Abstract
BACKGROUND
Covalent (irreversible) Bruton’s tyrosine kinase (BTK) inhibitors have transformed the treatment of multiple B-cell cancers, especially chronic lymphocytic leukemia (CLL). However, resistance can arise through multiple mechanisms, including acquired mutations in BTK at residue C481, the binding site of covalent BTK inhibitors. Noncovalent (reversible) BTK inhibitors overcome this mechanism and other sources of resistance, but the mechanisms of resistance to these therapies are currently not well understood.
METHODS
We performed genomic analyses of pretreatment specimens as well as specimens obtained at the time of disease progression from patients with CLL who had been treated with the noncovalent BTK inhibitor pirtobrutinib. Structural modeling, BTK-binding assays, and cell-based assays were conducted to study mutations that confer resistance to noncovalent BTK inhibitors.
RESULTS
Among 55 treated patients, we identified 9 patients with relapsed or refractory CLL and acquired mechanisms of genetic resistance to pirtobrutinib. We found mutations (V416L, A428D, M437R, T474I, and L528W) that were clustered in the kinase domain of BTK and that conferred resistance to both noncovalent BTK inhibitors and certain covalent BTK inhibitors. Mutations in BTK or phospholipase C gamma 2 (PLCγ2), a signaling molecule and downstream substrate of BTK, were found in all 9 patients. Transcriptional activation reflecting B-cell–receptor signaling persisted despite continued therapy with noncovalent BTK inhibitors.
CONCLUSIONS
Resistance to noncovalent BTK inhibitors arose through on-target BTK mutations and downstream PLCγ2 mutations that allowed escape from BTK inhibition. A proportion of these mutations also conferred resistance across clinically approved covalent BTK inhibitors. These data suggested new mechanisms of genomic escape from established covalent and novel noncovalent BTK inhibitors. (Funded by the American Society of Hematology and others.)
Covalent (irreversible) bruton’s tyrosine kinase (BTK) inhibitors bind to the C481 residue of BTK and block the ATP-binding pocket, thereby preventing catalytic activity. Despite excellent outcomes for patients with chronic lymphocytic leukemia (CLL) treated with covalent BTK inhibitors, resistance is ultimately acquired in many patients.1,2 Resistance to covalent BTK inhibitors is best understood in CLL, in which mutations at the BTK C481 amino acid residue impair drug binding and thereby restore the catalytic activity of BTK.3 In addition, activating mutations in phospholipase C gamma 2 (PLC0γ2), a direct substrate of BTK, render malignant cells less reliant on BTK.2
Noncovalent (reversible) BTK inhibitors were developed to improve on the pharmacologic properties of covalent BTK inhibitors while also maintaining potency against BTK C481 mutations. These agents do not require binding to the BTK C481 residue and effectively inhibit both wild-type and mutant BTK with C481 substitutions.4 In preclinical studies, noncovalent BTK inhibitors, including pirtobrutinib (LOXO-305), ARQ-351,4 fenebrutinib, and vecabrutinib, inhibited B-cell–receptor signaling in BTK C481–mutant cell and animal models. Moreover, the phase 1–2 clinical trial of pirtobrutinib showed promising efficacy for patients with B-cell cancer who had previously been treated with covalent BTK inhibitors (with 62% of patients with CLL having a response), including patients with or without BTK C481 mutations (with a response occurring in 71% and 66% of the patients, respectively).5
Because most patients with CLL receive multiple therapies over the course of their treatment, an understanding of the mechanisms of resistance to each class of agents is critical. However, published reports of resistance to noncovalent BTK inhibitors in patients have been lacking. We therefore set out to identify die genetic mechanisms and transcriptional characteristics of CLL in patients with clinical resistance to noncovalent BTK inhibitors.
METHODS
PATIENTS
We obtained peripheral blood specimens (and bone marrow aspirates and lymph node–biopsy specimens, when clinically indicated) before treatment and at the time of disease progression during treatment from patients at the Memorial Sloan Kettering Cancer Center with relapsed or refractory CLL treated with pirtobrutinib who were participating in the phase 1–2 BRUIN study (NCT037405295), The BRUIN study is an open-label study of pirtobrutinib in patients with relapsed or refractory B-cell cancers.6 In the phase 1 portion, patients were treated with pirtobrutinib monotherapy according to a standard dose-escalation study design in 28-day cycles. In the phase 2 dose-expansion cohort, patients were treated with pirtobrutinib at the recommended phase 2 dose of 200 mg once daily. Key eligibility criteria for the enrollment of patients with CLL in the BRUIN study Included two previous lines of therapy (later amended to one previous line of therapy if it included a covalent BTK inhibitor). All the patients provided written informed consent, and the institutional review board at Memorial Sloan Kettering Cancer Center approved the protocol.
Responses were assessed by the investigators in accordance with the criteria from the 2018 International Workshop on Chronic Lymphocytic Leukemia.7 Institutional review board–approved correlative studies conducted as part of a separate biospecimen protocol were performed with the use of specimens obtained from patients before pirtobrutinib treatment and at the time of the occurrence of resistance to pirtobrutinib during treatment. Clinical data were collected and collated by the Memorial Sloan Kettering Cancer Center without the involvement of Loxo Oncology and include clinical outcomes in this subgroup of patients as of May 1, 2021. The authors designed the correlative analysis, gathered the data, analyzed the data, and vouch for the accuracy and completeness of the data. The corresponding authors wrote the submitted manuscript without sponsor-funded editorial support.
ANALYSIS AND VALIDATION OF RESISTANCE MECHANISMS
Mutational analysis was performed with specimens from nine patients with resistance by means of targeted next-generation sequencing with the MSK-IMPACT Heme/HemePACT8 platform for all patients and by means of single-cell DNA sequencing with the Mission Bio Tapestri platform9 for two of the patients. Additional details are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org.
For the validation of resistance mutations, BTK and PLCG2 plasmids were transduced into TMD8 cells (a B-cell lymphoma line with constitutive Notch signaling) and OCl-LYlO cells (derived from an activated B-cell–type diffuse large B-cell lymphoma). Cells expressing mutant or control alleles were used for competition assays, cell-viability assays, immunoblots, and intracellular calcium-release assays. Further details regarding the methods of validation are provided in the Supplementary Appendix.
STRUCTURAL MODELING
Structural modeling of BTK was performed with UCSF ChimeraX.10 The published structures of ibrutinib (Protein Data Bank ID, 5P9J11) and ARQ-531 bound to BTK (Protein Data Bank ID, 6E4F4) were used to map mutations found in patients onto the BTK kinase domain. Pirtobrutinib was modeled into the BTK structure with the use of induced fit docking and binding pose nietadynamics12 with Schrödinger Suite software, version 2021–1 (Schrödinger).
SINGLE-CELL RNA SEQUENCING
Single-cell RNA sequencing and CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) were performed on specimens from five healthy donors between the ages of 46 and 58 years and from six patients with CLL treated with pirtobrutinib (four who had a relapse during treatment and two who had a sustained response to treatment), with the use of a panel of 277 antibodies.13 Derails of the CITE-seq assay and analyses are provided in the Supplementary Appendix.
RESULTS
BTK MUTATIONS FOUND AT CLINICAL RESISTANCE TO NONCOVALENT BTK INHIBITORS
A total of 55 patients with CLL who were treated with the noncovalent BTK inhibitor pirtobrutinib in the phase 1–2 BRUIN study were evaluated for inclusion in the current study.5 At the time of data cutoff, 12 patients had discontinued therapy because of progressive disease, 38 patients continued to receive therapy, and 5 patients had discontinued therapy for reasons other than disease progression. Of the 12 patients who had discontinued pirtobrutinib therapy because of progressive disease, 1 patient with primary refractory disease (after 31 days of pirtobrutinib treatment) was excluded from resistance analyses because of the short duration of drug exposure. In addition, 2 patients who had progressive disease while receiving pirtobrutinib did not have post-treatment specimens available. Specimens were therefore available for 9 patients with resistance to pirtobrutinib that developed during treatment. Detailed clinical characteristics and treatment histories for each of these 9 patients who discontinued pirtobrutinib treatment because of progressive disease are shown in Figure S1 and Tables S1, S2, and S3 in the Supplementary Appendix. The best overall response among these 9 patients was partial response in 4 patients (44%). Progression during treatment manifested with biopsy-confirmed Richter transformation to diffuse large B-cell lymphoma in 3 patients.
In the first 2 patients with disease progression during treatment with pirtobrutinib, sequencing at progression revealed mutations in the kinase domain of BTK outside the C481 residue (V416L and M437R) that had not been present before treatment (Fig. 1A and 1B). Additional acquired mutations were observed in the BTK kinase domain, including T474I (in 2 patients) and L528W (in 4 patients). In one of these patients, both T474I and L528W were found at progression (Fig. 1A). Overall, in this heavily pretreated group of patients with CLL with acquired resistance, 7 patients had acquired non-C481 BTK mutations and the remaining 2 had persistence of PLCγ2 mutations that had been identified before treatment (Fig. 1B). No patient had a new BTK C481 mutation arise during pirtobrutinib treatment, and no other recurrent mutation or copy-number alteration was identified at progression.
Figure 1. BTK Mutations in Patients with Chronic Lymphocytic Leukemia with Acquired Resistance to Noncovalent BTK Inhibitors.
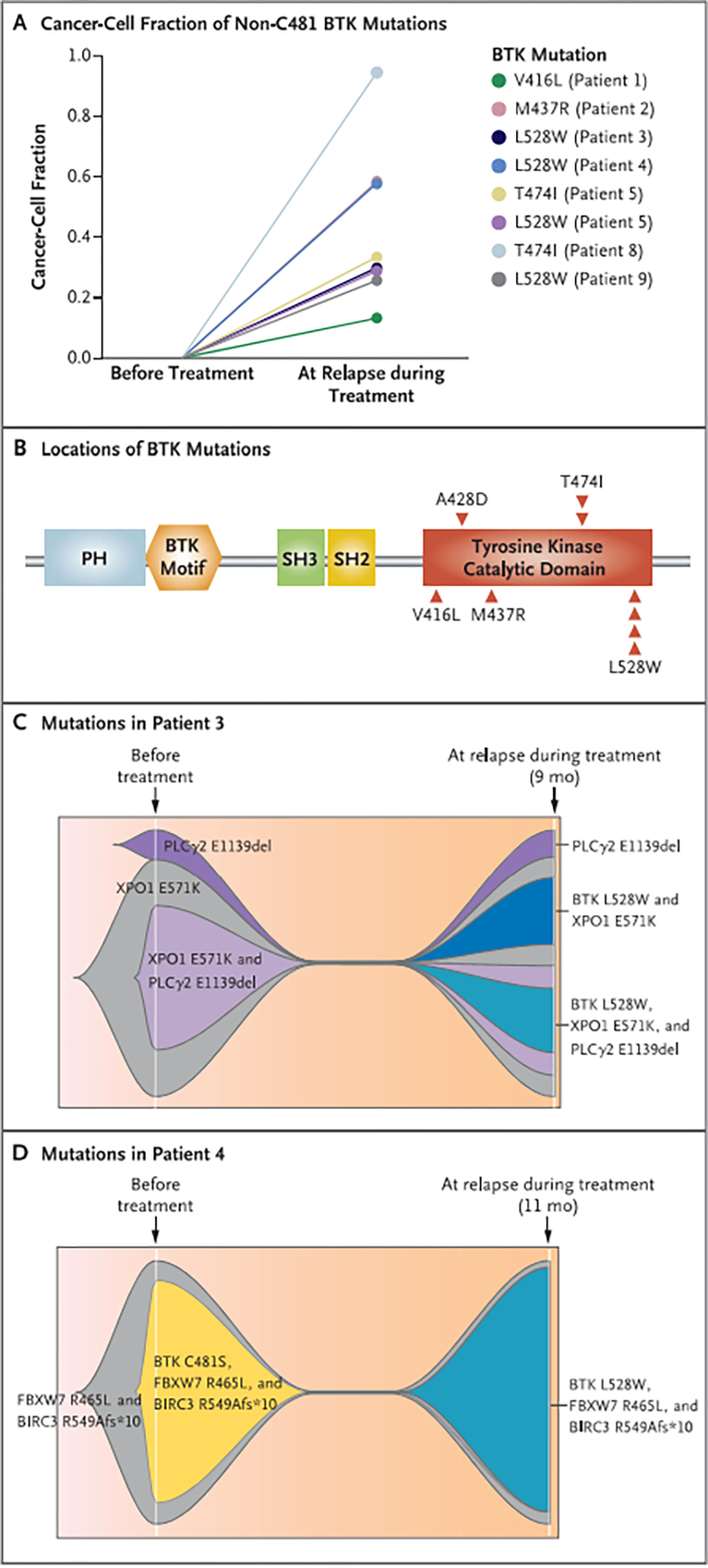
Panel A shows non-C481 Bruton’s tyrosine kinase (BTK) mutations found at the time of relapse during treatment, as revealed by serial targeted gene sequencing of specimens from patients with chronic lymphocytic leukemia treated with the noncovalent BTK inhibitor pirtobrutinib. The timing of specimen collection is shown on the x axis, and the cancer-cell fraction of the non-C481 BTK mutations is shown on the y axis. Panel B shows BTK mutations outside the BTK C481 residue found in patients with resistance to pirtobrutinib. Each individual occurrence of a mutation is depicted as an arrowhead. PH denotes pleckstrin homology domain, and SH Src homology domain. Panels C and D are fish-plot representations of single-cell mutational data from Patient 3 (Panel C) and Patient 4 (Panel D) before pirtobrutinib therapy and at relapse during treatment. Patient 3 had a phospholipase C gamma 2 (PLCγ2) mutation before pirtobrutinib therapy and was found to have acquired BTK L528W mutations at relapse after 9 months of treatment. Patient 4 had the BTK C481S mutation before pirtobrutinib therapy, and it was suppressed during treatment; this patient was found to have acquired BTK L528W at relapse after 11 months of treatment. Mutations in XPO1 (exportin 1), FBXW7 (F-box and WD repeat—containing protein 7), and BIRC3 (baculoviral IAP repeat–containing protein 3) were also found.
All 7 patients with newly acquired, non-C481 BTK mutations at clinical progression during pirtobrutinib treatment had received previous ibrutinib treatment and had discontinued ibrutinib for progressive CLL. In the 4 patients with preexisting BTK C481 mutations, BTK C481 clones were suppressed by pirtobrutinib in 2 patients, followed by acquisition of new non-C481 BTK mutations associated with clinical resistance to pirtobrutinib.
To understand the clonal architecture at resistance to pirtobrutinib, we performed single-cell mutational analysis of BTK, PLCG2, and 29 additional genes recurrently mutated in CLL before pirtobrutinib therapy and at the time of progression during treatment (Table S4). A total of 14,705 cells from two patients (Patients 3 and 4) were sequenced across these time points; each of these patients had BTK L528W mutations identified on the basis of bulk DNA sequencing at progression during pirtobrutinib treatment. In Patient 3, the BTK L528W mutation was not found within individual cells before treatment but was found in two subclones at progression during treatment, one of which also contained a PLCγ2 mutation within the same cells as the BTK L52SW mutation (Figs. 1C and S2A). In Patient 4, the BTK C481S mutation was suppressed by pirtobrutinib treatment, but BTK I.528W was found at progression during treatment, along with pathogenic mutations that had previously coexisted with BTK C481S (Figs. 1D and S2B).
FUNCTIONAL CHARACTERIZATION OF NON-C481 BTK MUTANTS
Each of the non-C481 BTK mutations identified in patients with pirtobrutinib resistance (V416L, A428D, M437R., T474I, and L528W) clustered within the kinase domain of BTK when mapped onto the three-dimensional structure of the protein (Fig. 2A). The published structure of ARQ-531 bound to BTK4 reveals a binding mode similar to that of ibrutinib. as does the pirtobrutinib–BTK interaction (Fig. 2B).
Figure 2. Resistance to BTK Inhibitors Conferred by BTK Mutations Outside the C481 Residue.
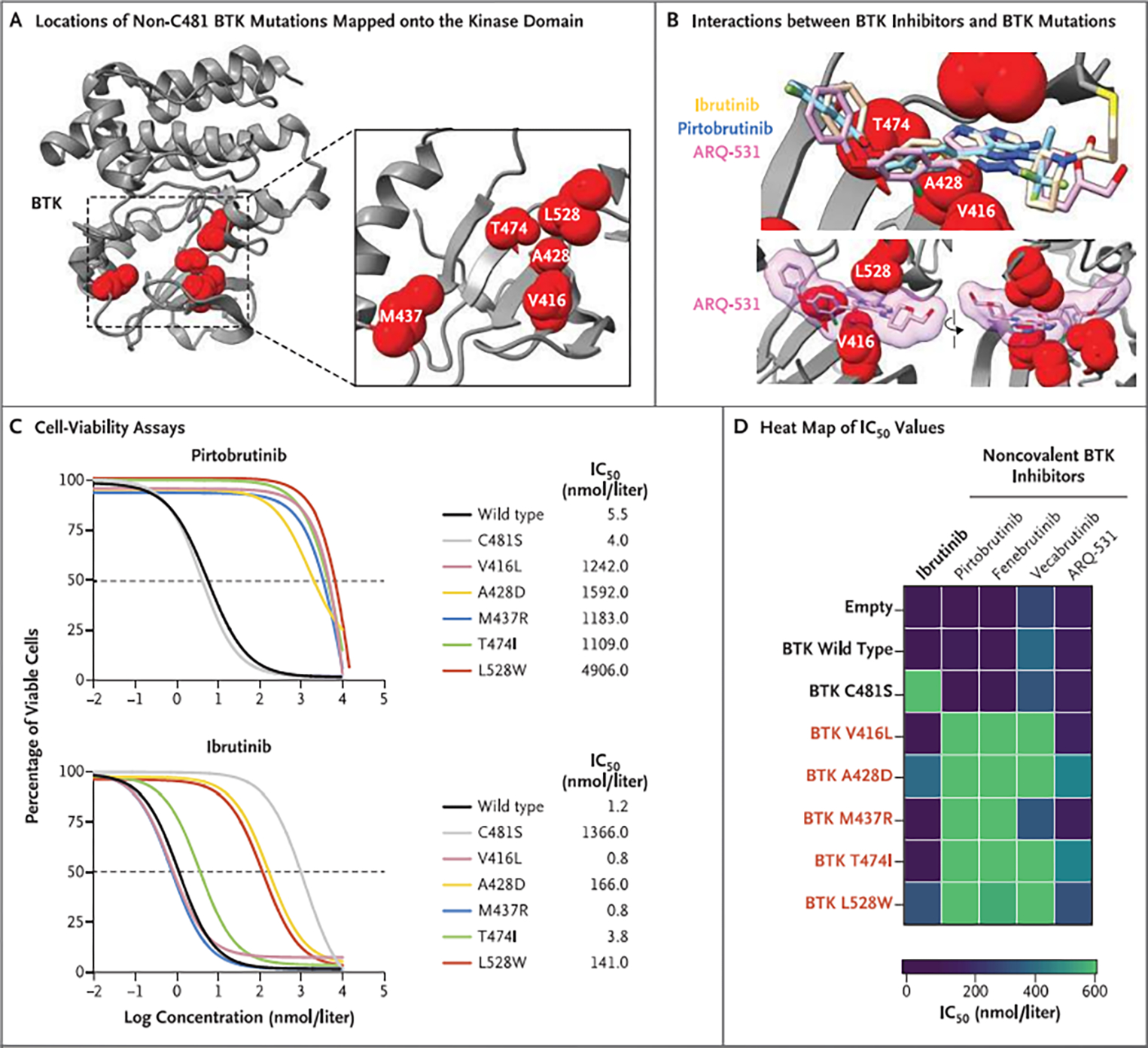
Panel A shows the locations of non-C481 BTK mutations (V416L, A428D, M437R, T474I, and L528W) identified in patients with pirtobrutinib resistance mapped onto the crystal structure of the BTK kinase domain (gray ribbon). Mutated amino acids are shown as red spheres. Panel B shows the chemical structures of ibrutinib, pirtobrutinib, and ARQ-531 overlaid onto the crystal structure of the BTK kinase domain to illustrate the interactions between noncovalent BTK inhibitors and the non-C481 BTK mutations identified in specimens from patients with pirtobrutinib resistance. Panel C shows the results of experiments in which TMD8 cells transduced with mutant BTK were treated with pirtobrutinib or ibrutinib for 72 hours and the half-maximal inhibitory concentrations (IC50) determined with the use of cell-viability assays. Panel D shows a heat map of IC50 values at 72 hours after treatment of TMD8 cells transduced with mutant BTK with ibrutinib or a panel of noncovalent BTK inhibitors (pirtobrutinib, fenebrutinib, vecabrutinib, or ARQ-531). These data show that BTK C481S is resistant to ibrutinib but sensitive to noncovalent BTK inhibitors, whereas cells expressing BTK mutants V416L, A428D, M437R, T474I. and L528W are less sensitive to noncovalent BTK inhibitors. The data in Panels C and D are from three independent replicates and have been normalized to values obtained with a vehicle control (dimethylsulfoxide).
To functionally characterize these BTK mutants, we introduced constructs expressing each mutant into the BTK-dependent human B-cell lymphoma cell line TMD8 and measured the half-maximal inhibitory concentration (IC50) on exposure to noncovalent BTK inhibitors (including pirtobrutinib, fenebrutinib, vecabrutinib, and ARQ-531) and ibrutinib (Figs. 2C, 2D, and S3A). Cells with BTK mutations that had been acquired in patients with resistance to pirtobrntinib (BTK V416L, M437R, T474I, and L528W) were much less sensitive to noncovalent BTK inhibitors than were cells expressing wild-type or C481S BTK. Measurement of the binding affinity of wild-type or mutant BTK to noncovalent BTK inhibitors indicated that each of the mutations tested impaired binding of both covalent and noncovalent inhibitors to BTK protein, a finding consistent with the cell-viability data (Fig. 3A). These drug-binding data also suggest that the in vitro sensitivity of some BTK-mutant–exptessiug cells to ibrutinib and ARQ-531 is most likely a result of the ability of these agents to inhibit numerous kinases beyond BTK with similar potency.4
Figure 3. Effect of BTK Resistance Mutations on Binding of Noncovalent and Covalent Inhibitors to BTK and on B-Cell–Receptor Signaling.
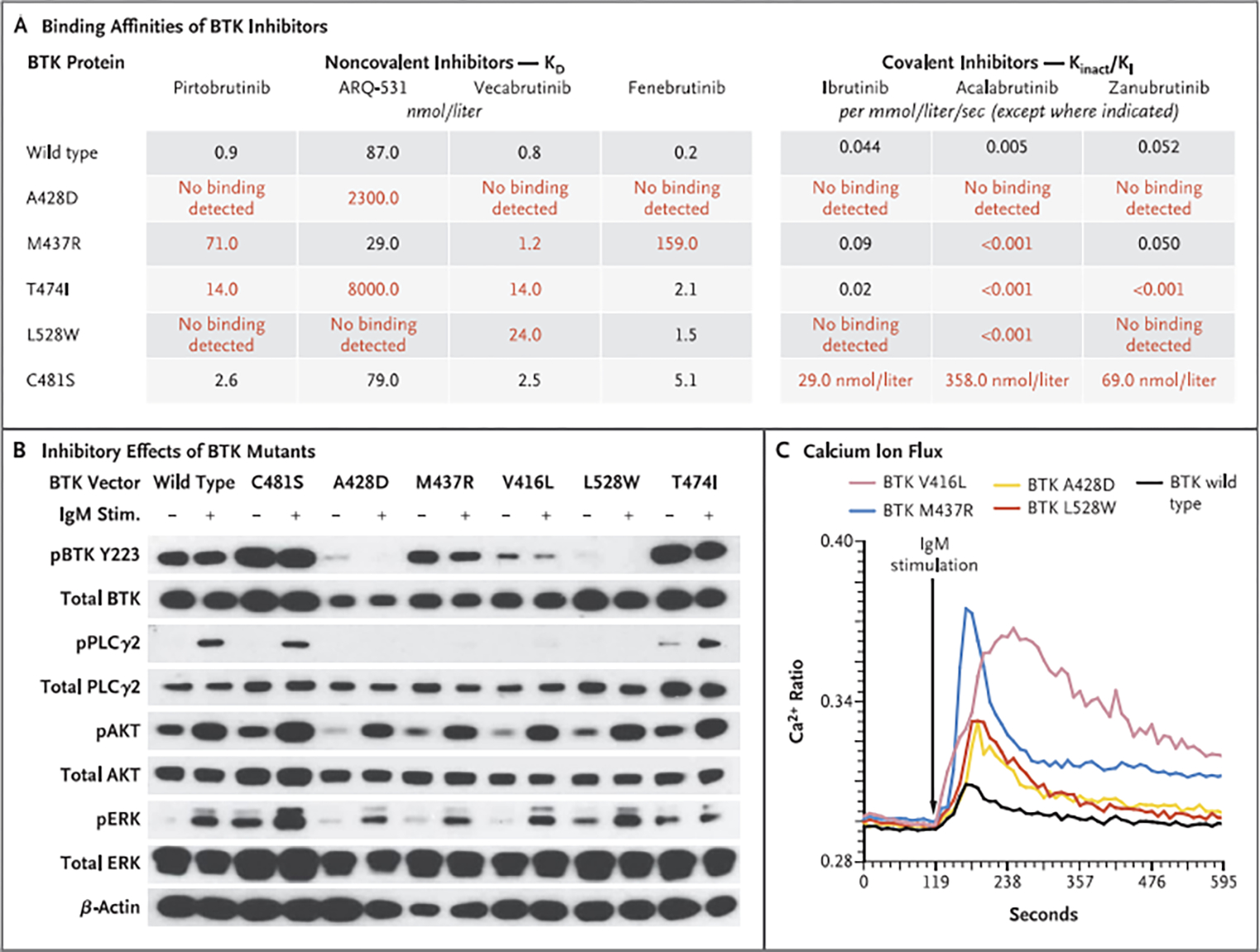
Panel A shows the binding affinities (KD) of each noncovalent BTK inhibitor to purified wild-type or mutant BTK protein, determined with the use of surface plasmon resonance technology. On the right are Kinact/K| values for covalent BTK inhibitors on the enzymatic activity of each wild-type or mutant BTK protein. The Kinact/K| value indicates the efficiency of covalent bond formation between each drug and BTK protein. Red values indicate mutants that decreased drug-binding efficiency by a factor of at least 10. The KD values for the covalent inhibitors to BTK C481S are reported. Panel B shows the inhibitory effects of TMD8 cells transduced with BTK mutants (C481S, A42ED, M437R, V416L, L528W, and T474I) on BTK autophosphorylation at Y223 and PLCγ2 phosphorylation at Y1217, On IgM stimulation of TMD8 cells, BTK mutants activated AKT (phosphorylated S473) and extracellular signal-related kinase (ERK; phosphorylated Y202 and Y204) signaling pathways similarly to wild-type BTK. (The prefix “p” indicates phosphorylation.) However, as shown in Panel C, intracellular calcium was elevated to higher levels in cells expressing BTK mutants than in those expressing wild-type BTK. The Ca2+ ratio is based on flow cytometry performed with lndo-1 (Life Technologies); higher values indicate greater concentrations of intracellular calcium.
Several mutations that were identified as conferring resistance to noncovalent BTK inhibitors also interfered with the ability of covalent BTK inhibitors to block BTK enzymatic activity (Fig. 3A). For example, BTK A428D and L528W mutations caused resistance to ibrutinib in vitro and prevented multiple covalent BTK inhibitors from inhibiting mutant BTK (Figs. 2D and 3A). Of note, several non-C481 BTK mutations and PLCγ2 mutations also conferred a growth advantage on exposure to noncovalent as well as covalent BTK inhibitors (Fig. S3B and S3C). Overall, these data indicate a causal link between BTK mutations identified here outside the C481 residue as well as PLCγ2 mutations and resistance to multiple noncovalent and covalent BTK inhibitors.
To examine the effects of BTK mutations on BTK autophosphorylation at Y223, a marker of BTK catalytic activity, HEK293T cells that lacked endogenous BTK were stably transfected with wild-type BTK, mutant BTK, or empty vector plasmids (Fig. S3D). Whereas wild-type BTK and several BTK mutants (C481S and T474I) increased BTK Y223 autophosphorylation, multiple BTK mutants that confer resistance to noncovalent and covalent BTK inhibitors greatly diminished BTK autophosphorylation of Y223. Similar results were seen in TMD8 cells, in which expression of BTK V416L, A428D, M437R, and L528W significantly reduced Y223 phosphorylated BTK and phosphorylation of PLCγ2 at tyrosine 1217, a catalytic substrate of BTK (Fig. 3B). The BTK residues at which mutations abrogate BTK catalytic activity (V416, A428, and L528) make up the ATP-binding interface of BTK and are thereby predicted to disrupt ATP binding and kinase activity (Fig. S3E), which would be consistent with the above findings.
Although noncovalent inhibitors suppressed BTK Y223 phosphorylation in cells expressing wild-type or C481S BTK, the resistance mutations identified here blocked the activity of multiple BTK inhibitors on BTK autophosphorylation (Fig. S4A). Furthermore, on IgM stimulation, catalytically inactive BTK mutants still enabled activation of AKT, extracellular signal-related kinase (ERK), and nuclear factor κB (Figs. 3B and S4B through S4E). Moreover, although AKT activation was abrogated by pittobrutinib in cells expressing wild-type BTK, AKT activation was not suppressed by pirtobrutinib in cells expressing BTK V416L, A428D, or L528W (Fig. S4F). Similarly, IgM stimulation of cells bearing these BTK mutations resulted in hyperactive calcium ion flux (Fig. 3C) and release of IP1 (inositol monophosphate) after B-cell–receptor stimulation (Fig. S4G), a reflection of intact downstream signaling. Overall, these data suggest that catalytically inactive BTK mutations acquired in patients with resistance to noncovalent BTK inhibitors allow persistent B-cell–receptor signaling despite ongoing treatment with the inhibitor.
TRANSCRIPTIONAL ACTIVATION OF B-CELL–RECEPTOR SIGNALING
To better understand the heterogeneity of CLL cells in response to BTK inhibition, we performed single-cell RNA sequencing and CITE-seq to simultaneously measure single-cell RNA and surface protein expression.14 CITE-seq was performed oil peripheral blood mononuclear cells (PBMCs) from five age-matched healthy controls and from six patients with CLL before pirtobrutinib treatment. CITE-seq was also performed at relapse during treatment for four of these six patients in whom resistance developed and at the time of ongoing response for two patients who had ongoing clinical responses after more than 1 year of pirtobrutinib therapy (Fig. S5A, S5B, and S5C and Table S5).
We mapped a total of 53,722 cells from 17 specimens to a previously described multimodal PBMC reference atlas15 and identified six broad hematopoietic cell types (Fig. S6A). We integrated and jointly analyzed B cells and CLL cells across patient specimens using LIGER (Linked Inference of Genomic Experimental Relationships).16 We observed reduced expression of the B-cell–receptor signaling pathway gene set (75 genes) in patients with ongoing response to pirtobrutinib (Figs. S6B, S7A, S7B, and S7C), a finding consistent with the effects of BTK inhibition. In contrast, transcriptional signatures reflecting activation of B-cell–receptor signaling increased in patients who had a relapse during treatment.
DISCUSSION
Resistance to covalent BTK inhibitors was first described in patients with CLL with acquired BTK C481 and PLCγ2 mutations.2,17 Here, we identified a cluster of mutations in BTK outside the C481 residue, as well as mutations in PLCγ2, that confer resistance to noncovalent BTK inhibitors in patients with CLL. Alrhough initially found in patients with acquired resistance to pirtobrutinib, the series of mutations described here confer resistance to a wide array of noncovalent BTK inhibitors in clinical development, including ARQ-531, fenebrutinib, and vecabrutinib, as well as to many existing covalent BTK inhibitors. These mutations impair the binding of a number of inhibitors to BTK. In so doing, several of these mutations (e.g., BTK V416L, A428D, M437R, and L528W) paradoxically hinder BTK catalytic activity yet allow effective B-cell–receptor signaling and AKT pathway activation despite exposure to covalent and noncovalent BTK inhibitors.
These findings of inactivating, disease-associated BTK mutations that augment AKT activation are reminiscent of kinase-inactivating BTK mutations in patients with previously untreated follicular lymphoma,18,19 a disease that is generally less sensitive to BTK inhibitors than CLL. Although it is known that BTK may serve functions independent of its kinase activity,20,21 the data presented here underscore the need to understand the link between catalytically inactive BTK and AKT activation. It will be important for future studies to determine how drug-resistant, catalytically inactive BTK mutations allow for persistent B-cell–receptor signaling. Of note, other well-established oncogenic mutations in kinases impair kinase activity but result in downstream pathway activation. For example, a series of BRAF mutations result in loss of RRAF kinase activity but enhance MAP kinase signaling.22 The non-C481 BTK mutations identified here might confer novel protein–protein interactions or release feedback inhibition of signaling pathways parallel to BTK–PLCγ2 signaling.
Our data suggest potential new therapeutic approaches to overcome the newly described BTK inhibitor resistance mechanisms. For example, these data provide a rationale for therapies aimed at addressing the potential scaffold function of BTK rather than inhibiting BTK kinase activity. Moreover, the finding that non-C481 BTK mutations allow persistent AKT signaling suggests that AKT inhibition might be another avenue to overcome resistance.
Despite these findings, it is important to note that we have assessed resistance in only 9 patients with relapse of CLL in an ongoing study that has enrolled more than 250 patients with CLL. Most patients in the overall study have not yet had a relapse, and therefore the present study has analyzed only the patients who have had the earliest relapses. It is therefore unclear whether similar mechanisms of resistance will be observed in patients who have had much longer disease control. Moreover, given that the mutations identified here all occurred in heavily pretreated patients (three of whom had progression with Richter transformation), it will be critical to evaluate whether similar resistance mechanisms arise in patients receiving noncovalent BTK inhibitors without any previous treatment with covalent BTK inhibitors in earlier lines of therapy.
Overall, the development of noncovalent BTK inhibitors represents a promising therapeutic advance for patients with CLL and other B-cell cancers that have previously been treated with covalent BTK inhibitors. We have identified a series of genetic mechanisms for acquired resistance to this new class of agents. More analyses of larger samples are necessary to characterize the frequency of these genetic events and how they will influence treatment options.
Supplementary Material
Acknowledgments
Supported by the American Society of Hematology (to Drs. Taylor, Mi, and Durham), the Robert Wood Johnson Foundation (to Dr. Taylor), the Doris Duke Charitable Foundation (to Dr. Taylor), the Edward P. Evans Foundation (to Drs. Taylor and Abdel-Wahab), the Society of MSK (to Dr. Durham), grants (2T32CA009207-36A1, to Dr. North 1K08CA230319-01, to Dr. Taylor; and 1P50 254838-01, to Dr. Abdel-Wahab) from the National Cancer Institute, the Lymphoma Foundation (to Dr. Thompson), Fondation de France (to Dr. Bourcier), the Lymphoma Research Foundation (to Dr. Mato), the CLL Society (to Dr. Mato), and Loxo Oncology. Dr. Aifantis’s work is supported by grants (R01CA216421, R01CA173636, R01CA228135, P01CA229086, and R01CA242020) from the National Cancer Institute and by the Edward P. Evans Foundation and the Leukemia and Lymphoma Society.
We thank Celina Komari, Yehudit Fox, and Kendall Meyer for their help in collecting the clinical data.
Footnotes
Contributor Information
Eric Wang, Human Oncology and Pathogenesis Program, New York
Xiaoli Mi, Human Oncology and Pathogenesis Program, New York
Meghan C. Thompson, Leukemia Service, New York
Skye Montoya, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Ryan Q. Notti, Memorial Sloan Kettering Cancer Center, the Laboratory of Molecular Electron Microscopy, Rockefeller University, New York
Jumana Afaghani, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Benjamin H. Durham, Human Oncology and Pathogenesis Program, New York Department of Medicine, and the Department of Pathology, New York.
Alex Penson, Human Oncology and Pathogenesis Program, New York
Matthew T. Witkowski, Department of Pathology and Laura and Isaac Perlmutter Cancer Center, New York University School of Medicine, New York
Sydney X. Lu, Human Oncology and Pathogenesis Program, New York
Jessie Bourcier, Human Oncology and Pathogenesis Program, New York
Simon J. Hogg, Human Oncology and Pathogenesis Program, New York
Caroline Erickson, Human Oncology and Pathogenesis Program, New York
Dan Cui, Human Oncology and Pathogenesis Program, New York
Hana Cho, Human Oncology and Pathogenesis Program, New York
Michael Singer, Human Oncology and Pathogenesis Program, New York
Tulasigeri M. Totiger, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Sana Chaudhry, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Mark Geyer, Leukemia Service, New York
Alvaro Alencar, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Adam J. Linley, Department of Molecular Physiology and Cell Signaling, Institute of Systems, Molecular, and Integrative Biology, University of Liverpool, Liverpool, United Kingdom
M. Lia Palomba, Lymphoma Service, New York
Catherine C. Coombs, University of North Carolina Medical Center, Chapel Hill
Jae H. Park, Leukemia Service, New York
Andrew Zelenetz, Lymphoma Service, New York
Lindsey Roeker, Leukemia Service, New York
Mary Rosendahl, Loxo Oncology at Lilly, Boulder, CO
Donald E. Tsai, Loxo Oncology at Lilly, Boulder, CO
Kevin Ebata, Loxo Oncology at Lilly, Boulder, CO
Barbara Brandhuber, Loxo Oncology at Lilly, Boulder, CO
David M. Hyman, Loxo Oncology at Lilly, Boulder, CO
Iannis Aifantis, Department of Pathology and Laura and Isaac Perlmutter Cancer Center, New’ York University School of Medicine, New York
Anthony Mato, Leukemia Service, New York
Justin Taylor, Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, Miami
Omar Abdel-Wahab, Human Oncology and Pathogenesis Program, New York Leukemia Service, New York.
REFERENCES
- 1.Woyach JA, Ruppert AS, Guinn D, et al. BTKC481S-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol 2017;35:1437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woyach JA. Furman RR, Liu T-M, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med 2014;370:2286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lampson BL, Brown JR. Are BTK and PLCG2 mutations necessary and sufficient for ibrutinib resistance in chronic lymphocytic leukemia? Expert Rev Hematol 2018:11:185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiff SD, Mantel R, Smith LL, et al. The BTK inhibitor ARQ 531 targets ibrutinib-resistant CLL and Richter transformation. Cancer Discov 2018;8:1300–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mato AR. Shah NN, Jurczak W, et al. Pirtobrutinib in relapsed or refractory B-cell malignancies (BRUIN): a phase 1/2 study. Lancet 2021;397:892–901. [DOI] [PubMed] [Google Scholar]
- 6.Mato A, Pagel JM, Coombs CC, et al. Pirtobrutinib, a next generation, highly selective, non-covalent BTK inhibitor in previously treated CLL/SLL: updated results from the phase 1/2 BRUIN study. Blood 2021;138:Suppl 1:391. abstract. [Google Scholar]
- 7.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018;131:2745–60. [DOI] [PubMed] [Google Scholar]
- 8.Durham BH, Getta R, Dietrich S, et al. Genomic analysis of hairy cell leukemia identifies novel recurrent genetic alterations. Blood 2017;130:1644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor J, Mi X, North K, et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 2020;136:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 2021;30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender AT, Gardberg A, Pereira A, et al. Ability of Bruton’s tyrosine kinase inhibitors to sequester Y551 and prevent phosphorylation determines potency for inhibition of Fc receptor but not B-cell receptor signaling. Mol Pharmacol 2017;91:208–19. [DOI] [PubMed] [Google Scholar]
- 12.Clark AJ, Tiwarjr P, Borrelli K, et al. Prediction of protein-ligand binding poses via a combination of induced fit docking and metadynamics simulations. J Chem Theory Comput 2016;12:2990–8. [DOI] [PubMed] [Google Scholar]
- 13.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184(13):3573.e29–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoeckius M, Zheng S, Houck-Loomis B, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 2018:19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 2019;177(7):1873.e17–1887.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman RR. Cheng S, Lu P, et al. Ibrutinib resistance in chronic lymphocytic leukemia. N Engl J Med 2014;370:2352–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu N, Wang F, Sun T, et al. Follicular lymphoma-associated BTK mutations are inactivating resulting in augmented AKT activation. Clin Cancer Res 2021;27:2301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krysiak K, Gomez F, White BS, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma, Blood 2017;129:473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middendorp S, Zijlstra AJE, Kersse-boom R, Dingjan GM, Jumaa H, Hendriks RW. Tumor suppressor function of Bruton tyrosine kinase is independent of its catalytic activity. Blood 2005;105:259–65, [DOI] [PubMed] [Google Scholar]
- 21.Middendorp S, Dingjan GM, Maas A, Dahlenborg K, Hendriks RW. Function of Bruton’s tyrosine kinase during B cell development is partially independent of its catalytic activity. J Immunol 2003;171:5988–96. [DOI] [PubMed] [Google Scholar]
- 22.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010;140:209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


