Abstract
Background
Programmed cell death ligand-1 (PD-L1) expression as a single biomarker for immune checkpoint blockade (ICB) was controversial. NKG2A was a PD1/PD-L1 axis-related immunity-dependent factor. NKG2A and PD-L1 expression as a combinatorial biomarker might improve the prediction of PD-L1 in patients with muscle-invasive bladder cancer (MIBC).
Methods
Three independent cohorts were enrolled in our study. 195 patients with bladder-derived metastatic urothelial carcinoma on PD-L1 inhibitor treatment from the IMvigor210 trial were enrolled. 124 MIBC patients from Zhongshan Hospital and 391 patients with MIBC from The Cancer Genome Atlas database were included in this study. The PD-L1/NKG2A-based risk stratification was validated in three independent cohorts, and its association with response to ICB and adjuvant chemotherapy (ACT), immune contexture and molecular features was evaluated. Histologic staining and genomic algorithm were performed to detect characteristics of NKG2A and PD-L1 expression and infiltration of immune cells.
Results
We identified NKG2AhiPD-L1hi patients could benefit more from cisplatin-based ACT and PD-L1 inhibitor. Further analyses revealed NKG2A and PD-L1 expression panel was linked to an immune-active tumor microenvironment with highly immune effector cells and effector molecules. In addition, NKG2A and PD-L1 expression panel was intrinsically correlated with genomic alterations related to therapeutic response in MIBC.
Conclusions
NKG2A and PD-L1 expression panel was associated with an immune inflamed microenvironment and acted as a combinatorial biomarker to predict the therapeutic response to ACT and PD-L1 blockade in MIBC.
Keywords: tumor microenvironment; urinary bladder neoplasms; immunotherapy; biomarkers, tumor
Key messages.
What is already known on this topic
Immune checkpoint blockade and adjuvant chemotherapy have shown quite durable antitumor responses, but lack of effective biomarkers limited the therapeutic benefits in patients with bladder cancer.
What this study adds
NKG2A and programmed cell death ligand-1 (PD-L1) expression panel ignited inflamed immune context in patients with bladder cancer.
NKG2A and PD-L1 expression panel predicted immunotherapeutic and chemotherapeutic benefit in patients with bladder cancer.
How this study might affect research, practice or policy?
NKG2A and PD-L1 expression panel could be a combinatorial biomarker for providing a precise decision in bladder cancer treatment.
NKG2A blockade combined with anti-PD-1/PD-L1 therapy is worth further investigation in bladder cancer.
Background
Bladder cancer is the 10th most prevalent cancer worldwide, with more than 573 000 new cases and approximately 213 000 deaths.1 About 25% of patients suffer from muscle-invasive bladder cancer (MIBC) at first diagnosis with poor clinical outcomes.2 For patients with MIBC, radical cystectomy (RC) complemented by cisplatin-based adjuvant chemotherapy (ACT) remains the mainstay treatment to improve survival.3
Recent studies showed that the immune checkpoint blockade (ICB) could improve the prognosis of patients resistant to ACT with advanced MIBC,4 especially programmed cell death-1 (PD-1)/ programmed cell death ligand-1 (PD-L1) targeting antibodies to reactivate antitumor immune response, representing a paradigm renovation for treatment of platinum-refractory advanced MIBC patients.5–7 However, immunotherapy remained poorly served by effective predictive biomarkers, and the response rates were suboptimal. Therefore, there was an urgent need to develop clinically useful predictive biomarkers to refine patient selection involving ACT and ICB.
Numerous researches suggested that PD-L1 was a predictive biomarker.8 However, using PD-L1 as a single biomarker was controversial because of the dynamic regulation of PD-L1 expression.9 10 PD-L1 expression was upregulated by both intracellular oncogenic variants such as loss of PTEN and ongoing immune responses.8 11 Immunity-dependent PD-L1 upregulation was more relevant for reactivating the tumor-killing activity of tumor infiltrating lymphocyte and had better predictive value.12 13 Therefore, it would be important to distinguish between immunity-dependent and immunity-independent PD-L1 expression. Recent studies provided a way that simultaneous measurement of PD1/PD-L1 axis-related immunity-dependent factors could potentially achieve this goal.14 To address this need, we integrated PD-L1 and classic immune checkpoint for identifying potential combinatorial biomarkers to predict ICB and ACT response.
As a classic immune checkpoint, NKG2A is mainly expressed on lymphocytes such as CD8+ αβ T cells and CD56hi natural killer (NK) cells. As an inhibitory member of the NKG2 family, NKG2A can dimerize with CD94 on the cell surface.15 HLAE is the primary ligand for NKG2A-CD94. NKG2A-CD94 engages with peptideloaded HLAE leading to the delivery of an inhibitory signal that suppresses competing signals from activated receptors and CD8+ T cell through SHP1/2 signaling.16
NKG2A is frequently coexpressed with PD-1 in patients. However, unlike PD-1 expression, which can be a hallmark of exhausted CD8+ T cells, CD8+ T cells expressing NKG2A might be activated T cells displaying superior antitumor properties.17 These results suggest NKG2A expression might be an additional brake for the PD-1/PD-L1 pathway to alter CD8+ T cells status. Recent trials have found that the combination of NKG2A and PD-L1 checkpoint blockade could enhance patients’ survival for microsatellite stabilized (MSS) colorectal cancer (NCT02671435) and non-small cell lung cancer (NCT03822351).18 19 However, the clinical value of the combination of NKG2A and PD-L1 expression remains scarcely explicit in MIBC.
Herein, our study constructed a classification of MIBC based on NKG2A and PD-L1 expression. We further examined their predictive value for ACT and ICB response through genomic and histological data in three large cohorts, including 195 patients on PD-L1 inhibitor from IMvigor210 trial and patients receiving ACT in The Cancer Genome Atlas (TCGA; n=391) and Zhongshan Hospital (ZSHS; n=124). Meanwhile, we correlated the stratification based on NKG2A and PD-L1 expression with immune contexture, key gene mutations and molecular subtypes.
Methods
Study cohort
Three independent cohorts were analyzed: IMvigor210 trial, TCGA cohort and ZSHS cohort.
Approved by the Clinical Research Ethics Committee of Zhongshan Hospital, the ZSHS cohort involved 215 patients with bladder cancer treated with RC. Patients received four cycles of platinum-based chemotherapy with gemcitabine/cisplatin or methotrexate/vinblastine/doxorubicin/cisplatin. Patients were given gemcitabine/carboplatin or gemcitabine/oxaliplatin when they were intolerant to cisplatin. A total of 91 patients were ineligible into this study under the following criteria: (1) 60 patients diagnosed as NMIBC; (2) 13 patients with non-urothelial carcinoma; (3) 18 patients with detachment in tissue microarray (TMA). Finally, 124 patients were included in the ZSHS cohort and 60 of them received platinum-based ACT treatment. Formalin-fixed, paraffin-embedded tissue blocks were available in all cases.
IMvigor210 trial was a single-arm phase II study investigating atezolizumab (1200 mg three times weekly) in patients with metastatic urothelial carcinoma (NCT02108652, NCT02951767).20 21 The therapeutic response was assessed by Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1 and immune modified RECIST. Patients who had achieved complete response (CR) and partial response (PR) to atezolizumab were defined as responders in this study, while patients with stable disease and progressive disease (PD) were defined as non-responders.22 All data of IMvigor210 trial were accessed through the IMvigor210CoreBiologies R package downloaded from http://research-pub.gene.com/IMvigor210CoreBiologies/. One hundred and fifty-three patients with non-bladder-derived metastasis urothelial carcinoma were excluded.
The clinicopathological characteristics and genomic data of patients in TCGA cohort were from TCGA-Assembler 2.0.5 in May 2018. The data regarding chemotherapeutic responsiveness were derived from TCGA-Assembler 2.0.5 in July 2021. Patients received chemotherapy as prescribed, but the specific chemotherapy regimen was unknown. A total of 21 patients were ineligible: 3 patients without survival information; 4 patients without RNA-seq data; 10 patients receiving neoadjuvant chemotherapy; and 4 patients with NMIBC. One hundred and seventy-five patients in TCGA cohort received ACT, while detailed information about the chemotherapy drugs was unavailable. Finally, TCGA cohort comprised 391 MIBC patients for further survival and bioinformatics analysis.
Overall survival (OS) considered as a clinical endpoint was calculated from the date of RC to the date of death or the last follow-up. Meanwhile, recurrence-free survival (RFS) was defined from the date of surgery to the date of recurrence or last follow-up. Objective response rate (ORR) was defined as the sum of CR and PR ratios. Clinical and pathological features of IMvigor210, ZSHS and TCGA cohorts were presented respectively in online supplemental tables 1–3.
jitc-2022-004569supp001.pdf (396.4KB, pdf)
RNA-seq data and processing
The RNA-seq expression data of TCGA cohort and IMvigor210 trial were retrieved along with the process of acquiring clinical information and were transformed as log2 (FPKM+1) for downstream analysis. Based on TCGA and IMvigor210 data, CIBERSORT was constructed to calculate the relative proportion of 22 immune cell types.23 The data of patient molecular subtype were retrieved through the BLCA subtyping R package from https://github.com/cit-bioinfo/BLCAsubtyping.24
Immunohistochemistry (IHC) and assay method
TMA was constructed by Shanghai Outdo Biotech Co, Ltd and was described in our previous study.25 This study enrolled 215 patients treated with RC from Zhongshan Hospital (surgery date: 2002–2014) of Fudan University. ZSHS samples were embedded in formalin-fixed paraffin and sectioned by TMA technique. All samples were stained with H&E for histological examination, and representative regions were marked on paraffin blocks. Duplicate 1.0-millimeter tissue cores from two different regions were used to construct TMAs. Single IHC staining was used for the following cells and their corresponding molecules: NKG2A+ cells (NKG2A), PD-L1, CD8+ cells (CD8), Th17 cells (IL-17), B cells (CD19), NK cells (CD56), PRF-1+ cells, IFN-γ+ cells and GZMB+ cells. For single staining, formalin-fixed MIBC tissue sections were paraffinized in xylene and hydrated in a diluted alcohol series. Then, antigen retrieval was accomplished with sodium citrate buffer (10 mM, pH 6.0) using a microwave for 15 min. Next, the sections were immersed in endogenous peroxidase blocking buffer (0.3%) for 30 min. After blocking with goat serum, the sections were incubated with corresponding primary antibodies at 4°C overnight. Then, after washing three times using Tris buffered saline with Tween 20 (TBST), the sections were subsequently incubated with secondary antibody for 30 min. The staining was performed with diaminobenzidine (DAB) stain system (ZLI-9019). Finally, the sections were counterstained with hematoxylin. All TMA slides were scanned digitally through Image Pro plus 6.0 and Nano-Zoomer-XR (Hamamatsu). Double IHC staining was used for the following molecules: CD4 and T-bet to detect Th1 cells, HLA-DR and CD11c to detect DCs, CD68 and HLA-DR to detect M1 cells. For double staining, after paraffin removal, antigen extraction and blocking, the first primary antibodies were incubated with slides overnight at 4°C. Subsequently, alkaline phosphatase-labeled secondary antibody was incubated, and Vector blue staining was performed. Then the second primary antibodies were incubated with TMA at 37°C for 2 hours, and TMA was incubated with horseradish peroxidase labeled antibody, before proceeding to staining using DAB reagent. In addition, the antibody information involved in this article was shown in online supplemental table 4.
Immune evaluation and cut-off value
Two independent pathologists unaware of clinical data evaluated TMA slides separately. The average evaluation of two pathologists was adopted. When variations in the enumeration exceeded five cells, two pathologists re-evaluated separately to reach a consensus. Immunohistochemistry (IHC) score was calculated by multiplication of the staining intensity and proportion. The staining intensity was graded as 0 (negative staining), 1 (weak staining), 2 (moderate staining) and 3 (strong staining), and the proportion was scored as a percentage of positive cells (0%–100%). The detailed evaluation method for stained cells was illustrated in online supplemental table 4. The kappa between the two pathologists’ evaluations for NKG2A and PD-L1 IHC reading from the same slide were 0.925 (95% CI 0.878 to 0.972, p<0.001) and 0.935 (95% CI 0.892 to 0.978, p<0.001), respectively. The cut-off values for NKG2A and PD-L1 were, respectively, 22.5 and 97.4 by the median value. The median of the mRNA expression of NKG2A and PD-L1 were 0.41 and 0.44 in IMvigor210 cohort and 0.11 and 0.94 in TCGA cohort, respectively.
Genomic analysis and variant assessment
The tumor mutation burden (TMB) was identified as the total non-silent somatic mutation counts in TCGA and IMvigor210 cohorts. For analysis, we defined TMB ≥10 mut/Mb as ‘TMB high’.23 TMB data were obtained from https://portal.gdc.cancer.gov/. TMB data for patients in TCGA cohort were calculated using results from whole-exome sequencing and maftools package, while TMB data for IMvigor210 trial were obtained by targeted large-panel sequencing.26 Gene alterations (GAs) were defined as the aggregation of gene mutation and copy number variation. Either nonsense, missense, frameshift, splice-site variants affecting consensus nucleotides, or deleterious homozygous deletions and amplifications were defined as GA.
Statistics
Kruskal-Wallis test was used to depict the diversity of immune infiltration in NKG2A/PD-L1-stratified groups. Kaplan-Meier method was used to assess 5-year OS and RFS, whereas log-rank test and Cox regression models were applied for the assessment of the prognostic and risk significance. Spearman correlation analysis was performed to investigate the bivariate correlation. The χ2 test was applied to depict the relation of clinicopathological parameters, molecular subtypes and GAs with NKG2A and PD-L1 expression. The kappa statistics was used to assess the consistency for IHC reading. The two-sided p≤0.05 was considered statistically significant. The data were analyzed through IBM SPSS Statistics V.20.0 and R software V.4.0.4.
Results
NKG2A and PD-L1 expression panel predicts PD-L1 blockade benefit in MIBC
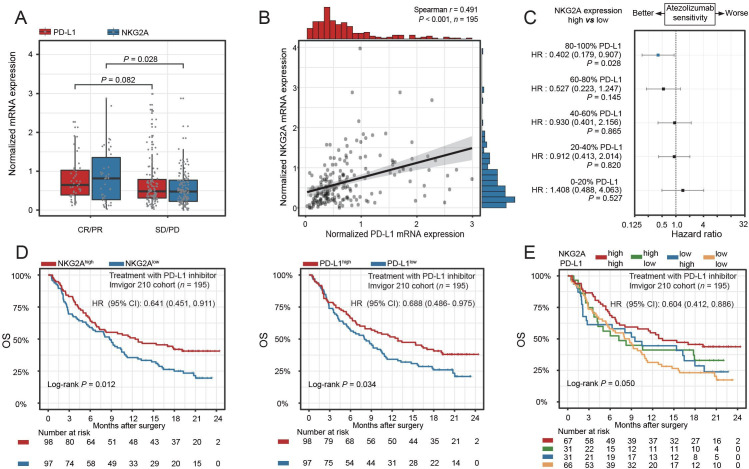
We noticed that NKG2A expression was significantly enriched in responders as compared with non-responders in the IMvigor210 cohort (p=0.028, figure 1A). However, PD-L1 expression merely indicated a trend toward improved immunotherapy response rate (figure 1A, p=0.082). Both NKG2A and PD-L1 expression in baseline tumor indicated favorable OS (p=0.012, p=0.034, figure 1D). Therefore, we tested whether the combination of NKG2A and PD-L1 expression would improve the prediction of therapeutic response compared with either single biomarker. In the IMvigor210 cohort, we observed a positive correlation between NKG2A expression and PD-L1 expression (p<0.001, figure 1B). Furthermore, analysis of the hazard curves for PD-L1 by NKG2A expression status showed that the positive association of high NKG2A expression with OS increased in magnitude as PD-L1 mRNA expression increased (figure 1C). In IMvigor210, we noted that the combination of NKG2A expression with PD-L1 expression was significantly associated with improved OS (p=0.050, figure 1E). In addition, the ORR for NKG2AhiPD-L1hi subgroup was 0.667 (95% CI 0.505 to 0.828, online supplemental table 5). Together, our study suggested that NKG2A expression plus PD-L1 expression could serve as a combinatorial biomarker to predict clinical response to ICB.
Figure 1.
NKG2A and PD-L1 expression panel predicts PD-L1 blockade benefit in MIBC. (A) NKG2A and PD-L1 mRNA expression between responder and non-responder group in IMvigor210 trial. Data were analyzed by Mann-Whiney U test and presented as median and IQR. (B) Spearman correlation between NKG2A and PD-L1. (C) HR estimates for OS in IMvigor210 cohort. HRs compare NKG2A high and low mRNA expression at different PD-L1 mRNA expression values. Plotting symbols give point estimates of HR; horizontal bars give 95% CIs. (D) Kaplan-Meier analyses of OS in patients in IMvigor210 cohort, stratified according to NKG2A mRNA expression values and PD-L1 mRNA expression values. Data were analyzed by log-rank test. (E) Kaplan-Meier analyses of OS in patients in the IMvigor210 trial stratified according to the combination of NKG2A and PD-L1. Data were analyzed by log-rank test. HRs with 95% CI were estimated by Cox proportional hazards model. MIBC, muscle-invasive bladder cancer; OS, overall survival; PD-L1, programmed cell death ligand-1; RFS, recurrence-free survival.
NKG2A and PD-L1 expression panel indicates ACT benefit in MIBC
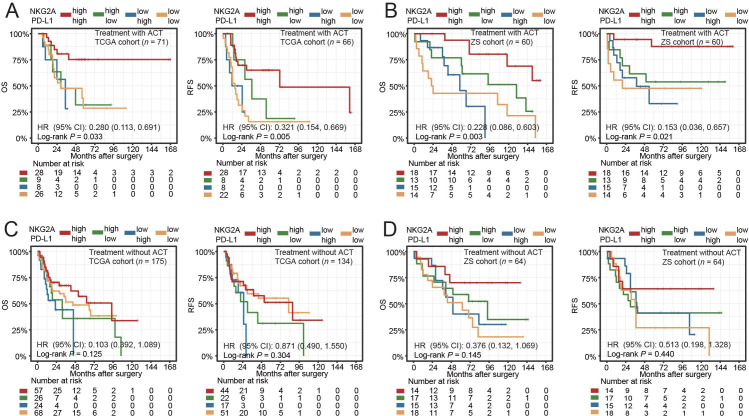
Furthermore, to investigate the predictive value of NKG2A and PD-L1 expression panel on the benefit of ACT, we developed a classification scheme based on NKG2A and PD-L1 expression using the median values in TCGA cohort and ZSHS cohort. Representative images of IHC staining for NKG2A and PD-L1 expression were shown in online supplemental figure 1B. NKG2A and PD-L1 expression could significantly lead to improved survival in patients receiving ACT in the TCGA cohort (OS: p=0.033; RFS: p=0. 005; figure 2A) and the ZSHS cohort (OS: p=0.003; RFS: p=0.021; figure 2B). Moreover, the ORR of NKG2AhiPD-L1hi patients receiving ACT in the TCGA cohort was 0.490 (95% CI 0.345 to 0.635, online supplemental table 5). However, for patients without ACT application, NKG2A and PD-L1 expression failed to reduce their risk of death and recurrence in the TCGA cohort (OS: p=0. 125; RFS: p=0. 304; figure 2C) and the ZSHS cohort (OS: p=0. 145; RFS: p=0. 440; figure 2D). Conclusively, we identified NKG2A and PD-L1 expression panel could be associated with superior therapeutic response to ACT.
Figure 2.
NKG2A and PD-L1 expression panel indicates adjuvant chemotherapy response in MIBC. (A and B) Kaplan-Meier survival curves for OS and RFS of patients with ACT treatment from TCGA cohort (A) and Zhongshan cohort (B). (C and D) Kaplan-Meier survival curves for OS and RFS of patients without ACT treatment from TCGA cohort (C) and Zhongshan cohort (D). Data were analyzed by log-rank test. HRs with 95% CI were estimated by Cox proportional hazards model. ACT, adjuvant chemotherapy; OS, overall survival; PD-L1, programmed cell death ligand-1; RFS, recurrence-free survival; TCGA, The Cancer Genome Atlas.
NKG2A and PD-L1 expression panel ignites inflamed immune context in MIBC
The tumor immune microenvironment was commonly considered to be associated with the efficacy of chemotherapy and immunotherapy.27 Therefore, we used CIBERSORT23 to calculate the infiltration of 22 immune cells in the TCGA and IMvigor210 cohorts to reveal the potential effect of NKG2A and PD-L1 expression on the immune microenvironment. NKG2Ahigh and PD-L1high subgroup was associated with higher levels of immune effector cells and immune effector molecules expression (figure 3A, B). These results illustrated high expression of NKG2A and PD-L1 was associated with the inflamed immune micro-environment in MIBC. To validate the results, we performed IHC to quantify immune cells and cytokines in the ZSHS cohort. Consistent with the results of the TCGA cohort, patients with high expression of NKG2A and PD-L1 had more immune effector cells (CD8+ T cells, M1 macrophages and NK cells, figure 3C) and effector molecules (IFN-γ, GZMB and PRF-1, figure 3D), indicating an immune-activated environment. Collectively, these results suggested that NKG2A and PD-L1 expression panel was associated with an inflamed immune microenvironment in MIBC.
Figure 3.
NKG2A and PD-L1 expression panel associates with inflamed immune context in MIBC. (A and B) Heatmap showing CIBERSORT analysis of 22 kinds of intratumoral infiltration immune cells across stratification in patients in TCGA cohort (A) and IMvigor210 trial (B). (C–E) Quantification analyses of effecter immune contexture including lymphoid cells (C), myeloid cells (D) and molecules (E) across stratification based on the evaluation of IHC staining. Data were analyzed by Kruskal-Wallis test, and presented as median and IQR. *P<0.05, **p<0.01, ***p<0.001. GZMB, granzyme B; IFN-γ, interferon-γ; IHC, immunohistochemistry; MIBC, muscle-invasive bladder cancer; PD-L1, programmed cell death ligand-1; PRF-1, perforin 1; TCGA, The Cancer Genome Atlas.
NKG2A and PD-L1 expression panel confers molecular alterations in MIBC
Remarkably, multiple studies have shown the impact of molecular features and genetic alteration on the tumor immune microenvironment and on ICB and ACT response heterogeneity.28 We found that patients characterized by basal squamous and high TMB were more distributed in NKG2Ahigh and PD-L1high patients. Furthermore, we selected several pathways associated with the therapeutic response to ACT and ICB (homologous recombination repair pathway, mismatch repair pathway, other DNA repair, histone modification, cell cycle, RAS/RAF pathway, PIK3CA/Akt pathway and receptor tyrosine kinase signaling) in the TCGA and IMvigor210 cohorts to explore the underlying genetic mechanism for the predictive value of our framework.29 We found that in both cohorts, more alterations in DNA repair, especially in the RB1 gene, were associated with the NKG2Ahigh and PD-L1high subgroup (figure 4A, B). Furthermore, we explored the predictive value of the NKG2Ahigh PD-L1high subgroup in the context of genetic alterations. We identified that in patients without selective pathway alterations, combined expression of NKG2A and PD-L1 could serve as a combined biomarker to predict response to ICB (figure 4C, D), which suggested NKG2AhiPD-L1hi patients without selected pathway alteration might benefit more from ICB therapy.
Figure 4.
NKG2A and PD-L1 expression panel correlates with molecular alterations in MIBC. (A and B) The landscape of genomic alterations according to NKG2A and PD-L1 expression in TCGA cohort (A) and IMvigor210 trial (B). (C and D) the HR for overall survival with NKG2Ahigh PD-L1high tumors versus others in the context of pathway alterations and molecular subtypes representing better responses in patients with NKG2Ahigh PD-L1high tumors. HRs were evaluated by univariate Cox analysis in ACT applied patients in TCGA cohort (C) and atezolizumab-applied patients in IMvigor210 cohort (D). *P<0.05, **p<0.01, ***p<0.001. ACT, adjuvant chemotherapy; GA, gene alteration; HRR, homologous recombination repair; MIBC, muscle-invasive bladder cancer; MMR, mismatch repair; PD-L1, programmed cell death ligand-1; RTK, receptor tyrosine kinase; TCGA, The Cancer Genome Atlas; TMB, tumor mutation burden; WT, wild type.
Discussion
Convincing evidence indicated that PD-L1 expression was upregulated by both intracellular oncogenic variants and ongoing immune responses, while immunity-dependent PD-L1 upregulation had better predictive value.8 Therefore, it would be of significance to distinguish immunity-dependent and immunity-independent PD-L1 expression. Recent studies provided a way to achieve this goal through simultaneous measurement of PD-1/PD-L1 axis-related immunity-dependent factors.14 NKG2A, as an additional brake for the PD-1/PD-L1 pathway, was regulated through inflammatory signaling.17 Meanwhile, inhibition of the NKG2A immune checkpoint restored the effector functions of CD8+ T cells and NK cells in preclinical cancer models.30 Overall, based on previous studies and our findings from three independent clinical cohorts, NKG2A and PD-L1 expression acted as a combinatorial biomarker to predict responses to ACT and PD-L1 blockade.
Tumor microenvironment (TME) status played an important role in tumor progression and therapeutic response.31 Inflamed TME was characterized by the infiltration of CD8+ T cells, myeloid cells and monocytic cells in the tumor parenchyma and the upregulation of effector cytokines. This profile suggested a pre-existing antitumor immune response in the microenvironment.32 Our study revealed that NKG2A and PD-L1 expression panel positively correlated with more immune cell infiltration (CD8+ T cells, B cells, M1 macrophages and NK cells) and elevated effector molecules level (IFN-γ, GZMB and PRF-1), implying that NKG2A and PD-L1 expression panel was associated with an inflamed microenvironment in MIBC. Furthermore, it was commonly reported that an inflamed microenvironment was associated with the efficacy of chemotherapy and immunotherapy,27 which was also supported in our previous studies.33–35 Consequently, the predictive value of NKG2A and PD-L1 expression panel was reasonable, reflecting the interaction between immunity and cancer.
Recent studies reported that combination immunotherapy might be more effective than single immune checkpoint inhibitors.36 37 Moreover, NKG2A was often coexpressed with PD-1 in tumors and NKG2A blockade combined with anti-PD-1/PD-L1 therapy improved tumor control.38 Consistent with these data, the combination of NKG2A and PD-L1 antibodies was more effective than single PD-L1 blockade in several targeted drug therapy trials for MSS colorectal cancer (NCT02671435) and non-small cell lung cancer (NCT03822351).18 19 However, the therapeutic responsiveness of the combination of NKG2A and PD-L1 antibodies in MIBC patients has not been reported.
Remarkably, GAs were the molecular drivers of tumorigenesis and led to transcriptional and translational variations that affected the dynamic balance between tumor cells and the microenvironment and response to ACT and ICB.39 40 In our study, we provided genomic alteration profiling and found that RB1 alterations were significantly much more frequent in NKG2AhiPD-L1hi patients. Previous studies illustrated that RB1 had a key role in restraining cell cycle and reflected activation of the IFN pathway.41 42 We also found that patients characterized by basal squamous and high TMB were more distributed in NKG2AhiPD-L1hi patients. Patients with basal squamous subtype and high TMB were generally sensitive to immunotherapy.43 These results indicated an underlying genetic mechanism for the predictive value of our framework.
Several limitations in our study should be mentioned. This was a retrospective analysis of three large sample cohorts that correlated the predictive value of the combined NKG2A and PD-L1 expression in patients with ACT and ICB. We would further validate our findings in a prospective, larger, multicentered randomized trial in the future. Moreover, although we found that high TMB was more distributed in NKG2AhiPD-L1hi patients, differences in calculation were between the TCGA cohort and IMvigor210 trial because of different sequencing technologies, which may have biased our analysis. In addition, patients in IMvigor210 cohort suffered not MIBC but metastasis urothelial carcinoma originating from the bladder. Therefore, the immune microenvironment and molecular features between the TCGA cohort and IMvigor210 cohort might be slightly different.
Our study identified for the first time the expression of NKG2A and PD-L1 as a combinatorial biomarker to predict the response to ACT and PD-L1 blockade in MIBC. We also confirmed that NKG2A and PD-L1 expression panel was associated with an inflamed microenvironment in MIBC. Cumulatively, this study provided insight into the predictive value of linking PD-L1 and NKG2A to precise screen sensitive patients for PD-L1 inhibitors and ACT.
Acknowledgments
We would like to thank Dr Lingli Chen (Department of Pathology, Zhongshan Hospital, Fudan University, Shanghai, China) and Dr Yunyi Kong (Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China) for their excellent pathological technology help.
Footnotes
SY, HZ, KJ and FS contributed equally.
Contributors: SY, HZ, KJ and FS for acquisition of data, analysis and interpretation of data, statistical analysis and drafting of the manuscript; ZL, YC, YW and YZ for technical and material support; ZW, LX and JX for study concept and design, analysis and interpretation of data, drafting of the manuscript, obtained funding and study supervision; JX is the guarantor for this study. All authors read and approved the final manuscript.
Funding: This study was funded by grants from National Natural Science Foundation of China (31770851, 81872082, 82002670, 82103408), Shanghai Municipal Natural Science Foundation (19ZR1431800, 22ZR1413400), Shanghai Sailing Program (21YF1407000) and Fudan University Shanghai Cancer Center for Outstanding Youth Scholars Foundation (YJYQ201802). All these study sponsors have no roles in the study design, in the collection, analysis and interpretation of data.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s)
Ethics approval
This study was approved by the Clinical Research Ethics Committee of Zhongshan Hospital, Fudan University (No. B2015-030). Written informed consent was obtained from each patient. Participants gave informed consent to participate in the study before taking part.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Smith AB, Deal AM, Woods ME, et al. Muscle-invasive bladder cancer: evaluating treatment and survival in the National Cancer Data Base. BJU Int 2014;114:719–26. 10.1111/bju.12601 [DOI] [PubMed] [Google Scholar]
- 3.Witjes JA, Bruins HM, Cathomas R, et al. European Association of Urology Guidelines on Muscle-Invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur Urol 2021;79:82–104. 10.1016/j.eururo.2020.03.055 [DOI] [PubMed] [Google Scholar]
- 4.Rijnders M, de Wit R, Boormans JL, et al. Systematic review of immune checkpoint inhibition in urological cancers. Eur Urol 2017;72:411–23. 10.1016/j.eururo.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 5.Apolo AB, Infante JR, Balmanoukian A, et al. Avelumab, an anti–programmed death-ligand 1 antibody, in patients with refractory metastatic urothelial carcinoma: results from a multicenter, phase Ib study. JCO 2017;35:2117–24. 10.1200/JCO.2016.71.6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balar AV, Galsky MD, Rosenberg JE, et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 2017;389:67–76. 10.1016/S0140-6736(16)32455-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Retz M, Siefker-Radtke A, et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol 2017;18:312–22. 10.1016/S1470-2045(17)30065-7 [DOI] [PubMed] [Google Scholar]
- 8.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity 2018;48:434–52. 10.1016/j.immuni.2018.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol 2016;17:e542–51. 10.1016/S1470-2045(16)30406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US food and drug administration (FDA) approvals of immune checkpoint inhibitors. J Immunother Cancer 2019;7:278. 10.1186/s40425-019-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu C, Fillmore CM, Koyama S, et al. Loss of LKB1 and PTEN leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 2014;25:590–604. 10.1016/j.ccr.2014.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HS, Lee JH, Nam SJ, et al. Association of PD-L1 expression with tumor-infiltrating immune cells and mutation burden in high-grade neuroendocrine carcinoma of the lung. J Thorac Oncol 2018;13:636–48. 10.1016/j.jtho.2018.01.008 [DOI] [PubMed] [Google Scholar]
- 13.Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 2018;17:129. 10.1186/s12943-018-0864-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seiwert TY, Burtness B, Mehra R, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1B trial. Lancet Oncol 2016;17:956–65. 10.1016/S1470-2045(16)30066-3 [DOI] [PubMed] [Google Scholar]
- 15.Petrie EJ, Clements CS, Lin J, et al. CD94-NKG2A recognition of human leukocyte antigen (HLA)-E bound to an HLA class I leader sequence. J Exp Med 2008;205:725–35. 10.1084/jem.20072525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viant C, Fenis A, Chicanne G, et al. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat Commun 2014;5:5108. 10.1038/ncomms6108 [DOI] [PubMed] [Google Scholar]
- 17.André P, Denis C, Soulas C, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by Unleashing both T and NK cells. Cell 2018;175:1731–43. 10.1016/j.cell.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho M, Bendell JC, Han S-W, et al. Durvalumab + monalizumab, mFOLFOX6, and bevacizumab in patients (PTS) with metastatic microsatellite-stable colorectal cancer (MSS-CRC). Annals of Oncology 2019;30:v490–1. 10.1093/annonc/mdz253.027 [DOI] [Google Scholar]
- 19.Martinez-Marti A, Majem M, Barlesi F, et al. LBA42 COAST: an open-label, randomised, phase II platform study of durvalumab alone or in combination with novel agents in patients with locally advanced, unresectable, stage III NSCLC. Ann Oncol 2021;32:S1320. 10.1016/j.annonc.2021.08.2121 [DOI] [Google Scholar]
- 20.Necchi A, Joseph RW, Loriot Y, et al. Atezolizumab in platinum-treated locally advanced or metastatic urothelial carcinoma: post-progression outcomes from the phase II IMvigor210 study. Ann Oncol 2017;28:3044–50. 10.1093/annonc/mdx518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariathasan S, Turley SJ, Nickles D, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 2016;387:1909–20. 10.1016/S0140-6736(16)00561-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamoun A, de Reyniès A, Allory Y, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol 2020;77:420–33. 10.1016/j.eururo.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L, Zhu Y, An H, et al. Clinical significance of tumor-derived IL-1β and IL-18 in localized renal cell carcinoma: associations with recurrence and survival1Contributed equally to this work. Urol Oncol 2015;33:68.e9–16. 10.1016/j.urolonc.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 26.Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fridman WH, Zitvogel L, Sautès–Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 28.Garnis C, Buys TPH, Lam WL. Genetic alteration and gene expression modulation during cancer progression. Mol Cancer 2004;3:9. 10.1186/1476-4598-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyamoto DT, Mouw KW, Feng FY, et al. Molecular biomarkers in bladder preservation therapy for muscle-invasive bladder cancer. Lancet Oncol 2018;19:e683–95. 10.1016/S1470-2045(18)30693-4 [DOI] [PubMed] [Google Scholar]
- 30.van Montfoort N, Borst L, Korrer MJ, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 2018;175:e1715:1744–55. 10.1016/j.cell.2018.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinshaw DC, Shevde LA. The tumor microenvironment Innately modulates cancer progression. Cancer Res 2019;79:4557–66. 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen DS, Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature 2017;541:321–30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 33.Zeng H, Zhou Q, Wang Z, et al. Stromal LAG-3+ cells infiltration defines poor prognosis subtype muscle-invasive bladder cancer with immunoevasive contexture. J Immunother Cancer 2020;8:e000651. 10.1136/jitc-2020-000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Zhou Q, Wang Z, et al. Intratumoral TIGIT+ CD8+ T-cell infiltration determines poor prognosis and immune evasion in patients with muscle-invasive bladder cancer. J Immunother Cancer 2020;8:e000978. 10.1136/jitc-2020-000978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu H, Zhu Y, Wang Y, et al. Identification and validation of stromal immunotype predict survival and benefit from adjuvant chemotherapy in patients with muscle-invasive bladder cancer. Clin Cancer Res 2018;24:3069–78. 10.1158/1078-0432.CCR-17-2687 [DOI] [PubMed] [Google Scholar]
- 36.McDermott DF, Huseni MA, Atkins MB, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24:749–57. 10.1038/s41591-018-0053-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 2015;161:205–14. 10.1016/j.cell.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Montfoort N, Borst L, Korrer MJ, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 2018;175:e1715:1744–55. 10.1016/j.cell.2018.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet 2021;22:3–18. 10.1038/s41576-020-0265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiter JG, Baretti M, Gerold JM, et al. An analysis of genetic heterogeneity in untreated cancers. Nat Rev Cancer 2019;19:639–50. 10.1038/s41568-019-0185-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamid AA, Gray KP, Shaw G, et al. Compound genomic alterations of TP53, PTEN, and Rb1 tumor suppressors in localized and metastatic prostate cancer. Eur Urol 2019;76:89–97. 10.1016/j.eururo.2018.11.045 [DOI] [PubMed] [Google Scholar]
- 42.Vidotto T, Nersesian S, Graham C, et al. DNA damage repair gene mutations and their association with tumor immune regulatory gene expression in muscle invasive bladder cancer subtypes. J Immunother Cancer 2019;7:148. 10.1186/s40425-019-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2018;174:1033. 10.1016/j.cell.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004569supp001.pdf (396.4KB, pdf)
Data Availability Statement
Data are available on reasonable request.