Abstract
The nature of the SHV-1 β-lactamase gene was analyzed in 97 epidemiologically unrelated Klebsiella pneumoniae strains isolated from clinical samples. β-Lactamase bands that focused at a pI of 7.6 (SHV-1-type) in 74 strains, at a pI of 7.1 (LEN-1-type) in 13 strains, and at a pI of 5.4 (TEM-1-type) in 10 strains were detected by analytical isoelectric focusing (IEF). Among the 74 SHV-1-producing strains, 40 had, in addition to the pI 7.6 band, an additional band on IEF: 20 had a band with a pI of 7.1 and 20 had a band with a pI of 5.4. Most of the 74 SHV-1-producing strains (76.7%) carried plasmids. Transfer of β-lactam resistance by conjugation was possible in only 9.3% of the strains tested. SHV-1 gene-specific PCR-restriction fragment length polymorphism (PCR-RFLP) analysis of the chromosomal DNA was positive for 93 of the 97 strains and negative for only 4 of the 10 samples with K. pneumoniae TEM-1 producers. In an attempt to approximate the location of the SHV gene locus by endonuclease restriction analysis, RFLP analysis with Southern blotting of chromosomal DNA with a labeled SHV-1 fragment as a probe was used to study the 97 strains. A trial with EcoRI showed at least one positive hybridization band for 96 strains; two bands were detected for 8 strains. The hybridization was negative for only one TEM-1 β-lactamase-producing strain. DNA sequence analysis showed no differences in promoter regions or extra stop-triplet sequences; only point mutations determined different allelic variants. The novel SHV-type variants are designated SHV-32 and SHV-33. As a result of the RFLP and sequencing analyses, it can be postulated that the loci for SHV-1 and LEN-1 genes are arranged in tandem. Our results strongly support the hypothesis that the ancestor of the SHV-1 β-lactamase originated from the K. pneumoniae chromosome.
The SHV-1 β-lactamase, first described by Pitton in 1972 as Pit-2 (19), is a class A, group 2b β-lactamase (according to the Bush-Jacoby-Medeiros classification [5]). Its hydrolytic spectrum of activity is similar to that of the TEM-1 β-lactamase, but it achieves better activity against ampicillin (5).
SHV-1 has been identified in several species of enterobacteria and is generally considered a plasmid-encoded enzyme (5). Nevertheless, this β-lactamase is found at a higher frequency (up to 80 to 90%) in Klebsiella pneumoniae (3, 9–12, 19). Thus, SHV-1 has putatively been considered chromosomally borne. In 1979, Matthew et al. (14) and Nugent and Hedges (17) demonstrated the extrachromosomal location of the blaSHV-1 gene in some K. pneumoniae strains. Other β-lactamases of putative chromosomal origin were occasionally found in K. pneumoniae strains, which were unable to transfer ampicillin resistance (18). Among them, LEN-1, a class A, group 2a β-lactamase, is the most frequently occurring and is clearly chromosomally encoded (2).
To obtain a better understanding of the structure and function of the SHV-1 family of β-lactamases, several groups of investigators have sought to detect differences among β-lactamases of the SHV-1 family in K. pneumoniae mainly by comparing the sequences of SHV-1 (4, 15), LEN-1 (2), and OHIO-1 (22). In the study described here we attempted to clarify the nature of the SHV-1 β-lactamase gene by analyzing 97 unrelated K. pneumoniae strains isolated from clinical samples.
MATERIALS AND METHODS
Bacterial strain characterization.
Ninety-seven K. pneumoniae strains were studied. All were isolated from pathological human products between 1979 and 1984 and were provided by five laboratories (laboratories L1 to L5). The strains were not epidemiologically related. Most of the strains were from laboratory L1 (49.5%). The other laboratories provided 18.5% (laboratory L2), 18.6% (laboratory L3), 7.2% (laboratory L4), and 6.2% (laboratory L5) of the strains. Urine was the most frequent type of sample (79.2%), followed by blood (4.1%), exudates (7.2%), respiratory specimens (1%), and other types of specimens (9.3%). Biotype characterization was performed by standard biochemical analyses. The reference strain used to extract the SHV-1 DNA probe was Escherichia coli HB101 (F− hsdS20 recA13 ara-14 proA2 leu lacY1 galK2 rpsL20 xyl-55 mtl-1 supE44) carrying plasmid pMON38.
Expression of the different β-lactamases was studied by previously described analytical isoelectric focusing (IEF) procedures (13) performed with samples of crude cell-free sonic extracts in Multiphor equipment (Pharmacia Biotech, Uppsala, Sweden). The pH gradients (pH 3.5 to 9.5, 4 to 6.5, and 5.5 to 8.5) were measured with a surface pH electrode device (LKB Multiphor electrode); enzyme bands were detected by nitrocefin staining (0.5 mmol of nitrocefin per liter in 0.1 mol of phosphate buffer per liter [pH 7.0]).
Antibiotic susceptibility and transfer of resistance.
Testing of the MICs of ampicillin, mezlocillin, carbenicillin, cephalothin, cefotaxime, ceftazidime, and aztreonam was performed by the agar dilution method. In addition, the MICs of ampicillin (2:1) and mezlocillin (4:1) in combination with clavulanic acid were also determined.
Conjugation experiments were performed by a two-step procedure with E. coli K-12 (C600; F− without the lac, thia, leu, and threo genes but with Nalr) as the recipient strain. From an overnight broth, culture recipient and donor strains were mixed at a proportion of 2:1 and were incubated at 37°C for 4 h. A sample of 0.1 ml was incubated overnight at 37°C on Mueller-Hinton agar; E. coli transconjugants were selected on agar plates (Diagnostics Pasteur, Marnes La Coquette, France) with nalidixic acid (40 μg/ml) containing ampicillin (60 μg/ml).
SHV-1 genetic analysis.
Plasmid DNA was isolated by a commercial method (Magic Miniprep DNA purification system; Promega Corporation, Madison, Wis.) based on the alkaline lysis method, followed by DNA purification with ion-exchange resins. Chromosomal DNA was isolated by the cetyltrimethylammonium bromide (CTAB) method (23). The size of the chromosomal DNA was estimated to be over 80 kb, and the 260/280 purity ratio was 1.8. Recombinant plasmid pMON38 carrying the SHV-1pMON38 gene was kindly provided by George A. Jacoby. Plasmid pMON38 was used as a template to obtain the 178-bp PCR product as a probe for Southern blot analysis and as a template to set up a specific PCR-restriction fragment length polymorphism (PCR-RFLP) analysis method. The probe was labeled with digoxigenin-dUTP with a PCR digoxigenin DNA labeling system (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's instructions.
A specific PCR-RFLP test was developed after the designation of primer sequences from the SHV-1 sequence (GenBank accession number AF148850) (4) by selecting the most specific coding area that distinguished SHV-1 from other β-lactamases, particularly LEN-1. The primer sequences were located at SHV-1 gene (GenBank accession number AF148850) (4) sequences 5′-T97AAGCGAAAGCCAGCTGTCG116-3′-OH (forward primer) and 5′-T274TTCGCTCCAGCTGTTCGTC255-3′-OH (backward primer); both primers detected a 178-bp fragment. The PCR amplification reaction mixture consisted of a 50-μl volume containing 50 mM Tris buffer (pH 8.0), 1.3 mM MgCl2, 5% dimethyl sulfoxide, 0.2 mM deoxynucleoside triphosphates, each primer at a concentration of 125 nM, and 1.25 U of Taq DNA polymerase (Amplitaq; Roche, Basel, Switzerland). The reaction was performed in a Perkin-Elmer 9600 thermocycler. The amplification method was as follows: the reaction was initiated by a denaturing step at 94°C for 1 min, followed by 30 cycles of denaturation at 92°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 10 s; the reaction was terminated with an extension step at 72°C for 1 min. The amplified fragment was electrophoresed in 1.2% agarose gels with TBE (Tris-borate-EDTA buffer, which consisted of 45 mM Trizma base, 45 mM boric acid, and 1 mM EDTA [pH 8.0]), yielding a fragment of 178 bp, which was a positive reaction. The PCR product was further digested with 3 U of NotI restriction endonuclease to test for specificity for SHV-1 and yielded specific fragments of 56 and 122 bp.
RFLP analysis with Southern blotting was performed with chromosomal DNA samples. Several endonucleases were tested at 3 U/μg of DNA overnight at the appropriate temperature for each endonuclease. Electrophoresis was done on a 0.5% agarose gel (20 by 20 cm) at 20 V for 24 h. Transfer to a positively charged nylon membrane (Boehringer Mannheim) was performed with a vacuum transfer system (Vacu-Gene XL; Pharmacia Biotech, Uppsala, Sweden). Conditions of stringency were taken into account as follows: the hybridization temperature was 68°C and the hybridization solution was 250 mM Na2HPO4, 1 mM EDTA, 20% sodium dodecyl sulfate, and 0.5% blocking reagent (Boehringer Mannheim). Detection of specific fragments was performed by chemiluminescence with the CSPD reagent (Boehringer Mannheim) at a dilution of 1:100 for 5 min. The sizes of the fragments obtained by RFLP analysis were determined with a digoxigenin-labeled bacteriophage lambda-HindIII DNA ladder marker that was also detected by chemiluminescence.
DNA sequencing analyses were performed with a Perkin-Elmer sequencing kit (ABI PRISM dye terminator cycle sequencing kit) adapted for an ABI PRISM 377 sequencer (Perkin-Elmer). The reactions were performed in a Perkin-Elmer 9600 thermocycler with 100 ng of chromosomal DNA from the tested strain as the template and according to the manufacturer's instructions; purified PCR products were electrophoresed and analyzed with an ABI PRISM 377 sequencer. Sequencing primers were defined to identify the whole sequence of the SHV-1 gene (a total of 1,120 bp; EMBL accession number M59181) (15) in two steps: both strands of the gene were sequenced, including 864 bp of the coding region, 117 bp upstream, and 139 bp downstream. The first pair of primers used was 5′-G8ATGAAAAATGATGAAGGAA27-3′-OH (forward primer set 1) and 5′-A576TCTGGCGCAAAAAGGCAGT557-3′-OH (backward primer set 1); the second pair of primers was 5′-C514GCCAATCTGCTACTGGCCA533-3′-OH (forward primer set 2) and 5′-G1127GAGGCCACGTTTATGGCGT1108-3′-OH (backward primer set 2). Sequencing data for 16 strains and pMON38, which was used as a probe, were compared to reported data for the blaSHV-1 gene listed in GenBank under accession number AF148850 (4). The sequences of the strains were translated and the amino acids were numbered as described by Ambler et al. (1).
Nucleotide sequence accession numbers.
The novel allelic variants reported in this study were designated SHV-32 and SHV-33 (http://www.lahey.org/studies/webt.html), and the nucleotide sequence data reported for the strains producing those variant enzymes and for K263 (designated as LEN-2) will appear in the GenBank nucleotide sequence database under accession numbers AY037778, AY037779, and AY037780, respectively.
RESULTS
Analytical IEF of sonic extracts yielded different β-lactamase bands that focused at a pI of 7.6 (SHV-1) in 74 strains, at a pI of 7.1 (LEN-1) in 13 strains, and at a pI of 5.4 (TEM-1) in 10 strains. Additionally, among the 74 SHV-1-type strains, 34 had a single band and 40 had, in addition to the pI 7.6 band, an additional band on IEF: 20 with a band at a pI of 7.1 and 20 with a band at a pI of 5.4. Antibiotic susceptibility assessed according to the MICs for the 97 K. pneumoniae strains tested indicated that the strains had broad-spectrum activities, showing both penicillinase and cephalosporinase activities, but the percentage of strains resistant to the different antibiotics studied tended to correlate with the enzyme produced (Table 1). Resistance to broad-spectrum cephalosporins or aztreonam was not observed. Nevertheless, for 33 strains that were not considered to have extended-spectrum β-lactamases, ceftazidime MICs ranged from ≥0.5 to 4 μg/ml.
TABLE 1.
Susceptibility profile relationship with type of β-lactamase
| pI by IEF (no. of strains) | No. (%) of strains resistant toa:
|
|||||
|---|---|---|---|---|---|---|
| AMP | AMP + CLA | MEZ | MEZ + CLA | CAR | CEF | |
| 5.4 (10) | 10 (100) | 6 (60) | 9 (90) | 0 (0) | 10 (100) | 6 (60) |
| 7.6 + 5.4 (20) | 20 (100) | 8 (40) | 11 (55) | 1 (5) | 20 (100) | 11 (55) |
| 7.6 (34) | 34 (100) | 6 (18) | 17 (50) | 0 (0) | 34 (100) | 19 (56) |
| 7.6 + 7.1 (20) | 20 (100) | 2 (10) | 9 (44) | 0 (0) | 20 (100) | 6 (30) |
| 7.1 (13) | 13 (100) | 0 (0) | 2 (15) | 0 (0) | 12 (92) | 2 (15) |
AMP, ampicillin; CLA, clavulanic acid; MEZ, mezlocillin; CAR, carbenicillin; CEF, cephalothin. The following breakpoints were used: for ampicillin, ampicillin-clavulanic acid, and cephalothin, ≥16 μg/ml; for mezlocillin and mezlocillin-clavulanic acid, ≥64 μg/ml; for carbenicillin, ≥32 μg/ml.
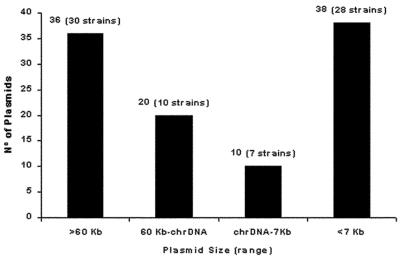
Plasmid analysis was performed with the 74 SHV-producing strains to study the frequency and distribution of plasmids. Most of the strains (76.7%) carried from one to five plasmids. The group of strains most frequently encountered was that with only one plasmid (37%), and 23% of the strains had no detectable plasmids. Plasmids were mainly distributed into two groups according to size: 54% were large (>30-kb) plasmids and 37% were very small (<7-kb) plasmids. Strains frequently had different combinations of plasmids by size and were compiled into different categories, as shown in Fig. 1. Among the 74 strains, 59 were studied in conjugation experiments; the strains excluded were the SHV-1 and TEM-1 producers because of their plasmid-encoded TEM-1 β-lactamase. Transfer of β-lactam resistance to the recipient strain, E. coli K-12 C600 F−, was positive for only 5 of the 59 (9.3%) strains tested. The five parental strains were mezlocillin resistant. The sizes of the plasmids detected in the transconjugant strains were >60 kb in three strains and 3.5 kb in two strains.
FIG. 1.
Analysis of the size ranges of plasmids among the 74 SHV-1-expressing strains by IEF. chrDNA, fragmented chromosomal DNA band (approximately 30 kb).
SHV-1 gene-specific PCR-RFLP analysis of the chromosomal DNA was positive for 93 of the 97 strains. PCR was negative for negative control strains (several E. coli clinical isolates that were nonproducers of the SHV-1 β-lactamase) and for only 4 of the 10 K. pneumoniae strains that produced the TEM-1 enzyme. A specific control analysis was performed with the restriction enzyme NotI, which has an SHV-1-specific restriction site. The NotI site is absent from the LEN-1 and OHIO genes, which have 90 and 95% nucleotide gene sequence homologies, respectively, with the SHV-1 sequence analyzed.
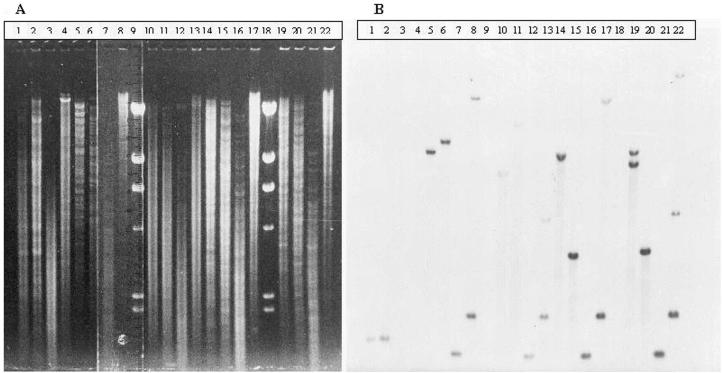
The 97 strains were studied by RFLP analysis with Southern blotting of chromosomal DNA. In an attempt to approximate the gene locus by endonuclease restriction analysis, several enzymes were tested (Fig. 2). The strategy used was to test the enzymes that did not cut the genes (EcoRI, HindIII) or that were specific for the SHV-1 gene (NotI) and/or the LEN-1 gene (BamHI, KpnI) (Table 2). In the trial with EcoRI, at least one positive hybridization band was found for 96 strains. The hybridization was negative for only one TEM-1 β-lactamase-producing strain (strain K534; Fig. 2, lane 3); PCR was also negative for this strain. Curiously, strains K251 and K273 (Fig. 2, lanes 1 and 2, respectively), which were also PCR negative (Table 2), were positive for a fragment; thus, further sequence analysis was suggested. Among the 96 positive strains, 88 had one EcoRI-specific hybridization band and two bands were detected for 8 strains. The fragments found were 8.5 kb (88.5%), 8 kb (6.7%), 7 kb (2.9%), and >23 kb (1.9%). The 8-kb fragment was always found together with the 8.5-kb fragment; the 7-kb fragment was always found alone; the >23-kb fragment was found alone in one strain and was combined with the 8.5-kb fragment in another strain. Among all strains that produced LEN-1, as determined by IEF, PCR analysis detected the SHV-1 gene and RFLP analysis detected one fragment of 8.5 kb (83% of strains) and two fragments of 8 and 8.5 kb (7% of strains). In addition, in the six strains that produced TEM-1 and in which the SHV-1 gene was detected by PCR, the fragment obtained by RFLP analysis was 8.5 kb.
FIG. 2.
RFLP analysis with Southern blotting with different chromosomal DNA samples. (A) Electrophoresis run. Lanes (K. pneumoniae strain tested and enzyme used for restriction, indicated by ⊥): 1, K251 ⊥ BamHI; 2, K273 ⊥ BamHI; 3, K534 ⊥ BamHI; 4, P111 (E. coli expressing TEM-1) ⊥ BamHI (negative control pattern); 5, K51 ⊥ EcoRI; 6, K51 ⊥ BamHI; 7, K51 ⊥ PstI; 8, K51 ⊥ NotI; 10, K48 ⊥ EcoRI; 11, K48 ⊥ BamHI; 12, K48 ⊥ PstI; 13, K48 ⊥ NotI; 14, K53 ⊥ EcoRI; 15, K53 ⊥ BamHI; 16, K53 ⊥ PstI; 17, K53 ⊥ NotI; 19, K75 ⊥ EcoRI; 20, K75 ⊥ BamHI; 21, K75 ⊥ PstI; 22, K75 ⊥ NotI; 9 and 18, unlabeled bacteriophage λ ⊥ HindIII (ladder marker). (B) Corresponding blot with labeled SHV-1 probe, as described in Materials and Methods.
TABLE 2.
SHV-1 gene locus approximation according to the profile obtained by RFLP analysis with Southern blottinga
| PCR result for SHV-1 gene | Strain | pI by IEF | Fragment size (kb) obtained with the following endonucleaseb:
|
|||||
|---|---|---|---|---|---|---|---|---|
| EcoRI | HindIII | NotI | BamHI | KpnI | PstI | |||
| Positive | K75 | 7.6 | 8.5 + 8 | 10 + 9 | >23 + 5 + 2.3 | 4 | 10 + 9 | 1.8 |
| K48 | 7.6 | 7 | ND | 5 + 2.3 | 16 | ND | 1.8 | |
| K53 | 7.6 + 7.1 | 8.5 | 10 | 23 + 2.3 | 4 | ND | 1.8 | |
| K51 | 7.1 | 8.5 | ND | 23 + 2.3 | 10 | ND | 1.8 | |
| K567 | 7.1 | 8.5 | 10 | ND | 4 | 9 | 1.8 | |
| Negative | K263c | 5.4 | 8.5 | 10 | 2.3 | 1.8 | 2.4 | ND |
| K251 | 5.4 | 8.5 | 10 | 2.3 | 1.8 | 4.5 | ND | |
| K273 | 5.4 | 8.5 | 10 | 2.3 | 1.8 | 4.5 | ND | |
Total chromosomal DNA was used for RFLP analysis.
EcoRI and HindIII did not cut the gene for SHV-1, LEN-1, or OHIO-1, whereas NotI, BamHI and KpnI are one-site gene-specific endonucleases for SHV-1 (NotI) and LEN-1 (BamHI, KpnI) genes. Finally, PstI has one site in all genes. ND, Not determined.
Sequencing analysis detected an allelic variant of LEN-1 (see Table 3).
DNA sequence analysis was performed with 17 DNA samples: pMON38 DNA; plasmid DNAs of the 5 transconjugant strains (strains TK75, TK136, TK167, TK175, and TK198); chromosomal DNAs of 3 SHV-1-type K. pneumoniae strains (strains K280, K48, and K237), with strain K280 being sensitive to mezlocillin and the other two strains being resistant to mezlocillin; and chromosomal DNAs of 3 LEN-1-producing K. pneumoniae strains (strains K103, K218, and K299) and 5 TEM-1-producing K. pneumoniae strains (strains K8, K39, K117, K154, and K263), as determined by IEF. Table 3 shows the results for both alignments for 16 different DNA sequences and their respective deduced amino acid sequences. For comparison purposes, the published sequences of the SHV-1 (GenBank accession number AF148850), LEN-1 (EMBL accession number X04515), and OHIO-1 (EMBL accession number M33655) genes were also considered. All matches were expressed relative to our SHV-1pMON38 sequence or its deduced amino acid sequence. As expected, the DNA and amino acid sequences of SHV-1pMON38 showed high degrees of homology (99.20 and 100%, respectively) with those of the SHV-1 enzyme from GenBank (accession number AF148850). Only 7 nucleotides were found to be different, and all of them were out of the coding region. The other sequences were also very homologous, with homologies varying between 98.8 and 100% for DNA sequence homology and 99.3 and 100% for amino acid sequence homology. Seven of the 15 strains that were sequenced and that were PCR positive for the SHV-1 gene showed amino acid changes (strains K48, K103, K218, K299, K8, K39, and K154) (Table 3). The amino acid sequences of isolates K48 and K39 were consistent with that of SHV-11 (a non-extended-spectrum β-lactamase) (16), and both showed a 7-kb fragment by RFLP analysis with EcoRI and Southern blotting. According to the β-lactamase nomenclature described on the World Wide Web (http://www.lahey.org/studies/webt.html), strains K48 and K39, strains K103 and K218, and strain K154 have already been designated producers of SHV-31, SHV-27 (6), and SHV-28, respectively. The remaining two strains (strains K299 and K8) produce new SHV variants designated SHV-32 and SHV-33, respectively, and their sequence data are included in the GenBank database under accession numbers AY037778 and AY037779, respectively.
TABLE 3.
DNA and deduced amino acid sequence alignmentsa
| Strain | % DNA homology | % Protein homology | Amino acid substitutionb | PCR result | Fragment size (kb) by RFLP analysis with EcoRI | β-Lactamase allelic variantc |
|---|---|---|---|---|---|---|
| TK75, TK136, TK167 | 100.00 | 100.00 | + | 8 | ||
| TK175 | 100.00 | 100.00 | + | 8.5 | ||
| TK198 | 99.91 | 100.00 | + | 8.5 | ||
| K280 (Mzs) | 99.54 | 100.00 | + | 8.5 | ||
| K237 (Mzr) | 100.00 | 100.00 | + | 8.5 | ||
| K48 (Mzr) | 98.80 | 99.65 | Leu35Gln | + | 7 | SHV-31 |
| K39 | 99.35 | 99.65 | Leu35Gln | + | 7 | SHV-31 |
| K117 | 99.53 | 100.00 | + | 8.5 | ||
| K103, K218 | 99.36 | 99.65 | Gly156Asp | + | 8.5 | SHV-27 |
| K154 | 99.34 | 99.65 | Tyr7Phe | + | 8.5 | SHV-28 |
| K299 | 99.18 | 99.30 | Ala126Val Gly156Asp | + | 8.5 | SHV-32 |
| K8 | 99.63 | 99.65 | Pro226Ser | + | 8.5 | SHV-33 |
| K263d | 90.92 | 90.91 | Val22Ala, Tyr24Asp, Asn53Ser, Leu88Val, 284AlaAlaLeulleGluHisTrp290 | − | 8.5 | LEN-2 |
| SHV-1e | 99.20 | 100.00 | SHV-1 | |||
| LEN-1f | 89.95 | 87.76 | ||||
| OHIO-1g | 94.88 | 94.41 |
Matches are expressed relative to the SHV-1pMON38 sequence used in this study. Mzr, mezlocillin resistant; Mzs, mezlocillin susceptible.
Amino acid sequence numbers according to Ambler et al. (1).
Sequence data for SHV-32, SHV-33, and LEN-2 will appear in the GenBank nucleotide sequence data under accession numbers AY037778, AY037779, and AY037780, respectively. Allele designations are reported at http://www.lahey.org/studies/webt.html.
Amino acid substitutions for K263 were determined by comparison with the LEN-1 consensus sequence.
Sequence from GenBank (accession number AF148150).
Sequence from EMBL data bank (accession number X04515).
Sequence from EMBL data bank (accession number M33655).
The enzyme from one of the TEM-1-type producer strains (strain K263) showed lower levels of DNA (90.9%) and amino acid (90.9%) sequence homology with the DNA and amino acid sequences of SHV-1pMON38 but had higher levels of DNA (98.7%) and amino acid (97.5%) sequence homology with the DNA and amino acid sequences of LEN-1. This strain can also be considered to produce a new variant, in this case, a LEN enzyme (EMBL accession number X04515). Strain K263 was PCR negative and showed an 8.5-kb fragment by RFLP analysis with Southern blotting. In view of these results, the sonic extracts were concentrated by lyophilization and further analyzed by IEF; a clear band with a pI of 7.1, similar to that for LEN-1, was detected. This LEN-1 and SHV-1 allelic variant (strain K263) may indicate that the chromosomal loci for SHV-1 and LEN-1 have a common ancestor; the sequence data for the enzyme from K263, designated LEN-2, will be reported in the GenBank database under accession number AY037780.
Neither differences in promoter regions nor extra triplet stop sequences were observed. Point mutations, which in some cases implied an amino acid change, were the only finding. These point mutations, when found, determined different allelic variants which, in an important number of cases, were the same for different strains (data not shown).
DISCUSSION
IEF clearly showed the high frequency of expression of the pI 7.6 β-lactamase in K. pneumoniae. Two β-lactamases combined, usually SHV-1 plus LEN-1 or TEM-1, were also frequently detected in this and other (7, 10, 20) studies. However, using a more highly sensitive method (PCR analysis), we demonstrated the chromosomal location of the SHV-1 gene in K. pneumoniae by detecting the gene in chromosomal DNA. The PCR method not only permits confirmation of IEF data but also improves the IEF technique by detecting those strains in which the gene was not expressed and, subsequently, in which the enzyme could not be detected. In some strains, poor enzyme expression can result in underestimation of the presence of the unaltered gene. However, PCR would not necessarily imply a functional expression of the gene. PCR analyses had high degrees of specificity for the detection of the SHV-1 gene and could be useful in studies that test the horizontal propagation of SHV-1.
Plasmids are frequently found in K. pneumoniae, and these are mainly self-transferable (>30 kb); however, SHV-1 is poorly transferred from K. pneumoniae to other strains, possibly because of its chromosomal location (8). In conjugation experiments we found that a plasmid-borne SHV-1 gene was transferred in a few cases, which confirmed our PCR results. The five donor strains (K75, K167, K136, K175, and K198) had MIC profiles indicating that they were highly resistant to mezlocillin, probably due to an extra copy of the gene carried on plasmids. Nevertheless, only two of these transconjugants—those carrying the smaller multicopy plasmids—were resistant to mezlocillin (TK75, TK167). The large plasmids were not identified in these particular transconjugant strains; this fact can probably be explained by the very small copy number of the large plasmids in the new recipient host. Moreover, three of the parental strains (strains K75, K167, and K136) had two large fragments, as determined by RFLP analysis with Southern blotting, indicating, in addition to the fact that the enzyme is plasmid borne, chromosomal gene duplication (8.5- and 8-kb EcoRI-specific fragments).
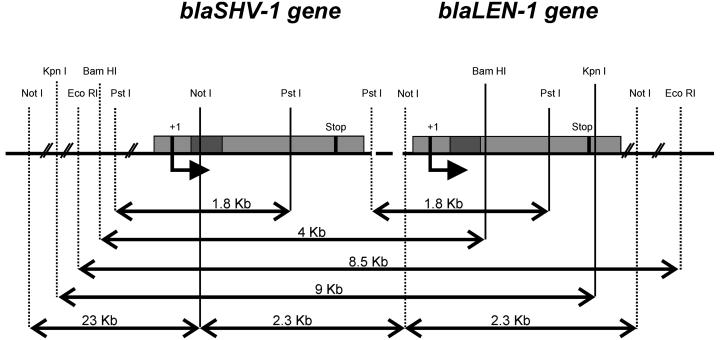
RFLP analysis with Southern blotting showed that the SHV-1 β-lactamase gene resides in a very conservative 8.5-kb EcoRI-EcoRI fragment. This region may include the LEN-1 β-lactamase gene in tandem, as deduced from the RFLP profile found in the TEM-1-producing strains, which were PCR negative, and sequencing analyses confirmed the presence of an allelic variant of the LEN-2 gene (Table 3). Additionally, on most occasions, only one hybridization fragment was found even in samples with strains expressing both enzymes. In eight strains, the genetic driving force of this particular region must have evolved to carry a double copy of the gene, as shown by the presence of two hybridization fragments by RFLP analysis with Southern blotting. As described above for the strains to which SHV-1 was transferable by plasmids, these strains with a double copy of the gene were also highly mezlocillin resistant.
DNA sequence analysis showed no differences in the promoter regions that would have explained the high or the low level of expression of the SHV-1 gene and the relationship to antibiotic resistance. Recently, a single A→C mutation at the second position of the −10 region has been reported; this mutation confers a high level of expression of the chromosomally encoded SHV-1 gene and, consequently, resistance to ceftazidime and piperacillin-tazobactam (21). No stop-codon sequences or other structures were observed that would have indicated why in some cases the enzyme is not detected by IEF. Only dispersed mutations were seen, indicative of an evolution in clusters (repetitive or conservative point mutations); in some cases, these mutations were translated into conservative amino acid changes (Table 3). Some of these allelic variants were also observed in other studies (6, 8, 16; http://www.lahey.org/studies/webt.html), but some others were new (those that produce the SHV-32, SHV-33, and LEN-2 enzymes).
In conclusion, our results strongly support the hypothesis that the ancestor of the SHV-1 β-lactamase originated from the K. pneumoniae chromosome. As a result of the RFLP and sequencing analyses, it can be postulated that the loci for the SHV-1 and LEN-1 genes are arranged in tandem (Fig. 3). However, to clearly confirm this finding, a specific analysis to test for the LEN-1 gene would clarify the occurrence of the gene and how close its locus is to that for the SHV-1 gene. It is possible that environmental antibiotic pressure selected some clustered point mutations and strains with a duplication of the gene inside the chromosome. This phenomenon was probably the first step toward transfer of the gene into a mobile element (a plasmid or transposon), where it acquired the potential for horizontal distribution to other species, in which it is plasmid encoded. In this new environment and with subsequent adaptations, the gene may confer high levels of resistance under antibiotic pressure.
FIG. 3.
Postulated gene loci for SHV-1 and LEN-1 β-lactamase genes.
ACKNOWLEDGMENTS
This work was supported by grants from the Spanish Fondo de Investigación Sanitaria (grants FIS 94/1829 and 92/170).
George A. Jacoby is acknowledged for kindly providing the pMON38 clone. We are also grateful to C. O'Hara for English revision of the manuscript.
Footnotes
Through this study, we wish to posthumously pay our last respects to Clara Roy, who was the head of the Microbiology Department for many years.
REFERENCES
- 1.Ambler R P, Coulson A F, Frére J M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tiraby G, Waley S G. A standard numbering scheme for the class A β-lactamase. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Ohta M, Kido N, Fujii Y, Komatsu T, Kato N. Close evolutionary relationship between the chromosomally encoded β-lactamase gene of Klebsiella pneumoniae and the TEM-1 β-lactamase gene mediated by R-plasmids. FEBS Lett. 1986;207:69–74. doi: 10.1016/0014-5793(86)80014-x. [DOI] [PubMed] [Google Scholar]
- 3.Babini G S, Livermore D M. Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob Agents Chemother. 2000;44:2230. doi: 10.1128/aac.44.8.2230-2230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford P A. Automated thermal cycling is superior to traditional methods for nucleic sequencing of blaSHV genes. Antimicrob Agents Chemother. 1999;43:2960–2963. doi: 10.1128/aac.43.12.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corkill J E, Cuevas L E, Gurgel R Q, Greensill J, Hart C A. SHV-27: a novel cefotaxime-hydrolysing beta-lactamase, identified in Klebsiella pneumoniae isolates from a Brazilian hospital. J Antimicrob Chemother. 2001;47:463–465. doi: 10.1093/jac/47.4.463. [DOI] [PubMed] [Google Scholar]
- 7.Fuster C, Roy C, Reig R, Raya C, Coira A. Frequency of plasmid-mediated β-lactamases in Enterobacteriaceae isolated in Spain. Eur J Clin Microbiol. 1993;12:67–69. doi: 10.1007/BF01997065. [DOI] [PubMed] [Google Scholar]
- 8.Haeggman S, Löfdahl L G, Burman S. An allelic variant of the chromosomal gene for class A β-lactamase K2, specific for Klebsiella pneumoniae, is the ancestor of SHV-1. Antimicrob Agents Chemother. 1997;41:2705–2709. doi: 10.1128/aac.41.12.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung M, Shannon K, French G. Rarity of transferable β-lactamase production by Klebsiella species. J Antimicrob Chemother. 1997;39:737–745. doi: 10.1093/jac/39.6.737. [DOI] [PubMed] [Google Scholar]
- 10.Liu P Y F, Gur D, Hall L M C, Livermore D M. Survey of the prevalence of β-lactamases amongst 1000 gram-negative bacilli isolated consecutively at the Royal London Hospital. J Antimicrob Chemother. 1992;30:429–447. doi: 10.1093/jac/30.4.429. [DOI] [PubMed] [Google Scholar]
- 11.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore D M, Williams J D. Betalactams: mode of action and mechanism of bacterial resistance. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 502–578. [Google Scholar]
- 13.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 14.Matthew M, Hedges R W, Smith J T. Types of β-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979;138:657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercier J, Levesque R C. Cloning of SHV-2, OHIO-1, and OXA-6 β-lactamases and cloning and sequencing of SHV-1 β-lactamase. Antimicrob Agents Chemother. 1990;34:1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nugent M F, Hedges R W. The nature of genetic determinants for the SHV-1 β-lactamase. Mol Gen Genet. 1979;175:239–243. doi: 10.1007/BF00397222. [DOI] [PubMed] [Google Scholar]
- 18.Petit A, Ben-Yaghlane-Bouslama H, Sofer L, Labia R. Characterization of chromosomally encoded penicillinases in clinical isolates of Klebsiella pneumoniae. J Antimicrob Chemother. 1992;29:629–638. doi: 10.1093/jac/29.6.629. [DOI] [PubMed] [Google Scholar]
- 19.Pitton J S. Mechanism of bacterial resistance to antibiotics. Rev Physiol. 1972;65:15–93. doi: 10.1007/3-540-05814-1_2. [DOI] [PubMed] [Google Scholar]
- 20.Reig R, Roy C, Hermida M, Teruel D, Coira A. A survey of β-lactamases from 618 isolates of Klebsiella spp. J Antimicrob Chemother. 1993;31:29–35. doi: 10.1093/jac/31.1.29. [DOI] [PubMed] [Google Scholar]
- 21.Rice L, Carias L, Hujer A, Bonafede M, Hutton R, Hoyen C, Bonomo R. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:362–367. doi: 10.1128/aac.44.2.362-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlaes D M, Currie-McCumber C, Hull A, Behlau I, Kron M. OHIO-1 β-lactamase is part of the SHV-1 family. Antimicrob Agents Chemother. 1990;34:1570–1576. doi: 10.1128/aac.34.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Wiley Interscience; 1988. pp. 241–245. [Google Scholar]