Abstract
Posttranslational modifications of antibody products affect their stability, charge distribution, and drug activity and are thus a critical quality attribute. The comprehensive mapping of antibody modifications and different charge isomers (CIs) is of utmost importance, but is challenging. We intended to quantitatively characterize the posttranslational modification status of CIs of antibody drugs and explore the impact of posttranslational modifications on charge heterogeneity. The CIs of antibodies were fractionated by strong cation exchange chromatography and verified by capillary isoelectric focusing-whole column imaging detection, followed by stepwise structural characterization at three levels. First, the differences between CIs were explored at the intact protein level using a top-down mass spectrometry approach; this showed differences in glycoforms and deamidation status. Second, at the peptide level, common modifications of oxidation, deamidation, and glycosylation were identified. Peptide mapping showed nonuniform deamidation and glycoform distribution among CIs. In total, 10 N-glycoforms were detected by peptide mapping. Finally, an in-depth analysis of glycan variants of CIs was performed through the detection of enriched glycopeptides. Qualitative and quantitative analyses demonstrated the dynamics of 24 N-glycoforms. The results revealed that sialic acid modification is a critical factor accounting for charge heterogeneity, which is otherwise missed in peptide mapping and intact molecular weight analyses. This study demonstrated the importance of the comprehensive analyses of antibody CIs and provides a reference method for the quality control of biopharmaceutical analysis.
Keywords: Antibody, Charge isomers, Mass spectrometry, Posttranslational modification, Glycopeptide
Graphical abstract
Highlights
-
•
A stepwise investigation on the diversity and dynamics of modifications of antibody charge isomers (CIs) was performed.
-
•
The results from SCX-HPLC and cIEF-WCID supported each other.
-
•
In-depth analysis on glycan variants of CIs was achieved by analyzing the enriched glycopeptides.
-
•
Sialic acid and deamidation modifications were critical factors for the charge heterogeneity.
1. Introduction
Charge heterogeneity of monoclonal antibodies (mAbs) is formed by enzymatic and nonenzymatic processes during the manufacture of biopharmaceuticals. The development and manufacture of therapeutic mAbs is a highly regulated process, and the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use proposed a level of heterogeneity allowed for biopharmaceutical products. Charge heterogeneity has an important influence on the stability of mAbs and their biological functions and has become a critical quality attribute [1] that must be tested in the processes of stability research, release, and approval [[2], [3], [4], [5]]. Cleavage of C-terminal lysine results in the loss of one or two positively charged lysine residues, leading to the formation of basic variants [6,7]. Deamidation of N- and O-linked glycans, glycation, and the presence of negatively charged sialic acid lead to an increase in negative charges and the appearance of acidic variants [[8], [9], [10]].
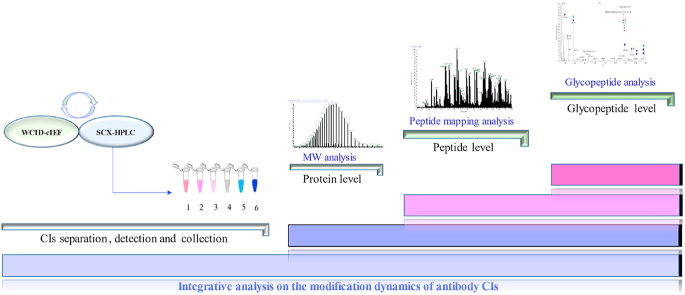
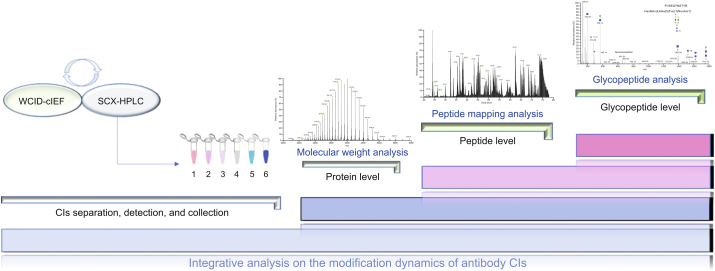
There are many methods to detect charge isomers (CIs), including polyacrylamide gel electrophoresis [11]. However, its low resolution and complicated operation results in fewer applications. Ion exchange chromatography uses either salt elution or pH gradient elution mode [[12], [13], [14], [15]], with the subsequent collection of the flow-through for the mass spectrometry (MS) analysis of protein modifications [16]. The online liquid chromatography (LC)-MS method was developed using an organic salt buffer that can be tolerated by MS to perform CI separation [17], but the quality of the mass spectrum and universality of the methods should be considered, and the detailed interpretation of site-specific modifications is difficult. This challenge also exists for the approach of coupling capillary electrophoresis with MS [18,19]. In this study, we aimed to characterize the global modification status of antibodies and reveal the dynamics of modifications along with different CIs of antibodies. The experimental design is shown in Fig. 1. The method for detecting and separating the CIs of the IgG1 antibody was established by strong cation exchange chromatography (SCX-HPLC) and capillary isoelectric focusing-whole column imaging detection (cIEF-WCID), and the former was followed by fractionation to collect CIs. Highly accurate and high-resolution MS detection was used to analyze the differences in the molecular weight of each CI at the intact protein level. We further investigated the effects of various modifications of CIs at the peptide level. Finally, the effects of glycans on CIs were explored by the hydrophilic interaction liquid chromatography (HILIC) enrichment of glycopeptides and LC-MS/MS analysis.
Fig. 1.
Experimental design for the in-depth structural analysis of charge isomers (CIs) of IgG antibody. SCX-HPLC: strong cation exchange chromatography; cIEF-WCID: capillary isoelectric focusing-whole column imaging detection.
2. Materials and methods
2.1. Materials
The IgG1 antibody used in this study was provided by the China National Institute of Metrology (Beijing, China), abbreviated as C_mAb, and its C-terminal lysine was knocked out at the gene level. RM8671 was purchased from National Institute of Standards and Technology (NIST) (Gaithersburg, MD, USA), named as NIST_mAb, formic acid (FA) was purchased from Fluka (Seelze, Germany), trifluoroacetic acid (TFA) was from ACROS (Walsham, MA, USA), and trypsin from Promega (Madison, WI, USA). Ampholyte (HR, AESlyte 3–10), SH AESlyte 2.5–5, isoelectric point standard 7.03, and isoelectric point standard 9.33 were purchased from CEInfinite (Ontario, Canada). Chemicals, including NaOH, H3PO4, NaH2PO4, Na2HPO4, and NaCl, were purchased from the Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Methyl cellulose (MC), NH4HCO3, tris (2-carboxyethyl) phosphine (TCEP), acetonitrile (ACN), PNGase F and iodoacetamide were purchased from Sigma (St. Louis, MO, USA).
Instruments used included L-3120 HPLC (Beijing, China), CEInfinite C01 WCID (Ontario, Canada), Thermo Fisher Exactive Plus EMR and Thermo Fisher Q-Exactive™ Plus (Walsham, MA, USA).
Columns used included size-exclusion chromatography column (SEC) (4.6 mm × 300 mm, 3 μm) and SCX (4.6 mm × 250 mm, 5 μm) purchased from Sepax (Newark, DE, USA). A pre-column C18 (150 μm × 2 cm, 3 μm) (Beijing, China) and C18 capillary column (150 μm × 15 cm, 1.9 μm) (Beijing, China) were also used.
2.2. Methods
2.2.1. Separation of CIs
cIEF-WCID: The samples were first diluted to 10 mg/mL with deionized water. Then, 5 μL of samples were mixed with 1 μL of HR AESlyte 3–10, 3 μL of SH AESlyte 2.5–5, 40 μL of 0.5% MC, 0.5 μL of isoelectric point standard (7.03), and 0.5 μL of isoelectric point standard (9.33), then were centrifuged at 10,000 g for 2 min. For each analysis, 5 μL of the mixed sample was injected. The instrument parameters used were as follows: column temperature, 25 °C; cathode electrode solution, 0.1 mol/L NaOH; anode electrode solution, 0.08 mol/L H3PO4; and detection wavelength, 280 nm. The focusing procedure was 1000 V for 1 min, 2000 V for 1 min, and 3000 V for 4 min. The current was below 15 μA.
For SCX-HPLC, the sample was diluted to 1 mg/mL with deionized water, and 10 μg was injected for each analysis. Then 10 mmol/L of phosphate (pH 6.0; buffer A) and 0.5 mol/L of NaCl in buffer A (pH 6.0; buffer B) at a flow rate of 0.6 mL/min were used for the separation with the gradient: 0–5.0 min, 0% B; 5.0–43.0 min, 0–25% B; 43.0–43.5 min, 25%–90% B; 43.5–48.5 min, 90% B; and 48.5–55 min, 90%–0% B.
2.2.2. Molecular weight detection
Proteins were separated isocratically on the SEC column (20% ACN and 0.1% FA in 79.9% H2O). The detection wavelength was 280 nm, and the flow rate was 0.2 mL/min. The mass spectrometer was operated in positive ion mode, with an acquisition range from m/z 1000–6000 and maximum IT of 200 ms. The molecular weight calculation was performed with Protein Deconvolution 4.0 with 99% confidence. The mass tolerance was set at 20 ppm, and the relative abundance threshold ranged from 0% to 1%.
2.2.3. Peptide mapping analysis
First, 5 μg of each CI fraction was diluted with 50 mM NH4HCO3 to a total volume of 50 μL. Then, TCEP and CAA were added to 10 and 20 mM final concentrations, and the sample was incubated at 25 °C for 0.5 h. Trypsin was added at a ratio of enzyme to protein 1:30 (μg:μg), and digestion was maintained for 16 h at 37 °C. The resultant peptides were desalted, and 500 ng was injected for nanoLC-MS/MS analysis [20]. The mobile phase consisted of 0.1% FA in ACN (A) and 0.1% FA in 80% ACN (B) at a flow rate of 600 nL/min. The gradient was 0–14 min, 8%–13% B; 14–51 min, 13%–25% B; 51–68 min, 25%–38% B; 68–69 min, 38%–95% B; and 69–75 min, 95% B.
MS parameters were as follows: for full MS, resolution 70000, AGC target value 3e6, maximum injection time 20 ms, and scan range 300–2000 m/z; for MS2, resolution 17500, AGC target value 1e6, maximum injection time 100 ms, loop count 5, isolation window 1.6 Da, scan range 200–2000 m/z, step NCE 20 and 30, and dynamic exclusion 18 ms.
The mAb sequence matching was run through pFind with the following parameters: fixed modification of carbamidomethylation (C); variable modifications: deamidation (N/Q) and oxidation (M); precursor tolerance 20 ppm, fragment tolerance 20 ppm, peptide mass range 100–10000 Da, and peptide length 3–100 amino acids.
MaxQuant search of the mAb sequence was based on the following parameters: fixed modification of carbamidomethylation (C); variable modification: deamidation (N/Q), oxidation (M), G0F, and G1F; min peptide length three amino acids; and match-between-run on.
For the Byonic search, the data were searched against the mAb sequence and 52 common N-glycan databases. The fixed modification was carbamidomethylation (C). The variable modifications were deamidation (N/Q) and oxidation (M).
2.2.4. Glycopeptide enrichment and detection
First, 2 mg of HILIC medium was activated in 200 μL of 0.1% TFA for 15 min at 25 °C, and the upper layer solution was discarded after centrifugation. Then, three replicate washings of the HILIC media were performed with 200 μL of 0.2% TFA in 80% ACN solution for 15 min. The digested 20 μg peptides were dissolved in 0.2% TFA in 80% ACN solution, mixed with HILIC media, and incubated for 2 h. The mixtures were then added to a tip filled with a C8 membrane at the end and washed twice with 80 μL of 0.2% TFA in 80% ACN solution (non-glycopeptide). Then, 80 μL of 0.1% TFA was added to elute the glycopeptide twice. The samples were then dried using a SpeedVac at 45 °C and analyzed by nanoLC-MS/MS. The MS parameters of N-glycopeptide were the same as those of the peptide mapping parameters.
2.2.5. Statistical analysis
Peptide mapping analysis: We calculated the fraction of total (FOT) for each peptide to normalize the intensity data. The detailed workflow is shown in Fig. S1.
N-Glycoform analysis: Data calculation and graphics processing were performed using R v.3.2.1. Bubble plots were generated using the ggplot2 package based on the PSMs of N-glycoforms.
3. Results and discussion
3.1. SCX-HPLC and cIEF-WCID separation
Two antibodies were used in the study. One was procured from the National Institute of Metrology, China (NIMC), named C_mAb. The other was purchased from the NIST (RM 8671), named NIST_mAb, which was used as a reference for necessary comparisons. The theoretical pI was 8.2 for C_mAb and 8.41 for NIST_mAb (by Compute pI/Mw tool; http://ca.expasy.org/tools/pi_tool.html). We used cIEF-WCID to measure the actual isoelectric point first and verified the method with NIST_mAb. This method was easy to achieve with a high-resolution view of CI distribution with only a small sample quantity. We performed SCX-HPLC using a phosphate buffer system [9] and verified this method with NIST_mAb. The gradient was optimized so that the CI peaks were separated with better resolution and facilitated the collection of the CIs.
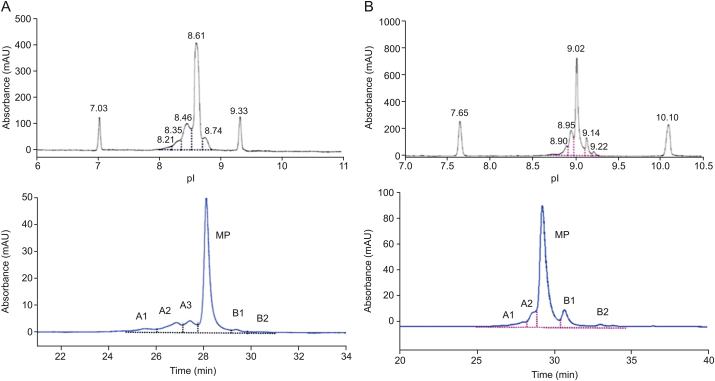
Figs. 2A and B show that the two methods generated similar profiles. For C_mAb, the SCX-HPLC acid peak proportion was 23.9%, the main peak was 71.9%, and the basic peak was 4.1%. The cIEF-WCID profile showed an acid peak of 31.08%, a main peak isoelectric point at 8.61 with a proportion of 61.43%, and a basic peak content of 7.48%. The results of the two methods were mutually verified, further illustrating their accuracy and feasibility. Both methods showed that C_mAb had a lower basic peak content, indicating that the C-terminal lysine knockout had a significant influence on the basic peak. For NIST_mAb, both results from cIEF-WCID and SCX-HPLC had two basic peaks, corresponding to 1 or 2 C- terminal lysines. This result showed that the C-terminal lysine had a great influence on the distribution of antibody CIs.
Fig. 2.
(A) Profiles from cIEF-WCID (upper panel) and SCX-HPLC (lower panel) separation of C_mAb; pI 7.03 and pI 9.33 denoted the pI markers. (B) Profiles from cIEF-WCID (upper panel) and SCX-HPLC (lower panel) separation of NIST_mAb; pI 7.65 and pI 10.10 denoted the pI markers. A1: acid peak1; A2: acid peak2; A3: acid peak3; MP: main peak; B1: basic peak1; B2: basic peak2.
3.2. SCX-HPLC separation of CI fractions
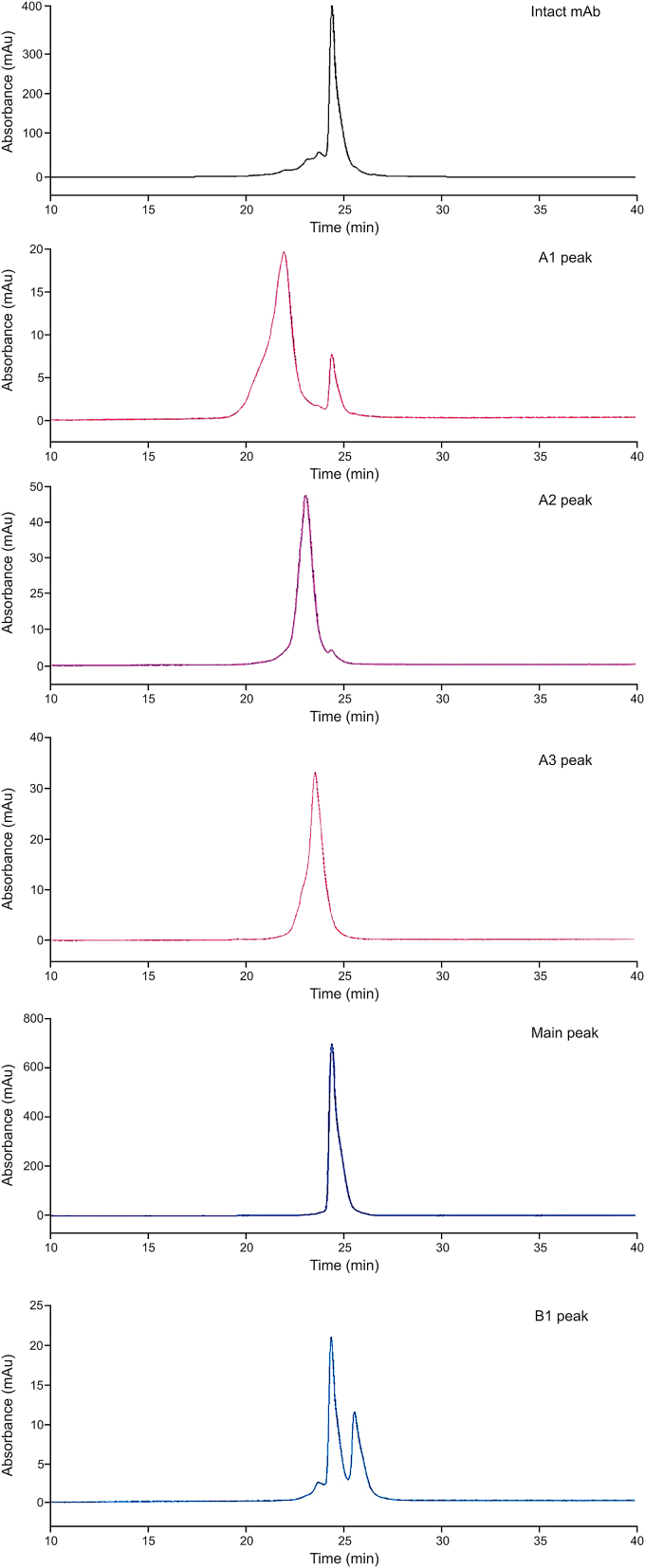
The collected CI fractions were reanalyzed by SCX-HPLC to evaluate their purity (Fig. 3). We compared the CIs with the intact C_mAb at the same elution time. The purities of the A1, A2, A3, and MP fractions were 96%, 98%, 99%, and 99%, respectively. This antibody had only a 4.1% basic peak, which was mainly due to the absence of lysine at the C-terminus. The purity of B1 was 49% (contained part of the composition of the main peaks). Although B2 could not be detected by UV, we still collected this and conducted MS analysis.
Fig. 3.
SCX-HPLC profiles of CI fractions. The purity of A1, A2, A3, MP, and B1 was 96%, 98%, 99%, 99%, and 49%, respectively.
3.3. Molecular weight detection
The reSpect™ algorithm was employed to resolve the spectra [21,22], which calculates the average mass using the form of the peak interval (used to represent the mass). In the current study, we optimized our data analysis strategy to confidently identify either high- or low-abundance species using a mass tolerance of 20 ppm. The molecular weight distributions are shown in Fig. 4, and the N-glycoform abundance (common component) is shown in Fig. S2.
Fig. 4.
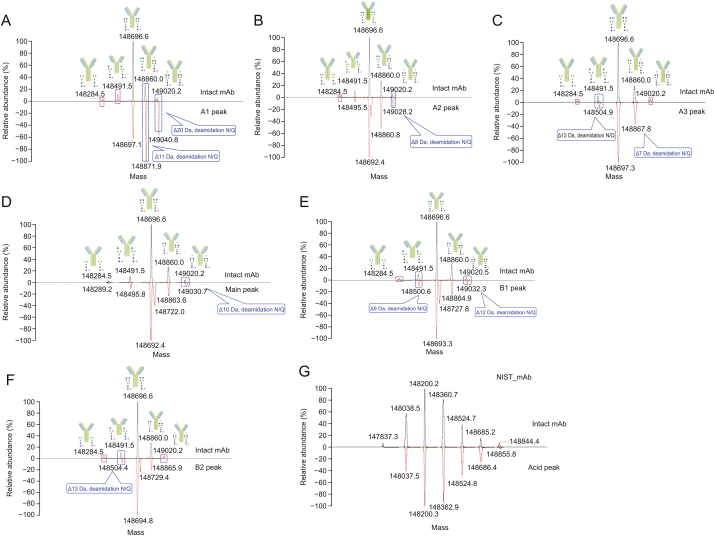
Molecular weight distribution analysis of CIs. Pink solid frames indicated the components that were not present in the CIs. Blue dotted boxes indicated a deamidation modification on the CIs. A1: acid peak1; A2: acid peak2; A3: acid peak3; MP: main peak; B1: basic peak1; B2: basic peak2. (A) A1 vs. intact C_mAb; (B) A2 vs. intact C_mAb; (C) A3 vs. intact C_mAb; (D) MP vs. intact C_mAb; (E) B1 vs. intact C_mAb; (F) B2 vs. intact C_mAb; (G) reference: acid peak of NIST_mAb vs. intact NIST_mAb.
The glycoform distribution of acid peak A1 differed significantly from that of the intact antibody (Figs. 4A and S2). The differences in the molecular weight were presumed to be a consequence of deamidation [23], and this was later confirmed in the peptide mapping analysis. The A2 glycoform distribution was also inconsistent with the intact antibody distribution (Fig. 4B). The A2 peak lacked the G0F/G0F-2HexNAc glycoform compared to the intact antibody; however, the difference was relatively small, and the deamidation modification also existed. For the A3 glycoform distribution, the G0F/G0F-2HexNAc and G1F/G1F glycoforms were absent (Fig. 4C). The MP fraction had glycoform distributions almost identical to that of the intact antibody (Fig. 4D). A small peak appeared in the deconvolution plot of MP, which was presumed to be oxidation modification [24] and confirmed in the following peptide mapping analysis. The distributions of the main peaks of A2, B1, and B2 were the same as those in the above results. Fig. 4E shows the basic B1 peak. Although it contained a component that was 51% of the main peak, the glycoform still showed difference from the intact antibody, and deamidation also occurred. The basic peak B2 was significantly different from that of the intact antibody with respect to the glycoform distribution, and the G0F/G0F-2HexNAc and G1F/G1F glycoforms were missing (Fig. 4F). We also presented the results of the acid peak of NIST_mAb (Fig. 4G), and these verified that glycoforms and modifications affected the molecular weight distribution. We concluded from molecular weight analyses that glycoforms might affect the distribution of CIs [25,26] as well as deamidation. Usually, deamidation occurs in the variable domains, especially in the exposed and flexible complementarity-determining regions as well as in the constant domains [27,28]. Detailed information on the deconvoluted molecular weight can be found in Tables S1–7.
3.4. Peptide mapping analysis
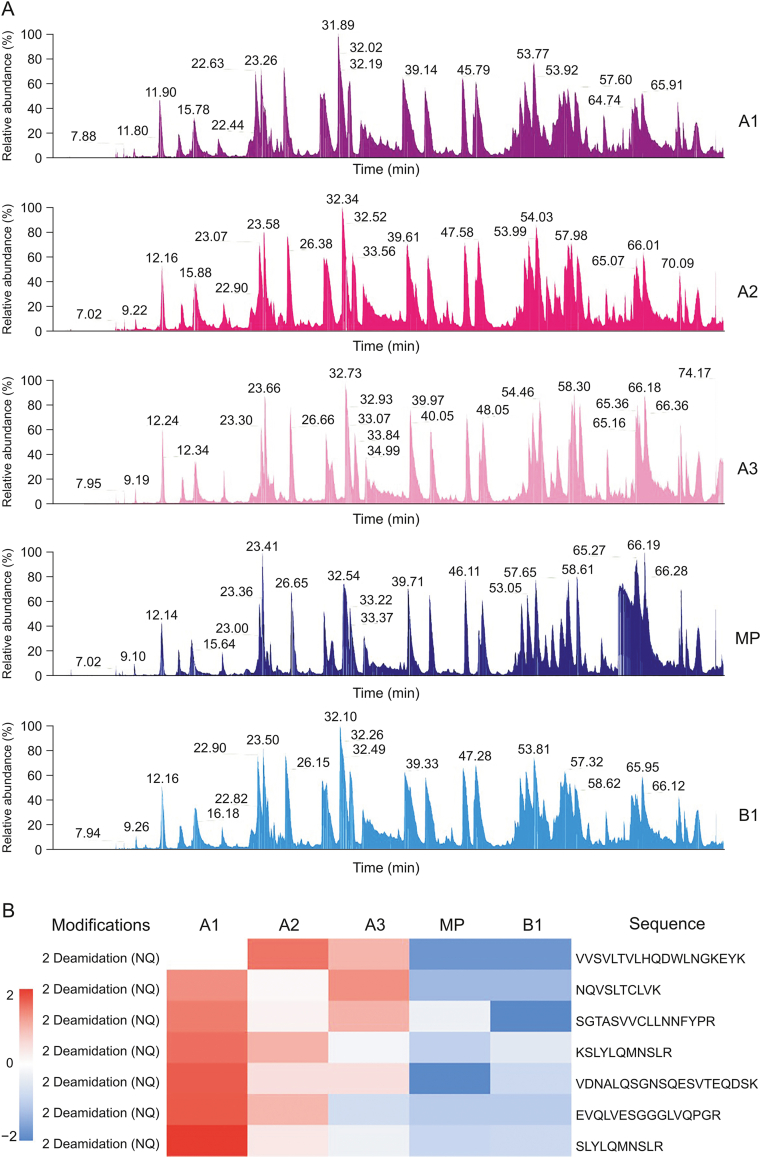
The total ion chromatogram (TIC) of peptide mapping is shown in Fig. 5A. The software pFind with the “Open Search” function [29,30] was first used to search the raw data to find peptide modifications. Next, the raw data were retrieved using MaxQuant software, and the “match-between-runs” function was used to extract the intensity of each identification [31]. The heatmap of peptide mapping results showed that the acid peak had more deamidation (N/Q) (Fig. 5B). Along with the weak acidic peaks, the intensity of the deamidation decreased. Other modifications are shown in Fig. S3. This result suggests that deamidation is an important factor that affects CIs and confirms the deamidation in the intact molecular weight analysis of CIs [32].
Fig. 5.
LC-MS/MS analysis of CIs. (A) TIC of peptide mapping of each CI and (B) heatmap view of the 2 deamidation (NQ) modification from peptide mapping. A1: acid peak1; A2: acid peak2; A3: acid peak3; MP: main peak; B1: basic peak1.
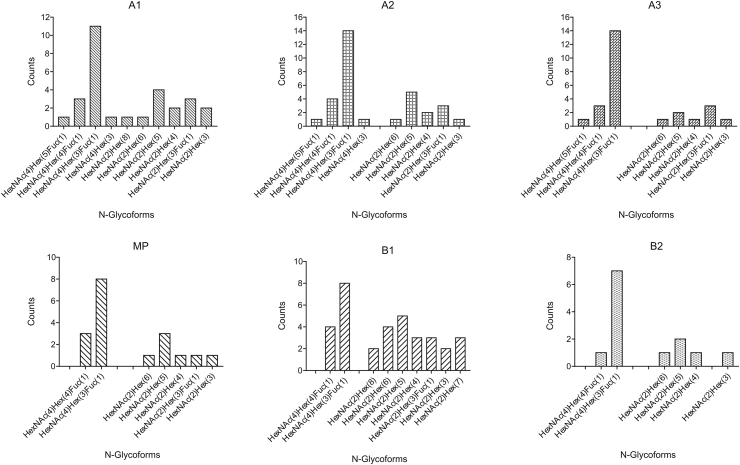
Next, we explored glycopeptide identifications in the peptide mapping analysis. Byonic software is a recommended tool for intact glycopeptide identification [33,34]. Both the spectrum counting method [35] (Fig. 6) and extraction peak area method (Fig. S4) were used for glycopeptide quantitation. Both methods indicated that G0F and G1F were the major glycoforms of the antibody, and all the 10 types of high mannose components not present in molecular weight analysis were detected in this analysis, further suggesting that glycoforms (non-major glycoforms) might affect the distribution of CIs.
Fig. 6.
Spectrum counts of N-Glycoforms for each CI.
3.5. Identification of enriched glycopeptides
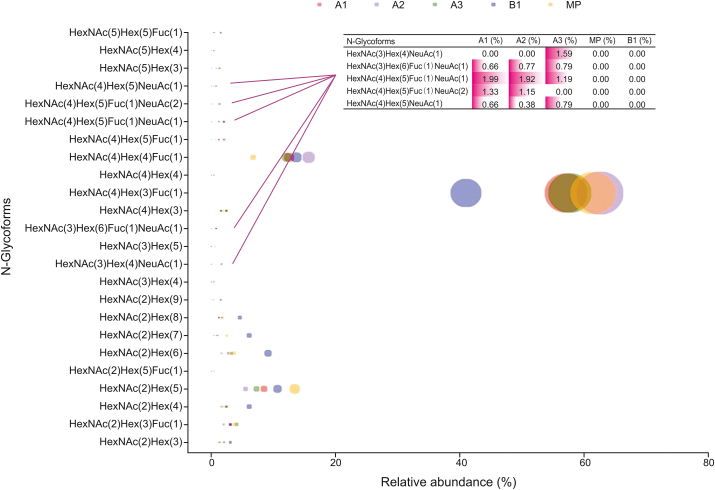
An important feature of therapeutic proteins is glycosylation modification as it is an important factor in protein activity and stability [36,37]. The above results indicated that N-glycoform variations might affect the distribution of CIs. To more accurately explain the effect of glycosylation on charge heterogeneity, it is necessary to carry out further exploration of the N-glycoform. Glycopeptides in each CI digest were enriched with HILIC media [36] and analyzed by nanoLC-MS/MS (Fig. S5). The mass spectrum signals for glycopeptides were significantly improved after HILIC enrichment. Spectra for one of the sialic acid-containing glycopeptides after HILIC enrichment are shown in Fig. S6. The results of the N-glycoforms and sialic acid content table are shown in a bubble plot (Fig. 7). The main N-glycoforms in each of the CIs were G0F and G1F. In total, 24 N-glycoforms were detected, which was higher than the number detected in the peptide mapping analysis. A fraction of sialic acid-containing glycans was detected in the A1, A2, and A3 CIs with a decreasing tendency, while none was detected in the B1 and MP fractions. This result suggested that although sialic acid-containing glycans were not the dominant N-glycoforms, they were still an important factor affecting the distribution of CIs.
Fig. 7.
Bubble plot of N-glycoform content after HILIC enrichment. The size of the bubble indicates the relative glycan content. Different colors represent different CIs.
4. Conclusion
In this study, we adopted two different chromatographic methods, SCX-HPLC and cIEF-WCID, for separation of the IgG antibody CIs in parallel. The two methods supported each other to provide a more complete view of the CIs. Based on proteomics technology, the effects of modifications on CIs were comprehensively characterized. Intact molecular weight distribution, peptide mapping, and in-depth glycopeptide analyses were performed. The results showed that at the protein level, the differences in N-glycoforms and abundances were the most important factors, followed by deamidation (N/Q) modification. At the peptide level, deamidation (N/Q) modification was more prominent. Upon further HILIC enrichment of glycopeptides, more N-glycoforms were detected, in which the sialylation modification appeared only in the acidic CIs. These results suggested that the sialic acid and deamidation modifications were the most critical factors affecting the charge heterogeneity. Therefore, we provide a stepwise and in-depth approach to study the influence of CIs on protein biopharmaceuticals.
CRediT author statement
Xinling Cui: Methodology, Validation, Formal analysis, Investigation, Writing - Original draft preparation, Writing - Reviewing and Editing; Wei Mi: Formal analysis, Investigation, Writing - Original draft preparation; Zhishang Hu, Xiaoyu Li, Bo Meng, and Xinyuan Zhao: Software, Formal analysis; Xiaohong Qian, Tao Zhu, and Wantao Ying: Conceptualization, Reviewing, Editing, Supervision.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We are grateful for the financial support from the National Key Program for Basic Research of China (Grant Nos.: 2018YFC0910302 and 2017YFF0205400), the National Natural Science Foundation of China (Grant No.: 81530021), and Innovation Foundation of Medicine (Grant Nos.: BWS14J052 and 16CXZ027).
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.11.006.
Contributor Information
Tao Zhu, Email: tao.zhu@cansinotech.com.
Wantao Ying, Email: yingwantao@ncpsb.org.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Torkashvand F., Vaziri B. Main quality attributes of monoclonal antibodies and effect of cell culture components. Iran. Biomed. J. 2017;21:131–141. doi: 10.18869/acadpub.ibj.21.3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reusch D., Haberger M., Maier B., et al. Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles--part 1: separation-based methods. mAbs. 2015;7:167–179. doi: 10.4161/19420862.2014.986000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parr M.K., Montacir O., Montacir H. Physicochemical characterization of biopharmaceuticals. J. Pharmaceut. Biomed. Anal. 2016;130:366–389. doi: 10.1016/j.jpba.2016.05.028. [DOI] [PubMed] [Google Scholar]
- 4.Leblanc Y., Ramon C., Bihoreau N., et al. Charge variants characterization of a monoclonal antibody by ion exchange chromatography coupled on-line to native mass spectrometry: case study after a long-term storage at +5 ℃. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1048:130–139. doi: 10.1016/j.jchromb.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz S.A., Engen J.R., Mazzeo J.R., et al. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 2012;11:527–540. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris R.J., Kabakoff B., Macchi F.D., et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J. Chromatogr. B Biomed. Sci. Appl. 2001;752:233–245. doi: 10.1016/s0378-4347(00)00548-x. [DOI] [PubMed] [Google Scholar]
- 7.Khawli L.A., Goswami S., Hutchinson R., et al. Charge variants in IgG1: isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs. 2010;2:613–624. doi: 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Gaza-Bulseco G., Faldu D., et al. Heterogeneity of monoclonal antibodies. J. Pharm. Sci. 2008;97:2426–2447. doi: 10.1002/jps.21180. [DOI] [PubMed] [Google Scholar]
- 9.Du Y., Walsh A., Ehrick R., et al. Chromatographic analysis of the acidic and basic species of recombinant monoclonal antibodies. mAbs. 2012;4:578–585. doi: 10.4161/mabs.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekete S., Beck A., Guillarme D. Characterization of cation exchanger stationary phases applied for the separations of therapeutic monoclonal antibodies. J. Pharmaceut. Biomed. Anal. 2015;111:169–176. doi: 10.1016/j.jpba.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 11.Takeo K., Tanaka T., Nakamura K., et al. Studies on the heterogeneity of anti-hapten antibodies by means of two-dimensional affinity electrophoresis. Electrophoresis. 1989;10:818–824. doi: 10.1002/elps.1150101204. [DOI] [PubMed] [Google Scholar]
- 12.Fekete S., Beck A., Veuthey J.L., et al. Ion-exchange chromatography for the characterization of biopharmaceuticals. J. Pharmaceut. Biomed. Anal. 2015;113:43–55. doi: 10.1016/j.jpba.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 13.Lingg N., Berndtsson M., Hintersteiner B., et al. Highly linear pH gradients for analyzing monoclonal antibody charge heterogeneity in the alkaline range: validation of the method parameters. J. Chromatogr., A. 2014;1373:124–130. doi: 10.1016/j.chroma.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Fekete S., Beck A., Fekete J., et al. Method development for the separation of monoclonal antibody charge variants in cation exchange chromatography, Part I: salt gradient approach. J. Pharmaceut. Biomed. Anal. 2015;102:33–44. doi: 10.1016/j.jpba.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Fekete S., Beck A., Fekete J., et al. Method development for the separation of monoclonal antibody charge variants in cation exchange chromatography, Part II: pH gradient approach. J. Pharmaceut. Biomed. Anal. 2015;102:282–289. doi: 10.1016/j.jpba.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Füssl F., Cook K., Scheffler K., et al. Charge variant analysis of monoclonal antibodies using direct coupled pH gradient cation exchange chromatography to high-resolution native mass spectrometry. Anal. Chem. 2018;90:4669–4676. doi: 10.1021/acs.analchem.7b05241. [DOI] [PubMed] [Google Scholar]
- 17.Füssl F., Trappe A., Cook K., et al. Comprehensive characterisation of the heterogeneity of adalimumab via charge variant analysis hyphenated on-line to native high resolution Orbitrap mass spectrometry. mAbs. 2019;11:116–128. doi: 10.1080/19420862.2018.1531664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyon A., Excoffier M., Janin-Bussat M.C., et al. Determination of isoelectric points and relative charge variants of 23 therapeutic monoclonal antibodies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1065–1066:119–128. doi: 10.1016/j.jchromb.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 19.Hirayama A., Abe H., Yamaguchi N., et al. Development of a sheathless CE-ESI-MS interface. Electrophoresis. 2018;39:1382–1389. doi: 10.1002/elps.201800017. [DOI] [PubMed] [Google Scholar]
- 20.Kulak N.A., Pichler G., Paron I., et al. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 21.Makarov A., Denisov E. Dynamics of ions of intact proteins in the Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 2009;20:1486–1495. doi: 10.1016/j.jasms.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Senko M.W., Beu S.C., McLaffertycor F.W. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 23.Chelius D., Rehder D.S., Bondarenko P.V. Identification and characterization of deamidation sites in the conserved regions of human immunoglobulin gamma antibodies. Anal. Chem. 2005;77:6004–6011. doi: 10.1021/ac050672d. [DOI] [PubMed] [Google Scholar]
- 24.Sokolowska I., Mo J., Dong J., et al. Subunit mass analysis for monitoring antibody oxidation. mAbs. 2017;9:498–505. doi: 10.1080/19420862.2017.1279773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jefferis R. Glyco-engineering of human IgG-Fc to modulate biologic activities. Curr. Pharmaceut. Biotechnol. 2016;17:1333–1347. doi: 10.2174/1389201017666161029225929. [DOI] [PubMed] [Google Scholar]
- 26.Gavrilov Y., Shental-Bechor D., Greenblatt H.M., et al. Glycosylation may reduce protein thermodynamic stability by inducing a conformational distortion. J. Phys. Chem. Lett. 2015;6:3572–3577. doi: 10.1021/acs.jpclett.5b01588. [DOI] [PubMed] [Google Scholar]
- 27.Lyubarskaya Y., Houde D., Woodard J., et al. Analysis of recombinant monoclonal antibody isoforms by electrospray ionization mass spectrometry as a strategy for streamlining characterization of recombinant monoclonal antibody charge heterogeneity. Anal. Biochem. 2006;348:24–39. doi: 10.1016/j.ab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Vlasak J., Bussat M.C., Wang S., et al. Identification and characterization of asparagine deamidation in the light chain CDR1 of a humanized IgG1 antibody. Anal. Biochem. 2009;392:145–154. doi: 10.1016/j.ab.2009.05.043. [DOI] [PubMed] [Google Scholar]
- 29.Chi H., Liu C., Yang H., et al. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018;36:1059–1061. doi: 10.1038/nbt.4236. [DOI] [PubMed] [Google Scholar]
- 30.Chi H., He K., Yang B., et al. pFind-Alioth, A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J. Proteomics. 2015;125:89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 32.Timm V., Gruber P., Wasiliu M., et al. Identification and characterization of oxidation and deamidation sites in monoclonal rat/mouse hybrid antibodies. J. Chromatog.r B Analyt. Technol. Biomed. Life Sci. 2010;878:777–784. doi: 10.1016/j.jchromb.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 33.Lee L.Y., Moh E.S., Parker B.L., et al. Toward automated N-glycopeptide identification in glycoproteomics. J. Proteome Res. 2016;15:3904–3915. doi: 10.1021/acs.jproteome.6b00438. [DOI] [PubMed] [Google Scholar]
- 34.Bern M., Kil Y.J., Becker C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics. 2012;40 doi: 10.1002/0471250953.bi1320s40. 13.20.1–13.20.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley N.M., Hebert A.S., Westphall M.S., et al. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 2019;10:1311. doi: 10.1038/s41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen P.H., Mysling S., Højrup P., et al. Glycopeptide enrichment for MALDI-TOF mass spectrometry analysis by hydrophilic interaction liquid chromatography solid phase extraction (HILIC SPE) Methods Mol. Biol. 2013;951:131–144. doi: 10.1007/978-1-62703-146-2_10. [DOI] [PubMed] [Google Scholar]
- 37.Dong Q., Yan X., Liang Y., et al. In-depth characterization and spectral library building of glycopeptides in the tryptic digest of a monoclonal antibody using 1D and 2D LC-MS/MS. J. Proteome Res. 2016;15:1472–1486. doi: 10.1021/acs.jproteome.5b01046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.