Abstract
Patients with ovarian cancer exhibit low response rates to anti-programmed cell death protein-1 (PD-1) based therapies, despite ovarian tumors demonstrating measurable immune responses. Therefore, the aim of the present study was to comparatively examine expression of notable immune co-stimulatory and co-inhibitory receptors in order identify the most abundant receptors that could potentially serve as therapeutic targets to enhance immunotherapy response in high grade serous ovarian cancer (HGSOC). The Cancer Genome Atlas (TCGA) was employed to compare levels of various HGSOC and pan-cancer cohorts. To confirm these findings at the protein level, immunofluorescence of select receptors was performed in 29 HGSOC patient tissue samples. TCGA and Kaplan Meier analysis was employed to determine the association of highly expressed immune receptors with clinical outcomes. TIM-3 and OX40 exhibited the highest expression in HGSOC at both the gene and protein level, with TIM-3 demonstrating highest levels on both CD8+ and CD4+ T cell subsets. Pan-cancer analysis determined that TIM-3 and OX40 levels were similar to those in immunotherapy-responsive cancers, while PD-1 exhibited much lower expression in HGSOC. Finally, OX40 was most strongly associated with improved patient survival. Overall, the current study suggested that TIM-3 and OX40 are frequently expressed intratumoral immune receptors in HGSOC and thus represent promising immune targets. Furthermore, the present analysis strongly suggested that OX40 was significantly associated with a longer survival and could potentially be utilized as a prognostic factor for improved patient outcomes in HGSOC.
Keywords: high grade serous ovarian cancer, OX40, TIM-3
Introduction
In the USA in 2022, there will be approximately 19,880 women newly diagnosed with epithelial ovarian cancer (EOC) and approximately 12,810 deaths attributed to this disease (1). Unfortunately, the five-year survival rate for EOC is only 48% (2), with minimal improvement in survival in the last 30 years (3). This is due to the fact that many patients develop a recurrence within 12-18 months of completion of their primary treatment regimen, at which time the cancer is less responsive to traditional platinum based-chemotherapeutics (4,5). In recent years, there have been approvals of new targeted therapies such as anti-angiogenic and poly (ADP-ribose) polymerase (PARP) inhibitors that modestly improve progression-free survival (PFS); however, neither have produced a substantial overall survival (OS) benefit for patients (6–8). Currently, the focus of EOC clinical trials is immune checkpoint inhibitors that restore the anti-tumor function of CD8+ T cells, such as monoclonal antibodies targeting programmed cell death protein-1 (PD-1) (9). While these PD-1 inhibitors have proven to be successful in cancers such as melanoma, non-small cell lung, and renal cell carcinoma, EOC clinical trials have shown that only a small portion of patients (10–33%) respond to anti-PD-1 therapy (10–12). Despite EOC patients exhibiting low responses to anti-PD-1 based therapies, ovarian tumors produce anti-tumor immune responses that can be detected in ascites, peripheral blood, and tumors (13). In addition, it has been well documented that higher intratumoral T cell numbers correlate to a better clinical prognosis for EOC patients (14,15). Therefore, there is a strong need for improved immunotherapeutic approaches for this patient population.
In recent years, a plethora of pre-clinical in vivo studies show that combinatorial targeting of alternative T cell co-receptors in combination with PD-1 increases tumor immune cell infiltration and improves survival in EOC patients (16). However, there have been no studies comparing expression levels of diverse intratumoral immune co-receptors in EOC tissue. The expression of different intratumoral immune co-receptors in EOC is of particular importance, since one potential reason that PD-1 based therapies have been unsuccessful in EOC is because programmed death-ligand 1 (PD-L1) often exhibits low expression in patient tumors, suggesting that not all ovarian tumors utilize the PD-1 signaling pathway to evade immune detection (17). To address this knowledge deficit, we performed immunohistochemistry analysis of high grade serous ovarian cancer (HGSOC) patient tissue along with analysis of The Cancer Genome Atlas (TCGA) ovarian cancer dataset to comprehensively examine intratumoral expression of PD-1 and seven of the most commonly investigated immune co-receptors in the field of oncology, which are summarized in Table I (18–38). Therefore, this analysis provides a thorough depiction of immune co-receptor composition in HGSOC for the first time.
Table I.
Commonly investigated immune receptors in the field of oncology.
| Immune receptor | Cellular expression (18) | Main ligand(s) (18) | Main Physiological roles | Phases in clinical development (18,22) |
|---|---|---|---|---|
| PD1 (CD279) | T cells, B cells, NK cells and tumor infiltrating lymphocytes | PD-L1 and PD-L2 | Inhibits T cell proliferation (23) and both adaptive and innate immune responses (24). Maintains immune tolerance (24) | FDA approved (pembrolizumab and nivolumab) |
| CTLA-4 (CD152) | T cells | B7-1 (CD80) B7-2 (CD286) | Inhibits T cell proliferation, differentiation and function (36) | FDA approved (ipilimumab) |
| BTLA (CD272) | cells, resting B cells, NK cells,macrophages, and dendritic cells | HVEM (TNFRSF14) | Inhibits T cell proliferation (19) Promotes CD8+ T cell and T regulation cell differentiation (20,21) | Pre-clinical |
| CD137 (4-1BB) | Dendritic cells, NK cells, adaptive/activated CD4+ and CD8+ T cells and T regulation cells | CD137L (4-1BBL) | Induces T cell proliferation and survival via production of INFy and IL-2 (25). Maintenance and memory of CD8+ T cells (26) | II (solid tumors, NHL, NSCLC, RCC, HNCC and HCC) |
| LAG-3 (CD223) | T cells, B cells, NK cells and dendritic cells | MHC Class II and HLA class II FGL1 (37) | Negatively regulated T cell activation and function (38) Controls memory T cell expansion (28). Promotes tolerance on CD8+ T cells (29). Required for maximal suppressive T regulation cell activity (27,29) | I/II (solid tumors, pancreatic and breast melanoma) |
| OX40 (TNFRSF4 and CD134) | T cells | OX40L (TNFSF4) | Enhances proliferation and survival of T cells, increasing effector molecule expression and cytokine secretion (30,31). Inhibits T regulation cell Function (30) | I (solid tumors, melanoma and NSCLC) |
| TIM-3 (HAVCR2 and CD366) | CD8+/CD4+ cells, T regulation cells, Th17 cells and NK cells | Galectin-9 (LGAL9) | Mediates T cell exhaustion during chronic viral infections (32). Promotes MDSCs (33). Regulates function of FOXP3+T regulation cells (33) | I/II (advanced malignances and solid tumors) |
| ICOS (CD278) | Activated T cells (34) | ICOSL (34) | Enhances broad cytokine production to enhance proliferation of effector and regulatory T cell populations and promote memory cell development (35) | I/II (advanced solid tumors) (35) |
Materials and methods
Patient samples
Formalin-fixed, paraffin embedded (FFPE) tumor blocks from 29 stage III, grade 3 serous EOC patients were obtained from Women and Infants Hospital's Pathology Department, under Institutional Review Board (IRB) approval. Summarized patient data is presented in Table II. Specimen processing and analysis of samples was performed with IRB approval and in compliance with Women and Infant's IRB HIPAA requirements. All patient tumor samples were representative of patients' primary debulking, and thus naïve to therapy.
Table II.
Patient clinical outcomes.
| Patient number | Age at diagnosis (years) | CA125 pre-op | HE4 pre-op | Stage (FIGO) | Grade (FIGO) | Debulking status | PFS (months) | OS (months) |
|---|---|---|---|---|---|---|---|---|
| 1 | 80-84 | >1,000 | 0-500 | IIIC | 3 | Optimal | 36 | 96+ |
| 2 | 65-69 | 0-500 | 0-500 | IIIC | 3 | Suboptimal | 6 | 10 |
| 3 | 60-64 | 0-500 | 0-500 | IIIC | 3 | Optimal | 8 | 18 |
| 4 | 55-59 | 500-1,000 | 500-1,000 | IIIC | 3 | Suboptimal | 18 | 55 |
| 5 | 60-64 | 0-500 | >1,000 | IIIA | 3 | Optimal | 14 | 29 |
| 6 | 80-84 | >1,000 | 500-1,000 | IIIC | 3 | Suboptimal | 21 | 30 |
| 7 | 55-59 | 1,000 | 500-1,000 | IIIC | 3 | Optimal | 23 | 78 |
| 8 | 80-84 | >1,000 | >1,000 | IIIC | 3 | Suboptimal | 38 | 84+ |
| 9 | 60-64 | 500-1,000 | >1,000 | IIIC | 3 | Optimal | 10 | 66 |
| 10 | 75-79 | 0-500 | 0-500 | IIIC | 3 | Optimal | 19 | 45 |
| 11 | 75-79 | 0-500 | N/A | IIIC | 3 | Optimal | 62 | 66+ |
| 12 | 70-74 | >1,000 | 500-1,000 | IIIC | 3 | Optimal | 7 | 28 |
| 13 | 50-54 | >1,000 | 0-500 | IIIC | 3 | Optimal | 20 | 50 |
| 14 | 50-54 | 0-500 | N/A | IIIC | 3 | Optimal | 90+ | 90+ |
| 15 | 65-69 | >1,000 | N/A | IIIC | 3 | Optimal | 20 | 67+ |
| 16 | 55-59 | 0-500 | N/A | IIIC | 3 | Suboptimal | 6 | 14 |
| 17 | 75-79 | 500-1,000 | 0-500 | IIIC | 3 | Optimal | 67+ | 67+ |
| 18 | 55-59 | 0-500 | 0-500 | IIIC | 3 | Optimal | 26 | 85+ |
| 19 | 65-69 | >1,000 | N/A | IIIC | 3 | Suboptimal | 7 | 15 |
| 20 | 70-74 | >1,000 | N/A | IIIC | 3 | Optimal | 16 | 38 |
| 21 | 55-59 | 0-500 | N/A | IIIC | 3 | Optimal | 33 | 84 |
| 22 | 65-69 | 0-500 | 0-500 | IIIC | 3 | Optimal | 45 | 49+ |
| 23 | 65-69 | N/A | N/A | IIIC | 3 | Optimal | 4 | 6 |
| 24 | 50-54 | 0-500 | 0-500 | IIIC | 3 | Optimal | 93+ | 93+ |
| 25 | 40-44 | 0-500 | 0-500 | IIIC | 3 | Optimal | 84+ | 84+ |
| 26 | 65-69 | 0-500 | >1,000 | IIIC | 3 | Optimal | 10 | 22 |
| 27 | 55-59 | 0-500 | N/A | IIIC | 3 | Optimal | 100+ | 100+ |
| 28 | 60-64 | 0-500 | N/A | IIIC | 3 | Optimal | 72+ | 72+ |
| 29 | 55-56 | 0-500 | N/A | IIIC | 3 | Suboptimal | 62 | 64 |
CA125, cancer antigen 125; HE4, Human epididymis protein 4; PFS, progression free survival; OS, overall survival.
Immunofluorescence
FFPE human ovarian cancer tissue slides were baked at two hours at 65°C. Slides were then washed in xylene, 100% ethanol, 95% ethanol, 70% ethanol, deoxygenated water, and FTA Hemagglutination Buffer. Antigen retrieval was then performed using Antigen Retrieval Solution (1X) (Vector Laboratories, H-3300) and heated to 95°C for 20 min. Slides were then blocked with 5% horse serum in FTA Hemagglutination buffer and incubated overnight in primary antibody at 4°C. Secondary antibody was then applied to slides following incubation in the dark at room temperature for one hour. Slides were washed between each step using FTA Hemagglutination buffer and cover-slipped with DAPI containing mounting medium (Vector Laboratories, H-1200). A representative image of immune receptor co-staining with DAPI can be seen in Fig. S1. Primary antibodies were all used at a [1:50] dilution, with vendor and catalog numbers as follows: CD8 (Origene TA802079 and Abcam ab4055), CD4 (Origene, UM800010 and Abcam ab133616), PD-1 (Origene, UM870089), OX40 (Origene UM870166), BTLA (Origene, TA505536), CD137 (Abcam, ab232990), TIM-3 (Origene, TA807034), LAG-3 (Origene, TA807082). The secondary antibodies used were either Anti-Rabbit IgG Dylight 488 or Anti-Mouse IgG Dylight 594 (Vector Laboratories, DI-1488 and DI-2594), each at a 1:1,000 dilution.
Microscopy
Representative confocal images were acquired with a Nikon C1si confocal (Nikon Inc.) or a Nikon Eclipse Ti Microscope (Nikon Inc.) using diode lasers 402, 488, and 561. To obtain images for cell counting, ten randomly selected fields per case were acquired with a Nikon C1si confocal (Nikon Inc.) or a Nikon Eclipse Ti Microscope (Nikon Inc.) using a 40X objective. Each wavelength was acquired separately and an RGB image was created.
Image analysis
Image processing and analysis were performed in Photoshop CS6 (Adobe) or GIMP image analysis software (GIMP, Bremen, Germany). For each of the ten randomly selected image fields, total numbers of positive immune cells were counted. Single positive CD8+, CD4+, PD-1+, OX40+, TIM-3+, LAG-3+, BTLA+, and CD137+ cells were counted, as well as double positive CD8+ and CD4+ cells with each of the six immune receptors. Where average T cell or immune receptor levels are reported, the total number of positive cells per field was averaged from each patient from the cohort stained. Immune receptor and T cell counts for each patient are summarized in Data S1.
The Cancer Genome Atlas (TCGA)
The ovarian cancer TCGA dataset with complete RNA-sequencing results (n=378) from The Cancer Genome Atlas was obtained using GenomicDataCommons (version 1.12.0) and RStudio (R version 4.0.0) (39,40) which can be found at http://github.com/Bioconductor/GenomicDataCommons. Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values were obtained for PD-1, CTLA-4, ICOS, LAG-3, BTLA, CD137, TIM-3, OX40, PD-L1, OX40L, and Galectin-9.
cBioPortal
Survival outcomes and residual disease related to mRNA expression (−log10) of either OX40, TIM-3, Galectin-9, or PD-L1 were obtained from TCGA ovarian serous cystadenocarcinoma cohort (Nature 2011) in cBioPortal (https://cbioportal.org). Data was available for 316 samples. Moreover, TCGA PanCancer Atlas Studies from cBioPortal was employed to determine expression levels of PD-1, OX40, and TIM-3 [mRNA expression RNA seq log2 (value+1)] in breast (n=1082), cervical (n=294), ovarian (n=300), uterine (n=527), clear cell renal cell carcinoma (n=510), papillary renal cell carcinoma (n=283), lung adenocarcinoma (n=510), squamous lung cancer (n=484), and melanoma (n=443) cohorts.
Kaplan-Meier plotter
The ovarian cancer Kaplan-Meier Plotter (https://kmplot.com/analysis/index.php?p=service&cancer=ovar), which compiles Gene Expression Omnibus (GEO) Series (GSE) and TCGA data for analysis (41), was employed to determine the association between TIM-3 and OX40 expression with PFS and OS, using median expression as a cutoff.
Statistical analysis
Kaplan-Meier survival curve analysis was used to compare survival in patients from our Women and Infants Hospital cohort with high and low levels of CD4+ OX40+ T cells in which the top and bottom quartiles of expression delineated the groups. Mann Whitney U test was employed to compare mean ranks of TIM-3, OX40, and PD-1 expression across various cancers as well as the %CD8+ OX40+ populations in patients with low or high median CA125 levels (U value and P-value reported). Two-tailed unpaired t-test was used to determine significant differences in ligand expression of OX40L, Galectin-9, and PD-L1, as well as expression of OX40, TIM-3, OX40L, Galectin-9, or PD-L1 according to survival and residual disease outcomes. Statistical analyses were performed in GraphPad Prism. P<0.05 was considered significant.
Results
Intratumoral composition of immune co-receptors in HGSOC
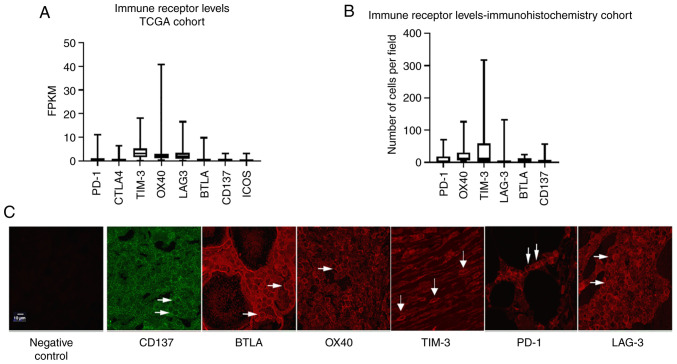
Analysis of ovarian cancer TCGA data was performed to compare mRNA levels (FKPM) of the top eight most commonly studied immune receptors in EOC: PD-1, cytotoxic T lymphocyte associated protein-4 (CTLA-4), T-cell immunoglobin and mucin domain containing 3 (TIM-3), OX40, CD137, B and T lymphocyte attenuator (BTLA), and inducible T cell co-stimulator (ICOS). The highest median transcript levels were observed in TIM-3 (3.13) and OX40 (2.02), followed by LAG-3 (1.86). PD-1 demonstrated a median expression level of 0.54, with all other immune co-receptors-CTLA-4, BTLA, CD137, and ICOS-exhibiting transcript levels of ≥0.28 (Fig. 1A). To confirm this data, immunohistochemistry was employed to determine protein levels of select immune co-receptors in a cohort of ten HGSOC patient tissues. Overall, there was a strong concordance between TCGA mRNA expression and protein expression by immunohistochemistry, with OX40 and TIM-3 immune co-receptors demonstrating the highest median expression (11.75 and 11.5 positive cells per field, respectively). Furthermore, in agreement with TCGA mRNA expression, PD-1, BTLA, and CD137 exhibited low intratumoral median expression with ≤1.5 positive cells per field. Interestingly, LAG-3+ cell numbers did not correlate with TCGA mRNA data, as immunohistochemical analysis revealed an average of only 0.5 positive cells per field (Fig. 1B), which was corroborated with two different LAG-3 antibodies (data not shown). A representative image of all co-receptors can be seen in Fig. 1C.
Figure 1.
Intratumoral composition of immune co-receptors in HGSOC. (A) Transcript expression (FKPM) of immune co-receptors in the TCGA ovarian cancer cohort. (B) Number of positive immune co-receptors per field from immunohistochemical analysis of a ten-patient HGSOC cohort. (C) Representative confocal images of positive immune co-receptor staining in patient tumors (magnification, ×40). Arrows indicate positive cells. HGSOC, high-grade serious ovarian cancer; TCGA, The Cancer Genome Atlas.
Characterization of TIM-3 and OX40 expression in helper and cytotoxic T cell population
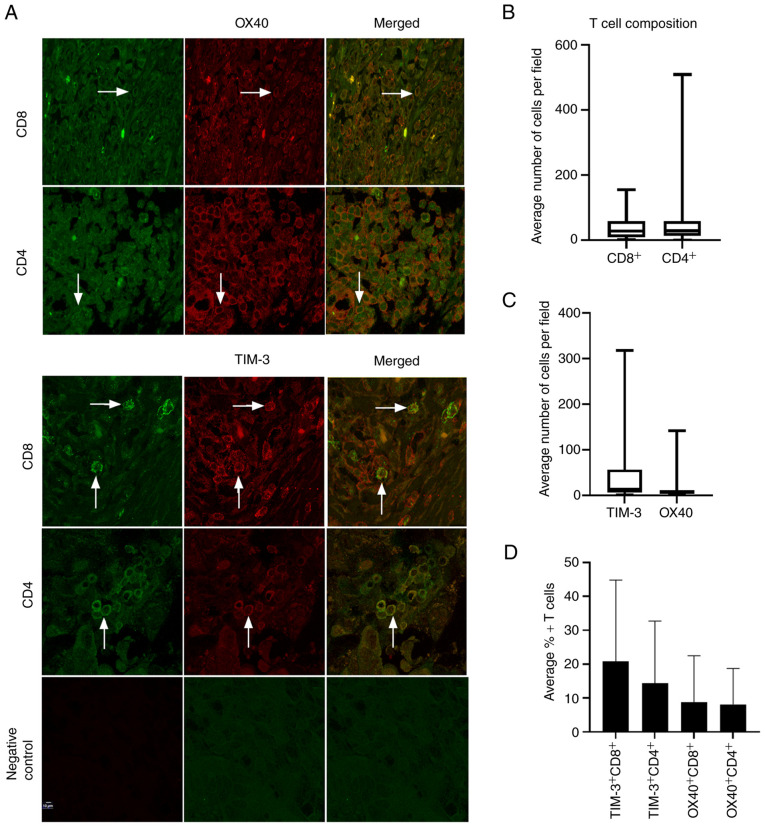
Next, since TIM-3 and OX40 were the most highly expressed immune co-receptors at both the gene and protein level, an additional 19 HGSOC patient tissues were stained for OX40 and TIM-3 in both CD4+ and CD8+ T cell populations via immunohistochemistry, for a total of 29 tissues (Fig. 2A). Median levels of positive cells per field were 31 for CD8+ cells and 29.1 for CD4+ cells (Fig. 2B). TIM-3 and OX40 demonstrated median average cells per field of 14.3 and 6.3, respectively (Fig. 2C). Examining co-expression of TIM-3 and OX40 in both cytotoxic and helper T cell populations revealed that TIM-3 was present on 20.8% of CD8+ T cells, and 14.0% of CD4+ T cells, while OX40 was expressed on 8.9% of CD8+ and 8.0% of CD4+ T cells (Fig. 2D), establishing that TIM-3 was more highly expressed on both CD8+ and CD4+ subsets.
Figure 2.
TIM-3 and OX40 expression in cytotoxic and helper T cell populations. (A) Representative confocal images of the double immunofluorescence staining of TIM-3 or OX40 with CD8 or CD4 from a 29-patient cohort (magnification, ×40). Arrows indicate double-positive cells. (B) Number of CD8+ and CD4+ T cells per field. (C) Number of TIM-3+ and OX40+ cells per field. (D) Average percentages of TIM-3 and OX40 positive CD8+ and CD4+ T cells.
TIM-3 and OX40 expression across select cancer subtypes
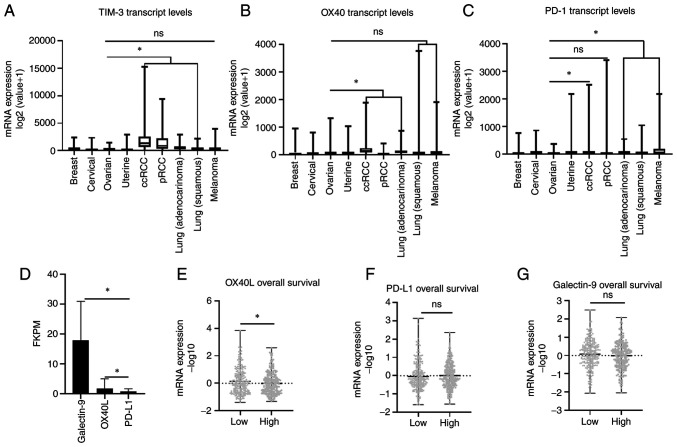
After examining the specific staining patterns of TIM-3 and OX40 in helper and cytotoxic T cells, transcript expression of both TIM-3 and OX40 along with PD-1 were compared across a Pan Cancer TCGA cohort that included breast, gynecologic malignancies, and immunotherapy responsive cancers such as renal cell carcinoma (RCC), lung cancer, and melanoma. There was no significant difference in TIM-3 expression between ovarian cancer and melanoma (U=64916, P=0.5632), while a significant lower expression was observed in ovarian cancer compared to clear cell RCC (ccRCC) (U=17157, P<0.0001), papillary RCC (pRCC) (U=18861, P<0.0001), lung adenocarcinoma (U=48751, P<0.0001), and squamous lung cancer (U=65968, P=0.0314) (Fig. 3A). A similar trend was observed with OX40 as transcript levels were not significantly different between ovarian cancer and melanoma (U=64329, P=0.4601), or squamous lung cancer (U=70892, P=0.5796) cohorts, while exhibiting significantly lower levels (P<0.0001) compared to ccRCC (U=35011) and lung adenocarcinoma (U=52535), and higher levels compared to pRCC (U=16069) (Fig. 3B). Finally, comparing PD-1 mRNA expression revealed significantly lower expression (P<0.0001) in ovarian cancer cohorts compared to melanoma (U=42864), ccRCC (U=41215), lung adenocarcinoma (U=33339), and squamous lung cancer (U=40306). No significant difference between PD-1 expression was observed between ovarian cancer and pRCC cohorts (U=38942, P=0.084). Moreover, the ovarian cancer cohort demonstrated the lowest median transcript expression of PD-1 out of all cancers analyzed (Fig. 3C). These results collectively suggest that OX40 and TIM-3 may represent more clinically relevant immune co-receptors for immunotherapy targeting than PD-1 in ovarian cancer.
Figure 3.
Comparative ligand expression and pan-cancer analysis for TIM-3 and OX40. TCGA Pan Cancer Atlas Cohort was employed to examine mRNA expression [log2(value+1)] of (A) TIM-3 (B) OX40 and (C) PD-1 across select cancer subtypes. (D) Galectin-9, OX40L and PD-L1 average transcript expression (FKPM) from the TCGA ovarian cancer cohort. TCGA ovarian cancer cohort analysis of (E) OX40L (F) PD-L1 and (G) Galectin-9 mRNA expression (−log10) stratified by median overall survival. *P<0.005 as indicated. NS, not significant; TCGA, The Cancer Genome Atlas; ccRCC, clear cell renal cell carcinoma; pRCC, papillary renal cell carcinoma; PD-1, programmed cell death protein-1.
TIM-3 and OX40 ligand expression
Ligand expression for TIM-3 (Galectin-9), OX40 (OX40L) and PD-1 (PD-L1) was compared in the ovarian cancer TCGA cohort. Significantly higher (P<0.001) transcript expression (FKPM) of Galectin-9 (22.16-fold) and OX40L (2.07-fold) was observed compared to PD-L1 levels (Fig. 3D). In addition, when ligand levels were stratified according to median OS, mean mRNA expression (−log10 value) of OX40L was significantly (P=0.048) elevated in patients with a shorter survival (Fig. 3E), while TIM-3 (P=0.4873) and PD-L1 (P=0.2221) expression was not significantly different when stratified by OS (Fig. 3F and G). Therefore, as was observed for their co-receptor levels, Galectin-9 and OX40L are more abundantly expressed than PD-L1 in HGSOC.
Clinical outcomes related to TIM-3 and OX40 expression
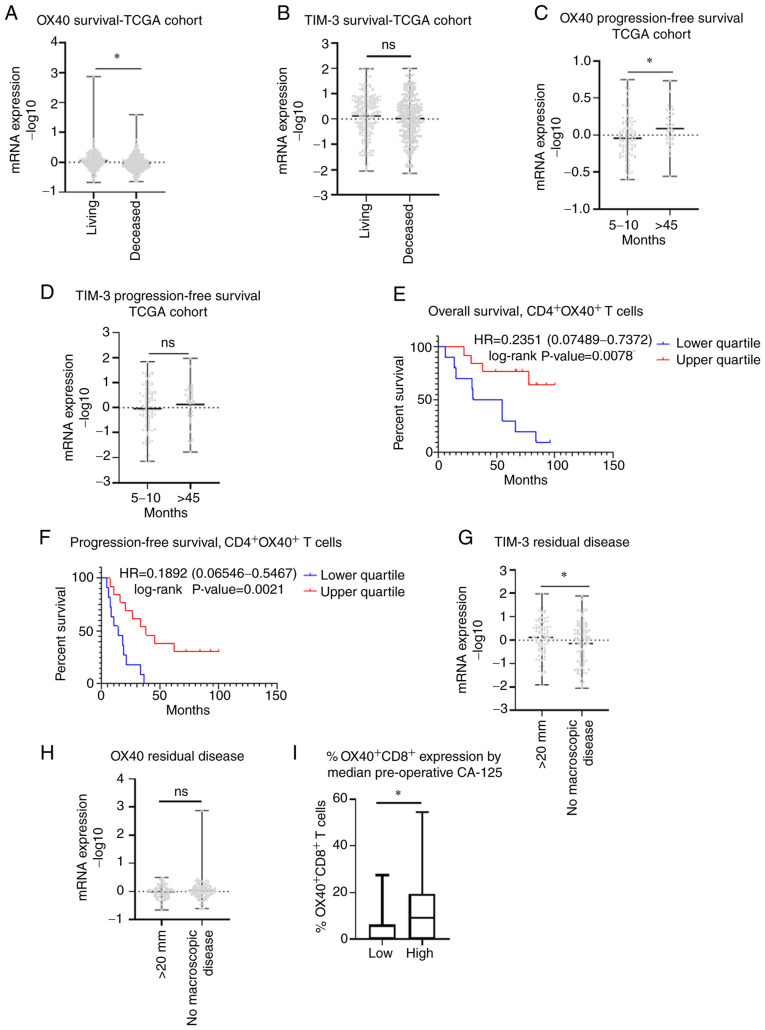
Next, we stratified TIM-3 and OX40 protein expression from our immunohistochemical analysis by median patient PFS and OS. No significant difference in expression between short and long median survival groups was observed for either receptor (Fig. S2A-D). This observation was confirmed at the genomic level utilizing TCGA data (Fig. S2E-H). Upon examining alternative survival cohorts from TCGA data, we observed that transcript expression (−log 10) of OX40 was 2.16-fold higher (P=0.0010) in patients who were living vs. deceased. Similarly, median TIM-3 levels were higher in living patients than deceased patients, however this association did not reach significance (P=0.1841) (Fig. 4A and B). Furthermore, comparing patients with a short (5–10 months) and an exceptionally long (>45 months) PFS, we saw that OX40 was significantly higher (P=0.0134) in patients with a longer PFS (Fig. 4C). While TIM-3 levels were also elevated in patients with a longer PFS, this trend was not significant (P=0.3271) (Fig. 4D). Moreover, Kaplan-Meier curve analysis from our immunohistochemical data demonstrated that higher levels of OX40 in CD4+ T cells was associated with an improved OS (HR=0.2351 [0.07489-0.7372], P=0.0078) (Fig. 4E) and PFS (HR=0.1892 [0.06546-0.5467], P-value=0.0021) (Fig. 4F). TCGA analysis was again applied to compare OX40 and TIM-3 expression when stratified according to residual disease outcomes following debulking surgery. TIM-3 levels were significantly higher in patients who exhibited >20 mm residual disease than in patients with no macroscopic disease (P=0.0471) (Fig. 4G). In contrast, there was no significant difference in OX40 levels between the cohorts with >20 mm residual disease vs. no macroscopic disease (P=0.3493) (Fig. 4H). Finally, as CA-125 is routinely used as a serum diagnostic marker in conjunction with radiology to detect ovarian disease, we sought to determine how OX40 expression related to these pre-operative levels. Our immunohistochemical data revealed a significantly higher percentage of OX40+ CD8+ cells in patients with higher median preoperative CA-125 levels (U=55, P=0.0345) (Fig. 4I), with no significant relationships detected between CA-125 and OX40+ cells alone or OX40+ CD4+ cells (data not shown).
Figure 4.
Clinical outcomes associated with TIM-3 and OX40 expression. TCGA ovarian cancer cohort analysis of mRNA expression (−log10) in (A) OX40 and (B) TIM-3 stratified according to living vs. deceased patients. Transcript expression (−log10) of (C) OX40 and (D) TIM-3 in patients with a low progression-free survival (5–10 months) and high (>45 months) from the TCGA ovarian cancer cohort. Kaplan-Meier survival curve analysis comparing (E) overall and (F) progression-free survival of upper and lower quartile expression of CD4+ OX40+ T cells from Women and Infants Hospital's immunohistochemical data in 29 patients with HGSOC. Log-rank hazard ratios and P-values are reported, with 95% confidence intervals in parentheses. mRNA expression (−log10) of (G) TIM-3 and (H) OX40 grouped according to no macroscopic disease and >20 mm of disease from the TCGA ovarian cancer dataset. (I) % OX40+ CD8+ T cells stratified according to median CA125 preoperative levels obtained from immunohistochemical staining in 29 HGSOC tumors. *P<0.005 as indicated. CA125, cancer antigen 125; NS, not significant; TCGA, The Cancer Genome Atlas.
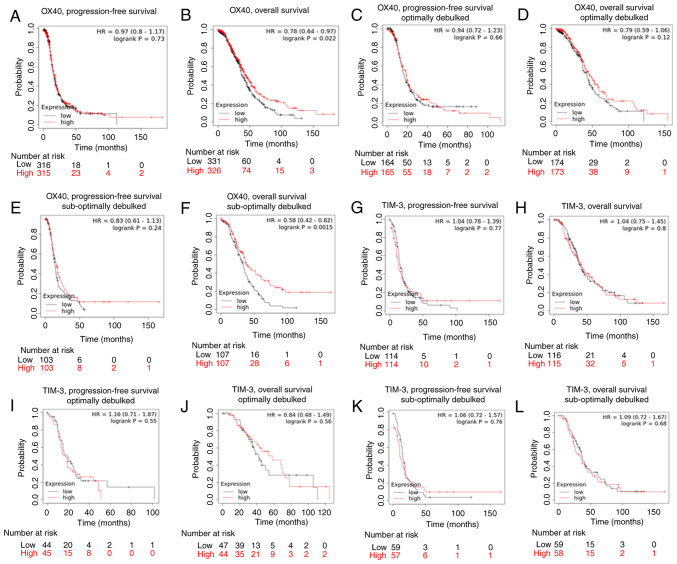
Lastly, we employed Kaplan-Meier survival curve analysis to examine the relationship between TIM-3 and OX40 and survival using TCGA and GSE data. Higher OX40 expression was significantly associated with a longer OS [HR=0.78 (0.64-0.97), P=0.022], but not PFS [HR=0.97 (0.8-1.17), P=0.73] in stage III grade 3 patients (Fig. 5A and B). Once restricting expression according to debulking status, it was discovered that while increased OX40 expression was not associated with improved OS or PFS in patients that were optimally debulked, in suboptimally debulked patients a significant association with improved OS [HR=0.58 (0.42-0.82), P=0.0.0015], and not PFS [HR=0.83 (0.61-1.13), P=0.24] was identified. (Fig. 5C-F). Conversely, no relationship was detected between TIM-3 expression and survival outcomes in stage III, grade 3 ovarian cancer patients (Fig. 5G-L). Taken as a whole, these survival analyses identified that high OX40 expression is most significantly and consistently associated with improved patient survival.
Figure 5.
Kaplan-Meier survival curve analysis of TIM-3 and OX40. Kaplan-Meier survival analysis was performed using The Cancer Genoma Atlas and Expression Omnibus Series data in patients with serous stage III, grade 3 ovarian cancer. Kaplan-Meier curves were generated to determine the association of OX40 with (A) progression-free survival, (B) overall survival, (C) progression-free survival (optimal debulking sub-cohort), (D) overall survival (optimal debulking sub-cohort), (E) progression-free survival (suboptimal debulking sub-cohort) and (F) overall survival (suboptimal debulking sub-cohort). Kaplan-Meier curves were generated depicting the association of TIM-3 with (G) progression-free survival, (H) overall survival, (I) progression-free survival (optimal debulking sub-cohort), (J) overall survival (optimal debulking sub-cohort), (K) progression-free survival (suboptimal debulking sub-cohort), and (L) overall survival (suboptimal debulking sub-cohort). Median expression of either OX40 or TIM-3 was used to delineate ‘low’ vs. ‘high’ expressing groups. Log-rank hazard ratios and P-values are reported, with 95% confidence intervals in parentheses.
Discussion
This comparative analysis revealed that TIM-3 and OX40 are two prominently expressed immune co-receptors in HGSOC, and consistently more highly expressed than PD-1 at the RNA and protein level. Furthermore, specific levels of both co-receptors were characterized in helper and cytotoxic T cell populations, revealing higher expression of TIM-3 in CD4+ and CD8+ T cells. While this T cell expression analysis was limited due to a small sample size, it will be necessary to verify these findings in a larger patient cohort, as well as expand our analyses to regulatory T cell populations. Nevertheless, to the best of our knowledge, there has been only one comprehensive intratumoral immune profiling paper in HGSOC. Rådestad et al similarly discovered high levels of TIM-3 in CD4+ and CD8+ populations and additionally found that that highest degree of co-expression out of all immune receptors analyzed existed on TIM-3+ PD-1+ CD8+ T cells (42). The naturally high expression of TIM-3, along with its high degree of co-expression with PD-1 indicates that TIM-3 could be an efficacious combinatorial partner to anti-PD-1 therapy. Moreover, a study by Fucikova et al found that TIM-3 was highly expressed on CD8+ populations, with PD-1+ TIM-3+ CD8+ T cells exhibiting all hallmarks of functional exhaustion and correlating with poor clinical outcomes in HGSOC (43). Conversely, neither CTLA-4, LAG-3, or the PD-1/PD-L1 axis alone were found to contribute to clinically meaningful immunosuppression, suggesting that TIM-3 might play a pivotal role in suppressing immune responses in HGSOC (43). Hence, the high degree of expression exhibited by TIM-3 in HGSOC discovered by our group and others coupled with its high degree of co-expression with PD-1 suggests that it may represent a viable treatment modality to increase ovarian cancer patient response to PD-1 based therapy.
Our data revealed that while TIM-3 exhibited higher levels on both helper and cytotoxic T cell populations, OX40 was most significantly associated with longer patient survival. Using ovarian cancer TCGA and GSE data, we found that higher OX40 transcript levels are consistently associated with a significantly longer survival. Furthermore, OX40′s ligand, OX40L was found to be significantly associated with improved survival. Finally, our original data suggests that intratumoral co-expression of CD4 and OX40 is significantly associated with improved HGSOC patient PFS and OS. Interestingly, a study by Ramser et al reported that levels of OX40 in ovarian cancer patient pre-treatment biopsies significantly correlated to chemosensitivity. In addition, patients who exhibited an increase in intratumoral OX40 expression within recurrent biopsies had improved recurrence free survival (RFS) (44). These studies corroborate our data showing that increased OX40 expression is associated with improved patient clinical outcomes both in the upfront and recurrent treatment setting, suggesting OX40′s utility as a prognostic factor for improved survival in HGSOC. In order to validate these results, a large prospective cohort should be initiated in addition to further investigations that examine if OX40 expression can be detected through non-invasive sampling to increase clinical utility of this potential immune prognostic marker.
There have been limited pre-clinical studies performed that have explored the efficacy of targeting either TIM-3 or OX40 with a PD-1 inhibitor. A study by Guo et al reported that targeting OX40 and PD-1 in combination significantly reduced murine tumor growth, increased levels of helper and cytotoxic T cells, promoted interferon-gamma (IFNγ) secretion, and decreased levels of regulatory T cells (Treg) and myeloid-derived suppressor cells (MDSCs) (45). Furthermore, an in vitro study that targeted TIM-3 and PD-1 in combination demonstrated an increase in the production of granzyme B, INFγ, and perforin in cytotoxic T cells, while inhibition of each immune receptor alone produced no such effects (43). In recent years, several early clinical trials have been initiated examining monoclonal antibodies against OX40 and TIM-3 in combination with anti-PD-1 based inhibitors. An OX40 agonist (PF-04518600) has been investigated in combination with the PD-1 inhibitor avelumab in a phase 2 triple negative breast cancer trial (NCT-3971407) and a phase 1b/2 trial in solid tumors that includes a platinum resistant ovarian cancer cohort (NCT02554812). Furthermore, a phase 1 trial evaluating a hexavalent OX40 agonist (INBR-X-106) in combination with another PD-1 inhibitor, pembrolizumab, in advanced or metastatic solid tumors has also been initiated (NCT041987). There is one TIM-3 inhibitor (TSR-022) currently being investigating in clinical trials. A phase 1 trial evaluating TSR-022 in combination with nivolumab (anti-PD-1 monoclonal antibody) in solid tumors (NCT02817633), as well as a phase 2 trial in melanoma in addition to the PD-1 inhibitor dostarlimab (NCT04139902) have commenced. Ultimately, results from these early clinical trial studies will provide insight into the effectiveness of OX40 and TIM-3 based immunotherapies.
This study represents an intratumoral expression analysis of several of the most commonly studied immune receptors in EOC, which identified TIM-3 and OX40 as having the highest distribution of expression. Therefore, these receptors represent potential therapeutic targets that in combination with current PD-1 based inhibitors could improve patient response to immunotherapy. Furthermore, our analyses suggest that OX40 is most significantly associated with improved survival compared to TIM-3, and may serve as a prognostic immune factor for improved outcomes in HGSOC.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Joselynn Wallace (COBRE Center for CBHD, Brown University) for assistance with acquiring TCGA data. The authors would also like to thank Dr Christina Raker (Division of Research, Women and Infants Hospital) for counseling on all statistical analyses performed.
Funding Statement
The present study was supported by the Rhode Island Hospital Center for Cancer Research Development COBRE Pilot Award supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (grant no. 5P30GM110759); the Division of Gynecologic Oncology, Program in Women's Oncology at Women & Infants Hospital; Swim Across America; the Kilguss Research Core of Women & Infants Hospital, and the COBRE Center for Computational Biology of Human Disease (CBHD) at Brown University. The Kilguss Research Core is supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (grant no. P30GM114750). The COBRE Center for CBHD is supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (grant no. P20GM109035).
Availability of data and materials
All data generated during this study are included in this published article.
Authors' contributions
NEJ and JRR were responsible for the conceptualization and design of the study. ADV, MW and NEJ performed immunohistochemical staining. Image acquisition and analysis was executed by VH, JRR and NEJ. ADV, JBE, and KM performed chart reviews. All statistical analyses were performed by NEJ and JRR. JO obtained all tissue samples utilized and assured they met minimum quality standards. JRR oversaw all aspects of the study. NEJ, JRR and ADV prepared the manuscript. NEJ and JRR confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patient tissue used for this study was obtained under the approval of the Women and Infants Hospital Institutional Review Board (protocol no. 1326537). Under this approved protocol, a waiver of consent and Health Insurance Portability and Accountability Act (HIPAA) was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.American Cancer Society, corp-author. https://www.cancer.org/cancer/ovarian-cancer/about/key-statistics.html. [ February 1; 2022 ];Key Statistics for Ovarian Cancer. [Google Scholar]
- 2.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, Bhattacharya R, Novak M, Jones S, Phallen J, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. 2017;8:1093. doi: 10.1038/s41467-017-00962-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan RD, Clamp AR, Evans DGR, Edmondson RJ, Jayson GC. PARP inhibitors in platinum-sensitive high-grade serous ovarian cancer. Cancer Chemother Pharmacol. 2018;81:647–658. doi: 10.1007/s00280-018-3532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvakumaran M, Pisarcik DA, Bao R, Yeung AT, Hamilton TC. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- 6.Rossi L, Verrico M, Zaccarelli E, Papa A, Colonna M, Strudel M, Vici P, Bianco V, Tomao F. Bevacizumab in ovarian cancer: A critical review of phase III studies. Oncotarget. 2017;8:12389–12405. doi: 10.18632/oncotarget.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu G, Yang D, Sun Y, Shmulevich I, Xue F, Sood AK, Zhang W. Differing clinical impact of BRCA1 and BRCA2 mutations in serous ovarian cancer. Pharmacogenomics. 2012;13:1523–1535. doi: 10.2217/pgs.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Earl H, Molica S, Rutkowski P. Spotlight on landmark oncology trials: The latest evidence and novel trial designs. BMC Med. 2017;15:111. doi: 10.1186/s12916-017-0884-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieber S, Reinartz S, Raifer H, Finkernagel F, Dreyer T, Bronger H, Jansen JM, Wagner U, Worzfeld T, Müller R, Huber M. Prognosis of ovarian cancer is associated with effector memory CD8+ T cell accumulation in ascites, CXCL9 levels and activation-triggered signal transduction in T cells. Oncoimmunology. 2018;7:e1424672. doi: 10.1080/2162402X.2018.1424672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamanishi J, Mandai M, Konishi I. Immune checkpoint inhibition in ovarian cancer. Int Immunol. 2016;28:339–348. doi: 10.1093/intimm/dxw020. [DOI] [PubMed] [Google Scholar]
- 11.Webb JR, Milne K, Kroeger DR, Nelson BH. PD-L1 expression is associated with tumor-infiltrating T cells and favorable prognosis in high-grade serous ovarian cancer. Gynecol Oncol. 2016;141:293–302. doi: 10.1016/j.ygyno.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Drakes ML, Mehrotra S, Aldulescu M, Potkul RK, Liu Y, Grisoli A, Joyce C, O'Brien TE, Stack MS, Stiff PJ. Stratification of ovarian tumor pathology by expression of programmed cell death-1 (PD-1) and PD-ligand-1 (PD-L1) in ovarian cancer. J Ovarian Res. 2018;11:43. doi: 10.1186/s13048-018-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez GM, Galpin KJC, McCloskey CW, Vanderhyden BC. The tumor microenvironment of epithelial ovarian cancer and its influence on response to immunotherapy. Cancers (Basel) 2018;10:242. doi: 10.3390/cancers10080242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 15.Santoiemma PP, Powell DJ., Jr Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol Ther. 2015;16:807–820. doi: 10.1080/15384047.2015.1040960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James NE, Woodman M, DiSilvestro PA, Ribeiro JR. The perfect combination: Enhancing Patient response to PD-1-based therapies in epithelial ovarian cancer. Cancers (Basel) 2020;12:2150. doi: 10.3390/cancers12082150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghisoni E, Imbimbo M, Zimmermann S, Valabrega G. Ovarian cancer immunotherapy: Turning up the heat. Int J Mol Sci. 2019;20:2927. doi: 10.3390/ijms20122927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torphy RJ, Schulick RD, Zhu Y. Newly emerging immune checkpoints: Promises for future cancer therapy. Int J Mol Sci. 2017;18:2642. doi: 10.3390/ijms18122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritthipichai K, Haymaker CL, Martinez M, Aschenbrenner A, Yi X, Zhang M, Kale C, Vence LM, Roszik J, Hailemichael Y, et al. Multifaceted role of BTLA in the control of CD8+ T-cell fate after antigen encounter. Clin Cancer Res. 2017;23:6151–6164. doi: 10.1158/1078-0432.CCR-16-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn R, Hutchinson T, Murphy KM, Ware CF, Croft M, Salek-Ardakani S. CD8 T cell memory to a viral pathogen requires trans cosignaling between HVEM and BTLA. PLoS One. 2013;8:e77991. doi: 10.1371/journal.pone.0077991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon T, Bromberg JS. BTLA+ dendritic cells: The regulatory T cell force awakens. Immunity. 2016;45:956–958. doi: 10.1016/j.immuni.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Marin-Acevedo JA, Dholaria B, Soyano AE, Knutson KL, Chumsri S, Lou Y. Next generation of immune checkpoint therapy in cancer: New developments and challenges. J Hematol Oncol. 2018;11:39. doi: 10.1186/s13045-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zak KM, Grudnik P, Magiera K, Dömling A, Dubin G, Holak TA. Structural biology of the immune checkpoint receptor PD-1 and its ligands PD-L1/PD-L2. Structure. 2017;25:1163–1174. doi: 10.1016/j.str.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Han Y, Liu D, Li L. PD-1/PD-L1 pathway: Current researches in cancer. Am J Cancer Res. 2020;10:727–742. [PMC free article] [PubMed] [Google Scholar]
- 25.Makkouk A, Chester C, Kohrt HE. Rationale for anti-CD137 cancer immunotherapy. Eur J Cancer. 2016;54:112–119. doi: 10.1016/j.ejca.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Snell LM, Lin GH, McPherson AJ, Moraes TJ, Watts TH. T-cell intrinsic effects of GITR and 4-1BB during viral infection and cancer immunotherapy. Immunol Rev. 2011;244:197–217. doi: 10.1111/j.1600-065X.2011.01063.x. [DOI] [PubMed] [Google Scholar]
- 27.Bauché D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, Ganal-Vonarburg SC, Wilson DC, McClanahan TK, Malefyt RW, et al. LAG3+ regulatory T cells restrain interleukin-23-Producing CX3CR1+ gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity. 2018;49:342–352.e5. doi: 10.1016/j.immuni.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Workman CJ, Cauley LS, Kim IJ, Blackman MA, Woodland DL, Vignali DAA. Lymphocyte activation gene-3 (CD223) regulates the size of the expanding T cell population following antigen activation in vivo. J Immunol. 2004;172:5450–5455. doi: 10.4049/jimmunol.172.9.5450. [DOI] [PubMed] [Google Scholar]
- 29.Grosso JF, Kelleher CC, Harris TJ, Maris CH, Hipkiss EL, De Marzo A, Anders R, Netto G, Getnet D, Bruno TC, et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest. 2007;117:3383–3392. doi: 10.1172/JCI31184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willoughby J, Griffiths J, Tews I, Cragg MS. OX40: Structure and function-What questions remain? Mol Immunol. 2017;83:13–22. doi: 10.1016/j.molimm.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Buchan SL, Rogel A, Al-Shamkhani A. The immunobiology of CD27 and OX40 and their potential as targets for cancer immunotherapy. Blood. 2018;131:39–48. doi: 10.1182/blood-2017-07-741025. [DOI] [PubMed] [Google Scholar]
- 32.Jayaraman P, Jacques MK, Zhu C, Steblenko KM, Stowell BL, Madi A, Anderson AC, Kuchroo VK, Behar SM. TIM3 Mediates T Cell Exhaustion during Mycobacterium tuberculosis infection. PLoS Pathog. 2016;12:e1005490. doi: 10.1371/journal.ppat.1005490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016;44:989–1004. doi: 10.1016/j.immuni.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Z, Mayer AT, Nobashi TW, Gambhir SS. ICOS Is an Indicator of T-cell-mediated response to cancer immunotherapy. Cancer Res. 2020;80:3023–3032. doi: 10.1158/0008-5472.CAN-19-3265. [DOI] [PubMed] [Google Scholar]
- 35.Solinas C, Gu-Trantien C, Willard-Gallo K. The rationale behind targeting the ICOS-ICOS ligand costimulatory pathway in cancer immunotherapy. ESMO Open. 2020;5:e000544. doi: 10.1136/esmoopen-2019-000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosseini A, Gharibi T, Marofi F, Babaloo Z, Baradaran B. CTLA-4: From mechanism to autoimmune therapy. Int Immunopharmacol. 2020;80:106221. doi: 10.1016/j.intimp.2020.106221. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, Chen L, Chen Y, Zhu G, Yin W, et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell. 2019;176:334–347.e12. doi: 10.1016/j.cell.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews LP, Marciscano AE, Drake CG, Vignali DA. LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev. 2017;276:80–96. doi: 10.1111/imr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Team RC. R: A language and environment for statistical computing. 2013. http://www.R-project.org/ [ June 15; 2020 ]; [Google Scholar]
- 40.Morgan M, Davis S. R: A language and environment for statistical computing. https://bioconductor.org/packages/GenomicDataCommons. [ June 15; 2020 ];NIH/NCI Genomic Data Commons Access. [Internet] [Google Scholar]
- 41.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 42.Rådestad E, Klynning C, Stikvoort A, Mogensen O, Nava S, Magalhaes I, Uhlin M. Immune profiling and identification of prognostic immune-related risk factors in human ovarian cancer. Oncoimmunology. 2019;8:e1535730. doi: 10.1080/2162402X.2018.1535730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fucikova J, Rakova J, Hensler M, Kasikova L, Belicova L, Hladikova K, Truxova I, Skapa P, Laco J, Pecen L, et al. TIM-3 dictates functional orientation of the immune infiltrate in ovarian cancer. Clin Cancer Res. 2019;25:4820–4831. doi: 10.1158/1078-0432.CCR-18-4175. [DOI] [PubMed] [Google Scholar]
- 44.Ramser M, Eichelberger S, Däster S, Weixler B, Kraljević M, Mechera R, Tampakis A, Delko T, Güth U, Stadlmann S, et al. High OX40 expression in recurrent ovarian carcinoma is indicative for response to repeated chemotherapy. BMC Cancer. 2018;18:425. doi: 10.1186/s12885-018-4339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Z, Wang X, Cheng D, Xia Z, Luan M, Zhang S. PD-1 blockade and OX40 triggering synergistically protects against tumor growth in a murine model of ovarian cancer. PLoS One. 2014;9:e89350. doi: 10.1371/journal.pone.0089350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article.