Abstract
Factor C protein isolated from the horseshoe crab, Carcinoscorpius rotundicauda, has endotoxin binding capability. Synthetic peptides of 34 amino acids based on the sequence of two regions of factor C (Sushi 1 and Sushi 3) as well as their corresponding mutants exhibited activities against 30 clinical isolates of Pseudomonas aeruginosa. Collectively, all four peptides demonstrated exceptionally effective bactericidal activity against P. aeruginosa with 90% minimal bactericidal concentrations (MBC90s) in the range of 0.06 to 0.25 μg/ml (16 to 63 nM). Viable bacteria were reduced by 90% after 7 min and were totally eradicated within 40 to 50 min. These peptides are minimally hemolytic against both rabbit and human erythrocytes even at concentrations up to 1,600-fold their MBC90s. Both in vitro and in vivo studies indicate that cytotoxic effects are small even at 1,000-fold their MBC90s. Furthermore, the Sushi peptides are tolerant of high-salt and adverse pH conditions. These findings demonstrate the promising therapeutic potential of the Sushi peptides.
In recent decades, nosocomial infection has drawn more attention from the medical community, owing to the ease of acquisition and lack of lasting effective clinical management. Pseudomonas aeruginosa is the epitome of an opportunistic human pathogen (17) causing infections of the urinary tract, respiratory system, and soft tissue. It also causes dermatitis, bacteremia, and a variety of systemic infections, particularly in victims of severe burns (38), patients with diabetes, and cancer and AIDS patients who are immunosuppressed. Hospitals and other medical facilities provide an immeasurable reservoir for pseudomonads to develop resistance to a variety of naturally occurring antibiotics (2, 17, 21). Many reports have shown that pseudomonads maintain antibiotic resistance plasmids, both R factors and resistance transfer factors, and are able to transfer these genes by conjugation and transformation. To date, only a few antibiotics remain effective against them. These include fluoroquinones, aminoglycosides, and imipenem (2, 5, 17, 21). However, resistance against these antibiotics is developing rapidly (2, 15, 17). The futility of treating Pseudomonas infections with antibiotics is most dramatically illustrated in cystic fibrosis (CF) (15, 34) and bronchiectasis (16) patients; virtually all of these patients eventually succumb to infection with multidrug-resistant strains that make treatment difficult, if not impossible. The pathogenesis of infections by Pseudomonas is multifactorial, as suggested by the wide array of its virulence determinants (9).
Pseudomonads are naturally resistant to many antibiotics, due to the permeability barrier afforded by their outer membrane, lipopolysaccharide (LPS). Furthermore, their tendency and ability to colonize the surfaces of biofilms (8) make them impervious to therapeutic concentrations of antibiotics. Recently, the concept of eradication via targeted disruption of bacterial LPS by cationic peptides and proteins was introduced (7, 33). These peptides and proteins, which are mainly α-helical or β-sheet in structure, assert their effect by disrupting the bacterial membrane, causing pore formation that eventually leads to osmotic imbalance and cell death (23). For effective antimicrobial therapy, such peptides and proteins need to satisfy several important criteria: (i) potent antimicrobial activity over a wide range of pH, (ii) rapid killing rate, (iii) low toxicity, (iv) low hemolytic activity, and (v) delivery to the target site of infection without degradation of the peptide. While numerous antimicrobial peptides like FALL-39 (1), SMAP-29 (33), lepidopteran cecropin (37), and magainin (40) have been reported, few display all of the above-mentioned attributes. Thus, the search for new, more-powerful, and yet safe antimicrobial peptides continues to be a priority.
In our laboratory, we have characterized the LPS binding region of factor C (28, 29, 35), the first enzyme in the endotoxin-induced coagulation cascade in the horseshoe crab (10, 11, 27). Four peptides derived from the Sushi 1 and 3 domains of the factor C sequence (12) (namely, S1, S1Δ, S3, and S3Δ) exhibited high affinity for LPS (36). These peptides are collectively termed Sushi peptides. Further analyses of these peptides showed them to have low cytotoxicity and the capability to neutralize LPS biotoxicity, to suppress LPS-induced cytokine production, and to confer protection against LPS-induced lethality in mice (36). Therefore, LPS toxicity, as seen during the course of antibiotic treatment, will be dramatically reduced. This property would provide an advantage over existing antibiotics and most other non-LPS-sequestering cationic antimicrobial peptides, in suppressing the adverse effects of LPS-induced septic shock during or after treatment. Septic shock is characterized by a drastic fall in blood pressure, cardiovascular collapse, and multiple organ failure (4, 19, 24) and is responsible for over 100,000 deaths a year in the United States alone (13). Septic shock often creates more complications than the actual infection itself when massive amounts of LPS are released by bacteria disintegrated by antibiotics (19, 26). This condition is especially pronounced in children, the elderly, and immunocompromised patients. This report documents factor C-derived Sushi peptides with potent activity against clinical isolates of multidrug-resistant P. aeruginosa.
MATERIALS AND METHODS
Bacterial cultures, antibiotics, and antimicrobial peptides.
Thirty clinical isolates of P. aeruginosa (obtained from the National University Hospital, Singapore) and the standard strain, ATCC 27853, were tested for antibiotic sensitivity by the disk diffusion method of Bauer et al. (3) against amikacin, aztreonam, cefoperazone, ceftazidime (CAZ), ciprofloxacin, gentamicin, imipenem, netilmicin, and piperacillin. The antibiotic disks were from Oxoid. Polymyxin B sulfate (PB) (500,000 U) and PB nonapeptide were obtained from Sigma. Colistimethate sulfate (here, designated colistin) was kindly provided by Alpharma. Clinical strain 3, which was the most resistant, was selected for further study, alongside with P. aeruginosa ATCC 27853 as the standard strain. All tests were carried out in triplicate.
Peptide synthesis and purification.
All peptides used in this study were synthesized and purified by Genemed Synthesis, Inc., San Francisco, Calif. The first peptide, N-GFKLKGMARISCLPNGQWSNFPPKCIRECAMVSS-C, corresponds to residues 171 to 204 of the Sushi 1 domain of CrFC (12) and is designated S1, with a molecular weight of 3,758. The second Sushi peptide, N-HAEHKVKIGVEQKYGQFPQGTEVTYTCSGNYFLM-C, corresponds to residues 268 to 301 of the factor C Sushi 3 domain and is designated S3, with an MW of 3,892. Two changes were made to introduce lysine residues into S1 and S3, resulting in S1Δ(171–204Δ177,179) (S1Δ) with an MW of 3,727 and S3Δ(268–301Δ276,278) (S3Δ) with an MW of 3,962. All four peptides, collectively referred to as Sushi peptides, were purified by high-performance liquid chromatography to >95% purity. The calculated pI values for peptides S1, S1Δ, S3, and S3Δ are 9.85, 10.08, 7.27, and 9.62, respectively.
Determination of MBC for peptides.
The minimum bacterial concentration (MBC) test was a modification of the MIC determination of cationic antimicrobial peptides by modified microtiter broth dilution method proposed by the Hancock laboratory (http://www.cmdr.ubc.ca/bdoh/MIC.htm). Test strains of P. aeruginosa were cultured in 10 ml of Mueller-Hinton broth (MHB) (Becton Dickinson) and shaken at 230 rpm overnight at 37°C with a shaker incubator (model 4536; Forma Scientific, Inc.). Overnight broth cultures were diluted to give a final cell population of 105 CFU/ml. One-hundred-microliter aliquots of the bacterial suspension were dispensed into sterile polypropylene eight-strip PCR tubes (Quality Scientific Plastics). Eleven microliters of serial twofold-diluted Sushi peptides, with final concentrations in the range of 0.03 to 4 μg/ml, was then added. The peptides were constituted at 10 times the required test concentrations in 0.01% acetic acid and 0.2% bovine serum albumin. Positive controls were cultures without test peptides. Uninoculated MHB was used as a negative control. Cultures were incubated at 37°C for 18 to 24 h, with the PCR tubes held in a horizontal position and shaken at 230 rpm. Cell counts were determined by a standard drop count method (21). Results were expressed as MICs and MBCs, whereby MIC is the lowest concentration of peptide that reduces growth by more than 50% and MBC is the lowest concentration of peptide that prevents any residual colony formation (http://www.cmdr.ubc.ca/bobh/MIC.htm). MIC90s and MBC90s are defined with respect to a collection of 30 different strains, representing the concentration at which 90% of the strains were inhibited or killed, respectively.
Killing rate of P. aeruginosa by Sushi peptides.
The killing rate assay was adapted from the MBC test. A fixed final concentration of 0.06 μg of the peptide ml was incubated with P. aeruginosa ATCC 27853 and clinical isolate 3; bacterial counts were performed at different time intervals. To test the limits of the bactericidal activities of the peptides, an initial density of 109 CFU/ml was used.
Effect of pH on Sushi peptides.
The effect of pH (6.0 to 8.0) on the peptides (from 0.03 to 1 μg/ml) was tested in the MBC assay with 105 CFU of P. aeruginosa ATCC 27853 or clinical isolate 3. The pH of MHB was adjusted with HCl or NaOH. MHB (pH 7.3) inoculated with bacteria was used as a negative control. Cultures were incubated at 37°C for 18 to 24 h and shaken at 230 rpm. The cultures were transferred into 96-well microtiter plates (Nunclon Δ surface; Nunc). Optical density at 595 nm (OD595) was measured with a SPECTRAmax 340 plate reader with SOFTmax PRO (version 1.2.0) software.
Effect of salt on Sushi peptides.
Peptides ranging in concentration from 0.03 to 1 μg/ml were added to MHB containing 50 to 300 mM NaCl. Incubation of P. aeruginosa ATCC 27853 or clinical isolate 3 was carried out at 37°C for 18 to 24 h. The overnight cultures were transferred into 96-well microtiter plates, and OD595 was measured.
Hemolysis assay.
The hemolysis assay was adapted from the method of Shin et al. (32). Human and rabbit erythrocytes were both used to test the hemolytic activities of the peptides. Whole blood was collected in a sterile, heparinized, borosilicate tube and centrifuged (3K10 centrifuge; Sigma) at 1,000 × g for 5 min at 4°C. The supernatant, including the leukocytes above the erythrocyte pellet, was removed carefully and discarded. Intact erythrocytes were washed three times with 3 volumes of prechilled pyrogen-free saline (PFS). Erythrocyte suspensions were adjusted to 0.8% for the hemolysis assay. Serial twofold dilutions of the peptides were prepared in PFS, and 100-μl aliquots were added to equal volumes of 0.8% erythrocyte suspension in sterile 96-well microtiter plates (Nunclon Δ surface; Nunc) in triplicate. The plates were incubated at 37°C for 1 h. Subsequently, intact erythrocytes were pelleted by centrifugation at 1,000 × g for 5 min at 4°C. One hundred microliters of supernatant from each well was transferred accordingly to a new 96-well microtiter plate, and the amount of hemoglobin released into the supernatant was determined by reading the absorbance at 414 nm against a reference wavelength of 490 nm. A positive control with 100 μl of 0.4% erythrocyte lysed in 1% Triton X-100 was taken as 100% lysis. The negative control was erythrocytes in PFS alone, which gave minimal lysis. This was taken as 0%.
RESULTS
The antibacterial activity of Sushi peptides against gram-negative bacteria.
The antimicrobial properties of the Sushi peptides have been tested on a range of gram-negative bacteria (Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 13883, Salmonella enterica serovar Typhimurium ATCC 14028, P. aeruginosa ATCC 27853, Vibrio parahaemolyticus, and Aeromonas hydrophila) with MICs ranging from ≤1.56 to 100 μg/ml. We pursued further investigations of a common gram-negative pathogen, P. aeruginosa. Table 1 summarizes the antibiogram of the 30 clinical isolates of P. aeruginosa and strain 3, which exhibited the broadest resistance against the antibiotics tested. Of particular interest is strain 3's resistance to antibiotics such as expanded-spectrum cephalosporins and aminoglycosides. Therefore, strain 3 was used in further studies with the Sushi peptides. P. aeruginosa ATCC 27853 was used as a standard control strain. The ATCC 27853 control strain was often more susceptible than the clinical strains.
TABLE 1.
Summary antibiogram of P. aeruginosa clinical isolates (n = 30)
| Antibiotic |
P. aeruginosa
|
No. of resistant clinical strains | |
|---|---|---|---|
| Strain 3 | ATCC 27853 | ||
| Amikacin | R | S | 3 |
| Aztreonam | R | S | 13 |
| Cefoperazone | R | S | 12 |
| CAZ | R | S | 4 |
| Ciprofloxacin | R | S | 3 |
| Gentamicin | R | S | 10 |
| Imipenem | S | S | 1 |
| Netilmicin | R | S | 3 |
| Piperacillin | R | S | 5 |
Sensitivity to antibiotics was tested by the method of Bauer et al. (3). S, susceptible; R, resistant.
Bactericidal concentration of Sushi peptides against P. aeruginosa.
The killing efficiency of the four Sushi peptides was calculated by enumerating surviving bacteria by the standard drop count method. All four peptides (S1, S1Δ, S3, and S3Δ) showed potent bactericidal activity against the 30 clinical strains (Table 2), with a rapid exponential killing effect within concentrations of only two- to fourfold. The killing curve of clinical isolate strain 3 is shown in Fig. 1. Although the isolates exhibited variable resistance to aminoglycosides and cephalosporins, the MBC90 for the peptides against them was 0.06 μg/ml (16 nM) for S1 and S3 and 0.25 μg/ml (63 nM) for S1Δ and S3Δ. These values are 32 to 133 times more potent than those of currently available antibiotics (for example, PB) and 64 to 267 times more effective than colistin (Table 2). Hence, these values are unsurpassed by those for any known peptides with activity against P. aeruginosa.
TABLE 2.
MIC, MBC, and hemolytic activities of Sushi peptides compared to other antibiotic peptides for clinical strains of P. aeruginosa
| Test peptide | MIC (μg/ml)
|
MBC (μg/ml)
|
% of hemolysis (at 100 μg/ml)a | ||||
|---|---|---|---|---|---|---|---|
| 50% | 90% | Range | 50% | 90% | Range | ||
| S1 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | 0.06 | ≤0.03–1 | 0 |
| S1Δ | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | 0.25 | ≤0.03–1 | 0 |
| S3 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | 0.06 | ≤0.03–0.25 | 8 |
| S3Δ | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | 0.25 | ≤0.03–0.5 | 37 |
| Colistin | 2 | 4 | 1–16 | 8 | 16 | 4–16 | ND |
| PB nonapeptide | ≥32 | ≥32 | ≥32 | ≥32 | ≥32 | ≥32 | ND |
| PB | 0.5 | 1 | 0.25–1 | 4 | 8 | 1–16 | ND |
For S1, S1Δ, and S3, less than 10% hemolysis was induced at 100 μg/ml. ND, not determined.
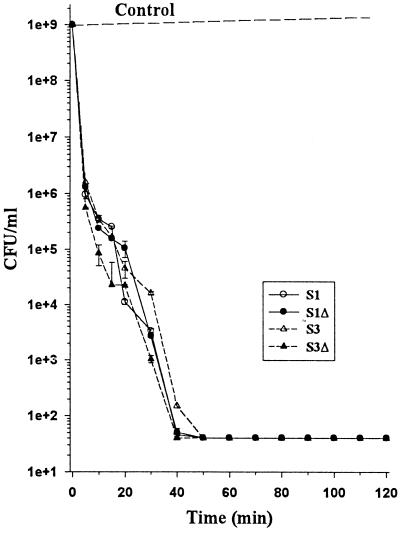
FIG. 1.
Bactericidal action of Sushi peptides against P. aeruginosa strain 3. Killing curve of P. aeruginosa 3 by Sushi peptides. The initial density of P. aeruginosa was 105 CFU/ml.
Sushi peptides exhibit rapid bactericidal action.
Rapid bactericidal action is one of the essential features of an effective therapeutic agent. With a low MBC90 concentration, we proceeded to investigate the killing time for the Sushi peptides with a much higher initial cell population of 109 CFU of P. aeruginosa 3 or ATCC 27853 per ml. At 0.06 μg/ml, all four peptides achieved 90% reductions in viable counts within 7 min for both strains tested (Fig. 2). By 40 to 50 min, the peptides had totally eradicated the bacteria (Fig. 2 and 3).
FIG. 2.
Time-dependent killing of P. aeruginosa 3. An initial cell density of 109 CFU of P. aeruginosa 3/ml was used in the assay. The effect of test peptides at 0.06 μg/ml was assessed by enumerating the viable cells (CFU per milliliter) at indicated time intervals after overnight incubation.
FIG. 3.
Eradication time course of P. aeruginosa 3 by S3Δ peptide by using a plate count assay. A plate count at 10-fold dilutions on Mueller-Hinton agar showed the effect of S3Δ peptide (0.06 μg/ml) on P. aeruginosa 3 with an initial cell count of 109 CFU/ml at indicated time intervals.
Sushi peptides show tolerance to pH range.
The functionality of the Sushi peptides was studied over pH 6.0 to 8.0 with P. aeruginosa 3 or ATCC 27853. S1, S1Δ, and S3 inhibited bacterial growth, whereas S3Δ showed reduced efficacy between pH 7.0 to 8.0. Figure 4 illustrates the effect of pH on the inhibitory ability of the four peptides against strain 3. Subsequent plate counts performed with the cultures revealed that over the pH range, S1 retained its MBC90 of 0.06 μg/ml against 105 CFU/ml. S1Δ, S3, and S3Δ changed from being bactericidal at pH 6.0 to 7.0 to being bacteriostatic at pH 7.5 to 8.0. Despite the transition from bactericidal to bacteriostatic activity, S1, S1Δ, and S3 were still able to reduce the initial concentration of 105 CFU/ml in the assays. However, S3Δ lost its bacteriostatic effect at a pH of > 7.0.
FIG. 4.
Effect of pH on Sushi peptides on P. aeruginosa 3. OD595 of different Sushi peptides was measured against P. aeruginosa 3 at their respective MBC90 concentrations. For each pH value, a positive control was set up with the respective MHB incubated with bacteria alone. The negative control was taken at OD595 of MHB alone.
Applicability of Sushi peptides at high-salt concentration.
The limit to salt tolerance of the Sushi peptides was analyzed at concentrations from 50 to 300 mM, although the osmolarity of body fluids ranges from 120 to 150 mM in a normal individual. All peptides transitioned from bactericidal to bacteriostatic activity, exerting an inhibitory effect over the range of salt concentrations tested. Even at 300 mM NaCl, all four Sushi peptides showed activities against P. aeruginosa 3 at 0.06 μg/ml (data not shown). All peptides maintained their antibacterial function over the salt concentrations tested, albeit transitioning from bactericidal to bacteriostatic activity.
Sushi peptides have low hemolytic activity.
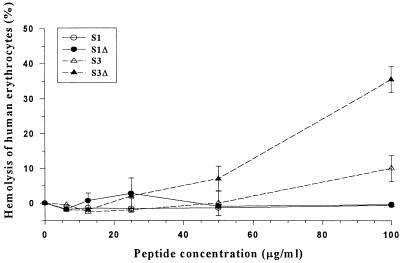
The absence or lack of hemolytic activity is crucial to the applicability of an antimicrobial agent for therapeutic use in humans and animals. The hemolytic activity of the peptides was determined with human erythrocytes. At 50 μg/ml (12.5 μM), the peptides showed minimum hemolytic activity ranging from 0 to 7%. Even at concentrations of 100 μg/ml (25 μM), up to 1,600-fold their MBC90s, the Sushi peptides showed minimal hemolytic activity (<5%) for S1, S1Δ, and S3, while S3Δ showed a higher hemolytic activity of 35% (Fig. 5). In a separate assay, the hemolytic activity of Sushi peptides was tested with rabbit erythrocytes. At the same concentration of 100 μg/ml, all the peptides showed hemolytic activity below 6% (data not shown). These results demonstrated the specificity of the peptides towards Pseudomonas spp., presumably the LPS layer, but not to human nor rabbit erythrocytes.
FIG. 5.
Percentage of hemolysis of human erythrocytes induced by the peptides. Erythrocytes at 0.4% were incubated with different concentrations of peptides (6 to 100 μg/ml). Erythrocytes lysed in 1% Triton X-100 were taken as 100% lysis. The negative control was 0.4% erythrocytes in PFS.
DISCUSSION
P. aeruginosa is a fast-replicating bacterium, which displays a short lag phase and doubling time. Owing to its pathogenicity and antibiotic-resistant nature, a bactericidal agent with rapid action will be the most effective and appropriate counter measure in controlling its spread from infected wounds. This is especially pronounced with secondary infections, like those in CF patients (2) and acute bacteremia in AIDS patients or those that occur near or in vital organs like the cornea, as well as in exposed skin on burn patients. Undesirably, pseudomonads have acquired a multitude of resistance mechanisms over the years against many of the antibiotics used to combat them. Pseudomonads that are resistant to first- and second-generation aminoglycosides, like gentamicin and amikacin (17, 38), cephalosporins, and imipenems (21) are rapidly increasing in number. New drugs that are effective against these emerging multidrug-resistant strains are urgently needed. A multifaceted approach to their eradication is essential to significantly reduce the possibility of the emergence of new resistant strains. Most antibiotics exert their bactericidal action by inhibiting a crucial biochemical enzyme (39). However, resistance can be attained through the acquisition of an antibiotic resistance plasmid, e.g., beta-lactamase, which expresses a new isoform of the targeted enzyme. Another mode of intervention is thus necessary to complement the current biochemical route.
The antimicrobial potency of the Sushi peptides was tested with 30 clinical isolates and a control strain of P. aeruginosa ATCC 27853. The resistance pattern of these strains gave a close representation of the resistant strains of P. aeruginosa found in Singapore (Table 1). The 30 clinical isolates showed very high resistance against most antibiotics used for the treatment of P. aeruginosa. Yet the Sushi peptides exhibited low MBC90s (0.06 to 0.25 μg/ml; 16–63 nM) for these multidrug-resistant strains of P. aeruginosa. A similar profile was observed for P. aeruginosa ATCC 27853. It is pertinent that the MBC90 range is narrow (two- to fourfold) for all isolates. These MBC90s obtained for the peptides are unsurpassed by any known antibiotics of metabolite or peptide origin. Comparatively, Sushi peptides are up to 1 to 2 orders of magnitude more effective than any other reported cationic antimicrobial peptides against P. aeruginosa (6, 14, 22, 30, 31). Although the peptides are probably targeted at the lipid A domain, different MBCs were observed for the 30 clinical isolates. The strong binding affinities of the Sushi peptides for lipid A (36) suggest an explanation for the susceptibility of the clinical strains tested. The Sushi peptides probably act by disrupting the LPS-lamellar organization in the bacterial cell membrane by physical means that eventually lead to osmotic imbalance and cell lysis.
At a concentration of 0.06 μg/ml, the Sushi peptides were able to eradicate 90% of 109 CFU of P. aeruginosa clinical strain 3 per ml (Fig. 2 and 3) and ATCC 27853 per ml within 7 min of incubation. A similar profile was observed for P. aeruginosa ATCC 27853. Complete eradication probably occurred within the first two generations of bacterial growth, which will reduce the possibility of mutation to resistance. Thus, this rapid killing rate should prevent the development of resistance, since it will require several precise mutations to occur at multiple enzymes along the LPS synthesis pathway to ultimately yield a modified LPS structure that is sufficiently different to evade Sushi peptide recognition. However, the possibility of developing or acquiring resistance cannot be precluded if some of these strains were allowed to mutate at sublethal peptide concentrations.
The effectiveness of the Sushi peptides was well maintained over a broad range of pH levels and salt concentrations. All the peptides were bactericidal from pH 6.0 to 7.0 at their respective MBC90s. S1, with a calculated pI of 9.85, is of particular interest, as it maintained its bactericidal potency across the pH range tested. It is observed that as the pH approached the pI of the peptides, the loss of most cationic charges on the peptides led to a loss of ionic interaction with LPS, which thus affected their bactericidal action. Surprisingly, both S1Δ (pI = 10.08) and S3Δ (pI = 9.62), with a pI relatively close to that of S1, did not perform as expected. They exhibited a bactericidal response at pH 6.0 to 7.0 and a bacteriostatic effect at pH 7.5 to 8.0.
The peptides were also resistant to high-salt concentrations. At up to 300 mM NaCl, Sushi peptides (≤0.03 μg/ml) inhibited Pseudomonas growth of an initial cell population of 105 CFU/ml (data not shown). Again, the transition from bactericidal to bacteriostatic activity was probably due to disruption of electrostatic interactions between the peptides and the bacterial LPS. Nevertheless, the peptides retained their bacteriostatic efficacy in controlling the proliferation of P. aeruginosa in a high-salt environment, similar to the lung fluids of CF patients where most antibiotics are inaccessible or unsuitable (15, 34). Hence, Sushi peptides can be developed for topical and aerosol applications.
The low MBC90 (0.06 to 0.25 μg/ml), rapid killing rate (40 to 50 min), versatility at high osmolarity (300 mM NaCl), tolerance of a broad pH range (6.0 to 8.0), and low or insignificant hemolytic activity as well as a lack of cytotoxic activity are excellent properties upon which Sushi peptides could be developed as highly effective and bactericidal candidate antibiotic against P. aeruginosa. The lack of cytotoxicity was confirmed both by in vitro and in vivo assays which showed minimal lysis of THP-I cells and also by the absence of aberrant behavior or death in C57BL/6J mice (36).
The LPS layer of gram-negative bacteria is essential to their growth and propagation. The LPS consists of a variable polysaccharide group and a lipid A moiety, which is the major trigger of a pathophysiological response. The massive release of LPS can be more deadly than the bacterial infection itself. The amounts of LPS released by antibiotics vary among different gram-negative bacterial strains. It is found that the amount of CAZ-induced release of bacterial LPS caused higher rates of lethality in mice than purified LPS alone (19). Moreover, the release of LPS is also shown to be associated with an increase in bacteria count (25). The mechanism of this phenomenon is still unknown. Perilously, antibiotic-induced LPS release occurs as early as 6 h after treatment (20).
The high affinity of Sushi peptides against E. coli lipid A also implies that the bactericidal potency of the peptides can be expanded to other gram-negative bacteria, without the risk of LPS release during the bactericidal action. Such peptides not only afford effective bactericidal action against P. aeruginosa but also encompass multidrug-resistant P. aeruginosa. The therapeutic indices (Table 2) of the Sushi peptides further illustrate their potential in controlling the emergence of such multidrug-resistant strains.
ACKNOWLEDGMENTS
We thank G. Kumarasinghe of the Department of Laboratory Medicine, National University Hospital, Singapore, Singapore, for providing the 30 clinical isolates of P. aeruginosa.
This work was supported by National Science and Technology Board grant LS/99/004.
REFERENCES
- 1.Agerberth B, Gunne H, Odeberg J, Kogner P, Boman H G, Gudmundsson G H. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc Natl Acad Sci USA. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda E A G, Marinho I S, Boulos M, Sinto S I, Caiaffa H H, Mendes C M, Oplustil C P, Sader H, Levy C E, Levin A S. Nosocomial infections caused by multi-resistant Pseudomonas aeruginosa. Infect Control Hosp Epidemiol. 1999;20:620–623. doi: 10.1086/501683. [DOI] [PubMed] [Google Scholar]
- 3.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–496. [PubMed] [Google Scholar]
- 4.Bone R C. The pathogenesis of sepsis. Ann Intern Med. 1991;115:457–460. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- 5.Bustamante C I, Wharton R C, Wade J C. In vitro activity of ciprofloxacin in combination with ceftazidime, aztreonam, and azlocillin against multiresistant isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:1814–1815. doi: 10.1128/aac.34.9.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catchpole C R, Andrews J M, Brenwald N, Wise R. A reassessment of the in-vitro activity of colistin sulphomethate sodium. J Antimicrob Chermother. 1997;39:255–260. doi: 10.1093/jac/39.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Chopra I. Research and development of antibacterial agents. Curr Opin Microbiol. 1998;1:495–501. doi: 10.1016/s1369-5274(98)80080-5. [DOI] [PubMed] [Google Scholar]
- 8.Costerton J W, Lewandowski Z, Caldwell D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 9.Danel F, Hall L M C, Gur D, Livermore D M. OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother. 1998;42:3117–3122. doi: 10.1128/aac.42.12.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding J L, Chai C, Pui A W M, Ho B. Expression of full length and deletion homologues of Carcinoscorpius rotundicauda factor C in Saccharomyces cerevisiae: immunoreactivity and endotoxin. J Endotoxin Res. 1997;4:33–43. [Google Scholar]
- 11.Ding J L, Navas III M A A, Ho B. Two forms of factor C from the amoebocytes of Carcinoscorpius rotundicauda: purification and characterisation. Biochim Biophys Acta. 1993;1202:149–156. doi: 10.1016/0167-4838(93)90076-4. [DOI] [PubMed] [Google Scholar]
- 12.Ding J L, Navas III M A A, Ho B. Molecular cloning and sequence analysis of factor C cDNA from the Singapore horseshoe crab, Carcinoscorpius rotundicauda. Mol Mar Biol Biotechnol. 1995;4:90–103. [PubMed] [Google Scholar]
- 13.Downey J S, Han J. Cellular activation mechanisms in septic shock. Front Biosci. 1998;3:468–476. doi: 10.2741/a293. [DOI] [PubMed] [Google Scholar]
- 14.Giacometti A, Cirioni O, Barchiesi F, Fortuna M, Scalise G. In-vitro activity of cationic peptides alone and in combination with clinically used antimicrobial agents against Pseudomonas aeruginosa. J Antimicrob Chemother. 1999;44:641–645. doi: 10.1093/jac/44.5.641. [DOI] [PubMed] [Google Scholar]
- 15.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 16.Hla S W H, Hui K P, Tan W C, Ho B. Genome macrorestriction analysis of sequential Pseudomonas aeruginosa isolates from bronchiectasis patients without cystic fibrosis. J Clin Microbiol. 1996;34:575–578. doi: 10.1128/jcm.34.3.575-578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horan T C, White J W, Jarvis W R, Emori T G, Culver D H, Munn V P, Thornsberry C, Olson D R, Hughes J M. Nosocomial infection surveillance, 1984. CDC surveillance summary. Morb Mortal Wkly Rep. 1986;35(SS1):17–29. [PubMed] [Google Scholar]
- 18.Karima R, Matsumoto S, Higashi H, Matsushima K. The molecular pathogenesis of endotoxic shock and organ failure. Mol Med Today. 1999;5:123–132. doi: 10.1016/s1357-4310(98)01430-0. [DOI] [PubMed] [Google Scholar]
- 19.Kirikae T, Kirikae F, Saito S, Tominaga K, Tamura H, Uemura Y, Yokochi T, Nakano M. Biological characterization of endotoxins released from antibiotic-treated Pseudomonas aeruginosa and Escherichia coli. Antimicrob Agents Chemother. 1998;42:1015–1021. doi: 10.1128/aac.42.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langevelde P, Kwappenberg K M C, Groeneveld P H P, Mattie H, Dissel J T. Antibiotic-induced lipopolysaccharide (LPS) release from Salmonella typhi: delay between killing by ceftazidime and imipenem and release of LPS. Antimicrob Agents Chemother. 1998;42:739–743. doi: 10.1128/aac.42.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorian V, editor. Antibiotics in laboratory medicine 4th ed. Baltimore, Md: The William & Wilkins Co.; 1996. pp. 902–1163. [Google Scholar]
- 22.Mosca D A, Hurst M A, So W, Viajar B S C, Fujii C A, Falla T J. IB-367, a protegrin peptide with in vitro and in vivo activities against the microflora associated with oral mucositis. Antimicrob Agents Chemother. 2000;44:1803–1808. doi: 10.1128/aac.44.7.1803-1808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oren Z, Shai Y. Mode of action of linear amphiphatic α-helical antimicrobial peptides. Biopolymers. 1998;47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Parrillo J E, Parker M M, Nathanson C, Cunnion A F, Ognibene F P. Septic shock in humans. Advances in the understanding of pathogenesis, cardiovascular, dysfunction and therapy. Ann Intern Med. 1990;113:227–237. doi: 10.7326/0003-4819-113-3-227. [DOI] [PubMed] [Google Scholar]
- 25.Porat R, Clark B D, Wolff S M, Diarello C A. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science. 1991;254:430–432. doi: 10.1126/science.1833820. [DOI] [PubMed] [Google Scholar]
- 26.Prins J M. Antibiotic induced release of endotoxin—clinical data and human studies. J Endotoxin Res. 1996;3:269–273. [Google Scholar]
- 27.Pui A W M, Ho B, Ding J L. Yeast recombinant factor C from horseshoe crab binds endotoxin and causes bacteriostasis. J Endotoxin Res. 1997;4:391–400. [Google Scholar]
- 28.Roopashree S D, Ho B, Ding J L. Recombinant COS-1 cells express Carcinoscorpius rotundicauda factor C. Biotechnol Lett. 1997;19:357–362. [Google Scholar]
- 29.Roopashree S D, Ho B, Ding J L. The Cys-rich and EGFP-like domains of Carcinoscorpius rotundicauda factor C yields soluble fusion protein with GFP. Biotechnol Lett. 1997;19:1147–1150. [Google Scholar]
- 30.Sawa T, Kurahashi K, Ohara M, Gropper M A, Doshi V, Larrick J W, Wiener-Kronish J P. Evaluation of antimicrobial and lipopolysaccharide-neutralizing effects of a synthetic CAP18 fragment against Pseudomonas aeruginosa in a mouse model. Antimicrob Agents Chemother. 1998;42:3269–3275. doi: 10.1128/aac.42.12.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwab U, Gilligan P, Jaynes J, Henke D. In vitro activities of designed antimicrobial peptides against multidrug-resistant cystic fibrosis pathogens. Antimicrob Agents Chemother. 1999;43:1435–1440. doi: 10.1128/aac.43.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin S Y, Kang J H, Hahm K S. Structure—antibacterial, antitumor and hemolytic activity relationships of cecropin A-magainin 2 and cecropin A-melittin hybrid peptides. J Peptide Res. 1999;53:82–90. doi: 10.1111/j.1399-3011.1999.tb01620.x. [DOI] [PubMed] [Google Scholar]
- 33.Skerlavaj B, Benincasa M, Risso A, Zanetti M, Gennaro R. SMAP-29: a potent antibacterial and antifungal peptide from sheep leukocytes. FEBS Lett. 1999;463:58–62. doi: 10.1016/s0014-5793(99)01600-2. [DOI] [PubMed] [Google Scholar]
- 34.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 35.Tan N S, Ho B, Ding J L. High-affinity LPS binding domain(s) in recombinant factor C of a horseshoe crab neutralizes LPS-induced lethality. FASEB J. 2000;14:859–870. doi: 10.1096/fasebj.14.7.859. [DOI] [PubMed] [Google Scholar]
- 36.Tan N S, Patricia N M L, Yau Y H, Chong P K W, Ho B, Ding J L. Definition of endotoxin-binding sites in horseshoe crab factor C recombinant sushi proteins and neutralization of endotoxin by sushi peptides. FASEB J. 2000;14:1801–1813. doi: 10.1096/fj.99-0866com. [DOI] [PubMed] [Google Scholar]
- 37.Teshima T, Ueky Y, Nakai T, Shiba T. Structure determination of lepidopteran, self-defense substance produced by silkworm. Tetrahedron. 1986;42:829–834. [Google Scholar]
- 38.Trafny E A. Susceptibility of adherent organisms from Pseudomonas aeruginosa and Staphylococcus aureus strains isolated from burn wounds to antimicrobial agents. Int J Antimicrob Agents. 1998;10:223–228. doi: 10.1016/s0924-8579(98)00042-9. [DOI] [PubMed] [Google Scholar]
- 39.Udo E E, Dashti A A. Detection of genes encoding aminoglycoside-modifying enzymes in staphylococci by polymerase chain reaction and dot blot hybridization. J Antimicrob Agents. 2000;13:273–279. doi: 10.1016/s0924-8579(99)00124-7. [DOI] [PubMed] [Google Scholar]
- 40.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]