19 autopsies as possible and to provide a central electronic hub for data curation and analysis,
-
2.
to support autopsy centres from pathology, neuropathology and forensic medicine,
-
3.
to support researchers and facilitate multicentre autopsy-based studies,
-
4.
to integrate the registry into the national and international research landscape and report results to policy makers, professional societies, and the public.
Methods
Ethical issues
Only cases that met the legal requirements, i.e., consent given by the deceased or next of kin for autopsy or request for autopsy by the health authorities or by the prosecutor's office were included in the registry. The registry was approved by the ethical committee of the medical faculty of the RWTH Aachen University (EK 092/20). Additionally, each participating centre had a local ethical approval. The registry was registered with the German Clinical Trials Registry (https://www.drks.de/drks_web/navigate.do?navigationId=trial.HTML&TRIAL_ID=DRKS00025136).
Participation and eligibility
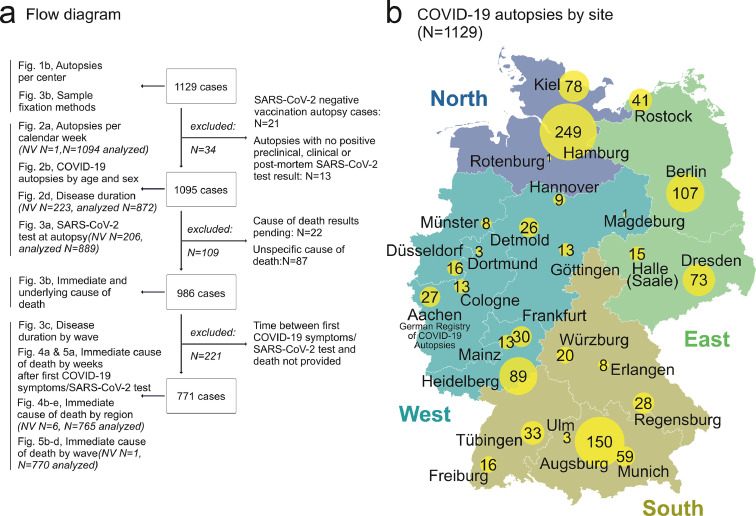
Participation in the registry is voluntary, and the registry was communicated via mailing lists of pathological societies (DGP - German Society for Pathology e.V., BDP Federal Association of German Pathologists e.V., DGNN - German Society for Neuropathology and Neuroanatomy e.V., DGRM - German Society for Forensic Medicine), at congresses and in the German pathology publication “Der Pathologe” (Springer Nature).16 The data were retrieved from the final autopsy report or the laboratory information system by a member of the autopsy team (physician or autopsy technician), scientific staff member or by medical documentation assistants. All cases with a positive SARS-CoV-2 test (usually antigen test from nasopharyngeal swab or PCR from nasopharyngeal swab or from tissue), either preclinical, clinical or at post-mortem, were eligible for registration and analysis. Accordingly, entries regarding post-SARS-CoV-2 vaccination autopsies without SARS-CoV-2 infection (N=21, Figure 1a) were not included in the present analyses. In Germany, vaccination became available for elderly persons during the 2nd wave, which is reflected in only very few cases post-vaccination. As we expect the highly important post-SARS-CoV-2 vaccination autopsy cohort to be further increased in the near future, it will be subject of subsequent analyses from the registry.
Figure 1.
a) Flow diagram of included and excluded cases. b) COVID-19 autopsies per site. From N=1129 autopsies, contributed by N=29 university and non-university autopsy centers in N=27 cities, N=1095 autopsy cases were eligible for analyses. (Map source: Map Data from OpenStreetMap. This data is available under the Open Database License and under Creative Commons Attribution-Share Alike 2.0 license.)
NV = no value
Besides full autopsies, minimal invasive autopsies were also eligible. In N=17 cases, biopsies were mentioned as sample type.
Data management
The data acquisition is performed with the electronic data capture system (EDC) LibreClinica (version 1.0.0rt snapshot). The purpose of the first stage of development of the registry is to improve the availability of cases and samples to enable research. Participation is voluntary and biomaterial remains at the respective site (decentralized biobanking). In order to keep the effort for the participating sites as low as possible, only a small data set on patients and autopsy findings and the number of available samples, are collected. The dataset is under continuous development. A detailed overview of the current data set version 4.0.0 is given in Supplementary Table 1.
Table 1.
Stratification of cohort by region.
| Region | N autopsies by postal code | % of total | N autopsies by regional centres | % of total |
|---|---|---|---|---|
| East (0-1) | 218 | 19 | 236 | 21 |
| North (2) | 313 | 28 | 328 | 29 |
| West (3-6) | 216 | 20 | 248 | 22 |
| South (7-9) | 344 | 30 | 317 | 28 |
| no value | 23 | 2 | 0 | 0 |
| Sum | 1129 | 100 | 1129 | 100 |
The cause of death data was taken from the final autopsy report after the histological examination. Entries without sequential cause of death reporting in combination with a nonspecific condition as the immediate cause of death or with missing data in line 1a (e.g., cardiovascular failure without underlying diseases) were excluded (Figure 1a). Nomenclature of diagnoses was curated following ICD-11 International Statistical Classification of Diseases and related health problems, eleventh revision, 1st ed. To determine the disease duration, the interval between the first symptoms of COVID-19, or, if not available, between the first positive SARS-CoV-2 test and death was calculated. Cause of death data was centrally reviewed and curated following the ICD-10: International statistical classification of diseases and related health problems, tenth revision, 2nd ed., Rules and guidelines for mortality and morbidity coding and International Guidelines for Certification and Classification (Coding) of COVID-19 as Cause of Death.17 Specifically, a chain of events leading directly to death was established, with the immediate cause of death (the final disease, injury, or complication directly causing death) in line 1a and the underlying cause of death (the disease or injury that initiated the chain of morbid events that led directly and inevitably to death) on the lowest used line (1b or 1c, depending on the number of underlying conditions, see Table 2 for an example). The sequence of cause of death was retrieved from the registry entries. In two instances, the sequence of cause of death was changed as suggested in the International Guidelines for the Classification (Coding) of COVID-19 as a Cause of Death: 1. when ischemic heart disease was reported as the cause of death in COVID-19-positive individuals, it was not classified as a COVID-19 death (N=7 cases). 2. in COVID-19 positive individuals, death due to diffuse alveolar damage/acute respiratory distress syndrome (DAD/ARDS), was always considered to be due to COVID-19 (following the WHO guideline, with the exception of aspiration pneumonia), even if COVID-19 had not been reported as the cause of death by the autopsy centre (N=19 cases). “Therapy-associated” deaths comprised N=6 bleeding complications of extracorporeal membrane oxygenation therapy, N=2 bleeding complications of intensive care therapy, N=2 complications of tracheotomy and N=1 bleeding complication after pulmonary embolism lysis therapy. The category “COVID-19 other” comprised N=11 spontaneous bleeding complications, N=2 cerebral infarction, N=2 disseminated intravasal coagulation, N=2 cardiac arrhythmia, N=2 myocarditis, N=1 acute renal failure, N=1 coronary thrombosis, N=1 mesenteric ischemia, N=1 bacterial endocarditis, N=1 hypoxic encephalopathy, N=1 other viral pneumonia, N =1 right ventricular failure without mention of pulmonary embolism and N=2 cardiac failure in patients with amyloidosis. The category “Other” comprised N=4 acute bacterial infection (extrapulmonary), N=1 acute kidney failure, N=1 thrombotic microangiopathy and N=1 hyponatraemia.
Table 2.
Example of how to certify the chain of events for deaths due to COVID-19, modified from.17
| 1 Report disease or condition directly leading to death in line “a” Report chain of events in “due to” order (if applicable) State the underlying cause on the lowest used line |
Cause of Death | ||
 |
a | Acute respiratory distress syndrome | |
 |
b | Due to: Diffuse alveolar damage |
|
| c | Due to: COVID-19 (SARS-CoV-2 test positive) |
||
| 2 Other significant conditions contributing to death | Coronary heart disease | ||
| Diabetes mellitus | |||
| Obesity | |||
Samples
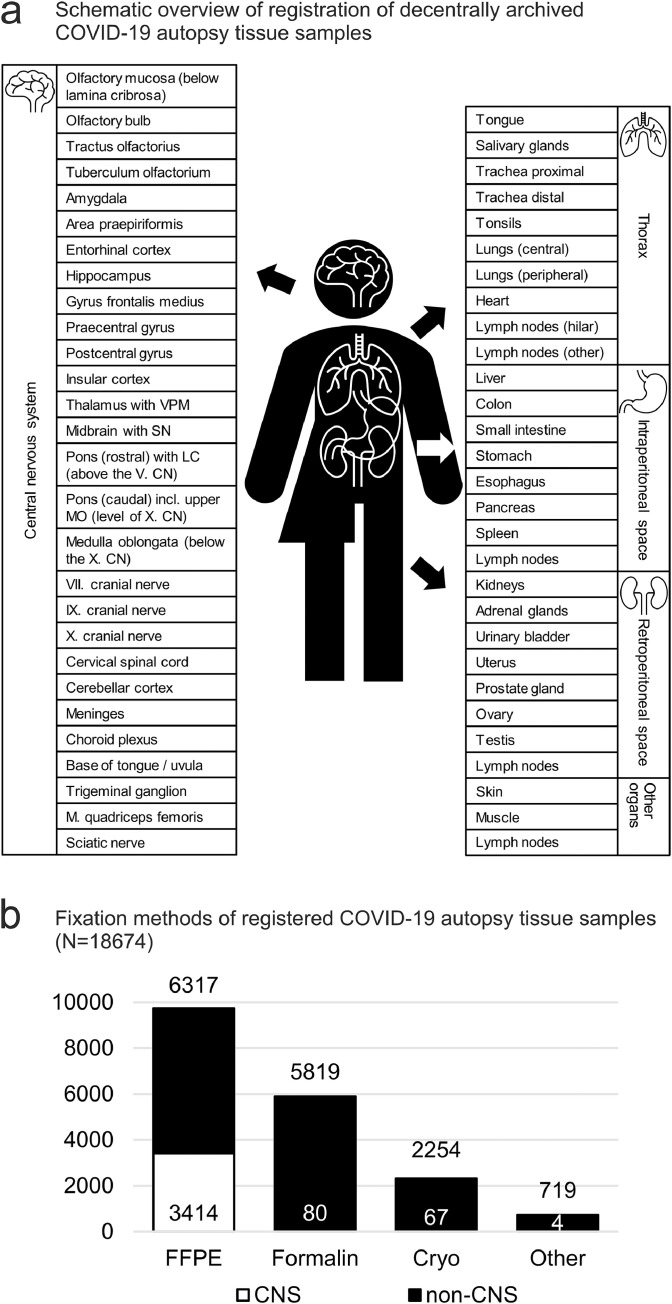
Decentrally archived samples were entered into the registry per tissue type/localization, but not as number of samples per tissue type/localization. Even if multiple lung samples were collected, to keep the data entry process simple, the lung was represented only twice in a list of organs and localizations (e.g., central and peripheral, Figure 6a, Supplementary Table 1). In comparison, the data model for central nervous system samples is very detailed.
Figure 6.
a) Schematic overview of registration of decentrally archived COVID-19 autopsy tissue samples. Samples are categorized into central nervous system, thorax, intraperitoneal space and retroperitoneal space. b) Available tissues from registered COVID-19 autopsies for different fixation methods (minimum N=18674 tissues from 1129 COVID-19 autopsies, numbers represent different organs/tissues, not total amount of samples). Decentrally archived samples from COVID-19 autopsy tissue comprise mainly formalin-fixed, paraffin-embedded samples, followed by formalin-fixed, cryopreserved and otherwise preserved biomaterials e.g., in gutaraldehyde for electron microscopy, or in RNA preserving fixation medium for RNA studies). CNS samples appear overrepresented in comparison to non-CNS samples due to numerous sampling localizations, compared to less detailed sampling localizations for non-CNS samples.
CN = cranial nerve; CNS = central nervous system; FFPE = formalin-fixed, paraffin embedded; LC = locus coeruleus; MO = medulla oblongata; SN = substantia nigra; VPM = nucleus ventralis posteromedialis
Cohort stratification by pandemic wave
Pandemic waves were defined based on COVID-19 death data from the Robert Koch Institute as follows: 1st wave, from calendar week 10 to 31, inclusive, in 2020. Calendar weeks 32 in 2020 to 11, inclusive, in 2021 were selected for the 2nd wave.18 The 3rd wave was defined as beginning calendar week 12 to 38, inclusive, in 2021.
Cohort stratification by region
The first digit of the German postal code of the deceased person's home address was used to stratify the cohort by region. Postal codes beginning with 0-1 were assigned to the "East" 2 to the "North", 3-6 to the "West", and 7-9 to the "South" (Table 1). It should be noted that stratification by postcode results in different numbers than adding the numbers of regional centres. This may be due to patients being transferred from one region to another due to ICU capacity, but also due to individuals residing in a region that is not identical to the region in which they are registered officially.
Statistics
2 × 2 contingency tables were used for association between sex and peaks in age groups (Figure 2b) with graphpad.com/quickcalcs/contingency1/ (Fisher's exact test, two tailed P value). Two-way ANOVA with Bonferroni post test was performed with GraphPad Prism. Graphs were created using GraphPad Prism, Microsoft Excel and R (for details, see Supplementary Table 2). Missing data were considered to be missing at random.
Figure 2.
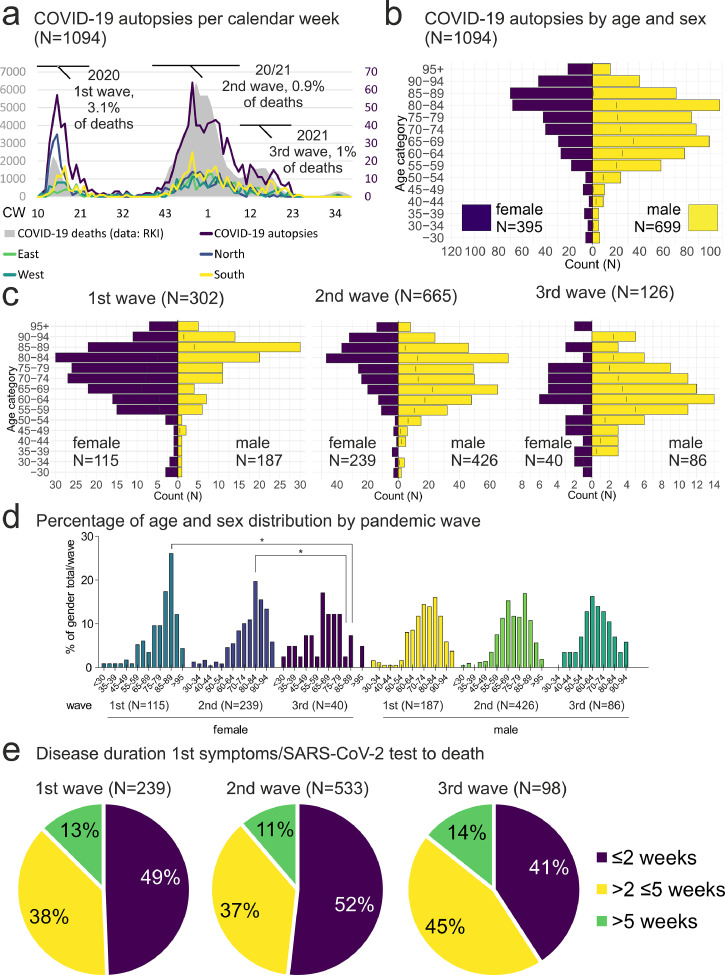
a) COVID-19 autopsies per calendar week (N=1094, 1·18% of all COVID-19 deaths). COVID-19 autopsies (purple line) follow the pandemic peaks of reported COVID-19 deaths (grey area, data: RKI). Note high autopsy rate in the Northern region during the 1st pandemic wave (blue line, one case excluded due to missing value). b) COVID-19 autopsies by age and sex (N=1094). Age and sex distribution of COVID-19 autopsy cases shows male predominance in patients from 50-80 years, but more female autopsy cases in patients older than 85 years. The age peaks ≥85 years and female sex and 65-69 years and male sex showed a formally significant association, while the age peak 80-84 years and male sex did not. (p < 0·001, Fisher's exact test, two tailed P value). c) Age and sex distribution of COVID-19 autopsy numbers by age and sex stratified by pandemic wave. Comparison of age groups between different waves in female or male sex showed no significant differences. d) The comparison of the percentage of age groups of female or male sex of the total number of female/male COVID-19 deceased persons per pandemic wave showed a significant decrease of female cases in the age group of 85-89 years from the 1st to the 3rd and 80-84 years from the 2nd to 3rd pandemic wave (p < 0·05). Comparison between male and female age groups during each wave showed no significant results (two-way ANOVA with Bonferroni post-test). d) Disease duration (N=870). The disease duration from first COVID-19 symptoms/Positive SARS-CoV-2 test to death was less than two weeks in 41-52% of the analysed cases, while it was between 2 and 5 weeks in 37-45% and longer than 5 weeks in 11-14% of cases.
CW = calendar week; RKI = Robert Koch-Institute
Role of the funding source
The funding source had no involvement in study design, data collection, data analysis, interpretation, or writing of the report.
Results
Participation and eligibility
As of October 2021, the registry included N=1129 autopsy cases. Of all N=1129 autopsy cases, N=771 to N=1129 records were eligible, depending on the analysis and missing data points, as shown in the flow diagram in Figure 1a. 69271 single data points, collected from 29 sites across Germany, were available for analyses (Figure 1b). Time points of death spanned calendar week 10, 2020, through calendar week 39, 2021.
Autopsy rates
The number of autopsies per calendar week reflected the pandemic peaks in Germany (Figure 2a). Autopsy rates averaged 1·18% from calendar week 10 in 2020 to calendar week 38 in 2021, but varied between pandemic waves. During the first wave, 3·13% of all reported COVID-19 deaths were autopsied (calendar week 10 to 22 in 2020, N=285 autopsies/ N=9104 reported COVID-19 deaths), whereas during the second and third waves, 0·96% of all reported COVID-19 deaths were autopsied (calendar week 39 in 2020 to calendar week 22 in 2021, N=784 autopsies/ N=82093 reported COVID-19 deaths). The distribution of the autopsy rates per week was not Gaussian, in 36 of 82 weeks from calendar week 10 in 2020 to calendar week 38 in 2021, the autopsy rate was 0-1% of all reported COVID-19 deaths. The interquartile range of the autopsy rates per week was 2·5%, with a mean absolute deviation of 2·2%. The difference in autopsy rate is likely due to much higher death counts in the 2nd and 3rd waves, due to the increased incidence. Given that the absolute numbers of autopsies per calendar week during these waves were comparable, this might suggest a “saturation” of autopsy capacity.
Age and sex distribution
The male-to-female ratio was 1·8:1, with peaks at 65-69 (formally p < 0·001) and 80-84 (ns) years in men and >85 years (formally p < 0·001, Fisher's exact test, two tailed p-value) in women (Figure 2b). Stratified by pandemic wave, the ratio of males to females was 1·6:1 during the 1st wave, 1·8:1 during the 2nd wave and 2·1:1 during the 3rd wave (Figure 2c). The decrease in female COVID-19 autopsies in the age groups of 85-89 years from the 1st to 2nd pandemic wave and 80-84 years from the 2nd to 3rd pandemic wave was statistically significant (Figure 2d, p < 0·05, two-way ANOVA with Bonferroni post test). Comparison of the male age groups between different pandemic waves and between female and male age groups within the pandemic waves showed no significant differences.
Disease duration
The disease duration from onset of first COVID-19 symptoms/positive SARS-CoV-2 test to death was less than two weeks in most cases (median 2 weeks, interquartile range 2·6 weeks). However, more than one third of cases showed a disease duration of more than two and less than five weeks and more than one in ten cases died more than five weeks after the first COVID-19 symptoms/positive SARS-CoV-2 test in each pandemic wave (Figure 2e).
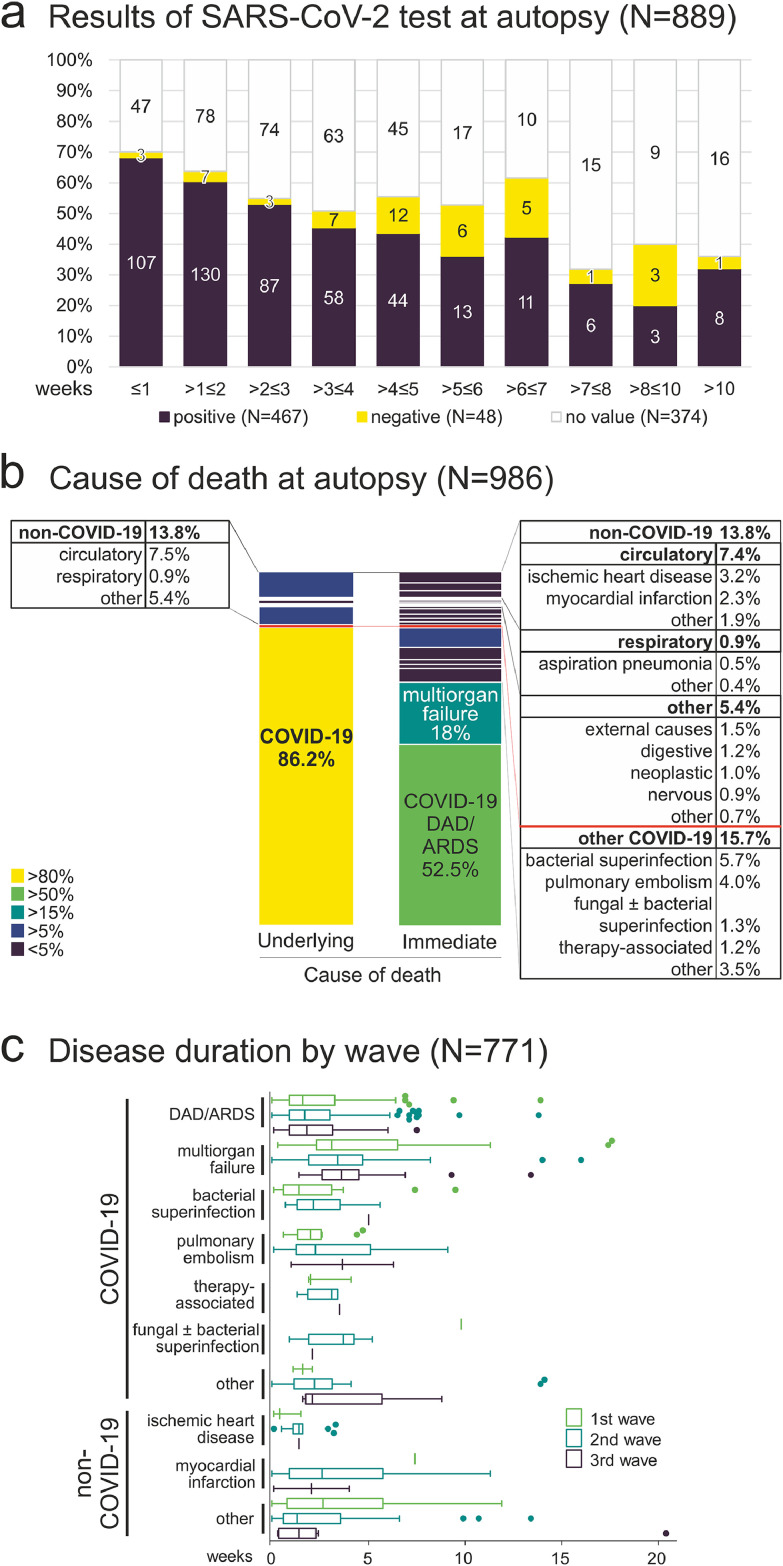
Post-mortem SARS-CoV-2 test
At autopsy, post-mortem SARS-CoV-2 tests were performed in 516 cases (Figure 3a). The ratio of positive to negative test results was 10:1, decreasing with increasing disease duration/postmortem interval. The longest interval between first COVID-19 symptoms/positive SARS-CoV-2 test and positive SARS-CoV-2 test result at autopsy was 18·7 weeks, disease duration 17·4 weeks, postmortem interval 9 days). The longest postmortem interval, after which SARS-CoV-2 RNA was detected at autopsy was 36 days. This finding can be explained by the fact that in Hamburg, a unique collaboration for systematic SARS-CoV-2 mortality monitoring has been pursued by the health authority and forensic medicine since March 2020. This enabled autopsies of non-hospitalized COVID-19 deceased persons, who had been found dead at their homes with sometimes long postmortem intervals.
Figure 3.
a) Results of SARS-CoV-2 test at autopsy (N=889). When autopsies took place within three weeks from the first COVID-19 symptoms/SARS-CoV-2 test, the test for SARS-CoV-2 at autopsy was positive in more than half of the cases. When the disease duration and post-mortem interval were longer than three weeks, the percentage of positive SARS-CoV-2 tests at autopsy decreased to less than 50%. b) Cause of death at autopsy (COVID-19 autopsies with positive clinical or post-mortem test for SARS-CoV-2, N=986). COVID-19 was the underlying cause of death in 86·2% of COVID-19 autopsies, whereas COVID-19 was a concomitant disease in 13·8% of COVID-19 autopsies. The most common immediate cause of death in COVID-19 deaths was diffuse alveolar damage/acute respiratory distress syndrome (DAD/ARDS), followed by multiorgan failure. c) Comparison of the disease duration of the most common immediate causes of death as shown in b) by pandemic wave (Tukey method for plotting the whiskers and outliers, no significant differences between waves, two way ANOVA with Bonferroni post test).
Cause of death
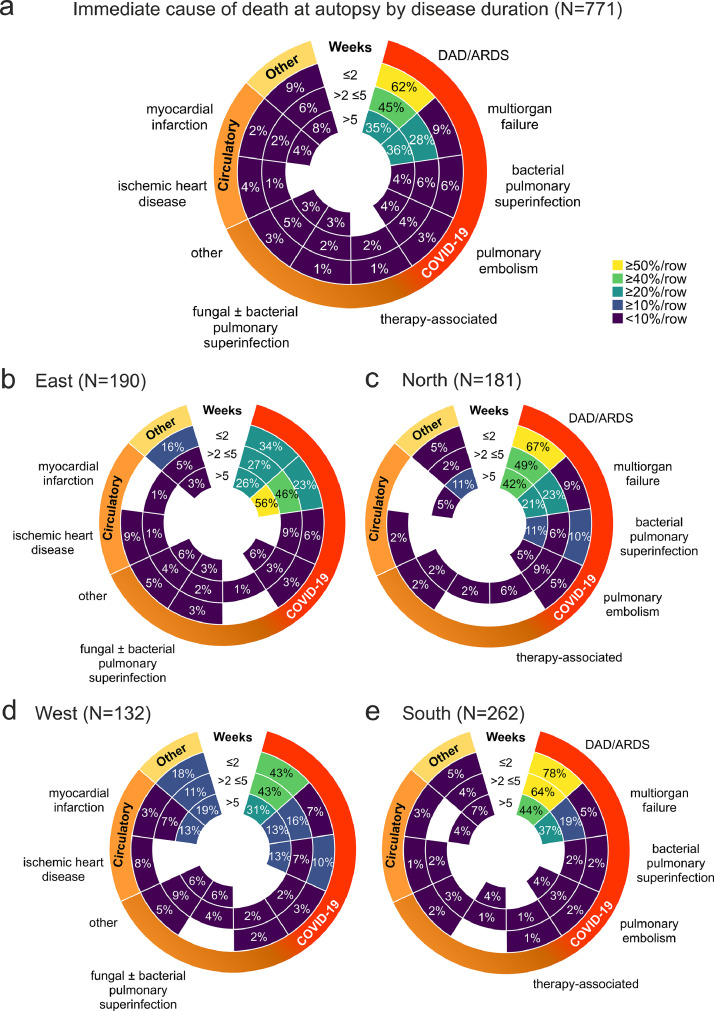
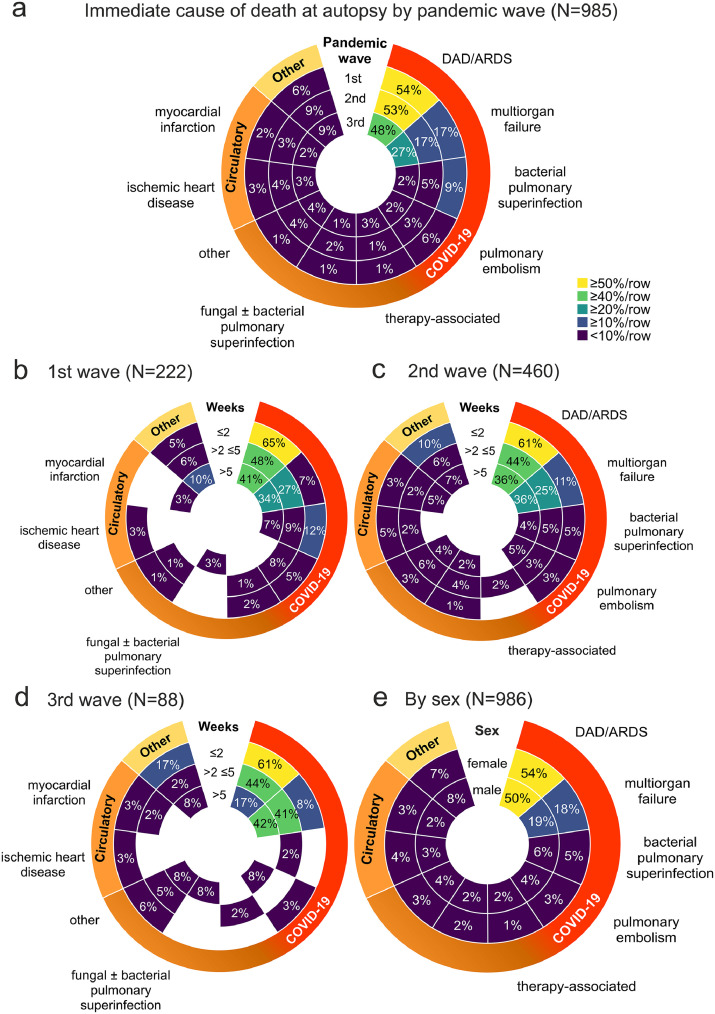
To analyse the cause of death in COVID-19, we examined the chain of events that directly led to death (Table 2). In 86% of autopsy cases, COVID-19 was the underlying cause of death, whereas in 14% of COVID-19 autopsies, COVID-19 was a concomitant disease (Figure 3b). Comparison of the immediate cause of death diagnoses between pandemic waves showed no significant differences in the disease duration (Figure 3c). Looking at percentage of immediate cause of death diagnoses by disease duration, the most common immediate cause of death was DAD/ARDS, followed by multiorgan failure (Figure 4a). Further stratification by region showed similar regional trends with DAD/ARDS as the leading cause of death, followed by multiorgan failure and bacterial pulmonary superinfection, with a significantly higher prevalence in the ≤2 weeks disease duration group in the South compared with the East and West and subsequent decrease in the South. The prevalence of multiorgan failure at >5 weeks disease duration was also higher in the East than in the North and West (Figure 4b-e). A stratification of immediate cause of death by pandemic wave showed a decrease in DAD/ARDS, bacterial pulmonary superinfection and pulmonary embolism from the 1st to the 3rd pandemic wave and an increase of multiorgan failure as an immediate cause of death (Figure 5a). Further stratification by disease duration during the pandemic waves showed a decreasing frequency of DAD/ARDS and increasing frequency of multiorgan failure as an immediate cause of death with increasing time interval between first COVID-19 symptoms/SARS-CoV-2 test in each pandemic wave (Figure 5b-d), which were statistically significant in the 3rd wave (Figure 5d). A stratification of cause of death by sex showed similar distributions of cause of death diagnoses between COVID-19 autopsies of male and female deceased persons without significant differences (Figure 5e).
Figure 4.
a) Immediate cause of death at autopsy (weeks after first COVID-19 symptoms/SARS-CoV-2 test, N=771). Radial heat map (scaled to rows) of the immediate cause of death sorted by interval between first COVID-19 symptoms/SARS-CoV-2 test and death shows highest prevalence of DAD/ARDS as immediate cause of death in the first two weeks of the disease, decreasing subsequently, and an inverse development for multiorgan failure as the second most prevalent immediate cause of death (no significant differences between disease duration intervals). b-e) A further stratification of a) by region shows similar regional trends for cause of death per week after first COVID-19 symptoms/SARS-CoV-2 test. The higher prevalence of DAD/ARDS in COVID-19 autopsies after a disease duration ≤2 (p < 0·01) and >2 ≤5 weeks in the South compared to the East (p < 0·05) and ≤2 weeks in the South compared to the West were significant (p < 0·05), as well as the decrease of DAD/ARDS from ≤2 weeks to >5 weeks in the South (p < 0·05) and higher prevalence of multiorgan failure after >5 weeks in the East compared to the North (p < 0·05) and to the West (p < 0·01), two-way ANOVA with Bonferroni post-test). All radial heatmaps are scaled to row, colour codes and labelling identical to a).
Figure 5.
a) Immediate cause of death at autopsy (weeks after first COVID-19 symptoms/SARS-CoV-2 test, N=985). Radial heat map (scaled to rows) of the immediate cause of death sorted by pandemic wave shows decreasing incidence of bacterial pulmonary superinfection and pulmonary embolism with each wave (no significant differences between the waves). b-d) A further stratification of a) by pandemic wave and interval from first COVID-19 symptoms/SARS-CoV-2 test shows similar percentages of persons deceased from DAD/ARDS during the three waves, but decreasing bacterial pulmonary superinfection with increasing disease duration. The decrease in DAD/ARDS from ≤2 to <5 weeks (p < 0·001) and the increase in multiorgan failure from ≤2 to subsequent weeks (p < 0·05) in the third wave d) were significant (two-way ANOVA with Bonferroni post test). e) The comparison between sex and cause of death regardless of disease duration showed similar distributions of cause of death diagnoses between COVID-19 autopsies of female and male deceased persons (no significant differences). All radial heat maps are scaled to row, colour codes and labelling identical to a).
Samples
The registry also records various anatomic localizations of available biospecimens (Figure 6a), that are frozen/cryopreserved, formalin-fixed and paraffin-embedded (FFPE) or otherwise fixed (e.g., in glutaraldehyde for electron microscopy or in RNase inactivating media for RNA analyses, Figure 6b).
Discussion
The first goal of the registry was to collect autopsy-derived data on as many COVID-19 autopsies as possible. This was achieved by close cooperation and support of all German professional societies for pathology, neuropathology and forensic medicine, which enabled a wide dissemination. Other promotional efforts included presentations at national congresses and the national medical journal “Der Pathologe”. Finally, the establishment of a German Network for Autopsies in Pandemics (DEFEAT PANDEMIcs) further strengthened the inclusion of centres, since the registry serves as the electronic backbone for centralized data reporting within the project. We were able to engage more than three quarters of all national university institutes of pathology, neuropathology, and forensic medicine to participate in the national network. The open and participatory design of the registry, allowing access to own data, and guaranteeing the rights on own data and its publication, was readily accepted by the community. There are several reasons for the relatively low number of autopsied cases, e.g., the early recommendation by the Robert Koch Institute in Germany not to perform COVID-19 autopsies (which, however, was corrected shortly thereafter), but also the overall situation in pathology with the declining interest of both pathologists and clinicians to perform autopsies. To assess the actual infectious potential of SARS-CoV-2 during autopsy, centres from the DEFEAT PANDEMIcs consortium performed a study on infectiousness of contaminated autopsy personal protective equipment.19 We believe that COVID-19 strongly documented and “rejuvenated” the interest in autopsies and that initiatives like the DeRegCOVID might further increase the value of autopsies as an important medical research tool.
Another aspect of the first goal is central data curation and analyses. Regarding curation, the first technical curation was realized by IT specialists, and the second curation of cause of death diagnoses was performed by the establishment of a sequential cause of death chain of events, with the underlying disease on the lowest line used, and the immediate cause of death on the first line. The central analyses allowed for the evaluation of the largest multi-centre cohort of COVID-19 autopsies currently available. A previous autopsy study found that 53% of the cohort studied died due to COVID-19, defined as a cause of death due to respiratory or cardiorespiratory failure with DAD.20 This is well in line with our data, with 52·5% of our cases meeting this study's definition of death due to COVID-19. However, because we also considered deaths due to events subsequent to COVID-19, such as a subsequent multiorgan failure, as "death due to COVID-19" according to the WHO recommendation, the proportion of COVID-19 deaths was higher, accounting for 86·2% of autopsies overall. Other previous studies reported “acute respiratory disease” or COVID-19 pneumonia as the immediate cause of death in 75-83% of COVID-19 autopsies and overall prevalence of DAD/ARDS, regardless of its relevance as a cause of death, was 80·9% of COVID-19 autopsies. 13,21,22 In a large single-centre Russian study, COVID-19 played a significant role in the cause of death in 57% of COVID-19 autopsies.3 Our finding that the prevalence of DAD/ARDS decreases from the 1st wave to subsequent waves is consistent with the result of a recent study from several European and non-European countries.14 Previous studies on COVID-19 autopsies performed very early during the 1st wave reported up to 20% of deaths in COVID-19 autopsies due to pulmonary embolism.23,24 This percentage is much higher compared to 6%, 3% and 2% in the 1st, 2nd and 3rd wave, respectively, in our cohort. The lower percentage of pulmonary embolism as immediate cause of death in our cohort can be explained by guidelines for anticoagulation, published early in the 1st pandemic wave.25 In a meta-analysis of mould disease in fatal COVID-19, invasive mould disease was found in 2% at autopsy, which is consistent with our analysis showing a prevalence of 1% to 3% with increasing disease duration. As for non-invasive mould disease, the prevalence at autopsy may be underestimated because it could be missed with standard histologic stains and moulds are a common contaminant.26 However, the studies remain difficult to compare, because existing guidelines on terminology of cause of death diagnoses do not specifically address autopsy reports. Also, most previous studies analysed autopsy cases regardless of the disease duration, making interpretation and direct comparison of the results difficult. Our finding of regional differences in the distribution of cause of death diagnoses over the different disease duration categories might be related to regional differences in infection dynamics, including ICU capacities and viral variants.27 Another consideration to keep in mind is that COVID-19 is an angiocentric disease, that may cause aggravation of cardiovascular or neurovascular diseases. If we would have considered COVID-19 as the underlying disease in immediate cardiovascular causes of death as well, the impact of COVID-19 as underlying cause of death would increase to more than 90% of our cohort. However, as this is currently not recommended in the World Health Organization's International Guidelines for Certification and Classification (Coding) of COVID-19 as Cause of Death, we did not include these cases into COVID-19 deaths during our analyses. The reasons for regional variability of the cause of death distribution and the contribution of COVID-19 to death from cardiovascular disease are important questions requiring further studies.
The second objective of the registry is to support the autopsy centres, by serving as a central information hub for practical aspects of COVID-19 autopsies, such as providing Standards of Practice (SOPs) for autopsies in pandemic situations. Such an SOP was prepared by the registry very early in the pandemic, and support in e.g., personal protective measures and sampling was provided.16 The registry also provides support on all aspects of the electronic system and data input process.
The third main objective of the DeRegCOVID is to act as an honest broker, i.e., to connect researchers with autopsy centres that can provide available material or data for the research question they would like to address. Research projects/inquiries can be submitted using a simple application form available on the website www.DeRegCOVID.ukaachen.de. The registry team reviews the applications and assists applicants with project planning, as appropriate, since there is often little experience with research on autopsy specimens. The registry team then checks the availability of the requested biomaterial and forwards the application to the respective autopsy centres. The decentralized biobanking concept, in which all samples remain at the respective site, leaves the rights to the samples and data with the individual centres, which autonomously decide on the suitability and availability of their resources. By December 2021, the registry has connected autopsy centres with researchers from various ongoing scientific projects, resulting in 23 publications in peer-reviewed journals. To date, several basic science topics have been addressed, e.g., the switch of macrophages driving profibrotic lung sequelae of COVID-19,2 T-cell cytotoxicity in COVID-19,28 imaging intact human organs with local resolution at the cellular level,29 and SARS-CoV-2 infection leading to viral infiltration of the pancreas,30 but clinical questions are also being addressed.
The fourth major objective of DeRegCOVID is to provide reports to the federal health authorities and the German Ministry of Health, considerably improving the information available for policy making, and communicating the essential role of autopsies in pandemic management to the public and policymakers through intensive public relations effort.31, 32, 33 An unexpected and important achievement of this close interaction was an adjustment of the legislation (German Infection Protection Act §25 and §60) to facilitate the performance of autopsies during pandemics.34
The registry currently has several limitations. One of them is the lack of centralized histology evaluation, thus relying on autopsy report data generated by each centre. However, because less than 10% of the data specifying the cause of death (N=109 of N=1095) could not be analysed due to missing or inconclusive data, this approach seems sufficiently feasible. Performing and reporting autopsies are a major aspect in pathology training and part of the daily routine at all German university hospitals and also at some, but not all, non-university pathology institutes. Almost one quarter of autopsy cases were contributed by contributors from forensic medicine, which also perform cause of death investigations as major part of their training and daily routine. In addition, a specific guideline on the certification of COVID-19 deaths was published by the WHO in April 2020,17 and early on during the pandemic, virtual congresses were held by the German (and European) professional pathology societies with COVID-19 sessions and specific virtual lectures on the pathologic features of COVID-19. From a macroscopic and microscopic point of view, the morphologic findings of COVID-19 DAD/ARDS are distinct, with heavy lungs void of air and diffuse alveolar damage at various stages.11 A special issue on COVID-19 has been published in the German pathology journal “Der Pathologe”35 with a special focus on SARS-CoV-2 detection and COVID-19 manifestations in routine pathology diagnostics. Collectively, these aspects are reassuring the validity of the data. In the future, support by specialized reference centres or consensus readings, particularly using digital pathology might facilitate centralized readings and further strengthen the data validity. Furthermore, our eCRF does not yet capture all information and metadata. In this first registry, we have tried to balance the amount of information with the amount of time needed for data input to assess whether and how such a registry can support research in general. Also, the current sample registration section in the eCRF is concise regarding non-CNS localizations, but much more granular regarding CNS localizations. This was due to the fusion of the DeRegCOVID with a previously separate registry for COVID-19 autopsy-derived CNS samples (CNS-COVID-19). Further extension of the depth of captured data, particularly combined with support systems for automated data input, are among the important next steps in the further development of the registry.
Another limitation is the variability in the nomenclature of autopsy-derived diagnoses. For neoplastic diseases, specific terminology has been developed and implemented in pathology since the 1960s to facilitate interdisciplinary communication, guideline-based decision making and cancer registration (International Classification of Diseases for Oncology, ICD-O). Only in 2021, with the release of ICD-11, that an integration of morphologic diagnoses such as “CB05.0 diffuse alveolar damage” has been achieved. Due to its recency, ICD-11 has not yet been introduced into autopsy reporting practice in Germany. Furthermore, although a German recommendation guideline on the sequential cause of death reporting was released in 2017,36 the structure of cause of death reports remains variable at different centres. Therefore, an electronic death certificate within the registry might be helpful to not only harmonize cause of death terminology, but also to speed up the registration of pandemic deaths in the future. As an example, in Portugal, a complete electronic death certification has been used since 1st January 2014.37,38
Another limitation is that our data do not provide a complete national coverage, because participation is voluntary. Based on our correspondence with other centres, we suspect that 10-20% of COVID-19 autopsies in Germany may not have been included in the registry. Furthermore, in most centres only hospitalized patients were autopsied, potentially leading to a selection bias. The age and sex characteristics of our cohort are consistent with data published by German national health authorities. The underlying cause of death was COVID-19 in 86% of autopsy cases (“dying of COVID-19"), while COVID-19 was a concomitant disease in 14% (“dying with COVID-19"), in agreement with data published by the German Federal Statistical Office (83% dying of COVID-19, 17% dying with COVID-19).18 Since epidemiological characteristics and the proportion of deaths attributable to COVID-19 in our cohort show high concordance with data on all COVID-19 deaths published by the German Federal Statistical Office, the results are comparable and likely representative.39
In conclusion, a centralized national autopsy registry can provide multicentre, quantitative information on COVID-19 deaths at the national level, providing information and facilitating medical research, policy making and public discussion. There are several limitations of the registry, which outline the future directions for further improving this infrastructure, not only for pandemic preparedness but also for other autopsy-based research studies. We hope that this report might spark interest in the development of similar national autopsy-registries, which could be potentially integrated on a European or even global level.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
Contributors
Drs von Stillfried and Boor had full access to all of the data in the study and take responsibility for the integrity of the data, which were provided by the study collaborators, and the accuracy of the data analysis. Concept and design: von Stillfried, Bülow, Röhrig, Boor. Acquisition, analysis, or interpretation of data: von Stillfried, Bülow, Röhrig, Boor. Drafting of the manuscript: von Stillfried, Bülow, Röhrig, Boor. Obtained funding: Boor. Administrative, technical, or material support: von Stillfried, Bülow, Röhrig, Boor. Supervision: Boor. All authors and collaborators approved of the manuscript.
Acknowledgements
The authors thank Benita Freeborn for proofreading of the manuscript.
Funding
This work was supported by the German Registry of COVID-19 Autopsies (www.DeRegCOVID.ukaachen.de), funded Federal Ministry of Health (ZMVI1-2520COR201), by the Federal Ministry of Education and Research within the framework of the network of university medicine (DEFEAT PANDEMIcs, 01KX2021).
Data availability statement
The current eCRF data set version 4.0.0 is available as Supplementary material. The autopsy-derived data that support the findings of this study will be made available to researchers on reasonable request after submitting a request to the DeRegCOVID registry (www.deregcovid.ukaachen.de).
# DeRegCOVID collaborators:
Jana Böckera, Jens Schmidta, Pauline Tholena, Raphael Majeedb, Jan Wienströerb, Joachim Weisc, Juliane Bremerc, Ruth Knücheld, Anna Breitbachd, Claudio Cacchid, Benita Freebornd, Sophie Wucherpfennigd, Oliver Springe, Georg Braunf, Christoph Römmelef, Bruno Märklg, Rainer Clausg, Christine Dhillong, Tina Schallerg, Eva Siposg, Klaus Hirschbühlh, Michael Wittmannh, Elisabeth Klingi, Thomas Krönckej, Frank L. Heppnerk,l,m, Jenny Meinhardtk, Helena Radbruchk, Simon Streitk, David Horstn, Sefer Elezkurtajn, Alexander Quaaso, Heike Göbelo, Torsten Hansenp, Ulf Titzep, Johann Lorenzenq, Thomas Reuterq, Jaroslaw Woloszynq, Gustavo Barettonr, Julia Hilsenbeckr, Matthias Meinhardtr, Jessica Pablikr, Linna Sommerr, Olaf Holotiuks, Meike Meinels, Nina Mahlket, Irene Espositou, Graziano Crudeleu, Maximilian Seidlu, Kerstin U. Amannv, Roland Corasw, Arndt Hartmannx, Philip Eichhornx, Florian Hallerx, Fabienne Langex, Kurt Werner Schmidy, Marc Ingenwerthy, Josefine Rawitzery, Dirk Theegarteny, Christoph G. Birngruberz, Peter Wildaa, Elise Gradhandaa, Kevin Smithaa, Martin Wernerab, Oliver Schillingab, Till Ackerac, Stefan Gattenlöhnerad, Christine Stadelmannae, Imke Metzae, Jonas Franzae, Lidia Storkae, Carolina Thomasae, Sabrina Zechelae, Philipp Ströbelaf, Claudia Wickenhauserag, Christine Fathkeag, Anja Harderag, Benjamin Ondruschkaah, Eric Dietzah, Carolin Edlerah, Antonia Fitzekah, Daniela Fröbah, Axel Heinemannah, Fabian Heinrichah, Anke Kleinah, Inga Kniepah, Larissa Lohnerah, Dustin Möbiusah, Klaus Püschelah, Julia Schädlerah, Ann-Sophie Schröderah, Jan-Peter Sperhakeah, Martin Aepfelbacherai, Nicole Fischerai, Marc Lütgehetmannai, Susanne Pfefferleai, Markus Glatzelaj, Susanne Krasemannaj, Jakob Matschkeaj, Danny Jonigkak, Christopher Werleinak, Peter Schirmacheral, Lisa Maria Domkeal, Laura Hartmannal, Isabel Madeleine Kleinal, Constantin Schwabal, Christoph Röckenam, Johannes Friemannan, Dorothea Langerao, Wilfried Rothap, Stephanie Stroblap, Martina Rudeliusaq, Konrad Friedrich Stockar, Wilko Weichertas, Claire Delbridgeas, Atsuko Kasajimaas, Peer-Hendrik Kuhnas, Julia Slotta-Huspeninaas, Gregor Weirichas, Peter Barthat, Eva Wardelmannat, Katja Evertau, Andreas Büttnerav, Johannes Manhartav, Stefan Nigburav, Iris Bittmannaw, Falko Fendax, Hans Bösmüllerax, Massimo Granaiax, Karin Klingelax, Verena Warmax, Konrad Steinestelay, Vincent Gottfried Umathumay, Andreas Rosenwaldaz, Florian Kurzaz, Niklas Vogtaz
a Center for Translational & Clinical Research (CTC-A), University Hospital RWTH Aachen, Aachen, Germany
b Institute for Medical Informatics, University Hospital RWTH Aachen, Aachen, Germany
c Institute of Neuropathology, University Hospital RWTH Aachen, Aachen, Germany
d Institute of Pathology, University Hospital RWTH Aachen, Aachen, Germany
e Anesthesiology and Intensive Care, University Hospital Augsburg, Augsburg, Germany
f Gastroenterology, University Hospital Augsburg, Augsburg, Germany
g General Pathology and Molecular Diagnostics, University Hospital Augsburg, Augsburg, Germany
h Hematology and Oncology, University Hospital Augsburg, Augsburg, Germany
i Laboratory Medicine and Microbiology, University Hospital Augsburg, Augsburg, Germany
j Radiology, University Hospital Augsburg, Augsburg, Germany
k Department of Neuropathology, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany
l German Center for Neurodegenerative Diseases (DZNE) Berlin, Berlin, Germany
m Cluster of Excellence, NeuroCure, Berlin, Germany
n Institute of Pathology, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany
o Department of Pathology, University Hospital Cologne, Cologne, Germany
p Institute of Pathology, University Hospital OWL of the Bielefeld University, Campus Lippe, Detmold, Germany
q Department of Pathology, Klinikum Dortmund, Dortmund, Germany
r Institute of Pathology, University Hospital Dresden, Dresden, Germany
s Gemeinschaftspraxis für Pathologie, Dresden, Germany
t Institute of Forensic Medicine, University Hospital Düsseldorf, Düsseldorf, Germany
u Institute of Pathology, University Hospital Düsseldorf, Düsseldorf, Germany
v Department of Nephropathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
w Institute of Neuropathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
x Institute of Pathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
y Institute of Pathology, University Hospital Essen, Essen, Germany
z Institute of Forensic Medicine, University Hospital Frankfurt, Frankfurt, Germany
aa Senckenberg Institute of Pathology, University Hospital Frankfurt, Frankfurt, Germany
ab Institut für Klinische Pathologie, Medical Center-University of Freiburg, Freiburg, Germany
ac Institute of Neuropathology, University Hospital Giessen and Marburg, Giessen, Germany
ad Institute of Pathology, University Hospital Giessen and Marburg, Giessen, Germany
ae Institute of Neuropathology, University Medical Center Göttingen, Göttingen, Germany
af Institute of Pathology, University Medical Center Göttingen, Göttingen, Germany
ag Institute of Pathology, University Hospital Halle (Saale), Halle (Saale), Germany
ah Institute of Legal Medicine, University Medical Center Hamburg- Eppendorf, Hamburg, Germany
ai Institute of Medical Microbiology, Virology, and Hygiene, University Medical Center Hamburg- Eppendorf, Hamburg, Germany
aj Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
ak Institute of Pathology, Hannover Medical School, Hannover, Germany
al Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany
am Department of Pathology, University Hospital Schleswig-Holstein, Kiel, Germany
an Department of Pathology, University Hospital Cologne, Lüdenscheid, Germany
ao Institute of Pathology, Klinikum Magdeburg, Magdeburg, Germany
ap Institute of Pathology, University Medical Center Mainz, Mainz, Germany
aq Institute of Pathology, Ludwig-Maximilians-Universität Munich, Munich, Germany
ar Department of Nephrology, TUM School of Medicine of Technical University of Munich, Munich, Germany
as Institute of Pathology, TUM School of Medicine of Technical University of Munich, Munich, Germany
at Gerhard Domagk Institute of Pathology, University Hospital Münster, Münster, Germany
au Institute of Pathology, University Hospital Regensburg, Regensburg, Germany
av Institute of Legal Medicine, Rostock University Medical Center, Rostock, Germany
aw Institute of Pathology, Agaplesion Diakonieklinikum Rotenburg, Rotenburg, Germany
ax Institute of Pathology and Neuropathology, University Hospital Tübingen, Tübingen, Germany
ay Department of Pathology, Bundeswehrkrankenhaus Ulm, Ulm, Germany
az Institute of Pathology, University of Würzburg, Würzburg, Germany
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.lanepe.2022.100330.
Contributor Information
Peter Boor, Email: pboor@ukaachen.de.
German Registry of COVID-19 Autopsies (DeRegCOVID), DeRegCOVID Collaborators:
Jana Böcker, Jens Schmidt, Pauline Tholen, Raphael Majeed, Jan Wienströer, Joachim Weis, Juliane Bremer, Ruth Knüchel, Anna Breitbach, Claudio Cacchi, Benita Freeborn, Sophie Wucherpfennig, Oliver Spring, Georg Braun, Christoph Römmele, Bruno Märkl, Rainer Claus, Christine Dhillon, Tina Schaller, Eva Sipos, Klaus Hirschbühl, Michael Wittmann, Elisabeth Kling, Thomas Kröncke, Frank L. Heppner, Jenny Meinhardt, Helena Radbruch, Simon Streit, David Horst, Sefer Elezkurtaj, Alexander Quaas, Heike Göbel, Torsten Hansen, Ulf Titze, Johann Lorenzen, Thomas Reuter, Jaroslaw Woloszyn, Gustavo Baretton, Julia Hilsenbeck, Matthias Meinhardt, Jessica Pablik, Linna Sommer, Olaf Holotiuk, Meike Meinel, Nina Mahlke, Irene Esposito, Graziano Crudele, Maximilian Seidl, Kerstin U. Amann, Roland Coras, Arndt Hartmann, Philip Eichhorn, Florian Haller, Fabienne Lange, Kurt Werner Schmid, Marc Ingenwerth, Josefine Rawitzer, Dirk Theegarten, Christoph G. Birngruber, Peter Wild, Elise Gradhand, Kevin Smith, Martin Werner, Oliver Schilling, Till Acker, Stefan Gattenlöhner, Christine Stadelmann, Imke Metz, Jonas Franz, Lidia Stork, Carolina Thomas, Sabrina Zechel, Philipp Ströbel, Claudia Wickenhauser, Christine Fathke, Anja Harder, Benjamin Ondruschka, Eric Dietz, Carolin Edler, Antonia Fitzek, Daniela Fröb, Axel Heinemann, Fabian Heinrich, Anke Klein, Inga Kniep, Larissa Lohner, Dustin Möbius, Klaus Püschel, Julia Schädler, Ann-Sophie Schröder, Jan-Peter Sperhake, Martin Aepfelbacher, Nicole Fischer, Marc Lütgehetmann, Susanne Pfefferle, Markus Glatzel, Susanne Krasemann, Jakob Matschke, Danny Jonigk, Christopher Werlein, Peter Schirmacher, Lisa Maria Domke, Laura Hartmann, Isabel Madeleine Klein, Constantin Schwab, Christoph Röcken, Johannes Friemann, Dorothea Langer, Wilfried Roth, Stephanie Strobl, Martina Rudelius, Konrad Friedrich Stock, Wilko Weichert, Claire Delbridge, Atsuko Kasajima, Peer-Hendrik Kuhn, Julia Slotta-Huspenina, Gregor Weirich, Peter Barth, Eva Wardelmann, Katja Evert, Andreas Büttner, Johannes Manhart, Stefan Nigbur, Iris Bittmann, Falko Fend, Hans Bösmüller, Massimo Granai, Karin Klingel, Verena Warm, Konrad Steinestel, Vincent Gottfried Umathum, Andreas Rosenwald, Florian Kurz, and Niklas Vogt
Appendix. Supplementary materials
References
- 1.Puelles VG, Lutgehetmann M, Lindenmeyer MT, et al. Multiorgan and Renal Tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wendisch D, Dietrich O, Mari T, et al. SARS-CoV-2 infection triggers profibrotic macrophage responses and lung fibrosis. Cell. 2021;184(26) doi: 10.1016/j.cell.2021.11.033. 6243-61 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rybakova MG, Karev VE, Kuznetsova IA. [Anatomical pathology of novel coronavirus (COVID-19) infection. First impressions] Arkh Patol. 2020;82(5):5–15. doi: 10.17116/patol2020820515. [DOI] [PubMed] [Google Scholar]
- 4.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosmuller H, Traxler S, Bitzer M, et al. The evolution of pulmonary pathology in fatal COVID-19 disease: an autopsy study with clinical correlation. Virchows Arch. 2020;477(3):349–357. doi: 10.1007/s00428-020-02881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elezkurtaj S, Greuel S, Ihlow J, et al. Causes of death and comorbidities in hospitalized patients with COVID-19. Sci Rep. 2021;11 doi: 10.1038/s41598-021-82862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evert K, Dienemann T, Brochhausen C, et al. Autopsy findings after long-term treatment of COVID-19 patients with microbiological correlation. Virchows Arch. 2021;479(1):97–108. doi: 10.1007/s00428-020-03014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzek A, Schadler J, Dietz E, et al. Prospective postmortem evaluation of 735 consecutive SARS-CoV-2-associated death cases. Sci Rep. 2021;11(1):19342. doi: 10.1038/s41598-021-98499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagiannis D, Umathum VG, Bloch W, et al. Antemortem vs Postmortem Histopathologic and Ultrastructural Findings in Paired Transbronchial Biopsy Specimens and Lung Autopsy Samples From Three Patients With Confirmed SARS-CoV-2. Am J Clin Pathol. 2021 doi: 10.1093/ajcp/aqab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschbuhl K, Dintner S, Beer M, et al. Viral mapping in COVID-19 deceased in the Augsburg autopsy series of the first wave: A multiorgan and multimethodological approach. PLoS One. 2021;16(7) doi: 10.1371/journal.pone.0254872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaller T, Hirschbuhl K, Burkhardt K, et al. Postmortem Examination of Patients With COVID-19. JAMA. 2020;323(24):2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong DWL, Klinkhammer BM, Djudjaj S, et al. Multisystemic Cellular Tropism of SARS-CoV-2 in Autopsies of COVID-19 Patients. Cells. 2021;10(8) doi: 10.3390/cells10081900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper JE, Padera RF, Dolhnikoff M, et al. A Postmortem Portrait of the Coronavirus Disease 2019 (COVID-19) Pandemic: A Large Multi-institutional Autopsy Survey Study. Arch Pathol Lab Med. 2021;145(5):529–535. doi: 10.5858/arpa.2020-0786-SA. [DOI] [PubMed] [Google Scholar]
- 14.Fortarezza F, Pezzuto F, Hofman P, et al. COVID-19 Pulmonary Pathology: The Experience of European Pulmonary Pathologists throughout the First Two Waves of the Pandemic. Diagnostics. 2022;12(1):95. doi: 10.3390/diagnostics12010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Stillfried S, Bulow RD, Rohrig R, Knuchel-Clarke R, Boor P, DeRegCovid Autopsy registry can facilitate COVID-19 research. EMBO Mol Med. 2020;12(8):e12885. doi: 10.15252/emmm.202012885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Stillfried S, Acker T, Aepfelbacher M, et al. Cooperative approach of pathology and neuropathology in the COVID-19 pandemic: German registry for COVID-19 autopsies (DeRegCOVID) and German network for autopsies in pandemics (DEFEAT PANDEMIcs) Pathologe. 2021 [Google Scholar]
- 17.World Health Organization. International Guidelines for Certification and Classification (Coding) of COVID-19 as Cause of Death. 16 April 2020. https://www.who.int/classifications/icd/Guidelines_Cause_of_Death_COVID-19.pdf. (Accessed 5 November 2021).
- 18.Statistisches Bundesamt (Destatis). Pressrelease #327 from 8 July. 8 July 2021 2021. https://www.destatis.de/EN/Press/2021/07/PE21_327_23211.html;jsessionid=4AC8A191948E0A5CC9E5A66CEEF29488.live732. (Accessed 3 November 2021).
- 19.Brandner JM, Boor P, Borcherding L, et al. Contamination of personal protective equipment during COVID-19 autopsies. Virchows Arch. 2022 doi: 10.1007/s00428-021-03263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haberecker M, Schwarz EI, Steiger P, et al. Autopsy-Based Pulmonary and Vascular Pathology: Pulmonary Endotheliitis and Multi-Organ Involvement in COVID-19 Associated Deaths. Respiration. 2021:1–11. doi: 10.1159/000518914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edler C, Schroder AS, Aepfelbacher M, et al. Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int J Legal Med. 2020;134(4):1275–1284. doi: 10.1007/s00414-020-02317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satturwar S, Fowkes M, Farver C, et al. Postmortem Findings Associated With SARS-CoV-2: Systematic Review and Meta-analysis. Am J Surg Pathol. 2021;45(5):587–603. doi: 10.1097/PAS.0000000000001650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabrese F, Pezzuto F, Fortarezza F, et al. Pulmonary pathology and COVID-19: lessons from autopsy. The experience of European Pulmonary Pathologists. Virchows Arch. 2020;477(3):359–372. doi: 10.1007/s00428-020-02886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wichmann D, Sperhake JP, Lutgehetmann M, et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann Intern Med. 2020;173(4):268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langer F, Kluge S, Klamroth R, Oldenburg J. Coagulopathy in COVID-19 and Its Implication for Safe and Efficacious Thromboprophylaxis. Hamostaseologie. 2020;40(3):264–269. doi: 10.1055/a-1178-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kula BE, Clancy CJ, Hong Nguyen M, Schwartz IS. Invasive mould disease in fatal COVID-19: a systematic review of autopsies. Lancet Microbe. 2021;2(8) doi: 10.1016/S2666-5247(21)00091-4. e405-e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuppert A, Polotzek K, Schmitt J, Busse R, Karschau J, Karagiannidis C. Different spreading dynamics throughout Germany during the second wave of the COVID-19 pandemic: a time series study based on national surveillance data. Lancet Reg Health Eur. 2021;6 doi: 10.1016/j.lanepe.2021.100151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georg P, Astaburuaga-Garcia R, Bonaguro L, et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell. 2021 doi: 10.1016/j.cell.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh CL, Tafforeau P, Wagner WL, et al. Imaging intact human organs with local resolution of cellular structures using hierarchical phase-contrast tomography. Nat Methods. 2021 doi: 10.1038/s41592-021-01317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steenblock C, Richter S, Berger I, et al. Viral infiltration of pancreatic islets in patients with COVID-19. Nat Commun. 2021;12(1):3534. doi: 10.1038/s41467-021-23886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledford H. Autopsy slowdown hinders quest to determine how coronavirus kills. 7 May 2020. https://www.nature.com/articles/d41586-020-01355-z. (Accessed 1 December 2021). [DOI] [PubMed]
- 32.Burkhardt M. [Corona knowledge from autopsies-Pathology shows Covid-19 long-term damage] Article in German. 18 December 2020. https://www.zdf.de/nachrichten/panorama/corona-pathologie-lunge-schaeden-100.html. (Accessed 1 December 2021).
- 33.dpa-infocom. [Pathologist: autopsies help better understand Corona] Article in German. 6 February 2020. https://www.sueddeutsche.de/gesundheit/gesundheit-kiel-pathologe-obduktionen-helfen-corona-besser-zu-verstehen-dpa.urn-newsml-dpa-com-20090101-210206-99-325754. (Accessed 1 December 2021).
- 34.German Federal Ministry of Justice and Consumer Protection. [Law on the Prevention and Control of Infectious Diseases in Humans (Infection Protection Act - IfSG) § 25 Investigations] German. http://www.gesetze-im-internet.de/ifsg/__25.html. (Accessed 6 December 2021).
- 35.Der Pathologe. COVID-19. 2021. https://www.springermedizin.de/der-pathologe-2-2021/18934598. (Accessed 11 January 2022).
- 36.Bundesverband Deutscher Pathologen e.V., e.V. DGfP. [S1- Guideline for the performance of autopsies in pathology] Article in German. August 2017. https://www.pathologie.de/?eID=downloadtool&uid=1667. (Accessed 3 November 2021).
- 37.Direção-Geral da Saúde. [Mortality in real time] Portuguese. 1 December 2021. https://evm.min-saude.pt/. (Accessed 1 December 2021).
- 38.Pinto CS, Anderson RN, Martins H, Marques C, Maia C, do Carmo Borralho M. Mortality Information System in Portugal: transition to e-death certification. Eurohealth (Lond) 2016;22(2):1–53. [PMC free article] [PubMed] [Google Scholar]
- 39.Robert Koch-Institut. [Deaths by date of death] Article in German. 28 October 2021. https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Projekte_RKI/COVID-19_Todesfaelle.html. (Accessed 3 November 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The current eCRF data set version 4.0.0 is available as Supplementary material. The autopsy-derived data that support the findings of this study will be made available to researchers on reasonable request after submitting a request to the DeRegCOVID registry (www.deregcovid.ukaachen.de).
# DeRegCOVID collaborators:
Jana Böckera, Jens Schmidta, Pauline Tholena, Raphael Majeedb, Jan Wienströerb, Joachim Weisc, Juliane Bremerc, Ruth Knücheld, Anna Breitbachd, Claudio Cacchid, Benita Freebornd, Sophie Wucherpfennigd, Oliver Springe, Georg Braunf, Christoph Römmelef, Bruno Märklg, Rainer Clausg, Christine Dhillong, Tina Schallerg, Eva Siposg, Klaus Hirschbühlh, Michael Wittmannh, Elisabeth Klingi, Thomas Krönckej, Frank L. Heppnerk,l,m, Jenny Meinhardtk, Helena Radbruchk, Simon Streitk, David Horstn, Sefer Elezkurtajn, Alexander Quaaso, Heike Göbelo, Torsten Hansenp, Ulf Titzep, Johann Lorenzenq, Thomas Reuterq, Jaroslaw Woloszynq, Gustavo Barettonr, Julia Hilsenbeckr, Matthias Meinhardtr, Jessica Pablikr, Linna Sommerr, Olaf Holotiuks, Meike Meinels, Nina Mahlket, Irene Espositou, Graziano Crudeleu, Maximilian Seidlu, Kerstin U. Amannv, Roland Corasw, Arndt Hartmannx, Philip Eichhornx, Florian Hallerx, Fabienne Langex, Kurt Werner Schmidy, Marc Ingenwerthy, Josefine Rawitzery, Dirk Theegarteny, Christoph G. Birngruberz, Peter Wildaa, Elise Gradhandaa, Kevin Smithaa, Martin Wernerab, Oliver Schillingab, Till Ackerac, Stefan Gattenlöhnerad, Christine Stadelmannae, Imke Metzae, Jonas Franzae, Lidia Storkae, Carolina Thomasae, Sabrina Zechelae, Philipp Ströbelaf, Claudia Wickenhauserag, Christine Fathkeag, Anja Harderag, Benjamin Ondruschkaah, Eric Dietzah, Carolin Edlerah, Antonia Fitzekah, Daniela Fröbah, Axel Heinemannah, Fabian Heinrichah, Anke Kleinah, Inga Kniepah, Larissa Lohnerah, Dustin Möbiusah, Klaus Püschelah, Julia Schädlerah, Ann-Sophie Schröderah, Jan-Peter Sperhakeah, Martin Aepfelbacherai, Nicole Fischerai, Marc Lütgehetmannai, Susanne Pfefferleai, Markus Glatzelaj, Susanne Krasemannaj, Jakob Matschkeaj, Danny Jonigkak, Christopher Werleinak, Peter Schirmacheral, Lisa Maria Domkeal, Laura Hartmannal, Isabel Madeleine Kleinal, Constantin Schwabal, Christoph Röckenam, Johannes Friemannan, Dorothea Langerao, Wilfried Rothap, Stephanie Stroblap, Martina Rudeliusaq, Konrad Friedrich Stockar, Wilko Weichertas, Claire Delbridgeas, Atsuko Kasajimaas, Peer-Hendrik Kuhnas, Julia Slotta-Huspeninaas, Gregor Weirichas, Peter Barthat, Eva Wardelmannat, Katja Evertau, Andreas Büttnerav, Johannes Manhartav, Stefan Nigburav, Iris Bittmannaw, Falko Fendax, Hans Bösmüllerax, Massimo Granaiax, Karin Klingelax, Verena Warmax, Konrad Steinestelay, Vincent Gottfried Umathumay, Andreas Rosenwaldaz, Florian Kurzaz, Niklas Vogtaz
a Center for Translational & Clinical Research (CTC-A), University Hospital RWTH Aachen, Aachen, Germany
b Institute for Medical Informatics, University Hospital RWTH Aachen, Aachen, Germany
c Institute of Neuropathology, University Hospital RWTH Aachen, Aachen, Germany
d Institute of Pathology, University Hospital RWTH Aachen, Aachen, Germany
e Anesthesiology and Intensive Care, University Hospital Augsburg, Augsburg, Germany
f Gastroenterology, University Hospital Augsburg, Augsburg, Germany
g General Pathology and Molecular Diagnostics, University Hospital Augsburg, Augsburg, Germany
h Hematology and Oncology, University Hospital Augsburg, Augsburg, Germany
i Laboratory Medicine and Microbiology, University Hospital Augsburg, Augsburg, Germany
j Radiology, University Hospital Augsburg, Augsburg, Germany
k Department of Neuropathology, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany
l German Center for Neurodegenerative Diseases (DZNE) Berlin, Berlin, Germany
m Cluster of Excellence, NeuroCure, Berlin, Germany
n Institute of Pathology, Charité – Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Berlin, Germany
o Department of Pathology, University Hospital Cologne, Cologne, Germany
p Institute of Pathology, University Hospital OWL of the Bielefeld University, Campus Lippe, Detmold, Germany
q Department of Pathology, Klinikum Dortmund, Dortmund, Germany
r Institute of Pathology, University Hospital Dresden, Dresden, Germany
s Gemeinschaftspraxis für Pathologie, Dresden, Germany
t Institute of Forensic Medicine, University Hospital Düsseldorf, Düsseldorf, Germany
u Institute of Pathology, University Hospital Düsseldorf, Düsseldorf, Germany
v Department of Nephropathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
w Institute of Neuropathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
x Institute of Pathology, University Hospital Erlangen-Nürnberg, Erlangen, Germany
y Institute of Pathology, University Hospital Essen, Essen, Germany
z Institute of Forensic Medicine, University Hospital Frankfurt, Frankfurt, Germany
aa Senckenberg Institute of Pathology, University Hospital Frankfurt, Frankfurt, Germany
ab Institut für Klinische Pathologie, Medical Center-University of Freiburg, Freiburg, Germany
ac Institute of Neuropathology, University Hospital Giessen and Marburg, Giessen, Germany
ad Institute of Pathology, University Hospital Giessen and Marburg, Giessen, Germany
ae Institute of Neuropathology, University Medical Center Göttingen, Göttingen, Germany
af Institute of Pathology, University Medical Center Göttingen, Göttingen, Germany
ag Institute of Pathology, University Hospital Halle (Saale), Halle (Saale), Germany
ah Institute of Legal Medicine, University Medical Center Hamburg- Eppendorf, Hamburg, Germany
ai Institute of Medical Microbiology, Virology, and Hygiene, University Medical Center Hamburg- Eppendorf, Hamburg, Germany
aj Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
ak Institute of Pathology, Hannover Medical School, Hannover, Germany
al Institute of Pathology, Heidelberg University Hospital, Heidelberg, Germany
am Department of Pathology, University Hospital Schleswig-Holstein, Kiel, Germany
an Department of Pathology, University Hospital Cologne, Lüdenscheid, Germany
ao Institute of Pathology, Klinikum Magdeburg, Magdeburg, Germany
ap Institute of Pathology, University Medical Center Mainz, Mainz, Germany
aq Institute of Pathology, Ludwig-Maximilians-Universität Munich, Munich, Germany
ar Department of Nephrology, TUM School of Medicine of Technical University of Munich, Munich, Germany
as Institute of Pathology, TUM School of Medicine of Technical University of Munich, Munich, Germany
at Gerhard Domagk Institute of Pathology, University Hospital Münster, Münster, Germany
au Institute of Pathology, University Hospital Regensburg, Regensburg, Germany
av Institute of Legal Medicine, Rostock University Medical Center, Rostock, Germany
aw Institute of Pathology, Agaplesion Diakonieklinikum Rotenburg, Rotenburg, Germany
ax Institute of Pathology and Neuropathology, University Hospital Tübingen, Tübingen, Germany
ay Department of Pathology, Bundeswehrkrankenhaus Ulm, Ulm, Germany
az Institute of Pathology, University of Würzburg, Würzburg, Germany