Abstract
Plant leucine-rich repeat (LRR) receptor-like kinases (RLKs) and LRR receptor-like proteins (RLPs) comprise a large family of cell surface receptors that play critical roles in signal perception and transduction. Both LRR-RLKs and LRR-RLPs rely on regulatory LRR-RLKs to initiate downstream signaling pathways. BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (BAK1/SERK3) and SUPPRESSOR OF BIR1-1 (SOBIR1) are important and extensively studied regulatory LRR-RLKs with distinct functions. Although the regulatory mechanism of BAK1 activation has been studied in detail, the activation mechanism of SOBIR1 remains poorly understood. Here, the crystal structures of the catalytically inactive kinase domain of SOBIR1 (SOBIR1-KD) from Arabidopsis thaliana were determined in complexes with AMP-PNP and Mg2+. The results show that SOBIR1-KD contains a uniquely long β3-αC loop and adopts an Src-like inactive conformation with an unusual architecture at the activation segment, which comprises three helices. Biochemical studies revealed that SOBIR1 is transphosphorylated by BAK1 following its autophosphorylation via an intermolecular mechanism, and the phosphorylation of Thr529 in the activation segment and the β3-αC loop are critical for SOBIR1 phosphorylation. Further functional analysis confirmed the importance of Thr529 and the β3-αC loop for the SOBIR1-induced cell death response in Nicotiana benthamiana. Taken together, these findings provide a structural basis for the regulatory mechanism of SOBIR1 and reveal the important elements and phosphorylation events in the special stepwise activation of SOBIR1-KD, the first such processes found in regulatory LRR-RLKs.
Key words: LRR-RLK, SOBIR1, crystal structure, unusual architecture, autophosphorylation, stepwise activation
SOBIR1 is an indispensable adaptor RLK for LRR-RLP-mediated signaling, and its phosphorylation status is positively linked with its constitutive immune activity in plants. This study proposes a model for the stepwise activation of SOBIR1 in which it is autophosphorylated via an intermolecular mechanism and subsequently transphosphorylated by BAK1. These results provide a framework for elucidating the activation mechanism of SOBIR1-RLP-dependent signaling cascades and offer new insights into mechanisms of kinase regulation.
Introduction
Plants use germline-encoded cell surface receptors to perceive various external cues and transmit signals to induce responses (Man et al., 2020). Such receptors include receptor-like kinases (RLKs) and receptor-like proteins (RLPs), which participate in the regulation of plant growth, development, immune responses, and many other essential processes (Liebrand et al., 2014; Man et al., 2020). A typical RLK frequently includes a leucine-rich repeat (LRR) extracellular domain that recognizes specific ligands, a single membrane-spanning transmembrane (TM) domain, and an intracellular kinase domain (KD) that is lacking in RLPs. Previous research has demonstrated that both LRR-RLKs and LRR-RLPs rely on regulatory LRR-RLKs to initiate downstream signaling pathways. The LRR-RLK BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE 3 (BAK1/SERK3) is essential for BRASSINOSTEROID INSENSITIVE 1 (BRI1)-mediated pathways and was demonstrated to be a general regulator in multiple signaling pathways, functioning as a ligand-binding co-receptor that influences development and immunity (Liebrand et al., 2014; Gao et al., 2018). The crystal structure of the BAK1KD was determined in the active state, providing the structural basis for the impact of phosphorylation on the activation process (Yan et al., 2012). In general, the perception of a specific ligand by the extracellular domain of an LRR-RLK recruits BAK1, and this is followed by the transphosphorylation and activation of their cytosolic KDs and subsequent initiation of downstream signaling (Chinchilla et al., 2007; Wang et al., 2008; Postma et al., 2016). LRR-RLPs lack the KD and are thought to rely on the regulatory LRR-RLK SUPPRESSOR OF BIR1-1 (SOBIR1, also called EVERSHED) to activate cytoplasmic signaling pathways. Although extensive genetic analyses have demonstrated the functional significance of SOBIR1, the molecular mechanism of its cytoplasmic activation is poorly understood.
SOBIR1 was initially identified in a suppressor screen of the BAK1-Interacting RLK 1 (bir1-1) mutant, which shows autoimmune responses (Gao et al., 2009). Overexpression of SOBIR1 induces cell death in both Arabidopsis and Nicotiana benthamiana, suggesting that SOBIR1 is a positive regulator of cell death (Gao et al., 2009). SOBIR1 homologs are transcriptionally upregulated by a broad range of pathogens and microbe-associated molecular patterns (MAMPs), often leading to rapid downstream responses, such as an oxidative burst and mitogen-activated protein kinase (MAPK) activation. Mutations in sobir1 can also suppress the autoimmunity induced by BAK1 overexpression (Dominguez-Ferreras et al., 2015). SOBIR1 has been reported to constitutively associate with a series of RLPs in a ligand-independent manner. Some examples of RLPs are Cf-2, Cf-4, Cf-9, Ve1, RLP23, RLP30, and RLP42, all of which are required for pathogen perception in tomato (Solanum lycopersicum) or Arabidopsis thaliana (Gao et al., 2009; Zhang et al., 2013, 2014; Albert et al., 2019). Furthermore, SOBIR1-RLP complexes such as SOBIR1-Cf-4 and SOBIR1-RLP23 generally require a ligand-dependent interaction with BAK1 or other SERKs for downstream signaling (Zhang et al., 2014; van der Burgh et al., 2019; Wu et al., 2019). Although SOBIR1 provides the intracellular KD to form the SOBIR1-RLP complex, which is structurally equivalent to bipartite RLKs, SOBIR1-RLP- and RLK-mediated immune responses have distinct requirements for BOTRYTIS-INDUCED KINASE 1 (BIK1), a receptor-like cytoplasmic kinase (RLCK), suggesting that they have distinct activation mechanisms (Wan et al., 2019). Consistently, it was shown that BIK1 negatively regulates RLP23/RLP42-mediated immunity but positively regulates flagellin-sensitive 2 (FLS2)- and elongation factor Tu receptor (EFR)-mediated immunity (Wan et al., 2019). SOBIR1's extracellular domain contains five LRRs, and its TM domain includes the GxxxG dimerization motif, which is necessary for interactions with several RLPs (Bi et al., 2016; Albert et al., 2019; Hohmann and Hothorn, 2019). The cytoplasmic portion of SOBIR1 contains a KD, the activity of which is essential for RLP signal transduction (Liebrand et al., 2013; Wu et al., 2019). Despite significant similarity in their protein domain architectures, the SOBIR1-KD shares only 34.6% sequence similarity with the BAK1 cytoplasmic domain (CD) and lacks a C-terminal tail essential for BAK1-induced immunity (Wu et al., 2018a).
SOBIR1 is an indispensable adaptor RLK for LRR-RLP-mediated signaling, and the phosphorylation status of SOBIR1 is positively linked to its constitutive immune activity in plants. However, less is known about the molecular basis for phosphorylation-dependent regulation of SOBIR1 activity. In this study, the crystal structures of catalytically inactive SOBIR1-KD provide new insights into its kinase regulation mechanism. The structures reveal that SOBIR1 adopts an Src-like inactive conformation with an unusual architecture at the activation segment, which comprises three helices. In vitro biochemical and in vivo functional analyses indicate that SOBIR1 is autophosphorylated via an intermolecular mechanism and subsequently transphosphorylated by BAK1. The phosphorylation of Thr529 in the activation segment, as well as the relatively long β3-αC loop, are essential for SOBIR1 activation. We thus propose a model for the stepwise activation of SOBIR1. Collectively, these results not only clarify the biochemical and structural characteristics of SOBIR1-KD but also provide the framework for elucidating the activation mechanism of SOBIR1-RLP-dependent signaling cascades.
Results
SOBIR1-KD is autophosphorylated via an intermolecular mechanism
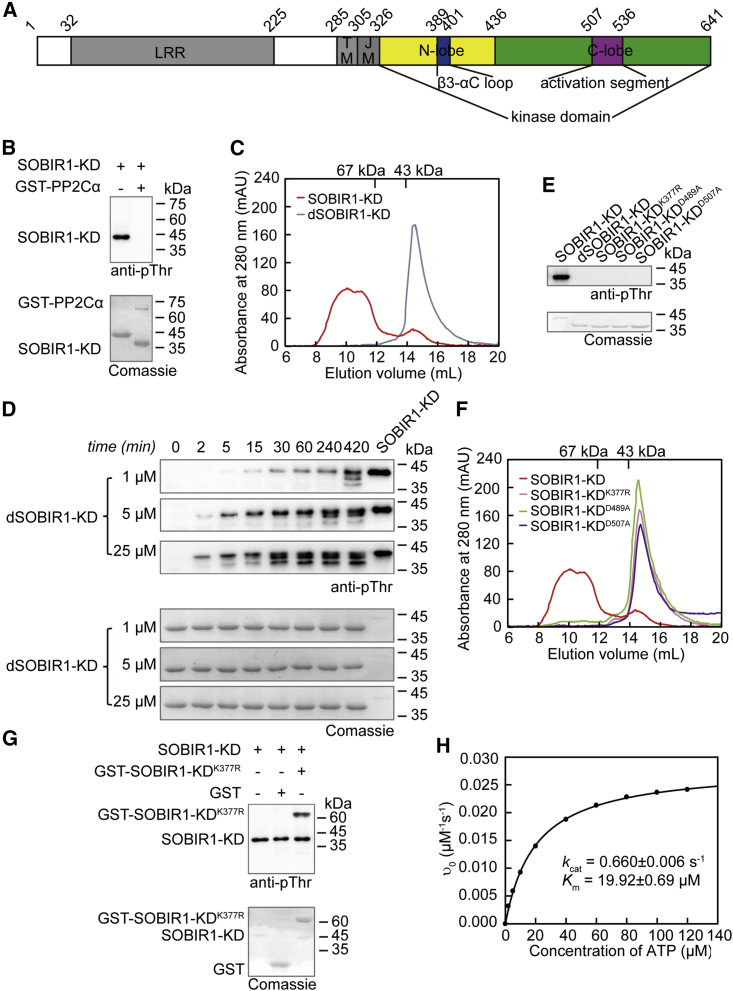
To illustrate the molecular mechanism underlying the activation of SOBIR1-KD, we expressed and purified SOBIR1-KD (residues 326–641) (Figure 1A). Consistent with a previous report, the bacterially expressed SOBIR1-KD was highly phosphorylated, implying that the protein can be autophosphorylated during expression and/or purification in vitro (Leslie et al., 2010) (Figure 1B, lane 1). The protein was completely dephosphorylated after incubation with protein phosphatase PP2Cα (Figure 1B, lane 2). Size-exclusion chromatography (SEC) analysis indicated that the wild-type SOBIR1-KD is heterogeneous with potentially different oligomeric forms (Figure 1C). However, the dephosphorylated recombinant SOBIR1-KD (dSOBIR1-KD hereafter) is primarily monomeric, consistent with the 35.6-kDa calculated monomeric molecular mass. These results suggest that the phosphorylation of SOBIR1-KD may influence its oligomerization state. To confirm that SOBIR1-KD can be autophosphorylated, we performed in vitro kinase assays with different concentrations of dSOBIR1-KD. After incubation with 1 mM ATP and 10 mM Mg2+ at 25°C, samples with equal loading were analyzed by western blotting using the nonspecific anti-phospho-Thr antibody. As shown in Figure 1D, dSOBIR1-KD was autophosphorylated in 2 min, and the autophosphorylation of dSOBIR1-KD at 1 μM was slower than that at 25 μM. The rate of autophosphorylation of SOBIR1-KD was highly dependent on protein concentration, suggesting that SOBIR1 autophosphorylation occurs by an intermolecular (in trans) rather than an intramolecular (in cis) mechanism (Wang et al., 2011).
Figure 1.
Biochemical properties of SOBIR1-KD
(A) Schematic diagram of AtSOBIR1 (Swiss-Prot ID: Q9SKB2). LRR domain (shown in gray); TM domain (shown in gray); juxtamembrane (JM) domain (shown in gray); N-lobe, N-terminal lobe (shown in yellow); C-lobe, C-terminal lobe (shown in green). The β3-αC loop and the activation segment are presented in blue and magenta, respectively.
(B) Phosphorylation states of the recombinant wild-type and PP2Cα-dephosphorylated SOBIR1-KD analyzed with a general anti-phospho-Thr antibody. The reaction mixture contained 10 μM SOBIR1-KD and 1 μM GST-PP2Cα.
(C) Determination of oligomerization states via SEC at a concentration of approximately 20 μM. Wild-type SOBIR1-KD eluted as a multimer, whereas the elution volume for the PP2Cα-dephosphorylated SOBIR1-KD corresponded to an apparent molecular mass of 36–40 kDa, which is consistent with the calculated monomeric molecular mass of 36 kDa. Elution volumes of the protein standards are indicated.
(D) Time-course analysis of SOBIR1-KD intermolecular autophosphorylation. Specifically, 1, 5, and 25 μM PP2Cα-dephosphorylated SOBIR1-KD samples were incubated with 1 mM ATP and 10 mM Mg2+ for the indicated period.
(E) Phosphorylation states of wild-type SOBIR1-KD, PP2Cα-dephosphorylated SOBIR1-KD, and the kinase-dead mutants SOBIR1-KDK377R, SOBIR1-KDD489A, and SOBIR1-KDD507A.
(F) Determination of oligomerization states via SEC at a concentration of approximately 20 μM. Wild-type SOBIR1-KD eluted as a multimer, whereas the elution volumes for kinase-dead mutants, SOBIR1-KDK377R, SOBIR1-KDD489A, and SOBIR1-KDD507A corresponded to an apparent molecular mass of 36–40 kDa, which is consistent with the calculated monomeric molecular mass of 36 kDa. Elution volumes of the protein standards are indicated.
(G) Western blotting analysis of the transphosphorylation of SOBIR1-KDK377R by wild-type SOBIR1-KD. The reaction mixture contained 10 μM GST-SOBIR1-KDK377R and 1 μM SOBIR1-KD. As a negative control, GST was incubated with wild-type SOBIR1-KD, and its phosphorylation signal was undetectable.
(H) Effects of ATP concentration on the initial velocity of the SOBIR1-KD-catalyzed reaction. The reaction mixture contained 50 nM wild-type SOBIR1-KD and 2, 5, 10, 20, 40, 60, 80, 100, or 120 μM ATP. The solid line represents the best-fitting result according to the Michaelis-Menten equation, with kcat and Km values of 0.660 ± 0.006 s−1 and 19.92 ± 0.69 μM, respectively.
To further investigate the intermolecular phosphorylation mechanism, we mutated SOBIR1-KD by individually replacing three catalytically important residues, Lys377Arg, Asp489Ala, and Asp507Ala (Taylor and Kornev, 2011). The purified SOBIR1-KDK377R, SOBIR1-KDD489A, and SOBIR1-KDD507A were analyzed by western blotting using the nonspecific anti-phospho-Thr antibody. Phosphorylation signals were not detectable for the mutated proteins, confirming the importance of these sites to the catalytic activity of SOBIR1-KD (Figure 1E). Moreover, SEC analysis showed that the three inactive SOBIR1-KD mutants were monomeric in solution, the same as dSOBIR1-KD (Figure 1F). We then performed in vitro kinase assays by incubating the phosphorylated wild-type SOBIR1-KD with the kinase-dead mutant SOBIR1-KDK377R and found that SOBIR1-KD could robustly phosphorylate both itself and SOBIR1-KDK377R (Figure 1G), demonstrating that in vitro autophosphorylation of SOBIR1-KD occurs in trans.
To further characterize the ATPase activity of SOBIR1-KD, we performed a hydrolysis reaction assay via enzyme-coupled spectrophotometry. A decrease in absorbance at 340 nm indicated that the consumption of the substrate was caused by a reaction that coupled the production of ADP to the oxidation of NADH (Roskoski, 1983). The kcat and Km values were determined to be 0.660 ± 0.006 s−1 and 19.92 ± 0.69 μM, respectively, suggesting that SOBIR1-KD could effectively hydrolyze ATP and providing evidence for its autophosphorylation (Figure 1H). These data indicated that the autophosphorylation of SOBIR1 is mediated by an intermolecular mechanism.
Overall structure of SOBIR1-KD
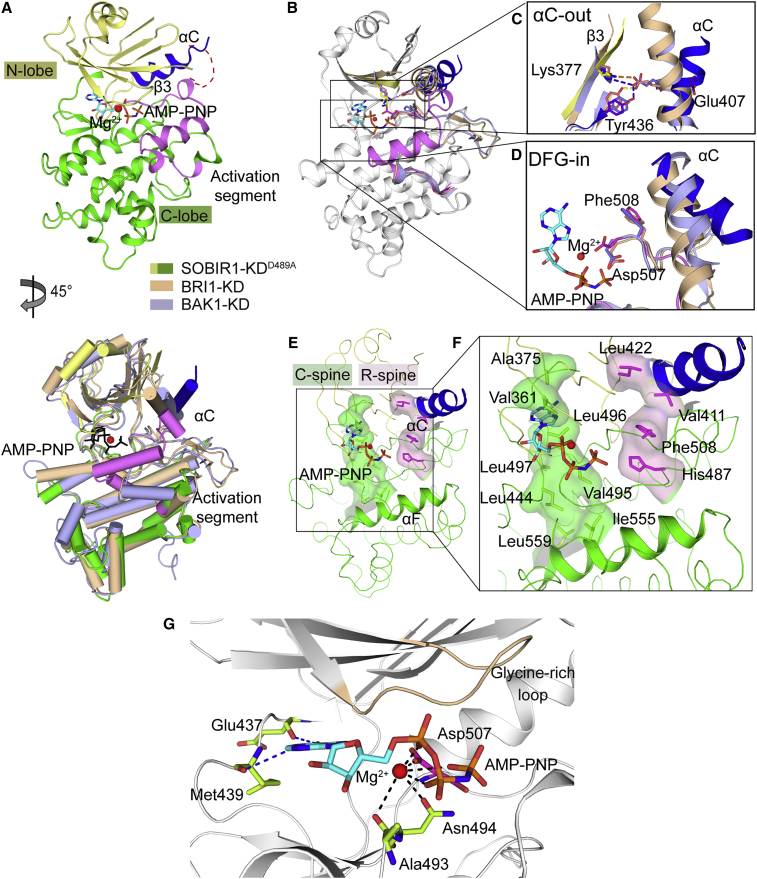
To gain more insights into the activation of SOBIR1 via phosphorylation regulation, we attempted to crystallize SOBIR1-KD. Although we were not able to culture crystals of the phosphorylated wild-type SOBIR1-KD, probably because the heterogeneity of the phosphorylated proteins impeded crystallization, we obtained the kinase-dead SOBIR1-KDD489A in complex with the non-hydrolyzable ATP analog AMP-PNP and Mg2+. This binary complex was determined at a 2.9-Å resolution via molecular replacement with the KD of BRI1 (PDB: 5LPY) as the search model (N- and C-terminal lobes replaced separately), as it has the highest sequence identity with SOBIR1 (37%) in the PDB database (Bojar et al., 2014). The final model was refined to an overall Rfree value of 23.9% (Rwork of 21.7%; Supplemental Table 1). The crystallographic asymmetric unit comprises two molecules (A and B) (Supplemental Figure 1A). Because SOBIR1-KDD489A is mostly monomeric in solution (Figure 1F), the crystal packing interactions formed by two independent molecules may be physiologically irrelevant. The overall structures of molecules A and B are similar, with a root-mean-square deviation (RMSD) value of 0.361 Å for 245 equivalent Cα positions (Supplemental Figure 1B). The structural analyses described hereafter are based on molecule A, which has a lower average temperature factor, indicating a more stable structure (Supplemental Figure 1C).
The structure of the unphosphorylated SOBIR1-KDD489A has a canonical bilobal kinase architecture comprising a small N-terminal lobe (residues 326–435) and a large C-terminal lobe (residues 439–641) connected by a flexible hinge region (residues 436–438) (Figure 2A). The N-terminal lobe includes a twisted, five-stranded antiparallel β sheet (β1–β5) as well as a prominent αC helix, and the C-terminal lobe consists mainly of α helices. The αC helix is preceded by a noncanonical α helix αB (residues 344–349), a short α helix, and an N-terminal four-residue β strand antiparallel to β4. The activation segments in both molecules A and B are traceable among all residues in the electron density map and are assembled into inhibitory α helices packed against the active site for substrate binding (Supplemental Figure 1A). An AMP-PNP bound to a single Mg2+ ion is observed to occupy the canonical ATP-binding pocket in the deep cleft between the two lobes of SOBIR1-KDD489A (Figure 2A). Notably, SOBIR1 has a long loop between β3 and αC in which some residues (383–399 in molecule A and 381–397 in molecule B) lack electron density, suggesting the high flexibility of this loop.
Figure 2.
Overall structure of SOBIR1-KDD489A complexed with the non-hydrolyzable ATP analog AMP-PNP
(A) Two views of the SOBIR1-KDD489A ribbon structure (45° rotation around a vertical axis). The SOBIR1-KDD489A color scheme is the same as that in the schematic diagram in Figure 1A. The bound AMP-PNP is presented as an orange stick, whereas Mg2+ is indicated by a red sphere. The disordered β3-αC loop is presented as a red dashed line. The same SOBIR1-KDD489A color scheme is used in the following figures, unless otherwise indicated. A structural comparison between SOBIR1-KD and BRI1-CD (PDB: 5LPY, in wheat) and BAK1-CD (PDB: 3UIM, in light blue) is presented in the right panel. The same BRI1-CD and BAK1-CD color scheme is used in the following figures, unless otherwise indicated. The AMP-PNP and Mg2+ in SOBIR1-KDD489A are presented as a black stick and a red sphere, respectively. The AMP-PNP and ions in BRI1-CD and BAK1-CD are not shown.
(B) Comparison between SOBIR1-KD and BRI1-CD and BAK1-CD after superimposing their active sites. The AMP-PNP and Mg2+ in SOBIR1-KDD489A are presented as an orange stick and a red sphere, respectively. The AMP-PNP and ions in BRI1-CD and BAK1-CD are not shown.
(C and D) Close-up views of the active sites with αC-out (C) and DFG-in (D). The essential residues are presented as sticks. Specifically, Lys377 in SOBIR1-KD corresponds to Lys911 in BRI1-CD and Lys317 in BAK1-CD; Glu407 in SOBIR1-KD corresponds to Glu927 in BRI1-CD and Glu334 in BAK1-CD; Tyr436 in SOBIR1-KD corresponds to Tyr956 in BRI1-CD and Tyr363 in BAK1-CD. The hydrogen bonds and salt bridges in BRI1-CD and BAK1-CD are presented as orange and blue dashed lines, respectively.
(E and F) C-spine and R-spine in the SOBIR1-KD-AMP-PNP-Mg2+ complex. For clarity, the αC and αF helices are depicted in cartoon form. The C-spine and R-spine are presented in green and magenta, respectively (E). Residues forming the C-spine and R-spine in the SOBIR1-KD-AMP-PNP-Mg2+ complex. The C-spine, comprising Val361, Ala375, Leu496, Val495, Leu497, Leu444, Leu555, and Leu559, is indicated by lemon sticks. The AMP-PNP associated with Val361, Ala375, and Leu496 is presented as an orange stick. The R-spine, consisting of Phe508, His487, Val411, and Leu422, is indicated by magenta sticks (F).
(G) View of the nucleotide-binding pocket of SOBIR1-KD occupied by AMP-PNP and a single Mg2+. The nucleotides and the nucleotide-interacting residues are presented as cyan and lemon sticks, respectively, whereas Mg2+ is indicated by a red sphere. Hydrogen bonds for the interaction with AMP-PNP and Mg2+ are presented in blue or black dashed lines.
SOBIR1-KDD489A adopts a DFG-in and αC-out Src-like inactive conformation
Protein kinases undergo large conformational changes when switching between active and inactive states (Johnson et al., 1996; Huse and Kuriyan, 2002). Crystal structures of active kinases suggested that they adopt similar conformations in which the αC helix rotates inward to prime the active site Lys from β3, and the activation loop adopts an extended conformation exposed to the solvent; however, inactive kinases take various conformations to impede the catalytically active center in various ways (Schwarz et al., 2019). The structures of the best-characterized plant RLKs, BRI1, and BAK1, were all determined in their active state with multiple phosphorylated sites (Yan et al., 2012; Bojar et al., 2014; Wang et al., 2014). Here, the structure of the inactive SOBIR1-KDD489A provides some new mechanistic insights into the phosphorylation-dependent activation of SOBIR1.
The regulatory αC helix possessing Glu407 of SOBIR1-KDD489A rotates away from the catalytic site, thereby breaking the critical salt bridge between Glu407 and Lys377 from strand β3 (αC-out), which is required for the catalytically competent KD conformation (Figure 2B and 2C). In contrast to other activated plant RLKs, the hydrogen bond between the gatekeeper Tyr436 and the conserved Glu407 in SOBIR1-KDD489A is disrupted because of the αC rotation (Bojar et al., 2014) (Figure 2C). Interestingly, the DFG motif of SOBIR1-KDD489A has a DFG-in conformation in which the side chain of Asp507 faces the active site but the side chain of Phe508 points away from this site, toward the αC helix (Figure 2D). Hence, the structure of SOBIR1-KDD489A adopts an Src-like inactive conformation, which was previously observed in the crystal structures of c-Src and other protein kinases, including IRAK4 and Abl (Levinson et al., 2006; Mikkola and Gahmberg, 2010; Wagner et al., 2015; Wang et al., 2019). Like most Src-like inactive kinases, SOBIR1 contains a basic residue (Lys512) at the third position downstream of the DFG motif (Roskoski, 2015) (Supplemental Figure 2). Unfortunately, the side chains of Lys512 and Lys377, which were supposed to form a salt bridge with the side chains of Glu407 and Asp507, respectively, in the αC-out configuration cannot be seen in the SOBIR1-KDD489A structure (Supplemental Figure 3A and 3C). In addition, the interaction between the main chain of Asp489 (replaced by Ala in our structure) and the side chain of Asn494 (Supplemental Figure 3B), detected in many Src-like kinases, helps to stabilize the inactive SOBIR1-KDD489A conformation (Roskoski, 2015; Wang et al., 2019).
To further dissect the inactive conformation of SOBIR1-KDD489A, we investigated the assembly of two functional protein kinase “spines”: the catalytic spine (C-spine) and the regulatory spine (R-spine) (Figure 2E and 2F). The integrity of these spines determines the active state of protein kinases and is also critical for the binding of ATP and the substrate (Kornev et al., 2006, 2008). The C-spine of SOBIR1-KDD489A consists of eight hydrophobic residues from the N- and C-lobes (Val361, Ala375, Leu496, Val495, Leu497, Leu444, Leu555, and Leu559) (Figure 2F). The DFG-in configuration of SOBIR1-KDD489A may facilitate the assembly of a C-spine with the AMP-PNP molecule. Specifically, the Val361, Ala375, and Leu496 side chains form a hydrophobic pocket that accommodates the adenine ring of AMP-PNP. By contrast, the R-spine of SOBIR1-KDD489A, comprising four non-consecutive hydrophobic residues, Phe508, His487, Val411, and Leu422, is broken by the outward-facing αC helix (Figure 2F). Unlike the other three residues that adopt a similar conformation in the active kinases, Val411 from αC is rotated away from the active site, disrupting the R-spine. These structural features confirm that SOBIR1-KDD489A adopts a catalytically inactive conformation.
The inactive SOBIR1-KD structure includes only one Mg2+ bound to the nucleotide-binding site, although the crystallization buffer contains a substantial amount of Mg2+ (Figure 2A). An active, phosphorylated protein kinase typically requires two Mg2+/Mn2+ ions to catalyze phosphoryl transfers (Bastidas et al., 2013; Jacobsen et al., 2012). The position of Mg2+ in the SOBIR1-KD structure is similar to the corresponding position in the inactive AMP-PNP-bound IRAK4 (PDB: 6EGF) and ATP-bound CDK2 (PDB: 1HCK) structures but differs from the corresponding position in BRI1 (PDB: 5LPV), which contains two Mn2+ (Schulze-Gahmen et al., 1996; Bojar et al., 2014; Wang et al., 2019) (Supplemental Figure 4). The Asp507 side chain faces the active site and interacts with Mg2+ in the structure, and the structure thus adopts a DFG-in conformation (Figure 2D). The Mg2+ ion associates with all three AMP-PNP phosphate groups, as well as the carbonyl group oxygen of the Ala493 main chain and the Asn494 side chain. The adenine purine group of AMP-PNP forms two hydrogen bonds with the main chains of the hinge residues Glu437 and Met439 (Figure 2G).
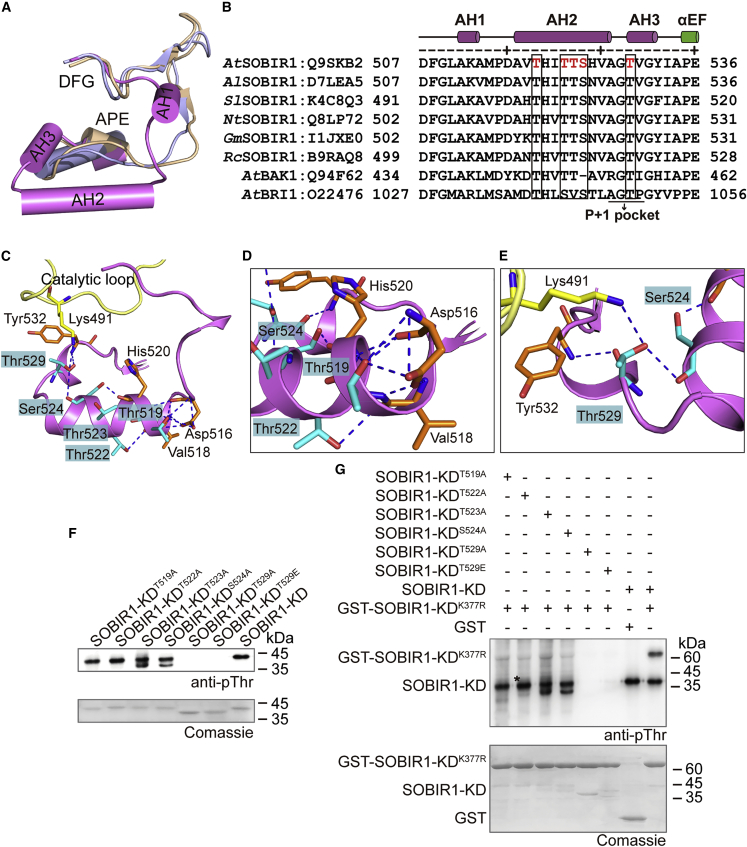
Thr529 is essential for SOBIR1 catalytic activity
In the SOBIR1-KDD489A structure, the residues of the activation segment (residues 507–536) are well defined in the electron density map and form three helices packed into the active site (Figure 3A and 3B). The region immediately downstream of the DFG motif is located at the beginning of the activation segment (residues 511–513) and assembles into a short α helix (activation segment helix 1 [AH1]), which is associated with the displaced αC helix, consistent with the Src-like inactive conformation of kinases (Levinson et al., 2006; Mikkola and Gahmberg, 2010; Roskoski, 2015; Wagner et al., 2015; Wang et al., 2019). Notably, the middle region of the SOBIR1-KD activation segment (residues 517–527) is compacted into a long α helix (activation segment helix 2 [AH2]), a conformation that is rarely observed in inactive kinase structures. To the best of our knowledge, only the KD of maize sucrose-induced receptor kinase 1 (PDB: 5UV4) has a similar activation loop conformation (Aquino et al., 2017). The last part of the activation segment (residues 529–532) also forms a short α helix (activation segment helix 3 [AH3]), corresponding to the part of the P+1 loop involved in substrate recognition (Madhusudan et al., 1994). Using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of wild-type SOBIR1-KD, we identified five phosphorylation sites in the activation segment (Thr519, Thr522, Thr523, Ser524, and Thr529) (Figure 3B, Supplemental Figure 5B and 5C). According to the structure, the first four sites are sequestered in AH2 and interact with the neighboring residues from the activation loop, and Thr519 is clamped by a pyramid-shaped interaction network formed by Asp516, Val518, and His520 (Figure 3C and 3D). This network is extended by Thr522 interacting with Val518 and Ser524 and by Ser524 forming hydrogen bonds with His520 and Thr529. The hydroxyl group of the Thr529 residue forms a hydrogen bond with the side-chain amino group of Lys491, which is located immediately downstream from the catalytic loop, indicating that Thr529 is probably involved in bridging the catalytic and activation loops to promote kinase activity. Thr529 also forms a polar contact with Tyr532 through its backbone amino group, which is located at the C terminus of AH3 (Figure 3C, 3E, and Supplemental Figure 2). It is worth noting that Tyr1052 in BRI1, corresponding to Tyr532 in SOBIR1, is essential for kinase activity in vitro and in vivo (Oh et al., 2009). These residues and interaction networks may be critical for stabilizing the inhibitory helical conformation of the activation loop, whereas the phosphorylation of the activation segment sites may disrupt these interactions, promoting an extended open-loop, the correct orientation of the catalytic bases, and the proper conformation of the αC helix.
Figure 3.
Analyses of the phosphorylation sites in the activation segment of SOBIR1-KD
(A) Comparison between the SOBIR1-KD activation segment and the BRI1 and BAK1 structures. SOBIR1, BRI1, and BAK1 are presented in magenta, beige, and light blue, respectively.
(B) Sequence alignment of the activation segments of SOBIR1 orthologs from different species as well as AtBAK1 and AtBRI1. The Swiss-Prot ID is provided after each protein name. The secondary structure elements of SOBIR1-KD are indicated above the sequences. Phosphorylation sites identified in AtSOBIR1-KD are highlighted in red. Conserved phosphorylation sites in the kinases are indicated in a black box. The P+1 pocket is labeled accordingly. At, A. thaliana; Al, Arabidopsis lyrata; Sl, S. lycopersicum; Nt, Nicotiana tabacum; Gm, Glycine max; Rc, Ricinus communis.
(C–E) View of the interaction networks of Thr519, Thr522, Ser524, and Thr529 in the SOBIR1-KD activation segment. The activation segment is presented in magenta, whereas the catalytic loop is in yellow. The phosphorylation sites identified by LC-MS/MS are labeled and presented in cyan, and the interacting residues are highlighted in orange (in the activation segment) or yellow (in the catalytic loop). Hydrogen bonds are presented as blue dashed lines. Detailed interactions of Thr519, Thr522, Ser524 (D), and Thr529 (E) are presented.
(F) Phosphorylation states of the activation segments of the SOBIR1-KD mutants.
(G) Western blot analysis of the transphosphorylation of SOBIR1-KDK377R by the SOBIR1-KD mutants. The reaction mixture contained 10 μM GST-SOBIR1-KDK377R and 1 μM SOBIR1-KD mutants. As a negative control, GST was incubated with wild-type SOBIR1-KD, and its phosphorylation signal was undetectable. All proteins in this assay were recombinantly expressed in E. coli. Asterisk (∗) represents a nonspecific band.
To investigate the functional significance of the identified phosphorylation sites, we conducted a series of site-directed mutagenesis experiments in which Thr519, Thr522, Thr523, Ser524, and Thr529 of SOBIR1-KD were individually replaced by Ala (phosphor-dead mutants) or by Glu (phosphor-mimic mutants). Immunoblotting using the anti-phospho-Thr antibody revealed that the autophosphorylation levels of SOBIR1-KDT519A, SOBIR1-KDT522A, SOBIR1-KDT523A, and SOBIR1-KDS524A were similar to that of the wild-type SOBIR1-KD. By contrast, replacement of Thr529 with either Ala or Glu produced a dominant-negative effect on the kinase activity of SOBIR1-KD (Figure 3F). To evaluate the contribution of individual phosphorylated residues to SOBIR1-KD kinase activity, we performed in vitro phosphorylation assays by incubating the phosphor-mutants with the kinase-dead SOBIR1-KDK377R in the presence of ATP-Mg2+, and the samples were further analyzed by immunoblotting using the anti-phospho-Thr antibody. The substitutions of Thr529 resulted in the complete loss of kinase activity and were not able to phosphorylate the kinase-dead SOBIR1-KDK377R, whereas the kinase activity of the other four substitutions was slightly lower than that of the wild-type SOBIR1-KD (Figure 3G), suggesting that phosphorylation at Thr529 is required for SOBIR1 kinase activity. Consistent with this result, the LC-MS/MS data revealed that only Thr529 in the activation segment was phosphorylated during the autophosphorylation of dSOBIR1-KD (Supplemental Table 2). The phosphorylation/dephosphorylation of Thr529 in the conserved P+1 pocket may modulate substrate specificity (Figure 3B). Previous sequence alignments indicated that this phosphorylation site may be highly conserved among the Arabidopsis LRR-RLKs, and 72.8% of LRR-RLKs have a Ser or Thr at the position equivalent to Thr529 in SOBIR1 (Wang et al., 2005). These results suggest that the phosphorylation of the highly conserved Thr529 is critical for SOBIR1 activation.
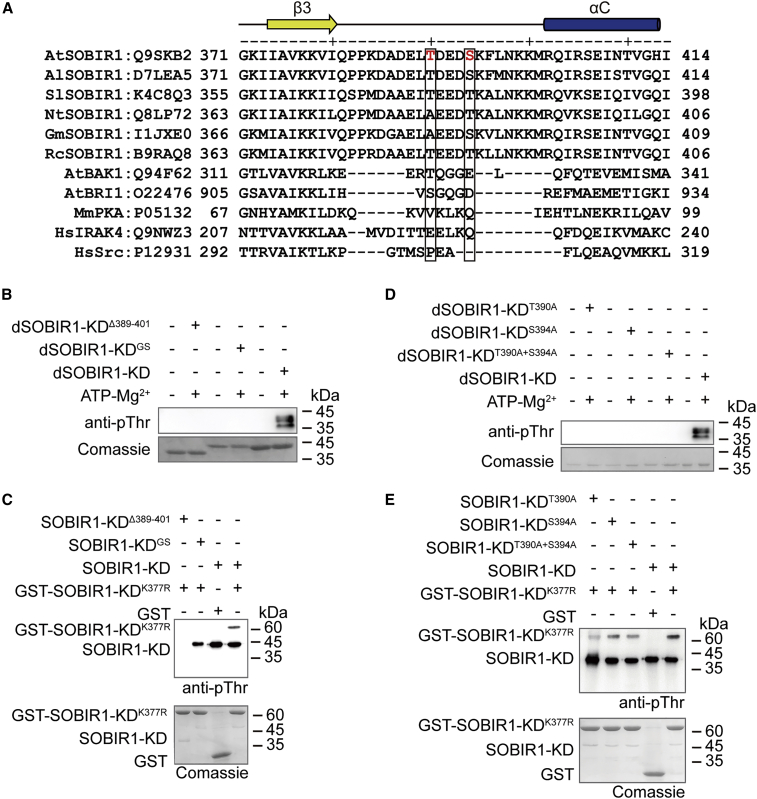
Thr390 and Ser394 in the β3-αC loop are important for SOBIR1 autophosphorylation
In the SOBIR1-KDD489A structure, the lack of electron density in the region between β3 and αC (383–399 in molecule A and 381–397 in molecule B) indicated the disordered nature of this segment (Supplemental Figure 1A). We therefore aligned the sequences of SOBIR1 orthologs, typical RLKs, and Src-like inactive kinases. As shown in Figure 4A, the length of the loop between β3 and αC (hereafter called the β3-αC loop) is conserved among the SOBIR1 orthologs, and this loop is significantly longer than that of the other kinases, suggesting that this region may have a specific function in the regulation of SOBIR1 kinase activity.
Figure 4.
The β3-αC loop of SOBIR1-KD is important for SOBIR1 autophosphorylation
(A) Sequence alignment of the β3-αC loop of SOBIR1 orthologs from different species and other kinases. The Swiss-Prot ID is provided after each protein name. The secondary structure elements of SOBIR1-KD are indicated above the sequences. Phosphorylation sites identified in AtSOBIR1-KD are highlighted in red. At, A. thaliana; Al, A. lyrata; Sl, S. lycopersicum; Nt, N. tabacum; Gm, G. max; Rc, R. communis; Mm, Mus musculus; Hs, Homo sapiens.
(B) Autophosphorylation of PP2Cα-dephosphorylated SOBIR1-KDΔ389-401 and SOBIR1-KDGS. Specifically, 25 μM PP2Cα-dephosphorylated proteins were incubated in the presence or absence of 1 mM ATP and 10 mM Mg2+ for 1 h.
(C) Western blotting analysis of the transphosphorylation of SOBIR1-KDK377R by the SOBIR1-KD mutants. The reaction mixture contained 10 μM GST-SOBIR1-KDK377R and 1 μM SOBIR1-KD mutants. As a negative control, GST was incubated with wild-type SOBIR1-KD, and its phosphorylation signal was undetectable.
(D) Autophosphorylation of PP2Cα-dephosphorylated SOBIR1-KDT390A, SOBIR1-KDS394A, and SOBIR1-KDT390A+S394A. Specifically, 25 μM PP2Cα-dephosphorylated proteins were incubated in the presence or absence of 1 mM ATP and 10 mM Mg2+ for 1 h. Reactions were terminated by boiling in 2× loading buffer, and samples were then analyzed by SDS-PAGE and western blotting.
(E) Western blotting analysis of the transphosphorylation of SOBIR1-KDK377R by the SOBIR1-KD mutants. The reaction mixture contained 10 μM GST-SOBIR1-KDK377R and 1 μM SOBIR1-KD mutants. As a negative control, GST was incubated with wild-type SOBIR1-KD, and its phosphorylation signal was undetectable.
To characterize the biochemical function of the β3-αC loop, we purified two recombinant mutants of SOBIR1-KD: one with the loop region deleted (residues 389–401) (SOBIR1-KDΔ389-401) and the other with the loop region replaced by 13 GS repeats (SOBIR1-KDGS). SEC analysis indicated that SOBIR1-KDΔ389-401 existed predominantly as a monomer, whereas SOBIR1-KDGS was heterogeneous, similar to the wild-type SOBIR1-KD (Supplemental Figure 6A). This result suggested that the β3-αC loop may affect the oligomerization state of SOBIR1. Moreover, phosphorylation of SOBIR1-KDΔ389-401 was completely abolished (Supplemental Figure 6B, lane 2), whereas the phosphorylation of SOBIR1-KDGS (Supplemental Figure 6B, lane 3) was slightly weaker than that of the wild-type SOBIR1-KD (Supplemental Figure 6B, lane 1). These results suggested that the β3-αC loop is required for the autophosphorylation of SOBIR1.
To further elucidate the functional importance of the β3-αC loop, we performed crystallization with recombinant SOBIR1-KDΔ389-401. The SOBIR1-KDΔ389-401 structure was determined at a 2.9-Å resolution by molecular replacement with SOBIR1-KDD489A as the search model (Supplemental Figure 6C and 6D). Compared with SOBIR1-KDD489A, SOBIR1-KDΔ389-401 had a similar overall structure (RMSD = 0.442 Å), suggesting that deletion of the β3-αC loop did not significantly perturb the overall structure of SOBIR1-KD (Supplemental Figure 6E). Next, we performed in vitro autophosphorylation assays of PP2Cα-dephosphorylated SOBIR1-KDΔ389-401 (dSOBIR1-KDΔ389-401) and SOBIR1-KDGS (dSOBIR1-KDGS). The samples were then analyzed by western blotting using the general anti-phospho-Thr antibody. Unlike the wild-type dSOBIR1-KD (Figure 4B), neither dSOBIR1-KDΔ389-401 (Figure 4B, lane 2) nor dSOBIR1-KDGS (Figure 4B, lane 4) was able to undergo ATP-initiated autophosphorylation. In addition, SOBIR1-KDΔ389-401 and SOBIR1-KDGS were unable to phosphorylate the kinase-dead SOBIR1-KDK377R, suggesting that the kinase activity of SOBIR1 requires the β3-αC loop (Figure 4C).
LC-MS/MS analysis detected two phosphorylated residues, Thr390 and Ser394, in the β3-αC loop of wild-type SOBIR1-KD but not in the dephosphorylated protein (Supplemental Table 2; Supplemental Figure 7A and 7B). Similar to Thr529, both Thr390 and Ser394 were phosphorylated when dSOBIR1-KD was incubated with ATP/Mg2+ (Supplemental Table 2; Supplemental Figure 7C and 7D). We speculated that one or both of Thr390 and Ser394 might participate in the activation of SOBIR1. To verify this hypothesis, we generated SOBIR1-KDT390A, SOBIR1-KDS394A, and SOBIR1-KDT390A+S394A mutants and dephosphorylated them with PP2Cα. We then performed in vitro autophosphorylation assays by incubating dephosphorylated dSOBIR1-KDT390A, dSOBIR1-KDS394A, and dSOBIR1-KDT390A+S394A in the presence of 1 mM ATP and 10 mM Mg2+. Western blotting analysis indicated that the Thr390 and Ser394 mutations resulted in a complete loss of autophosphorylation activity (Figure 4D). Interestingly, the kinase-dead SOBIR1-KDK377R was still phosphorylated by SOBIR1-KDT390A, SOBIR1-KDS394A, and SOBIR1-KDT390A+S394A, although to a lesser extent than the wild-type SOBIR1-KD (Figure 4E). These observations demonstrated that Thr390 and Ser394 in the β3-αC loop are essential for SOBIR1 autophosphorylation but are not crucial for the kinase activity of SOBIR1-KD.
Reciprocal phosphorylation between SOBIR1 and BAK1 can enhance SOBIR1 activity
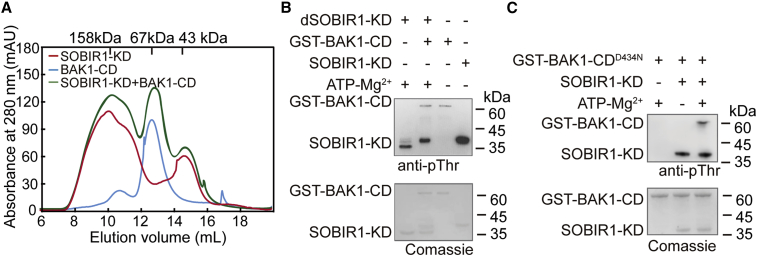
Previous research has shown that kinase-active BAK1 is essential for many SOBIR1-induced constitutive immune responses and contributes to the transphosphorylation of SOBIR1 in vivo (Albert et al., 2019; van der Burgh et al., 2019; Wu et al., 2019). To further investigate the mechanism underlying the reciprocal phosphorylation of SOBIR1-KD and BAK1-CD, we constructed the cytosolic domain of AtBAK1 (BAK1-CD, residues 255–615) with an N-terminal glutathione S-transferase (GST) tag and the corresponding kinase-dead mutant BAK1-CDD434N. SOBIR1-KD was incubated with BAK1-CD to investigate whether BAK1-CD could influence the heterogeneous status of SOBIR1-KD. SEC analysis showed that SOBIR1-KD was still able to form oligomers in the presence of BAK1-CD in vitro, suggesting that the oligomerization of SOBIR1 was due to the unique β3-αC loop and that ligand-induced BAK1 recruitment may have little effect on the formation of SOBIR1 oligomers (Figure 5A). We next incubated BAK1-CD with dSOBIR1-KD in the presence of 1 mM ATP and 10 mM Mg2+. Compared with SOBIR1 autophosphorylation (Figure 5B, lane 1), adding BAK1-CD increased the phosphorylation level of SOBIR1-KD (Figure 5B, lane 2), indicating that transphosphorylation by BAK1-CD was capable of increasing SOBIR1 phosphorylation. In addition to the autophosphorylation site Thr529, the other four residues in the activation segment of SOBIR1-KD (Thr519, Thr522, Thr523, and Ser524) could also be phosphorylated, as demonstrated by LC-MS/MS analysis (Supplemental Table 2; Supplemental Figure 5E and 5F). These results suggested that BAK1-CD could increase the phosphorylation of SOBIR1-KD, consistent with the importance of BAK1 for SOBIR1-induced cell death and constitutive immunity in planta (Albert et al., 2015; Albert et al., 2019; Postma et al., 2016; van der Burgh et al., 2019; Wu et al., 2019; Zhang et al., 2013). We also examined the phosphorylation of BAK1-CD by SOBIR1-KD by incubating the wild-type SOBIR1-KD with the kinase-dead BAK1-CDD434N in the presence of 1 mM ATP and 10 mM Mg2+. Western blotting and LC-MS/MS data indicated that SOBIR1-KD could phosphorylate BAK1-CD at Thr446, Thr449, Thr450, and Thr455, which are located in the activation segment and had previously appeared to be crucial for BAK1 kinase activity (Figure 5C and Supplemental Figure 8; Supplemental Table 2). Thus, consistent with previous reports, our data suggested that the reciprocal phosphorylation of SOBIR1 and BAK1 is important for their activation.
Figure 5.
Reciprocal phosphorylation between SOBIR1 and BAK1
(A) Determination of the oligomerization states of SOBIR1-KD, BAK1-CD, and SOBIR1-KD incubated with BAK1-CD via SEC. The reaction mixture contained 30 μM SOBIR1-KD and 10 μM GST-BAK1-CD. Elution volumes of the protein standards are indicated.
(B) Western blotting analysis of the transphosphorylation of PP2Cα-dephosphorylated SOBIR1-KD by wild-type BAK1-CD. The reaction mixture contained 10 μM dSOBIR1-KD and 1 μM GST-BAK1-CD.
(C) Western blotting analysis of the transphosphorylation of BAK1-CDD434N by wild-type SOBIR1-KD. The reaction mixture contained 10 μM GST-BAK1-CDD434N and 1 μM SOBIR1-KD.
Thr529 and the αC-βC loop are indispensable for SOBIR1 activity in planta
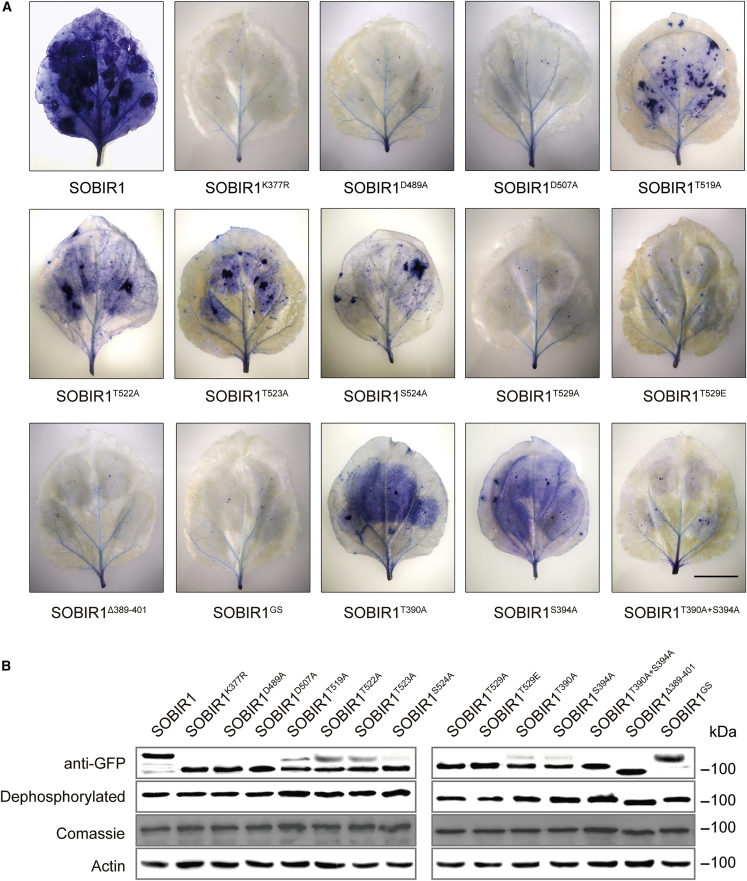
SOBIR1 functions as a positive regulator of immune responses, and overexpression of AtSOBIR1 triggers constitutive immune induction, including cell death (Wu et al., 2018b; van der Burgh et al., 2019). To assess the functional importance of critical segments and phosphorylation sites identified from the SOBIR1-KD crystal structure, we generated eGFP-tagged wild-type SOBIR1 from A. thaliana and various mutants, including SOBIR1K377R, SOBIR1D489A, SOBIR1D507A, SOBIR1T519A, SOBIR1T522A, SOBIR1T523A, SOBIR1S524A, SOBIR1T529A, SOBIR1T529E, SOBIR1T390A, SOBIR1S394A, SOBIR1T390A+S394A, SOBIR1Δ389-401, and SOBIR1GS. These constructs were transiently expressed in N. benthamiana using Agrobacteria-mediated transformation.
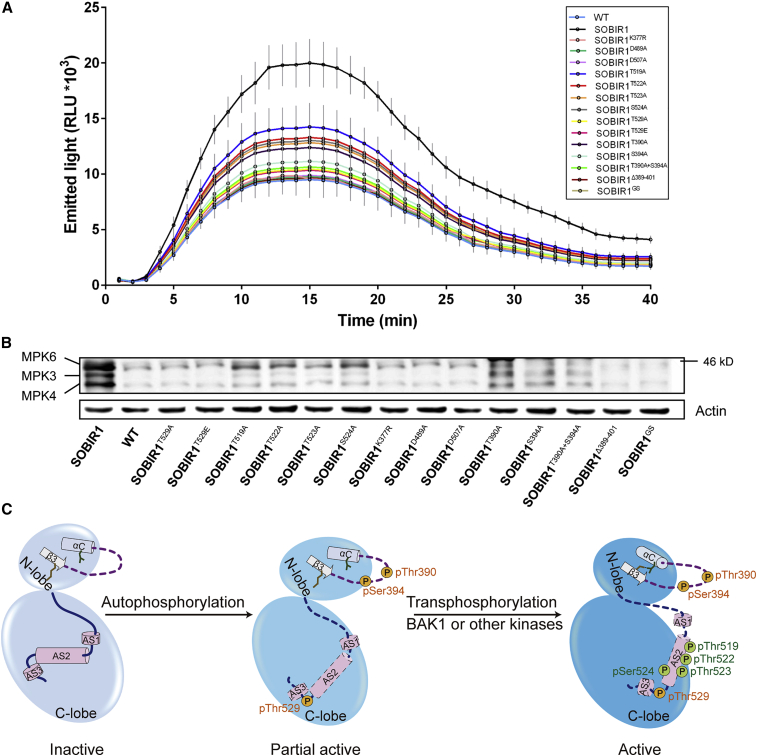
We found that obvious cell death could be induced by expressing the wild-type SOBIR1 in N. benthamiana but not by expressing the kinase-dead SOBIR1K377R, SOBIR1D489A, and SOBIR1D507A (Figure 6A and Supplemental Figure 9). Western blotting analyses indicated that the expression level of each mutant in N. benthamiana leaves was comparable to that of the wild-type SOBIR1 (Figure 6B). To further assess whether mutations in SOBIR1 affected downstream immune responses, we analyzed the reactive oxygen species (ROS) production and MAPK activation elicited by flg22, a peptide derived from bacterial flagellin that acts as a pathogen-associated molecular pattern (PAMP). As shown in Figure 7, flg22-induced ROS production and activation of MPK3, MPK4, and MPK6 were significantly increased in SOBIR1-overexpressing N. benthamiana. By contrast, flg22-induced immune responses were compromised when the catalytically inactive variants SOBIR1K377R, SOBIR1D489A, and SOBIR1D507A were expressed in N. benthamiana. This observation was consistent with the fact that SOBIR1 kinase activity is essential for its function (van der Burgh et al., 2019). More importantly, it validates the functional importance of Lys377, Glu489, and Glu507 in SOBIR1-induced cell death and flg22-induced immune activation.
Figure 6.
Trypan blue staining of SOBIR1 variants expressed in N. benthamiana
(A)AtSOBIR1 and various mutants fused to C-terminally enhanced green fluorescent protein were transiently expressed in N. benthamiana by Agrobacterium-mediated transient expression (agroinfiltrations). Pictures of representative leaves stained with trypan blue exhibit levels of cell death. All assays were performed three times, and a representative photograph is shown. Scale bar, 2 cm.
(B) All infiltrated N. benthamiana leaves were collected after phenotype analysis, mixed with 2× loading buffer (v/v), and boiled at 95°C for 15 min. Expression levels of all proteins were then checked by western blotting with anti-GFP antibody.
Figure 7.
Analysis of ROS production and MAPK activation mediated by SOBIR1 and its mutant variants using transient expression assays in N. benthamiana
(A and B) flg22-induced ROS burst in N. benthamiana expressing SOBIR1 and its mutant variants. AtSOBIR1 and all mutant variants were transiently expressed in leaves of N. benthamiana by agroinfiltration. After 2 days, oxidative burst and MAPK activation were induced by treatment with 300 nM flg22. ROS production over time was measured by a luminol-based assay after flg22 treatment (A). flg22-induced MAPK activation was measured by immunoblotting with anti-ERK 15 min after flg22 treatment (B). Error bars, ±SD of six replicates. Wild-type represents N. benthamiana without SOBIR1 expression and was used as a negative control. The actin level was used as the loading control. All experiments were performed in triplicate with similar results.
(C) Model of SOBIR1-KD stepwise activation. Inactive SOBIR1 is not phosphorylated. The β3-αC loop is disordered and the Lys-Glu salt bridge is broken because of the outward-facing αC helix (αC-out). The activation segment uniquely forms three helices without any phosphorylated sites (left). Ligand recognition induces conformational changes to SOBIR1, leading to SOBIR1 autophosphorylation. In addition, Thr390 and Ser394 in the βC-αC loop and Thr529 in the activation segment are phosphorylated during this process, and SOBIR1 is partially active (middle). BAK1 or other kinases then transphosphorylate SOBIR1 at Thr519, Thr522, Thr523, and Ser524 to fully activate SOBIR1. During the activation, these phosphorylation events re-position the helices within the activation segment and enable the αC helix to swing inward toward the ATP-binding site (right). Finally, the SOBIR1-KD adopts a fully active conformation. Specifically, the activation segment is extended with phosphorylated sites to provide a platform for protein substrate binding and the induction of downstream signaling.
We also investigated the functional significance of the activation segment residues Thr519, Thr522, Thr523, Ser524, and Thr529. Consistent with our structural and phosphorylation analyses, amino acid substitutions in Thr529 that essentially disrupted SOBIR1 activation resulted in a complete loss of cell death, accompanied by marked reductions in ROS production and MAPK activation after flg22 treatment, similar to observations for SOBIR1K377R, SOBIR1D489A, and SOBIR1D507A (Figures 6A and 7). However, overexpression of SOBIR1T519A, SOBIR1T522A, SOBIR1T523A, and SOBIRS524A resulted in reduced cell death compared with wild-type SOBIR1 but higher flg22-triggered ROS production and MAPK activation compared with SOBIR1T529A. To test the importance of the β3-αC loop, we overexpressed SOBIR1Δ389-401 and SOBIR1GS and found that they were also essential for SOBIR1-induced cell death and immune signaling, supporting the functional importance of this segment for SOBIR1 activation (Figures 6A and 7). Individually mutating the two phosphorylation sites (Thr390 and Ser394) in the β3-αC loop still caused constitutive cell death, ROS burst, and MAPK activation, although these responses were compromised compared with those obtained with wild-type SOBIR1. However, the double mutant SOBIR1T390A+S394A exhibited almost complete loss of SOBIR1 activity. Therefore, these results further support the essential role of Thr529 and the β3-αC loop in SOBIR1-mediated cell death and PAMP-triggered immune response (Figure 6A).
Discussion
Plant RLK SOBIR1 is essential for plant signaling pathways (Albert et al., 2019; Domazakis et al., 2018; Takahashi et al., 2018; van der Burgh et al., 2019; Wu et al., 2019). Overexpression of SOBIR1 induces cell death in both Arabidopsis and N. benthamiana, suggesting that SOBIR1 is a positive regulator of cell death (Gao et al., 2009). Previous research showed that the kinase-dead AtSOBIR1D498N suppresses autoimmunity, indicating that SOBIR1 kinase activity is required for downstream signaling (Liebrand et al., 2013). Similar to many kinases, SOBIR1-KD is activated by phosphorylation of the activation segment, and the initial phosphorylation events involve autophosphorylation (Beenstock et al., 2016; Ferrao et al., 2014; Lavoie et al., 2014; Mayo et al., 2019; Steichen et al., 2012). AtSOBIR1 is able to constitutively form homodimers independent of its kinase activity in planta, suggesting that AtSOBIR1 may undergo autophosphorylation via an intermolecular mechanism (van der Burgh et al., 2019). Consistent with this notion, the biochemical data presented here confirm that AtSOBIR1 trans-autophosphorylates in vitro. However, western blotting analysis showed that the autophosphorylation of dSOBIR1-KD was always weaker than the phosphorylation of wild-type SOBIR1-KD, suggesting that full activation of SOBIR1-KD cannot be achieved by autophosphorylation alone and may require the involvement of other kinases (Figure 1D). In many RLP/SOBIR1-mediated signaling pathways, BAK1 and other SERK family members are also required (Albert et al., 2015; Albert et al., 2019; Liebrand et al., 2014; Postma et al., 2016; van der Burgh et al., 2019; Wu et al., 2019). For example, during RLP Cf-4-mediated immunity, transient overexpression of the AtBAK1 variants AtBAK1C408Y and AtBAK1D416N, which contain mutations in the BAK1-KD, impaired AtSOBIR1 activity, suggesting that SOBIR1 functions depend on kinase-active BAK1 (van der Burgh et al., 2019). Interestingly, SOBIR1-KD still forms oligomers in the presence of BAK1-KD in vitro, suggesting that ligand-induced BAK1 recruitment may have little effect on the formation of SOBIR1 oligomers (Figure 5A). On the other hand, our study verified the reciprocal phosphorylation between SOBIR1 and BAK1 in vitro and further identified the reciprocal phosphorylation sites on both kinases. Based on our results and previous studies, we propose that SOBIR1 is activated via autophosphorylation and sequential transphosphorylation by BAK1 (Figure 7C). In the absence of ligands, SOBIR1 forms hetero-multimers with RLPs through the GxxxG dimerization motif in the TM domain. The cytoplasmic KD adopts an Src-like inactive conformation, and the activation segment forms impact helices without any phosphorylation. The perception of ligands induces conformational changes that initiate the autophosphorylation of SOBIR1. Specifically, Thr390 and Ser394 in the β3-αC loop and Thr529 in the activation segment are phosphorylated during this process. Then the low phosphorylation status of SOBIR1 that has limited kinase activity recruits BAK1 and possibly also other kinases to form complexes in which reciprocal phosphorylation occurs, resulting in a fully activated SOBIR1. These phosphorylation events re-position the helices of the activation segment and enable the αC helix to swing inward toward the active site. Finally, the fully phosphorylated SOBIR1-KD adopts an active conformation in which the activation segment is extended to provide a platform for protein substrate binding and the induction of downstream signaling.
Eukaryotic protein kinases operate in a large number of distinct signaling pathways, where the tight regulation of their activation and inactivation are crucial to both normal cellular functions and abnormal phenotypes (Johnson et al., 1996; Huse and Kuriyan, 2002; Kornev et al., 2006; Rabiller et al., 2010; Beenstock et al., 2016). Kinases are usually kept off, and the acquisition of catalytic activity is often buried under multiple layers of control. It is necessary to understand both the active and the inactive states to unravel the phosphorylation-dependent regulation mechanism of protein kinases. Many kinase structures have now been solved, suggesting that the overall protein kinase fold is extremely well conserved. However, protein kinases are highly dynamic molecules that undergo large conformational changes within the active site when toggling between active and inactive states. It is worth noting that all active kinases converge to a relatively conserved conformation and possess conserved structural features, including DFG-in and αC-in configurations, assembled hydrophobic R- and C-spines, and similar conformations of the activation segment (Kornev et al., 2006, 2008). By contrast, the conformations of inactive kinases can vary significantly, as they are not subject to the chemical constraints that the active states must satisfy. Until recently, the structures of important plant RLKs such as BRI1 and BAK1 were all determined in their active conformations, which can be regarded as prototypes to illustrate SOBIR1 structure following activation (Gao et al., 2012; Bojar et al., 2014; Wang et al., 2014). Compared with those of other kinases, the unique sequential and structural features of inactive SOBIR1 revealed that the regulation of its activation may depend on phosphorylation of the β3-αC loop and the activation segment, which probably results in a switch from αC-out to αC-in conformations and an extended open activation loop. Hence, the structure of the kinase-dead version of SOBIR1 may deepen our understanding of the ligand-free inactive state of this important kinase and provide a basis for studying the phosphorylation-dependent regulation of SOBIR1 activity.
Unphosphorylated SOBIR1-KD adopts the Src-like inactive conformation observed in the structures of several mammalian kinases, including Src and IRAK4 (Wang et al., 2019; Xu et al., 1999). Similar to the AMP-PNP-bound inactive IRAK4 structures, the non-hydrolyzable ATP analog AMP-PNP in our structures adopts a catalytically incompetent configuration (Wang et al., 2019). A comparison between unphosphorylated SOBIR1-KD and Src-like inactive kinases revealed a common outward-facing αC helix (αC-out) and side-chain movement of the Asp507 residue toward the single Mg2+ ion (DFG-in) (Figure 2C and 2D). The conformation of Src-like inactive kinases is stabilized by conserved interaction networks. For example, in the structure of inactive Src kinase complexed with AMP-PNP (PDB: 2SRC), salt bridges are formed between Lys295 in β3 and Asp404 in the DFG motif, Glu310 in αC and Arg409 in the activation segment, and Asp386 in the HRD motif and Asn391 at the loop before β7 (Xu et al., 1999). Similarly, Asp489 interacts with Asn494, and the interaction between these residues is maintained even though Asp489 in the HRD motif is replaced by Ala in our structure (Supplemental Figure 3B). In our structural analysis, this interaction was confirmed to help stabilize the inactive conformation of SOBIR1-KD. The side chains of the corresponding residues Lys377 and Lys512 are not built in the SOBIR1-KDD489A structure because the resolution is too low (Supplemental Figure 3A and 3C). To the best of our knowledge, almost all resolved plant receptor kinase CDs (e.g., BRI1 and BAK1) are in the active conformation (i.e., αC-in and DFG-in) (Yan et al., 2012; Bojar et al., 2014; Wang et al., 2014). Unlike our inactive structures, the activation loops in BRI1 and BAK1 adopt extended configurations and possess multiple phosphorylated residues (Yan et al., 2012; Bojar et al., 2014; Wang et al., 2014). The Src-like kinases have a conserved basic residue at the third position after the DFG motif, and this residue is thought to interact with the Glu in the αC helix to stabilize the inactive conformation (Roskoski, 2015; Wang et al., 2019). Similarly, BRI1 and BAK1 also possess a basic residue (Arg1032 in BR1 and Lys439 in BAK1) (Supplemental Figure 2). Thus, we speculate that they also adopt an Src-like conformation in their inactive states.
In contrast to other reported structures, the activation segments in our structures adopted an unusual architecture comprising three helices (AH1, AH2, and AH3) (Figure 3C). Of these helices, AH1 is conserved in the Src-like inactive conformational kinases but is converted to a β sheet in the active structure of BRI1-CD (Bojar et al., 2014). In addition, AH2 adopts an unusual helical architecture with four potential phospho-residues followed by Thr529 in the conserved P+1 pocket, which is important for substrate recognition and forms a part of AH3. None of these active sites is fully formed in SOBIR1-KD. The activation segment helices occlude the substrate-binding sites and are stabilized by the interaction networks formed by the potential phospho-residues (Thr519, Thr522, Ser524, and Thr529) and other interacting residues (Lys419, Asp516, Val518, His520, and Tyr532) (Figure 3C–3E). Among the plant RLKs, Ser524 is conserved in SOBIR1 orthologs. The corresponding site (Ser346) also exists in the mammalian IRAK4 activation loop but contributes little to maintain the active conformation of phosphorylated IRAK4-KD (Ferrao et al., 2014). In the active BRI1, the corresponding site (Ser1044) is phosphorylated and inserted into the positively charged phosphate-binding pocket formed by Arg922, Arg1008, and Arg1032 (Wang et al., 2014). In the BAK1 crystal structure, this phosphate-binding pocket formed by positively charged Arg415, Lys439, and Arg453 is associated with the phosphate group of Thr450, which corresponds to Thr523 in SOBIR1 (Yan et al., 2012). In the SOBIR1-KD structure, the electrostatic potentials indicate that there is no positively charged pocket surrounding potential phospho-residues, probably because of the inactive conformation and the helical activation segment. Sequence alignments revealed that two corresponding sites in SOBIR1 are conserved basic residues (Arg488 in the HRD motif and Lys512 in the activation segment) (Supplemental Figure 2), implying that SOBIR1-KD probably forms a positively charged pocket to stabilize the phosphate group in the active conformation. The Thr529 residue is locked by the Lys491-Ser524-Tyr532 network, whereas the corresponding phosphorylated Thr455 in BAK1 interacts with Gly457, His458, and Lys418 through its phosphate group in the active conformation to further stabilize the activation segment and enhance kinase activity (Yan et al., 2012). Interestingly, the corresponding Thr1049 in BRI1 is not phosphorylated in the active conformation, but it is phosphorylated in planta, suggesting that phosphorylation/dephosphorylation of this site may control the substrate specificity of RLKs (Wang et al., 2008; Bojar et al., 2014). These structural features stabilize the inactive conformation, and the phosphorylation of these potential phospho-residues may promote the formation of a catalytically active structure and thus activate SOBIR1.
In this study, the β3-αC loop was shown to be important for SOBIR1 activation, a feature unique among known plant RLKs. Sequence alignment analysis showed that the β3-αC loop is longer than those in other RLKs but is conserved among SOBIR1 orthologs from various plant species. In some mammalian kinases, the β3-αC loop is a crucial determinant of basal kinase activity. For example, in BRAF, EGFR, and HER2, shortening the β3-αC loop activated these kinases to induce cancers; this activation may be due to the αC helix favoring the αC-in conformation (Foster et al., 2016). However, compared with other family members, ERK1/2 has a relatively short β3-αC loop, resulting in limited autophosphorylation. Accordingly, the activation of ERK1/2 relies on the MEK kinase as an upstream activator (Sang et al., 2019). Inserting an amino acid in the loop of ERK1/2 resulted in full autoactivation in the absence of MEK. In addition, the intramolecular binding between the β3-αC loop residue Ala228 and the hydrophobic pocket of the SH2 domain in Csk is essential for protein activation (Mikkola and Gahmberg, 2010). These observations suggest that the β3-αC loop has an essential role in the regulation of kinase activity. We determined that Thr390 and Ser394 in the β3-αC loop are essential for SOBIR1 autophosphorylation (Figure 4D and 4E). In addition, functional studies also revealed that phosphorylation of both Thr390 and Ser394 is important for SOBIR1 activity (Figure 6A and Supplemental Figure 9), suggesting that the autophosphorylation of SOBIR1-KD may be the first step in SOBIR1 activation. Thr390 in SOBIR1 is conserved in BAK1 but is replaced by a Ser in BRI1, whereas Ser394 is replaced by the phospho-mimicking residues Asp in BAK1 and Glu in BRI1 (Figure 4A).
Sequence alignments and structural comparisons among SOBIR1-KD, BRI1-CD, and BAK1-CD suggested that their regulatory mechanisms of kinase activation may differ significantly. For example, Thr872 and Thr880 in BRI1, which correspond to Thr278 and Ser286 in BAK1, are highly conserved among Arabidopsis LRR-RLKs. Previous studies have shown that phosphorylation of these residues adversely affects the activity of BAK1 and BRI1 (Wang et al., 2005, 2008). Notably, the corresponding residues in SOBIR1 are Glu336 and Glu344, which generally mimic phosphorylation (Supplemental Figure 2). Sequence alignments also revealed that the Tyr463 in BAK1 and the Tyr1057 in BRI1 are conserved, but they are replaced by Phe537 in SOBIR1 (Supplemental Figure 2). Tyr463 in BAK1 is essential for the catalytic activity of the enzyme, and the substitution of this residue by Phe substantially inhibited phosphorylation (Oh et al., 2010). Therefore, the regulatory mechanism of SOBIR1 and the means by which it activates plant immunity may differ significantly from those of the well-studied BAK1, thus offering new mechanistic insights into phosphorylation-dependent RLK activation.
In summary, we propose a model for phosphorylation-dependent SOBIR1-KD activation in which SOBIR1-KD is autophosphorylated via an intermolecular mechanism, then transphosphorylated by activated BAK1. Our structural and functional studies determined that Thr529 in the activation segment and the β3-αC loop are indispensable for the activation of SOBIR1. Our data may be useful for further elucidating the regulatory mechanisms of SOBIR1 and clarifying how SOBIR-type regulators contribute to LRR-RLP-mediated cell death and signaling pathways associated with immunity. In the future, it will be interesting to determine the mechanism by which the β3-αC loop and activation segment stabilize the active conformation of SOBIR1-KD and to further investigate how RLPs trigger the activation of SOBIR1.
Methods
Cloning, expression, and purification
The coding sequences of SOBIR1-KD (residues 326–641) and BAK1-CD (residues 255–615) were amplified from A. thaliana cDNA. The SOBIR1-KD coding sequence was cloned into the pET15b vector (Novagen, Temecula, CA, USA) for the production of a fusion protein with an N-terminal His6-tag or into a modified pGEX-6P-1 vector (Novagen, Temecula, CA, USA) for the production of a fusion protein with an N-terminal GST-tag. The BAK1-CD coding sequence was cloned into the pGEX-6P-1 vector. All mutations in SOBIR1-KD and BAK1-CD were generated by the standard Quickchange PCR procedure and verified by DNA sequencing. All proteins were overexpressed in Escherichia coli BL21(DE3) cells (TransGen Biotech, Beijing, China) for 14 h at 20°C and then purified at 4°C by affinity chromatography with Ni-NTA (Qiagen, Hilden, Germany) and GST (Qiagen) columns, followed by ion-exchange chromatography (Source 15Q, GE Healthcare Life Sciences, Issaquah, WA, USA). Proteins were further concentrated and subjected to gel filtration chromatography (Superdex 200 HR 10/30 column, GE Healthcare Life Sciences). They were eluted in a buffer containing 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 2 mM dithiothreitol (DTT). The purified proteins were stored at −80°C. The proteins for the kinase assays were supplemented with 20% (v/v) glycerol. Protein concentrations were determined with a spectrophotometer (SpectraMax 190, Molecular Devices) and the theoretical molar extinction coefficients at 280 nm (Gill and von Hippel, 1989).
SEC
The homogeneity of wild-type SOBIR1-KD, dephosphorylated SOBIR1-KD, and mutated SOBIR1-KDK377R, SOBIR1-KDD489A, and SOBIR1-KDD507A were evaluated by gel filtration using a Superdex 200 column that was pre-equilibrated with a buffer comprising 10 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 2 mM DTT. Specifically, 1-ml aliquots (approximately 20 μM) of the isolated proteins were loaded onto the column sequentially and then monitored based on the absorbance at 280 nm, with an elution flow rate of 0.5 ml min−1. SOBIR1-KD, SOBIR1-KDΔ389-401, and SOBIR1-KDGS were analyzed similarly. All proteins were diluted to 2 μM and then mixed with 2× loading buffer (v/v) (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 200 mM DTT, and 0.1% bromophenol blue), boiled at 95°C for 5 min, and then analyzed by western blotting with an anti-phospho-Thr antibody to assess the phosphorylation state of each protein.
Kinase assays and western blotting
PP2Cα-dephosphorylated SOBIR1-KD was generated by incubating 20 μM wild-type SOBIR1-KD and 2 μM GST-PP2Cα in kinase reaction buffer (50 mM MOPS [pH 7.0], 100 mM NaCl, 10 mM MgCl2, and 0.2 mM EDTA) at 25°C for 1 h. The samples were then mixed with 2× loading buffer (v/v), boiled at 95°C for 5 min, and analyzed by western blotting with an anti-phospho-Thr antibody to assess the phosphorylation state of each protein.
The autophosphorylation of PP2Cα-dephosphorylated SOBIR1-KD was evaluated by incubating 1, 5, and 25 μM dephosphorylated SOBIR1-KD in reaction buffer (50 mM MOPS [pH 7.0], 100 mM NaCl, 10 mM MgCl2, 0.2 mM EDTA, and 1 mM ATP) at 25°C for 2, 5, 15, 30, 60, 240, and 420 min. All reagents were obtained from Sigma (St Louis, MO, USA). Samples were collected at specific time points, mixed with 2× loading buffer (v/v), and boiled at 95°C for 5 min. All samples were then analyzed by western blotting with anti-phospho-Thr antibody.
In addition, 10 μM GST-SOBIR1-KDK377R was incubated with 1 μM wild-type SOBIR1-KD or its variants in the same reaction buffer at 25°C for 1 h. The phosphorylation of SOBIR1-KDK377R was analyzed by western blotting with anti-phospho-Thr antibody.
The transphosphorylation of SOBIR1-KD by BAK1-CD was analyzed by incubating 10 μM kinase-dead SOBIR1-KDK377R and 1 μM BAK1-CD in the same reaction buffer at 25°C for 1 h. The reaction was terminated by adding 2×loading buffer (v/v) and boiling at 95°C for 5 min. The transphosphorylation of BAK1-CDD434N by SOBIR1-KD was examined by incubating 10 μM kinase-dead BAK1-CDD434N and 1 μM SOBIR1-KD in the same reaction buffer at 25°C for 1 h. The samples were then analyzed by western blotting with anti-phospho-Thr antibody to examine the phosphorylation state of each protein.
All protein samples were separated in a 12% SDS-PAGE gel and stained with Coomassie Brilliant Blue. For western blotting, proteins were transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA), which were then immersed in a 5% BSA (prepared in TBST buffer) blocking solution. The membranes were incubated with anti-phospho-Thr antibody (1:5000; Cell Signaling Technology, Beverly, MA, USA) at 4°C for 12 h, and then they were incubated with a peroxidase-conjugated goat anti-mouse antibody (1:2000; Abcam, Cambridge, UK) at room temperature for 1 h. Finally, the phosphorylation states of the proteins were monitored with the ECL chemiluminescence blotting substrate (Millipore).
ATPase activity
The ATPase activity of SOBIR1-KD was measured with a spectrophotometer, using ATP as the substrate. The assay, which coupled the production of ADP to the oxidation of NADH (Roskoski, 1983), was performed by adding 50 nM SOBIR1-KD to 1.8 ml of reaction solution (50 mM MOPS [pH 7.0], 100 mM NaCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mM NADH, 1.0 mM phosphoenolpyruvate, 15 units ml−1 pyruvate kinase, 20 units ml−1 lactate dehydrogenase, and different amounts of ATP) and incubating at 25°C for 5 min. The reaction was monitored continuously at 340 nm with a Lambda 45 spectrophotometer (PerkinElmer Life Sciences, Boston, MA, USA). The formation of ADP was measured based on an extinction coefficient of 6220 cm−1 M−1 for NADH, and the initial reaction rates were determined according to the linear slope of the progress curves. The kcat and Km values were determined with the Michaelis-Menten equation and a nonlinear regression analysis program.
Crystallization, X-ray diffraction, data collection, and structural characterization
Both SOBIR1-KDD489A (6 mg ml−1) and SOBIR1-KDΔ389-401 (8 mg ml−1) were incubated with 1 mM AMP-PNP and 10 mM MgCl2 at a 10:1 molar ratio on ice for 1 h and then subjected to crystallization trials. Crystals were grown according to the hanging drop vapor diffusion method. Specifically, the proteins were mixed with an equal volume of crystallization buffer at 20°C. The SOBIR1-KDD489A-AMP-PNP-Mg2+ crystals were obtained from crystallization buffer containing 0.1 M Bis-tris propane (pH 8.15), 18% PEG 8000, 3% MPD, and 8% glycerol. The SOBIR1-KDΔ389-401-AMP-PNP-Mg2+ crystals were obtained from crystallization buffer containing 0.1 M Hepes (pH 7.0) and 20% PEG 8000. All reagents were purchased from Hampton Research (Aliso Viejo, CA). The crystals were instantaneously frozen in liquid nitrogen. The diffraction datasets for SOBIR1-KDD489A and SOBIR1-KDΔ389-401 were collected at beamlines 19U and 17U, respectively, at the Shanghai Synchrotron Radiation Facility (Shanghai, China) and then processed with HKL-2000 (Otwinowski and Minor, 1997). The SOBIR1-KDD489A structure was solved by molecular replacement using Phaser (McCoy et al., 2007), with the BRI1 KD (PDB: 5LPY) structure as the search model for SOBIR1-KDD489A and SOBIR1-KDD489A as the search model for SOBIR1-KDΔ389-401. Phenix (Adams et al., 2010) and Coot (Emsley et al., 2010) were used for the standard refinement. The structure validation was completed with MolProbity (Williams et al., 2018). The data processing and refinement statistics are summarized in Supplemental Table 1. All structures visualized in this article were prepared with PyMOL (http://www.pymol.org).
In-gel protein digestion and peptide recovery
The wild-type SOBIR1-KD analyzed by LC-MS/MS was expressed in E. coli and purified as described above. PP2Cα-dephosphorylated SOBIR1-KD was generated by incubating 10 μM SOBIR1-KD and 1 μM GST-PP2Cα in kinase reaction buffer at 25°C for 1 h. The mixture was then loaded onto GST-NTA columns, and dSOBIR1-KD was eluted. Autophosphorylated SOBIR1-KD was generated by incubating 10 μM dSOBIR1-KD with 1 mM ATP and 10 mM Mg2+ at 25°C for 3 h. BAK1-phosphorylated SOBIR1-KD was generated by incubating 10 μM dSOBIR1-KD and 1 μM GST-BAK1-CD in kinase reaction buffer at 25°C for 1 h.
After proteins were separated by SDS-PAGE, the stained protein bands were digested after overnight incubation in 50 mM NH4HCO3/acetonitrile (1:1, v/v). Samples were treated with 75% acetonitrile and equilibrated in 20 mM ammonium bicarbonate. Acetonitrile was added after removing the liquid, and protein bands were dehydrated until they turned completely white; they were then placed in a vacuum. Protein bands were reduced with 10 mM DTT at 56°C for 1 h, then alkylated in 55 mM iodoacetamide at room temperature for 1 h in the dark. After removing the liquid, acetonitrile was added to dehydrate the protein samples, which were then digested with trypsin (1 μg μl−1 in 20 mM ammonium carbonate, pH 8.9). The ammonium carbonate buffer was added until the protein bands returned to their original size. The trypsin digestion was completed overnight at 37°C. The resulting peptides were recovered by two extractions (20 min each) with 100 μl of 60% acetonitrile acid. Once successful extraction of very hydrophobic peptides was achieved, a third extraction buffer containing 60% acetonitrile and 0.1% formic acid was added to resuspend the extracts, which were then concentrated. Each extraction was finally resuspended in 2% acetonitrile containing 0.1% formic acid and then centrifuged at 20,000 g for 15 min.
Determination of in vitro phosphorylation sites by LC-MS/MS analysis
The Acclaim PepMap C18-reversed-phase column (75 μm × 2 cm, 3 μm, 100 Å; Thermo Fisher Scientific, San Jose, CA) and the reversed-phase C18 column (75 μm × 10 cm, 5 μm, 300 Å; Agela Technologies, Tianjin, China) were used to separate peptides via gradient elution at a flow rate of 400 nl min−1. The peptides were then analyzed with the Q-Exactive MS system (Thermo Fisher Scientific) with the following settings: positive ion mode; data-dependent manner with a full MS scan from 350–2000 m/z; full scan resolution at 70 000; and MS/MS scan resolution at 17 500. The minimum signal threshold value for the MS/MS scanning was 1E+5, with an isolation width of 2 Da. To analyze the labeled samples, high-energy collision dissociation was employed in two MS/MS acquisition modes with normalized collision energies of 20% and 28%.
Analysis of LC-MS/MS data
Peptides were identified and quantified with Mascot software (version 2.3.01) using The Arabidopsis Information Resource (TAIR) database search algorithm and the integrated false discovery rate analysis function. These data were used to screen the TAIR10_pep_20101214 protein sequence database (35 386 sequences; 14 482 855 residues).
Plant growth and transient expression
N. benthamiana was grown under 16 h of light at 25°C and 8 h of darkness at 21°C in 75% relative humidity. The AtSOBIR1 coding sequence was cloned into the pROK2-eGFP vector to generate a 35S promoter-driven construct and verified by DNA sequencing. All mutations of AtSOBIR1 were generated by the standard Quickchange PCR procedure, and the primers used for gene cloning are shown in Supplemental Table 3. Agrobacterium tumefaciens infiltrations were performed as previously described (Van der Hoorn et al., 2000) with appropriate modification. All binary vectors were transformed into A. tumefaciens strain C58C1 and grown in LB medium with appropriate antibiotics (20 mg/L rifampicin and 50 mg/L kanamycin), then shaken (200 rpm) at 28°C for about 12–14 h. After the optical density at 600 nm (OD600) value reached 0.6–0.8, the cultures were centrifuged at 4000 g for 10 min at room temperature and then resuspended in MES buffer (10 mM MES-KOH [pH 5.6], 10 mM MgCl2, and 100 μM acetosyringone) to a final optical density of 1. All constructs were infiltrated into 4-week-old N. benthamiana leaves, and samples were collected 7 d after agroinfiltration for phenotype analysis. SOBIR1- and mutant-induced cell death phenotypes in N. benthamiana were detected by trypan blue staining, which was performed as described previously (Fernández-Bautista et al., 2016).
ROS production and MAPK activation analysis
For measurement of ROS burst, leaf discs (0.125 cm2) of N. benthamiana were incubated overnight in water in a 96-well titer plate using one leaf disc per well. ROS produced by the leaf discs were measured by a luminol-based assay (Felix et al., 1999). Luminescence, shown as relative light units (RLU), was measured using a Tecan Infinite M200 microplate reader (Austria). AtSOBIR1 and all mutants were transiently expressed in leaves of N. benthamiana by agroinfiltration. After 2 days, oxidative burst and MAPK activation were elicited with 300 nM flg22. Samples for the measurement of MAPK activation were harvested 15 min after flg22 treatment. Detection of the phosphorylation of N. benthamiana MAPKs was performed according to the method described by Flury et al. (2013) with slight modification. MAPK activation was detected by immunoblotting using anti-ERK antibody (Rabbit mAb #4370, Cell Signaling Technology). All experiments were performed in triplicate.
Funding
This work was supported by the National Natural Science Foundation of China (31571963).
Author contributions
X.W. and T.G. performed protein purification, prepared crystal samples, and collected the diffraction data. X.W., Y.W. and H.Y. performed the biochemical experiments. S.Z. performed the in vivo functional experiments. Y.W. processed the diffraction data and solved the structures. X.W., Y.W., S.Z. and G.S. performed data analysis and wrote the manuscript. G.G., X.L., S.F., Y.G., and F.W. gave important suggestions for the project. F.X. supervised the project. All authors approved the manuscript.
Acknowledgments
We thank the Shanghai Synchrotron Radiation Facility (SSRF) and the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing, China) for providing facility support for crystallographic data collection and processing, as well as BGI Tech Solutions (Beijing, China) for performing the LC-MS/MS experiments and data analyses. No conflict of interest is declared.
Published: January 19, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Data and code availability
The PDB accession numbers for the AMP-PNP-bound SOBIR1-KDD489A and AMP-PNP-bound SOBIR1-KDΔ389-401 structures reported herein are 7CTV and 7CTX, respectively.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I., Bohm H., Albert M., Feiler C.E., Imkampe J., Wallmeroth N., Brancato C., Raaymakers T.M., Oome S., Zhang H., et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants. 2015;1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- Albert I., Zhang L., Bemm H., Nurnberger T. Structure-function analysis of immune receptor AtRLP23 with its ligand nlp20 and coreceptors AtSOBIR1 and AtBAK1. Mol. Plant Microbe Interact. 2019;32:1038–1046. doi: 10.1094/MPMI-09-18-0263-R. [DOI] [PubMed] [Google Scholar]
- Aquino B., Counago R.M., Verza N., Ferreira L.M., Massirer K.B., Gileadi O., Arruda P.F. Structural characterization of maize SIRK1 kinase domain reveals an unusual architecture of the activation segment. Front. Plant Sci. 2017;8:852. doi: 10.3389/fpls.2017.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas A.C., Deal M.S., Steichen J.M., Guo Y., Wu J., Taylor S.S. Phosphoryl transfer by protein kinase A is captured in a crystal lattice. J. Am. Chem. Soc. 2013;135:4788–4798. doi: 10.1021/ja312237q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenstock J., Mooshayef N., Engelberg D. How do protein kinases take a selfie (autophosphorylate)? Trends Biochem. Sci. 2016;41:938–953. doi: 10.1016/j.tibs.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Bi G., Liebrand T.W., Bye R.R., Postma J., van der Burgh A.M., Robatzek S., Xu X., Joosten M.H. SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor-like proteins. Mol. Plant Pathol. 2016;17:96–107. doi: 10.1111/mpp.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojar D., Martinez J., Santiago J., Rybin V., Bayliss R., Hothorn M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014;78:31–43. doi: 10.1111/tpj.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J.D., Felix G., Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Domazakis E., Wouters D., Visser R.G.F., Kamoun S., Joosten M., Vleeshouwers V. The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol. Plant Microbe Interact. 2018;31:795–802. doi: 10.1094/MPMI-09-17-0217-R. [DOI] [PubMed] [Google Scholar]
- Dominguez-Ferreras A., Kiss-Papp M., Jehle A.K., Felix G., Chinchilla D. An overdose of the Arabidopsis coreceptor BRASSINOSTEROID INSENSITIVE1-ASSOCIATED RECEPTOR KINASE1 or its ectodomain causes autoimmunity in a SUPPRESSOR OF BIR1-1-dependent manner. Plant Physiol. 2015;168:1106–1121. doi: 10.1104/pp.15.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin.pdf. The Plant Journal. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Bautista N., Domínguez-Núñez J., Moreno M.M., Berrocal-Lobo M. Plant tissue trypan blue staining during Phytopathogen infection. Bio-Protocol. 2016;6 doi: 10.21769/BIOPROTOC.2078. [DOI] [Google Scholar]
- Ferrao R., Zhou H., Shan Y., Liu Q., Li Q., Shaw D.E., Li X., Wu H. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Mol. Cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flury P., Klauser D., Boller T., Bartels S. MAPK phosphorylation assay with leaf disks of Arabidopsis. Bio-Protocol. 2013;3:e929. [Google Scholar]
- Foster S.A., Whalen D.M., Ozen A., Wongchenko M.J., Yin J., Yen I., Schaefer G., Mayfield J.D., Chmielecki J., Stephens P.J., et al. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell. 2016;29:477–493. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Gao J., Ma Y., Sun Y., Zhao H., Hong D., Yan L., Lou Z. Crystallization and preliminary crystallographic analysis of Arabidopsis thaliana BRI1-associated kinase 1 (BAK1) cytoplasmic domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:340–342. doi: 10.1107/S1744309112004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., Bi D., Cheng Y.T., Chen S., Li X., et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe. 2009;6:34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Gao X., Ruan X., Sun Y., Wang X., Feng B. BAKing up to survive a battle: functional dynamics of BAK1 in plant Programmed cell death. Front. Plant Sci. 2018;9:1913. doi: 10.3389/fpls.2018.01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hohmann U., Hothorn M. Crystal structure of the leucine-rich repeat ectodomain of the plant immune receptor kinase SOBIR1. Acta Crystallogr. D Struct. Biol. 2019;75:488–497. doi: 10.1107/S2059798319005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Jacobsen D.M., Bao Z.Q., O'Brien P., Brooks C.L., 3rd, Young M.A. Price to be paid for two-metal catalysis: magnesium ions that accelerate chemistry unavoidably limit product release from a protein kinase. J. Am. Chem. Soc. 2012;134:15357–15370. doi: 10.1021/ja304419t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.N., Noble M.E., Owen D.J. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Kornev A.P., Haste N.M., Taylor S.S., Eyck L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev A.P., Taylor S.S., Ten Eyck L.F. A helix scaffold for the assembly of active protein kinases. Proc. Natl. Acad. Sci. U S A. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H., Li J.J., Thevakumaran N., Therrien M., Sicheri F. Dimerization-induced allostery in protein kinase regulation. Trends Biochem. Sci. 2014;39:475–486. doi: 10.1016/j.tibs.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Leslie M.E., Lewis M.W., Youn J.Y., Daniels M.J., Liljegren S.J. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development. 2010;137:467–476. doi: 10.1242/dev.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson N.M., Kuchment O., Shen K., Young M.A., Koldobskiy M., Karplus M., Cole P.A., Kuriyan J. A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand T.W., van den Berg G.C., Zhang Z., Smit P., Cordewener J.H., America A.H., Sklenar J., Jones A.M., Tameling W.I., Robatzek S., et al. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U S A. 2013;110:10010–10015. doi: 10.1073/pnas.1220015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand T.W., van den Burg H.A., Joosten M.H. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014;19:123–132. doi: 10.1016/j.tplants.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Madhusudan T.,E.A., Xuong N.-H., Adams J.A., Eyck L.F.T., Taylor S.S., Sowadski J.M. cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man J., Gallagher J.P., Bartlett M. Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 2020;226:1492–1505. doi: 10.1111/nph.16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo C.B., Erlandsen H., Mouser D.J., Feinstein A.G., Robinson V.L., May E.R., Cole J.L. Structural basis of protein kinase R autophosphorylation. Biochemistry. 2019;58:2967–2977. doi: 10.1021/acs.biochem.9b00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola E.T., Gahmberg C.G. Hydrophobic interaction between the SH2 domain and the kinase domain is required for the activation of Csk. J. Mol. Biol. 2010;399:618–627. doi: 10.1016/j.jmb.2010.04.045. [DOI] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Kota U., Goshe M.B., Clouse S.D., Huber S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2009;106:658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Wu X., Zhao Y., Clouse S.D., Huber S.C. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. U S A. 2010;107:17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Postma J., Liebrand T.W., Bi G., Evrard A., Bye R.R., Mbengue M., Kuhn H., Joosten M.H., Robatzek S. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 2016;210:627–642. doi: 10.1111/nph.13802. [DOI] [PubMed] [Google Scholar]
- Rabiller M., Getlik M., Kluter S., Richters A., Tuckmantel S., Simard J.R., Rauh D. Proteus in the world of proteins: conformational changes in protein kinases. Arch. Pharm. (Weinheim) 2010;343:193–206. doi: 10.1002/ardp.201000028. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr. Assays of protein kinase. Methods Enzymol. 1983;99:3–6. doi: 10.1016/0076-6879(83)99034-1. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol. Res. 2015;94:9–25. doi: 10.1016/j.phrs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Sang D., Pinglay S., Wiewiora R.P., Selvan M.E., Lou H.J., Chodera J.D., Turk B.E., Gumus Z.H., Holt L.J. Ancestral reconstruction reveals mechanisms of ERK regulatory evolution. eLife. 2019;8:e38805. doi: 10.7554/eLife.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Gahmen U., De Bondt H.L., Kim S.H. High-resolution crystal structures of human cyclin-dependent kinase 2 with and without ATP: bound waters and natural ligand as guides for inhibitor design. J. Med. Chem. 1996;39:4540–4546. doi: 10.1021/jm960402a. [DOI] [PubMed] [Google Scholar]
- Schwarz D., Merget B., Deane C., Fulle S. Modeling conformational flexibility of kinases in inactive states. Proteins. 2019;87:943–951. doi: 10.1002/prot.25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J.M., Kuchinskas M., Keshwani M.M., Yang J., Adams J.A., Taylor S.S. Structural basis for the regulation of protein kinase A by activation loop phosphorylation. J. Biol. Chem. 2012;287:14672–14680. doi: 10.1074/jbc.M111.335091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Murano T., Ishikawa A. SOBIR1 and AGB1 independently contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2018;82:1922–1930. doi: 10.1080/09168451.2018.1498727. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Kornev A.P. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burgh A.M., Postma J., Robatzek S., Joosten M. Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol. 2019;20:410–422. doi: 10.1111/mpp.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn R.A., Laurent F., Roth R., De Wit P.J. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 2000;13:439–446. doi: 10.1094/MPMI.2000.13.4.439. [DOI] [PubMed] [Google Scholar]
- Wagner T., Alexandre M., Duran R., Barilone N., Wehenkel A., Alzari P.M., Bellinzoni M. The crystal structure of the catalytic domain of the ser/thr kinase PknA from M. tuberculosis shows an Src-like autoinhibited conformation. Proteins. 2015;83:982–988. doi: 10.1002/prot.24754. [DOI] [PubMed] [Google Scholar]
- Wan W.L., Zhang L., Pruitt R., Zaidem M., Brugman R., Ma X., Krol E., Perraki A., Kilian J., Grossmann G., et al. Comparing Arabidopsis receptor kinase and receptor protein-mediated immune signaling reveals BIK1-dependent differences. New Phytol. 2019;221:2080–2095. doi: 10.1111/nph.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiang J., Wang J., Chen L., Fan S.L., Wu J.W., Wang X., Wang Z.X. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014;24:1328–1341. doi: 10.1038/cr.2014.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wu J.W., Wang Z.X. Mechanistic studies of the autoactivation of PAK2: a two-step model of cis initiation followed by trans amplification. J. Biol. Chem. 2011;286:2689–2695. doi: 10.1074/jbc.M110.156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ferrao R., Li Q., Hatcher J.M., Choi H.G., Buhrlage S.J., Gray N.S., Wu H. Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4) J. Biol. Chem. 2019;294:4511–4519. doi: 10.1074/jbc.RA118.005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B., et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Liu Y., Xu F., Zhang Y. Differential requirement of BAK1 C-terminal tail in development and immunity. J. Integr. Plant Biol. 2018;60:270–275. doi: 10.1111/jipb.12623. [DOI] [PubMed] [Google Scholar]
- Wu J., Reca I.B., Spinelli F., Lironi D., De Lorenzo G., Poltronieri P., Cervone F., Joosten M., Ferrari S., Brutus A. An EFR-Cf-9 chimera confers enhanced resistance to bacterial pathogens by SOBIR1- and BAK1-dependent recognition of elf18. Mol. Plant Pathol. 2019;20:751–764. doi: 10.1111/mpp.12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., van der Burgh A.M., Bi G., Zhang L., Alfano J.R., Martin G.B., Joosten M. The bacterial effector AvrPto targets the regulatory coreceptor SOBIR1 and suppresses defense signaling mediated by the receptor-like protein Cf-4. Mol. Plant Microbe Interact. 2018;31:75–85. doi: 10.1094/MPMI-08-17-0203-FI. [DOI] [PubMed] [Google Scholar]
- Xu W., Doshi A., Lei M., Eck M.J., Harrison S.C. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol. Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- Yan L., Ma Y., Liu D., Wei X., Sun Y., Chen X., Zhao H., Zhou J., Wang Z., Shui W., et al. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 2012;22:1304–1308. doi: 10.1038/cr.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kars I., Essenstam B., Liebrand T.W., Wagemakers L., Elberse J., Tagkalaki P., Tjoitang D., van den Ackerveken G., van Kan J.A. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 2014;164:352–364. doi: 10.1104/pp.113.230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Fraiture M., Kolb D., Loffelhardt B., Desaki Y., Boutrot F.F., Tor M., Zipfel C., Gust A.A., Brunner F. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell. 2013;25:4227–4241. doi: 10.1105/tpc.113.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PDB accession numbers for the AMP-PNP-bound SOBIR1-KDD489A and AMP-PNP-bound SOBIR1-KDΔ389-401 structures reported herein are 7CTV and 7CTX, respectively.