Summary
Background
Evolutionary pressure has led to the emergence of SARS-CoV-2 variants, with the most recent Omicron variant containing an unparalleled 30 mutations in the spike protein. Many of these mutations are expected to increase immune evasion, thus making breakthrough cases and re-infection more common.
Methods
From June 2020 to December 2021 serial blood samples (initial post recovery, 6 months, 12 months) were collected from a COVID-19 convalescent cohort in Boston, MA. Plasma was isolated for use in Mesoscale Discovery based antibody binding assays. Unvaccinated donors or those vaccinated prior to the primary blood draw were excluded from this analysis, as were those who did not have at least two blood draws. Wilcoxon signed rank tests were used to compare pre- and post-vaccination titers and antibody response against different variants, while McNemar tests were used to compare the proportions of achieving ≥ 4 fold increases against different variants.
Findings
Forty-eight COVID convalescent donors with post-infection vaccination (hybrid immunity) were studied to evaluate the levels of cross-reactive antibodies pre- and post- vaccination against various SARS-CoV-2 Spike and RBD proteins. Vaccination with BNT162b2, mRNA-1273 or Ad26.COV2.S led to a 6.3 to 7.8 fold increase in anti-Spike antibody titers and a 7·0 to 7·4 fold increase in anti-WT, Alpha and Delta RBD antibody. However, a lower response was observed for Beta and Omicron RBDs with only 7/48 (15%) and 15/48 (31%) donors having a ≥4 fold increase in post-vaccination titers against Beta and Omicron RBDs. Structural analysis of the Beta and Omicron RBDs reveal a shared immune escape strategy involving residues K417-E484-N501 that is exploited by these variants of concern.
Interpretation
Through mutations of the K417-E484-N501 triad, SARS-CoV-2 has evolved to evade neutralization by the class I/II anti-RBD antibody fraction of hybrid immunity plasma as the polyclonal antibody response post-vaccination shows limitations in the ability to solve the structural requirements to bind the mutant RBDs.
Funding
Massachusetts Consortium on Pathogen Readiness (280870.5116709.0016) and the National Institute of Allergy and Infectious Diseases (1R01AI161152-01A1).
Keywords: SARS-CoV-2, COVID-19, Hybrid immunity, Omicron variant, Vaccine induced antibody titers, Neutralization escape mutations
Research in context.
Evidence before this study
As SARS-CoV-2 continues to infect people around the globe, naturally occurring mutations allow for viral evolution and the generation of numerous variants, some of which remain in circulation today. While vaccination is a critical step to controlling viral spread and limiting the severity of infection, current vaccines rely on the ancestral SARS-CoV-2 Wuhan strain. The emergence of the Omicron variant in November 2021 exposed the risk associated with using ancestral strains, as the unprecedented number of spike mutations lead to concerns regarding the efficacy of vaccines as well as other therapeutic measures. As concerns began to mount, it was important to determine if vaccination led to a protective immune response against circulating variants of concern. We searched PubMed for articles published up to December 15, 2021 using the search terms ("SARS-CoV- 2"[All Fields] AND "vaccine"[All Fields] AND "plasma"[All Fields] AND "Omicron"[All Fields]). Our search yielded two preprint studies of vaccine effectiveness against Omicron, one from South Africa that assessed the neutralization potential of plasma from BNT162b2 vaccinated individuals to neutralize Omicron virus and a second from the United States that compared neutralization potency of plasma from individuals vaccinated with either mRNA or Ad26.COV2.S. The study from South Africa first demonstrated that Omicron continues to infect cells via the ACE2 receptor and then showed that while neutralization was higher in donors with hybrid immunity, all vaccinated groups tested exhibited a 22-fold Omicron escape versus the D614G variant. The United States study similarly demonstrated a 30-60-fold loss of neutralization in a pseudovirus based assay when compared to WT Wuhan virus in vaccinated donors, however in donors who received an additional mRNA booster dose or were vaccinated post infection, a 38- and 154-fold increase in neutralization was observed. Due to the emergent nature of the Omicron strain, studies assessing plasma neutralization of this variant were scarce at the time this study was initiated.
Added value of this study
While this is not the first study to analyze plasma antibody titers in vaccinated individuals to Omicron and other variant spike proteins, to the best of our knowledge this is the first study that performs a structural analysis regarding the convergent immune evading mutations that have been discovered. By analyzing 21 previously reported anti-SARS-CoV-2 spike monoclonal antibodies, we are able to discern structural interactions of class I and II RBD antibodies with residues K417-E484-N501 on the RBD, and further identify how mutations at these residues can effectively abrogate monoclonal antibody binding and neutralization. Further applying this logic to polyclonal antibody sera, we provide a mechanistic understanding as to why infection followed by vaccination with ancestral spike strain induced a lower binding ability to Beta and Omicron RBDs than WT, Alpha, and Delta RBDs.
Implications of all evidence available
All donors tested in this study mounted an immune response to the variant Spike proteins following infection and vaccination, however, the increased antibody titers were not persistent, providing evidence that booster doses of vaccines are required to maintain immunity. Structural analysis of previously reported antibodies allows for a mechanistic view of SARS-CoV-2 immune evasion of the most potent class I and II antibodies through RBD mutations in the K417-E484-N501 triad and provides a rational for optimizing future vaccines to improve breadth of neutralization and epitope diversity.
Alt-text: Unlabelled box
Introduction
Coronaviruses contain the largest known viral RNA genome and replication is mediated by a viral RNA polymerase and other replicative enzymes in ORF1b. Though coronaviruses contain a unique proofreading enzyme, the inherently low fidelity of the RNA polymerase allows for mutations to arise naturally during replication.1 While many of these mutations decrease viral fitness and lead to extinction, on rare occasion these mutations can provide an evolutionary advantage, such as allowing for cross-species transmission, increased infectivity, or improved receptor binding.2, 3, 4, 5, 6 When combined with directed evolution via selective pressure from natural immunity or therapeutic intervention, this pathway can also lead to mutations that provide a means for the virus to evade anti-viral therapies and infection or vaccination based immunity.
Since SARS-CoV-2 first began circulating in late 2019, the World Health Organization has identified five variants of concern, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529). Recent surges have been caused by Delta and Omicron, each of which contains a unique set of spike mutations that creates an evolutionary advantage and enabled them to rapidly spread across the globe. With high infectivity and an unprecedented 30 nonsynonymous amino acid mutations in the spike protein, the Omicron variant is of particular concern as many of the mutations have been previously reported to enable immune evasion and reduce vaccine effectiveness.7 These mutations combined with naturally waning immunity following both infection and vaccination creates the potential for re-infection of convalescent patients and increased breakthrough infections in vaccinated individuals, however the vaccination of COVID-19 convalescent individuals, known as “hybrid immunity,” has been demonstrated to generate a more diverse and cross-reactive antibody pool.8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Here we investigated the breadth of antibody responses to both the SARS-CoV-2 Spike and RBD in a cohort of individuals with hybrid immunity. By comparing the titers against the ancestral strain and variants of concern, we have made observations that suggest a convergent strategy has been used by the SARS-CoV2 to achieve immune escape.
Methods
Ethics
This study was approved by the Dana-Farber Cancer Institute Institutional Review Board (IRB protocol #20-201). This protocol was prepared during the beginning of the COVID-19 pandemic specifically for the collection of COVID-19 convalescent donors. Therefore, we did not observe any potential impacts from COVID-19 and no study adaptations nor special methodologies were utilized.
Donor recruitment and inclusion criteria
Starting in June 2020, COVID-19 convalescent individuals in the Boston area were invited to participate in the COVID-19 Protective Immunity Study which involved completing a demographic/infection history survey and providing three staggered blood samples (initial post infection, 6 months, 12 months). Recruitment efforts included digital posters around the DFCI/Longwood Medical Center campus, messages posted on internal websites, and a link on the Marasco Lab's website. DFCI patients may have been informed of the study by their treating physician, however all other participants were passively recruited via digital study fliers and advertisements. Participants were required to be greater than 18 years of age and fully recovered from COVID-19 with two negative nasopharyngeal PCR tests. However, a time-based resolution of infection was later allowed following DFCI Infection Control policies. Interested donors were instructed to contact DFCI Research Nursing for consenting and enrollment.
Sample collection
Sequential samples (initial draw post infection with optional blood draws at 6 and 12 months) were collected from enrolled donors using BD K2EDTA vacutainers (lavender) via venipuncture by qualified nursing staff. After brief mixing in the vacutainer, blood samples were diluted 1:1 with autoMACS Rinsing solution (Miltenyi Biotec) + 2% heat inactivated FBS (Gibco). Diluted samples were carefully layered on 15 ml of GE Ficoll Paque Plus then centrifuged, allowing for the isolation of plasma and PBMCs which were aliquoted and stored at -20 C and -80 C, respectively. Sample collection from human subjects was performed under a protocol approved by the DFCI Institutional Review Board (IRB# 20-201).
RBD expression and purification
WT (Wuhan), Alpha, Beta, and Delta RBD DNA was amplified via PCR and cloned into a mammalian expression vector with a C-terminal 6xHis-Avi tag. Cloned plasmids were expressed using Expifectamine and the Expi293F (Life Technologies) platform following manufacturer protocols. Proteins were purified from supernatant via Ni-NTA Superflow (Qiagen) and dialyzed into PBS using Amicon Ultra-15 Centrifugal Filter units (Millipore Sigma). Purified proteins were confirmed via SDS-PAGE and ELISA binding assay with recombinant ACE2. Omicron RBD was purchased from AcroBiosystems.
The following plasmids were used as PCR templates: pCAGGS-S-B.1.617.2-FLAG was a gift from Daniel Conway (Addgene plasmid #177097; http://n2t.net/addgene:177097; RRID:Addgene_177097); Vector pCMV/R Containing the SARS-Related Coronavirus 2, Spike Glycoprotein Gene, Lineage B.1.1.7, Alpha Variant, NR-55304 (BEI Resources); Vector pCMV/R Containing the SARS-Related Coronavirus 2, Spike Glycoprotein Gene, Lineage B.1.351, Beta Variant, NR-55305 (BEI Resources); pCMV3-SARS-CoV-2 (2019-nCoV) Spike RBD ORF from SinoBiologics.
Mesoscale discovery platform (MSD)
Antibody titers were measured by MSD assay and arrayed such that all samples could be tested against an individual Spike or RBD protein on a single plate. 384 well High Bind plates (MSD) were coated with 25 ul of protein at 1 ug ml−1 protein in PBS overnight at 4 C. The next morning, plates were washed with PBS and blocked with PBS + 1% BSA (assay dilutant) for 3 h at room temperature (RT). During this time, plasma samples were diluted 1:100 with assay dilutant (final dilution of 1:200 when taking into account the initial dilution during cell isolation). Blocked plates were washed with PBS and 25 ul of diluted plasma sample was added to each well. Plates were incubated at RT for 2 h, followed by 4 rounds of washing with PBS. 25 ul of diluted (1:500) Sulfo-TAG labeled polyclonal anti-human IgG secondary (Mesoscale Discovery) was then added to each well. Plates were incubated for 2 h, then washed 6x with PBS, followed by the addition of 35 ul MSD Read Buffer with Surfactant. To ensure plates did not sit without buffer after washing, plates were washed and read on an MSD S600 before moving on to the next plate.
Data handling
Study data was collected and managed using REDCap electronic data capture tools hosted at DFCI and de-identified information was provided for research use. REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies, providing 1) an intuitive interface for validated data capture; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for data integration and interoperability with external sources.19, 20 Study size was determined by the number of donors with hybrid immunity and both pre and post vaccination samples collected. Samples from donors with only one bleed (n = 44) did not match the criteria of this study and thus were not tested in these assays. Titer results for all samples (N = 48) that were tested are included in this manuscript, however one donor with inconclusive vaccination dates was excluded from assignment to long (final draw >6 months post vaccination) or short (final draw <6 months post vaccination) interval groups for secondary analysis. An increase of 4 fold from pre-vaccination titers was used as the threshold to identify donors with increased titers post vaccination. Samples were not randomized nor blinded during analysis. For monoclonal antibody analysis (Table 1), all the RBD-Antibody structures were obtained from PDB (https://www.rcsb.org, Date of last access: Jan 07, 2022) and analyzed in Pymol (https://pymol.org/2/, Date of last access: Jan 07, 2022).
Table 1.
Relationship of anti-RBD monoclonal antibodies with the K417-E484-N501 triad. Molecular interactions of 21 anti-RBD monoclonal antibodies with published structures were analyzed to characterize binding interactions with the K417-E484-N501 triad.
| Table 1. SARS-CoV-2 RBD antibody relationship to K417-E484-N501 triad | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antibody | Epitope Class | Heavy Chain | Light Chain | Contacts (Yes/No); Bond |
PDB | Refs. | ||
| K417 | E484 | N501 | ||||||
| C102 | I | IGHV3-53 | IGKV3-20 | Pi-Alkyl, H-bond | No | 2 H-bonds | 7K8M | Barnes et al.52 |
| C105 | I | IGHV3-53 | IGLV2-8 | Yes | No | Yes | 6XCA | Barnes et al.53 |
| B38 | I | IGHV3-53 | IGKV1-9 | H-bond | No | H-bond | 7BZ5 | Wu et al.54 |
| CC12.3 | I | IGHV3-53 | IGKV3-20 | Salt Bridge | No | No | 6XC4 | Yuan et al.55 |
| COVOX-222 | I | IGHV3-53 | IGKV3-20 | Salt Bridge, Two H-bonds | No | Yes | 7Q9G | Liu et al.56 |
| COVOX-253 | I | IGHV1-58 | IGKV3-20 | No | H-Bond | No | 7BEN | Dejnirattisai et al.57 |
| Fab 1-57 | I | IGHV3-72 | IGKV3-20 | H-Bond | No | No | 7LS9 | Cerutti et al.58 |
| Fab 2-7 | I | IGHV2-5 | IGLV2-14 | No | No | H-bond | 7LSS | Cerutti et al.58 |
| BD629 | I | IGHV3-53 | IGKV3-20 | H-bond | No | H-bond | 7CH5 | Du et al.59 |
| C002 | II | IGHV3-30 | IGKV1-39 | No | Salt Bridge | No | 7K8S | Barnes et al.52 |
| LY-COV555 | II | IGHV1-69 | IGKV1-39 | No | Salt Bridge | No | 7KMG | Jones et al.26 |
| REGN10933 | II | IGHV3-15 | IGKV1-33 | No | H-bond | No | 6XDG | Hansen et al.60 |
| BD-368-2 | II | IGHV3-23 | IGKV2-28 | No | Salt Bridge | No | 7CHC | Du et al.59 |
| C104 | II | IGHV4-34 | IGKV3-20 | No | H-bond | No | 7K8U | Barnes et al.52 |
| LY-CoV1404 | III | IGVH2-5 | IGLV2-14 | No | No | H-bond | 7MMO | Westendorf et al.61 |
| S2M11 | III | IGHV1-2 | IGKV3-20 | H-bond | Salt Bridge, H-bond | No | 7K43 | Tortorici et al.62 |
| C135 | III | IGHV3-30 | IGKV1-5 | No | No | No | 7K8Z | Barnes et al.52 |
| S309 | III | IGHV1-18 | IGKV3-20 | No | No | No | 7R6X | Starr et al.63 |
| S2×259 | IV | IGHV1-69 | IGLV1-40 | No | No | H-bond to backbone | 7RAL | Tortorici et al.64 |
| CR3022 | IV | IGHV5-51 | IGKV4-1 | No | No | No | 7JN5 | Wu et al.65 |
| C022 | IV | IGHV4-39 | IGKV1-5 | No | No | No | 7RKU | Jette et al.66 |
Statistical analysis
MSD samples were performed in triplicate and the average values of the triplicate were analyzed. Geometric mean titers (GMTs) and ratio of post- vs pre-vaccination GMTs (GMTRs) were summarized for each strain. Continuous variables, such as GMTs and GMTRs, were compared between different subgroups using Wilcoxon rank-sum tests, while binary variables, such as achieving ≥ 4 fold increase in titers, were compared between different subgroups using Fisher's exact tests. Wilcoxon signed rank tests were used to compare pre- and post-vaccination titers and antibody response against different variants, while McNemar tests were used to compare the proportions of achieving ≥ 4 fold increase against different variants. Analysis was performed in GraphPad Prism version 9·2·0 and R version 4·1·3 (http://www.R-project.org). P values <0·05 were considered statistically significant and no adjustments were made for multiple comparisons.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Cohort description
An observational cohort of 48 COVID-19 convalescent volunteers with hybrid vaccination was established in Boston, MA. Serial blood samples were collected between June 2020 and December 2021 amid the ongoing pandemic and represented post recovery, 6, and 12 months time points. Cohort size was determined by the number of donors with hybrid immunity who provided both pre- and post- vaccination samples. The majority of these donors received mRNA vaccines from Pfizer or Moderna (18/49 BNT162b2, 23/49 mRNA-1273, 1/49 undisclosed) following their first blood collection with the remaining (7/49) receiving Johnson and Johnson's adenovirus-based vaccine (Ad26.COV2.S). 71% (35/49) of the donors were female and the medium participant age is 50 years (range 22–73 years) (Tables S1, S2). As all donors were infected prior to February 2021 (Table S2), the timing of collections was such that contemporaneous variants of concern constituted a small fraction of cases in the Boston area.21 As such, it is presumed that all donors were infected with Wuhan or early D614G strains. The majority of donors had PCR confirmed case of COVID-19, however that was not a condition of enrollment in the study and self-reported cases were also accepted.
Plasma antibody binding to spike and RBD proteins
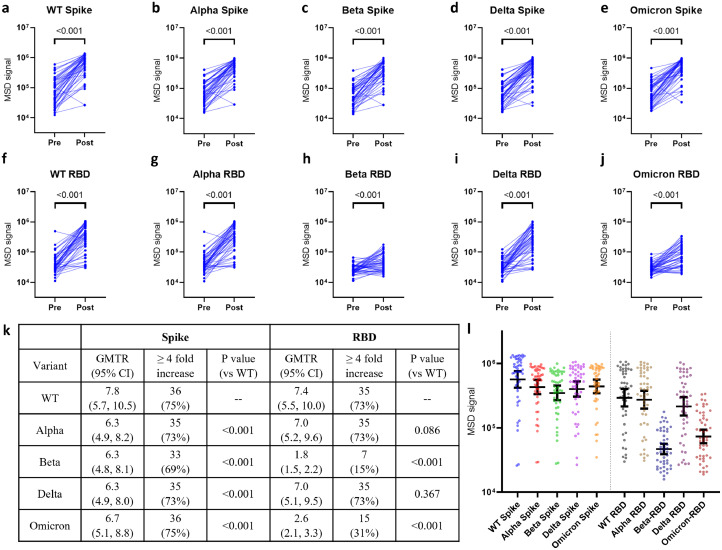
Plasma titers against the spike protein of WT, Alpha, Beta, Delta, and Omicron variants (Figure1a–e) were measured via the Mesoscale Discovery (MSD) platform. As expected, vaccination with BNT162b2, mRNA-1273, or Ad26.COV2.S led to a statistically significant increase of anti-Spike antibodies against the variant spikes tested, with the highest and lowest fold rise in titers observed in WT (GMTR: 7·8 [95% CI: 5·7, 10·5]) and Beta (GMTR: 6·3 [95% CI: 4·8, 8·1]), respectively (Figure 1k). Though vaccination leads to an increase of anti- WT, Alpha, and Delta RBD antibody titer (Figure 1f,g,i) with a GMTR of 7·4 (95% CI: 5·5, 10·0), 7·0 (95% CI: 5·2, 9·6), and 7·0 (95% CI: 5·1, 9·5), respectively, the GMTRs of vaccine-mediated increase against Beta and Omicron were only 1·8 (95% CI: 1·5, 2·2) and 2·6 (95% CI: 2·1, 3·3). Moreover, 7/48 (15%) and 15/48 (31%) of the donors had at least 4 fold increase in titer against Beta or Omicron RBDs, respectively (Figure 1k), which represents a statistically significant decrease (both p < 0·001) compared to WT RBD (Figure 1l).
Figure 1.
Antibody responses to full-length Spike and RBD proteins from various SARS-CoV-2 variants of concern using samples from 48 donors. Titers were measured by MSD ELISA at 1:200 dilution against full length spike (a–e) or RBD (f–j) proteins. (k) GMTR for each variant with 95% CI and number of donors who displayed a ≥ 4 fold increase in antibody titer post vaccination. P values represent a comparison of GMTR between WT and the indicated variants using Wilcoxon signed rank test. (l) Plasma titers against indicated Spike or RBD protein post vaccination. P values for statistical comparison in (l) is found in Table S6. Dots indicate individual donors and error bar represents overall Geometric Mean with 95% CI. All samples were performed in triplicate. Statistical analysis for a–j was performed using Wilcoxon Signed-Rank tests for paired data.
Similar to data published by van Gils et al., we observed that mRNA vaccination leads to a greater increase in plasma anti-Spike antibodies, with mRNA-1273 fold-changes generally higher than BNT162b2 (Table S7).22 This is most clearly demonstrated with the WT, Alpha, and Omicron spike GMTR, though the trend can also be seen with Beta and Delta. Interestingly, in our cohort the mRNA vaccines did not elicit the highest anti-RBD response, as the donors vaccinated with a single dose of Ad26.COV2.S generally had higher titers against the variant and WT RBDs (Table S7). This trend was also evident for the Beta and Omicron RBDs, however the small sample size of Ad26.COV2.S donors precluded us from further exploring the differential effect of vaccines.
Vaccination results in a decreased long-term antibody response to Beta and Omicron RBDs
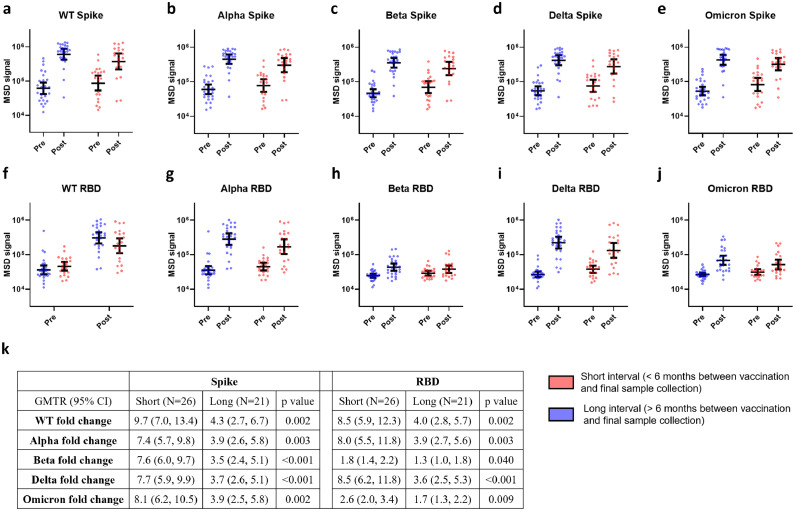
To further analyze the temporal dependency of vaccination on plasma antibody titers, 47 donors (one was excluded due to inconclusive vaccination dates) were grouped into two categories: “Short Interval” donors (n = 26) with final blood draw <6 months post vaccination and “Long Interval” donors (n = 21) with final blood draw >6 months post vaccination (Figure S2). Figure 2a–e provides evidence that regardless of the duration between vaccination and blood collection, the anti-Spike response displayed strong cross-reactivity with all variants tested. Though at the final blood draw the “Short Interval” group tended to have higher anti-Spike titers than the “Long interval” group, the difference between them did not reach statistical significance for any variants (Table S8). We next looked at the effect of vaccination induced antibody production by comparing post-vaccination anti-Spike GMTs to pre-vaccination anti-Spike GMTs. For all variants, the difference between “Short Interval” and “Long Interval” donors in fold-rise of anti-Spike titer is statistically significant, with the GMTR ranging from 7·4 to 9·7 in “Short Interval” donors and from 3·5 to 4·3 in “Long Interval” donors, a finding consistent with the well-established waning of anti-Spike antibody titers (Figure 2k).
Figure 2.
COVID-19 vaccination induced waning antibody responses to the RBD domains of Beta and Omicron spike variants using samples from 47 donors. The intervals between the completion of vaccination and final sample collection were calculated and segregated as < 6 months (short interval, red bars) and > 6 months (long interval, blue bars). Full length spike (a–e) and RBD (f–j) antibody titers for donors in different interval groups is shown. The titers were measured by MSD ELISA at 1:200 dilution, each sample was performed in triplicate. k) GMTR with 95% CI comparing pre and post vaccination for each group is shown. Dots represent individual donors and error bars indicate Geometric Mean with 95% CI. Statistical analysis was performed by Kruskal-Wallis rank sum test.
Similar to the results seen with anti-Spike antibodies, a statistically significant difference is seen when comparing the fold-rise of anti-RBD titers in “Short Interval” donors when compared to “Long Interval” donors (Figure 2k). When focusing on vaccination induced RBD-specific antibodies in the “Short Interval” group, a statistically significant greater fold increase in antibody titer was observed against WT (GMTR: 8·5 [95% CI: 5·9, 12·3]), Alpha (GMTR: 8·0 [95% CI: 5·5, 11·8]) and Delta (GMTR: 8·5 [95% CI: 6·2, 11·8]) compared to the Beta (GMTR: 1·8 [95% CI: 1·4, 2·2]) or Omicron (GMTR: 2·6 [95% CI: 2·0, 3·4]) RBDs (Figure 2k), indicating that vaccination elicited lower levels of antibodies cross-reactive to the Beta and Omicron RBDs. A statistically significant rise in cross-reactive anti-RBD GMTs in “Long Interval” donors was also observed except for against Beta RBD (GMTR: 1·3 [95% CI: 0·97, 1·8], p = 0·24), and while a statistically significant rise in Omicron RBD titers was observed (GMTR: 1·7 [95% CI: 1·3, 2·2]), they tended to be lower compared to WT, Alpha, and Delta RBDs (Figure 2f–j). As a whole, Figure 2 provides evidence to suggest that vaccination of COVID-19 convalescent individuals elicits a lower titer of Beta and Omicron RBD cross-reactive antibodies and these antibodies represent a reduced fraction of the plasma antibody pool after 6 months.
Identification of K417-E484-N501 triad in the RBD as a common viral escape strategy
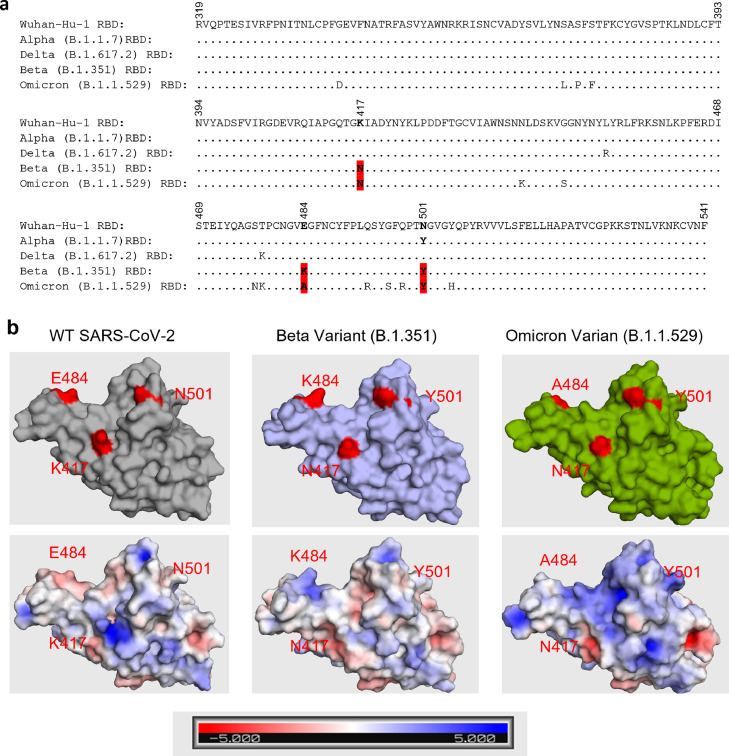
The finding that Beta and Omicron RBDs escaped binding from a fraction of plasma antibodies (Figs. 1 & 2) lead us to hypothesize that these variants share a similar escape strategy that separates them from the other variants. Therefore, we aligned the amino acid sequences of the RBDs and confirmed that both Beta and Omicron contain mutations at residues K417 (to N), E484 (to K for Beta, A for Omicron), and N501 (to Y) (Figure 3a, Table S9). While these mutations have been observed previously, with K417 occurring in Beta, Gamma, Delta, and Omicron, E484 occurring in Beta, Gamma, Zeta, Eta, Theta, Iota, Mu, and Omicron, and N501 occurring in Alpha, Beta, Gamma, Eta, Mu, and Omicron, only Beta, Gamma, and Omicron have mutations at all three residues in the K417-E484-N501 triad.21,23 These three residues lie on the ACE2 binding motif and are also involved in critical interactions with RBD class I and II monoclonal antibodies. A table summarizing these interactions for 21 antibodies covering the four RBD classes is presented in Table 1. K417, located in the middle of the class I antigenic site forms key molecular interactions (π-alkyl, hydrogen-bonds (H-bonds), or salt bridges) with 7/9 class I antibodies (Figure 3, Table 1) and mutations at this residue have been shown to reduce neutralization potency of class I and II antibodies.24,25 E484 sits at the boundary of class I and II antigenic sites and serves as a key source of H-bonds and electrostatic interactions for various neutralizing antibodies (Figure 3, Table 1), leading to critical energy forming contacts with 5/5 class II antibodies. Mutations at this site can severely dampen binding and neutralization for some of the most potent neutralizing antibodies including bamlanivimab and casirivimab, as well as reduce vaccine-elicited neutralizing antibody titers.25, 26, 27, 28, 29, 30, 31 N501 makes key interactions via hydrogen bonding with 4/9 class I antibodies (Figure 3, Table 1) and interestingly, the N501Y mutation has been reported to increase the binding affinity to ACE2.23,32,33 These observations demonstrate that class I and II antibodies frequently target the K417-E484-N501 triad, suggesting that reduced titers observed against the Beta and Omicron RBDs are likely a direct result of escape from class I and II antibodies. Taken together, the K417-E484-N501 triad in the RBD region is commonly used by SARS-CoV-2 variants to escape neutralization while simultaneously enhancing binding affinity to the ACE2 receptor.
Figure 3.
RBD sequence alignment of Wuhan-Hu-1 (WT), Alpha (B.1.1.7), Delta (B.1.617.2), Beta (B.1.351), and Omicron (B.1.1.529). (a) Amino acid sequence alignment is shown with K417-E484-N501 highlighted. (b) Surface representation of WT, Beta, and Omicron RBD is shown with their respective residues 417, 484, and 501 highlighted in red (top). Electrostatic potential of surface residues for each RBD is also shown (bottom). PDB files used WT: 7CH5, Beta: 7LYN, Omicron: 7T9L.
Discussion
In this manuscript we study a cohort of COVID convalescent individuals with hybrid immunity to characterize the plasma antibody response against Spike and RBD proteins of the WT, Alpha, Beta, Delta, and Omicron variants. As discussed in Schmidt et al., individuals with hybrid immunity can develop higher, more diverse antibody titers compared to individuals whose only exposure came from vaccination.12,13 However, recent strains including Beta and Omicron have shown to be particularly resistant to neutralization by both monoclonal antibodies and convalescent/vaccinated sera.34,35 Our cohort studies are focused on determining polyclonal anti-Spike and RBD titers and making observations on the effects of vaccine type as well as the breadth and durability of the antibody response post-vaccination. Structural and modeling studies of mutations allowed us to identify a critical triad at K417-E484-N501 as a common escape pathway from the anti-RBD antibody response.
With our cohort we are able to evaluate the differential response elicited by the three vaccines currently administered in the United States. Ad26.COV2.S is known to elicit a low anti-Spike/RBD antibody titer while mRNA vaccines rapidly induce high antibody titers that decay post vaccination.36 Interestingly in our cohort, while vaccination with either mRNA vaccine induced a greater anti-Spike antibody response, vaccination with Ad26.COV2.S consistently led to a greater increase in anti-RBD antibodies for all variants tested (Table S4). The difference in time between vaccination and sample collection could be a factor in the greater number of anti-RBD antibodies, as samples from Ad26.COV2.S vaccinated donors were collected on average 88·6 days post-vaccination, whereas mRNA-1273 and BNT162b2 vaccinated donors were collected 121·5 and 139·3 days post-vaccination, respectively, however anti-RBD titers in individuals vaccinated with Ad26.COV2.S have been shown to remain stable over 8 months.36
With an extensive list of mutations, the Omicron variant possesses the potential for enhanced transmissibility, decreased effectiveness of vaccines and therapeutic monoclonal antibody therapies, and an increased risk of re-infection in convalescent patients. Serological analysis by our lab demonstrates that vaccination effectively boosts plasma antibody titers against the ancestral spike protein and the spike proteins of Alpha, Beta, Delta, and Omicron (Figure 1a–e). Though vaccine induced anti-Spike antibodies are generally cross-reactive with the different variants, the ability of these antibodies to target divergent RBDs is more restrictive, with the Alpha and Delta RBDs possessing similar binding patterns to that of the WT, while the mutations in Beta and Omicron lead to statistically significantly (p < 0·001) lower levels of plasma antibody binding compared to WT (Figure 1f–j,k). The difference observed between Spike and RBD titers is not unexpected, as the spike protein contains a greater number of epitopes than the RBD and thus the loss of binding to the mutated epitopes correlates to a smaller overall change.
To examine the temporal relationship between vaccination and the cross-reactive antibody pool, donors in our cohort were divided into short and long interval groups depending on the time between vaccination and final blood draw. As shown in Figure 2a–e, the time between vaccination and the final sample collection does not impact the anti-Spike antibody titers, as both groups display a substantial increase in plasma antibody titer regardless of variant tested. However, this timeframe plays an important role in the vaccine's ability to elicit anti-RBD antibodies cross-reactive with the immune evading Beta and Omicron RBDs. Though anti-RBD titers in our cohort were comparatively lower for these two RBDs, a stark difference can be seen for those who were vaccinated early in the pandemic versus later, with early vaccinees failing to display a sustained cross-reactive plasma antibody population to Beta and Omicron RBDs >6 months post vaccination (Figure 2f–j). With the observation that vaccination of COVID-19 convalescent donors induced a lower fold change of cross-reactive antibodies against Beta and Omicron RBDs (Figure 2k), we provide evidence to suggest that these variants evaded vaccine elicited antibodies more effectively than other variants tested in this study.
The mutations at K417, E484, and N501 in the ACE2 binding domain observed in both Beta and Omicron have independently arisen in multiple SARS-CoV-2 variants, suggesting a selective advantage to these substitutions. Gobeil et al. demonstrated that in the D614G variant, ∼56% of the spikes are observed in the 3-RBD-down or closed position, however the K417N, E484K, N501Y mutations seen in Beta destabilize RBDs in the down position, leading to ∼88% of the spikes adopting an open conformation (≥ 1 RBD-up), increasing viral attachment potential and transmissibility.37 Further indicating the importance in these mutations to the transmissibility of the variants, Barton et al. demonstrated in vitro that the Beta variant RBD has 3·7-fold increased affinity for ACE2 compared to the WT RBD.38 In addition to their impact on transmissibility, E484 and K417 reside on epitopes commonly targeted by class I and II antibodies, with numerous reports demonstrating decreased binding and neutralization for variants containing mutations at these residues.39,40 The importance of escaped RBD immunity compared to full-length spike immunity is exemplified in work published by Garcia-Beltran et al. who utilized pseudovirus neutralization assays to demonstrate that the neutralization resistance of Beta is mainly due to mutations in the RBD and not mutations in the NTD or S2 domains.41
The preferential targeting of the triad residues following vaccination or infection with ancestral strains is in part due to the fact that antibodies preferentially bind charged or polar antigen residues.42 As glutamate and lysine both contain solvent exposed, electrically charged side chains (Figure 3b, Table 1), antibodies raised against the WT Spike protein, are drawn to these residues and rely on the π-alkyl, H-bonds, or salt bridge interactions as a critical source of binding energy. A great example of this is bamlanivimab which was isolated from a convalescent individual early in the pandemic and has sub-nanomolar affinity to the WT RBD.43 Structural analysis indicates that the bamlanivimab heavy and light chains combine to form three salt bridges with E484 and mutation to lysine (E484K) completely abolishes RBD binding.44 While the E484K charge reversal seen in Beta is quite drastic and likely results in the statistically significant (p < 0·0001) decrease in plasma titers, E484A seen in Omicron is a subtler change and likely accommodated by a greater number of plasma antibodies leading to a less severe drop in titer (Figure 1k). As the Alpha and Delta RBDs do not contain mutations at these residues, plasma titers remain similar to that of the WT RBD. Though our results show a greater decrease in antibody titer of Beta compared to Omicron, other studies have demonstrated that Omicron possesses greater neutralization escape than Beta.9,44,45 As the binding assays in this study are not focused on specific neutralization epitopes, higher binding titers are not necessarily correlated with neutralization potential. This contradiction could also arise as a result of mechanistic differences in these assays, as the MSD is an equilibrium binding assay where even low affinity antibodies bind and contribute to the overall signal. Conversely, neutralization is a dynamic assay where affinity and target epitope play a major role, with lower affinity antibodies often displaying limited neutralization efficacy.46
For a more focused structural analysis, we further analyzed 21 published monoclonal antibodies, including nine class I, five class II, four class III, and three class IV antibodies, and their interactions with the triad residues (Table 1). We found that at least one residue in the triad interacts with 9/9 (100%) class I, 5/5 (100%) class II, 2/4 (50%) class III, and 1/3 (33·3%) class IV antibodies. This suggests that the triad is preferentially targeted by class I and II antibodies, and thus these two classes of antibodies are most susceptible to triad mutations. As class I/II antibodies are known to have more potent neutralization than class III/IV antibodies, mutations in the triad such as those seen in Beta and Omicron would be expected to reduce plasma neutralization potential, an effect confirmed in multiple studies.10,34,47 Here we provide evidence to suggest that due to antibody epitope bias arising from ancestral strain infection or vaccination, hybrid immunity elicited antibodies have a lower probability of binding variants with triad mutations. What is not known is whether this binding limitation is the result of original antigenic sin where antibodies against conserved epitopes are preferentially elicited on repeat exposure, or are there limitations within immune repertoires of antibodies that bind to the RBD class I and II epitopes without depending on these triad residues for energetically favorable interactions.48 Either way, these mutations represent a convergent strategy for SARS-CoV-2 to evolve and escape immunologic pressure exerted by prior infection and/or vaccination, without sacrificing viral fitness or affinity to ACE2. In addition, while vaccine boosters are an effective short-term solution, our study suggests that optimizing vaccine designs to mitigate against immune evading mutations adopted by the virus should be explored as a means to elicit a more “universal” neutralizing antibody response.
Limitations
The strategy of utilizing triad mutations to escape antibody binding observed in the Beta and Omicron variants was identified by serological and structural analysis described in this manuscript. As mentioned in the results section, the Gamma (P.1) variant also includes triad mutations (K417T-E484K-N501Y) that likely disrupt immune recognition. However, due to the similarities seen in Beta and Gamma, and the fact that, global circulation of both strains was negligible compared to Delta and Omicron at the time of this study, only Beta was chosen for analysis.19 Additionally, studies by Wanwisa et al. and Uriu et al. demonstrated that compared to ancestral strains, Beta and Gamma display reduced neutralization by convalescent or vaccinated plasma, while Liang et al. showed similar results with a panel of neutralizing antibodies.49, 50, 51 Taken together, this suggests that the Gamma variant also utilizes triad mutations to evade the immune system, further supporting our hypothesis.
Another limitation of this study is the small size of the cohort and lack of infection only and vaccine only control cohorts for comparison. Though enrollment began in summer 2020 prior to widespread vaccine administration, the majority of our donors are healthcare workers who were among the first groups vaccinated. As such, the majority of our donors received a vaccine between the first and final sample collection and our IRB protocol was not designed to recruit vaccinated, non-infected volunteers. Therefore, we could not compare the potential differential effects of vaccine only, infection only, and infection followed by vaccination. This study is also limited to vaccines approved and administered in the United States as of December 2021 (BNT162b2, mRNA-1273, and Ad26.COV2.S), however the results are broadly applicable as these vaccines have been widely approved and distributed around the world. Due to the observational nature of the study design, confounding factors could adversely impact our findings. For example, those who received mRNA vaccines tended to be younger and more likely to be female, possibly leading to a greater vaccination response. However, within the mRNA vaccine group those who received mRNA-1273 tended to be older and more likely to be male, suggesting that differences in mRNA vaccine may have a greater impact on antibody titers than observed in this study. Other potential confounding variables in this study included the high prevalence of healthcare workers and donor distribution in regard to ethnicity and interval between infection and vaccination. Additional bias is our cohort arises from the lack of a fixed window between vaccination and sample collection.
Contributors
MRC, HK, and WAM conceptualized and designed the study. MRC, HK, CDC, YW, KM, and GMK isolated samples and executed experiments reported in this manuscript. MRC, HK, and CDC analyzed the data. CYH performed statistical analysis of the data and interpreted the results. MRC, HK, and WAM drafted the original manuscript. MRC and HK had full access to and verified all data. All authors had full access to study results, reviewed the manuscript, and approved the final version.
Data sharing
Data generated by this study is available upon request to the corresponding author.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
The authors would like to recognize and thank DFCI Research Nursing for their contributions to this work in consenting donors, completing enrollment surveys, and for providing the clinical expertise to collect samples used in this study. This work was supported by grants 280870.5116709.0016 from the Massachusetts Consortium on Pathogen Readiness (MassCPR) and 1R01AI161152-01A1 from the National Institute of Allergy and Infectious Diseases (NIAID); these funds were used for salary support of research staff and the purchasing of reagents.
The following reagents were obtained through BEI Resources, NIAID, NIH: Vector pCMV/R Containing the SARS-Related Coronavirus 2, Spike Glycoprotein Gene, Lineage B.1.1.7, Alpha Variant, NR-55304; Vector pCMV/R Containing the SARS-Related Coronavirus 2, Spike Glycoprotein Gene, Lineage B.1.351, Beta Variant, NR-55305; Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, Wuhan-Hu-1 with C-Terminal Histidine and Avi Tags, Recombinant from HEK293F Cells, NR-53524; Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, B.1.1.7 Lineage with C-Terminal Histidine and Avi Tags, Recombinant from HEK293 Cells, NR-55311; Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, B.1.351 Lineage with C-Terminal Histidine and Avi Tags, Recombinant from HEK293 Cells, NR-55310; Spike Glycoprotein (Stabilized) from SARS-Related Coronavirus 2, Delta Variant with C-Terminal Histidine and Avi Tags, Recombinant from HEK293 Cells, NR-55614.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104025.
Appendix. Supplementary materials
References
- 1.Robson F., Khan K.S., Le T.K., et al. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholz S., Pokhrel S., Kraemer B.R., et al. Paired SARS-CoV-2 spike protein mutations observed during ongoing SARS-CoV-2 viral transfer from humans to minks and back to humans. Infect Genet Evol. 2021;93 doi: 10.1016/j.meegid.2021.104897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ilmjärv S., Abdul F., Acosta-Gutiérrez S., et al. Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant. Sci Rep. 2021;11:1–13. doi: 10.1038/s41598-021-91662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ou J., Zhou Z., Dai R., et al. V367F mutation in SARS-CoV-2 spike RBD emerging during the early transmission phase enhances viral infectivity through increased human ACE2 receptor binding affinity. J Virol. 2021;95 doi: 10.1128/jvi.00617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun F., Wang X., Tan S., et al. SARS-CoV-2 quasispecies provides an advantage mutation pool for the epidemic variants. Microbiol Spectr. 2021;9:1–16. doi: 10.1128/spectrum.00261-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J Med Virol. 2021 doi: 10.1002/jmv.27526. n/a. [DOI] [PubMed] [Google Scholar]
- 8.Basile K, Rockett RJ, Mcphie K, et al. Improved neutralization of the SARS-CoV-2 Omicron variant after Pfizer-BioNTech BNT162b2 COVID-19 vaccine boosting. BioRxiv. 2021 doi: 10.1101/2021.12.12.472252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemet I., Kliker L., Lustig Y., et al. Third BNT162b2 Vaccination neutralization of SARS-CoV-2 Omicron infection. N Engl J Med. 2021:2008–2009. doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022 doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt F., Muecksch F., Weisblum Y., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2021:2008–2009. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt F., Weisblum Y., Rutkowska M., et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamatatos L., Czartoski J., Wan Y.H., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotty S. Hybrid immunity. Science. 2021;372:1392–1393. doi: 10.1126/science.abj2258. [DOI] [Google Scholar]
- 15.Seow J., Graham C., Merrick B., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terpos E., Stellas D., Rosati M., et al. SARS-CoV-2 antibody kinetics eight months from COVID-19 onset: persistence of spike antibodies but loss of neutralizing antibodies in 24% of convalescent plasma donors. Eur J Intern Med. 2021;89:87–96. doi: 10.1016/j.ejim.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., et al. Rapid decay of anti–SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muecksch F., Wise H., Batchelor B., et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. 2021;223:389–398. doi: 10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodcroft E.B. CoVariants: SARS-CoV-2 mutations and variants of interest 2021. https://covariants.org/.
- 22.van Gils MJ, Lavell AHA, van der Straten K, et al. Four SARS-CoV-2 vaccines induce quantitatively different antibody responses against SARS-CoV-2 variants. MedRxiv. 2021 doi: 10.1101/2021.09.27.21264163. [DOI] [Google Scholar]
- 23.Cameroni E., Saliba C., Bowen J.E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021 doi: 10.1101/2021.12.12.472269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tegally H., Wilkinson E., Giovanetti M., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z., Schmidt F., Weisblum Y., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones B.E., Brown-Augsburger P.L., Corbett K.S., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med. 2021;13:1–17. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen J., Baum A., Pascal K.E., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. Partial resistance of SARS-CoV-2 Delta variants to vaccine-elicited antibodies and convalescent sera. IScience. 2021;24 doi: 10.1016/j.isci.2021.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls A.C., Miranda M.C., Schäfer A., et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell. 2021;184:5432–5447. doi: 10.1016/j.cell.2021.09.015. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K., Werner A.P., Koch M., et al. Serum neutralizing activity elicited by mRNA-1273 vaccine. N Engl J Med. 2021;384:1468–1470. doi: 10.1056/NEJMc2102179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier D.A., De Marco A., Ferreira I.A.T.M., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starr T.N., Greaney A.J., Hilton S.K., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carreño J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against a B.1.1.529 variant SARS-CoV-2 isolate. Nature. 2021 doi: 10.1101/2021.12.20.21268134. 2021.12.20.21268134. [DOI] [PubMed] [Google Scholar]
- 35.Lu L., Mok B.W.Y., Chen L.L., et al. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. 2021:1–28. doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collier A.Y., Yu J., McMahan K., et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N Engl J Med. 2021;385:2010–2012. doi: 10.1056/NEJMc2115596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobeil S.M.C., Janowska K., McDowell S., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science. 2021;373 doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barton M.I., Macgowan S., Kutuzov M., Dushek O., Barton G.J., Anton Van Der Merwe P. Effects of common mutations in the SARS-CoV-2 spike RBD and its ligand the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10:1–19. doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greaney A.J., Starr T.N., Barnes C.O., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun. 2021;12 doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendy M., Kaufman S., Ponga M. Molecular strategies for antibody binding and escape of SARS-CoV-2 and its mutations. Sci Rep. 2021;11:1–11. doi: 10.1038/s41598-021-01081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Beltran W.F., Lam E.C., St. Denis K., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M., Zhu D., Zhu J., Nussinov R., Ma B. Local and global anatomy of antibody-protein antigen recognition. J Mol Recognit. 2018;31:e2693. doi: 10.1002/jmr.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H., Wei P., Zhang Q., et al. 501Y.V2 and 501Y.V3 variants of SARS-CoV-2 lose binding to bamlanivimab in vitro. MAbs. 2021;13:1–6. doi: 10.1080/19420862.2021.1919285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers B.L., Chakraborty S., Xue Y., et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci Transl Med. 2022;7842:1–12. doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.GeurtsvanKessel CH, Geers D, Schmitz KS, et al. Divergent SARS CoV-2 Omicron-specific T- and B-cell responses in COVID-19 vaccine recipients. MedRxiv 2021;2202:2021.12.27.21268416. [DOI] [PMC free article] [PubMed]
- 46.Zost S.J., Gilchuk P., Case J.B., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cele S., Jackson L., Khan K., et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. MedRxiv Prepr Serv Heal Sci. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 48.Gostic K.M., Ambrose M., Worobey M., Lloyd-Smith J.O. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dejnirattisai W., Huo J., Zhou D., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2021 doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uriu K., Kimura I., Shirakawa K., et al. Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N Engl J Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang K.H., Chiang P.Y., Ko S.H., et al. Antibody cocktail effective against variants of SARS-CoV-2. J Biomed Sci. 2021;28:1–12. doi: 10.1186/s12929-021-00777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barnes C.O., Jette C.A., Abernathy M.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnes C.O., West A.P., Huey-Tubman K.E., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Y., Wang F., Shen C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan M., Liu H., Wu N.C., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C., Zhou D., Nutalai R., et al. The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dejnirattisai W., Zhou D., Ginn H.M., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200. doi: 10.1016/j.cell.2021.02.032. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerutti G., Rapp M., Guo Y., et al. Structural basis for accommodation of emerging B.1.351 and B.1.1.7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29:655–663. doi: 10.1016/j.str.2021.05.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du S., Cao Y., Zhu Q., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020 doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hansen J., Baum A., Pascal K.E., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westendorf K, Žentelis S, Foster D, et al. LY-CoV1404 potently neutralizes SARS-CoV-2 variants. BioRxiv Prepr Serv Biol 2021:2021.04.30.442182. 10.1101/2021.04.30.442182.
- 62.Tortorici M.A., Beltramello M., Lempp F.A., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;957:eabe3354. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Starr T.N., Czudnochowski N., Liu Z., et al. Springer US; 2021. SARS-CoV-2 RBD antibodies that Maximize Breadth and Resistance to Escape. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tortorici M.A., Czudnochowski N., Starr T.N., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu N.C., Yuan M., Bangaru S., et al. A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. PLOS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jette C.A., Cohen A.A., Gnanapragasam P.N.P., et al. Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.