Abstract
As the understanding of cancer grows, new therapies have been proposed to improve the well-known limitations of current therapies, whose efficiency relies mostly on early detection, surgery and chemotherapy. Mesenchymal stem cells (MSCs) have been introduced as a promissory and effective therapy. This fact is due to several useful features of MSCs, such as their accessibility and easy culture and expansion in vitro, and their remarkable ability for ‘homing’ towards tumors, allowing MSCs to exert their anticancer effects directly into tumors. Additionally, MSCs offer the practicability of being genetically engineered to carry anticancer genes, increasing their specificity and efficacy for fighting tumors. In the present study, the antitumoral efficacy and post-implant survival of mice bearing lymphomas implanted intratumorally were determined using mouse bone marrow-derived (BM)-MSCs transduced with soluble TRAIL (sTRAIL), full length TRAIL (flTRAIL), or interferon β (IFNβ), naïve BM-MSCs, or combinations of these. The percentage of surviving mice was determined once all not-implanted mice succumbed. It was found that the percentage of surviving mice implanted with the combination of MSCs-sTRAIL and MSCs-IFN-β was 62.5%. Lymphoma model achieved 100% fatality in the non-treated group by day 41. On the other hand, the percentage of surviving mice implanted with MSCs-sTRAIL was 50% and with MSCs-INFβ 25%. All the aforementioned differences were statistically significant (P<0.05). In conclusion, all implants exhibited tumor size reduction, growth delay, or apparent tumor clearance. MSCs proved to be effective anti-lymphoma agents; additionally, the combination of soluble TRAIL and IFN-β resulted in the most effective antitumor and life enlarging treatment, showing an additive antitumoral effect compared with individual treatments.
Keywords: mesenchymal stem cells, TNFα-related apoptosis-inducing ligand, interferon β, lymphoma, immunocompetent, lentivirus, cancer treatment
Introduction
Cancer is one of the most lethal diseases. Conventional treatment consists of surgery, chemotherapy and radiotherapy. This therapeutic scheme, based on 5FU and oxaliplatin is useful only when cancer is detected early, but it lacks specificity and is not practical in the presence of metastases (1). Moreover, this therapy has remained mostly unchanged for decades. Besides, all the aforementioned treatments, and particularly radiotherapy and chemotherapy, produce severe harmful effects (2). Recently, new approaches that focus on cancer mechanisms to increase specificity in cancer cell elimination efficacy and safety are being intensively analyzed. Some of these new therapies introduce the use of nanoparticles, monoclonal antibodies, miRNAs, gene and cell therapy with the use of cells as delivery vectors for antitumoral proteins (3–10). All promise to be more specific, efficacious and safer than conventional treatments.
In the present study, engineered mesenchymal stem cells (MSCs) were used as antitumoral therapy, considering that MSCs gather several desirable features (11) as anticancer weapons, such as their abundance in tissues, particularly in bone marrow (BM) (12) and adipose tissue (13). Besides, these cells have the outstanding ability for ‘homing’ to the tumor microenvironment. Homing allows MSCs to exert their anticancer effects directly towards tumors (14). Nevertheless, a serious limitation for using MSCs as anticancer therapy is that it is hard to predict the naïve MSCs behavior in the tumor, as these cells, under the influence of the tumor microenvironment, can improve tumor growth (15–17). Thus, an alternative is needed that increases the anticancer potency of MSCs and reduces the risk of favoring cancer progression instead of fighting it. Achieving this goal is possible since MSCs offer the practicability of being genetically engineered to overexpress coded proteins and find and attack malignant cells. Therefore, engineering MSCs can ensure that these cells specifically attack malignant cells and overcome the naïve MSCs efficacy for fighting tumors. To transduce murine BM-MSCs, the murine soluble fraction of the TNFα-related apoptosis-inducing ligand (sTRAIL) and interferon β (INFβ) was chosen, considering the following features of each transgene: sTRAIL is a powerful anticancer protein extensively used due to the selective apoptotic effect that this protein induces in malignant cells. TRAIL targets the death receptors (DR)4 and DR5, which activate apoptosis by a caspase-8 derived process; the aforementioned receptors are mainly overexpressed by malignant cells (18,19). sTRAIL exerts its function once it has been trimerized and bound with cell receptors. However, Wong et al (19) pointed out that full-length (fl)TRAIL can induce apoptosis more efficiently than soluble (s)TRAIL. Interferon beta (IFNβ) is a type I member of the interferon family with pleiotropic roles, including immunomodulator, antiproliferative and cancer-inhibitory activity (20). IFNβ has already been examined and reported as an effective gene therapy agent (21); however, it was hypothesized in the present study that the combination of both proteins could increase the reach of monotherapy efficiency. To achieve this goal, a murine syngeneic model of lymphoma was used, with an immunocompetent strain (BALB/c) and BM-MSCs freshly obtained from mice belonging to the same BALB/c strain. The purpose of the present study was to determine the percentage of surviving mice and lymphoma reduction in mice implanted with MSCs carrying sTRAIL or IFNβ transgenes and their combinations.
Materials and methods
Mice
BALB/c mice were born, grown and maintained in the Laboratory of Animal Experimentation of the Autonomous University of Nuevo Leon. Male mice aged 4 to 8 weeks were used to isolate BM-MSCs. A total of 72 female BALB/c mice (12 weeks old) weighing 19–22 g were used to induce solid lymphomas and evaluate the in vivo anti-lymphoma efficacy. From mice birth and throughout the experiments until the last mouse was euthanized, all animals were kept in individually ventilated cages at 25°C with sterilized air with EPA filters (UREAtac; LAB&Bio), with 50% humidity, ≤500 ppm of CO2, ≤15 ppm of NH3 and a light/dark cycle of 12/12 h. Access to food (Labdiet 5001; LabSupply) and purified sterile water was provided ad libitum.
Lymphoma cells
The thymic lymphoma cell line L5178Y was used to produce lymphomas; the clone that was used (3.7.2C) was derived from the L5178YS cell line. In vitro, these cells grow in suspension; in vivo, when inoculated intramuscularly, they produce solid lymphomas, and when injected intraperitoneally, they grow abundantly and produce ascites. The L5178Y cell line was purchased from the American Type Culture Collection (cat. no. CRL-9518™; ATCC®) and cultured as instructed by the manufacturer. The culture medium was Dulbecco's modified Eagles medium (DMEM) high glucose medium (4.5 g/l), 0.1% pluronic, 100 µg gentamycin/ml, 2.5 µg amphotericin B/ml, and 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.).
Lentiviral vectors
Lentiviral vectors were produced and provided by Cyagen Biosciences, Inc. as a gene delivery tool, named as follows: pLV[Exp]-EGFP/Neo-EF1A>{sMurTRAIL}, pLV[Exp]-EGFP/Neo-EF1A>mIfnb1[NM_010510.1], pLV[Exp]-EGFP/Neo-EF1A>{MurIL12-70p}. The lentiviral vectors were of 3rd generation and also included an integration cassette for G-418 (geneticin; Gibco; Thermo Fisher Scientific, Inc.) resistance for cell selection and a green fluorescent protein (GFP) gene to evaluate transgene integration.
MSC isolation and cell culture
BM-MSCs were obtained and characterized as previously described (22). Briefly, six mice were placed inside a CO2 chamber to sacrifice them (70% vol/min); immediately after, their femurs and tibias were dissected under aseptic conditions. Their epiphysis was removed, and using a hypodermic needle (27 gauge), their BM was obtained by perfusing the shafts inner barrel with 500-1,000 µl DMEM/Nutrient Mixture F-12 (DMEM F-12) medium supplemented (s) with 10% FBS, 100 µg gentamicin/ml, and 2.5 µg amphotericin B/ml (Gibco; Thermo Fisher Scientific, Inc.) and poured into T-25 cell culture flasks (Corning, Inc.). The first cell passage was accomplished as follows: The aforementioned preparations were incubated for 24 h at 37°C in a 5% CO2 humid atmosphere. The expended medium and the non-adherent cells were discarded. The culture flasks with the adherent cells were replenished with 4 ml fresh medium and incubated as before until the cell monolayer reached 80% confluence (7–10 days). The second passage was performed by discarding the spent medium and incubating the cell monolayer with a solution of 0.25% trypsin and 0.1% EDTA (Gibco; Thermo Fisher Scientific, Inc.) in sterile phosphate-buffered saline (PBS), washing trice with PBS and inoculating 5×105 cells in a new culture flask with fresh 5 ml of supplemented DMEM F-12 and incubating it as before. The procedure described for the second passage was repeated in each of the following passages.
MSC characterization
The characterization of MSCs and the assays performed with them were accomplished with cell cultures from passage 3 or 4. MSCs were characterized by immunocytochemistry and their multipotency was evaluated (Fig. 1). Cells were fixed with methanol-acetone (1:1) at 4°C per 10 min. Regarding immunocytochemistry, CD90 and CD105 surface markers were investigated using specific monoclonal antibodies included in the mouse and rabbit specific HRP/DAB (ABC) detection IHC kit (cat. no. ab64264; Abcam). Antibodies were diluted in PBS to 1:200 (CD90) and 1:25 (CD105). MSC multipotency was examined by inducing the cells to differentiate to osteoblasts or chondroblasts, using the appropriate reagents in the Mouse Mesenchymal Stem Cell Functional Identification kit (cat. no. SC010; R&D Systems, Inc.) and following the manufacturer's instructions. The osteoblasts were identified by staining with the Von Kossa technique and chondroblasts with Alcian blue. Cells were fixed with 4% paraformaldehyde for 20 min at ambient temperature. In Von Kossa staining, slides were submerged in saturated lithium carbonate solution for 20 min, and then washed with water and submerged in a 5% silver nitrate solution. Slides were exposed to bright light for 60 min and then were submerged in 5% sodium thiosulfate solution for 5 min. Finally, the slides were counterstained with neutral red. In Alcian blue staining, the colorant was added at ambient temperature for 10 min, and then the slides were washed with water and counterstained with neutral red at ambient temperature for 2 min.
Figure 1.
BM-MSCs characterization. (A) BM-MSCs were adherent to the culture flask and presented elongated morphology (scale bar, 100 µm). They also expressed high mesenchymal markers, such as (B) CD90 and (C) CD105 (scale bar, 100 µm). In addition, they could differentiate to chondrogenic by (D) Alcian blue staining and calcified areas generated by osteogenic cells with (E) Von Kossa stain. BM-MSCs, bone marrow mesenchymal stem cells.
Construct design
All transgene constructs contained murine sequences of the genes of interest, a promotor cassette for Elongation Factor 1-Alpha (EF1-α) to allow a constitutive, high protein expression, and the Kozak consensus sequence (GCC GCC ACC) (23). The constructs used in the present study were designed with Vector Builder software (Cyagen Biosciences, Inc.). The design of the sTRAIL transgene is presented in Fig. 2A. This design contained the sTRAIL nucleotide sequence codifying the amino acid fragment 118 to 291; an isoleucine zipper sequence to facilitate in situ TRAIL's trimerization (24,25), a furin specific cleavage site (CGCACCAAACGC), a signal sequence (SS) derived from Gaussia sp. (a copepod) luciferase (TGG GAG TCAAAGTTCTGTTTGCCCTGATCTGCATCGCTG-TGGCCGAGGCC) to conduct protein secretion (26) and Kozak's sequence. As revealed in Fig. 2B, besides EF1-α and Kozak's sequence, the construct used to perform the transduction of BM-MSCs with the flTRAIL transgene contained a nucleotide sequence coding its 1–291 amino acids. The entire flTRAIL was retracted from GeneBank (accession number U37522, URL: http://www.ncbi.nlm.nih.gov/nuccore/U37522). Regarding IFNβ, the murine IFNβ gene sequence was retracted from GeneBank (accession number NM_010510, URL: http://www.ncbi.nlm.nih.gov/nuccore/NM_010510). As demonstrated in Fig. 2C, the scheme of the IFNβ transgene design was as follows: The complete IFNβ nucleotide sequence was included and added with Kozak's sequence.
Figure 2.
Lentiviral transduction of BM-MSC. (A-C) Lentiviral vectors were designed to contain (A) sTRAIL, (B) flTRAIL and (C) IFNβ. (D) Representative fluorescent images of BM-MSCs transduced with lentiviral vectors; nucleus stained with DAPI (blue) and transduced BM-MSCs expressing GFP protein (green signal). (E and F) Western blot analysis confirming protein expression of (E) flTRAIL and sTRAIL (F) and IFNβ in untransfected MSCs and MSCs transduced with a lentivirus that does not contain any of the genes of interest (empty lentiviral vector pLV[Exp]-EGFP:T2A:Puro-EF1A> mCherry, Vector Builder). MSCs express each of the genes flTRAIL, sTRAIL and IFN and as positive control the recombinant protein. Actin was used as a loading control. BM-MSCs, bone marrow mesenchymal stem cells; sTRAIL, TRAIL soluble; flTRAIL, TRAIL full length; IFNβ, interferon β.
Transduction of murine BM-MSC
BM-MSCs (5×105) were seeded into T-25 culture flasks (Corning, Inc.) containing 5 ml DMEM F-12 supplemented with 10% FBS and incubated overnight under standard conditions, 50% confluency. Then, protamine sulfate (Sigma-Aldrich), 5 µg/ml were added to the BM-MSC cultures. Each lentivirus preparation carrying sTRAIL, flTRAIL, or INFβ transgenes was added to separate BM-MSC cultures. The multiplicity of infection (MOI) employed was two viral particles/one BM-MSC (27). The BM-MSCs infected with the lentivirus vectors were incubated overnight, to allow viral infection. Then, spent media was discarded and replenished with fresh DMEM-F12 and incubated 48 h before fluorescence analysis. Lentivirus integration was validated with fluorescence emitted by GFP, which was visualized with a fluorescence microscope (Eclipse 10i; Nikon Corporation) and a representative field photographed with a Sight DS-2MV (Nikon Corporation) digital camera. The selection of transduced BM-MSCs was accomplished by adding 200 µg of geneticin/ml to the culture medium. Transduction efficiency was calculated by applying the following equation: % Transduction efficiency=BM-MSC-GFP/total BM-MSC ×100. Where BM-MSC-GFP are transduced cells that exhibit a green fluorescent signal under the epifluorescence microscope. Total BM-MSC is the total number of cells detected by counting nuclei by fluorescence with DAPI (blue signal). Mounting medium Vectashield with DAPI (7 µl) per slide was applied (Vecta Mount; Vector Laboratories, Inc.).
Protein extraction
Gene expression was validated by western blot analysis. BM-MSCs (5×105) transduced with sTRAIL, flTRAIL or INFβ were placed, separately, into T-75 flasks (Corning, Inc.), containing 10 ml of DMEM-F12 and incubated at 37°C with 5% CO2 for 48 h and allowing 70% confluency. The spent medium was discarded, and then the cell culture flasks were replenished with 10 ml fresh supplemented media and incubated at 37°C with 5% CO2 for 48 h. For soluble proteins (sTRAIL and IFNβ), the spent medium was centrifuged at 4,000 × g for 5 min, concentrated 40 times with an Amicon filtration unit of 10 kDa MW cut-off (Merck KGaA). FlTRAIL is a transmembrane protein that was extracted and purified. FlTRAIL-BM-MSCs were cultured as aforementioned with the sTRAIL-BM-MSCs or IFNβ-BM-MSCs until flTRAIL-BM-MSCs culture reached 80% confluency. Then, flTRAIL-BM-MSCs were harvested and washed twice with PBS. Subsequently, 1×106 cells were resuspended in 50 µl of lysis buffer [Triton X-100 (50 ml), Tris-HCl 1 M pH 7.5 (5 ml), KCl 2 M (12.5 ml), MgCl2 1 M (1 ml), DTT (1 µl, Invitrogen; Thermo Fisher Scientific, Inc.) and a protease inhibitor cocktail (5 µl; Sigma-Aldrich; Merck KGaA)]. The mixture was adjusted at 500 µl with ultrapure sterile water, chilled in ice for 20 min, centrifuged at 16,000 × g at 4°C for 5 min, and the supernatant was collected.
Western blot analysis
The aforementioned protein concentrates were quantified with Bradford reagent (Bio-Rad Laboratories, Inc.). After, 40 µg protein concentrate (5 µl) was submitted to 2% sodium dodecyl sulfate (SDS), 4% stacking and 12% resolving polyacrylamide gel electrophoresis (PAGE) at 100 V for 120–150 min and the protein bands were electro-transferred to a 0.45-µm mesh polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc.) at 100 V for 60 min. Protein was Electro-transfer to a PVDF membrane. The membrane was blocked for 1 h with 5% skimmed milk in Tris-buffered saline with Tween-20 (TBST) at 4°C. The protein bands of sTRAIL, flTRAIL, or IFNβ were identified with rabbit monoclonal antibodies anti-murine TRAIL (1:200; cat. no. ab10516; Abcam) or 1:500 rabbit polyclonal anti-murine IFNβ (cat. no. ab85803; Abcam), as primary antibodies, incubated overnight at 4°C. The secondary antibody was an HRP-conjugated anti-rabbit polyclonal antibody (1:10,000; cat. no. W4011; Promega Corporation), incubated for 2.5 h at 4°C. Dilutions of all antibodies were performed with Tris-buffered saline (TBS buffer) in 5% skimmed milk.
The marked protein bands were revealed with the luminol kit Clarity Western ECL blotting substrate (Bio-Rad Laboratories, Inc.) following the manufacturer's protocol and analyzed with the Molecular Imager Chemidoc XRS+ Imaging System (Bio-Rad Laboratories, Inc.) equipped with the Image Lab software.
Commercial recombinant proteins were used as positive controls in all western blot assays; soluble TRAIL recombinant protein (cat. no. 315-19; PeproTech, Inc.) which consists in the extracellular fraction of murine TRAIL, weighing 20 kDa; ~200 ng of recombinant protein was loaded into the gel well. IFNβ standard (cat. no. IF011; Merck KGaA) consisted of recombinant murine IFNβ weighing 22 kDa; ~2 kU was loaded into the gel well.
Induction of lymphomas
L5178Y cells (5×105) were inoculated in the right gastrocnemius with a 27-gauge syringe. The cells were suspended in 100 µl of saline solution for a total of 72 female BALB/c mice. The mice exhibited mild discomfort immediately after inoculation; however, no sign of discomfort was observed overnight. The implanted leg of each mouse was measured with a Vernier calibrator to calculate the gastrocnemius volume (mm3), using the following equation: Vol=π/6 (LxWxh); where Vol is the volume of the leg, expressed in mm3, π is a radius constant (3.1416), L is the leg's length, measured from the ankle to the knee. W is the width of the thickest portion of the leg, and h is the height, measuring from the rear leg section (gastrocnemius) to the front (tibialis anterior). These measurements were performed every third day throughout the experiment. When legs carrying lymphomas showed an increment in the leg volume of 1–2 mm3, mice were implanted with the transduced BM-MSCs.
Implantation of transduced BM-MSCs
Mice with lymphomas were divided into nine groups of eight mice each. Transduced BM-MSCs were implanted to one of these groups (G) as follows: G1, BM-MSCs-sTRAIL; G2, BM-MSCs-IFNβ plus-flTRAIL; G3, BM-MSCs-IFNβ; G4, flTRAIL; G5, sTRAIL plus IFNβ, G6, not transduced or naïve BM-MSC plus-IFNβ plus-flTRAIL; G7, naïve BM-MSC; G8, mice not implanted with BM-MSCs but injected with isotonic saline; G9, mice not implanted with BM-MSCs. In addition, the experiment was repeated with groups G5, sTRAIL plus IFNβ, and G6, not transduced BM-MSC to confirm results. However, mice were reduced to five animals per group with the same characteristics. In all implanted mice, 1×106 viable BM-MSCs, suspended in 100 µl isotonic sterile saline, were distributed into the entire volume of each tumor. The number of BM-MSCs transduced separately with two transgenes or two transgenes plus not transduced MSCs were inoculated with an appropriate proportion of BM-MSCs to sum a total of 1×106 cells. For example, G5 mice were implanted with 5×105 BM-MSCs-sTRAIL and 5×105-INFβ and mice from G6 were implanted with 3.33×105 not transduced BM-MSC, plus 3.33×105 BM-MSCs IFNβ plus 3.33×105 flTRAIL. All inoculations were performed in aseptic conditions with a new, sealed sterile 27-gauge needle for each mouse. Before and throughout the experiments, mice behavior, their ability to eat and drink, and pain indicators were monitored. When a mouse showed intense suffering as previously described (28), these signs included bent down low ears, orbital tightening, bristly hair, sluggishness, or a lack of response to external stimuli. When all signs were detected, the mouse was euthanized. These animals were considered deceased due to their induced lymphoma. Euthanasia was practiced by cervical dislocation. Tumor volume and mouse weight measuring were performed thrice a week, as at the beginning of the experiment.
Tumor biopsies of ~5 mm3 were dissected from recently deceased animals. The samples from each lymphoma were preserved with Tissue-Tek optimum cutting temperature (O.C.T.) (Sakura Finetek Europe B.V.), freezing the blocks with liquid nitrogen and stored at −80°C.
Histopathological analysis
Frozen samples were cut with a cryostat (model 2230; Shenyang Roundfin Trade Co., Ltd.), stained with hematoxylin and eosin, and observed with a clear field using a light microscope (model DM IL LED; Leica Microsystems GmbH) at 100–400 amplification diameters. Images were captured with a camera (DFC295; Leica Microsystems GmbH) attached to the microscope.
Analysis of results
The number of deceased mice from all groups was monitored daily. When the last not-implanted mouse with BM-MSCs carrying lymphomas succumbed, the percentage of surviving mice from each group and the number of days elapsed since the lymphoma cells were inoculated were registered.
Statistical analysis
The survival rate was analyzed with a Kaplan-Meier estimator with a Mantel-Cox test to assess statistical significance with SPSS software (SPSS Statistics for Windows; version 22.0; IBM Corp.). Statistical analysis was conducted using GraphPad Prism version 7.00 for Windows (GraphPad Software, Inc.). Differences between the analyzed groups were determined with the t-test and one-way ANOVA (followed by Tukey's post hoc test). P<0.05 was considered to indicate a statistically significant difference.
Ethical considerations
The present study was approved (approval no. BI15-005) by the Scientific Research and Bioethics Committee and the Institutional Animal Care Committees of the School of Medicine of the Autonomous University of Nuevo Leon (Monterrey, Mexico). The mice were handled according to the Mexican Standard NOM-062-ZOO-1999. The mice were kept in the animal facility of the Department of Immunology in individually ventilated cages (Lab & Bio) that maintain filtered air flow and a barrier that protects them from contact with external agents. The system records pressure, oxygen, CO2, humidity, temperature, and ammonia concentration. Furthermore, this system maintains all these parameters in uniform conditions. The cages were cleaned every 2–3 days by the technical personnel of the vivarium. The model was euthanized if the tumor exceeded 10% of the normal mouse body weight according to the international manual Institutional Animal Care and Use Committee (2002) and would be euthanized by inhalation of CO2 with subsequent cervical dislocation and monitoring of vital signs. They were also euthanized if they showed signs of severe pain according to the Rodent Management Laboratory Manual accepted by the American Association for the Accreditation of Animal Care Laboratories (29): decreased intake of water and food, weight loss (>20%), bristly coat, dry skin, lacerations, posture, abnormal gait, head tilt, lethargy, bloating, diarrhea, seizures, discharge from body orifices and dyspnea. The experimental groups were kept under close care from the day of inoculation until the last mouse in the control group succumbed. The mice succumbed due to tumor growth. However, the mice in the experimental groups that survived until the last mouse in the untreated group were sacrificed on the same day that the last untreated mouse lived.
Results
BM-MSCs express TRAIL and IFNβ
BM-MSC isolated from mice presented fibroblast morphology in culture and were characterized by expression of CD90 and CD105 superficial markers that presented >95% positive cell staining (Fig. 1A-C). In addition, BM-MSC differentiated to chondroblasts and osteoblasts, showing positivity to Alcian blue and Von Kossa stains, respectively (Fig. 1D and E). A representative field of transduced BM-MSCs is shown in Fig. 2D. All cells fluoresced intensely in green due to expression of the GFP gene and geneticin selection, denoting an efficient integration of each gene construct. The percentage of transduction efficiency of BM-MSC-sTRAIL and -flTRAIL was 90% and for BM-MSC-IFNβ was 95%. For protein expression, western blot results are shown in Fig. 2E and F. The protein bands, whose images are demonstrated in Fig. 2E, correspond to sTRAIL and an sTRAIL standard, respectively, with both weighing ~20 kDa (control). The image of flTRAIL, revealing a ladder of five bands; the smallest one weighs ~20 kDa, which matches the 2 soluble TRAIL standard control; however, 4 heavier bands of ~30, ~39, ~45, and ~60 kDa were also revealed. In Fig. 2F, two IFNβ bands are demonstrated, one weighing ~22 kDa and a slightly heavier ~24 kDa band. Alongside as control, the recombinant IFNβ standard shows a single band weighing ~22 kDa.
BM-MSC expressing sTRAIL and IFNβ reduce solid lymphoma engraftment
On day 7 post-inoculation (PI), a slight inflammation was observed in all inoculated subjects. PI leg volumes on day 9, comparing the non-inoculated and inoculated groups are revealed in Fig. 3; considering one standard deviation as a reference, it displays the totality of the study subjects with tumor engraftment.
Figure 3.

Leg tumor volumes at day 9. Figure shows differences between leg volumes of inoculated and not inoculated mice. *P<0.001 vs. W/O inoculum.
First, deceased mice from the untreated group were registered by day 26 PI; therefore, the last tumor measure with full subjects' groups was on day 23 PI. Fig. 4A highlights the most relevant results: the untreated group shows the largest tumor (average 2.5 cm3) and first deceased mouse at day 26 PI; on the other hand, the group with combined treatment of IFNβ and sTRAIL showed the smallest tumor (1.2 cm3) by day 23, and the first mouse succumbed by day 29 PI. Notably, separated treatments of both sTRAIL and IFNβ also showed considerably smaller tumors (compared with the untreated group) but slightly larger than the combined one (1.3 and 1.5 cm3, respectively).
Figure 4.
Murine tumor model. (A) Tumor mass showed differences between groups, with the ones treated with combination of sTRAIL and IFNβ exhibiting the smallest volumes. (B) Mice weight remained unaffected during the experiment. sTRAIL, TRAIL soluble; flTRAIL, TRAIL full length; IFNβ, interferon β.
BM-MSC expressing sTRAIL, flTRAIL and IFNβ treatments do not affect mice weight
During the study, mice weights were documented. Any mouse that exhibited a loss of ≥20% of the baseline weight was withdrawn from the study and euthanized. The weight of all groups during the study is revealed in Fig. 4B; mice showed stable weight by day 19 PI; however, important weight variability was registered thereafter. All mice were carefully observed to detect a disability to eat or drink; however, despite massive tumors, mice were eating and drinking; thus, weight variability was associated with tumor growth. All subjects gained weight according to the developed tumor mass.
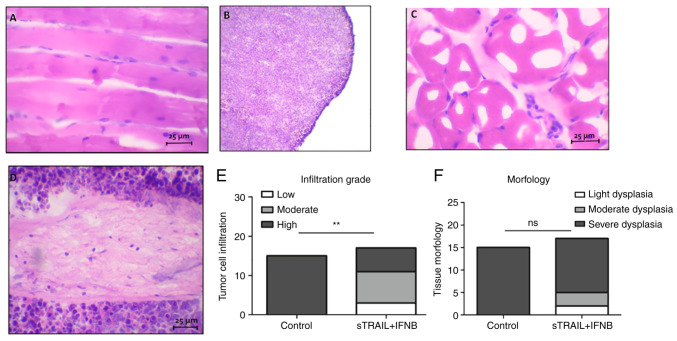
In addition, tumor tissues were analyzed with a light microscope. A section of the tumor of untreated mice was compared with tissue of the inoculated leg of surviving mice. Images of histological sections are revealed in Fig. 5. It was demonstrated how the tumor tissue almost completely displaced muscle tissue (Fig. 5B and D), exhibiting basophilic predominance of lymphocyte (tumoral cell) nuclei, which in contrast to a surviving mouse, most of the tissue was acidophilic with skeletal muscle tissue (Fig. 5A and C). It was also observed that a few nuclei dispersed over the muscle tissue, indicating an active immune reaction. The statistical analysis of the infiltration rate and morphology observed in treated tissues is revealed in Fig. 5E and F. Chi-square showed differences between the control and the treatment group (P<0.001); however, in morphology, no statistical differences were identified.
Figure 5.
Tumor histological sections. (A and C) Tumors treated with sTRAIL and IFNβ. (B and D) Tumors without treatment. (E and F) Semiquantitative analysis according to (E) cellular infiltration grade and (F) morphology in treated tissues and control. Scale bar, 25 µm. **P<0.001. sTRAIL, TRAIL soluble; IFNβ, interferon β.
BM-MSC expressing sTRAIL and IFNβ increase survival of mice with solid lymphoma engraftment
The last untreated mouse died at day 41 PI; thus, a cut-off point was established. All groups comparing each treated group against the untreated group were analyzed. In Fig. 6A, a Kaplan-Meier estimator with the most relevant results is demonstrated: 100% of untreated mice succumbed due to the tumor by day 41 PI. On the other hand, the combined treatment of IFNβ and sTRAIL showed a 62.5% survival rate. Additionally, the separated groups of IFNβ and sTRAIL showed 25 and 50% survival, respectively, in addition to initial tumor mass, which showed a similar result, suggesting an additive effect. The log-rank test showed significant differences between the groups (P<0.05). All P-values are included in Table I.
Figure 6.

Kaplan-Meier curves. (A) First model with all treatments. (B) Second model with sTRAIL and IFNβ treatment and MSC naïve. *P<0.05 and **P<0.001 compared with the untreated group. sTRAIL, TRAIL soluble; IFNβ, interferon β; MSC, mesenchymal stem cell.
Table I.
Survival significant differences between treated and non-treated groups.
| Treatment | P-value |
|---|---|
| sTRAIL | 0.017 |
| flTRAIL + IFNβ | 0.068 |
| IFNβ | 0.170 |
| flTRAIL | 0.072 |
| sTRAIL + IFNβ | 0.008 |
| MSC naïve + IFNβ + flTRAIL | 0.042 |
| MSC naïve | 0.038 |
| Saline solution | 0.309 |
MSC, mesenchymal stem cell; sTRAIL, TRAIL soluble; flTRAIL, TRAIL full length; IFNβ, interferon β.
To confirm these results, the combination treatment of BM-MSC expressing sTRAIL and IFNβ was repeated. The results are presented in Fig. 6B. The control treatment of naïve BM-MSCs was added. At first, both treatments reduced tumor growth in the right gastrocnemius; however, near day 20, mice of the groups treated with naïve BM-MSCs presented an increase in tumor volume. On day 36 PI, the last mouse of the naïve BM-MSC group was euthanized, and 40% of survivors remained in the group treated with sTRAIL plus IFNβ. The log-rank test showed significant differences between the groups (P<0.001). Mice were kept monitored until day 50; nevertheless, mice did not present visually solid tumors in the right gastrocnemius.
All treatments showed anti-tumorigenic activity to some extent. Independent treatments showed poorer antitumor activity than combined ones. IFNβ exhibited the lowest therapeutic activity with a 25% survival rate, sTRAIL showed 37.5%. Combined treatment showed 37.5% survival rate, indicating a non-additive effect since it remained nearly the same as when treatment was performed with sTRAIL alone. Notably, saline solution had a 25% survival rate. A combined treatment of MSC naïve, IFNβ and flTRAIL showed a 50% survival rate, indicating a slight improvement over flTRAIL and IFNβ alone, suggesting MSC antitumoral effect.
Discussion
Cancer xenograft in vivo murine models have been a frequent approach in numerous research groups; indeed, the chimeric models of human tissue in immunodeficient mice have allowed achievement of tremendous advances and testing several cancer models and treatments (30). The current landscape deems murine models as the cornerstone in biomedical research (31).
MSC approaches in murine cancer models involve the following criteria: first, a cancer model; that is, a tumor-inducing cell line in vivo of certain tissue; second, determination of the source of the MSCs, as there are significant phenotypic and genotypic differences among the cells in terms of regenerative capacity, multipotency, isolation yield, and overall behavior within the tumor; third, one or more anticancer transgenes to be expressed and delivered; and finally, fourth, a mouse strain according to the aims of the study (32). CD105, CD90 and CD73 are main markers highly expressed by BM-MSCs (33). In the present study, CD105 and CD90 were only evaluated. CD73 expression on MSCs was reported heterogeneity from different sources as adipose tissue, BM, amniotic membrane (34). However, previous research has shown that CD73 has the power to promote cell proliferation, tumor angiogenesis, and has the ability to make tumor cells escape immunological recognition (35), so its evaluation would be very interesting for future studies.
MSCs have been reported to possess homing ability led by chemokines and growth factors from the tumor site. Moreover, this therapeutic effect depends on the capacity of reaching the tumor site, by migration, expression of adhesion molecules, and engraftment in the tumor microenvironment. It has been previously reported that MSCs decrease the incidence of lymphoma tumors and improve survival of C57BL/6 mice when intravenously administered (36). In addition, intraperitoneal administration of MSCs in a disseminated non-Hodgkin's lymphoma murine model increases the overall survival of treated mice (37). Both studies demonstrated MSC homing to tumoral cells. Nevertheless, there is a lack of enough lymphoma studies that determine the complete mechanisms of antitumoral activity of MSC, which opens an important field to research. In the present study, a murine, syngeneic, immunocompetent, lymphoma model treated with BM-MSCs overexpressing TRAIL (soluble fraction or full-length protein) and IFNβ by intratumoral inoculation was employed to elucidate if unmodified MSC would exert a pro-tumoral or anti-tumoral effect and if such an effect would be reverted or enhanced, respectively by the transgenes alone or combined.
The tumor microenvironment is determinant in tumor progression as T and B lymphocytes are strongly influenced by it and vice versa, affecting other cells. T-cells are the most important line of defense regarding cancer development; nonetheless, tumors can produce TGF-β, which favors T cell conversion to regulatory T-cell, which exhibits protumor effects by immunosuppression (38).
B-cell subsets have demonstrated cooperative or opposite roles within the tumors. Immunosuppressive regulatory B cells or tumor-infiltrating lymphocytes have protumor effects as they tend to be up to 25% of cells within the tumor mass, can produce lymphotoxin, an angiogenesis-inducing protein (39), and secrete antibodies against tumor-specific antigens (such as p53) (40) with variable outcomes. On the other hand, there are also effector B-cells with antitumor effects by antigen presentation, CD4 T-cell activation, and macrophage conversion to antitumoral M1 phenotype (41). In aims for an improved understanding of cancer development and treatments, in our opinion, a homogenous, syngeneic model provides substantial advantages.
To the best of our knowledge, there are no reports of lymphoma immunocompetent models with combined TRAIL (soluble fraction and complete) and IFNβ treatments. Soluble TRAIL required several accessories to be secreted extracellularly to exert its intended function: an isoleucine zipper to facilitate ligand trimerization (25), a signal sequence-derived Gaussia princeps luciferase (26) to conduct protein extraction, and a furin cleavage site to eliminate all upstream accessory sequences (24); all transgenes proved to be recognizable as they were easily detected by western blotting. FlTRAIL showed 5 bands (weighing ~20, ~30, ~39, ~45 and ~60 kDa), which differs from a 24–25 kDa expected weight; however, similar reports suggested a glycosylated version of the protein (27) or possible oligomerization of the ligand. Additionally, a ~20 kDa band suggests the expression of soluble TRAIL as well. IFNβ showed 2 bands (~22 and ~24 kDa), similar to flTRAIL; the smaller band matched the recombinant standard, and the observed ~24 kDa band is probably a glycosylated version of the protein.
TRAIL has been examined in numerous studies and clinical trials due to its unique property of targeting transformed cells, but its short half-life and toxicity make it impractical; thus, the need for a dose-based therapy by cell delivery is plausible. Soluble TRAIL fraction is normally secreted by the cell to procure DR4+ or DR5+ cells more easily than the complete protein; thus, flTRAIL is anchored to the cell membrane. Therefore, it requires a markedly closer cell-cell approach to trigger apoptosis. Notably, Yuan et al (27) demonstrated that flTRAIL exhibited a more efficient cell-killing effect in TRAIL sensitive and resistant cell lines in vitro. Conversely, in the present in vivo study, sTRAIL resulted in the best single-gene treatment in tumor-reducing effects (with an average volume of 1.4 cm3 leg-tumor size as opposed to flTRAIL with 1.5 cm3) and survivability (50% compared with 37.5% for flTRAIL). This finding could be due to inherent differences between in vitro and in vivo models and protein presentation, as multimers of flTRAIL are more likely as they are suspended in a controlled environment. MSCs administered intravenously or in another form of distant administration require a large number of cells due to loss and death on the way to the tumor site (4,11); the majority get trapped in capillaries and in the lungs owing to their large size and expression of adhesion molecules (2). Despite this, the ones that arrive at the tumor site could develop activity, which in turn is enhanced through genetic modification to express cytokines and pro-apoptotic proteins (11–13). By contrast, intratumoral-inoculated cells are significantly more restricted, relying only on cells close to the tumor parenchyma puncture site; additionally, the soluble TRAIL construct was designed to be secreted abundantly and perfused more efficiently through the tumor. Furthermore, to reduce variation during the administration of the MSCs in each of the experimental groups, the inoculation of cells was performed less than 1 h after cell counting further avoiding any reduction in viability.
IFNβ has been intensively studied in several models (21,42,43) with abundant evidence of its anticancer activity, namely, immune system stimulation, angiogenesis, and malignant cell inhibition using S-phase accumulation (44–46). In the present study, IFNβ treatment dealt relatively poor tumor-reducing activity while administered alone, as it exhibited only 25% of survival rate. Notably, combined treatment with sTRAIL showed a 62.5% survival rate, which surpasses both single-gene treatments (sTRAIL and IFNβ), suggesting an additive effect.
IFNβ was unexpectedly ineffective. Interferons type I have direct effects on T-cells; for instance, IFNβ-based treatment is the first choice to treat multiple sclerosis by modulation of T cell activity (47); therefore, malignant T cells are inherently influenced by it. However, the specific nature of such effects is unclear as current information is notably paradoxical due to the complex effect IFNβ exerts on T cells (48). To elucidate this subject goes beyond the aim of the present study. On the other hand, IFNβ can induce apoptosis by upregulation and enhanced sensitivity of endogenous TRAIL and upregulation of TRAIL-R1 and TRAIL-R2 in cancer cell lines; moreover, there is evidence that IFNβ induces a lengthened S-phase which also potencies TRAIL's apoptosis-inducing effect in nasopharyngeal (49), and cervical carcinomas (50).
It is well known that more than caspase cascade activation, TRAIL can also modulate NF-κB signaling (51,52), which is controversial as NF-κB can upregulate the expression of survival factors such as members of the apoptosis family inhibitor and Bcl-xL (53). Conversely, TRAIL-dependent NF-κB activation triggers apoptosis rather than survival in epithelial cell lines and T cells (54,55). In addition, TRAIL sensitivity can be upregulated by IFNs by overexpression of the TRAIL-R2 DR5 (56–58). IFNβ induces TRAIL-R2 expression in ME-180 cells in the S phase (50), and in melanoma and breast cancer cells, IFNβ induces apoptosis by an extrinsic pathway dependent on TRAIL expression. TRAIL overexpression is a Stat1 dependent process associated with activation of the TRAIL promoter (59).
Related to the MSC treatment, first, 50% survival was observed, but in a second experiment, recurrence and tumor growth after day 26 was identified. This finding showed that MSCs present immunoregulatory properties, as they can inhibit T-cell proliferation and migratory properties and are resistant to natural killer cell-mediated cytolysis. Those attributes contribute to the application of MSC-based delivery of therapeutic genes to solid tumors (60). However, MSC could improve cancer development by increasing the expression of immunosuppressive cytokines such as IL-10 when activated with signals from the tumor microenvironment (22,61,62). In addition, it was observed that the saline solution (placebo) group showed a 25% survival rate, which was reasonably expected considering all mice were immunocompetent. Mice reached immune maturity, which could be explained as a local immunological reaction due to intramuscular solution inoculation that resulted in mild bleeding and muscle inflammation, acting as a local adjuvant.
By day 41 PI, the surviving mice reduced the tumor to the point of being imperceptive. The histological analysis of the untreated mice showed almost no proper muscle tissue but instead was completely saturated with lymphoid cells; by contrast, treated mice showed seemingly healthy muscle tissue, with histologically normal muscle fibers and few scattered lymphoid cells. This finding bifurcates into two rather simplistic yet actual possibilities: i) The main component of the tumor microenvironment is tumor infiltrating lymphocytes of different phenotypes as helper and cytotoxic T cells; these cells could be normal, healthy lymphoid cells due to the immune surveillance induced by the malignant cells, which in this scenario, are seemingly depleted; or ii) Remaining transformed lymphoma cells within the muscle tissue represent an ongoing immunological process rather than a terminated one (63,64). Both scenarios could be elucidated by finer cell characterization, a longer-term observation period to detect relapse, and larger subject groups. In addition, the combined treatment of sTRAIL and IFNβ did recover the tissue morphology mainly due to extended tumor damage. Thus, the mechanism of tissue repair after cancer elimination needs improvement.
In conclusion, the present findings demonstrated that MSCs are effective anti-lymphoma vehicles, as non-treated mice developed massive tumor masses and succumbed. Moreover, in this model, sTRAIL had improved tumor-reducing effects, and IFNβ enhanced it.
Acknowledgements
The authors would like to thank Dr Sergio Lozano (Autonomous University of Nuevo Leon, Monterrey, Mexico) for reviewing the manuscript.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AGQR and CAGV contributed to designing, performing experiments, analyis and discussion of results. HMR helped to design of experiments, wrote, analyzed and corrected the manuscript. SSF designed the experiments, performed literature analysis and discussion of results. MCSC and AYLF helped in the management and in the care with murine model. ASD helped in the standardization of immunohistochemistry and with the software of the fluorescence microscope. GPR helped with statistical analysis. RMDOL contributed to designing of transgenes of lentivirus. JFI edited the text, analyzed the results and reviewed the final manuscript. ENGT contributed to the design of experiments, analysis of the results, discussion and correction of the manuscript. AGQR, CAGV and ENGT confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved (approval no. BI15-005) by the Scientific Research and Bioethics Committee and the Institutional Animal Care Committees of the School of Medicine of the Autonomous University of Nuevo Leon (Monterrey, México).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Urrutiocoechea A, Alemany R, Balart J, Villanueva A, Viñals F, Capellá G. Recent advances in cancer: An Overview. Curr Pharm Des. 2010;16:3–10. doi: 10.2174/138161210789941847. [DOI] [PubMed] [Google Scholar]
- 2.Herskovic A, Martz K, Al-Sarraf M, Leichman L, Brindle J, Vaitkevicius V, Cooper J, Byhardt R, Davis L, Emami B. Combined chemotherapy and radiotherapy compared with radiotherapy alone in patients with cancer of the esophagus. N Engl J Med. 1992;326:1593–1598. doi: 10.1056/NEJM199206113262403. [DOI] [PubMed] [Google Scholar]
- 3.Singh P, Pandit S, Mokkapati VRSS, Garg A, Ravikumar V, Mijakovic I. Gold nanoparticles in diagnostics and therapeutics for human cancer. Int J Mol Sci. 2018;19:1979. doi: 10.3390/ijms19071979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baetke SC, Lammers T, Kiessling F. Applications of nanoparticles for diagnosis and therapy of cancer. Br J Radiol. 2015;88:20150207. doi: 10.1259/bjr.20150207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–221. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;176:139–148. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loebinger M, Sage E, Davies D, Janes S. TRAIL-expressing mesenchymal stem cells kill the putative cancer stem cell population. Br J Cancer. 2010;103:1692–1697. doi: 10.1038/sj.bjc.6605952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Hu J, Li Y, Cao W, Wang Y, Ma Z, Li F. Mesenchymal stem cells expressing interleukin-18 inhibit breast cancer in a mouse model. Oncol Lett. 2018;15:6265–6274. doi: 10.3892/ol.2018.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matosevic S. Viral and nonviral engineering of natural killer cells as emerging adoptive cancer immunotherapies. J Immunol Res. 2018;2018:4054815. doi: 10.1155/2018/4054815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedenstein AJ, Gorskaja JF, Kulagina NN. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol. 1976;4:267–274. [PubMed] [Google Scholar]
- 12.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 13.Igura K, Zhang X, Takahashi K, Mitsuru A, Yamaguchi S, Takahashi TA. Isolation and characterization of mesenchymal progenitor cells from chorionic villi of human placenta. Cytotherapy. 2004;6:543–553. doi: 10.1080/14653240410005366-1. [DOI] [PubMed] [Google Scholar]
- 14.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: The devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 16.Shinagawa K, Kitadai Y, Tanaka M, Sumida T, Kodama M, Higashi Y, Tanaka S, Yasui W, Chayama K. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int J Cancer. 2010;127:2323–2333. doi: 10.1002/ijc.25440. [DOI] [PubMed] [Google Scholar]
- 17.Yu JM, Jun ES, Bae YC, Jung JS. Mesenchymal stem cells derived from human adipose tissues favor tumor cell growth in vivo. Stem Cells Dev. 2008;17:463–473. doi: 10.1089/scd.2007.0181. [DOI] [PubMed] [Google Scholar]
- 18.Pan G, O'Rourke K, Chinnaiyan A, Gentz R, Ebner R, Ni J, Dixit V. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 19.Wong SHM, Kong WY, Fang CM, Loh HS, Chuah LH, Abdullah S, Ngai SC. The TRAIL to cancer therapy: Hindrances and potential solutions. Crit Rev Oncol Hematol. 2019;143:81–94. doi: 10.1016/j.critrevonc.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Nicolini A, Carpi A, Rossi G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006;17:325–337. doi: 10.1016/j.cytogfr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Studeny M, Marini FC, Champlin RE, Zompetta C, Fidler IJ, Andreeff M. Bone marrow-derived mesenchymal stem cells as vehicles for interferon-β delivery into tumors. Cancer Res. 2002;62:3603–3608. [PubMed] [Google Scholar]
- 22.Gonzalez-Villarreal C, Rodriguez M, Said-Fernández S. How desirable and undesirable features of naïve or genetically reengineered mesenchymal stem cells are being considered in preclinical or clinical assays. J BUON. 2017;22:812–830. [PubMed] [Google Scholar]
- 23.Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:3–6. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MH, Billiar TR, Seol DW. The secretable form of trimeric TRAIL, a potent inducer of apoptosis. Biochem Biophys Res Commun. 2004;321:930–935. doi: 10.1016/j.bbrc.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 25.Yan C, Li S, Li Z, Peng H, Yuan X, Jiang L, Zhang Y, Fan D, Hu X, Yang M, Xiong D. Human umbilical cord mesenchymal stem cells as vehicles of CD20-specific TRAIL fusion protein delivery: A double-target therapy against non-Hodgkin's lymphoma. Mol Pharm. 2013;10:142–151. doi: 10.1021/mp300261e. [DOI] [PubMed] [Google Scholar]
- 26.Stern B, Olsen LC, Tröße C, Ravneberg H, Pryme IF. Improving mammalian cell factories: The selection of signal peptide has a major impact on recombinant protein synthesis and secretion in mammalian cells. Trends Cell Mol Biol. 2007;10:19–24. [Google Scholar]
- 27.Yuan Z, Kolluri KK, Sage EK, Gowers KH, Janes SM. Mesenchymal stromal cell delivery of full-length tumor necrosis factor-related apoptosis-inducing ligand is superior to soluble type for cancer therapy. Cytotherapy. 2015;17:885–896. doi: 10.1016/j.jcyt.2015.03.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langford DJ, Bailey AL, Chanda ML, Clarke SE, Drummond TE, Echols S, Glick S, Ingrao J, Klassen-Ross T, Lacroix-Fralish ML, et al. Coding of facial expressions of pain in the laboratory mouse. Nat Methods. 2010;7:447–449. doi: 10.1038/nmeth.1455. [DOI] [PubMed] [Google Scholar]
- 29.Committee on Rodents, Institute of Laboratory Animal Resources, Commission on Life Sciences and the National Research Council, corp-author. Laboratory Animal Managent Rodents. National Academies Press; Washington, D.C., USA: 1996. p. 99. [Google Scholar]
- 30.Bankert RB, Egilmez NK, Hess SD. Human-SCID mouse chimeric models for the evaluation of anti-cancer therapies. Trends Immunol. 2001;22:386–393. doi: 10.1016/S1471-4906(01)01943-3. [DOI] [PubMed] [Google Scholar]
- 31.Haouzi P. Murine models in critical care research. Crit Care Med. 2011;39:2290–2293. doi: 10.1097/CCM.0b013e3182227550. [DOI] [PubMed] [Google Scholar]
- 32.Frese K, Tuveson D. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 33.Boxall SA, Jones E. Markers for characterization of bone marrow multipotential stromal cells. Stem Cells Int. 2012;2012:975871. doi: 10.1155/2012/975871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darzi S, Werkmeister JA, Deane JA, Gargett CE. Identification and characterization of human endometrial mesenchymal stem/stromal cells and their potential for cellular therapy. Stem Cells Transl Med. 2016;5:1127–1132. doi: 10.5966/sctm.2015-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Hou H, Li M, Yu X, Zuo H, Gao J, Zhang M, Li Z, Guo Z. CD73+ Mesenchymal stem cells ameliorate myocardial infarction by promoting angiogenesis. Front Cell Dev Biol. 2021;9:637239. doi: 10.3389/fcell.2021.637239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song N, Gao L, Qiu H, Huang C, Cheng H, Zhou H, Lv S, Chen L, Wang J. Mouse bone marrow-derived mesenchymal stem cells inhibit leukemia/lymphoma cell proliferation in vitro and in a mouse model of allogeneic bone marrow transplant. Int J Mol Med. 2015;36:139–149. doi: 10.3892/ijmm.2015.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Secchiero P, Zorzet S, Tripodo C, Corallini F, Melloni E, Caruso L, Bosco R, Ingrao S, Zavan B, Zauli G. Human bone marrow mesenchymal stem cells display anti-cancer activity in SCID mice bearing disseminated non-hodgkin's lymphoma xenografts. PLoS One. 2010;5:e11140. doi: 10.1371/journal.pone.0011140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu E, Chia PZC, Chen W. TGFβ in T cell biology and tumor immunity: Angel or devil? Cytokine Growth Factor Rev. 2014;25:423–435. doi: 10.1016/j.cytogfr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuen GJ, Demissie E, Pillai S, Burnet M. B lymphocytes and cancer: A love-hate relationship. Trends Cancer. 2017;2:747–757. doi: 10.1016/j.trecan.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: A review. Cancer Res. 2000;60:1777–1788. [PubMed] [Google Scholar]
- 41.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;20:294–307. doi: 10.1038/s41577-019-0257-x. [DOI] [PubMed] [Google Scholar]
- 42.Lei H, Furlong PJ, Jin HR, Mullins D, Cantor R, Fraker DL, Spitz FR. AKT activation and response to interferon-β in human cancer cells. Cancer Biol Ther. 2005;4:709–715. doi: 10.4161/cbt.4.7.1767. [DOI] [PubMed] [Google Scholar]
- 43.Ren C, Kumar S, Chanda D, Kallman L, Chen J, Mountz JD, Ponnazhagan S. Cancer gene therapy using mesenchymal stem cells expressing interferon-β in a mouse prostate cancer lung metastasis model. Gene Ther. 2008;15:1446–1453. doi: 10.1038/gt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Z, Greene G, Pettaway C, Dinney CP, Eue I, Lu W, Bucana CD, Balbay MD, Bielenberg D, Fidler IJ. Suppression of angiogenesis, tumorigenicity, and metastasis by human prostate cancer cells engineered to produce interferon-beta. Cancer Res. 1999;59:872–879. [PubMed] [Google Scholar]
- 45.Pestka S, Langer JA. Interferons and their actions. Ann Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 46.Qin X, Runkel L, Deck C, Dedios C, Barsoum J. Interferon-beta induces S phase accumulation selectively in human transformed cells. J Interf Cytokine Res. 1997;17:355–367. doi: 10.1089/jir.1997.17.355. [DOI] [PubMed] [Google Scholar]
- 47.Kavrochorianou N, Markogiannaki M, Haralambous S. IFN-β differentially regulates the function of T cell subsets in MS and EAE. Cytokine Growth Factor Rev. 2016;30:47–54. doi: 10.1016/j.cytogfr.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Teige I, Liu Y, Issazadeh-Navikas S. IFN-β Inhibits T cell activation capacity of central nervous system APCs. J Immunol. 2006;177:3542–3553. doi: 10.4049/jimmunol.177.6.3542. [DOI] [PubMed] [Google Scholar]
- 49.Makowska A, Wahab L, Braunschweig T, Kapetanakis NI, Vokuhl C, Denecke B, Shen L, Busson P, Kontny U. Interferon beta induces apoptosis in nasopharyngeal carcinoma cells via the TRAIL-signaling pathway. Oncotarget. 2018;9:14228–14250. doi: 10.18632/oncotarget.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vannucchi S, Chiantore MV, Fiorucci G, Percario ZA, Leone S, Affabris E, Romeo G. TRAIL is a key target in S-phase slowing-dependent apoptosis induced by interferon-β in cervical carcinoma cells. Oncogene. 2005;24:2536–2546. doi: 10.1038/sj.onc.1208403. [DOI] [PubMed] [Google Scholar]
- 51.Yang J, LeBlanc FR, Dighe SA, Hamele CE, Olson TL, Feith DJ, Loughran TP. TRAIL mediates and sustains constitutive NF-kB activation in LGL leukemia. Blood. 2018;131:2803–2815. doi: 10.1182/blood-2017-09-808816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mühlenbeck F, Schneider P, Bodmer JL, Schwenzer R, Hauser A, Schubert G, Scheurich P, Moosmayer D, Tschopp J, Wajant H. The tumor necrosis factor-related apoptosis-inducing ligand receptors TRAIL-R1 and TRAIL-R2 have distinct cross-linking requirements for initiation of apoptosis and are non-redundant in JNK activation. J Biol Chem. 2000;275:32208–32213. doi: 10.1074/jbc.M000482200. [DOI] [PubMed] [Google Scholar]
- 53.Wang CY, Guttridge DC, Mayo MW, Baldwin AS. NF-κB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19:5923–5929. doi: 10.1128/MCB.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baetu TM, Kwon H, Sharma S, Grandvaux N, Hiscott J. Disruption of NF-kappaB signaling reveals a novel role for NF-kappaB in the regulation of TNF-related apoptosis-inducing ligand expression. J Immunol. 2001;167:3164–3173. doi: 10.4049/jimmunol.167.6.3164. [DOI] [PubMed] [Google Scholar]
- 55.Shetty S, Brown Gladden J, Henson ES, Hu X, Villanueva J, Haney N, Gibson SB. Tumor necrosis factor-related apoptosis inducing ligand (TRAIL) up-regulates death receptor 5 (DR5) mediated by NFkappaB activation in epithelial derived cell lines. Apoptosis. 2002;7:413–420. doi: 10.1023/A:1020031023947. [DOI] [PubMed] [Google Scholar]
- 56.Sedger LM, Shows DM, Blanton RA, Peschon JJ, Goodwin RG, Cosman D, Wiley SR. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol. 1999;163:920–926. [PubMed] [Google Scholar]
- 57.Meng RD, El-Deiry WS. P53-independent upregulation of KILLER/DR5 TRAIL receptor expression by glucocorticoids and interferon-gamma. Exp Cell Res. 2001;262:154–169. doi: 10.1006/excr.2000.5073. [DOI] [PubMed] [Google Scholar]
- 58.Shigeno M, Nakao K, Ichikawa T, Suzuki K, Kawakami A, Abiru S, Miyazoe S, Nakagawa Y, Ishikawa H, Hamasaki K, et al. Interferon-alpha sensitizes human hepatoma cells to TRAIL-induced apoptosis through DR5 upregulation and NF-kappa B inactivation. Oncogene. 2003;22:1653–1662. doi: 10.1038/sj.onc.1206139. [DOI] [PubMed] [Google Scholar]
- 59.Choi EA, Lei H, Maron DJ, Wilson JM, Barsoum J, Fraker DL, El-Deiry WS, Spitz FR. Stat1-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand and the cell-surface death signaling pathway by interferon beta in human cancer cells. Cancer Res. 2003;63:5299–5307. [PubMed] [Google Scholar]
- 60.Bao Q, Zhao Y, Niess H, Conrad C, Schwarz B, Jauch KW, Huss R, Nelson PJ, Bruns CJ. Mesenchymal stem cell-based tumor-targeted gene therapy in gastrointestinal cancer. Stem Cells Dev. 2012;21:2355–2363. doi: 10.1089/scd.2012.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taraballi F, Pasto A, Bauza G, Varner C. Immunomodulatory potential of mesenchymal stem cell role in diseases and therapies: A bioengineering prospective. J Immunol Regen Med. 2019;4:100017. [Google Scholar]
- 62.Monteran L, Erez N. The Dark side of fibroblasts: Cancer-Associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front Immunol. 2019;10:1835. doi: 10.3389/fimmu.2019.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu P, Fu YX. Tumor-infiltrating T lymphocytes: Friends or foes? Lab Investig. 2006;86:231–245. doi: 10.1038/labinvest.3700389. [DOI] [PubMed] [Google Scholar]
- 64.Badalamenti G, Fanale D, Incorvaia L, Barraco N, Listì A, Maragliano R, Vincenzi B, Calò V, Iovanna JL, Bazan V, Russo A. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell Immunol. 2019;343:103753. doi: 10.1016/j.cellimm.2018.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.