Abstract
The SARS-CoV-2 Omicron variant emerged in South Africa in November 2021, and has later been identified worldwide, raising serious concerns.
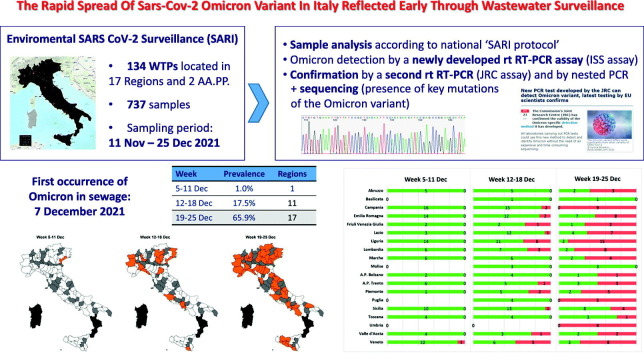
A real-time RT-PCR assay was designed for the rapid screening of the Omicron variant, targeting characteristic mutations of the spike gene. The assay was used to test 737 sewage samples collected throughout Italy (19/21 Regions) between 11 November and 25 December 2021, with the aim of assessing the spread of the Omicron variant in the country. Positive samples were also tested with a real-time RT-PCR developed by the European Commission, Joint Research Centre (JRC), and through nested RT-PCR followed by Sanger sequencing.
Overall, 115 samples tested positive for Omicron SARS-CoV-2 variant. The first occurrence was detected on 7 December, in Veneto, North Italy. Later on, the variant spread extremely fast in three weeks, with prevalence of positive wastewater samples rising from 1.0% (1/104 samples) in the week 5–11 December, to 17.5% (25/143 samples) in the week 12–18, to 65.9% (89/135 samples) in the week 19–25, in line with the increase in cases of infection with the Omicron variant observed during December in Italy. Similarly, the number of Regions/Autonomous Provinces in which the variant was detected increased from one in the first week, to 11 in the second, and to 17 in the last one. The presence of the Omicron variant was confirmed by the JRC real-time RT-PCR in 79.1% (91/115) of the positive samples, and by Sanger sequencing in 66% (64/97) of PCR amplicons.
In conclusion, we designed an RT-qPCR assay capable to detect the Omicron variant, which can be successfully used for the purpose of wastewater-based epidemiology. We also described the history of the introduction and diffusion of the Omicron variant in the Italian population and territory, confirming the effectiveness of sewage monitoring as a powerful surveillance tool.
Keywords: SARS-CoV-2, Omicron, Variant, RT-qPCR, Sewage, Wastewater-based epidemiology
Graphical abstract
1. Introduction
The SARS-COV-2 Omicron variant emerged in South Africa on 24 November 2021 and has later been identified in numerous countries worldwide. On 26 November 2021, WHO designated B.1.1.529 as a Variant of Concern, named Omicron, asking to enhance surveillance and sequencing efforts to better understand circulation of SARS-CoV-2 variants.
As of 20 January 2022, Omicron has been identified in all EU/EEA countries, and as on 30 January it was the dominant variant (accounting for >50% of sequenced viruses) in 19 of the 22 EU/EEA countries with adequate sequencing volume, with 268.835 Omicron cases reported (https://www.ecdc.europa.eu/en/covid-19/country-overviews).
In Italy, the first Omicron clinical case was described on 22 November in Milan, in an Italian man who had returned on 11 November from Mozambique (first Omicron sequence submitted in GISAID with accession ID EPI_ISL_6777160). Later on, the highly contagious Omicron variant spread quickly. As on 8 February 2022, a total of 8588 omicron sequences have been submitted to GISAID from Italy, from 16 of the 19 Italian Regions: namely Abruzzo (576), Basilicata (10), Calabria (3), Campania (1319), Emilia Romagna (664), Friuli Venezia Giulia (528), Lazio (730), Liguria (60), Lombardia (639), Piemonte (582), Puglia (280), Sicilia (2007), Toscana (58), Trentino Alto Adige (152), Umbria (425), Valle d'Aosta (16), Veneto (538), and from the two autonomous provinces (AA.PP.) of Bolzano and Trento. No sequences were available for the Regions Marche, Molise, and Sardegna.
Wastewater testing for SARS-CoV-2 has emerged as a valuable warning system for its circulation in the population and tracking the presence of novel variants in sewage is recognized as a key tool to control the spread of SARS-CoV-2 and ensure public health response. The European Commission has recommended to the EU member states to set up the monitoring of SARS-CoV-2 in wastewater by 1st October 2021 (Rec. 2021/472 of 17 March 2021), focusing on the emergence and spread of SARS-CoV-2 variants. Italy has been monitoring urban sewage since July 2020, at the beginning as a pilot study, and then, since October 2021, in the framework of the EU wastewater surveillance. The periodic detection of SARS-CoV-2 variants and the analysis of genetic diversity is part of the surveillance. In March and May 2021 Variants of Concern (VoCs) and Variants of Interest (VoI) have been identified in urban sewage in Italy (La Rosa et al., 2021a; La Rosa et al., 2021b). Since October 2021 regular national “flash surveys” - i.e. periodic sampling campaigns, held simultaneously in all regions over a short period of time - are conducted in Italy, aimed at assessing the diversity of SARS-CoV-2 in wastewater (see Acque reflue - ISS for the published monthly reports). The reports related to samples collected between 1 and 5 November (https://doi.org/10.5281/zenodo.5985196) and 28 November and 3 December 2021 (https://doi.org/10.5281/zenodo.5985276) showed, as expected, the predominance of the Delta variant associated to significant variability, and no presence of the Omicron variant by analysis of raw data of Next Generation amplicon sequencing.
As soon as the detection of the SARS-CoV-2 Omicron variant was first reported in Italy, we designed a real-time RT-PCR assay to enable rapid identification of the novel VoC, targeting characteristic mutations in the spike gene, and evaluated its specificity and sensitivity against other SAR-CoV-2 variants. In parallel, we tested an assay developed by the European Commission, Joint Research Centre (JRC) designed in silico for the detection of Omicron targeting a spike region with a unique cluster of mutations (Petrillo et al., 2021). The JRC invited control laboratories worldwide to validate it in vivo on clinical samples (https://joint-research-centre.ec.europa.eu/jrc-news/efficient-tracing-omicron-new-pcr-test-2021-12-13_en). Subsequently, both the assays were used to test sewages collected throughout Italy in the period between 11 November (date of entry in Italy of the first Omicron case in Italy) and 25 December 2021, to investigate how the Omicron variant spread over time and geographically. Moreover, Sanger sequencing was performed on positive samples, to validate results and study sequence variability.
2. Material and methods
2.1. Real-time RT-PCR design and evaluation of specificity and sensitivity
A new real-time RT-PCR assay targeting the spike region of the Omicron variant was designed using the Primer3Plus software (Primer3Plus (bioinformatics.nl). The spike region of Omicron harbours a number of mutations, some of which are in common with other variants (e.g. the deletion 69/70 shared with Alpha, T95I and G142D shared with Delta, N501Y shared with Alpha, Beta and Gamma), while other are unique for Omicron, and therefore suitable for the design of specific assays. We selected the region harbouring mutations H655Y, N679K and P681H, which are each present in >98.9% of the Omicron variant sequences (Supplementary materials, Fig. S1) as on 8 January 2022. Only mutation P681H is in common with the Alpha variant which was de-escalated by the European Centre for Diseases Control on 23 September 2021 due its drastically reduced circulation in Europe.
For the optimization of the assay different reaction conditions were evaluated: primer/probe concentrations, annealing temperatures (58 °C and 60 °C), and real time RT-PCR reagents (UltraSense One-Step qRT-PCR System by Invitrogen and AgPath-ID One-Step RT-PCR Reagents by Applied Biosystems). The newly designed assay was tested on a RNA sample extracted from a nasopharyngeal swab of a patient resident in the region of Apulia, collected on 7 December 2021, and kindly provided by Dr. Maria Chironna, Department of Biomedical Sciences and Human Oncology, School of Medicine and Surgery, University of Bari Aldo Moro. This sample had been previously characterized as Omicron variant by whole genome sequencing and submitted to GISAID with ID EPI_ISL_7565149. Viral RNA was quantified using a previously described real-time RT-qPCR (La Rosa et al., 2021c; Petrillo et al., 2021) and was standardized at 6 × 102 genome copies (g.c.)/μl for the use as a positive control in PCR runs and for sensitivity assessment. For the latter, the standardized RNA was used to prepare serial dilutions which were analysed in eight replicates to calculate the limit of detection (LOD50) of the assay on pure target. The same dilutions were also used to spike nucleic acids extracted from wastewater samples collected in July 2021 (i.e. before the emergence of the Omicron variant) to assess the real-time assay LOD50 on environmental samples. Calculation were performed according to Wilrich and Wilrich (2009), with the spreadsheet available at https://www.wiwiss.fu-berlin.de/fachbereich/vwl/iso/ehemalige/wilrich/index.html.
The specificity of the assay was evaluated using RNAs extracted from clinical samples characterized as Alpha, Beta, Gamma, and Delta variants (sample IDs 7652, 7584, 7654 and 8019, respectively), kindly provided by Dr. Paola Stefanelli at the Department of Infectious Disease at the Italian National Institute for Health (ISS). All RNAs had been previously standardized at a concentration of 1 × 103 genome copies (g.c.)/μl. Moreover, the in-house assay was evaluated for specificity using the European Virus Archive – EVA GLOBAL (EVAg) panel, consisting of RNAs from different Alfa- and Beta- coronaviruses, namely HCoV-NL63, HCoV-229E, HCoV-OC43, MERS-CoV, SARS-CoV and the prototype strain of SARS-CoV-2 (kindly provided by the Erasmus University Medical Center (Rotterdam, The Netherlands).
Another real-time RT-qPCR assay, developed by the European Commission, Joint Research Centre (JRC) (https://doi.org/10.5281/zenodo.5747872), and found to be able to successfully detect the Omicron variant in silico, was tested to confirm results and compare the performance of the two assays as the JRC invited control laboratories worldwide to validate the assay in vivo on field samples.
Primers and probes used in the present study are shown in Table 1 .
Table 1.
Primer and probes used in the present study, targeting the spike region.
| PCR ID | Primer/probe name and orientation | Nucleotide sequence | Position in the SARS-CoV-2 Omicron genomea | Amplicon size (bps) | Targeted mutations |
|---|---|---|---|---|---|
| 999 | 2356_Omicron_fw | 5′-GGCTGTTTAATAGGGGCTGAATA-3′ | 23,429–23,451 | 141 | H655Y, N679K, P681H |
| 2357_Omicron_rev | 5′-GGCAATGATGGATTGACTAGC-3′ | 23,569–23,549 | |||
| 2358_Omicron_probe | 5′-FAM-TCAGACTCAGACTAAGTCTCATCGG-BHQ1-3′ | 23,509–23,533 | |||
| OmMetb | Omt-F | 5′-AACAAACCTTGTAATGGTGTTGC-3′ | 22,916–22,938 | 138 | S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H |
| Omt-R | 5′-TGCTGGTGCATGTAGAAGTTC-3′ | 23,053–23,033 | |||
| Omt-P | 5′-FAM-GATCATATAGTTTCCGACCCACTTATGGTG TTGGTC-BHQ1-3′ | 22,965–23,000 | |||
| 995 (1st cycle) |
2351_fw | 5′-CACGCCTATTATAGTGCGTGA-3′ | 22,102–22,122 | 693 | G339D, S371L, S373P, S375Fc |
| 2347_rev | 5′-CAAGCTATAACGCAGCCTGT-3′ | 22,794–22,775 | |||
| 996 (nested) |
2352_fw | 5′-TTCGGCTTTAGAACCATTGGTAG-3′ | 22,147–22,169 | 577 | |
| 2349_rev | 5′-TGGAGCGATTTGTCTGACTTC-3′ | 22,723–22,703 |
Position related to GISAID sequence accession ID: EPI_ISL_6913995.
Petrillo et al., 2021 (https://doi.org/10.5281/zenodo.5747872); probe quencher QSY was replaced with BHQ1.
List restricted to mutations with a prevalence in the lineage of at least 75%, data from https://outbreak.info/compare-lineages?pango=Omicron&gene=S&threshold=75&nthresh=1&sub=false&dark=true
2.2. Real-time RT-PCR assay on wastewater samples
Real-Time RT-PCR was performed on 737 wastewater samples (see Supplementary Material, Table S2) collected throughout Italy from 134 wastewater treatment plants – WTPs – located in 17 Regions and 2 AA.PP. (Supplementary materials, Fig. S2). These samples were collected between 11 November - the day of the incoming flight of the first recognized case of Omicron variant in Italy - and 25 December 2021, as a part of the SARS-CoV-2 surveillance established under Rec. 2021/472 of 17 March 2021. Of these, 123 samples collected in the period 28 November – 3 December 2021, had already been analysed in the monthly “flash surveys” on SARS-CoV-2 variants by Sanger and NGS (Covid-19: flash survey sulle acque reflue - dicembre 2021). Samples were collected, registered, processed for virus concentration and subjected to RNA extraction by the members of the SARS-CoV-2 environmental surveillance network in Italy (SARI network). The same reference analytical protocol (SARI protocol rev. 3, https://doi.org/10.5281/zenodo.5758725) was adopted by all network members. Briefly, 24 h composite sewage sample was concentrated by PEG precipitation followed by centrifugation following the method described by Wu and collaborators (Wu et al., 2020) with minor modifications (sample pasteurization at 56 °C per 30 min and removal of solid debris before precipitation through centrifugation at 4500 ×g per 30 min). RNA was extracted by magnetic silica beads and subsequently purified by the OneStep PCR Inhibitor Removal Kit (Zymo Research, CA, USA). RT-qPCR testing for SARS-CoV-2 was conducted according to previously published protocols (La Rosa et al., 2021c; Pierri et al., 2021) and quality insurance controls (process control virus, inhibition control) were included in the process to assess viral recovery and PCR inhibition. Purified RNAs were then shipped in dry ice to the Department of Environment and Health at ISS. Detection of the Omicron variant by real-time PCR was carried out with two different protocols:
-
a)
Newly designed assay (PCR ID_999 assay)
Following optimization of conditions, the RT-PCR mix (25 μL total volume) was prepared using the AgPath-ID One-Step RT-PCR Reagents (Applied Biosystems, MA, USA), and 5 μL of sample RNA were added to reactions containing 1× buffer, 1 μL of RT-PCR enzyme mix, 1.67 μL of detection enhancer. Primer/probe (Table 1) concentrations were as follow: 500 nM, 900 nM, and 250 nM of primer 2356_Omicron_fw, 2357_Omicron_rev, and probe 2358_Omicron_probe, respectively. Amplification conditions included reverse transcription for 30 min at 50 °C, inactivation for 5 min at 95 °C and 45 cycles of 15 s at 95 °C and 30 s at 60 °C.
-
b)
JRC assay (OmMet assay)
As no PCR conditions were specifically provided in the reference publication, the reaction mix was prepared as for the PCR ID_999 and the same thermal profile was adopted. Amplification with the JRC assay was performed for confirmation purpose on samples testing positive by the real-time PCR assay ID_999.
All real-time reactions were run on a QuantStudio 12 K Flex (Applied Biosystems). Samples with Ct values <40 were considered positive; samples with Ct values >40 were considered uncertain but were retained for nested RT-PCR testing.
A summary of the characteristics of the samples testing positive by the two real-time PCRs is shown in Table 2 .
Table 2.
Summary of the positive samples, including geographic region and sampling date, WTPs data, SARS-CoV-2 concentrations, results of the two real-time assays and availability of sequencing data.
| ID SARI | Region | Province | Sampling Date | WTP | Equivalent inhabitants | SARS-CoV-2 c.g./L | rt RT-PCR ISS (Ct) |
rt RT-PCR JRC (Ct) | Sequencing data |
|---|---|---|---|---|---|---|---|---|---|
| 6440 | Abruzzo | AQ | 21/12/21 | L'Aquila - Pile | 48,000 | <LOD | 37,29 | 38,70 | No |
| 6437 | PE | 21/12/21 | Montesilvano - Villa Carmine | 140,000 | <LOD | >40 | 39,64 | No | |
| 6439 | TE | 21/12/21 | Teramo - Villa Pavone | 41,824 | <LOD | 35,82 | 37,08 | No | |
| 6407 | Campania | AV | 21/12/21 | Manocalzati | 140,000 | 4,4E+03 | 37,63 | – | Yesa |
| 6351 | CS | 16/12/21 | Villa Literno | 631,714 | <LOD | 38,68 | 38,98 | Yes | |
| 6402 | CS | 21/12/21 | Area Casertana | 370,769 | 2,0E+03 | 38,55 | 39,57 | Yes | |
| 6409 | CS | 21/12/21 | Villa Literno | 631,714 | 1,2E+04 | 35,30 | 37,86 | Yesa | |
| 6348 | NA | 16/12/21 | Napoli OVEST - ex ingr. Camaldoli | 250,000 | 6,8E+03 | 36,13 | 39,46 | Yes | |
| 6401 | NA | 21/12/21 | Area Nolana | 400,000 | 1,3E+04 | 34,14 | 37,72 | No | |
| 6404 | NA | 21/12/21 | Napoli EST | 1,750,000 | 1,3E+04 | 34,78 | – | Yesa | |
| 6406 | NA | 21/12/21 | Napoli OVEST - ex ingr. Camaldoli | 250,000 | 2,4E+04 | 34,78 | 36,19 | Yes | |
| 6405 | NA | 21/12/21 | Napoli OVEST - ingr. Principale | 950,000 | 2,4E+04 | 33,88 | 36,42 | Yesa | |
| 6403 | SA | 21/12/21 | Nocera Superiore | 299,121 | 2,2E+04 | 35,91 | 36,93 | Yes | |
| 6408 | SA | 21/12/21 | Salerno | 700,000 | 8,3E+03 | 35,89 | 37,59 | Yesa | |
| 6187 | Emilia-Romagna | BO | 13/12/21 | IDAR Bologna | 800,000 | 2,5E+05 | 38,01 | 38,51 | Yes |
| 6244 | BO | 15/12/21 | IDAR Bologna | 800,000 | 1,1E+05 | 37,24 | 38,95 | Yes | |
| 6372 | BO | 20/12/21 | IDAR Bologna | 800,000 | 2,3E+05 | 34,95 | 37,04 | Yesa | |
| 6453 | BO | 22/12/21 | IDAR Bologna | 800,000 | 1,4E+04 | 32,99 | 34,89 | Yesa | |
| 6375 | FC | 21/12/21 | Forlì | 250,000 | 1,2E+05 | 37,31 | – | No | |
| 6373 | MO | 20/12/21 | Naviglio | 500,000 | 4,1E+04 | 38,02 | 39,84 | Yes | |
| 6454 | MO | 22/12/21 | Naviglio | 500,000 | 4,0E+04 | 36,48 | 36,62 | Yesa | |
| 6383 | PR | 22/12/21 | Parma Ovest | 168,000 | 9,0E+03 | 36,29 | – | Yesa | |
| 6382 | RE | 22/12/21 | Reggio Emilia Mancasale | 280,000 | 1,1E+03 | 36,21 | 38,64 | No | |
| 6456 | RN-FC | 22/12/21 | S. Giustina | 560,000 | 8,2E+04 | 34,72 | 36,76 | Yesa | |
| 6449 | Friuli-Venezia Giulia | PN | 21/12/21 | Cordenons | 15,000 | 2,3E+05 | 35,81 | 37,75 | Yesa |
| 6312 | UD | 14/12/21 | Udine | 200,000 | 8,4E+04 | >40 | – | Yes | |
| 6450 | UD | 21/12/21 | Udine | 200,000 | 4,9E+04 | 37,43 | – | No | |
| 6259 | Lazio | RM | 14/12/21 | Roma Est (linea 1 + linea 2) | 900,000 | 4,0E+04 | 37,91 | 39,31 | Yes |
| 6280 | RM | 20/12/21 | Anzio Colle Cocchino | 75,000 | 9,4E+03 | 37,50 | 39,57 | Yes | |
| 6283 | RM | 20/12/21 | Pomezia Capoluogo | 60,000 | 4,4E+03 | 37,56 | 37,97 | Yes | |
| 6285 | RM | 20/12/21 | Ponte Lucano di Guidonia | 50,000 | 9,3E+03 | 35,86 | 39,78 | Yesa | |
| 6284 | RM | 20/12/21 | Velletri La Chiusa | 36,700 | 1,1E+04 | 34,24 | 37,37 | Yesa | |
| 6732 | RM | 22/12/21 | Fregene | 76,000 | 1,5E+05 | 37,57 | – | Yes | |
| 6731 | RM | 22/12/21 | Roma Ostia | 350,000 | 1,3E+06 | 33,26 | 34,99 | Yesa | |
| 6730 | RM | 22/12/21 | Roma Sud | 1,100,000 | 1,1E+05 | 35,06 | 36,94 | Yes | |
| 6410 | Liguria | GE | 22/12/21 | Darsena | 160,000 | 1,2E+05 | 35,59 | 36,01 | No |
| 6234 | GE | 14/12/21 | Genova | 60,000 | 4,9E+04 | 34,41 | 38,57 | Yesa | |
| 6387 | GE | 21/12/21 | Pegli | 40,000 | 2,2E+05 | 32,58 | 35,88 | Yesa | |
| 6386 | GE | 21/12/21 | Punta Vagno | 250,000 | 1,0E+05 | 36,00 | 37,85 | Yes | |
| 6400 | GE | 22/12/21 | Punta Vagno | 250,000 | 1,1E+05 | 34,04 | 37,61 | No | |
| 6389 | GE | 21/12/21 | Quinto | 60,000 | 2,5E+05 | 31,87 | 34,69 | Yesa | |
| 6390 | GE | 21/12/21 | Rapallo | 90,000 | 1,8E+05 | 33,26 | 36,27 | Yesa | |
| 6239 | GE | 14/12/21 | Sestri Ponente | 130,000 | 4,1E+04 | 36,10 | 39,49 | No | |
| 6391 | GE | 21/12/21 | Sestri Ponente | 130,000 | 1,1E+05 | 35,61 | 37,84 | Yes | |
| 6240 | GE | 14/12/21 | Sturla | 60,000 | 1,1E+05 | 38,04 | 38,77 | Yes | |
| 6392 | GE | 21/12/21 | Sturla | 60,000 | 2,6E+05 | 33,60 | 35,19 | Yesa | |
| 6385 | GE | 16/12/21 | Valpolcevera | 160,000 | 5,7E+04 | 38,11 | 39,17 | No | |
| 6397 | GE | 22/12/21 | Valpolcevera | 160,000 | 2,0E+05 | 33,39 | 35,82 | No | |
| 6388 | GE | 21/12/21 | Voltri | 62,000 | 9,4E+04 | 34,19 | 38,89 | Yesa | |
| 6396 | IM | 21/12/21 | Camisano | 40,000 | 1,1E+05 | 36,83 | 37,86 | Yesa | |
| 6393 | IM | 21/12/21 | Sanremo - Capo Verde | 75,000 | 2,5E+05 | 36,47 | 38,56 | Yesa | |
| 6242 | SP | 14/12/21 | Camisano | 40,000 | 1,5E+04 | 36,76 | – | No | |
| 6243 | SP | 14/12/21 | Silea | 17,500 | 3,1E+04 | 34,76 | 38,29 | Yes | |
| 6398 | SP | 21/12/21 | Silea | 17,500 | 1,5E+05 | 32,82 | 34,29 | Yesa | |
| 6399 | SP | 21/12/21 | Stagnoni | 82,000 | 1,9E+05 | 34,44 | 36,18 | Yesa | |
| 6394 | SV | 20/12/21 | Savona | 280,000 | 3,0E+05 | 32,19 | 34,98 | Yesa | |
| 6430 | Lombardia | BS | 21/12/21 | Verziano | 296,000 | 9,5E+02 | 36,15 | – | Yes |
| 6431 | BS | 22/12/21 | Verziano | 296,000 | 3,5E+02 | 37,37 | 39,52 | No | |
| 6327 | CO-LC-MI-MB | 20/12/21 | Monza San Rocco | 600,000 | 5,3E+04 | 34,48 | 38,94 | Yes | |
| 6371 | CO-LC-MI-MB | 22/12/21 | Monza San Rocco | 600,000 | 3,5E+03 | 35,15 | – | Yes | |
| 6659 | MI | 12/12/21 | Milano Nosedo | 1,250,000 | 5,1E+04 | 37,04 | – | Yesa | |
| 6664 | MI | 14/12/21 | Milano San Rocco | 1,036,000 | 5,7E+04 | 37,43 | 37,07 | Yesa | |
| 6661 | MI | 19/12/21 | Milano Nosedo | 1,250,000 | 4,5E+04 | 35,69 | 37,18 | Yesa | |
| 6665 | MI | 19/12/21 | Milano San Rocco | 1,036,000 | 2,2E+04 | 37,28 | 38,60 | No | |
| 6251 | MI-VA | 14/12/21 | Canegrate | 137,950 | 6,9E+03 | 36,36 | 37,58 | Yesa | |
| 6445 | MI-VA | 21/12/21 | Canegrate | 137,950 | 3,3E+04 | 31,54 | 34,67 | Yesa | |
| 6446 | VA | 21/12/21 | Varese | 74,402 | 2,0E+04 | 33,43 | 36,91 | Yesa | |
| 6358 | Marche | AN | 21/12/21 | Camerano | 33,000 | 6,4E+03 | 38,81 | 39,54 | Yes |
| 6356 | AN | 21/12/21 | Zipa | 100,000 | 8,3E+03 | 37,49 | – | Yesa | |
| 6354 | PU | 21/12/21 | Ponte Metauro | 60,000 | 1,1E+04 | 37,61 | 39,64 | No | |
| 6355 | PU | 21/12/21 | Ponte Sasso | 18,000 | 4,9E+03 | 35,91 | – | Yes | |
| 6325 | P.A. Bolzano | BZ | 20/12/21 | IDA Merano | 356,520 | 6,5E+03 | 37,62 | – | Yes |
| 6117 | P.A. Trento | TN | 13/12/21 | Rovereto | 95,000 | 2,1E+05 | 36,58 | – | Yesa |
| 6314 | TN | 20/12/21 | Trento sud | 100,000 | 2,2E+05 | 36,22 | 39,94 | Yesa | |
| 6316 | TN | 22/12/21 | Trento nord | 120,000 | 1,5E+05 | 34,86 | 36,95 | Yesa | |
| 6317 | TN | 22/12/21 | Trento sud | 100,000 | 1,6E+05 | 34,95 | 36,65 | Yesa | |
| 6148 | Piemonte | AL | 15/12/21 | Alessandria | 110,000 | 1,3E+02 | 38,53 | – | No |
| 6334 | AL | 24/12/21 | Alessandria | 110,000 | 8,5E+03 | 33,46 | 36,99 | Yesa | |
| 6289 | BI | 20/12/21 | Biella Sud | 53,000 | 2,8E+03 | 37,20 | – | Yes | |
| 6333 | CN | 23/12/21 | Cuneo | 185,000 | 6,3E+03 | 35,93 | 36,54 | Yesa | |
| 6287 | NO | 20/12/21 | Novara | 184,000 | 1,2E+03 | 37,52 | 38,61 | Yesa | |
| 6144 | TO | 15/12/21 | Castiglione Torinese | 1,934,099 | 2,9E+02 | >40 | – | Yesa | |
| 6332 | TO | 22/12/21 | Castiglione Torinese | 1,934,099 | 9,1E+03 | 34,98 | 36,70 | Yesa | |
| 6424 | Puglia | BA | 20/12/21 | Bari Est | 461,394 | 3,8E+03 | 37,25 | – | Yes |
| 6425 | BA | 20/12/21 | Bari Ovest | 360,000 | 2,4E+03 | 36,50 | 39,46 | Yes | |
| 6426 | BA | 20/12/21 | Bitonto | 79,332 | 3,8E+03 | 37,22 | 39,78 | Yes | |
| 6427 | FG | 21/12/21 | Cerignola | 56,355 | 4,7E+04 | 37,36 | 38,64 | Yesa | |
| 6428 | FG | 21/12/21 | Foggia | 208,000 | 7,9E+03 | >40 | 39,88 | Yesa | |
| 6306 | Sicilia | AG | 20/12/21 | Agrigento | 55,000 | 4,1E+04 | 36,52 | – | Yes |
| 6328 | CL | 21/12/21 | Caltanissetta | 76,700 | 1,1E+05 | 38,02 | >40 | Yesa | |
| 6308 | PA | 21/12/21 | Acqua dei Corsari | 314,973 | 3,5E+04 | 38,16 | 37,07 | Yesa | |
| 6310 | PA | 21/12/21 | Bagheria | 75,000 | 3,7E+04 | 34,90 | 37,74 | Yes | |
| 6112 | RG | 14/12/21 | Vittoria C.da Mendolilli | 55,000 | 2,9E+03 | 39,95 | – | Yesa | |
| 6364 | Toscana | PI | 20/12/21 | Pisa Nord (San Jacopo) | 52,000 | 2,8E+04 | 37,73 | 39,50 | Yesa |
| 6290 | Umbria | PG | 20/12/21 | Pian della Genna | 90,000 | 4,0E+04 | 34,20 | 36,00 | Yesa |
| 6330 | PG | 22/12/21 | Foligno Casone | 90,000 | 1,3E+04 | 34,92 | 37,10 | Yesa | |
| 6329 | PG | 22/12/21 | Pian della Genna | 90,000 | 6,8E+04 | 32,78 | 34,84 | Yesa | |
| 6331 | TR | 22/12/21 | Terni | 150,000 | 2,5E+04 | 33,41 | 35,33 | Yes | |
| 6113 | Valle d'Aosta | AO | 12/12/21 | La Salle | 60,000 | 8,6E+03 | 37,73 | 39,60 | No |
| 6319 | AO | 19/12/21 | La Salle | 60,000 | 8,1E+03 | 35,89 | 37,11 | Yesa | |
| 6413 | AO | 21/12/21 | La Salle | 60,000 | 1,1E+04 | 35,99 | 39,21 | Yesa | |
| 6302 | Veneto | PD | 21/12/21 | Padova Ca' Nordio - zip | 98,500 | 4,1E+04 | 37,62 | – | Yesa |
| 6339 | TV | 21/12/21 | Treviso | 70,000 | 3,0E+04 | 34,24 | 39,53 | Yesa | |
| 6007 | VE | 07/12/21 | Venezia Fusina | 400,000 | 1,1E+04 | 38,70 | – | Yes | |
| 6146 | VE | 14/12/21 | Venezia Fusina | 400,000 | 2,3E+04 | 39,01 | 37,60 | Yesa | |
| 6340 | VE | 21/12/21 | Venezia Fusina | 400,000 | 2,6E+04 | 34,08 | 36,21 | Yesa | |
| 6432 | VE | 23/12/21 | Venezia Fusina | 400,000 | 1,5E+04 | 33,43 | 35,70 | Yesa | |
| 6235 | VR | 16/12/21 | Verona-collettore 1 M | 82,000 | 4,7E+04 | 32,04 | 35,89 | Yesa | |
| 6236 | VR | 16/12/21 | Verona-collettore 3 M | 102,000 | 2,4E+04 | 34,41 | 37,03 | Yesa | |
| 6238 | VR | 16/12/21 | Verona-collettore 8 M | 226,000 | 1,5E+04 | 34,82 | 37,94 | Yesa | |
| 6433 | VR | 23/12/21 | Verona-collettore 1 M | 82,000 | 2,0E+04 | 31,95 | 34,47 | Yesa | |
| 6434 | VR | 23/12/21 | Verona-collettore 3 M | 102,000 | 2,4E+04 | 31,84 | 34,82 | Yesa | |
| 6435 | VR | 23/12/21 | Verona-collettore 8 M | 226,000 | 1,7E+04 | 33,63 | 37,70 | Yesa | |
| 6147 | VI | 15/12/21 | Vicenza Casale | 92,000 | 1,6E+04 | 37,01 | – | Yes | |
| 6341 | VI | 21/12/21 | Vicenza Casale | 92,000 | 7,9E+03 | 34,12 | 37,30 | Yesa |
Presence of mutations characteristic of the Omicron variant.
2.3. Nested RT-PCR and sequencing
A newly designed nested RT-PCR assay targeting the spike protein and designed PCR ID_995/996 (first cycle/nested reaction), was used for further confirmation and to study viral genetic diversity though sequencing. The Omicron's Spike protein has several amino acid mutations (substitutions and deletions) in the selected region that are distinct compared to other VoCs (Kannan et al., 2022). Specifically, the nested assay amplifies a region of 577 bps, covering the amino acids 220 to 412 of the spike protein.
First-strand cDNA was synthesized using Super Script IV Reverse Transcriptase (ThermoFisher Scientific) according to the manufacturer's instructions, using reverse primer 2347. First PCR reaction was performed with 4 μL of cDNA and 1 μL of each primer (10 μM) in a final volume of 25 μL (Kit Platinum SuperFi Green PCR Master Mix, Thermo). The PCR conditions were as follows: 98 °C for 2 min, followed by 35 cycles at 98 °C for 10 s, 62 °C for 30 s, and 72 °C for 1 min and a final extension at 72 °C for 5 min. Nested PCR was performed using 2 μL of the first PCR product under the same conditions, with the exception of the annealing temperature raised at 63 °C. To avoid false-positive results, standard laboratory precautions were taken.
The PCR products were visualised by gel electrophoresis purified using a Montage PCRm96 Microwell Filter Plate (Millipore, Billerica, MA, USA), and were then sequenced on both strands (BioFab Research, Rome, Italy). Mutations were detected using the CoVsurver mutation Analysis of hCoV-19 implemented in GISAID (GISAID - CoVsurver mutations App).
Sequences were submitted to GenBank under the accession numbers ON196927-ON197019.
3. Results
3.1. Real-time RT-PCR design and evaluation of specificity and sensitivity
The optimization of PCR conditions showed that the RT-PCR reagents used had a significant impact on the performance of both real-time PCR assays, with ΔCt values between the results provided by the different amplification systems equal or above 5 cycles (see Supplementary Material, Fig. S3). Following optimization, the standardized Omicron RNA (6 × 102 g.c./μl) was successfully amplified by both the newly designed PCR ID_999 (ISS assay) and the OmMet PCR (JRC assay), although higher Ct values were obtained for the latter (30.15 ± 0.40 vs. 27.38 ± 0.66, Supplementary Material, Table S1). The ISS real-time RT-PCR provided a LOD50 of 0.28 g.c./μl of tested nucleic acids (1.4 g.c./reaction) on pure Omicron RNA, and of 2.67 g.c./μl (13.3 g.c./reaction) on nucleic acids extracted from sewage samples and spiked with the Omicron variant. As for the specificity of the assays, no amplification was obtained for the six Coronavirus RNAs included in the EVAg Coronavirus panel, but a partial cross-reactivity was observed with Alpha and Gamma variants which were however amplified at a lower efficiency (ΔCt of 5.02 and 3.78, respectively, compared to Omicron; Supplementary Material, Fig. S4).
3.2. Omicron detection by real-time RT-PCR in wastewater samples
None of the 355 samples collected between 11 November and 4 December tested positive for the Omicron variant. Of the 382 samples collected from 5 to 25 December, 111 tested positive (Ct values <40) for Omicron with the ISS assay (PCR ID_999) and 4 provided weak amplification signals (Ct values >40). An example of the results obtained is reported in Supplementary Materials, Fig. S5.Of the 115 positive samples, 91 (79.1%) were confirmed positive for the Omicron variant using the JRC assay (OmMet). Positive samples displayed an average Ct value (±SD) of 35.97 ± 2.17 (range 31.54–42.76) and 37.59 ± 1.58 (range 34.29–40.26) in the ISS and JRC assay, respectively, with a statistically significant difference of the two values (paired t-test, p = 0.043; Supplementary Materials, Fig. S6). Most of the discrepant results were associated to samples with high Ct values and, presumably, a lower viral concentration.
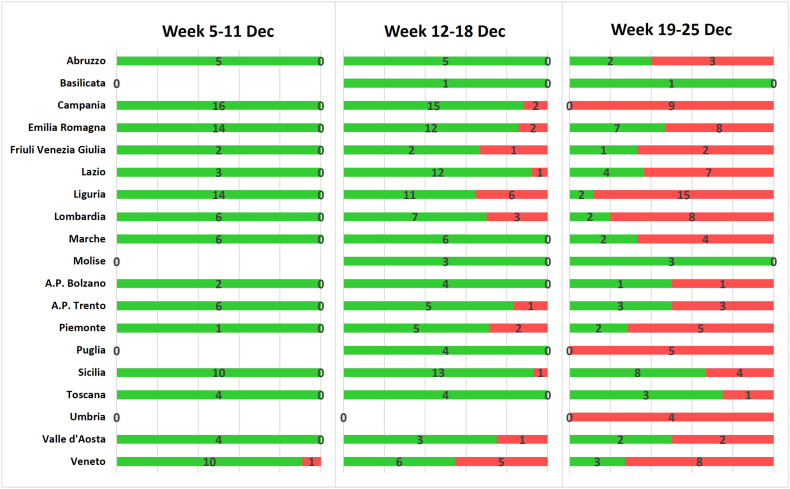
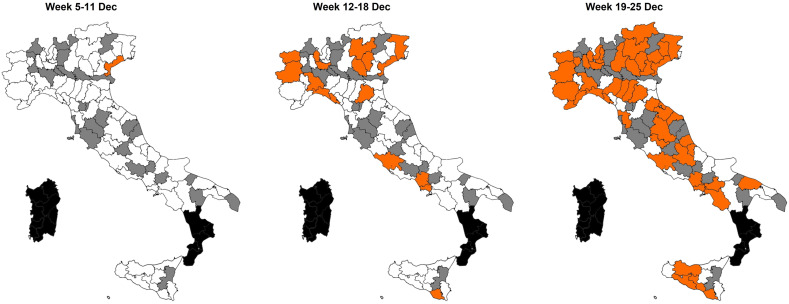
The first occurrence of the Omicron variant in wastewater in Italy was detected in a sample collected in Venice (Region of Veneto) on 07 December 2021. Later on, a rapid spread of the variant was observed (Fig. 1 ) as follow: 1/104 samples (1.0%) and one Region in the week 5–11 December; 25/143 samples (17.5%) and 11 Regions/AA.PP. in the week 12–18 December; and 89/135 samples (65.9%) and 17 Regions/AA.PP. in the week 19–25 December (Supplementary Material, Fig. S7). In the last week, the Omicron variant was detected in 100% of the samples collected in the Regions of Campania, Puglia, Umbria (Fig. 2 ) and in the majority of the samples in other Regions (e.g. Lazio, Liguria, Lombardia, Piemonte, Veneto).
Fig. 1.
Number of Omicron positive samples detected in the three weeks of December (05.12.2021–25.12.2021) divided by Region/A.P.
Samples negative for Omicron variant are reported in green; positive samples are represented in red.
Fig. 2.
Geographic spread of the Omicron positive samples during the three weeks of December (05.12.2021–25.12.2021) by province
Regions in which wastewater surveillance was not yet activated at the time of the study are reported in black. Provinces for which no samples were tested in the study are reported in grey. Detection of the Omicron variant is reported in orange.
3.3. Characterization of Omicron-positive samples by Sanger sequencing
Overall, 97 of the 115 Omicron-positive wastewater samples (84.3%) were successfully amplified with the nested RT-PCR assay targeting the S protein, and subsequently sequenced by Sanger sequencing. Of these, 64 samples showed mutations associated with the Omicron variant, three samples showed amino acid substitutions not discriminatory for variant assignment, and 4 were unreadable due to mixed electropherograms (Table 3 ). Moreover, 26 sequences showed no mutations.
Table 3.
Mutations detected by nested RT-PCR amplification followed by Sanger sequencing.
| ID SARI | Mutations | Suggested variant |
|---|---|---|
| 6236 | G339D | Omicron |
| 6383 | S375F | |
| 6339, 6408 | G339D, S371P | |
| 6333, 6404 | S371L, S375F | |
| 6330 | T323I, G339D | |
| 6393 | G257V, G339D, S371L, S373P, S375F | |
| 6234, 6235, 6372, 6387, 6388, 6394, 6407, 6409, 6433, 6435, 6449 | G339D, R346K, S371L, S373P, S375F (BA.1.1) | |
| 6117, 6144, 6238, 6251, 6284, 6285, 6314, 6316, 6317, 6340, 6356, | G339D, S371F, S373P, S375F | |
| 6146, 6287, 6290, 6308, 6319, 6328, 6329, 6334, 6341, 6364, 6389, 6390, 6392, 6396, 6398, 6399,6405, 6413,6427, 6432, 6434, 6445, 6446, 6453, 6454, 6456,6659, 6661, 6664 | G339D, S371L, S373P, S375F | |
| 6428 | G339D, S371F, S375F | |
| 6112, 6302, 6731 | G339D, S371P, S375F | |
| 6332 | G339D, S373P, S375F | |
| 6007, 6147, 6240, 6243, 6244, 6259, 6280, 6283, 6289, 6306, 6310, 6312, 6325, 6348, 6351, 6358, 6373, 6386, 6391, 6403, 6406, 6425, 6426, 6430, 6730, 6732 | No mutations | Unassigned |
| 6187, 6327, 6331, 6371 | Mixed electropherogram | |
| 6355, 6424 | P251L | |
| 6402 | P251L, S371P |
Sequence analysis showed a high degree of sequence variability within the Omicron variant sequences. In total, 10 amino acid substitutions were detected in the 577-bps fragment, with 12 different combinations (Table 3). The most frequent combination of mutations included the panel “G339D, S371L, S373P, S375F” (29 samples) followed by the panels “G339D, S371F, S373P, S375F” and “G339D, R346K, S371L, S373P, S375F” (11 samples each). This last combination of mutation is associated with sublineage BA1.1.
4. Discussion
The SARS-CoV-2 Omicron variant has rapidly replaced SARS-CoV-2 Delta variant in most European Union/European Economic Area (EU/EEA) countries (European Centre for Disease Prevention and Control, 2022). Immunity acquired through previous infection seems to be less effective against Omicron than against other variants, but the risk of severe COVID-19 remains low (Mallapaty 2022).
Omicron belongs to the Pango lineage B.1.1.529 which, compared to the Wuhan strain, is characterized by several amino acid changes in the spike protein, including A67V, DEL69/70, T95I, G142D, DEL143/145, N 211I, DEL212, G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F (list restricted to mutations with a prevalence in the lineage of at least 75%; data from https://outbreak.info/compare-lineages?pango=Omicron&gene=S&threshold=75&nthresh=1&sub=false&dark=true). This lineage has recently been partitioned into four sub-lineages BA.1 (B.1.1.529.1), BA.1.1 (B.1.1.529.1.1), BA.2 (B.1.1.529.2) and BA.3 (B.1.1.529.3).
The present study describes the development of a real time RT-PCR to differentiate the Omicron variant from other SARS-CoV-2 VoCs in environmental samples and its successful use for the purpose of Wastewater-Based Epidemiology (WBE). The spike protein was selected for assay design since Omicron harbours more than 32 mutations in the Spike protein alone, 15 of these mutations reside in the receptor-binding domain (Shah et al., 2022). Another assay, also targeting the S-region and developed at JRC, was tested for comparison and for results confirmation. Since this assay has been only tested in silico, JRC invited control laboratories worldwide to validate it in vivo on clinical samples (https://joint-research-centre.ec.europa.eu/jrc-news/efficient-tracing-omicron-new-pcr-test-2021-12-13_en). The validation of the ISS assay on clinical samples confirmed the ability of the test to detect the Omicron variant, though we found that it may have cross-reaction with the Alpha and Gamma variants to a minor extent. However, since the circulation of both Alpha and Gamma has drastically reduced in the EU/EEA (https://www.ecdc.europa.eu/en/covid-19/situation-updates/variants-dashboard) and the amplification efficiency is significantly lower than Omicron, this should not currently present any major problem for the use of the assay on wastewater samples.
The newly designed PCR ID_999 assay targets amino acid substitutions H655Y, N679K, and P681H of the S-region. As on 8 February 2022, a total 1,081,970 Omicron sequences have been deposited in GISAID, 1,072,798 of which (99.1%) show the combination of amino acid substitutions Spike_H655Y, Spike_N679K, Spike_P681H. These mutations have not been detected in non-Omicron variants worldwide and are reported in 8284/8588 (96.5%) of the Omicron sequences submitted to GISAID from Italy, thus far (as on 8 February 2022). Moreover, the three mutations are present in all Omicron sublineages, therefore the assay is potentially able to detect BA.1, BA.1.1, BA.2, and BA.3, which may be useful in light of the rapid evolution of the variant's epidemiology. The Omicron subvariant BA.2, indeed, is more transmissible than BA.1. Researchers in Denmark have found that BA.2 is about 1.5 times more transmissible than BA.1, currently the dominant version of omicron worldwide, and will likely become more common in the near future; moreover, it is more adept at infecting people who are vaccinated and even boosted (Lyngse et al., 2022). Early studies suggest that the BA.2 lineage might prolong the Omicron wave, but won't necessarily cause a fresh surge of COVID-19 infections (Callaway 2022). However, until more research is performed to reveal the epidemiological characteristics of this VoC, we should remain cautious regarding Omicron BA.2 (Huang et al., 2022).
The developed assay was successfully used to detect the Omicron variant in sewage samples, demonstrating the first occurrence in Italy in the first week of December 2021 and its rapid countrywide diffusion within three weeks. In only 21 days, the prevalence of Omicron-positive wastewater samples increased from 1.0% to 65.9%, and the Regions/AA.PP. detecting its presence from one to 17. This trend reflected the progressive diffusion of the variant in the population. Indeed, as on 20 December 2021, the clinical “flash survey” conducted in Italy showed that the Delta variant was still predominant, and the prevalence of the Omicron variant was 21.0% (Stima della prevalenza delle varianti VOC - Indagine 20.12.2021). On 3 January 2022 the prevalence of the Omicron variant in nasopharyngeal swabs increased at 80.8% (Stima della prevalenza delle varianti VOC - Indagine 03.01.2022), and on 17 January to 95.8% (Stima della prevalenza delle varianti VOC - Indagine 17.01.2022). Due to the nature of wastewater surveillance, which monitors clusters of the population and not individuals, the sharp increase of the Omicron variant and its path towards the replacement of the Delta variant, were captured earlier in this study compared to the clinical data. Indeed, the regular flash surveys on wastewater performed in January (Covid-19: flash survey sulle acque reflue - gennaio 2022) and February (Covid-19: flash survey sulle acque reflue - febbraio 2022), confirm the predominance of the Omicron variant throughout Italy, with only a residual presence of the Delta variant. The Delta variant has completely disappeared in the flash survey performed in March (Covid-19: flash survey sulle acque reflue - marzo 2022) confirming that, since December 2021, Omicron has taken over in the whole country.
Positive samples by the newly designed ISS real-time RT-PCR were also tested with another assay (JRC OmMet) to further confirm the results, and compare the performances of the two assays. Overall, 79.1% of positive samples were also confirmed by the second assay, discrepancies being mostly associated with samples with Ct values higher than the average and, presumably, lower concentration of the target. Indeed, considering the results of the optimization tests and of the analysis on wastewater samples, the JRC assay seems to provide higher amplification stringency (no cross-reactivity with other variants), but lower sensitivity (Ct values on average higher than the ISS assay). Since a protocol for the OmMet assay was not given in the published article (Petrillo et al., 2021), the OmMet assay was run at the same condition of the newly designed assay, as specific optimization of the assay was outside the scope of the work. Increase of the sensitivity of the assay could be incremented by further optimize reaction conditions.
It should be noted that only qualitative real-time RT-PCR assays were used in this study, aiming at evaluating presence/absence of the target; we therefore did not exactly measure the Omicron template by comparison of the Ct values to a standard curve. Indeed the aim of the present study was not comparing Omicron SARS-CoV-2 concentrations, but describing the history of the introduction and diffusion of the Omicron variant in the Italian population. However, by comparing absolute Ct values, we were able to show that they (and consequently the variant concentration) varied considerably. It is known that many factors impact the absolute value of Ct besides the concentration of the target, however a comparison can be done for data obtained in experiments using the same reaction conditions (instruments, reagents and assays). Therefore, the observation that the Ct value from one sample is higher than that of the other, could be valuable in concluding that the amount of template is lower in the first sample. Ct values ranged from 31,5 up to >40, that is more than 2.5 log difference, showing significant differences in sample concentrations. Lower Ct values (and therefore higher concentrations) were detected in North Italy in Veneto (where the first occurrence was also documented), Liguria and Emilia Romagna, while higher Ct values (lower concentrations) were documented in South Italy (Sicilia, Campania, and Puglia).
Further confirmation of the results obtained by the ISS assay was obtained by sequencing of positive samples. Overall, 55.6% of the positive samples (64 of 115) showed mutations characteristic of the Omicron variant. Interestingly, a considerable variability was observed within the Omicron sequences with 10 different amino acid substitutions detected in 12 combinations over a relatively short fragment (192 aa), the majority of positive samples harbouring the panel “G339D, S371L, S373P, S375F”. The amino acid substitutions found by Sanger sequencing suggest the presence of the sublineage BA.1, with a subset of samples having a panel of mutation associated to BA1.1 (G339D, R346K, S371L, S373P, and S375F). Characteristic mutations of BA.2 were not detected. Indeed, the first Omicron BA.2 Italian sequence deposited in GISAID (EPI_ISL_11191121) was obtained from a nasopharyngeal swab collected on 2021-03-06 in the Region of Emilia Romagna.
Twenty-four sequences showed no mutations in the amplified fragment, which could be consistent with the presence of the Delta variant, which was still the prevalent one at the beginning of December (Covid-19: flash survey sulle acque reflue - dicembre 2021). Indeed, in the portion of the spike protein spanning amino acids 220 to 412, the Delta variant harbours few mutations with low or very low prevalence in the lineage: S221L (0.3%), A222V (10%), T250I (0.5%), P251L (0.9%), S255F (0.2%), W258L (0.2%), A262S (0.2%), V289I (0.9%), T299I (0.3%) (https://outbreak.info/compare-lineages?pango=Delta&gene=S&threshold=0.2). We can therefore assume that these samples contained both the Omicron and the Delta variants, the latter being quantitatively prevalent and therefore amplified more efficiently by the nested PCR.
A few studies have successfully detected the Omicron variant in sewage since its introduction in the population. Ahmed and co-workers published on the first detection of SARS-CoV-2 Omicron VOC in an aircraft wastewater sample from a flight arriving to Australia from South Africa on the 25th of November 2021 by RT-qPCR assays followed by sequencing (Ahmed et al., 2022). Agravall and collaborators found all characteristic mutations of Omicron in wastewater originating from Frankfurt Airport before the first confirmed clinical case (Agravall et al., 2022). In the United States, early evidence of the SARS-CoV-2 Omicron variant was documented in the community in November–December 2021 by detecting key mutations associated with the variant in wastewater (Kirby et al., 2022). Finally, SARS-CoV-2 Omicron Variant mutations were detected in wastewater samples from Denmark using Nanopore Sequencing (Rasmussen et al., 2022).
This study described the history of the introduction and diffusion of the Omicron variant in the Italian population and territory. The rapid spread of the variant was captured, confirming the effectiveness of sewage monitoring as a powerful surveillance tool. From a public health point of view, when a new variant arises policymakers need a rapid assessment of whether the new variant impacts the transmissibility of the virus, severity, and/or immunity that are likely to have an impact on the epidemiological situation. WBE can be used to study the spread of the variants of SARS-CoV-2 in the general population, and it can therefore be considered as a valid and efficient alternative method to test campaigns for surveillance purposes.
CRediT authorship contribution statement
| Conceptualization | Ideas; formulation or evolution of overarching research goals and aims | GLR, ES |
| Methodology | Development or design of methodology; creation of models | GLR, ES |
| Software | Programming, software development; designing computer programs; implementation of the computer code and supporting algorithms; testing of existing code components | DB, MR, MG |
| Validation | Verification, whether as a part of the activity or separate, of the overall replication/ reproducibility of results/experiments and other research outputs | GLR, ES |
| Formal analysis | Application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data | GLR, ES, DB, MR, MG |
| Investigation | Conducting a research and investigation process, specifically performing the experiments, or data/evidence collection | GLR, MI, CV, PM, GBF, ES, the SARI Network |
| Resources | Provision of study materials, reagents, materials, patients, laboratory samples, animals, instrumentation, computing resources, or other analysis tools | GLR, ES, the SARI Network |
| Data Curation | Management activities to annotate (produce metadata), scrub data and maintain research data (including software code, where it is necessary for interpreting the data itself) for initial use and later reuse | GLR, ES, MR, MG, the SARI Network |
| Writing - Original Draft | Preparation, creation and/or presentation of the published work, specifically writing the initial draft (including substantive translation) | GLR, ES |
| Writing - Review & Editing | Preparation, creation and/or presentation of the published work by those from the original research group, specifically critical review, commentary or revision – including pre-or postpublication stages | GLR, ES, LB, LL, the SARI Network |
| Visualization | Preparation, creation and/or presentation of the published work, specifically visualization/ data presentation | GLR, ES, CV, PM, GBF |
| Supervision | Oversight and leadership responsibility for the research activity planning and execution, including mentorship external to the core team | GLR, ES |
| Project administration | Management and coordination responsibility for the research activity planning and execution | GLR, ES |
| Funding acquisition | Acquisition of the financial support for the project leading to this publication | GLR, ES, LB, LL |
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We wish to thank Dr. Maria Chironna, Department of Biomedical Sciences and Human Oncology, School of Medicine and Surgery, University of Bari Aldo Moro for sharing clinical omicron samples for testing the newly designed assay, Dr. Bernd Manfred Gawlik, European Commission's Joint Research Centre, for his coordination on building the EU Sewage Sentinel System for SARS-CoV-2 and its variants. This publication was supported by the European Virus Archive Global (EVA-GLOBAL) project, which provided the coronavirus RNA panel for the assessment of our in-house real time PCR.
We wish to acknowledge the financial support on the Italian Government (Decree of the Ministry of Health 30.10.2021, GU Serie Generale n.294 del 11-12-2021), of Ministry of Health (program CCM2020, project “Epidemiologia delle acque reflue: implementazione del sistema di sorveglianza per l'identificazione precoce di agenti patogeni, con particolare riferimento al SARS-CoV-2” [Wastewater based epidemiology: implementation of a surveillance system for the early detection of pathogens, and specifically SARS-CoV-2]) and of EU Commission (Gran Agreement 060701/2021/864481/SUB/ENV.C2).
We wish to thank Francesca Rita Iaia and Marta Vullo (Università degli Studi di Palermo), Lorenzo Dondero and Francesca Rispo (UNIGE - DISTAV), Rosa Maria Bertolotto, Marta Bellisomi, and Irene Tomesani (ARPAL), Daniele Colobraro (Regione Liguria), Silvia Scattolini, Ilaria Rosa, Daniela D'Angelantonio, and Giacomo Migliorati (Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise “G. Caporale”); Lorenzo Dondero, Francesca Rispo (UNIGE - DISTAV), Maria Bertolotto, Stefano Rosatto, Marta Bellisomi, and Irene Tomesani (ARPAL), Micaela Tiso (MICAMO srl);
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.155767.
Contributor Information
SARI network:
Achille Palma, Alberta Stenico, Alberto Antonelli, Alberto Izzotti, Alessandra Schiavuzzi, Alessandra Tosco, Amalia Porta, Andrea Franzetti, Andrea Turolla, Angela Costa, Angelo D’Argenzio, Angelo Romano, Anna Pariani, Annalaura Carducci, Anna-Maria Prast, Antonella Agodi, Antonella Cersini, Antonio Pizzolante, Arianna Azzellino, Barbara Bertasi, Bartolomeo Griglio, Carla Ancona, Carmelo Massimo Maida, Carmen Montanaro, Claudio Ottaviano, Clementina Cocuzza, Cristina Pignata, Daniele Nasci, Danilo Cereda, Desdemona Oliva, Doriana Antonella Giorgi, Elena Grasselli, Elena Mengon, Elena Nicosia, Elisabetta Carraro, Emanuela Ammoni, Enrica Ricci, Eric Grange, Ermanno Federici, Ettore Zuccato, Fabio Filippetti, Fabio Zuccon, Flavia Guarneri, Florida Damasco, Franca Palumbo, Francesca Apollonio, Francesca Borney, Francesca Ciuti, Francesca Cutrupi, Francesca Malpei, Francesca Pennino, Francesca Russo, Francesco Pizzo, Francesco Triggiano, Franco Rigoli, Gabriella Trani, Giancarlo Cecchini, Gianluca Borlone, Giorgia Allaria, Giorgio Bertanza, Giovanna Fusco, Giovanna La Vecchia, Giovanni Alborali, Giovanni Giammanco, Giovanni Santoro, Gisella Pitter, Giulia Lauretani, Giulia Nani, Giuseppa Purpari, Giuseppe Aprea, Giuseppe Bucciarelli, Giuseppe Di Vittorio, Giuseppe Lauria, Ileana Federigi, Irene Amoruso, Irene Ferrante, Laura De Lellis, Lisa Gentili, Lorella Zago, Lucia Decastelli, Luigi Bolognini, Luigi Cossentino, Manila Bianchi, Manuela Antonelli, Marco Guercio, Marco Verani, Marco Zampini, Margherita Ferrante, Maria Cadonna, Maria Grazia Cerroni, Maria Luisa Callegari, Maria Teresa Montagna, Maria Teresa Scicluna, Mariaconcetta Arizzi, Marika Mariuz, Marina Nadia Losio, Mario Palermo, Marta Paniccià, Maria Triassi, Martina Barchitta, Matteo Ramazzotti, Mattia Postinghel, Mauro Cravero, Mauro Ruffier, Maya Petricciuolo, Michele La Bianca, Michele Colitti, Monica Monfrinotti, Nadia Fontani, Nicola Ungaro, Nicoletta Formenti, Onofrio Mongelli, Osvalda De Giglio, Paola Angelini, Paola Foladori, Paolo Torlontano, Patrizia Montenegro, Pellegrinelli Laura, Piergiuseppe Calà, Renato Olivares, Renza Berruti, Rosa Anna Cifarelli, Rosanna Brienza, Sandro Binda, Sara Briscolini, Sara Castiglioni, Sara Muzio, Serena Manara, Silvia Bonetta, Silvia Magi, Silvia Riosa, Silvia Schiarea, Simona De Grazia, Sofia Barigelli, Stefano Rosatto, Tatjana Baldovin, Valeria Capparuccini, Valeria Primache, Vanessa Groppi, and Walter Mazzucco
Appendix A. The SARI network (“Sorveglianza Ambientale di SARS-CoV-2 attraverso i Reflui urbani in Italia”)
Abruzzo: Giuseppe Bucciarelli, Paolo Torlontano (Regione Abruzzo); Giuseppe Aprea (Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise “G. Caporale”);
Basilicata: Michele La Bianca (Regione Basilicata); Rosa Anna Cifarelli,
Achille Palma, Giovanna La Vecchia e Giuseppe Lauria (Agenzia Regionale
per la Protezione dell'Ambiente Basilicata – ARPAB); Rosanna Brienza e
Patrizia Montenegro (Acquedotto Lucano-AQL);
Campania: Angelo D'Argenzio (Regione Campania); Luigi Cossentino, Renato Olivares (Arpac - Agenzia Regionale per la Protezione Ambientale in Campania); Antonio Pizzolante, Giovanna Fusco (Istituto Zooprofilattico Sperimentale del Mezzogiorno); Alessandra Tosco, Amalia Porta (Università degli Studi di Salerno); Francesca Pennino, Triassi Maria (Università degli Studi di Napoli “Federico II”);
Emilia Romagna: Paola Angelini (Regione Emilia – Romagna); Laura De Lellis, Daniele Nasci (HERATech); Giovanni Alborali, Nicoletta Formenti, Flavia Guarneri (Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia-Romagna); Nadia Fontani, Giulia Nani, Franca Palumbo, Gianluca Borlone, Marco Guercio (IREN); Lisa Gentili (Arpae Emilia- Romagna);
Friuli Venezia Giulia: Marika Mariuz, Gabriella Trani, (Direzione Centrale Salute FVG); Anna Pariani (LABORATORIO HERAtech di Sasso Marconi –BO);
Lazio: Carla Ancona (DEPLAZIO - Dipartimento di Epidemiologia del Servizio Sanitario Regionale - Regione Lazio), Doriana Antonella Giorgi, Irene Ferrante, Monica Monfrinotti, Silvia Riosa, Valeria Capparuccini (ARPA Lazio - Agenzia Regionale per la Protezione Ambientale del Lazio), Maria Teresa Scicluna, Antonella Cersini (IZSLT - Istituto Zooprofilattico Sperimentale del Lazio e della Toscana), Mariaconcetta Arizzi, Giancarlo Cecchini, Claudio Ottaviano (Acea Elabori);
Liguria: Elena Nicosia (Regione Liguria settore tutela della salute negli ambienti di vita e di lavoro); Nadia Fontani, Giulia Nani, Franca Palumbo, Gianluca Borlone, Marco Guercio (Iren); Elena Grasselli, Giorgia Allaria (UNIGE - DISTAV); Alberto Izzotti (UNIGE – DIMES); Elena Nicosia, Stefano Rosatto, (ARPAL);
Lombardia: Emanuela Ammoni, Danilo Cereda (Regione Lombardia); Marina Nadia Losio, Barbara Bertasi (IZSLER - Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia); Andrea Aliscioni, Desdemona Oliva (CAP Holding); Sara Castiglioni; Silvia Schiarea, Ettore Zuccato (Istituto Mario Negri IRCCS); Manuela Antonelli, Arianna Azzellino, Francesca Malpei, Andrea Turolla (POLIMI); Sandro Binda, Pellegrinelli Laura, Valeria Primache (Univeristà degli Studi di Milano, Dipartimento di Scienze Biomediche per la Salute); Clementina Cocuzza, Andrea Franzetti (Università di Milano-Bicocca); Giorgio Bertanza (Università di Brescia); Maria Luisa Callegari (Università Cattolica del Sacro Cuore);
Marche: Luigi Bolognini, Fabio Filippetti (Regione Marche); Marta Paniccia’, Francesca Ciuti, Sara Briscolini (IZSUM - Istituto Zooprofilattico Sperimentale Umbria Marche); Silvia Magi (ARPAM);
Molise: Michele Colitti (Regione Molise); Carmen Montanaro (ASReM); Giuseppe Aprea (Istituto Zooprofilattico Sperimentale dell'Abruzzo e del Molise “G. Caporale”); Maria Grazia Cerroni (Arpa Molise);
Piemonte: Bartolomeo Griglio; Renza Berruti, Mauro Cravero, Angela Costa (Regione Piemonte); Manila Bianchi, Lucia Decastelli, Angelo Romano, Fabio Zuccon (IZSTO - Istituto Zooprofilattico Sperimentale del Piemonte Liguria e Valle d'Aosta SC Sicurezza e Qualità degli Alimenti); Elisabetta Carraro, Cristina Pignata (Dipartimento di Scienze della Sanità Pubblica e Pediatriche, Università di Torino), Silvia Bonetta (Dipartimento di Scienze della Vita e Biologia dei Sistemi, Università di Torino);
Puglia: Giuseppe Di Vittorio, Onofrio Mongelli (Regione Puglia); Osvalda De Giglio, Francesca Apollonio, Francesco Triggiano, Maria Teresa Montagna (Laboratorio di Igiene dell'Ambiente e degli Alimenti, Università degli Studi di Bari Aldo Moro - Dipartimento Scienze Biomediche e Oncologia Umana, Università degli Studi di Bari Aldo Moro); Nicola Ungaro (ARPA Puglia);
Sicilia: Mario Palermo (Regione Sicilia); Carmelo Massimo Maida, Walter Mazzucco (Università degli Studi di Palermo-Dipartimento PROMISE - sezione di Igiene); Simona De Grazia, Giovanni Giammanco (Centro di Riferimento Regionale per la Sorveglianza delle Paralisi Flaccide Acute (PFA) e ambientale della circolazione di poliovirus in Sicilia - AOUP Palermo); Giuseppa Purpari (IZS - Istituto Zooprofilattico Sperimentale della Sicilia), Margherita Ferrante, Antonella Agodi, Martina Barchitta (Università degli Studi di Catania - Dipartimento “G. F. Ingrassia”);
Toscana: Piergiuseppe Cala’ (Regione Toscana); Annalaura Carducci, Marco Verani, Ileana Federigi, Giulia Lauretani, Sara Muzio (Laboratorio di Igiene e Virologia Ambientale - Dipartimento di Biologia Università di Pisa); Matteo Ramazzotti, Alberto Antonelli (SOD microbiologia e virologia, azienda ospedaliera universitaria Careggi, Firenze);
Umbria: Enrica Ricci, Giovanni Santoro (Regione Umbria); Ermanno Federici, Maya Petricciuolo, Sofia Barigelli (Laboratorio Microbiologia Applicata e Ambientale, DCBB Università di Perugia);
Valle D'Aosta: Mauro Ruffier (Regione Valle d'Aosta); Francesca Borney, Eric Grange, Florida Damasco (Laboratorio chimico biologico microbiologico Arpa Valle d'Aosta);
Veneto: Francesca Russo, Gisella Pitter, Vanessa Groppi (Regione Veneto); Franco Rigoli, Marco Zampini (ARPAV - Agenzia Regionale per la Prevenzione e Protezione Ambientale del Veneto); Tatjana Baldovin, Irene Amoruso (Universita di Padova);
A.P. Trento: Elena Mengon (P.A. Trento); Maria Cadonna, Mattia Postinghel (ADEP SGI PAT); Francesco Pizzo, Alessandra Schiavuzzi (P. A. Trento); Francesca Cutrupi, Paola Foladori, Serena Manara (UNITN – Università di Trento);
A.P. Bolzano: Lorella Zago (P.A. Bolzano), Alberta Stenico, Anna-Maria Prast (Laboratorio biologico - Agenzia provinciale per l'ambiente e la tutela del clima (A.P.P.A.).
Appendix B. Supplementary data
Supplementary material
References
- Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 omicron variant at Frankfurt airport but limited spread in the City of Frankfurt, Germany, in November 2021. Microbiol. Resour. Announc. 2022 Feb 17;11(2) doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Smith W.J.M., Metcalfe S., Stephens M., Jennison A.V., Moore F.A.J., Bourke J., Schlebusch S., McMahon J., Hewitson G., Nguyen S., Barcelon J., Jackson G., Mueller J.F., Ehret J., Hosegood I., Tian W., Wang H., Yang L., Bertsch P.M., Tynan J., Thomas K.V., Bibby K., Graber T.E., Ziels R., Simpson S.L. Detection of the omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci. Total Environ. 2022 May;10(820) doi: 10.1016/j.scitotenv.2022.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Why does the Omicron sub-variant spread faster than the original? Nature. 2022 Feb 16 doi: 10.1038/d41586-022-00471-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . ECDC; Stockholm: 2022. Assessment of the further spread and potential impact of the SARS-CoV-2 Omicron variant of concern in the EU/EEA. 19th update - 27 January 2022. [Google Scholar]
- Huang J., Zeng G. Letter to the editor: epidemiology of the SARS-CoV-2 variant omicron BA.2 - vigilance needed. Euro Surveill. 2022 Mar;27(13) doi: 10.2807/1560-7917.ES.2022.27.13.2200254. PMID: 35362407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan S.R., Spratt A.N., Sharma K., Chand H.S., Byrareddy S.N., Singh K. Omicron SARS-CoV-2 variant: unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022;126 doi: 10.1016/j.jaut.2021.102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A.E., Welsh R.M., Marsh Z.A., Yu A.T., Vugia D.J., Boehm A.B., Wolfe M.K., White B.J., Matzinger S.R., Wheeler A., Bankers L., Andresen K., Salatas C., New York City Department of Environmental Protection. Gregory D.A., Johnson M.C., Trujillo M., Kannoly S., Smyth D.S., Dennehy J.J., Sapoval N., Ensor K., Treangen T., Stadler L.B., Hopkins L. Notes from the field: early evidenceof the SARS-CoV-2 B.1.1.529 (Omicron) variant in community wastewater - United States, November-December 2021. MMWR Morb Mortal Wkly Rep. 2022 Jan 21;71(3):103–105. doi: 10.15585/mmwr.mm7103a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Brandtner D., Mancini P., Veneri C., Bonanno Ferraro G., Bonadonna L., Lucentini L., Suffredini E. Key SARS-CoV-2 mutations of alpha, gamma, and eta variants detected in urban wastewaters in Italy by long-read amplicon sequencing based on nanopore technology. Water. 2021;13:2503. doi: 10.3390/w13182503. [DOI] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Bonadonna L., Lucentini L., Suffredini E. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ. 2021 Jan;1(750) doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Bonanno Ferraro G., Veneri C., Iaconelli M., Lucentini L., Bonadonna L., Brusaferro S., Brandtner D., Fasanella A., Pace L., Parisi A., Galante D., Suffredini E. Rapid screening for SARS-CoV-2 variants of concern in clinical and environmental samples using nested RT-PCR assays targeting key mutations of the spike protein. Water Res. 2021 Jun;1(197) doi: 10.1016/j.watres.2021.117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 2022 2022.01.28.22270044. [Google Scholar]
- Mallapaty S. COVID reinfections surge during Omicron onslaught. Nature. 2022 Feb 16 doi: 10.1038/d41586-022-00438-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Petrillo M., Querci M., Corbisier P., Marchini A., Buttinger G., Van den Eede G. n Silico Design of Specific Primer Sets for the Detection of B.1.1.529 SARS-CoV-2 Variant of Concern (Omicron) Zenodo. 2021;300 https://zenodo.org/record/5747872#.YbnC6HyZOUk. [Google Scholar]
- Pierri B., Mancusi A., YTR Proroga, Capuano F., Cerino P., Girardi S., Vassallo L., Lo Conte G., Tafuro M., Cuomo M.C., Di Concilio D., Vicenza T., Cozzi L., Di Pasquale S., La Rosa G., Beikpour F., Suffredini E. SARS-CoV-2 detection in nasopharyngeal swabs: performance characteristics of a real-time RT-qPCR and a droplet digital RT-PCR assay based on the exonuclease region (ORF1b, nsp 14) J Virol Methods. 2021 Dec 10;300 doi: 10.1016/j.jviromet.2021.114420. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L.D., Richter S.R., Midgley S.E., Franck K.T. Detecting SARS-CoV-2 Omicron B.1.1.529 variant in wastewater samples by using nanopore sequencing. Emerg. Infect. Dis. 2022 Apr 1;28(6) doi: 10.3201/eid2806.220194. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2022 Jan;24(12) doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilrich C., Wilrich P.T. Estimation of the POD function and the LOD of a qualitative microbiological measurement method. J. AOAC Int. 2009 Nov-Dec;92(6):1763–1772. [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., et al. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4) doi: 10.1128/mSystems.00614-20. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material