Abstract
In recent years, circular RNAs (circRNAs/circs) have attracted significant attention due to their potentially important functions in a variety of human cancer types. circ_0067934 is a newly identified circRNA, the role of which in gastric cancer (GC) has yet to be reported, to the best of our knowledge. In the present study, the expression levels of circ_0067934, microRNA (miR)-1301-3p and kinesin family member 23 (KIF23) in GC cells were detected via reverse transcription-quantitative PCR. Cell proliferation was measured using Cell Counting Kit-8 assays and EdU staining. Wound healing and Transwell assays were performed to assess cell migration and invasion, respectively. Western blotting was performed to measure the protein expression levels of Ki67, proliferating cell nuclear antigen, MMP2, MMP9 and KIF23. The starBase database and luciferase reporter assays were used to predict and verify the binding between circ_0067934 and miR-1301-3p, as well as KIF23, in GC cells. The results demonstrated that circ_0067934 expression was upregulated in GC cells, and circ_0067934 silencing significantly inhibited GC cell proliferation, migration and invasion. In addition, miR-1301-3p was regulated by circ_0067934, and miR-1301-3p overexpression suppressed GC cell migration, invasion and proliferation. miR-1301-3p was found to target KIF23, and KIF23 overexpression reversed the effects of circ_0067934 silencing and miR-1301-3p overexpression on cell proliferation, migration and invasion. In conclusion, circ_0067934 may regulate the proliferation, invasion and migration of GC cells via the miR-1301-3p/KIF23 signaling axis, which may represent a novel therapeutic target for GC metastasis.
Keywords: circular RNA, gastric cancer, circular RNA 0067934, microRNA-1301-3p/kinesin family member 23, migration, invasion
Introduction
Gastric cancer (GC) is a common malignant tumor with high morbidity and mortality rates (1). The radical treatment for advanced GC is surgical resection. However, 40–60% of patients with GC develop recurrence and metastasis after surgery and the 5-year overall survival rate of patients with advanced GC is only 10–21% (2,3). Therefore, the molecular mechanisms underlying the occurrence and metastasis of GC, as well as the search for potential diagnostic and therapeutic targets, have become hotspots in the basic research field of the molecular diagnosis and treatment of GC.
Circular RNAs (circRNAs/circs) are a class of non-coding RNAs, which have a special closed ring structure and are widely present in most organisms (4). circRNAs can regulate gene function at several levels, such as epigenetic, transcriptional and post-transcriptional levels (5). MicroRNAs (miRNAs/miRs) comprise a group of non-coding RNA molecules, 20-25 nucleotides in length, which can bind to the 3′-untranslated region (UTR) of target genes, causing their degradation or inhibiting their translation (6). Some studies have revealed that circRNAs and miRNAs are abnormally expressed in various tumor tissues and cells, and they may participate in the whole process of tumor development (7–9). For example, circ_BTG3 associated nuclear protein-mediated miR-503/La ribonucleoprotein 1, translational regulator signaling was shown to promote lung cancer development and progression (10). Moreover, circ_5692 targets miR-328-5p to inhibit the occurrence of liver cancer (11). Another study reported that circ_homeodomain interacting protein kinase 3 promoted colorectal cancer development and metastasis by sponging miR-7 (12). circ_0067934 is a newly identified circRNA. A systematic meta-analysis based on an integrated dataset pre-processed from three microarrays suggested that circ_0067934 may represent an authentic biomarker for GC screening (13). Previous studies have shown that circ_0067934 may be involved in several types of cancer, including breast cancer, non-small cell lung cancer (NSCLC) and bladder cancer (14–16). However, the role of circ_0067934 in GC remains elusive.
Therefore, the aim of the present study was to investigate the possible role of circ_0067934 in the proliferation, invasion and migration of GC cells, as well as elucidate its underlying mechanism.
Materials and methods
Bioinformatic analysis
The starBase database (version 3.0; http://starbase.sysu.edu.cn/index.php/) was used to predict the binding site of circ_0067934 and miR-5047, miR-1301-3p, miR-670-5p, miR-345-3p, miR-545-3p and miR-3605-5p. In addition, the binding site of miR-1301-3p and KIF23 was predicted using the starBase database.
Cell culture
The GES-1 normal gastric mucosal cell line, and the GC cell lines, AGS and MKN-45 (China Infrastructure of Cell Line Resources, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences), were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 11 mM glucose, 10% (v/v) FBS (cat. no. 10099141; Gibco; Thermo Fisher Scientific, Inc.), 50 µM β-mercaptoethanol (cat. no. M3148; MilliporeSigma) and 10 mM HEPES (cat. no. H1090; Beijing Solarbio Science & Technology Co., Ltd.) at 37°C with 5% CO2.
Cell transfection
Short hairpin RNA (shRNA/sh)-circ_0067934-1/2 and corresponding negative control (NC) shRNA (sh-NC), miR-1301-3p mimic and its NC (mimic-NC), a pcDNA3.1 expression vector containing full-length human KIF23 (Ov-KIF23) and corresponding NCs (Ov-NC) were obtained from Shanghai GenePharma Co., Ltd. The sequence for the miR-1301-3p mimic was 5′-UUGCAGCUGCCUGGGAGUGACUUC-3′. The NC shRNA sequence was 5′-UCACAACCUCCUAGAAAGAGUAGA-3′. The AGS cell suspension was inoculated into a 6-well plate and cultured in an incubator with 5% CO2 at 37°C for 24 h. The cultured cells were transfected with 100 nM of these recombinants using 2.5 µl/ml Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc.) at 37°C for 48 h in accordance with the manufacturer's instructions. After transfection, the transfection efficacy of the cells was detected via reverse transcription-quantitative (RT-q)PCR.
Cell proliferation assay
AGS cells from each group were collected after transfection for 48 h. After being counted, cells (1×103 cells/well) were seeded into a 96-well cell culture plate and cultured in an incubator with 5% CO2 at 37°C. After 24, 48 and 72 h, 10 µl Cell Counting Kit-8 (CCK-8) reagent (Beyotime Institute of Biotechnology) was added to each well in accordance with the manufacturer's instructions. Then, the cells were incubated at 37°C in the dark for 2 h. The absorbance of each well was measured at 450 nm with a microplate reader (Bio-Rad Laboratories, Inc.). Each experiment was repeated ≥3 times. In total, six duplicate wells were examined in each group.
EdU cell proliferation assay
AGS cells were seeded into 6-well plates (500 cells/well) and incubated at room temperature overnight. EdU solution (Beyotime Institute of Biotechnology) was added and the cells were incubated for 4 h at 37°C. After the working fluid was removed, the cells were fixed with 4% paraformaldehyde for 15 min at room temperature. The cells were then permeated with 0.5% Triton X-100 for 15 min. After the addition of Click reaction solution, the cells were incubated in the dark for 30 min at 37°C and imaged under a fluorescence microscope (Leica Microsystems GmbH; magnification, ×200).
Western blotting
AGS cells were harvested and total protein was extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology). Total protein was quantified using a protein concentration determination kit (cat. no. P0012; Beyotime Institute of Biotechnology). Proteins (30 µg/lane) were separated via SDS-PAGE (15%) and were subsequently transferred onto PVDF membranes (MilliporeSigma). After blocking with 5% BSA (Beyotime Institute of Biotechnology) at room temperature for 2 h, the membranes were then incubated with the following primary antibodies (Abcam) at 4°C overnight: Anti-Ki67 (cat. no. ab15580; 1:1,000), anti-proliferating cell nuclear antigen (anti-PCNA; cat. no. ab92552; 1:1,000), anti-MMP2 (cat. no. ab92536; 1:1,000), anti-MMP9 (cat. no. ab76003; 1:1,000) and anti-KIF23 (cat. no. ab174304; 1:1,000). After the incubation with the primary antibodies, the membranes were washed with TBS containing Tween-20 (0.1%) and incubated at room temperature for 1.5 h with HRP-conjugated secondary antibodies [Goat Anti-Mouse IgG(H+L), 1:2,000, cat. no. SA00001-1; Goat Anti-Rabbit IgG(H+L), 1:2,000, cat. no. SA00001-2; ProteinTech Group, Inc.]. Protein bands were visualized using the Odyssey Western Blot Analysis system (LI-COR Biosciences) and semi-quantified using ImageJ software, version 7.6.5 (National Institutes of Health).
Transwell assay
The invasive ability of the cells was examined using a Transwell invasion assay. The Transwell chambers (Costar; Corning, Inc.) were first coated with 0.1 ml Matrigel (Becton-Dickinson and Company) at 37°C for 1 h. AGS cells (5×105 cells/well) were collected and suspended at 2×105 cells/ml in serum-free DMEM. Cell suspensions were placed into the upper compartment of a Transwell chamber and cultured in 5% CO2 at 37°C. Medium supplemented with 10% FBS was added into the lower compartment of the Transwell chamber. After 24 h, the non-invaded cells on the upper face of the Transwell membrane were wiped off with a cotton swab. The cells invading to the lower surface of the filter were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution for 30 min at room temperature. Images were captured using a light microscope at ×100 magnification.
Wound healing assay
AGS cells were cultured in 6-well plates (6×104 cells/well) and incubated to 80–90% confluence in RPMI-1640 medium with 10% FBS at 37°C (17). A linear scratch was created in the cell monolayer using a 200-µl pipette tip. The cells were subsequently cultured under standard conditions in RPMI-1640 medium containing 2% FBS (Procell Life Science & Technology Co., Ltd.) for 48 h at 37°C in 5% CO2. The area occupied by cells migrating into the scratch was evaluated using an inverted microscope at ×100 magnification. The migration rate was calculated based on the formula: (Wound width at 0 h - wound width at 24 h)/wound width at 0 h × 100%. In total, five fields were randomly selected for analysis in each well.
Nuclear and cytoplasmic separation experiment
AGS cells were lysed and centrifuged for 5 min at 500 × g at 4°C. Then, the cytoplasmic components retained in the supernatant were collected. The precipitated nuclear components were further cleaved. The collected cytoplasmic components and nuclear lysates were mixed with 2X lysis/binding solution and then pumped through a filter tube. The extracted cytoplasmic and nuclear RNA were reverse-transcribed, and the expression level of circ_0067934 was detected via RT-qPCR. U6 was the positive control for the detection of nuclear RNA expression, and GAPDH was the positive control for the detection of cytoplasmic RNA expression (18).
Luciferase reporter assay
The circ_0067934 3′-UTR, containing wild-type (WT) or mutant (MUT) target sites for miR-1301-3p, was amplified via PCR and inserted into a pGL3 vector (Promega Corporation) to form the reporter vector circ_0067934-WT or circ_0067934-MUT. Then, the DNA sequence of KIF23 3′-UTR that included the putative binding sites of miR-1301-3p was subcloned into a pGL3 vector (Promega Corporation) to create a luciferase plasmid containing the WT sequence of KIF23 3′-UTR. The miR-1301-3p binding sites were mutated, and the MUT KIF23 3′-UTR sequence was subcloned into the pGL3 vector so as to create a mutant reporter vector. AGS cells were co-transfected with luciferase reporter vectors by using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. At 48 h after transfection, the relative luciferase activities were measured by using a Dual-Luciferase Reporter Assay (Promega Corporation) and luciferase activities were normalized against Renilla luciferase.
RT-qPCR analysis
Total RNA from cell samples was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and reverse-transcribed into cDNA using a commercial RevertAid™ cDNA Synthesis kit (Takara Bio, Inc.) at 42°C for ~1 h and 90°C for 5 min. The SYBR Premix Ex Taq™ II kit (Thermo Fisher Scientific, Inc.) was applied for qPCR. The PCR reaction mixture contained 3 mM MgCl2, 0.5 µM forward and reverse primers, 2 µl SYBR Green PCR master mix and 2 µl cDNA on a QuantStudio 3 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers used are shown in Table I. The thermocycling conditions of the qPCR were as follows: Initial denaturation for 5 min at 95°C, followed by 40 cycles for 30 sec at 95°C, annealing at 65°C for 45 sec and extension at 72°C for 1 min. A final extension step at 72°C for 7 min was performed in each PCR assay. The results were measured using the 2−ΔΔCq method (19). U6 served as an endogenous control for miRNAs and GAPDH was used as an endogenous control for circ_0067934 and KIF23.
Table I.
Sequences of primers for reverse transcription-quantitative PCR.
| Gene | Sequence |
|---|---|
| circ_0067934 | F: 5′-TAGCAGTTCCCCAATCCTTG-3′ |
| R: 5′-CACAAATTCCCATCATTCCC-3′ | |
| miR-5047 | F: 5′-GCCTAGACGAGACACAGTGC-3′ |
| R: 5′-GCCAAGACCTTACAACCGCA-3′ | |
| miR-1301-3p | F: 5′-GCCCGCTTGCAGCTGCCTGGGAG-3′ |
| R: 5′-GTGCAGGGTCCGAGGT-3′ | |
| miR-670-5p | F: 5′-AAACCCATGACACAGCACAA-3′ |
| R: 5′-ATTGCTTTGCCTCCAGAAGA-3′ | |
| miR-345-3p | F: 5′-GCCGAGAGGGGTCTGGAGA-3′ |
| R: 5′-CTCAACTGGTGTCGTGGA-3′ | |
| miR-545-3p | F: 5′-TGGCTCAGTTCAGCAGGAAC-3′ |
| R: 5′-CATTACTGGATCTATCAACAGG-3′ | |
| KIF23 | F: 5′-AGACAGAAGGCGAGGGATG-3′ |
| R: 5′-GGAGACGAATTGGTGGTGC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACATATA-3′ |
| R: 5′-ACGCTTCACGAATTTGAGTGTC-3′ | |
| GAPDH | F: 5′-GTCAAGGCTGAGAACGGGAA-3′ |
| R: 5′-AAATGAGCCCCAGCCTTCTC-3′ |
miR, microRNA; circ, circular RNA; KIF23, kinesin family member 23; F, forward; R, reverse.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, Inc.). All data are presented as the mean ± SD of three independent experiments. Statistical differences between two groups were determined using an unpaired two-tailed Student's t-test, while one-way followed by Tukey's post hoc test was used to analyze data in >2 groups. P<0.05 was considered to indicate a statistically significant difference.
Results
circ_0067934 silencing inhibits AGS cell proliferation
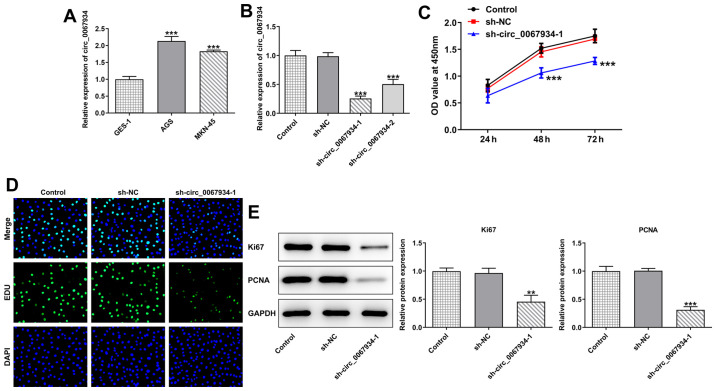
To investigate the role of circ_0067934 in GC cells, the expression level of circ_0067934 in GC cells was firstly detected via RT-qPCR. The results demonstrated that the expression level of circ_0067934 was upregulated in the GC cell lines, AGS and MKN-45, compared with the normal gastric mucosa cells, GES-1 (Fig. 1A). AGS cells showed the highest expression of circ_0067934, so these cells were selected for subsequent experiments.
Figure 1.
Effects of circ_0067934 on proliferation of gastric cancer cells. (A) The expression level of circ_0067934 in gastric cancer cell lines was detected via RT-qPCR. ***P<0.001 vs. GES-1 cells. (B) Transfection efficiency of sh-circ_0067934 was measured via RT-qPCR. (C) Cell proliferation was evaluated using a Cell Counting Kit-8 assay. (D) EdU staining was used to assess cell proliferative capacity. (E) Western blotting was performed to detect the protein expression levels of Ki67 and PCNA. Data are presented as the mean ± SD. Original magnification, ×200. **P<0.01, ***P<0.001 vs. sh-NC. RT-qPCR, reverse transcription-quantitative PCR; circ, circular RNA; sh, short hairpin RNA; NC, negative control; PCNA, proliferating cell nuclear antigen; OD, optical density.
Cells were then transfected with sh-circ_0067934-1/2 to knock down the expression of circ_0067934 (Fig. 1B). Compared with the sh-NC group, the cell proliferation ability of the sh-circ_0067934-1 group was significantly decreased (Fig. 1C and D). In addition, the results of western blotting revealed that the expression levels of Ki67 and PCNA were downregulated following transfection with sh-circ_0067934 (Fig. 1E).
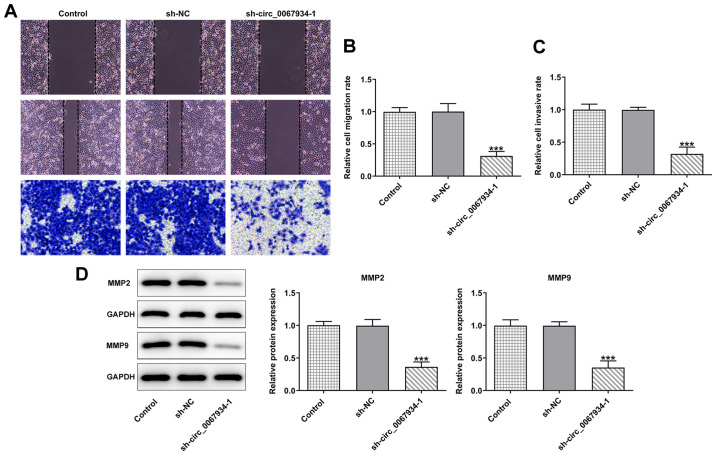
circ_0067934 silencing inhibits AGS cell migration and invasion
As shown in Fig. 2A-C, following transfection with sh-circ_0067934, the migratory and invasive abilities of AGS cells were decreased compared with the control and sh-NC groups. Additionally, western blotting revealed that the expression levels of the migration-related proteins, MMP2 and MMP9, were significantly downregulated compared with those in the control and sh-NC groups (Fig. 2D), indicating that circ_0067934 silencing inhibited AGS cell migration and invasion.
Figure 2.
Effects of circ_0067934 silencing on AGS cell migration and invasion. (A) Transwell and wound healing assay were used to measure (B) cell migration and (C) invasion. Original magnification, ×100. (D) Western blotting was conducted to detect the expression levels of MMP2 and MMP9. Data are expressed as the mean ± SD. ***P<0.001 vs. sh-NC. circ, circular RNA; sh, short hairpin RNA; NC, negative control
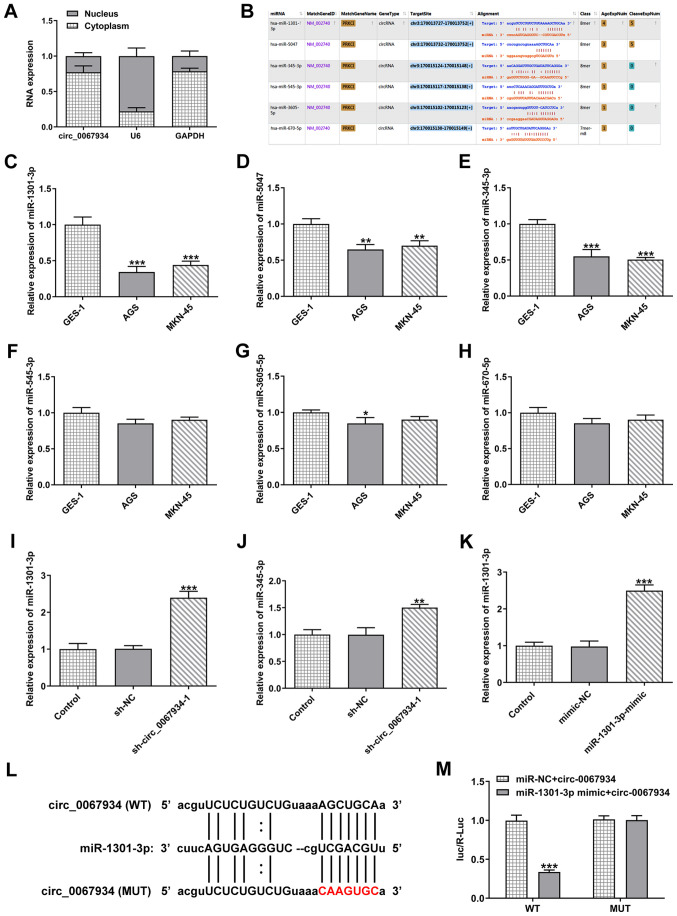
circ_0067934 targets miR-1301-3p
Next, the expression level of circ_0067934 in the cytoplasm and nucleus of GC cells was examined via RT-qPCR. The results demonstrated that circ_0067934 was mainly present in the cytoplasm of AGS cells (Fig. 3A). The binding sequences of circ_0067934 with miR-5047, miR-1301-3p, miR-670-5p, miR-345-3p, miR-545-3p and miR-3605-5p were predicted using the starBase database (Fig. 3B). Moreover, the RT-qPCR results demonstrated that the expression levels of miR-1301-3p and miR-345-3p were significantly decreased in GC cells compared with control cells (Fig. 3C-H). As shown in Fig. 3I and J, transfection with sh-circ_0067934 promoted the expression levels of miR-1301-3p more significantly than miR-345-3p. Thus, miR-1301-3p was selected for the subsequent experiments.
Figure 3.
circ_0067934 targets miR-1301-3p. (A) Expression level of circ_0067934 in the cytoplasm and nucleus of AGS cells was detected via RT-qPCR. (B) The starBase database was used to predict the binding site of circ_0067934 with miR-5047, miR-1301-3p, miR-670-5p, miR-345-3p, miR-545-3p and miR-3605-5p. The expression levels of (C) miR-1301-3p, (D) miR-5047, (E) miR-345-3p, (F) miR-545-3p, (G) miR-3605-5p and (H) miR-670-5p in gastric cancer cell lines was identified via RT-qPCR. The expression levels of (I) miR-1301-3p and (J) miR-345-3p were detected after transfection with sh-circ_0067934. (K) miR-1301-3p mimics increased miR-1301-3p expression in AGS cells. (L) The predictive binding sequence between circ-0067934 and miR-1301-3p. The red letters in the circ-0067934 showed the mutated sites. (M) Luciferase activity was determined using a luciferase reporter assay. Data are expressed as the mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs. GES-1 cells, sh-NC or mimic-NC. RT-qPCR, reverse transcription-quantitative PCR; circ, circular RNA; sh, short hairpin RNA; NC, negative control; miR, microRNA; WT, wild-type; MUT, mutant.
To determine the effect of miR-1301-3p on AGS cells, miR-1301-3p overexpression vector was transfected into AGS cells (Fig. 3K). Furthermore, the luciferase vectors containing the WT and MUT sequences (shown in red letters) of circ-0067934 were constructed (Fig. 3L) and the results indicated that miR-1301-3p was a direct target of circ_0067934 (Fig. 3M).
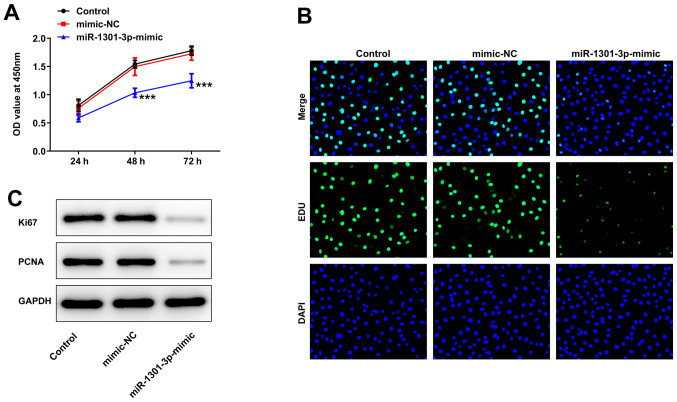
miR-1301-3p overexpression suppresses AGS cell proliferation
As presented in Fig. 4A, the optical density (OD) value of miR-1301-3p mimic-transfected cells was significantly lower compared with that of the mimic NC and control groups. Moreover, EdU staining revealed that transfection with miR-1301-3p mimic markedly inhibited cell proliferation (Fig. 4B). Furthermore, the protein expression levels of Ki67 and PCNA were notably suppressed by miR-1301-3p mimic in comparison with those in the NC and control groups (Fig. 4C).
Figure 4.
Effects of miR-1301-3p on proliferation of AGS cells. (A) Cell proliferation after transfection with miR-1301-3p mimic was measured using a Cell Counting Kit-8 assay. (B) An EdU assay was utilized to calculate cell proliferative capacity. Original magnification, ×200. (C) The protein expression levels of Ki67 and PCNA were estimated using western blotting. Data are expressed as the mean ± SD. ***P<0.001 vs. mimic-NC. NC, negative control; miR, microRNA; PCNA, proliferating cell nuclear antigen; OD, optical density.
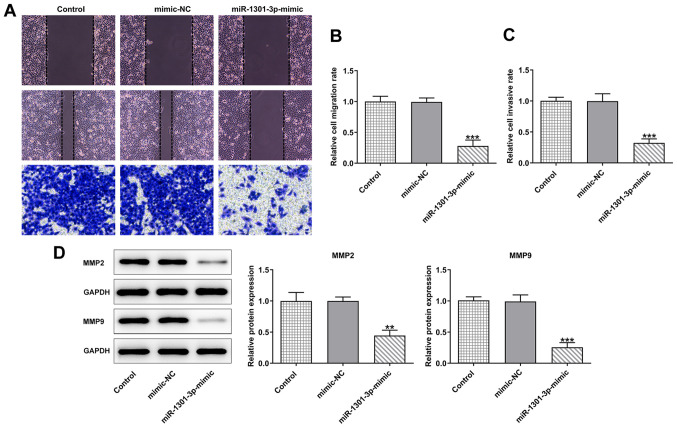
miR-1301-3p mimic represses the migration and invasion of AGS cells
To further investigate the role of miR-1301-3p in AGS cells, wound healing and Transwell assays were conducted to measure cell migration and invasion. As shown in Fig. 5A-C, the cell migratory and invasive abilities were weakened following transfection with miR-1301-3p mimic compared with that in the mimic NC group. In addition, according to the results of the western blotting assay, the expression levels of the migration-related proteins, MMP2 and MMP9, were significantly decreased following transfection with miR-1301-3p mimic compared with the mimic NC or control groups (Fig. 5D).
Figure 5.
Effects of miR-1301-3p mimic on AGS cell migration and invasion. (A) Transwell and wound healing assay were used to measure (B) cell migration and (C) invasion. Original magnification, ×100. (D) Western blotting was performed to detect the protein expression levels of MMP2 and MMP9. Data are expressed as the mean ± SD. **P<0.01, ***P<0.001 vs. mimic-NC. NC, negative control; miR, microRNA.
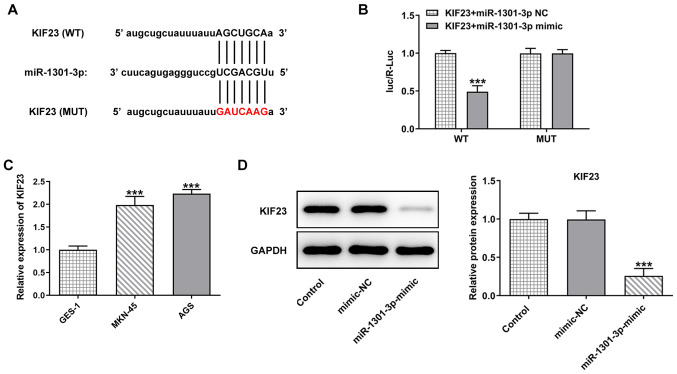
miR-1301-3p targets KIF23
The starBase database predicted the targeted binding of miR-1301-3p to KIF23 (Fig. 6A), and luciferase reporter assay verified the association between miR-1301-3p and KIF23 (Fig. 6B). Moreover, it was found that KIF23 expression was significantly upregulated in GC cells lines compared with the normal gastric mucosa cells (Fig. 6C). The results of western blotting identified that the expression of KIF23 was decreased following transfection with miR-1301-3p mimic compared with the mimic NC group (Fig. 6D). These results indicated that miR-1301-3p targets KIF23 in AGS cells.
Figure 6.
miR-1301-3p targets KIF23. (A) The starBase database was used to predict the binding sequence of miR-1301-3p with KIF23. (B) The luciferase reporter assay predicted the binding of miR-1301-3p to KIF23. (C) The expression level of KIF23 in gastric cancer cell lines was detected via reverse transcription-quantitative PCR. (D) Western blotting was employed to examine the expression level of KIF23 in AGS cells after transfection of miR-1301-3p mimic. Data are expressed as the mean ± SD. ***P<0.001 vs. mimic-NC. NC, negative control; miR, microRNA; KIF23, kinesin family member 23; WT, wild-type; MUT, mutant.
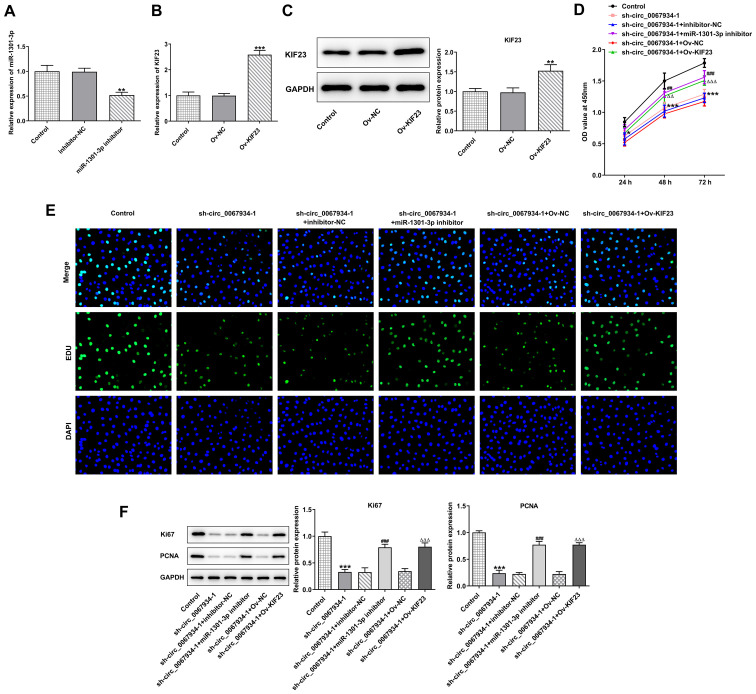
KIF23 overexpression promotes AGS cell proliferation
To determine the role of KIF23 in GC cells, the miR-1301-3p inhibitor and KIF23 overexpression vector were transfected into AGS cells (Fig. 7A-C). The level of cell proliferation was detected using a CCK-8 assay and the results revealed that the OD value was significantly enhanced following transfection with miR-1301-3p inhibitor or Ov-KIF23 in circ_0067934-silenced cells compared with their NC groups (Fig. 7D). In addition, compared with the sh-circ_0067934 + inhibitor-NC group and sh-circ_0067934 + Ov-NC group, miR-1301-3p inhibitor and Ov-KIF23 markedly increased the percentage of living cells (Fig. 7E). Consistently, the western blotting results demonstrated that miR-1301-3p silencing and KIF23 overexpression increased the protein expression levels of Ki67 and PCNA in cells transfected with sh-circ_0067934 (Fig. 7F).
Figure 7.
Effects of KIF23 on proliferation of AGS cells. (A) miR-1301-3p expression was detected via reverse transcription-quantitative PCR in cells transfected with miR-1301-3p inhibitor. KIF23 (B) mRNA and (C) protein expression was measured after transfection with Ov-KIF23. (D) A Cell Counting Kit-8 assay was applied for investigating cell proliferation. (E) Cell proliferative capacity was evaluated using an EdU assay. Original magnification, ×200. (F) Western blotting was conducted to measure the protein expression levels of Ki67 and PCNA. Data are expressed as the mean ± SD. **P<0.01, ***P<0.001 vs. Control or corresponding NCs; ##P<0.01, ###P<0.001 vs. sh-circ_0067934-1 + inhibitor-NC groups; △△P<0.01, △△△P<0.001 vs. sh-circ_0067934-1 + Ov-NC. NC, negative control; miR, microRNA; KIF23, kinesin family member 23; OD, optical density; Ov, overexpression; sh, short hairpin RNA; circ, circular RNA; PCNA, proliferating cell nuclear antigen.
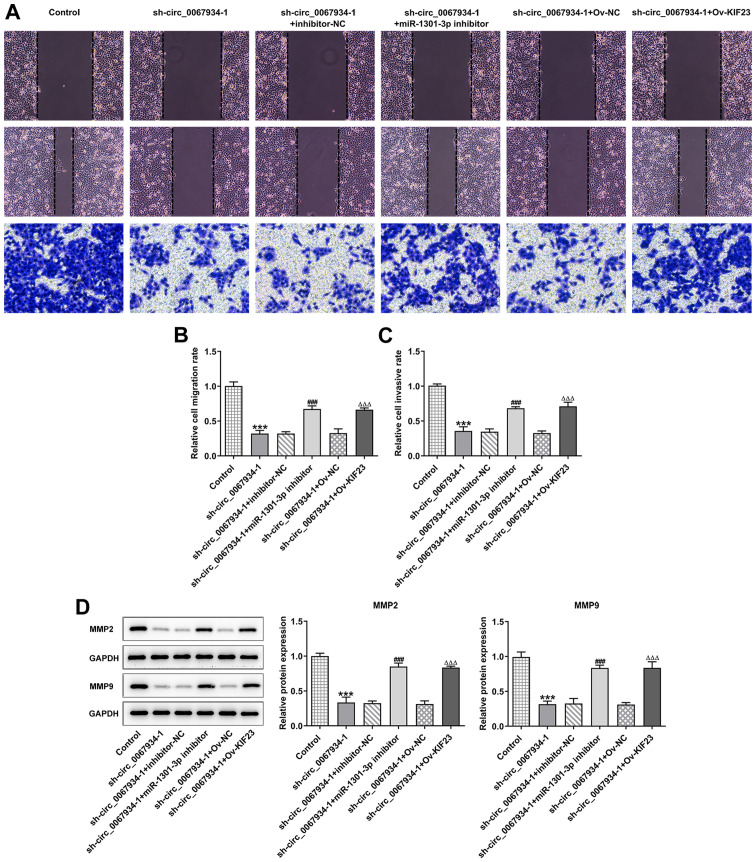
KIF23 overexpression facilitates AGS cell migration and invasion
As shown in Fig. 8A-C, KIF23 overexpression or miR-1301-3p knockdown promoted cell migration and invasion compared with the sh-circ_0067934-1 + Ov-NC and sh-circ_0067934 + inhibitor-NC groups. In addition, the western blotting results demonstrated that the protein expression levels of MMP2 and MMP9 were significantly increased following transfection with miR-1301-3p inhibitor or Ov-KIF23 in circ_0067934-silenced cells compared with their corresponding NC (Fig. 8D).
Figure 8.
Effects of KIF23 on AGS cell migration and invasion. (A) Transwell and wound healing assay were used to measure (B) cell migration and (C) invasion after transfection with Ov-KIF23. Original magnification, ×100. (D) Western blotting was used to measure the protein expression levels of MMP2 and MMP9 after transfection with Ov-KIF23. Data are expressed as the mean ± SD. ***P<0.001 vs. control; ###P<0.001 vs. sh-circ_0067934-1 + inhibitor-NC groups; △△△P<0.001 vs. sh-circ_0067934-1 + Ov-NC. NC, negative control; miR, microRNA; KIF23, kinesin family member 23; OD, optical density; Ov, overexpression; sh, short hairpin RNA; circ, circular RNA.
Discussion
GC is the fourth most common type of cancer, and its fatality rate is the second highest worldwide (20). The prognosis of patients with GC is poor and the 5-year relative survival rate for patients with advanced-stage disease is only 20% (21). Therefore, it is crucial to study the mechanisms underlying the pathogenesis and development of GC, as well as to identify novel therapeutic targets for this type of cancer.
circ_0067934 has been shown to act as an oncogene in a variety of cancer types (22–24). Although it has been reported that the expression of circ_0067934 is increased in GC tissues (13), its biological effect on GC cells is yet to be elucidated. In the present study, the effect of circ_0067934 on the proliferation, migration and invasion of GC cells was investigated by silencing circ_0067934. It was recently revealed that circRNAs may serve as key regulators in the development of human diseases, including cancer (25). It was also reported that downregulation of circ_0067934 decreased the proliferative ability of NSCLC cells and, thus, it may serve as a predictive marker for the prognosis of NSCLC and a target for the treatment of this disease (26).
In the present study, following transfection with sh-circ_0067934, CCK-8 assays and EdU staining were used to evaluate cell proliferation, and western blotting was used to detect the protein expression levels of Ki67 and PCNA, which are known to be closely associated with cell proliferation (27). Ki67 is a common cell proliferation status marker, which has high sensitivity in determining the proliferative activity of tumor cells and is associated with the occurrence, development, proliferation and metastasis of tumors (28). PCNA is synthesized in and localizes to the nucleus, and it is closely involved in the proliferation and invasion of tumor cells. PCNA has also been found to participate in the synthesis and metabolism of RNA and DNA in tumor cells, and it is closely associated with the differentiation and invasion of tumor cells, as well as with tumor metastasis, recurrence and prognosis (29). The current results demonstrated that circ_0067934 expression was higher in GC cells, and the expression levels of Ki67 and PCNA were downregulated after silencing of circ_0067934, which was consistent with previous findings (16,22,30). The wound healing assay demonstrated that the knockdown of circ_0067934 could reduce the migratory ability of GC cells, and the expression levels of the migration-related proteins, MMP2 and MMP9, were also downregulated. MMP2 and MMP9 are closely associated with cell migration (31). Overexpression of MMP2 and MMP9 can promote the degradation of the extracellular matrix and improve the cell migratory ability (32,33).
Recently, circRNAs have been considered to be members of the competing endogenous RNA family due to their abundant conserved miRNA response elements (34). Some studies have shown that miR-1301-3p was associated with a variety of cancer types, including prostate (35), breast (36) and colon cancer (37). According to data from the starBase database, the current study predicted that circ_0067934 targeted miR-1301-3p. The present study identified that the expression level of miR-1301-3p was significantly increased after circ_0067934 was silenced, while cell proliferation and migration were decreased.
KIF23 is a kinesin-like motor protein that serves crucial roles in cytokinesis (38). Previous studies have reported the oncogenic roles of KIF23 in several types of cancer, including GC (39–41). In the present study, the starBase database also predicted that miR-1301-3p could bind to KIF23, and when KIF23 was overexpressed, the cell proliferation, migratory and invasive abilities were enhanced. However, the current study primarily focused on the role of circ_0067934 and the mechanism involved in circ_0067934/miR-1301-3p/KIF23 in GC cells, and thus, did not investigate the regulation network of circ_0067934/miR-1301-3p/KIF23 in normal gastric mucosal cells. Future studies will further investigate the relationship of the three genes in normal gastric mucosal cells. In addition, due to the lack of samples from patients with GC, this study did not include clinical sample testing. Moreover, only one GC cell line, AGS, was used to examine the role of circ_0067934, miR-1301-3p and KIF23, and so, this study cannot determine the correlation of these circRNA, miRNA and KIF23 gene expressions in GC cells or patient samples.
In conclusion, the present study provided evidence that circ_0067934 can inhibit the proliferation, migration and invasion of GC cells via the miR-1301-3p/KIF23 axis, which may provide a theoretical and factual basis for the targeted therapy of GC.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JX, NS and JZ designed the experiments and made considerable contributions to the manuscript writing. JX, NS, WH, NZ and XL performed the experiments and analyzed the data. JX and NS revised the manuscript and guided the experiments. JX and NS confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017 Jul 3; doi: 10.1177/1010428317714626. (Epub ahead of print). doi: 10.1177/1010428317714626. [DOI] [PubMed] [Google Scholar]
- 2.Li JH, Zhang SW, Liu J, Shao MZ, Chen L. Review of clinical investigation on recurrence of gastric cancer following curative resection. Chin Med J (Engl) 2012;125:1479–1495. [PubMed] [Google Scholar]
- 3.Suzuki H, Mori H. Gastric cancer after Helicobacter pylori eradication. Gan To Kagaku Ryoho. 2018;45:1123–1127. (In Japanese) [PubMed] [Google Scholar]
- 4.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Zhang W, Nian X, Lu X, Li Y, Liu F, Wang F, He B, Zhao L, Zhu Y, et al. The previously uncharacterized lncRNA APP promotes prostate cancer progression by acting as a competing endogenous RNA. Int J Cancer. 2020;146:475–486. doi: 10.1002/ijc.32422. [DOI] [PubMed] [Google Scholar]
- 6.Khan S, Ayub H, Khan T, Wahid F. MicroRNA biogenesis, gene silencing mechanisms and role in breast, ovarian and prostate cancer. Biochimie. 2019;167:12–24. doi: 10.1016/j.biochi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, Tai S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M, Wang G, Tian W, Deng Y, Xu Y. MiRNA-based therapeutics for lung cancer. Curr Pharm Des. 2018;23:5989–5996. doi: 10.2174/1381612823666170714151715. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, Cui J. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503:2429–2435. doi: 10.1016/j.bbrc.2018.06.172. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao W, Quan J, Fan X. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10:900. doi: 10.1038/s41419-019-2089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B, Wang S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ding HX, Xu Q, Wang BG, Lv Z, Yuan Y. MetaDE-cased analysis of circRNA expression profiles involved in gastric cancer. Dig Dis Sci. 2020;65:2884–2895. doi: 10.1007/s10620-019-06014-6. [DOI] [PubMed] [Google Scholar]
- 14.Wang JM, Li XJ, Wang J. Circular RNA circ_0067934 functions as an oncogene in breast cancer by targeting Mcl-1. Eur Rev Med Pharmacol Sci. 2019;23:9499–9505. doi: 10.26355/eurrev_201911_19444. [DOI] [PubMed] [Google Scholar]
- 15.Zou Q, Wang T, Li B, Li G, Zhang L, Wang B, Sun S. Overexpression of circ-0067934 is associated with increased cellular proliferation and the prognosis of non-small cell lung cancer. Oncol Lett. 2018;16:5551–5556. doi: 10.3892/ol.2018.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Zhou Q, Zhong P. circ_0067934 increases bladder cancer cell proliferation, migration and invasion through suppressing miR-1304 expression and increasing Myc expression levels. Exp Ther Med. 2020;19:3751–3759. doi: 10.3892/etm.2020.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Liu T, Wang X, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46:2586–2594. doi: 10.3892/ijo.2015.2976. [DOI] [PubMed] [Google Scholar]
- 18.Yu F, Pillman KA, Neilsen CT, Toubia J, Lawrence DM, Tsykin A, Gantier MP, Callen DF, Goodall GJ, Bracken CP. Naturally existing isoforms of miR-222 have distinct functions. Nucleic Acids Res. 2017;45:11371–11385. doi: 10.1093/nar/gkx788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GL, Xia XL, Li XL, He FH, Li JL. Identification and expression analysis of the MSP130-related-2 gene from Hyriopsis cumingii. Genet Mol Res. 2015;14:4903–4913. doi: 10.4238/2015.May.11.23. [DOI] [PubMed] [Google Scholar]
- 20.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 21.Sitarz R, Skierucha M, Mielko J, Offerhaus GJA, Maciejewski R, Polkowski WP. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag Res. 2018;10:239–248. doi: 10.2147/CMAR.S149619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C, Wang Y, Li A, Zhang J, Xue F, Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2019;234:9225–9232. doi: 10.1002/jcp.27601. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D, Ni Y. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. 2018;497:626–632. doi: 10.1016/j.bbrc.2018.02.119. [DOI] [PubMed] [Google Scholar]
- 24.Cui XL, Wang XD, Lin SK, Miao CM, Wu M, Wei JG. Circular RNA circ_0067934 functions as an oncogene in glioma by targeting CSF1. Eur Rev Med Pharmacol Sci. 2019;23:8449–8455. doi: 10.26355/eurrev_201910_19157. [DOI] [PubMed] [Google Scholar]
- 25.Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060. doi: 10.26355/eurrev_201805_15063. [DOI] [PubMed] [Google Scholar]
- 27.Juríková M, Danihel L, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118:544–552. doi: 10.1016/j.acthis.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Stevanovic L, Choschzick M, Moskovszky L, Varga Z. Variability of predictive markers (hormone receptors, Her2, Ki67) and intrinsic subtypes of breast cancer in four consecutive years 2015-2018. J Cancer Res Clin Oncol. 2019;145:2983–2994. doi: 10.1007/s00432-019-03057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehm EM, Gildenberg MS, Washington MT. The many roles of PCNA in eukaryotic DNA replication. Enzymes. 2016;39:231–254. doi: 10.1016/bs.enz.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao M, Ma W, Ma C. Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed Pharmacother. 2020;129:110461. doi: 10.1016/j.biopha.2020.110461. [DOI] [PubMed] [Google Scholar]
- 31.Simbulan-Rosenthal CM, Dougherty R, Vakili S, Ferraro AM, Kuo LW, Alobaidi R, Aljehane L, Gaur A, Sykora P, Glasgow E, et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers (Basel) 2019;11:E1490. doi: 10.3390/cancers11101490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui F, Wang S, Lao I, Zhou C, Kong H, Bayaxi N, Li J, Chen Q, Zhu T, Zhu H. miR-375 inhibits the invasion and metastasis of colorectal cancer via targeting SP1 and regulating EMT-associated genes. Oncol Rep. 2016;36:487–493. doi: 10.3892/or.2016.4834. [DOI] [PubMed] [Google Scholar]
- 33.Park GB, Chung YH, Kim D. Induction of galectin-1 by TLR-dependent PI3K activation enhances epithelial-mesenchymal transition of metastatic ovarian cancer cells. Oncol Rep. 2017;37:3137–3145. doi: 10.3892/or.2017.5533. [DOI] [PubMed] [Google Scholar]
- 34.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song XL, Huang B, Zhou BW, Wang C, Liao ZW, Yu Y, Zhao SC. miR-1301-3p promotes prostate cancer stem cell expansion by targeting SFRP1 and GSK3β. Biomed Pharmacother. 2018;99:369–374. doi: 10.1016/j.biopha.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 36.Peng X, Yan B, Shen Y. MiR-1301-3p inhibits human breast cancer cell proliferation by regulating cell cycle progression and apoptosis through directly targeting ICT1. Breast Cancer. 2018;25:742–752. doi: 10.1007/s12282-018-0881-5. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Zhao Y, Xu M, Zhou F, Yan J. Serum miR-1301-3p, miR-335-5p, miR-28-5p, and their target B7-H3 may serve as novel biomarkers for colorectal cancer. J BUON. 2019;24:1120–1127. [PubMed] [Google Scholar]
- 38.Sun X, Jin Z, Song X, Wang J, Li Y, Qian X, zhang Y, Yin Y. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer. 2015;15:961. doi: 10.1186/s12885-015-1987-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Chen H, Dong P, Xie G, Zhou Y, Ma Y, Yuan X, Yang J, Han L, Chen L, et al. KIF23 activated Wnt/β-catenin signaling pathway through direct interaction with Amer1 in gastric cancer. Aging (Albany NY) 2020;12:8372–8396. doi: 10.18632/aging.103146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato T, Wada H, Patel P, Hu HP, Lee D, Ujiie H, Hirohashi K, Nakajima T, Sato M, Kaji M, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer. 2016;92:53–61. doi: 10.1016/j.lungcan.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 41.Li XL, Ji YM, Song R, Li XN, Guo LS. KIF23 promotes gastric cancer by stimulating cell proliferation. Dis Markers. 2019;2019:9751923. doi: 10.1155/2019/9751923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.