Abstract
Anaplastic lymphoma kinase (ALK) inhibitors have been shown to be effective in treating patients with ALK-positive non-small cell lung cancer (NSCLC), and crizotinib, ceritinib and alectinib have been approved as clinical first-line therapeutic agents. The availability of these inhibitors has also largely changed the treatment strategy for advanced ALK-positive NSCLC. However, patients still inevitably develop resistance to ALK inhibitors, leading to tumor recurrence or metastasis. The most critical issues that need to be addressed in the current treatment of ALK-positive NSCLC include the high cost of targeted inhibitors and the potential for increased toxicity and resistance to combination therapy. Recently, it has been suggested that the serine/threonine kinase 11 (STK11) mutation may serve as one of the biomarkers for immunotherapy in NSCLC. Therefore, the main purpose of this review was to summarize the role of STK11 in ALK-positive NSCLC. The present review also summarizes the treatment and drug resistance studies in ALK-positive NSCLC and the current status of STK11 research in NSCLC.
Keywords: non-small cell lung cancer, anaplastic lymphoma kinase-positive, STK11, drug resistance, targeted therapies
1. Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, with non-small cell lung cancer (NSCLC) being the most common subtype (1). The majority of patients with NSCLC are diagnosed at an advanced, inoperable stage (2) and have an overall 5-year survival rate of just 5% (3). This poor prognosis may be associated with tumor heterogeneity, acquisition and intrinsic resistance to therapeutic agents in NSCLC (4). If the most appropriate treatment is identified early and drug resistance is addressed to a satisfactory degree, patient survival can be significantly improved. Current non-surgical treatments for NSCLC in clinical practice include systemic chemotherapy, radiotherapy, targeted therapy and immunotherapy (IO). In recent years, rapid developments have been made in cellular and molecular biotechnology, and targeted gene therapy and IO are gradually gaining traction (5). Molecular testing is commonly used in NSCLC, and the detection of epidermal growth factor receptor (EGFR), B-Raf proto-oncogene, serine/threonine kinase (BRAF) and MET proto-oncogene, receptor tyrosine kinase (MET) mutations, as well as anaplastic lymphoma kinase (ALK), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), ret proto-oncogene (RET) and neurotrophic receptor tyrosine kinase 1 (NTRK1) translocations have been incorporated into the diagnostic criteria for NSCLC, and inhibitors of these kinases are now routinely used in the clinic (6). An increasing number of signaling pathways and driver genes are being identified, and therapeutic drugs for NSCLC are emerging (7,8). Targeted drugs can bind specifically to the oncogenic site and induce cancer cell-specific death (9). Targeted drugs have a higher efficacy and fewer side effects than chemotherapy (10,11). However, each generation of targeted drugs shows different degrees of resistance; therefore, identifying new therapeutic targets following resistance is crucial. Multiple mutations are not uncommon in clinical practice in recent years, and exploring serine/threonine kinase 11 (STK11) co-mutations in ALK-positive NSCLC patients is important.
A number of recent studies have linked the presence of STK11 mutations to the lack of response to IO in NSCLC (12–16). In addition, several clinical studies have further elucidated the biological role of STK11 mutations leading to primary resistance to IO (17–19). The implementation of STK11 mutations as a routine biomarker in NSCLC remains controversial and is not performed in daily practice (20).
Therefore, the aim of the present study was to investigate the role of STK11 in ALK-positive NSCLC, review the treatment of patients with ALK-positive NSCLC, and compare the clinical efficacy, resistance mutations and appropriate resistance solutions of three generations of ALK inhibitors.
2. Current research advances in ALK-positive NSCLC
ALK and NSCLC
As a receptor tyrosine kinase of the insulin receptor (IR) subfamily, ALK has been found to play an important role in various types of cancer, particularly in anaplastic large cell lymphoma (ALCL), NSCLC and neuroblastoma (21). In 2007, the echinoderm microtubule-associated protein like protein 4 (EML4)-ALK fusion gene was identified in a group of NSCLC patients (22). This fusion is the result of an inversion of the short arm of chromosome 2, where the human EML4 and ALK genes are present (23). EML4 contains a coiled oligomeric structural domain, which mediates the dimerization and structural activation of ALK. Like in ALCL, many different ALK fusions have been identified, but EML4-ALK is the most common variant (24). ALK has been reported to regulate several different pathways involved in cell proliferation and survival, such as the phosphatidylinositol-3-kinase (PI3K)/AKT/mammalian targets of rapamycin (mTOR), RAS/RAF/MAP kinase-extracellular signal-regulated kinase (ERK) kinase (MEK)/ERK and JAK/STAT pathways (Fig. 1), once it is dimerized and activated by autophosphorylation upon binding to its ligands, pleiotrophin and midkine (25,26). Direct evidence for the oncogenic potential of EML4-ALK in lung carcinogenesis has been found in mice. The transgenic overexpression of EML4-ALK in type II alveolar cells via the surface activated protein-c or Clara cell secretory protein promoter leads to the rapid development of tumors with features of lung adenocarcinoma (27,28). A total of 3–7% of NSCLC (mainly adenocarcinoma subtypes) cases are characterized by ALK rearrangements, which occur in a mutually exclusive manner with KRAS and EGFR mutations (29,30). Of note, a previous study developed in vivo induction models by inducing EML4-ALK rearrangement, which leads to lung carcinogenesis. These models have shown a sensitivity to ALK inhibition, thus serving as valuable tools to explore the mechanisms of EML4-ALK-induced lung cancer and response to ALK-targeted therapy (31). Of note, ALK-positive NSCLC patients have a healthy weight, are non-smokers or are young (32).
Figure 1.
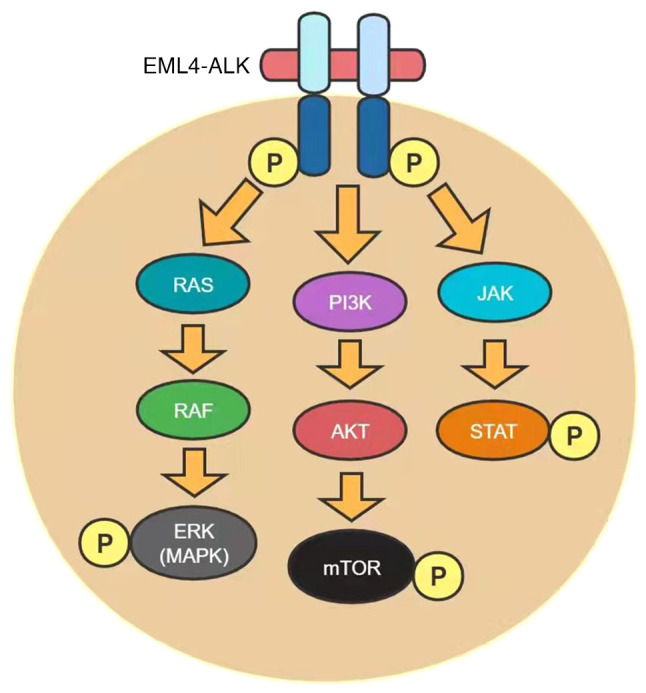
Signaling pathway of EML4-ALK activation. EML4, echinoderm microtubule-associated protein like protein 4; ALK, anaplastic lymphoma kinase; ERK, extracellular signal-regulated kinase; PI3K, phosphatidylinositol-3-kinase; mTOR, mammalian targets of rapamycin; JAK, Janus kinase; STAT, signal transducer and activator of transcription; P, phosphorylated.
Current targeted therapies for patients with ALK-positive NSCLC
Prior to the discovery of the EML4-ALK fusion protein, chemotherapy was the first-line treatment option (33). ALK inhibitors have shown potent clinical activity in patients with NSCLC (34). Three generations of ALK inhibitors have been approved for clinical use (2). Ten years ago, the first-generation ALK inhibitor crizotinib was approved for clinical treatment. Table I documents clinical studies associated with ALK inhibitors (35–46). In a series of subsequent clinical trials, crizotinib demonstrated good clinical efficacy, including in 149 patients with NSCLC and ALK mutations (PROFILE1001), with an objective remission rate (ORR) of 60.8% and progression-free survival (PFS) of 9.7 months in 143 patients evaluated; several other phases of clinical trials also achieved useful results (35) (Table I). However, since crizotinib cannot easily cross the blood-brain barrier, it leads to brain metastases and resistance in patients (24). The rapid development of resistance during the treatment cycle is the main limiting factor associated with crizotinib (47). Since then, progeny ALK inhibitors have demonstrated significant efficacy and better central nervous system (CNS) activity compared with crizotinib (48–50). Among the clinical studies of second-generation ALK inhibitors, the ASCEND series was a series of studies evaluating the safety and efficacy of ceritinib. Based on this series, ceritinib was approved by the food and drug administration (FDA) for clinical treatment in 2014, mainly for patients who were intolerant to crizotinib or whose disease progressed after taking crizotinib in ALK-positive patients (33). As a result, the FDA approved brigatinib for patients who had failed prior ALK inhibitor therapy (33). In the J-Alex study in Japan, alectinib was compared head-to-head with crizotinib. The latest data from this trial demonstrated a PFS of 34.1 and 10.2 months in the alectinib and crizotinib groups, respectively (42). In the global ALEX study, a new record was set with a PFS of 34.8 months for first-line treatment with alectinib (Table I), which was approved for the treatment of pure ALK-positive patients based on the results of the randomized phase III ALEX trial. In addition, alectinib was effective in preventing the development of brain metastasis and significantly reduced the risk of CNS progression in patients by 84% (43,44). The ALTA-1L study was a study comparing the efficacy and safety of brigatinib with those of crizotinib as first-line treatment for patients with ALK-positive metastatic NSCLC (51). The final results showed that when multiple targeted drugs cause resistance, patients can still benefit from brigatinib. Thus, brigatinib has a unique advantage in follow-up therapy. The third-generation ALK inhibitor lorlatinib was found to inhibit both the ALK and ROS1 pathways and overcome the multiresistance associated with first- and second-generation ALK inhibitors, while also crossing the blood-brain barrier (45). Regarding IO, irrespective of the histological type, patients treated with atezolizumab showed a significantly longer overall survival than platinum-based chemotherapy in patients with NSCLC with a high programmed cell death-ligand 1 (PD-L1) expression, with a significantly superior efficacy (52).
Table I.
Clinical studies associated with ALK inhibitors.
| Clinical study | Drug | mPFS (month) | ORR (%) |
|---|---|---|---|
| PROFILE 1001 (35) | Crizotinib | 9.7 | 60.8 |
| PROFILE 1005 (36) | Crizotinib | 8.4 | 54.0 |
| PROFILE 1007 (37) | Crizotinib | 7.7 | 65.0 |
| PROFILE 1014 (38) | Crizotinib | 10.9 | 74.0 |
| ASCEND-1 (39) | Ceritinib | 18.4 | 72.0 |
| ASCEND-2 (40) | Ceritinib | 5.7 | 38.6 |
| ASCEND-4 (41) | Ceritinib | 16.6 | 72.5 |
| J-ALEX (42) | Alectinib | 34.1 | 92.0 |
| ALEX (43,44) | Alectinib | 34.8 | 82.9 |
| ALTA-1L (45) | Brigatinib | 24.0 | 71.0 |
| B7461001 (46) | Lorlatinib | 9.6 | 46.0 |
ALK, anaplastic lymphoma kinase; mPFS, median progression-free survival; ORR, objective response rate.
Mechanisms of ALK inhibitor resistance
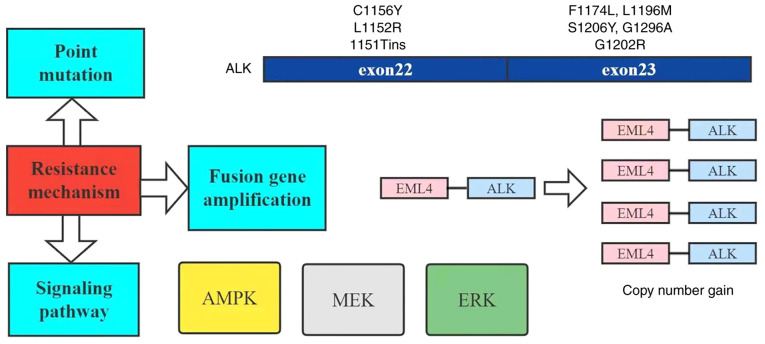
Although ALK inhibitors are clinically effective, patients still experience various types of drug resistance. This usually occurs in the form of ALK kinase structural domain mutations, ALK site amplification or activation of ‘bypass’ signaling pathways, ~1/3 of which are ALK kinase structural domain mutations (53). The first mutations identified were the L1196M and C1156Y (54), and L1196M is a residue known as the ‘gatekeeper’ that controls the entry of small-molecule ALK inhibitors into a hydrophobic pocket within the catalytic site and can spatially block inhibitor binding (55). Meanwhile, L1196M is the most common mutation in which patients develop resistance to crizotinib (47), while C1156Y develops resistance through other different mechanisms. Cysteine is similar to the catalytically important αC-helix within the structural domain of ALK tyrosine kinase, so its substitution for tyrosine is thought to prevent inhibitor binding by stabilizing the activity of ALK (47). Other resistant mutations that map to the same region with the same mechanism of resistance include 1151Tins, F1174C/L, L1198P, L1152R/P (47,56,57) and L1171T (12,13,58). Acquired resistance to crizotinib usually emerges after 1 year of treatment. ALK-E1210K mutations have been detected in patients treated with crizotinib (14). Other secondary mutations that occur following crizotinib treatment include L1196M, G1269A, G1202R, S1206Y, G1269A, L1152R, D1203N, I1171T, V1180L and C1156Y (13,47). The second-generation ALK inhibitors ceritinib, alectinib and brigatinib were found to show a stronger anti-ALK activity compared with crizotidnib (16). In addition, they exhibited greater CNS permeability and the ability to target multiple secondary ALK mutations. The direct application of second-generation ALK tyrosine kinase inhibitors has been shown to result in better therapeutic outcomes. In addition to ALK, ceritinib inhibits insulin-like growth factor 1 (IGF1), ROS1 and the IR (17,18). In addition, ceritinib inhibits multiple ALK mutations resistant to crizotinib, including L1196M, G1269A and S1206Y (47). However, C1156Y/T, I1151Tins and L1152P/R mutations have been associated with the emergence of ceritinib resistance (19). Alectinib has a better efficacy against G1269A, L1196M, F1174L and C1156Y mutations (20); however, alectinib treatment has been shown to cause the emergence of resistance mutations (I1171T/N/S, V1180L and G1202R) (14,58), and the activated bypass signaling pathway to be mediated by hepatocyte growth factor (HGF) and MET (59). Epithelial-mesenchymal transition (EMT) is a potential mechanism of alectinib resistance, characterized by the loss of E-cadherin and increased expression of waveform proteins (60). Brigatinib is a novel inhibitor of ALK, ROS1 and EGFR. Brigatinib inhibits crizotinib-resistant mutations, including ALK L1196M and EGFR T790M (51). Lorlatinib can be used to treat all known ALK inhibitor-induced resistance mutations and is the treatment of choice for patients with alectinib resistance (61). The L1198F mutation was recently reported to exhibit resistance to lorlatinib mainly by interfering with drug binding through spatial site block. It has also been reported that L1198F mutation enhances the binding of crizotinib, reduces the effect of C1156Y and enhances susceptibility to crizotinib resistance (62). However, as for how to overcome L1198F mutation, the clinical efficacy and resistance mechanisms of lorlatinib need to be elucidated.
Known mechanisms of resistance include point mutations, fusion gene amplification and bypass signaling through the activation of other oncogenes (Fig. 2) (63), and AMPK, which is closely associated with STK11 mutation and is one of the important pathways in the mechanism of ALK inhibitor resistance (30).
Figure 2.
Mechanisms of resistance to ALK inhibitors. ALK, anaplastic lymphoma kinase; EML4, echinoderm microtubule-associated protein like protein 4; AMPK, AMP-activated protein kinase; MEK, MAP kinase-ERK kinase; ERK, extracellular signal-regulated kinase.
3. Current research progress on STK11
Function of STK11
STK11 is considered an important tumor-suppressor gene with a wide range of metabolic functions (64). It encodes the serine/threonine kinase liver kinase B1 (LKB1), which activates a family of 12 downstream kinases, including AMPK, and plays a role in essential biological functions, including cellular energy regulation (65). It is also involved in several physiological processes, including the regulation of cellular metabolism, cell polarity and DNA damage response (65,66). Tumor cells with inactivated or lost STK11 are unable to activate adenosine monophosphate-activated protein kinases, and are therefore particularly vulnerable to energy stress states (67). STK11 inactivation was initially identified in human tumors associated with Peutz-Jeghers genetic syndrome (68). STK11 also negatively regulates mTOR signaling through its substrate AMPK, and STK11 loss leads to the aberrant activation of mTOR in a variety of tissues. mTOR inhibitor everolimus has been shown to be effective (69,70). STK11/LKB1 loss of function has been found in several cancer types, mainly through somatic alterations in the STK11 gene, such as nonsense mutations, loss of heterozygosity, insertions, intragenic deletions or chromosomal deletions (71–79), while in Asian populations, STK11 is mainly inactivated through focal deletions (80,81). Although STK11 is inactivated by a large spectrum of truncating mutations and behaves like a tumor-suppressor gene in different tumor models through mTOR repression (77,82,83), recent studies have shown that STK11 may also acquire oncogenic properties. Subsequently, somatic STK11 mutations have been reported in other cancer types (69), including NSCLC (71,84,85). It has now been demonstrated that the loss of LKB1 affects tumor progression through energy metabolism, cytokine inhibition, tumor immunosuppression and altered cell viability (86–90). Several types of tumors exhibit aberrant mutations in the STK11 gene. For example, STK11 deletion in cervical cancer and melanoma is associated with extensive and high-grade metastasis, and a heterozygous deletion of the STK11 locus in primary breast cancer is associated with metastasis (91,92). STK11-mutant lung cancer constitutes a genetic subgroup of aggressive NSCLC with an in vitro inhibition of mitogen-activated protein kinase and mTOR signaling-increased sensitivity (93). STK11 abnormalities have also been associated with cancer-related immune dysfunction. For example, STK11-mutant lung cancer suppresses immune surveillance responses (94) and STK11 deficiency decreases PD-L1 expression (95).
STK11 and NSCLC
STK11 is one of the most commonly mutated genes in lung adenocarcinoma, and the LKB1 protein encoded by the STK11 gene is the second most common tumor suppressor in NSCLC, with mutations or genomic loss occurring in 17–23% of NSCLC cases (84,96,97). In lung cancer, the short STK11 isoform, lacking 124 N-terminal amino acids, is defined as an oncogene (98). Indeed, it has been shown that cytoplasmic STK11 interacts with estrogen receptor (ER)α/SRC/PI3K to stimulate the AKT pathway and is associated with a shorter survival (99). These findings suggested that STK11 may play a tumor-suppressor or oncogene function. This dual mechanism may explain the lack of a clear association between STK11 alterations and prognosis in lung cancer (100). In vitro studies have shown that STK11 inactivation increases the motility and invasiveness of lung cancer, and facilitates epithelial-to-mesenchymal transition (EMT) in lung cancer, thus enhancing metastatic potential (101,102). In addition, it was shown that STK11/LKB1 inactivation promotes cancer cell growth and survival through the upregulation of hypoxia-inducible factor 1 (HIF-1). The inactivation of STK11/LKB1 in lung cancer cells leads to the upregulation of mTOR signaling, providing a growth advantage associated with mitochondrial dysfunction, due to autophagic injury (103,104). The aberrant activation of the PI3K/AKT/mTOR pathway has been identified in 90% of lung adenocarcinomas and 40% of squamous cell carcinomas (105). The PI3K/AKT/mTOR signaling pathway is mainly activated by receptor tyrosine kinases [e.g. epidermal growth factor receptor (EGFR), insulin-like growth factor receptor 1 (IGFR1), vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PGFR)] and is involved in a variety of biological functions, such as proliferation, differentiation, survival, adhesion, motility, invasion and cellular metabolism (106,107). In addition to the activation of the PI3K/AKT/mTOR pathway by growth factors and insulin, different nutritional and environmental signals, such as high levels of adenosine triphosphate (ATP), oxygen and elevated serum amino acid levels can also increase the activity of mTOR complex 1 (mTORC1). By contrast, intracellular and environmental stress signals, such as low ATP levels, hypoxia and DNA damage, inhibit mTORC1 activity mainly through AMPK activation (106). mTOR pathway activation has been found to be associated with poor clinical outcomes, invasiveness and metastasis (108–111). It is important to highlight that LKB1, encoded by STK11, is also associated with AMPK, and that LKB1/AMPK may counteract oxidative stress by inhibiting the synthesis of nicotinamide adenine dinucleotide phosphate (NADPH)-consuming fatty acids and increasing the oxidation of NADPH-producing fatty acids (112). At the same time, activated AMPK phosphorylates and activates the transcription factor nuclear related factor 2 (NRF2) (113). NRF2 then activates the transcription of antioxidant genes involved in NADPH production. High NADPH levels, together with autophagy, protect LKB1-savvy cancers from oxidative stress and reactive oxygen species (ROS)-induced chemotherapy (cisplatin, paclitaxel and adriamycin) (114). Thus, the activation of NRF2 is associated with more aggressive lung cancers and reduced patient survival (115). Of note, the activation of the LKB1/autophagy pathway enables circulating tumor cells to resist loss-of-nest apoptosis (116). Thus, cells lacking LKB1 undergo apoptosis in response to metabolic stress because they are unable to respond to energy deficiency and restore homeostasis in vivo (117). The regulation of STK11 expression and its role in cancer cell proliferation remains highly complex (118). For example, recent research has shown that asparagine and aspartic acid can regulate AMPK-mediated p53 activation by physically binding to LKB1 and regulating LKB1 activity. P53 has been reported to control cell survival by generating an auto-amplifying loop through asparagine-aspartate-mediated LKB1-AMPK signaling to regulate asparagine metabolism (118). In lung cancer, STK11/LKB1 alterations are the only marker significantly associated with PD-L1 negativity in patients with high/medium tumor mutation burden (TMB) (119). Both elevated TMB and increased PDL1 expression are associated with IO response (120,121). Kelch like ECH associated protein 1 (KEAP1) mutations or double allelic deletions are enriched in patients with LKB1-mutant NSCLC tumors. NRF2 is a transcriptional factor and KEAP1 is a negative regulator of NRF2 that binds to the antioxidant response element on DNA and initiates the transcription of several genes involved in the regulation of redox homeostasis and cellular detoxification (122). Clinical studies have shown that in KRAS-driven NSCLC, STK11 mutations leading to loss of function are associated with resistance to anti-programmed death 1 (PD-1) monoclonal antibody therapy, but the molecular mechanisms of pathogenesis are not yet clear (123). Despite some uncertainties, STK11 functional status is emerging as a reliable biomarker for predicting a lack of response to anti-PD-1 therapy in NSCLC patients. It has been reported in the literature that STK11 is significantly and significantly associated with decreased survival in meningiomas (124). Although the evidence on the biological role of STK11 is not sufficient, its prognostic significance in advanced NSCLC needs to be confirmed; clarifying the role of STK11 will facilitate the analysis of STK11 mutational status, which may also provide more options for targeted therapy and IO (125). The role and significance of STK11 in NSCLC needs to be further explored.
Current STK11 mutation therapies for NSCLC
Cancer typically evades immune surveillance by aberrantly expressing immune checkpoints (e.g. PD-1) that isolate tumor cells from the host immune system. Immune blockade using monoclonal antibodies against the immune checkpoint PD-1 and its primary ligand PD-L1 can greatly improve survival in advanced NSCLC, with the greatest impact in patients with stage III and first-line stage IV lung cancer (126–128). However, in patients with other types, the response rate was just 20% (129). In a retrospective cohort study, data from the Clinico-Genomic Database were used to identify patients with metastatic NSCLC who received first-line IO (alone or in combination) or chemotherapy in routine clinical practice. The results suggested that in NSCLC, patients with STK11 mutation (STK11m) exhibit poorer overall survival (OS) and PFS compared with patients with STK11 wild-type (STK11wt) receiving IO or chemotherapy. Survival outcomes analyzed by treatment line and type showed that OS and PFS were worse in the IO treatment group for STK11m vs. STK11wt (Table II) (130). The results of the study were not optimistic, which further suggested that STK11 mutations reduce the survival rate of patients with NSCLC. Most importantly, it is unclear whether STK11 can be used as a predictive biomarker to guide treatment selection, and prospective evaluation is still lacking. Therefore, immunotherapy should not be administered to patients with STK11-mutated tumors at the present time (131). At the same time, it has also been reported that LKB1 encoded by the STK11 gene may be associated with radioresistance in patients, and several previous studies have shown the role of LKB1 expression in regulating the response to radiotherapy, based on preclinical experiments (132–134). However, to the best of our knowledge, there are no clinical trials on STK11 mutations, so a comprehensive evaluation of patients with STK11 mutations in NSCLC could not be performed.
Table II.
Chemotherapy and immunotherapy for STK11 mutation status in NSCL.
| OS (month) | PFS (month) | |||
|---|---|---|---|---|
|
|
|
|||
| Variable | STK11m | STK11wt | STK11m | STK11wt |
| Chemotherapy (130) | ||||
| First-line | 11.7 | 18.9 | 4.5 | 6.1 |
| Second-line | 13.1 | 15.2 | 4.2 | 4.5 |
| Immunotherapy (131) | ||||
| First-line | 14.2 | 20.1 | 4.1 | 5.4 |
| Second-line | 6.6 | 13.6 | 2.2 | 3.1 |
NSCLC, non-small cell lung cancer; OS, overall survival; PFS, progression-free survival; STK11, serine/threonine kinase 11; STK11m, STK11 mutation; STK11wt, STK11 wild-type.
4. Conclusions and perspectives
Both STK11 and ALK can regulate tumor proliferation and growth through the mTOR pathway, and STK11 can be oncogenic in NSCLC through the AMPK pathway, which is included in the mechanism of drug resistance in patients with ALK-positive NSCLC. mTOR signaling is known to be a master regulator of homeostasis and to integrate various environmental signals to regulate cell growth, proliferation and metabolism. The deregulation of mTOR signaling, particularly its overactivation, is frequent in human cancer. Recent advances in molecular profiling have identified certain genes involved in encoding the mTOR pathway, including STK11, PIK3CA, PTEN and RPTOR independent companion of MTOR complex 2 (RICTOR), whose amplification or mutation induces mTOR pathway activation. AMPK is a central metabolic sensor that coordinates cell growth and energy balance. In terms of oncogenesis, LKB1 (mentioned previously) can directly phosphorylate and activate AMPK; therefore, in cells with LKB1 mutations, the AMPK protein is oxidized and inactivated, leading to cell death. In terms of drug resistance, if the AMPK pathway is abnormally regulated, patients with ALK-positive NSCLC then become resistant to the drug.
In conclusion, STK11 plays an important role in the treatment and drug resistance of patients with ALK-positive NSCLC. Although there is no definitive evidence on how STK11 affects the prognosis of ALK-positive patients with NSCLC, the results of this review showed that STK11 mutations may reduce the survival of ALK-positive NSCLC patients.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- ALK
anaplastic lymphoma kinase
- IR
insulin receptor
- ALCL
anaplastic large cell lymphoma
- EML4
echinoderm microtubule-associated protein like protein 4
- CNS
center nervous system
- FDA
food and drug administration
- AMPK
amp-activated protein kinase
- EGFR
epidermal growth factor receptor
- MEK
MAP kinase-Erk kinase
- ERK
extracellular signal-regulated kinase
- NRF2
nuclear related factor 2
- NADPH
nicotinamide adenine dinucleotide phosphate
- PFS
progression-free survival
- OS
overall survival
- ORR
objective remission rate
- LKB1
liver kinase B1
- TMB
tumor mutation burden
- ATP
adenosine triphosphate
- IO
immunotherapy
- STK11
serine/threonine kinase 11
- STK11m
STK11 mutation
- STK11wt
STK11 wild-type
- PD-1
programmed death 1
- PD-L1
programmed cell death-ligand 1
Funding Statement
The present study was supported by the Hubei Province Health and Family Planning Scientific Research Project (grant nos. WJ2019-21) to NL.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XCP and JC contributed to the design of this review. WZ, MW, MXM and ZQY reviewed the references. WZ, JC and XCP wrote the manuscript. WZ, LDY, YYC and NL designed and produced the tables and figures. YYC and NL revised the manuscript critically for important intellectual content. NL acquired the funding. YHY analyzed the data and designed the figures. LDY put a lot of effort into revising the manuscript and is listed as co-first author. Data authentication is not applicable. All authors read and approved the manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 2.Ye Z, Huang Y, Ke J, Zhu X, Leng S, Luo H. Breakthrough in targeted therapy for non-small cell lung cancer. Biomed Pharmacother. 2021;133:111079. doi: 10.1016/j.biopha.2020.111079. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Li N, Wang M, Zhang YH, Yan LD, Zhou W, Yu ZQ, Peng XC, Cai J. Tumorigenic effect of TERT and its potential therapeutic target in NSCLC (Review) Oncol Rep. 2021;46:182. doi: 10.3892/or.2021.8133. [DOI] [PubMed] [Google Scholar]
- 5.Brueckl WM, Ficker JH, Zeitler G. Clinically relevant prognostic and predictive markers for immune-checkpoint-inhibitor (ICI) therapy in non-small cell lung cancer (NSCLC) BMC Cancer. 2020;20:1185. doi: 10.1186/s12885-020-07690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. doi: 10.1016/j.critrevonc.2020.103194. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Li D, Luo H, Zhu X. Circular RNAs: The star molecules in cancer. Mol Aspects Med. 2019;70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Guo B, Li D, Du L, Zhu X. piRNAs: Biogenesis and their potential roles in cancer. Cancer Metastasis Rev. 2020;39:567–575. doi: 10.1007/s10555-020-09863-0. [DOI] [PubMed] [Google Scholar]
- 9.Gerlinger M. Targeted drugs ramp up cancer mutability. Science. 2019;366:1452–1453. doi: 10.1126/science.aaz9900. [DOI] [PubMed] [Google Scholar]
- 10.Liang G, Fan W, Luo H, Zhu X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomed Pharmacother. 2020;128:110255. doi: 10.1016/j.biopha.2020.110255. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Zhang Z, Lai WF, Cui L, Zhu X. How to overcome the side effects of tumor immunotherapy. Biomed Pharmacother. 2020;130:110639. doi: 10.1016/j.biopha.2020.110639. [DOI] [PubMed] [Google Scholar]
- 12.Ceccon M, Mologni L, Bisson W, Scapozza L, Gambacorti-Passerini C. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated Alk inhibitors. Mol Cancer Res. 2013;11:122–132. doi: 10.1158/1541-7786.MCR-12-0569. [DOI] [PubMed] [Google Scholar]
- 13.Toyokawa G, Hirai F, Inamasu E, Yoshida T, Nosaki K, Takenaka T, Yamaguchi M, Seto T, Takenoyama M, Ichinose Y. Secondary mutations at I1171 in the ALK gene confer resistance to both Crizotinib and Alectinib. J Thorac Oncol. 2014;9:e86–e87. doi: 10.1097/JTO.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 14.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, et al. Molecular mechanisms of resistance to first- and second-generation ALK Inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016;6:1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyokawa G, Seto T. Updated evidence on the mechanisms of resistance to ALKInhibitors and strategies to overcome such resistance: Clinical and preclinical data. Oncol Res Treat. 2015;38:291–298. doi: 10.1159/000430852. [DOI] [PubMed] [Google Scholar]
- 16.Costa DB. Clinical development and approval of second generation ALK inhibitors for ALKrearranged lung cancer. Transl Lung Cancer Res. 2014;3:373–375. doi: 10.3978/j.issn.2218-6751.2014.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roskoski R., Jr ROS1 protein-tyrosine kinase inhibitors in the treatment of ROS1 fusion protein-driven non-small cell lung cancers. Pharmacol Res. 2017;121:202–212. doi: 10.1016/j.phrs.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothschild SI. New treatment options for ALK+ advanced non-small-cell lung cancer: Critical appraisal of ceritinib. Ther Clin Risk Manag. 2016;12:735–741. doi: 10.2147/TCRM.S87876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi A. Alectinib for ALK-positive non-small-cell lung cancer. Expert Rev Clin Pharmacol. 2016;9:1005–1013. doi: 10.1080/17512433.2016.1195262. [DOI] [PubMed] [Google Scholar]
- 21.Kong X, Pan P, Sun H, Xia H, Wang X, Li Y, Hou T. Drug discovery targeting anaplastic lymphoma kinase (ALK) J Med Chem. 2019;62:10927–10954. doi: 10.1021/acs.jmedchem.9b00446. [DOI] [PubMed] [Google Scholar]
- 22.Qian M, Zhu B, Wang X, Liebman M. Drug resistance in ALK-positiveNon-small cell lungcancer patients. Semin Cell Dev Biol. 2017;64:150–157. doi: 10.1016/j.semcdb.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 24.Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: A paradigm for precision cancer medicine. Clin Cancer Res. 2015;21:2227–2235. doi: 10.1158/1078-0432.CCR-14-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales La Madrid A, Campbell N, Smith S, Cohn SL, Salgia R. Targeting ALK: A promising strategy for the treatment of non-small cell lung cancer, non-Hodgkin's lymphoma, and neuroblastoma. Target Oncol. 2012;7:199–210. doi: 10.1007/s11523-012-0227-8. [DOI] [PubMed] [Google Scholar]
- 26.Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res. 2011;17:2081–2086. doi: 10.1158/1078-0432.CCR-10-1591. [DOI] [PubMed] [Google Scholar]
- 27.Chen Z, Sasaki T, Tan X, Carretero J, Shimamura T, Li D, Xu C, Wang Y, Adelmant GO, Capelletti M, et al. Inhibition of ALK, PI3K/MEK, and HSP90 in murine lung adenocarcinoma induced by EML4-ALK fusion oncogene. Cancer Res. 2010;70:9827–9836. doi: 10.1158/0008-5472.CAN-10-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyo KH, Lim SM, Kim HR, Sung YH, Yun MR, Kim SM, Kim H, Kang HN, Lee JM, Kim SG, et al. Establishment of a conditional transgenic mouse model recapitulating EML4-ALK-positive human non-small cell lung cancer. J Thorac Oncol. 2017;12:491–500. doi: 10.1016/j.jtho.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y, et al. Multiplex reverse transcription-PCR screening for EML4-ALKfusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 30.Sharma GG, Mota I, Mologni L, Patrucco E, Gambacorti-Passerini C, Chiarle R. Tumor resistance against ALKTargeted therapy-where it comes from and where it goes. Cancers (Basel) 2018;10:62. doi: 10.3390/cancers10030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodig SJ, Mino-Kenudson M, Dacic S, Yeap BY, Shaw A, Barletta JA, Stubbs H, Law K, Lindeman N, Mark E, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gristina V, La Mantia M, Iacono F, Galvano A, Russo A, Bazan V. The emerging therapeutic landscape of ALK inhibitors in non-small cell lung cancer. Pharmaceuticals (Basel) 2020;13:474. doi: 10.3390/ph13120474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du X, Shao Y, Qin HF, Tai YH, Gao HJ. ALK-rearrangement in non-small-cell lung cancer (NSCLC) Thorac Cancer. 2018;9:423–430. doi: 10.1111/1759-7714.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Camidge DR, Bang YJ, Kwak EL, Iafrate AJ, Varella-Garcia M, Fox SB, Riely GJ, Solomon B, Ou SH, Kim DW, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: Updated results from a phase 1 study. Lancet Oncol. 2012;13:1011–1019. doi: 10.1016/S1470-2045(12)70344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackhall F, Ross Camidge D, Shaw AT, Soria JC, Solomon BJ, Mok T, Hirsh V, Jänne PA, Shi Y, Yang PC, et al. Final results of the large-scale multinational trial PROFILE 1005: Efficacy and safety of crizotinib in previously treated patients with advanced/metastatic ALK-positive non-small-cell lung cancer. ESMO Open. 2017;2:e000219. doi: 10.1136/esmoopen-2017-000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishio M, Kim DW, Wu YL, Nakagawa K, Solomon BJ, Shaw AT, Hashigaki S, Ohki E, Usari T, Paolini J, et al. Crizotinib versus chemotherapy in Asian patients with ALK-positive advanced non-small cell lung cancer. Cancer Res Treat. 2018;50:691–700. doi: 10.4143/crt.2017.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon BJ, Kim DW, Wu YL, Nakagawa K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, Tang Y, et al. Final overall survival analysis from a study comparing first-line crizotinib versus chemotherapy in ALK-mutation-positive non-small-cell lung cancer. J Clin Oncol. 2018;36:2251–2258. doi: 10.1200/JCO.2017.77.4794. [DOI] [PubMed] [Google Scholar]
- 39.Kim DW, Mehra R, Tan DSW, Felip E, Chow LQM, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, Riely GJ, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): Updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol. 2016;17:452–463. doi: 10.1016/S1470-2045(15)00614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crinò L, Ahn MJ, De Marinis F, Groen HJ, Wakelee H, Hida T, Mok T, Spigel D, Felip E, Nishio M, et al. Multicenter phase ii study of whole-body and intracranial activity with ceritinib in patients With ALK-rearranged non-small-cell lung cancer previously treated with chemotherapy and crizotinib: Results from ASCEND-2. J Clin Oncol. 2016;34:2866–2873. doi: 10.1200/JCO.2015.65.5936. [DOI] [PubMed] [Google Scholar]
- 41.Soria JC, Tan DSW, Chiari R, Wu YL, Paz-Ares L, Wolf J, Geater SL, Orlov S, Cortinovis D, Yu CJ, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): A randomised, open-label, phase 3 study. Lancet. 2017;389:917–929. doi: 10.1016/S0140-6736(17)30123-X. [DOI] [PubMed] [Google Scholar]
- 42.Hida T, Nokihara H, Kondo M, Kim YH, Azuma K, Seto T, Takiguchi Y, Nishio M, Yoshioka H, Imamura F, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): An open-label, randomised phase 3 trial. Lancet. 2017;390:29–39. doi: 10.1016/S0140-6736(17)30565-2. [DOI] [PubMed] [Google Scholar]
- 43.Pérol M, Pavlakis N, Levchenko E, Platania M, Oliveira J, Novello S, Chiari R, Moran T, Mitry E, Nüesch E, et al. Patient-reported outcomes from the randomized phase III ALEX study of alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer. Lung Cancer. 2019;138:79–87. doi: 10.1016/j.lungcan.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Camidge DR, Dziadziuszko R, Peters S, Mok T, Noe J, Nowicka M, Gadgeel SM, Cheema P, Pavlakis N, de Marinis F, et al. Updated efficacy and safety data and impact of the EML4-ALK fusion variant on the efficacy of alectinib in untreated ALK-positive advanced non-small cell lung cancer in the global phase III ALEX study. J Thorac Oncol. 2019;14:1233–1243. doi: 10.1016/j.jtho.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Camidge DR, Kim HR, Ahn MJ, Yang JCH, Han JY, Hochmair MJ, Lee KH, Delmonte A, García Campelo MR, Kim DW, et al. Brigatinib versus crizotinib in advanced ALK inhibitor-naive ALK-positive non-small cell lung cancer: Second interim analysis of the phase III ALTA-1L Trial. J Clin Oncol. 2020;38:3592–3603. doi: 10.1200/JCO.20.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solomon BJ, Besse B, Bauer TM, Felip E, Soo RA, Camidge DR, Chiari R, Bearz A, Lin CC, Gadgeel SM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: Results from a global phase 2 Study. Lancet Oncol. 2018;19:1654–1667. doi: 10.1016/S1470-2045(18)30649-1. [DOI] [PubMed] [Google Scholar]
- 47.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, Michellys PY, Awad MM, Yanagitani N, Kim S, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–673. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia B, Nagasaka M, Zhu VW, Ou SI, Soo RA. How to select the best upfront therapy for metastatic disease? Focus on ALK-rearranged non-small cell lung cancer (NSCLC) Transl Lung Cancer Res. 2020;9:2521–2534. doi: 10.21037/tlcr-20-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner MT, Zhao C, Zhang Q, Wasik MA. Nucleophosmin-anaplastic lymphoma kinase: The ultimate oncogene and therapeutic target. Blood. 2017;129:823–831. doi: 10.1182/blood-2016-05-717793. [DOI] [PubMed] [Google Scholar]
- 50.Tse BC, Said BI, Fan ZJ, Hueniken K, Patel D, Gill G, Liang M, Razooqi M, Brown MC, Sacher AG, et al. Longitudinal health utilities, symptoms and toxicities in patients with ALK-rearranged lung cancer treated with tyrosine kinase inhibitors: A prospective real-world assessment. Curr Oncol. 2020;27:e552–e559. doi: 10.3747/co.27.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castellanos EH, Horn L. Re-Evaluating progression in an era of progress: A review of first- and second-line treatment options in anaplastic lymphoma kinase-positive non-small cell lung cancer. Oncologist. 2016;21:755–761. doi: 10.1634/theoncologist.2015-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herbst RS, Giaccone G, de Marinis F, Reinmuth N, Vergnenegre A, Barrios CH, Morise M, Felip E, Andric Z, Geater S, et al. Atezolizumab for first-line treatment of PD-L1-selected patients with NSCLC. N Engl J Med. 2020;383:1328–1339. doi: 10.1056/NEJMoa1917346. [DOI] [PubMed] [Google Scholar]
- 53.Hallberg B, Palmer RH. The role of the ALK receptor in cancer biology. Ann Oncol. 2016;27((Suppl 3)):iii4–iii15. doi: 10.1093/annonc/mdw301. [DOI] [PubMed] [Google Scholar]
- 54.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 55.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heuckmann JM, Hölzel M, Sos ML, Heynck S, Balke-Want H, Koker M, Peifer M, Weiss J, Lovly CM, Grütter C, et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res. 2011;17:7394–7401. doi: 10.1158/1078-0432.CCR-11-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, Lathan C, Marcoux JP, Du J, Okuda K, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, Iafrate AJ, Takeuchi K, Taiji M, Okuno Y, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20:5686–5696. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isozaki H, Hotta K, Ichihara E, Takigawa N, Ohashi K, Kubo T, Ninomiya T, Ninomiya K, Oda N, Yoshioka H, et al. Protocol design for the bench to bed trial in alectinib-refractory non-small-cell lung cancer patients harboring the EML4-ALK fusion gene (ALRIGHT/OLCSG1405) Clin Lung Cancer. 2016;17:602–605. doi: 10.1016/j.cllc.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 60.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, Tang RW, Wang H, Tsaparikos K, Wang J, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, Burke BJ, Deng YL, Liu W, Dardaei L, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374:54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu YL, Lu S, Lu Y, Zhou J, Shi YK, Sriuranpong V, Ho JCM, Ong CK, Tsai CM, Chung CH, et al. Results of PROFILE 1029, a phase iii comparison of first-line crizotinib versus chemotherapy in East Asian patients with ALK-positive advanced non-small cell lung cancer. J Thorac Oncol. 2018;13:1539–1548. doi: 10.1016/j.jtho.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 64.Krishnamurthy N, Goodman AM, Barkauskas DA, Kurzrock R. STK11 alterations in the pan-cancer setting: Prognostic and therapeutic implications. Eur J Cancer. 2021;148:215–229. doi: 10.1016/j.ejca.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wohlhieter CA, Richards AL, Uddin F, Hulton CH, Quintanal-Villalonga À, Martin A, de Stanchina E, Bhanot U, Asher M, Shah NS, et al. Concurrent mutations in STK11 and KEAP1 promote ferroptosis protection and SCD1 dependence in lung cancer. Cell Rep. 2020;33:108444. doi: 10.1016/j.celrep.2020.108444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, Schrock AB, Hartmaier RJ, Trabucco SE, Gay L, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18:556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Höglund P, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 69.Mahoney CL, Choudhury B, Davies H, Edkins S, Greenman C, Haaften G, Mironenko T, Santarius T, Stevens C, Stratton MR, Futreal PA. LKB1/KRAS mutant lung cancers constitute a genetic subset of NSCLC with increased sensitivity to MAPK and mTOR signalling inhibition. Br J Cancer. 2009;100:370–375. doi: 10.1038/sj.bjc.6604886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parachoniak CA, Rankin A, Gaffney B, Hartmaier R, Spritz D, Erlich RL, Miller VA, Morosini D, Stephens P, Ross JS, et al. Exceptional durable response to everolimus in a patient with biphenotypic breast cancer harboring an STK11variant. Cold Spring Harb Mol Case Stud. 2017;3:a000778. doi: 10.1101/mcs.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG, Sidransky D. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–3662. [PubMed] [Google Scholar]
- 72.Hezel AF, Gurumurthy S, Granot Z, Swisa A, Chu GC, Bailey G, Dor Y, Bardeesy N, Depinho RA. Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol Cell Biol. 2008;28:2414–2425. doi: 10.1128/MCB.01621-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wingo SN, Gallardo TD, Akbay EA, Liang MC, Contreras CM, Boren T, Shimamura T, Miller DS, Sharpless NE, Bardeesy N, et al. Somatic LKB1 mutations promote cervical cancer progression. PLoS One. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill RK, Yang SH, Meerzaman D, Mechanic LE, Bowman ED, Jeon HS, Roy Chowdhuri S, Shakoori A, Dracheva T, Hong KM, et al. Frequent homozygous deletion of the LKB1/STK11gene in non-small cell lung cancer. Oncogene. 2011;30:3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SM, Choi JE, Na YK, Lee EJ, Lee WK, Choi YY, Yoon GS, Jeon HS, Kim DS, Park JY. Genetic and epigenetic alterations of the LKB1 gene and their associations with mutations in TP53 and EGFR pathway genes in Korean non-small cell lung cancers. Lung Cancer. 2013;81:194–199. doi: 10.1016/j.lungcan.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 76.Tanwar PS, Mohapatra G, Chiang S, Engler DA, Zhang L, Kaneko-Tarui T, Ohguchi Y, Birrer MJ, Teixeira JM. Loss of LKB1 and PTEN tumor suppressor genes in the ovarian surface epithelium induces papillary serous ovarian cancer. Carcinogenesis. 2014;35:546–553. doi: 10.1093/carcin/bgt357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Liu J, Li P, Mao X, Li W, Yang J, Liu P. Loss of LKB1 disrupts breast epithelial cell polarity and promotes breast cancer metastasis and invasion. J Exp Clin Cancer Res. 2014;33:70. doi: 10.1186/s13046-014-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang JY, Jiang SH, Liu DJ, Yang XM, Huo YM, Li J, Hua R, Zhang ZG, Sun YW. Decreased LKB1 predicts poor prognosis in pancreatic ductal adenocarcinoma. Sci Rep. 2015;5:10575. doi: 10.1038/srep10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W, Yin L, Song G, Han X, Yin Z, Luo D. LKB1 loss cooperating with BRAFV600E promotes melanoma cell invasion and migration by up-regulation MMP-2 via PI3K/Akt/mTOR pathway. Oncotarget. 2017;8:113847–113857. doi: 10.18632/oncotarget.22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, Suzuki K, Nakamoto M, Shimizu E, Minna JD, Yokota J. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–5918. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fang R, Zheng C, Sun Y, Han X, Gao B, Li C, Liu H, Wong KK, Liu XY, Chen H, Ji H. Integrative genomic analysis reveals a high frequency of LKB1 genetic alteration in Chinese lung adenocarcinomas. J Thorac Oncol. 2014;9:254–258. doi: 10.1097/JTO.0000000000000056. [DOI] [PubMed] [Google Scholar]
- 82.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 83.Liang J, Mills GB. AMPK: A contextual oncogene or tumor suppressor? Cancer Res. 2013;73:2929–2935. doi: 10.1158/0008-5472.CAN-12-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baas AF, Smit L, Clevers H. LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol. 2004;14:312–319. doi: 10.1016/j.tcb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Forcet C, Etienne-Manneville S, Gaude H, Fournier L, Debilly S, Salmi M, Baas A, Olschwang S, Clevers H, Billaud M. Functional analysis of Peutz-Jeghers mutations reveals that the LKB1 C-terminal region exerts a crucial role in regulating both the AMPK pathway and the cell polarity. Hum Mol Genet. 2005;14:1283–1292. doi: 10.1093/hmg/ddi139. [DOI] [PubMed] [Google Scholar]
- 88.Galan-Cobo A, Sitthideatphaiboon P, Qu X, Poteete A, Pisegna MA, Tong P, Chen PH, Boroughs LK, Rodriguez MLM, Zhang W, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res. 2019;79:3251–3267. doi: 10.1158/0008-5472.CAN-18-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koyama S, Akbay EA, Li YY, Aref AR, Skoulidis F, Herter-Sprie GS, Buczkowski KA, Liu Y, Awad MM, Denning WL, et al. STK11/LKB1 deficiency promotes neutrophil recruitment and proinflammatory cytokine production to suppress T-cell activity in the lung tumor microenvironment. Cancer Res. 2016;76:999–1008. doi: 10.1158/0008-5472.CAN-15-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The tumor suppressor gene LKB1 is associated with prognosis in human breast carcinoma. Clin Cancer Res. 2002;8:2085–2090. [PubMed] [Google Scholar]
- 92.Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, Roadcap DW, Ollila DW, Thomas NE, Castrillon DH, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer Cell. 2012;21:751–764. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–7832. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 94.Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, Engel BE, Xie M, Berglund AE, Creelan BC, et al. Differential association of STK11 and TP53 with KRASmutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene. 2016;35:3209–3216. doi: 10.1038/onc.2015.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lamberti G, Sisi M, Andrini E, Palladini A, Giunchi F, Lollini PL, Ardizzoni A, Gelsomino F. The mechanisms of PD-L1 regulation in non-small-cell lung cancer (NSCLC): Which are the involved players? Cancers (Basel) 2020;12:3129. doi: 10.3390/cancers12113129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roosan MR, Mambetsariev I, Pharaon R, Fricke J, Husain H, Reckamp KL, Koczywas M, Massarelli E, Bild AH, Salgia R. Usefulness of circulating tumor DNA in identifying somatic mutations and tracking tumor evolution in patients with non-small cell lung cancer. Chest. 2021;160:1095–1107. doi: 10.1016/j.chest.2021.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dahmani R, Just PA, Delay A, Canal F, Finzi L, Prip-Buus C, Lambert M, Sujobert P, Buchet-Poyau K, Miller E, et al. A novel LKB1 isoform enhances AMPK metabolic activity and displays oncogenic properties. Oncogene. 2015;34:2337–2346. doi: 10.1038/onc.2014.182. [DOI] [PubMed] [Google Scholar]
- 99.Bouchekioua-Bouzaghou K, Poulard C, Rambaud J, Lavergne E, Hussein N, Billaud M, Bachelot T, Chabaud S, Mader S, Dayan G, et al. LKB1 when associated with methylatedERα is a marker of bad prognosis in breast cancer. Int J Cancer. 2014;135:1307–1318. doi: 10.1002/ijc.28781. [DOI] [PubMed] [Google Scholar]
- 100.Koivunen JP, Kim J, Lee J, Rogers AM, Park JO, Zhao X, Naoki K, Okamoto I, Nakagawa K, Yeap BY, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99:245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roy BC, Kohno T, Iwakawa R, Moriguchi T, Kiyono T, Morishita K, Sanchez-Cespedes M, Akiyama T, Yokota J. Involvement of LKB1 in epithelial-mesenchymal transition (EMT) of human lung cancer cells. Lung Cancer. 2010;70:136–145. doi: 10.1016/j.lungcan.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 102.Yao YH, Cui Y, Qiu XN, Zhang LZ, Zhang W, Li H, Yu JM. Attenuated LKB1-SIK1 signaling promotes epithelial-mesenchymal transition and radioresistance of non-small cell lung cancer cells. Chin J Cancer. 2016;35:50. doi: 10.1186/s40880-016-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Momcilovic M, Shackelford DB. Targeting LKB1 in cancer-exposing and exploiting vulnerabilities. Br J Cancer. 2015;113:574–584. doi: 10.1038/bjc.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dobashi Y, Watanabe Y, Miwa C, Suzuki S, Koyama S. Mammalian target of rapamycin: A central node of complex signaling cascades. Int J Clin Exp Pathol. 2011;4:476–495. [PMC free article] [PubMed] [Google Scholar]
- 106.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 108.Chen B, Tan Z, Gao J, Wu W, Liu L, Jin W, Cao Y, Zhao S, Zhang W, Qiu Z, et al. Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J Exp Clin Cancer Res. 2015;34:126. doi: 10.1186/s13046-015-0239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krencz I, Sebestyén A, Fábián K, Márk Á, Moldvay J, Khoor A, Kopper L, Pápay J. Expression of mTORC1/2-related proteins in primary and brain metastatic lung adenocarcinoma. Hum Pathol. 2017;62:66–73. doi: 10.1016/j.humpath.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 110.Seki N, Takasu T, Mandai K, Nakata M, Saeki H, Heike Y, Takata I, Segawa Y, Hanafusa T, Eguchi K. Expression of eukaryotic initiation factor 4E in atypical adenomatous hyperplasia and adenocarcinoma of the human peripheral lung. Clin Cancer Res. 2002;8:3046–3053. [PubMed] [Google Scholar]
- 111.Yoshizawa A, Fukuoka J, Shimizu S, Shilo K, Franks TJ, Hewitt SM, Fujii T, Cordon-Cardo C, Jen J, Travis WD. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res. 2010;16:240–248. doi: 10.1158/1078-0432.CCR-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joo MS, Kim WD, Lee KY, Kim JH, Koo JH, Kim SG. AMPK facilitates nuclear accumulation of Nrf2 by phosphorylating at serine 550. Mol Cell Biol. 2016;36:1931–1942. doi: 10.1128/MCB.00118-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ciccarese F, Zulato E, Indraccolo S. LKB1/AMPK pathway and drug response in cancer: A therapeutic perspective. Oxid Med Cell Longev. 2019;2019:8730816. doi: 10.1155/2019/8730816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Singh A, Daemen A, Nickles D, Jeon SM, Foreman O, Sudini K, Gnad F, Lajoie S, Gour N, Mitzner W, et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin Cancer Res. 2021;27:877–888. doi: 10.1158/1078-0432.CCR-20-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Trapp EK, Majunke L, Zill B, Sommer H, Andergassen U, Koch J, Harbeck N, Mahner S, Friedl TWP, Janni W, et al. LKB1 pro-oncogenic activity triggers cell survival in circulating tumor cells. Mol Oncol. 2017;11:1508–1526. doi: 10.1002/1878-0261.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deng L, Yao P, Li L, Ji F, Zhao S, Xu C, Lan X, Jiang P. p53-mediated control of aspartate-asparagine homeostasis dictates LKB1 activity and modulates cell survival. Nat Commun. 2020;11:1755. doi: 10.1038/s41467-020-15573-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goodman AM, Piccioni D, Kato S, Boichard A, Wang HY, Frampton G, Lippman SM, Connelly C, Fabrizio D, Miller V, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid Tumors. JAMA Oncol. 2018;4:1237–1244. doi: 10.1001/jamaoncol.2018.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Donnelly LL, Hogan TC, Lenahan SM, Nandagopal G, Eaton JG, Lebeau MA, McCann CL, Sarausky HM, Hampel KJ, Armstrong JD, et al. Functional assessment of somatic STK11variants identified in primary human non-small cell lung cancers. Carcinogenesis. 2021;42:1428–1438. doi: 10.1093/carcin/bgab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gill CM, Loewenstern J, Rutland JW, Arib H, Pain M, Umphlett M, Kinoshita Y, McBride RB, Bederson J, Donovan M, et al. STK11 mutation status is associated with decreased survival in meningiomas. Neurol Sci. 2020;41:2585–2589. doi: 10.1007/s10072-020-04372-y. [DOI] [PubMed] [Google Scholar]
- 125.Facchinetti F, Bluthgen MV, Tergemina-Clain G, Faivre L, Pignon JP, Planchard D, Remon J, Soria JC, Lacroix L, Besse B. LKB1/STK11 mutations in non-small cell lung cancer patients: Descriptive analysis and prognostic value. Lung Cancer. 2017;112:62–68. doi: 10.1016/j.lungcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 126.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols MC, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Herbst RS, Baas P, Perez-Gracia JL, Felip E, Kim DW, Han JY, Molina JR, Kim JH, Dubos Arvis C, Ahn MJ, et al. Use of archival versus newly collected tumor samples for assessing PD-L1 expression and overall survival: An updated analysis of KEYNOTE-010 trial. Ann Oncol. 2019;30:281–289. doi: 10.1093/annonc/mdy545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zugazagoitia J, Molina-Pinelo S, Lopez-Rios F, Paz-Ares L. Biological therapies in nonsmall cell lung cancer. Eur Respir J. 2017;49:1601520. doi: 10.1183/13993003.01520-2016. [DOI] [PubMed] [Google Scholar]
- 130.Shire NJ, Klein AB, Golozar A, Collins JM, Fraeman KH, Nordstrom BL, McEwen R, Hembrough T, Rizvi NA. STK11 (LKB1) mutations in metastatic NSCLC: Prognostic value in the real world. PLoS One. 2020;15:e0238358. doi: 10.1371/journal.pone.0238358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mograbi B, Heeke S, Hofman P. The Importance of STK11/LKB1 assessment in non-small cell lung carcinomas. Diagnostics (Basel) 2021;11:196. doi: 10.3390/diagnostics11020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Herter-Sprie GS, Korideck H, Christensen CL, Herter JM, Rhee K, Berbeco RI, Bennett DG, Akbay EA, Kozono D, Mak RH, et al. Image-guided radiotherapy platform using single nodule conditional lung cancer mouse models. Nat Commun. 2014;5:5870. doi: 10.1038/ncomms6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.He Q, Li J, Dong F, Cai C, Zou X. LKB1 promotes radioresistance in esophageal cancer cells exposed to radiation, by suppression of apoptosis and activation of autophagy via the AMPK pathway. Mol Med Rep. 2017;16:2205–2210. doi: 10.3892/mmr.2017.6852. [DOI] [PubMed] [Google Scholar]
- 134.Wang Y, Li N, Jiang W, Deng W, Ye R, Xu C, Qiao Y, Sharma A, Zhang M, Hung MC, et al. Mutant LKB1 confers enhanced radiosensitization in combination with trametinib in KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2018;24:5744–5756. doi: 10.1158/1078-0432.CCR-18-1489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.