Abstract
The interaction of the SARS CoV2 spike glycoprotein with two sialic acid‐containing trisaccharides (α2,3 and α2,6 sialyl N‐acetyllactosamine) has been demonstrated by NMR. The NMR‐based distinction between the signals of those sialic acids in the glycans covalently attached to the spike protein and those belonging to the exogenous α2,3 and α2,6 sialyl N‐acetyllactosamine ligands has been achieved by synthesizing uniformly 13C‐labelled trisaccharides at the sialic acid and galactose moieties. STD‐1H,13C‐HSQC NMR experiments elegantly demonstrate the direct interaction of the sialic acid residues of both trisaccharides with additional participation of the galactose moieties, especially for the α2,3‐linked analogue. Additional experiments with the spike protein in the presence of a specific antibody for the N‐terminal domain and with the isolated receptor binding and N‐terminal domains of the spike protein unambiguously show that the sialic acid binding site is located at the N‐terminal domain.
Keywords: 13C-Labelling, Molecular Recognition, NMR Spectroscopy, SARS-Cov2, Sialic Acid
NMR experiments using 13C‐labelled sialyl‐containing trisaccharides unequivocally demonstrate that the N‐terminal domain of the Spike (S) SARS CoV‐2 glycoprotein directly binds sialic acid residues, highlighting the possibility of additional cellular receptor loci for the viral S protein at our cells.
SARS‐CoV‐2 virus uses the heavily glycosylated Spike (S) protein to promote cell attachment and fusion of the viral and cellular membrane and enter the host cell. The C‐terminal domain of the S promoter contains the receptor binding domain (RBD) that specifically recognizes angiotensin‐converting enzyme 2 (ACE2) as main host receptor.[ 1 , 2 , 3 , 4 , 5 ] However, different studies have shown limited ACE2 expression in the human respiratory system,[ 6 , 7 , 8 , 9 ] suggesting that, besides ACE2, additional receptors may also contribute to viral attachment and infection. [10] For instance, some lectins, such as DC‐SIGN, L‐SIGN and Siglec‐1, have been identified as enhancers of trans‐infection.[ 11 , 12 , 13 ] The binding of some of these lectins to the S glycoprotein has been also demonstrated. [14] At a different level, it has been described that several members of the Coronavirus family use sialylated glycans, which are abundantly expressed on the host cell surface of the respiratory tract, as attachment points.[ 15 , 16 ] The S glycoproteins of the human HCoV‐OC43[ 17 , 18 ] and HCoV‐HKU1 [19] viruses recognize sialic acid as sole receptor, while MERS‐CoV utilizes sialosides as co‐receptors in addition to DPP4 enzyme,[ 15 , 16 ] which brings the advantage of a two‐step attachment process. Sialic acids (Sia) are a class of acidic sugars often found at the termini of glycans in mammalian tissues.[ 20 , 21 ] 5‐N‐Acetyl neuraminic acid (Neu5Ac) is the most common form in humans, usually attached to galactose (Gal) through either α2,3‐ or α2,6‐linkages, to N‐acetyl galactosamine (GalNAc) in α2,6‐fashion or to another Neu5Ac moiety via α2,8‐linkage. Further structural diversity can arise from modification of the hydroxyl and N‐acetyl groups.[ 20 , 21 , 22 ] This structural diversity is crucial for coronavirus infection and for zoonosis to human hosts.[ 16 , 23 ]
Within the current pandemic due to SARS‐CoV‐2, [24] much controversy has been generated around the possibility that Sia containing glycoconjugates on the host cell surface could serve as attachment factors or auxiliary/co‐receptors.[ 10 , 25 ] Indeed, although a point‐of‐care device, based on a nanoparticle decorated with Neu5Ac, [26] has been demonstrated to be useful to detect the presence of the virus, other authors have neglected a major role for the participation of Sia. Alternatively, it has been recently proposed that the virus uses gangliosides as co‐receptors to facilitate viral entry. [27] In any case, no direct experimental evidence of the interaction of the S glycoprotein with Sia has been presented to the scientific community. Based on homology, in silico studies have suggested that the N‐terminal domain (NTD) of the S glycoprotein may contain a Sia binding site. [28] In any case, no experimental proof was presented.
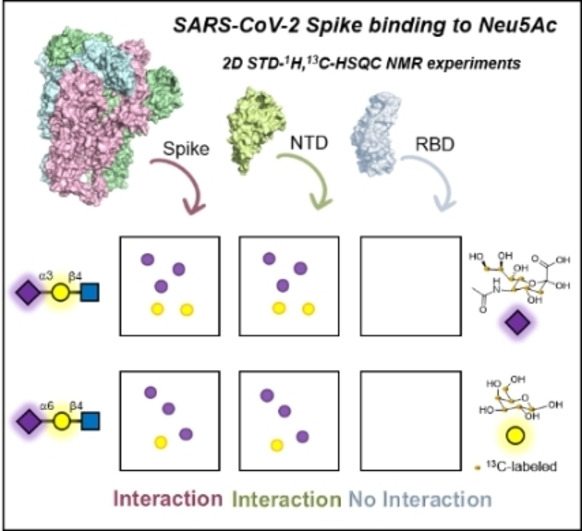
Herein, we experimentally demonstrate that the S glycoprotein of SARS‐CoV‐2 effectively binds Neu5Ac in both α2,3 and α2,6 sialyl N‐acetyllactosamine (SLN), using NMR spectroscopy. The S glycoprotein is highly glycosylated. These glycans on the protein represent a hurdle for NMR interaction studies with oligosaccharides, especially if they contain terminal sialylation,[ 14 , 29 , 30 , 31 ] as the distinction of the Neu5Ac moieties from the own glycoprotein from those that are exogeneous represents a major challenge. Therefore, we have synthesised and employed α2,3 and α2,6 SLN trisaccharides, 13C‐labelled at the Neu5Ac and Gal moieties, allowing the use of 2D STD‐1H,13C‐HSQC NMR experiments to detect only those STD NMR signals that correspond to protons attached to 13C atoms. [32] Therefore, the unequivocal identification of the Neu5Ac glycan binders is achieved, even for such heavily glycosylated protein. Furthermore, we have defined the glycan binding epitope, showing a higher contribution of the Gal ring in α2,3 than in α2,6 SLN to the binding. Finally, by using different protein constructs, such as the sole RBD, the NTD and a mAb specific for the NTD, we have also demonstrated that the Sia binding site is, in fact, located at the N‐terminal domain of the S glycoprotein, as previously suggested.
Saturation Transfer Difference (STD NMR) experiment is a robust method to detect ligand‐receptor interactions.[ 33 , 34 , 35 , 36 , 37 ] There is a long history on the use of 1H‐STD NMR to characterize protein‐glycan recognition events.[ 35 , 38 , 39 , 40 ] However, when the receptor is a glycoprotein, such as the S protein of SARS‐CoV‐2, this strategy fails since STD NMR signals arising from the glycans covalently linked to the protein also appear, as demonstrated in the blank STD spectrum of the S glycoprotein alone (Figure S6). Those NMR signals overlap with the expected 1H‐STD NMR signals of a putative glycan ligand, thus, precluding data analysis. Recently, we presented the application of 2D STD‐1H,13C‐HSQC experiments using specifically 13C‐labelled glycans to pinpoint glycan binding epitopes within repeating oligomers. [41] The introduction of 13C labels at specific position breaks the NMR chemical shift degeneracy that occurs in the non‐labelled compound, without introducing any artificial probe. Herein, the use of the 13C‐labels in the Neu5Ac moiety of the ligand molecules brings the additional advantage that the resulting 2D STD‐1H,13C‐HSQC NMR spectra are not disturbed by the signals from the glycans attached to the protein, which are not labelled, allowing for the specific identification of the binder (Figure S7 in Supporting Information for the blank 2D STD‐1H,13C‐HSQC experiments of the S glycoprotein, and 3′SLN* and 6′SLN*). To that scope, we first carried out the synthesis of the sialyl N‐acetyllactosamine trisaccharides with 13C‐labels at the Neu5Ac and Gal residues.
α2,3‐ and α2,6‐ SLN trisaccharides with 13C‐labelled Gal and Neu5Ac (3′SLN* and 6′SLN*) were prepared by one pot chemo‐enzymatic synthesis (Scheme 1). Neu5Ac with 13C labelled C3−C9 was prepared by coupling sodium pyruvate, 13C labelled at C3, with N‐acetyl mannosamine, 13C labelled at C1−C6, in the presence of the enzyme aldolase. The sugar donor was obtained by reacting the prepared 13C labelled sugar with cytidine 5′‐triphosphate (CTP) in the presence of cytidine‐5′‐monophosphate (CMP)‐Synthetase and magnesium chloride. [42] Next, commercial N‐acetyllactosamine uniformly 13C‐labelled at the Gal residue was treated with the synthetized CMP‐Neu5Ac in the presence of either bacterial PmST1 [43] or mammalian ST6Gal1, [44] providing the labelled α2,3‐SLN and α2,6‐SLN trisaccharides, respectively (see Supporting Information for details of the synthesis).
Scheme 1.
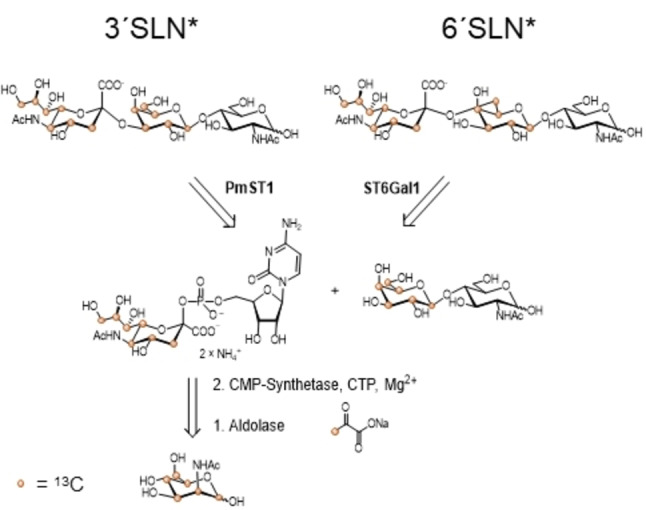
Retrosynthesis of α2,3 and α2,6 SLN with 13C‐labels on the NeuAc (C3 to C9) and Gal (C1 to C6) residues.
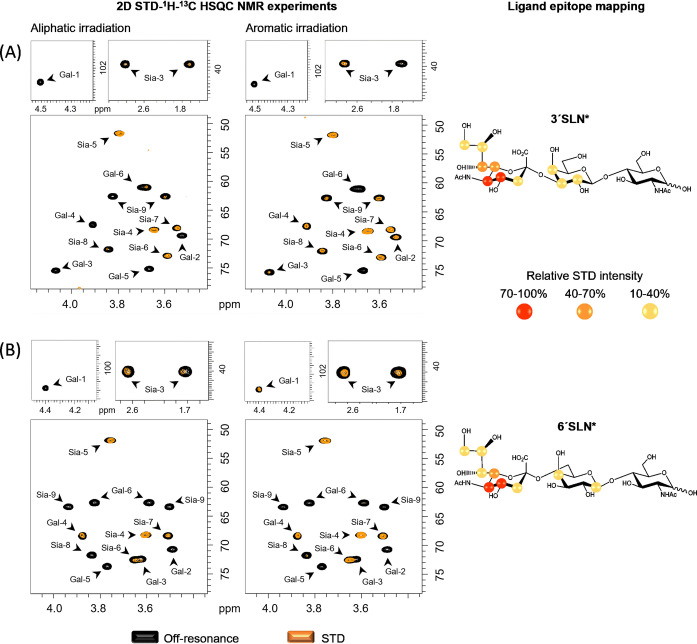
Binding of the 13C‐labelled α2,3‐ and α2,6‐SLN* to the extracellular domain of S protein was monitored through 2D STD‐1H,13C‐HSQC NMR experiments. These experiments provide STD signals only for those 1H covalently bonded to 13C‐labelled nuclei of the glycans, that is, in our case, all the protons of the Neu5Ac and Gal residues, excluding the N‐acetyl group of Neu5Ac, which is not 13C‐labelled. Two different frequencies were used to irradiate the protein, one at the aliphatic region (δ=0.8 ppm), and a second one at the aromatic region (δ=7.0 ppm). The resulting STD HSQC spectra showed the presence of selected STD signals, clearly indicating binding (Figure 1). For both compounds, the main STD signals belong to Neu5Ac residue, with the strongest ones belonging to C4−H4, C5−H5, C6−H6, and C7−H7 cross peaks. Weaker STD signals were detected for the C3−H3, C8−H8 and C9−H9 of Neu5Ac. Additional STD signals belong to the Gal ring, especially in the aromatic irradiation experiments, which are stronger for 3′SLN* than for 6′SLN* (Figure S10, Table S2). These data suggest that the Gal ring is closer to the S protein in the α2,3‐ than α2,6‐linked sialoside. Additionally, the comparison of the STD HSQC spectra obtained at the two different protein irradiation frequencies showed stronger STD intensities for the aromatic irradiation experiment (Tables S1–S2). These results strongly suggest that the Sia binding site of the S protein contains aromatic residues. [37]
Figure 1.
2D STD‐1H‐13C HSQC NMR experiments for the interaction of the S protein of SARS‐CoV‐2 with A) 3′SLN* and B) 6′SLN*. On the left: NMR spectra with aliphatic and aromatic protein irradiation. In black, the off‐resonance spectrum and superimposed in orange, the STD‐HSQC spectrum. On the right: Ligand epitope mapping presented as relative STD intensities and refers to the aromatic irradiation.
The ectodomain of the S glycoprotein on SARS‐CoV‐2 is rather large (426 kDa). It is composed of a stalk region (S2‐subunit) and a head domain (S1‐subunit), which contains the RBD at the C‐terminal domain and a N‐terminal domain.[ 45 , 46 ] Within the controversy mentioned above, it has been proposed that the RBD contains a Sia binding site with preference for gangliosides. [27] On the contrary, in silico studies predicted the N‐terminal domain as Neu5Ac locus.[ 28 , 47 , 48 ] Thus, once demonstrated that the S protein indeed binds Sia moieties, the NMR binding studies were carried out with the sole RBD and NTD, independently.
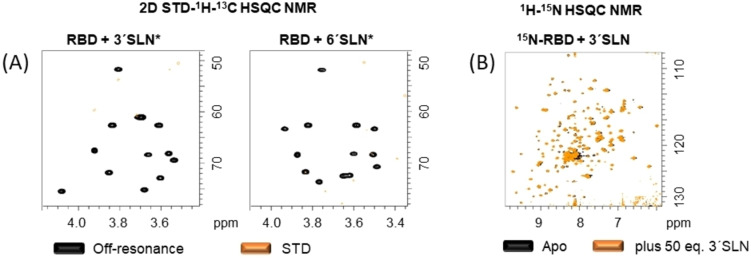
The RBD of the S glycoprotein was mixed with 3′SLN* and 6′SLN* and 2D STD‐1H,13C‐HSQC NMR experiments were acquired (Figure 2A). The resulting spectra did not show any STD NMR signals, clearly suggesting no binding of the RBD to the sialoglycans. It can be argued that the efficacy of the magnetization transfer from any protein to its ligand in STD NMR experiments depends, among other factors, on the size of the protein. The receptor binding domain is relatively small (24 kDa) which could result in less effective STD compared to the large S trimer. Thus, as a complementary strategy, a protein observed NMR approach was employed. Fittingly, the RBD has been recently produced with 15N‐labeling from E. coli, providing a NMR fingerprint of the protein. [49] However, the RBD has two N‐glycosylation sites which contain complex type N‐glycans when the protein is produced in mammalian cell culture, [34] but not in E. coli. These glycans may contribute to shape the protein 3D structure and influence the binding to sialoglycans. Thus, we generated the 1H‐15N labelled RBD in mammalian cell culture to ensure the complete post‐translational modification (see Supporting Information, section 5). The resulting 1H‐15N HSQC NMR spectrum is fairly similar to that obtained for the bacterial‐produced protein, allowing for mapping 60 % of the 1H‐15N cross peaks. Thus, the RBD was titrated with increasing amounts of both 3′SLN and 6′SLN up to 50 equivalents. No chemical shift perturbations were observed for any of the 1H‐15N cross peaks (Figure 2B and Figure S11, for detailed view of the 1H‐15N HSQC of 15N‐RBD in absence and presence of 3′SLN). Therefore, it can be concluded that, under these experimental conditions, the RBD of the S protein of SARS‐CoV‐2 does not bind to sialyl N‐acetyllactosamine.
Figure 2.
A) 2D STD‐1H‐13C HSQC NMR experiments of 3′SLN* and 6′SLN* with RBD. Note the absence of signals in the STD spectra. B) 1H‐15N HSQC of 15N‐RBD in absence (black) and presence (orange) of 50 equivalents of 3′SLN. No chemical shift perturbations were observed.
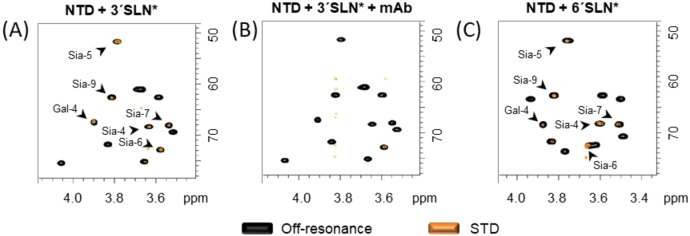
Next, the N‐terminal domain of the S glycoprotein was interrogated. The NTD was mixed with 3′SLN* and 6′SLN*, and 2D STD‐1H,13C‐HSQC NMR experiments were acquired (Figure 3). The resulting spectra showed STD NMR signals, demonstrating that the NTD contains a Sia binding site. Comparison of the STD results between the S protein ectodomain and the NTD revealed that the two protein constructs recognize both α2,3‐ and α2,6‐ SLN in a similar way, with a major Neu5Ac epitope and additional contributions from the Gal residue, which are again more pronounced for the α2,3‐ than the α2,6‐linked SLN (see Supporting Information section 6.3 and Figure S8). In general, lower STD intensities were observed with the sole NTD than with the much larger trimeric S protein.
Figure 3.
2D STD‐1H‐13C HSQC NMR experiments of NTD of S protein of SARS‐CoV‐2 with 3′SLN* (A) and 6′SLN (C). Note the presence of signals in the STD spectra. B) 2D STD‐1H‐13C HSQC NMR experiment of the SARS‐CoV‐2 spike protein with 3′SLN* and 3 equivalents of mAb against NTD. Note the dramatic reduction of the STD signals.
The sialic acid binding sites in the N‐terminal domain of the MERS, HCoV‐OC43 and HCoV‐HKU1 coronaviruses spike protein are defined by loops. Based on homology studies, it has been proposed that the sialic acid binding site of the S protein of SARS‐CoV‐2 presents similar structural elements. Thus, it can be hypothesized that the shape and accessibility of the Sia binding site depends on the employed protein constructs (ectodomain vs. NTD). To address this issue, one additional STD HSQC experiment was carried out by adding a mAb, which targets the NTD, to the NMR tube containing the S glycoprotein ectodomain and 3′SLN* (Figure 3B). Fittingly, a major decrease of the glycan STD NMR signals was observed, revealing that the mAb masks the sialoglycan binding site. This result clearly demonstrates that the sialic acid binding site in the NTD is functional and accessible under the S protein ectodomain presentation. Still, although the STD‐HSQC experiments with the NTD showed clear STD signals, their intensity is certainly lower than those with the larger trimeric S ectodomain. While this could be attributed to the different protein size as mentioned above, we may speculate that the Sia binding site, presumably localized in a loop region, is stabilized in the trimeric form of the S‐glycoprotein, minimizing dynamics and providing a more efficient interaction with respect to a looser binding locus in the single NTD.
These results provide a clear and non‐ambiguous experimental demonstration of the existence of direct binding between Sia containing oligosaccharides to the NTD of the S‐glycoprotein. The implications of this binding event to promote infection needs to be demonstrated. In any case, our results contribute to the further understanding of the SARS‐CoV‐2 infection mechanism and spread and may open new opportunities for the development of glycan‐based inhibitors to interfere with viral infection and ameliorate SARS‐CoV‐2 disease.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This research was funded by the European Research Council (ERC‐2017‐AdG, project number 788143‐RECGLYCA NMR to J.J.B.) and Agencia Estatal de Investigación (Spain), projects RTI2018‐094751‐B‐C21 to J.J.B. & A.A. and PID2019‐107770RA‐I00 to J.E.O., and by the Human Frontier Science Program (HFSP; grant LT000747/2018‐C to L.U.) and CIBER, an initiative of Instituto de Salud Carlos III (ISCIII), Madrid, Spain.
Dedicated to Professor Joan Bosch on the occasion of his 75th birthday.
L. Unione, M. J. Moure, M. P. Lenza, I. Oyenarte, J. Ereño-Orbea, A. Ardá, J. Jiménez-Barbero, Angew. Chem. Int. Ed. 2022, 61, e202201432; Angew. Chem. 2022, 134, e202201432.
Contributor Information
Dr. June Ereño‐Orbea, Email: jereno@cicbiogune.es.
Dr. Ana Ardá, Email: aarda@cicbiogune.es.
Prof. Dr. Jesús Jiménez‐Barbero, Email: jjbarbero@cicbiogune.es.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Cell 2020, 181, 281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Letko M., Marzi A., Munster V., Nat. Microbiol. 2020, 5, 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., Cell 2020, 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M., Nature 2003, 426, 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zamorano Cuervo N., Grandvaux N., eLife 2020, 9, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harmer D., Gilbert M., Borman R., Clark K. L., FEBS Lett. 2002, 532, 107–110. [DOI] [PubMed] [Google Scholar]
- 7. Lukassen S., Chua R. L., Trefzer T., Kahn N. C., Schneider M. A., Muley T., Winter H., Meister M., Veith C., Boots A. W., Hennig B. P., Kreuter M., Conrad C., Eils R., EMBO J. 2020, 39, e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C., Mol. Syst. Biol. 2020, 16, e9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Soni S., Jiang Y., Tesfaigzi Y., Hornick J. L., Çataltepe S., PLoS One 2021, 16, e0247510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun X.-L., Glycobiology 2021, 31, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lempp F. A., Soriaga L. B., Montiel-Ruiz M., Benigni F., Noack J., Park Y. J., Bianchi S., Walls A. C., Bowen J. E., Zhou J., Kaiser H., Joshi A., Agostini M., Meury M., Dellota E., Jaconi S., Cameroni E., Martinez-Picado J., Vergara-Alert J., Izquierdo-Useros N., Virgin H. W., Lanzavecchia A., Veesler D., Purcell L. A., Telenti A., Corti D., Nature 2021, 598, 342–347. [DOI] [PubMed] [Google Scholar]
- 12. Perez-Zsolt D., Muñoz-Basagoiti J., Rodon J., Elosua-Bayes M., Raïch-Regué D., Risco C., Sachse M., Pino M., Gumber S., Paiardini M., Chojnacki J., Erkizia I., Muñiz-Trabudua X., Ballana E., Riveira-Muñoz E., Noguera-Julian M., Paredes R., Trinité B., Tarrés-Freixas F., Blanco I., Guallar V., Carrillo J., Blanco J., Telenti A., Heyn H., Segalés J., Clotet B., Martinez-Picado J., Vergara-Alert J., Izquierdo-Useros N., Cell. Mol. Immunol. 2021, 18, 2676–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., Schoehn G., Bernardi A., Delgado R., Fieschi F., PLoS Pathog. 2021, 17, e1009576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lenza M. P., Oyenarte I., Diercks T., Quintana J. I., Gimeno A., Coelho H., Diniz A., Peccati F., Delgado S., Bosch A., Valle M., Millet O., Abrescia N. G. A., Palazón A., Marcelo F., Jiménez-Osés G., Jiménez-Barbero J., Ardá A., Ereño-Orbea J., Angew. Chem. Int. Ed. 2020, 59, 23763–23771; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23971–23979. [Google Scholar]
- 15. Li W., Hulswit R. J. G., Widjaja I., Raj V. S., McBride R., Peng W., Widagdo W., Tortorici M. A., Van Dieren B., Lang Y., Van Lent J. W. M., Paulson J. C., De Haan C. A. M., De Groot R. J., Van Kuppeveld F. J. M., Haagmans B. L., Bosch B. J., Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alejandra Tortorici M., Walls A. C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G. J., Bosch B. J., Rey F. A., de Groot R. J., Veesler D., Nat. Struct. Mol. Biol. 2019, 26, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hulswit R. J. G., Lang Y., Bakkers M. J. G., Li W., Li Z., Schouten A., Ophorst B., Van Kuppeveld F. J. M., Boons G. J., Bosch B. J., Huizinga E. G., De Groot R. J., Proc. Natl. Acad. Sci. USA 2019, 116, 2681–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vlasak R., Luytjes W., Spaan W., Palese P., Proc. Natl. Acad. Sci. USA 1988, 85, 4526–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q. K., Marasco W. A., Baric R. S., Sims A. C., Pyrc K., Li W., Sui J., J. Virol. 2015, 89, 7202–7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh S., in Sialic Acids Sialoglycoconjugates Biol. Life, Heal. Dis., Elsevier, Amsterdam, 2020, pp. 1–61. [Google Scholar]
- 21. Schauer R., Kamerling J. P., in Adv. Carbohydr. Chem. Biochem., Academic Press Inc., San Diego, 2018, pp. 1–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varki A., Trends Mol. Med. 2008, 14, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wasik B. R., Barnard K. N., Parrish C. R., Trends Microbiol. 2016, 24, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou P., Lou Yang X., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., Nature 2020, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dhar C., Sasmal A., Diaz S., Verhagen A., Yu H., Li W., Chen X., Varki A., Glycobiology 2021, 31, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baker A. N., Richards S., Guy C. S., Congdon T. R., Hasan M., Zwetsloot A. J., Gallo A., Lewandowski J. R., Stansfeld P. J., Straube A., Walker M., Chessa S., Pergolizzi G., Dedola S., Field R. A., Gibson M. I., ACS Cent. Sci. 2020, 6, 2046–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen L., McCord K. A., Bui D. T., Bouwman K. M., Kitova E. N., Elaish M., Kumawat D., Daskhan G. C., Tomris I., Han L., Chopra P., Yang T. J., Willows S. D., Mason A. L., Mahal L. K., Lowary T. L., West L. J., Hsu S. T. D., Hobman T., Tompkins S. M., Boons G. J., de Vries R. P., Macauley M. S., Klassen J. S., Nat. Chem. Biol. 2022, 18, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Awasthi M., Gulati S., Sarkar D. P., Tiwari S., Kateriya S., Ranjan P., Verma S. K., Viruses 2020, 12, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shajahan A., Supekar N. T., Gleinich A. S., Azadi P., Glycobiology 2020, 30, 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe Y., Allen J. D., Wrapp D., McLellan J. S., Crispin M., Science 2020, 369, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bagdonaite I., Thompson A. J., Wang X., Søgaard M., Fougeroux C., Frank M., Diedrich J. K., Yates J. R., Salanti A., Vakhrushev S. Y., Paulson J. C., Wandall H. H., Viruses 2021, 13, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fehér K., Groves P., Batta G., Jimenez-Barbero J., Muhle-Goll C., Kover K. E., J. Am. Chem. Soc. 2008, 130, 17148–17153. [DOI] [PubMed] [Google Scholar]
- 33. Mayer M., Meyer B., Angew. Chem. Int. Ed. 1999, 38, 1784–1788; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 1999, 111, 1902–1906. [Google Scholar]
- 34. Meyer B., Peters T., Angew. Chem. Int. Ed. 2003, 42, 864–890; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2003, 115, 890–918. [Google Scholar]
- 35. Ardá A., Jiménez-Barbero J., Chem. Commun. 2018, 54, 4761–4769. [DOI] [PubMed] [Google Scholar]
- 36. Viegas A., Manso J., Nobrega F. L., Cabrita E. J., J. Chem. Educ. 2011, 88, 990–994. [Google Scholar]
- 37. Monaco S., Tailford L. E., Juge N., Angulo J., Angew. Chem. Int. Ed. 2017, 56, 15289–15293; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 15491–15495. [Google Scholar]
- 38. Gimeno A., Valverde P., Ardá A., Jimenez-Barbero J., Curr. Opin. Struct. Biol. 2020, 62, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marchetti R., Perez S., Arda A., Imberty A., Jimenez-Barbero J., Silipo A., Molinaro A., ChemistryOpen 2016, 5, 274–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gimeno A., Reichardt N. C., Cañada F. J., Perkams L., Unverzagt C., Jiménez-Barbero J., Ardá A., ACS Chem. Biol. 2017, 12, 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moure M. J., Gimeno A., Delgado S., Diercks T., Boons G. J., Jiménez-Barbero J., Ardá A., Angew. Chem. Int. Ed. 2021, 60, 18777–18782; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 18925–18930. [Google Scholar]
- 42. Yu H., Yu H., Karpel R., Chen X., Bioorg. Med. Chem. 2004, 12, 6427–6435. [DOI] [PubMed] [Google Scholar]
- 43. Yu H., Chokhawala H., Karpel R., Yu H., Wu B., Zhang J., Zhang Y., Jia Q., Chen X., J. Am. Chem. Soc. 2005, 127, 17618–17619. [DOI] [PubMed] [Google Scholar]
- 44. Moremen K. W., Ramiah A., Stuart M., Steel J., Meng L., Forouhar F., Moniz H. A., Gahlay G., Gao Z., Chapla D., Wang S., Yang J. Y., Prabhakar P. K., Johnson R., Dela Rosa M., Geisler C., Nairn A. V., Seetharaman J., Wu S. C., Tong L., Gilbert H. J., Labaer J., Jarvis D. L., Nat. Chem. Biol. 2018, 14, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Science 2020, 367, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai Y., Zhang J., Xiao T., Peng H., Sterling S. M., Walsh R. M., Rawson S., Rits-Volloch S., Chen B., Science 2020, 369, 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bò L., Miotto M., Di Rienzo L., Milanetti E., Ruocco G., Front. Med. Technol. 2021, 2, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Milanetti E., Miotto M., Di Rienzo L., Nagaraj M., Monti M., Golbek T. W., Gosti G., Roeters S. J., Weidner T., Otzen D. E., Ruocco G., Front. Mol. Biosci. 2021, 8, 690655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schoenle M. V., Li Y., Yuan M., Clarkson M. W., Wilson I. A., Peti W., Page R., J. Am. Chem. Soc. 2021, 143, 7930–7934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.