SUMMARY
Unicellular eukaryotes have been suggested as undergoing self-inflicted destruction. However, molecular details are sparse compared with the mechanisms of programmed/regulated cell death known for human cells and animal models. Here, we report a molecular cell death pathway in Saccharomyces cerevisiae leading to vacuole/lysosome membrane permeabilization. Following a transient cell death stimulus, yeast cells die slowly over several hours, consistent with an ongoing molecular dying process. A genome-wide screen for death-promoting factors identified all subunits of the AP-3 complex, a vesicle trafficking adapter known to transport and install newly synthesized proteins on the vacuole/lysosome membrane. To promote cell death, AP-3 requires its Arf1-GTPase-dependent vesicle trafficking function and the kinase Yck3, which is selectively transported to the vacuole membrane by AP-3. Video microscopy revealed a sequence of events where vacuole permeability precedes the loss of plasma membrane integrity. AP-3-dependent death appears to be conserved in the human pathogenic yeast Cryptococcus neoformans.
Graphical abstract
In brief
Details about how mammalian cells die have yielded effective cancer therapies. Similarly, details about fungal cell death may explain failed responses to anti-fungal agents and inform next-generation anti-fungal strategies. Stolp et al. describe a potential mechanism of yeast cell death subversion, by inhibiting AP-3 vesicle trafficking to block vacuole/lysosome permeability.
INTRODUCTION
Long-standing conventions appear to challenge the existence of programmed or regulated cell death (PCD/RCD) mechanisms in unicellular eukaryotes. The orderly patterns of cell death observed in developing animals, which gave rise to the term, programmed cell death (Lockshin and Williams, 1965), also gave rise to the assumption that programmed death arose during the evolution of multicellular organisms. Additionally, for decades, the prevailing evolution theories rejected the possibility of cell suicide as an adaptation (the ultimate altruistic behavior) in unicellular organisms because this conflicted with the concept of individual-level theory of selection (versus multi- or group-level selection) (Durand, 2020; Nowak et al., 2010). Consistent with these conventions, unicellular eukaryotes and bacteria lack key molecular players of classical apoptosis, the best-studied cell death pathway in mammals (BAX-induced, caspase-3-mediated cell fragmentation, and engulfment by neighboring cells) (Nagata and Segawa, 2021).
Accumulating evidence has prompted reappraisal of long-held assumptions about the origins of PCD. The newly emerging concept is that programmed unicellular death predates and was required for multicellular organisms to arise, rather than the inverse (Huettenbrenner et al., 2003; Iranzo et al., 2014; Vanchurin et al., 2022). While classic apoptosis (caspase-3-mediated cell death) likely arose with metazoans, where it plays a key role in ontogeny, its absence in unicellular organisms does not preclude the concept of self-inflicted unicellular death, because the molecular details of cell death may be as diverse as the organisms themselves (Ameisen, 2002; Teng and Hardwick, 2015).
However, parallels between microorganisms and mammals continue to emerge, particularly for non-apoptotic RCD mechanisms. Pore-forming gasdermin proteins that mediate mammalian pyroptosis (a form of programmed necrosis) are found in some bacteria and multicellular/filamentous fungi (Daskalov et al., 2020; Johnson et al., 2022; Liu et al., 2016). Importantly, despite low sequence similarity, these diverse gasdermins share striking structural and mechanistic similarities to mediate cell suicide. Diverse cell death mechanisms with compelling empirical evidence have been partially delineated in bacteria and filamentous fungi, with and without parallels in animals (Heller et al., 2018; Ofir et al., 2021; Veneault-Fourrey et al., 2006). However, an equivalently advanced understanding of the dying processes in unicellular eukaryotes has not yet been achieved. Early evidence suggesting that yeast metacaspases induce apoptosis-like death resembling mammalian apoptosis was impactful in stimulating the field but has not garnered molecular support (Aouacheria et al., 2018; Minina et al., 2020). Thus, despite the arguments for altruistic death in unicellular eukaryotes, the question is unsettled.
Yeasts are widely used genetic models for a range of disciplines (e.g., autophagy, vesicle trafficking, cancer, and neurodegeneration) and are ideal models for dissecting unicellular eukaryotic cell death. Many Saccharomyces cerevisiae genes have been reported to enhance or suppress cell death (Chaves et al., 2021), including genes involved in developmental processes, fitness selection, and control of yeast viruses, some with semblance to mammals (Eastwood et al., 2012; Fannjiang et al., 2004; Gao et al., 2019; Ivanovska and Hardwick, 2005; Pozniakovsky et al., 2005). A common theme shared by the four best-characterized mammalian cell death pathways is membrane permeabilization carried out either by pore-forming proteins, such as BAX (apoptosis), MLKL (necroptosis), and gasdermins (pyroptosis), or by lipid peroxidation (ferroptosis), which represent commitment points or final steps toward cell suicide. In contrast, direct effectors of death in yeast are not yet identified. Permeabilization of the yeast vacuole/lysosome has been implicated as a final step in yeast cell death, potentially analogous to cell death by lysosomal membrane permeabilization (LMP) in mammals, but the details are unknown in both yeast and mammals (Eastwood et al., 2013; Kim and Cunningham, 2015; Kim et al., 2012; Watson and Khaled, 2020).
Surprisingly few yeast genetic screens have been aimed at identifying yeast genes that contribute to cell suicide following stress (Dong et al., 2017; Jarolim et al., 2013; Kim et al., 2012; Sousa et al., 2013; Teng et al., 2011, 2013). Instead, more efforts have focused on yeast genes that promote survival and drug resistance, seeking to control pathogens and support wine-making industries (Todd and Selmecki, 2020; Velazquez et al., 2016). Thus, the death of drug-treated fungal pathogens is generally not studied from the perspective of understanding the dying processes in fungal cells despite public health relevance. Cryptococcus neoformans is a major world threat, especially for individuals who are immunocompromised and recipients of organ transplants, worsened by increasing fluconazole resistance accelerated by the advent of prophylactic usage (Stott et al., 2021).
Yeasts may undergo multiple unconventional types of cell death. Whether these were selected as true adaptations during evolution or can be harnessed for therapeutic benefit, analogous to the anti-cancer BCL-2 inhibitor venetoclax (Roberts et al., 2016), is not known. Here, we provide evidence for an RCD pathway leading to vacuole/lysosome permeabilization in S. cerevisiae that appears to be conserved in C. neoformans.
RESULTS
Protracted time to death following heat-ramp stress
Non-programmed, unregulated cell death is defined by the Nomenclature Committee on Cell Death as a sudden catastrophic assault (Galluzzi et al., 2018). In this case, the cell does not contribute to its own death, and death is not preventable by any action from the cell or by any therapeutic treatment. To minimize death by assault, small volumes of yeast cells were treated with a near-lethal heat-ramp of 30°C–51°C delivered over 18 min using a programmable thermocycler, avoiding death by sudden heat shock (Figure 1A) (Teng et al., 2011; Teng and Hardwick, 2013). Our previous results suggested that yeast die slowly following a heat-ramp, based on vital dye staining at early times compared with clonogenic survival (colony-forming units [cfus]) after 2-days, with the caveat that these assays measure different cell properties. To validate vital dyes as useful proxies for impending cell death, two dyes were compared over an extended time course following heat-ramp, revealing that both dyes approximate loss of clonogenic survival at 48 h (Figure 1B). Trending differences are likely attributed to uncounted microscopic colonies and a few non-proliferative cells that may remain viable for longer periods. In addition, phloxine B, which is suggested to be pumped out of living cells (Kwolek-Mirek and Zadrag-Tecza, 2014), stains dying yeast cells several hours before propidium iodide (PI), a field standard that enters cells upon loss of plasma membrane integrity, marking definitive cell death (Figure 1B). Using the sensitive phloxine assay, median time to 50% death under these conditions was ~11.5 h (range 9–14 h) after heat-ramp for five wild-type strains (BY4709, BY4741, BY4742, W303, and SEY6210), determined by video microscopy of cells immobilized on agar (Figure 1C). Video frames illustrate the delay in phloxine staining (white arrows) and delayed proliferation of survivors (yellow arrows), and no phloxine-positive cells were observed to recover during the 16 h observation, although rare occurrences are not ruled out (Figure 1D). Death could occur first in the older mother or younger daughter cell (Figure 1E). Thus, yeast appear to die slowly, suggestive of a molecular dying process following stress and before death. Although this is consistent with gene-dependent cell suicide, it does not eliminate the possibility that cell damage incurred during the heat-ramp could take many hours to passively deteriorate. Therefore, we took a genetic approach to identify yeast genes that promote self-destruction following stress.
Figure 1. Protracted time to death following heat-ramp stress.
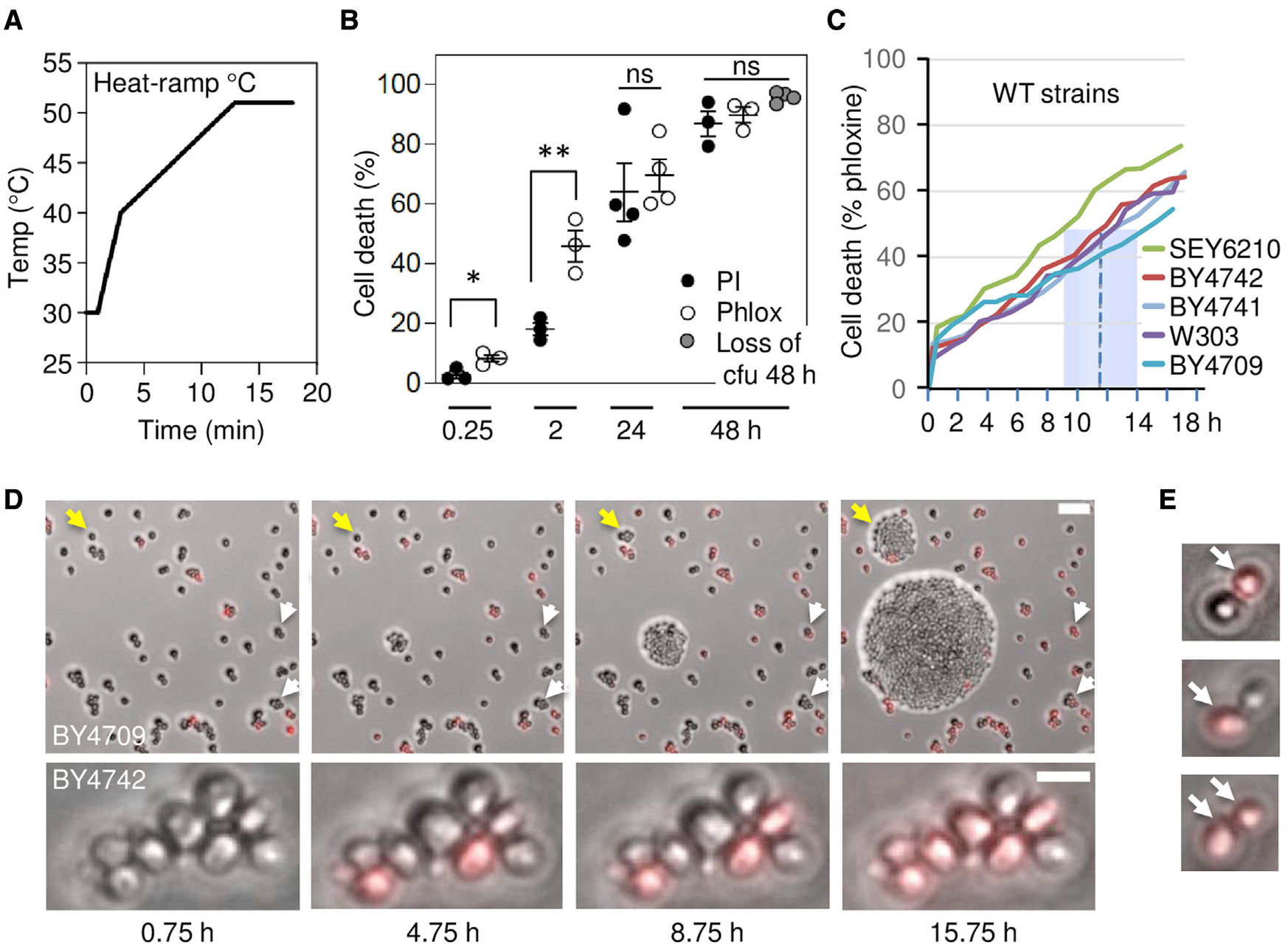
(A) Temperature plot for the 18 min, 30°C–51°C heat-ramp cell death stimulus delivered to log phase yeast throughout this study, except where noted (Teng et al., 2011).
(B) Cell death of wild-type yeast (BY4709) after heat-ramp determined as percent of starting cell number by propidium iodide (PI) or phloxine B (Phlox) staining in liquid phase (≤2 h post-heat-ramp) or in solid phase to avoid counting progeny of survivors (≥24 h), and by visible colony forming units (cfus, 48 h only). Mean ± SD plotted for 3–4 independent experiments per condition, counting ~200 per sample. Two-tailed t test, *p = 0.0157, **p = 0.0122, and two-way ANOVA with Tukey’s honest significant difference (HSD); p = 0.1006 to compare all three assays at 48 h.
(C) Time to death (phloxine+) determined by video microscopy of ~300 cells per strain after heat-ramp determined in solid phase. Median time to death (dashed line); range (blue box) for five wild-type strains under these growth and media conditions.
(D) Video microscopy frames from an independent time-lapse experiment, as described in (C). Example cells dying 9–16 h post-heat-ramp (white arrows) and early or delayed proliferation/clonogenicity (yellow arrows). Scale bar, 25 μm.
(E) Mother (lower larger) or daughter (upper smaller) cells may die first (phloxine+, arrows).
Genome-wide screen identifies death-resistant AP-3 deletion strains
To identify yeast genes that promote cell death following stress, we reanalyzed our previous genome-wide screen (Teng et al., 2011, 2013), this time to identify death-resistant deletion strains among ~5000 S. cerevisiae knockouts (BY4741) treated with a heat-ramp. We focused on pro-death genes (death-resistant knockouts) rather than anti-death genes, because deletion of any gene could potentially reduce fitness despite having no role in regulating cell death. Raw images of microscopic colonies in eight replicates were reacquired from a BioSpot Reader, visually inspected to remove artifacts, and colony counts were systematically corrected for undercounting at higher colony densities to identify death-resistant strains. A stringent initial cutoff yielded a hit rate of 1.84% (Figure 2A; Table S1). Gene ontology function analyses of these 84 hits readily identified all four knockouts of the heterotetrameric AP-3 adaptor complex (Figure 2B). AP-3 is composed of a small (Aps3), a medium (Apm3), and two large subunits (Apl5 and Apl6) and is responsible for trafficking newly synthesized proteins to their destination on the vacuole/lysosome membrane. In contrast, no components of the related AP-1 or AP-2 complexes, two other characterized vacuole trafficking pathways (CPY [Gga1 and Gga2] and CVT [Ape1, Ams1, and Atg19]), or Ent3 and Ent5 (Casler and Glick, 2020; Daboussi et al., 2012; Lynch-Day and Klionsky, 2010; Robinson et al., 1988) were among the top 200 hits, suggesting an AP-3-specific function in cell death.
Figure 2. Genome-wide screen identifies death-resistant AP-3 deletion strains.
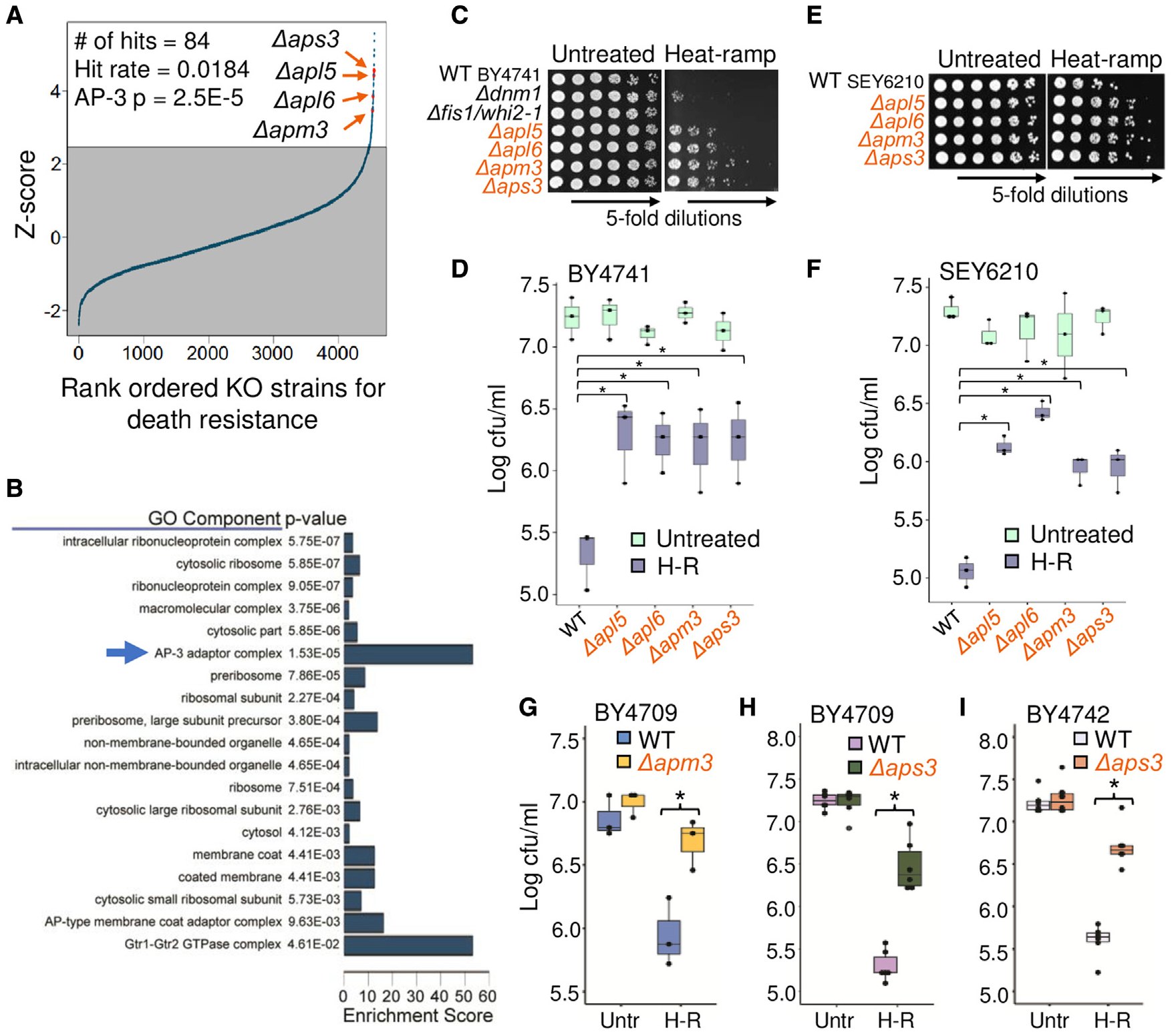
(A) Rank ordered survival of yeast knockout strains (BY4741) from genome-wide screen after post-diauxic-phase 20 min, 30°–62°C linear heat-ramp. Initial hit cutoff set at 1.5× interquartile range above 75th percentile of Z scores, ~2.47.
(B) Gene ontology component enrichment analysis of the 84 screen hits compared with all strains analyzed with Bonferroni correction for multiple hypothesis testing.
(C) Low throughput cell death assay of log phase optical density 600 (OD600)-adjusted cultures (BY4741) spotted on plates pre- and post-heat-ramp.
(D) Quantification for (C) from 3 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p ≤ 0.0048.
(E and F) As described for (C) and (D), except for SEY6210 background strains from 3 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p ≤ 0.0043.
(G) Survival (cfus) of newly generated CRISPR knockouts of APM3 in the amino acid prototroph BY4709. Quantified for 3 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 0.0068. Genome sequence and trafficking function tests in Figures S1A–S1D.
(H) As in (G), for new CRISPR knockout of APS3 (BY4709). Quantified for 6 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 1.24 × 10−8. Genome sequence and trafficking function tests in Figures S1E–S1H.
(I) As in (H), for new APS3 knockout generated by conventional recombination (BY4742). Quantified for 6 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 2.4 × 10−8.
Three AP-1, AP-2, and AP-3 adaptor complexes in yeast (five in mammals) sort, transport, and deliver membrane-associated proteins to their respective subcellular destinations with minimal overlap. Similar to mammals, yeast AP-1 and AP-3 recognize amino acid sequence motifs in their respective proteins protruding from late/post-Golgi or endosome membranes (Karim et al., 2018; Renard et al., 2010; Schoppe et al., 2020; Vowels and Payne, 1998). After engaging their cargo, AP-1 and AP-3 plus other proteins generate vesicles that transport and deliver their cargo by fusing to a target membrane with the aid of additional factors. Yeast AP-3 traffics from late/post-Golgi membranes directly to the vacuole/lysosome membrane, while AP-1 is important for Golgi and endosome recycling pathways (Casler and Glick, 2020; Cowles et al., 1997; Daboussi et al., 2012; Panek et al., 1997; Stepp et al., 1997).
The striking death resistance of AP-3 deletion strains was confirmed in small-scale heat-ramp tests and was more robust than our previous death-resistant landmark Δdnm1 (Fannjiang et al., 2004) (Figures 2C and 2D). Similar results were observed in the SEY6210 background strains (Figures 2E and 2F) and for several newly constructed knockouts generated by CRISPR and/or conventional recombination for APM3 (Figures 2G and S1A–S1D) and APS3 in different backgrounds (Figures 2H, 2I, and S1E–S1H). In contrast, knockouts of AP-1 (Δapl2, Δapl4, Δapm1, Δapm2, and Δaps1), AP-2 (Δapl1, Δapl3, Δapm4, and Δaps2) and the gamma-adaptin-like adaptors (Δgga1 and Δgga2) were sensitive to death, consistent with screen results (Figure S2).
Cell death resistance was not dependent on cell proliferation, because AP-3 knockouts were distinguishable from wild type, within ~15 min after heat-ramp by phloxine staining (Figure 3A), and to a lesser degree by PI staining (Figure 3B). Video microscopy revealed that AP-3-deficient yeast begin to proliferate sooner and die more slowly (time to death 50%, ~20.3 h) compared with wild type (10.3 h) (Figures 3C and 3D; Video S1). These findings are consistent with a genetically encoded cell suicide process promoted by the AP-3 complex in response to stress.
Figure 3. Rescue of AP-3 restores cell death induced by multiple stimuli.
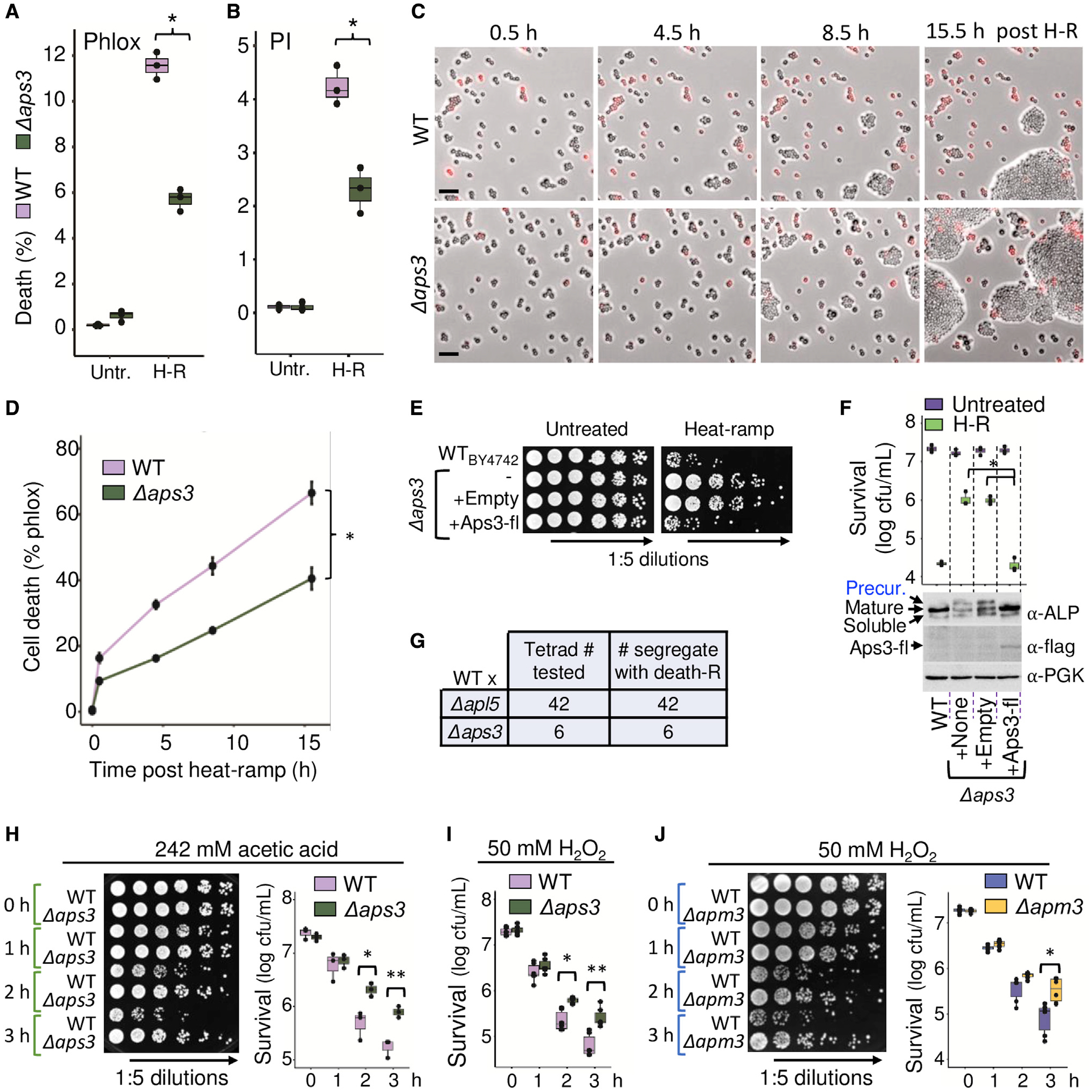
(A) Impending death of wild-type and Δaps3 in liquid cultures assessed as percent phloxine-positive before and ~15 min after heat-ramp. N = 3 independent experiments per condition, two-way ANOVA with Tukey’s HSD test post hoc, *p = 4.77 × 10−7.
(B) Same samples in (A) stained with propidium iodide (PI), *p = 1.54 × 10−4.
(C) Images from video microscopy of phloxine-stained wild-type (WT) and Δaps3 after heat-ramp. Scale bars, 25 μm.
(D) Quantification for (C) from 3 independent experiments; time to death (50%) for Δaps3 estimated by linear regression. One-way ANOVA with repeated measures, *p = 1.5 × 10−7.
(E) Rescue of Δaps3 cell death by C-terminal flag-tagged (fl) APS3 and native promoter. Viability determined by cfus on selection medium before and after heat-ramp (or acetic acid or H2O2, Figure S3).
(F) Quantification for (E), N = 3 independent experiments, two-way ANOVA with Tukey’s HSD test post hoc for Δaps3 + Aps3-flag rescue versus Δaps3 (*p = 1.78 × 10−7) or Δaps3 + control vector (*p = 1.53 × 10−7), respectively. WT versus Δaps3 (p = 1.95 × 10−7) or Δaps3 + control vector (p = 2.29 × 10−7). Western blot of unheated samples with anti-ALP/Pho8, anti-flag, and anti-PGK loading control.
(G) Summary of tetrad analysis for sporulated heterozygous diploids of WT crossed to Δapl5 (all 42 validated tetrads tested) and Δaps3 (all 6 validated tetrads tested). Death resistance uniformly segregated 2:2 with KanMX knockout marker.
(H) Resistance of new CRISPR Δaps3 strain to 242 mM acetic acid. N = 3 independent experiments, two-way ANOVA with Tukey’s HSD test post hoc, *p = 0.0064, **p = 0.0043.
(I) Resistance of new Δaps3 to 50 mM H2O2-induced death. N = 6 biological replicates from 3 independent experiments, two-way ANOVA with Tukey’s HSD test post hoc, *p = 0.0014, **p = 7.95 × 10−6.
(J) Resistance of new CRISPR knockout (KO) Δapm3 to 50 mM H2O2-induced death. N = 6 biological replicates from 3 independent experiments, two-way ANOVA with Tukey’s HSD test post hoc, *p = 3.25 × 10−5.
Rescue of AP-3 restores cell death induced by multiple stimuli
We previously demonstrated that 60%–70% of yeast knockout strains have evolved a second gene mutation strongly affecting cell death/survival following stress and that independent knockouts of the same gene (or genes of the same protein complex) tend to acquire mutations in a shared second gene (Cheng et al., 2008; Teng et al., 2013), analogous to Δsod1 strains that repeatedly develop mutations in PMR1 (Lapinskas et al., 1995). Therefore, death resistance of AP-3 knockouts could potentially be explained by natural selection for secondary gene mutations that arose as a consequence of deleting any one of the AP-3 genes.
However, this was not the case, because cell death susceptibility was restored by reinserting APS3 with its native promoter into the Δaps3 deletion strain (Figures 3E and 3F). To validate that rescue of APS3 also restored AP-3 vesicle trafficking function, we monitored trafficking of the endogenous AP-3 cargo protein alkaline phosphatase (ALP/Pho8), which defines the AP-3 pathway (also known as the ALP pathway). In the absence of AP-3, ALP/Pho8 is shunted to the vacuole by an alternate pathway, impairing its normal maturation on the vacuole membrane by luminal proteases that cleave the ALP/Pho8 precursor into mature and soluble forms (Cowles et al., 1997). Using this traditional assay, we found that reinsertion of APS3 prevented the accumulation of unprocessed ALP/Pho8 precursor (Figure 3F). In further support of AP-3-dependent cell death, analysis of tetrad spore sets derived from Δapl5 and Δaps3, each back-crossed to wild type (BY4742 or other strains), demonstrated 100% co-segregation of death resistance with the AP-3 locus (KanMX replacing AP-3 genes) (Figure 3G). We conclude that AP-3, and not genetic confounders, can promote yeast cell death.
Resistance to cell death was not limited to thermal stress because disruption of AP-3 also conferred resistance to acetic acid, a by-product of alcohol fermentation known to trigger yeast cell death, unlike other acids (Sousa et al., 2013; Vilela-Moura et al., 2011) (Figure 3H), and to hydrogen peroxide, a mimic of oxidative bursts produced by phagocytic host immune cells (Figures 3I and 3J). Rescue of Δaps3 with APS3-FLAG restored acetic-acid- and H2O2-induced death (Figure S3). Consistent with our findings, published supplemental tables of yeast cell death screens, using acetic acid (Sousa et al., 2013), endoplasmic reticulum (ER) stress (Kim et al., 2012), and thiosemicarbazone Ni(S-tcitr)2 (Baruffini et al., 2020), scored several AP-3 deletion strains as death resistant. Thus, AP-3 appears to contribute to cell death induced by multiple stimuli.
Vesicle trafficking function of AP-3 is required for cell death
Our genome-wide screen, plus additional analyses of Δapl5, Δapm3, and Δaps3 described thus far, imply that the intact AP-3 complex, rather than its individual components, promotes yeast cell death. This further implicates its vesicle trafficking function in the dying process. To test this more directly, we investigated the requirement for Arf1, a small GTPase required by human/yeast AP-1 and AP-3 complexes (inferred for yeast AP-3) for docking onto donor membranes to collect their cargo proteins (Anand et al., 2009; Nie et al., 2003; Ooi et al., 1998; Schoppe et al., 2020; Seaman et al., 1996). To address the role of yeast Arf1 in AP-3-dependent death without disrupting many other Arf1 functions (e.g., AP-1, GGA1/2, and exomer trafficking), we designed point mutations in AP-3 to prevent binding to Arf1. Mutations were selected based on sequence homology, crystal structures, and biochemical studies of mammalian AP-1 bound to human Arf1 (PDB: 6DFF and 4HMY). In these structures, an Arf1-GTP molecule is bound to the outer face of each of the two large AP-1 subunits (Morris et al., 2018; Ren et al., 2013). Two amino acid changes in human AP-1β1 (corresponding to yeast Apl6 L117D/I120D in AP-3) were shown to inhibit Arf1 binding and membrane association without affecting AP-1 complex assembly (Ren et al., 2013). The corresponding Apl6 L117D/I120D changes were inserted into the yeast genome and sequence validated (Figures 4A and S4).
Figure 4. AP-3 promotes cell death via its canonical vesicle trafficking functions.
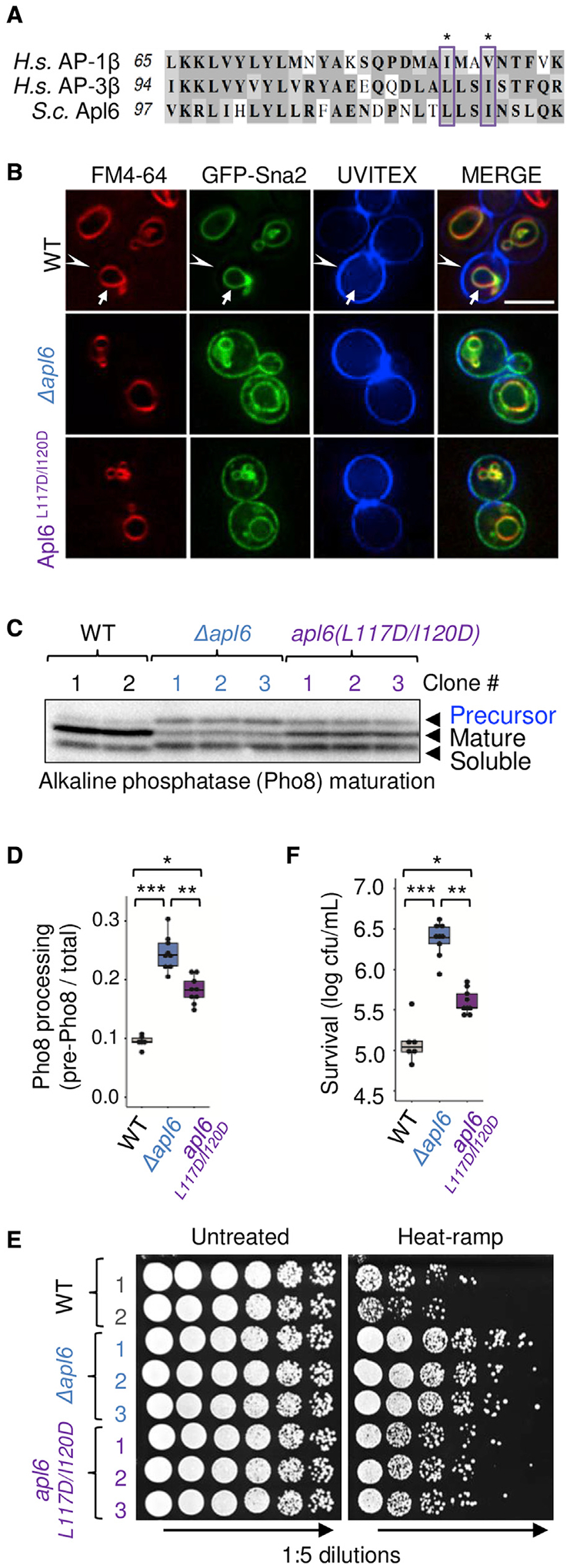
(A) Alignment of AP-3 orthologues, yeast Alp6, human AP-1 (AP1β), and human AP-3 (AP3β); conserved Arf1-binding contact sites (boxed).
(B) Fluorescent microscopy of GFP-Sna2 localization in yeast stained with FM4–64 (red) to mark vacuole membranes, and cell wall stain UVITEX (blue) to approximate the plasma membrane. Scale bar, 5 μm.
(C) Immunoblot of endogenous alkaline phosphatase (ALP/Pho8) to detect AP-3/vacuole-dependent processing of the precursor to proteolytically matured and soluble forms.
(D) Quantification for (C), calculated as Pho8 precursor relative to total (precursor + mature + soluble) for 3 independent experiments after pooling results for 2–3 independent strains per genotype. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 1.56 × 10−6, **p = 3.23 × 10−5, ***p = 1.57E-10.
(E) Viability of sequence-verified APL6-modified yeast strains (Figure S4) following heat-ramp and spotted on agar plates.
(F) Quantification for (E), plotted as log10 (cfus/mL) for 3 independent experiments after pooling results for 2–3 independent strains per genotype. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 2.67 × 10−6, **p = 3.98 × 10−12, ***p = 1.06 × 10−12.
As predicted, the apl6L117D/I120D mutant strain is defective for trafficking. GFP-Sna2, an AP-1/AP-3 cargo reporter (Renard et al., 2010) that normally resides on the vacuole membrane (marked with FM4–64), was mislocalized to the plasma membrane (marked by cell wall stain UVITEX) in the apl6L117D/I120D mutant, similar to the knockout Δapl6 (Figure 4B). Defective trafficking was confirmed for the AP-3-specific cargo protein ALP/Pho8 in apl6L117D/I120D, which had significant accumulation of its precursor, although less than Δapl6, indicating an obvious but partial defect in AP-3 trafficking (Figures 4C and 4D). Importantly, three apl6L117D/I120D strains lost ~85% of their pro-death effects (Figures 4E and 4F). Additional Arf1 binding sites on the Apl5 subunit may contribute to the incomplete effects of Apl6(L117D/I120D) (Morris et al., 2018; Ren et al., 2013). These findings indicate that a conserved Arf1 binding site on yeast AP-3 is required both for normal vesicle trafficking and for stress-induced cell death.
AP-3 is required shortly before the death stimulus
We considered how the AP-3 complex might promote cell death. We disfavor the possibility that AP-3 gains a new pro-death function under stress conditions, because thermostability of this large 275,000 kDa tetramer under heat-ramp conditions appears unlikely. Instead, we favor a model where AP-3 is the delivery vehicle for one or more cargo proteins that exhibit stress-induced toxicity, resulting in vacuole membrane damage. In this model, AP-3 is predicted to deliver its death-promoting payload to the vacuole prior to a death stimulus.
To test this idea, the auxin-induced degron (AID) system (Nishimura and Kanemaki, 2014) was used to determine the effects of rapid AP-3 inactivation on cell death. Strains were engineered by fusing the AID-6xflag cassette to the C terminus of knockin TAP-tag collection (Snyder et al., 2019). Of the four AID-tagged AP-3TAP subunits, Apl5 (Apl5TAP-AID-flag) was the most stably expressed and was used thereafter. The AID cassette also encodes the auxin-responsive OsTIR1 cullin adaptor needed to recruit AID-tagged proteins to the yeast cullin-1 E3 ligase for ubiquitination and degradation. As expected, the AID-tagged Apl5TAP strain was protected relative to Apl5TAP control from heat-ramp-induced cell death when pre-treated 4 h with auxin (50 μM indole-3-acetic acid [IAA]) to degrade Apl5, restoring survival similar to the apl5 knockout (Figures 5A and 5B).
Figure 5. AP-3 is required shortly before the cell death stimulus.
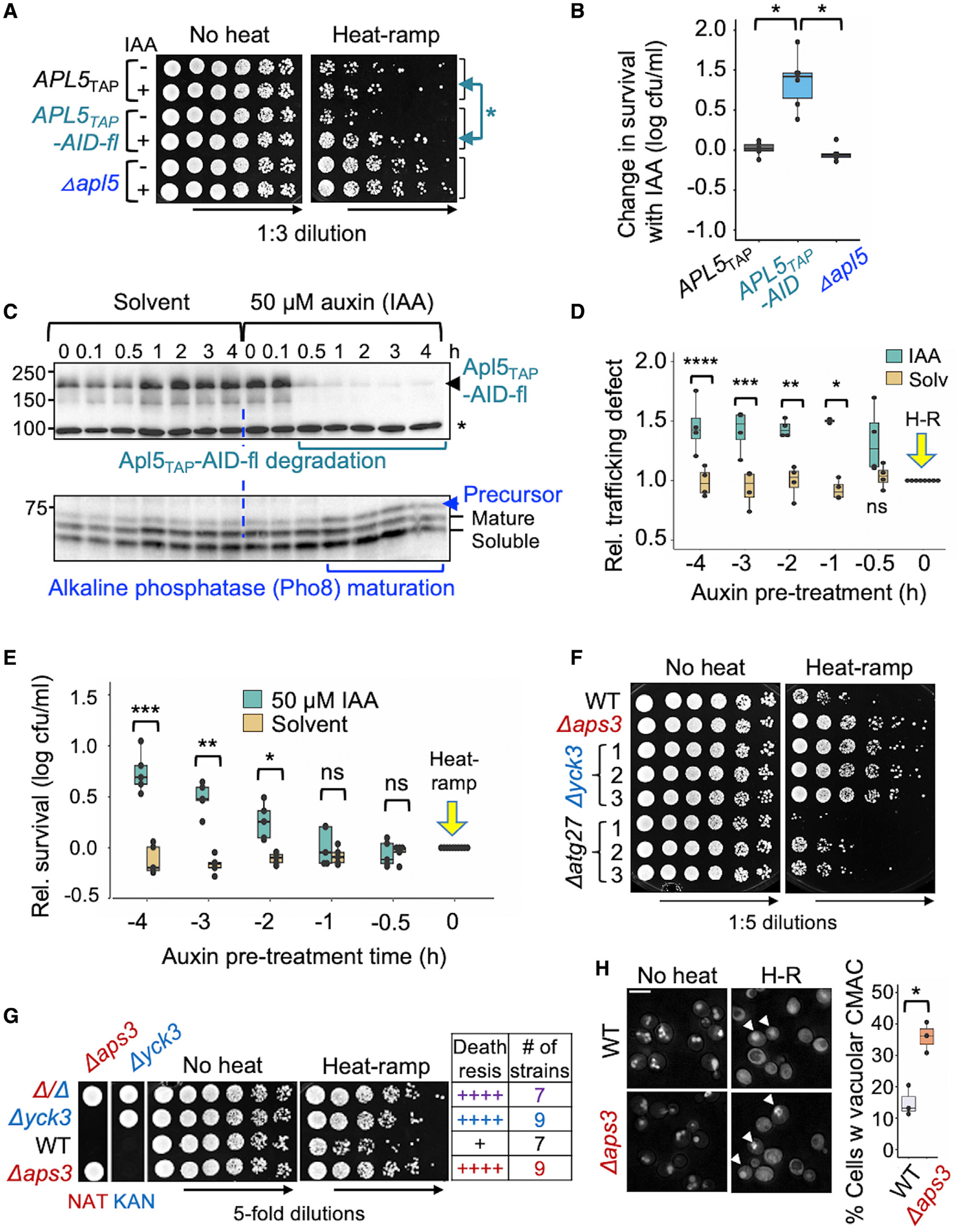
(A) Viability of yeast pre-treated 4 h with either solvent control (ethanol) or 50 μM auxin (IAA) to degrade Apl5TAP-AID-6xflag prior to heat-ramp and plated on agar in 3-fold serial dilutions.
(B) Quantification for (A); 6 independent experiments plotted as [(log10 cfus/mL with auxin) − (log10 cfus/mL with ethanol)]. Two-way ANOVA with Tukey’s HSD test post hoc: APL5TAP versus APL5TAP-AID (*p = 6.81 × 10−7); Δapl5 versus APL5TAP-AID (*p = 2.2 × 10−7); and Δapl5 versus APL5TAP (p = 0.931, ns).
(C) Immunoblots (upper) of yeast with anti-flag to monitor degradation of Apl5-TAP-AID-6xflag during 0–4 h with solvent control (ethanol) or 50 μM auxin (IAA). Immunoblot (lower) with anti-ALP/Pho8 to monitor AP-3 trafficking function based on maturation status of Pho8. *Non-specific band.
(D) Quantified AP-3 function for (C), calculated from densitometry as amount of precursor protein relative to total ALP/Pho8, and relative ALP defect is plotted as auxin/ethanol versus untreated for 4 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, ****p = 9.5 × 10−4, ***p = 5.7 × 10−4, **p = 0.00201, *p = 2.09 × 10−5, ns = 0.128.
(E) Relative survival of WT yeast pre-incubated with solvent alone or 50 μM auxin (IAA) prior to heat-ramp at time = 0, calculated as [(log10 cfus/mL with auxin or ethanol) − (log10 cfus/mL untreated)] for 5 independent experiments. Two-way ANOVA with Tukey’s HSD test post hoc, *p = 0.0016, **p = 5.2 × 10−9, ***p = <1.0 × 10−13.
(F) Heat-ramp cell death assay for three single-cell-derived substrains (BY4741) corresponding to AP-3 cargo proteins Yck3 and Atg27, and other cargo proteins (Figure S5).
(G) Heat-ramp cell death assay and corresponding genotypes verified by NAT or KAN (G418) drug selectivity for four spore-derived strains from a tetratype tetrad produced by sporulating heterozygous diploids (Δaps3:NatMx6 × Δyck3:KanMx4) and summary of death resistance scores for 32 spore-derived strains tested (8 tetrads).
(H) Fluorescent microscopy of WT and Δaps3 pre-stained with CMAC and imaged before and 30 min after heat-ramp. Arrowheads mark examples of heat-treated cells with retained vacuolar CMAC, quantified as percent of cells. Two-tailed t test for 3 fields from one experiment, counting >1,500 cells per genotype, *p = 0.0063.
To determine how long before the death stimulus AP-3 is needed to effect cell death, a time course of progressively shorter auxin pre-incubation times was tested. Apl5-AID protein was maximally degraded by ~30 min after auxin addition (Figure 5C, top), and AP-3 trafficking function was impaired by ~1 h based on ALP/Pho8 precursor accumulation (Figures 5C and 5D). For cell death, AP-3 was required between 1 and 2 h prior to heat-ramp, because degradation of Apl5 during the hour before a death stimulus did not significantly impair cell death (Figure 5E). This supports the model that AP-3 delivers sufficient amounts of its latent deadly cargo to the vacuole by 2 h before the death stimulus to initiate cell death. Thus, AP-3 is a critical component of this death pathway, but is not necessarily a direct effector of death.
AP-3 cargo protein Yck3 is a death effector of AP-3 at the vacuole
AP-3 cargo proteins are prime candidates for mediating cell death. AP-3 is likely a substantial contributor to the vacuole membrane proteome, yet relatively few AP-3 cargo proteins have been confirmed, and none had significant Z scores in our screen. However, most yeast knockouts contain a secondary mutation in at least 20% of their cell population that strongly affects cell death, typically increasing death (Teng et al., 2013). Therefore, we tested three single-cell-derived colonies/substrains of knockouts for 12 reported AP-3 cargo: ALP/Pho8, membrane-anchored protein kinase Yck3 (Sun et al., 2004), tSNARE Vam3 (Cowles et al., 1997), vSNARE Nyv1 (Darsow et al., 1998), amino acid transporters Ypq1-3 (Llinares et al., 2015), ESCRT-0 component Atg27 (Segarra et al., 2015), predicted zinc transporters Cot1 and Zrt3 (Yang et al., 2020), and the AP-3 reporter proteins Sna2 and Sna4 (Pokrzywa et al., 2009; Renard et al., 2010) of unclarified functions.
Among the 12 candidates, only the Δyck3 substrains were strikingly death-resistant, similar to AP-3 knockouts in validation tests (Figures 5F, S1, and S5), but not under the screen conditions (post-diauxic phase, 62°C heat-ramp), explaining why Δyck3 was missed in the screen Figure S6A). However, the screen phenotype is due to a second gene mutation in Δyck3 revealed only in post-diauxic phase, because segregation of the two genes by tetrad analysis confirmed that the pro-death effects in both log and post-diauxic phases are attributed to YCK3 (Figure S6B). Consistent with Yck3 working in the same AP-3 cell death pathway, double knockouts (Δaps3/Δyck3) phenocopy single knockouts (Figures 5G and S6C). Interestingly, endogenous and expressed Yck3 kinase were reported to promote loss of yeast viability after heat stress (Karim et al., 2018).
AP-3-dependent vacuole membrane permeabilization occurs before cell death
The release of vacuole contents into the cytoplasm could contribute to cell death, but the mechanisms of vacuole membrane permeabilization are not known. To determine if AP-3 promotes vacuole membrane permeabilization, cells were stained with the thiol-reactive dye CMAC-blue (210 molecular weight [mol wt]), which accumulates in the vacuole lumen and leaks into the cytoplasm if the vacuolar membrane is compromised (Karim et al., 2018; Llinares et al., 2015). Although both genotypes appear to undergo homotypic vacuole fusion, resulting in a single large vacuole following heat-ramp, fewer wild-type cells retained CMAC in the vacuole compared with Δaps3 (Figure 5H). Thus, AP-3 appears to promote vacuole membrane permeabilization.
To determine if vacuole membrane permeabilization is an early or late event in the dying process, CMAC release was monitored relative to the early cell death marker phloxine. Analogous to stochastic activation of apoptosis by mammalian cells treated with a death receptor agonist (Spencer et al., 2009), the onset of CMAC release under these conditions varied greatly, from 2 to 8.5 h (294 ± 131 min, n = 16 cells, excluding cells that released CMAC in the first 30 min after heat-ramp during set-up). Importantly, the time between CMAC release and the first hints of phloxine staining was 97 ± 48 min (range ~5–180 min, n = 18 cells from two independent experiments). In the example cell, CMAC begins to leak at specific points along the vacuole membrane ~6 h after heat-ramp (Figure 6A, arrows in heatmap) and gradually diffuses throughout the cell (gray scale). However, phloxine is not detectable until ~9 h after heat-ramp (arrowheads) and gradually intensifies over the next 30 min while CMAC dissipates (Figures 6A and S7A; Video S2). From Figure 1, the plasma membrane is not expected to become permeable until minutes to hours after phloxine staining. Our results imply that cells are alive when vacuole contents begin leaking into the cytoplasm. Thus, vacuole membrane permeabilization could represent a commitment point in the cell death pathway. Alternatively, mild vacuole permeability may be repairable, facilitated by delayed cell death.
Figure 6. AP-3-dependent vacuole membrane permeabilization.
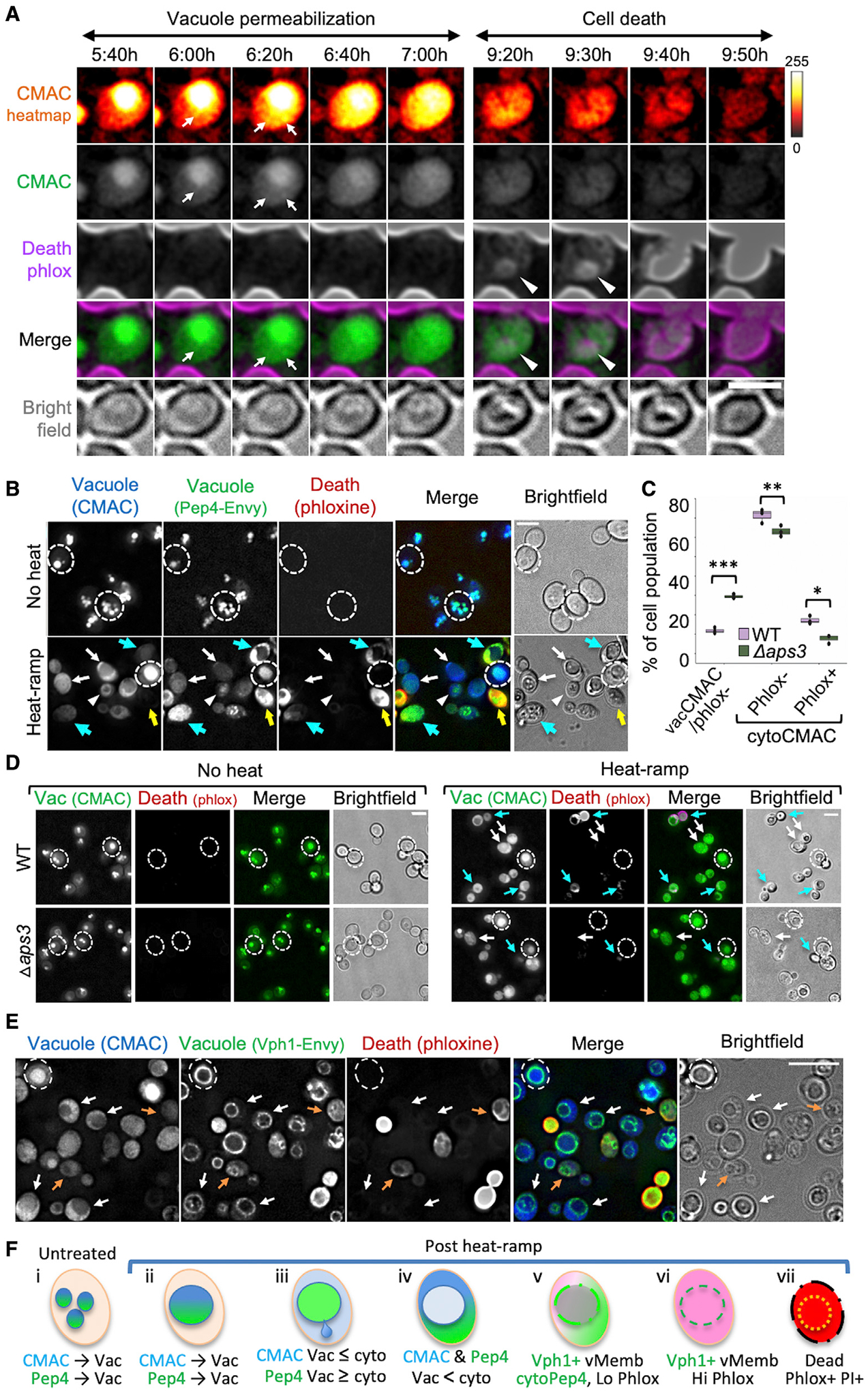
(A) Video microscopy images of wild-type yeast pre-stained with CMAC and imaged in phloxine at the indicated time stamps after heat-ramp. CMAC is presented as pixel intensity heatmap (color key, right), gray scale, and green fluorescence. Arrows mark spots where CMAC first emerges from the vacuole (depicted in Figure 6Fiii); arrowheads mark earliest phloxine staining. Scale bar, 2.5 μm.
(B) WT yeast expressing Pep4-Envy pre-stained with CMAC and imaged in phloxine ~20 min after heat-ramp. Dashed circles surround perimeter of example live (phlox−) cells that retain CMAC in the vacuole (Figure 6Fi and ii); arrowheads: example live (phlox−) cell with cytoplasmic CMAC but vacuolar Pep4 (Figure 6Fiii); white arrows: example live (phlox−) cells with partial or complete release of both CMAC and Pep4-Envy (Figure 6Fiii and iv); blue arrows: dying/dead cells (phlox+) with released Pep4 and fading/released CMAC (Figure 6Fv); and yellow arrows: dead cells (high phlox+) with released Pep4 and loss of CMAC (Figure 6Fvi). Scale bar, 5 μm.
(C) Quantification for (D) two-way ANOVA with Tukey’s HSD test post hoc for 3 fields from one experiment, counting >300 cells per genotype, *p = 0.0035, **p = 0.012, ***p = 1.05 × 10−5.
(D) WT and Δaps3 pre-stained with CMAC and imaged in phloxine before or ~30 min after heat-ramp. Dashed circles: surrounds perimeter of live (phlox−) cells with retained CMAC (Figure 6Fi and ii); white arrows: live (phlox−) cells with released CMAC (Figure 6Fiii and iv); and blue arrows: dead/dying (low or high phlox+) and released CMAC. Scale bar, 5 μm.
(E) WT yeast expressing Vph1-Envy pre-stained with CMAC and imaged in phloxine ~30 min after heat-ramp. Dashed circle: surrounds live (phlox−) cell with vacuolar CMAC (Figure 6Fii); white arrows: live (phlox−) with released CMAC and retained vacuole organelle structure (Vph1-Envy) (Figure 6Fv); and orange arrows: dying/dead cells (low phlox+) with released CMAC and indistinct vacuole membranes (Figure 6Fvi). Bright phlox+ cells depicted in Figure 6Fvii. Brightness of phloxine images in (B–E) was increased 20%. Scale bar, 5 μm.
(F) Diagram of the observed sequence of events described in (B–E).
To determine if vacuole membranes become permeable to molecules larger than CMAC, we monitored the localization of endogenous vacuolar protease Pep4 (human cathepsin D) with a fluorescent C-terminal tag (Gourlay and Ayscough, 2006; Mason et al., 2005; Pereira et al., 2010). Following heat-ramp, both Pep4-Envy (71.5 kDa) and CMAC were released from the vacuole lumen into the cytoplasm of live phloxine-negative cells (Figure 6B, white arrows). CMAC may leak more readily than Pep4 (arrowheads) and appears to diffuse away as dying cells accumulate phloxine (Figure 6B, blue/yellow arrows). Thus, the pores that develop in the vacuole membrane prior to staining for cell death are sufficiently large to pass intact proteins. Quantified data indicate that fewer wild-type cells retain vacuolar CMAC (Figures 6C and 6D), and those with released CMAC were more likely to be phloxine-stained compared with Δaps3 (Figure 6D, blue arrows). This supports the model that AP-3 promotes vacuole membrane permeabilization during the dying process.
Vacuole organelles appear to remain intact after CMAC release. To assess the structural integrity of vacuole membranes more directly, vacuole morphology was monitored in yeast expressing Envy-tagged endogenous Vph1, a subunit of the vacuole membrane-embedded Vo domain of the vacuolar V1Vo-ATPase. Rings of Vph1-Envy mark the vacuole membrane and remain clearly visible in cells with partially or fully released CMAC after heat-ramp (Figures 6E, white arrows, and 6F), although Vph1-Envy rings appear more uniform before CMAC release (dashed circle). However, vacuole membrane shapes are reorganized in phloxine-positive cells, possibly reflecting organelle fragmentation/fission or dissolution (Figure 6E, orange arrows, and Figure S7B). Thus, vacuole membranes become permeable before loss of organelle integrity.
Conservation of an AP-3-dependent cell death pathway
To test the possibility that AP-3 may promote cell death in a pathogenic yeast species, we tested the deletion strain of C. neoformans Ap3d1 (CNAG_02468), homolog of S. cerevisiae AP-3 subunit Apl5. Using a modified heat-ramp assay to accommodate the known heat-sensitivity of Cryptococcus species, we found that C. neoformans Δapl5, was strikingly death resistant compared with the wild-type control (Figures 7A and 7B). Thus, the AP-3 cell death pathway may extend to other fungal species. Interestingly, heat resistance of C. neoformans at 37°C body temperature is associated with virulence (Robert and Casadevall, 2009). Analysis of 4,800 fungal species for thermal tolerance revealed that a majority grow well below 30°C, but there was a rapid decline in the number of thermotolerant species as temperatures increase, and only a minority of fungal species survive at 37°C (Robert and Casadevall, 2009). Consequently, mammalian body temperatures create a thermal restriction zone for many potential fungal pathogens and provide considerable protection against mycoses. Thus, we also uncovered a new mechanism for thermal killing of fungi.
Figure 7. Cryptococcus neoformans AP-3 deficiency enhances survival.
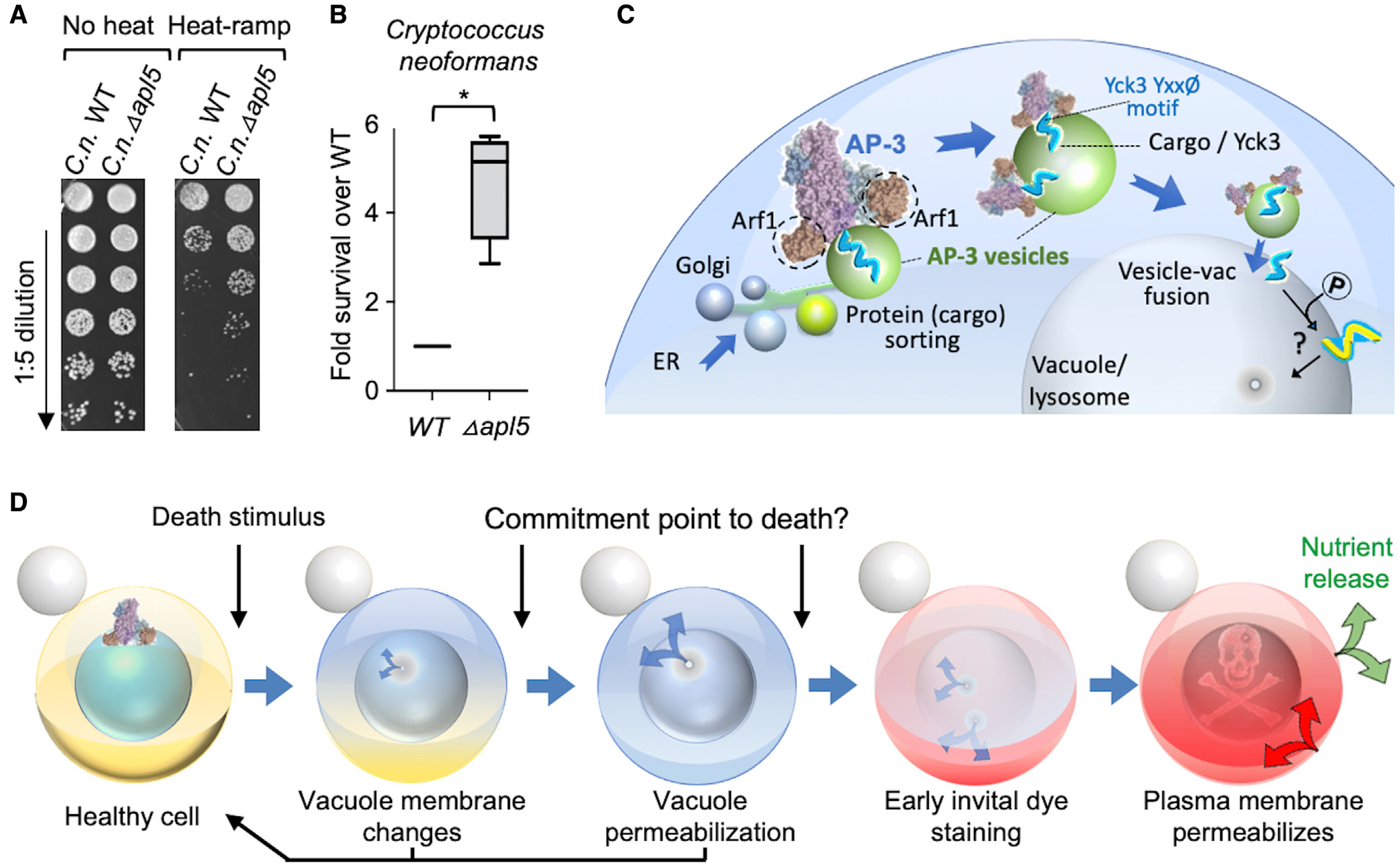
(A) Heat-ramp cell death assay adapted for Cryptococcus neoformans, comparing wild type and knockout of AP-3 subunit Apl5 homolog (CNAG_02468).
(B) Quantification for (A), boxplot of pooled results from 2 independent experiments testing the knockout for CNAG_02468 and two single-colony-derived knockout substrains normalized to wild-type KN99 control. Paired, two-tailed t test comparing log2 (cfu/mL) WT versus log2 (cfu/mL) ΔC.n. apl5 for heat-treated samples, *p = 0.0024; untreated samples are not different, p = 0.599.
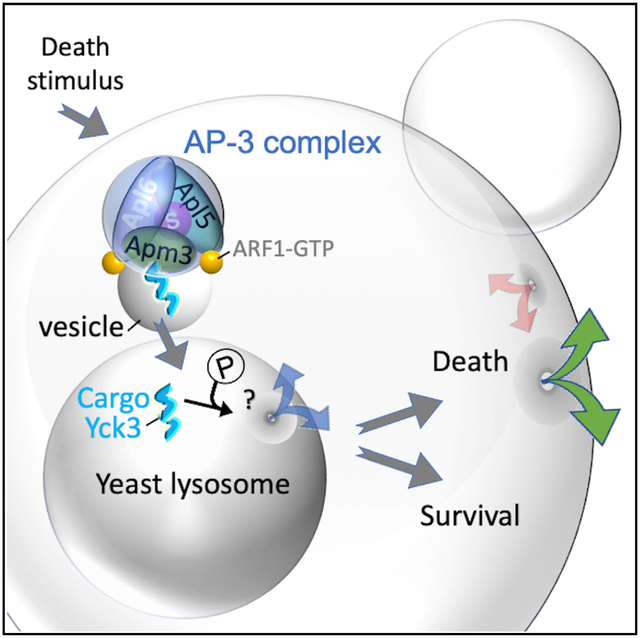
(C) Proposed yeast AP-3 vesicle trafficking cell death pathway depicts Arf1-dependent docking of AP-3 complex (based on PDB: 6DFF) at late/post-Golgi membranes (based on Video S3, in Levi et al., 2010); AP-3 recognition of Tyr motif in membrane-anchored casein kinase Yck3, which phosphorylates other proteins to promote vesicle fusion, vacuole membrane permeabilization, and cell death.
(D) Order of events and potential commitment points in the AP-3 cell death pathway.
DISCUSSION
Using genome-wide and targeted screens, we identified five cell-death-promoting genes in yeast, the four subunits of the AP-3 vesicle trafficking complex and Yck3 kinase, which is shuttled to the vacuole membrane by AP-3. We propose a model where AP-3 cargo proteins and potentially other factors contribute directly to vacuole membrane permeabilization, resulting in self-induced yeast cell death (Figure 7C). Other reports of vacuole membrane permeabilization (Mason et al., 2005; Pereira et al., 2013), together with our findings, imply that the AP-3 pathway responds to diverse death stimuli (heat, H2O2, and acetic acid). AP-3 potentially contributes to other death stimuli where vacuole membrane permeabilization occurs, such as hyperactivated Ras and spore death during nutrient limitation (Eastwood et al., 2012; Gourlay and Ayscough, 2006). This death pathway may be beneficial to survivors by providing nutrients liberated from dying cells, such as during starvation or yeast colony differentiation (Figure 7D) (Cap et al., 2012; Kireeva et al., 2021).
While a cell destined to die can be considered as dead from the point of insult, trivializing the mechanisms of death, it is important that not all cells die, and at the outset we cannot distinguish which cells will die or survive (e.g., Video S1). Similarly, we have shown that a fraction of animal cells exhibiting hallmarks of classic apoptosis in vivo (e.g., caspase activation) will ultimately survive (Tang et al., 2015). While unequal delivery of a death stimulus is one explanation for survival of a subset of cells, another explanation is the status of the death machinery within individual cells receiving equal doses, best demonstrated in cancer cells.
Like cancer cells, yeast cell death mechanisms may be subverted in those yeast that survive stress and regrow. We uncovered a potential mechanism of subversion–by reducing the AP-3 vesicle trafficking pathway or parts thereof. This is supported by evidence from genetic ablation, induced protein degradation (auxin), targeted disruption of AP-3 binding to its obligatory GTPase Arf1, and the faster recovery of AP-3-deficient cells following heat-ramp. Subversion of the AP-3 or other death pathway could potentially contribute to the pathogenicity of the heat-intolerant C. neoformans, which is responsible for 181,000 deaths per year worldwide (Rajasingham et al., 2017).
How do yeast die in the AP-3 pathway?
Our evidence suggests that AP-3 contributes more than other vacuole trafficking pathways to yeast cell death, implicating AP-3 cargo proteins as downstream death effectors. By unknown mechanisms, the contents of the vacuole lumen begin to emerge at distinct locations along the membrane (Figure 6A), possibly at unrepaired bilayers or through channels in the vacuole membrane. Several minutes to hours later, cells begin to exhibit marks of impending cell death detected by phloxine staining, which is followed by loss of plasma membrane integrity detected by PI minutes to hours afterward (Figure 7D).
The mechanism of vacuole membrane permeabilization during yeast cell death could be non-specific, such as lack of membrane repair in stressed cells or leakage caused by unfolded integral membrane proteins. However, the general trend in the mammalian cell death field is that specific death-promoting, pore-forming molecules are responsible for membrane permeabilization. Until recently, the large balloons emerging from dying mammalian cells were assumed to passively rupture. To the contrary, 16 kDa human/mouse NINJ1 oligomerizes to rupture these balloons during pyroptosis (Kayagaki et al., 2021).
Vacuolar proteases released into the cytoplasm have been prime candidates for yeast death effectors. However, reports are mixed on their roles in cell death (Chaves et al., 2021). Conceivably, vacuolar proteases could have a lesser role in cell death and a greater role in nutrient recycling by digesting cellular contents for survivors. This is somewhat analogous to mammalian apoptosis, where many genes participate in recognition and disposal of apoptotic corpses.
How might Yck3 kinase promote cell death?
AP-3 cargo proteins are obvious candidate cell death effectors. We identified one of these, the Yck3 protein kinase, and a second AP-3 cargo protein Zrt3 was reported to enhance yeast cell death (Terra-Matos et al., 2022). Yck3 interacts with AP-3 via a consensus tyrosine motif Yxxφ (444-YDSI) (Sun et al., 2004). Like ALP/Pho8, some Yck3 can reach the vacuole by default pathways, which may contribute to vacuole membrane permeabilization in AP-3-deficient cells.
By definition, both AP-3 and its cargo are present on both the membranes where vesicles are first formed (where Arf1 is required) and also on the destination membrane where cargo are delivered (vacuole). Thus, it is challenging to distinguish which of these sites is the nexus for cell death. However, an enforced retargeting strategy recently confirmed that Yck3 is dispensable for vesicle formation at origin membranes and instead functions at the vacuole membrane where Yck3 promotes tethering and SNARE-mediated fusion of AP-3 vesicles with vacuole membranes (Cabrera et al., 2010; Schoppe et al., 2020). This supports our model that Yck3 kinase promotes cell death by acting at the vacuole membrane.
Proteins that are phosphorylated by Yck3 kinase are also candidate effectors of cell death, including Vps41 in the HOPS complex that tethers AP-3 vesicles to the vacuole and orchestrates subsequent fusion (Cabrera et al., 2010; LaGrassa and Ungermann, 2005). Other targets of Yck3 include the vacuole membrane kinase Env7 (Manandhar et al., 2020); the guanine nucleotide exchange factor (GEF) for Ypt7/Rab7, Mon1-Ccz1 (Lawrence et al., 2014); and the SNARE Vam3 (Brett et al., 2008). Our cell death screen identified the RAG-like GTPases Gtr1 and Gtr2 (Figure 2B), which are known to modulate the activity of the target of rapamycin complex 1 (TORC1) in response to nutrients and are anchored to the vacuole membrane by Ego1/Meh1, a cargo protein of AP-3 (Hatakeyama et al., 2019). Ego1, Gtr1, and Gtr2 were previously reported to promote rapid cell death by activating TORC1 kinase in yeast cells experiencing ER stress (Kim and Cunningham, 2015). Consistent with these findings, yeast Whi2, a novel TORC1 inhibitor required to suppress TORC1 activity in low amino acid conditions, is also important to prevent cell death following stress (Chen et al., 2018; Teng and Hardwick, 2019; Teng et al., 2018).
Delineation of molecular cell dying processes in mammalian cells simultaneously revealed how tumor cells subvert normal dying processes, which in turn yielded newer and better therapies. These concepts may extend to microorganisms and to their failed responses to anti-fungal therapies. If borne out, we expect that knowledge of molecular fungal cell dying mechanisms will inform next-generation anti-fungal strategies to combat the rise in antibiotic-resistant fungal infections (Kulkarni et al., 2019).
Limitations of the study
A potential caveat to our model is that Yck3 kinase, in addition to being an AP-3 cargo protein, is also critical for the fusion of AP-3 vesicles to the vacuole. Thus, it is challenging to distinguish vesicle trafficking functions from death functions of Yck3. However, a role of Yck3 in vacuole fusion further supports our model that AP-3 contributes importantly to cell death via trafficking to the vacuole. A second limitation is that important yeast cell death genes are likely missed in our screen due to confounding secondary mutations, as reported here for Δyck3, or because their cell death functions are masked by their alternative day jobs in healthy cells, as in mammals (Chen et al., 2011; Fannjiang et al., 2003).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Dr JM Hardwick (hardwick@jhu.edu).
Materials availability
New strains and plasmids generated for this study will be made available upon request to the lead contact.
Data and code availability
Cell death screen data are reported in Table S1 (table key on line 4559). All Original western blot images have been deposited at Mendeley Data: https://doi.org/10.17632/h8mvg73p9t. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains, plasmids and media
Yeast strains and plasmids used in this work are listed in the key resources table, and were derived from BY, SEY, or W303 background strains of Saccharomyces cerevisiae, and KN99 background for Cryptococcus neoformans yeast strains. Frozen S. cerevisiae yeast stocks were streaked onto YPD (1% yeast extract, 2% peptone, and 2% dextrose) agar plates for 2 days at 30°C before amplifying in liquid cultures for experiments. Frozen stocks of Cryptococcus neoformans were streaked onto Sabouraud dextrose (SAB) agar plates for 2 days at 30°C before amplifying in SAB broth. YPD (S.c.) or SAB (C.n.) media were used throughout this study except where noted. Plasmids and linear DNAs were transformed into yeast by a high efficiency lithium acetate-carrier-polyethylene glycol method (Gietz and Schiestl, 2007). All yeast incubations were at 30°C.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG M2 (1:5000) | Sigma-Aldrich | Cat# F3165; RRID:AB_259529 |
| Mouse monoclonal anti-PGK 22C5D8 (1:5000) | Abcam | Abcam Cat# ab197960; RRID:AB_2756444 |
| Purified rabbit polyclonal anti-Pho8 (anti-CPY) (1:1000) | Gregory Payne; (Anand et al., 2009) | N/A |
| Purified rabbit polyclonal anti-Apm3 (1:5000) | Sandra Lemmon; (Panek et al., 1997) | N/A |
| Sheep anti-mouse HRP secondary (1:5000) | GE Healthcare | Cat# NA931; RRID:AB_772210 |
| Donkey anti-rabbit HRP secondary (1:5000) | GE Healthcare | Cat# NA934; RRID:AB_772206 |
| Chemicals, peptides, and recombinant proteins | ||
| CMAC (7-amino-4-chloromethycoumarin) | ThermoFisher Scientific | Cat#C2110 |
| Nourseothricin sulfate (NAT) | GoldBio | Cat#N500; CAS RN#96736-11-7 |
| Phloxine B | FisherScientific | Cat#P387-25 |
| Propidium Iodide | Invitrogen | Cat#P3566 |
| FM4-64 (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide) | ThermoFisher Scientific | Cat#T13320 |
| UVITEX 2B | Fisher Scientific (Polysciences, Inc.) | Cat#NC9859220 |
| IAA (3-indoleacetic acid) | Millipore Sigma | Cat#I3750 |
| Sabouraud dextrose (SAB) medium | BD Difco | BD238230; Cat#DF0382-17-9 |
| Deposited data | ||
| Uncropped western blots for all figures | Mendeley Data | https://doi.org/10.17632/h8mvg73p9t.1 |
| Experimental models: Organisms/strains | ||
| BY4709 [MATα ura3Δ0] | J. Boeke; (Brachmann et al., 1998) | N/A (Figure 1B–D) |
| BY4741 [MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0] | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 1C; 2D; S2) |
| BY4742 [MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0] | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 1C–D; 2–2I; 3E–3G; 5F; S5: S6; 5H) |
| BY4741-JMH [MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0] replicative aged | This paper | N/A (Figure 2C) |
| BY4741 Δdnm1 (Δdnm1::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figure 2C) |
| BY4741 Δfis1/whi2-1 (Δfis1::KanMX4, whi2-1) | (Cheng et al., 2008) | N/A (Figure 2C) |
| BY4741 Δapl5 (Δapl5::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 2C–D; 3G; 5A and 5B) |
| BY4741 Δapl6 (Δapl6::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 2C and 2D; Figure S2) |
| BY4741 Δapm3 (Δapm3::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 2C and 2D) |
| BY4741 Δaps3 (Δaps3::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 2C and 2D; Figure 3G) |
| BY4741 Δyck3 (Δyck3::KanMX4) | J. Boeke; (Brachmann et al., 1998) | N/A (Figures 5F and 5G; S6) |
| SEY6210 (MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801; GAL) | SusanMichaelis; (Robinson et al., 1988) | N/A |
| SEY6210 (CCY254 Δapl6::HIS3) | Scott Emr (Cowles et al., 1997) | N/A (Figures 2E and 2F) |
| SEY6210 (GOY4 Δapl5::HIS3) | Scott Emr (Cowles et al., 1997) | N/A (Figure 2E and 2F) |
| SEY6210 (GOY3 Δapm3::HIS3) | Scott Emr (Cowles et al., 1997) | N/A (Figure 2E and 2F) |
| SEY6210 (GOY5 Δaps3::HIS3) | Scott Emr (Cowles et al., 1997) | N/A (Figure 2E and 2F) |
| W303a [MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15] | Susan Michaelis, Johns Hopkins University | N/A (Figure 1C) |
| ZSY200 (BY4742 Δaps3::NatMX6) (homologous recombination) | This paper | N/A (Figures 2I; 5F; S5: S6; 5H) |
| ZSY201 (BY4709 pCRCT control colony #1 (D1) for APS3/APM3 CRISPR strains) | This paper | N/A (Figure S1G) |
| ZSY202 (BY4709 pCRCT control colony #2 (E1) for APS3/APM3 CRISPR strains) | This paper | N/A (Figures 2G–2H; S1C; 2A–2D; 3H–3J; 6A, 6C; S7) |
| ZSY203 (BY4709 Δaps3 CRISPR colony #1 (D2); sequence confirmed) | This paper | N/A (Figures S1E–S1H) |
| ZSY204 (BY4709 Δaps3 CRISPR colony #2 (E2); sequence confirmed) | This paper | N/A (Figures 2I; S1E–S1H, 3A–3D, 3H–3I; 6C) |
| ZSY205 (BY4709 Δaps3 CRISPR colony #3 (F2); sequence confirmed) | This paper | N/A (Figures S1E–S1H) |
| ZSY206 (BY4709 Δapm3 CRISPR colony #1 (D4); sequenced as WT) | This paper | N/A (Figures S1A–S1D) |
| ZSY207 (BY4709 Δapm3 CRISPR colony #2 (E4); sequenced as WT) | This paper | N/A (Figures S1A–S1D) |
| ZSY208 (Δapm3 CRISPR colony #3 (F2); sequence confirmed) | This paper | N/A (Figures 2G; S1A–S1D, 3J) |
| ZSY243 (ZSY200 + pRS303) [Δaps3::NatMX6, HIS3] | This paper | N/A (Figures 3E and 3F, S3) |
| ZSY248 (ZSY200 + APS3-flag) [Δaps3::NatMX6, APS3-Flag::HIS3] | This paper | N/A Figures 3E and 3F, S3) |
| ZSY209 (BY4709 pCRCT control colony #1 (B8) for APL6 CRISPR strains; sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY210 (BY4709 pCRCT control colony #2, B9, for APL6 CRISPR strains; sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY211 (BY4709 Δapl6 CRISPR colony #1 (C8); sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY212 (BY4709 Δapl6 CRISPR colony #2 (C9); sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY213 (BY4709 Δapl6 CRISPR colony #3 (C10); sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY214 (BY4709 apl6L117D/I120D CRISPR colony #1, D8; sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY215 (BY4709 apl6L117D/I120D CRISPR colony #2, D9; sequence confirmed) | This paper | N/A (Figures 4C–4F; Figure S4) |
| ZSY216 (BY4709 apl6L117D/I120D CRISPR colony #3, D10; sequence confirmed) | This paper | N/A (Figures 4B–4F; Figure S4) |
| ZSY217(Δyck3::KanMX4, Δaps3::NatMX6) [spore-derived from ZSY200 × Δyck3] | This paper | N/A (Figure 5G) |
| ZSY218 (Δyck3::KanMX4) [spore-derived from ZSY200 × Δyck3] | This paper | N/A (Figure 5G) |
| ZSY219 WT [spore-derived from ZSY200 × Δyck3] | This paper | N/A (Figure 5G) |
| ZSY220 (Δaps3::NatMX6) [spore-derived from ZSY200 × Δyck3] | This paper | N/A (Figure 5G) |
| APL5-TAP (BY4741 APL5-TAP::HIS3) | (Snyder et al., 2019) | N/A (Figures 5A and 5B) |
| APL5-AID (APL5-TAP-AID-6XFLAG::HIS3; OsTIR1::URA3) | Kyle Cunningham; (Snyder et al., 2019) | N/A Figures 5A–5E) |
| ZSY223 (BY4741 PEP4-ENVY::SpHIS5) | This paper | N/A (Figure 6B) |
| ZSY221 (BY4741 VPH1-ENVY::SpHIS5) | This paper | N/A (Figure 6D) |
| C. neoformans KN99alpha | FGSC 2016 Madhani plates, NIH R01AI100272 | http://www.fgsc.net/crypto/crypto.htm (Figures 7A and 7B) |
| C. neoformans Δapl5 (CNAG_02468) | FGSC 2016 Madhani plates, NIH R01AI100272 | http://www.fgsc.net/crypto/crypto.htm (Figures 7A and 7B) |
| Oligonucleotides | ||
| Sequences of Primers and CRISPR gene blocks are in Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: p41Nat 1-F GW | Leonid Kruglyak, UCLA (unpublished) | Addgene #58546 |
| Plasmid: pCRCT (encodes iCas9 + tracrRNA) | (Bao et al., 2015) | Addgene #60621 |
| Plasmid: pCRCT-APL6 disruption | This paper | N/A |
| Plasmid: pCRCT-APL6 L117D/I120D | This paper | N/A |
| Plasmid: pCRCT-APM3 disruption | This paper | N/A |
| Plasmid: pCRCT-APS3 disruption | This paper | N/A |
| Plasmid: pFA6A-link-GFPEnvy-SpHis5 | Linda Huang (Slubowski et al., 2015) | Addgene #60782 |
| Plasmid: pRS303 | Susan Michaelis, Johns Hopkins University | https://www.ncbi.nlm.nih.gov/nuccore/U03435 |
| Plasmid: pZDS100 (pRS303 + PAPS3-APS3-FLAG) | This paper | N/A |
| Plasmid: pRS416 + PSNA2-GFP-SNA2 | Pierre Morsomme; (Renard et al., 2010) | N/A |
| Other | ||
| Preprint of earlier version of this manuscript | BioRxiv | https://www.biorxiv.org/content/10.1101/2021.08.02.454728v1.full |
Yeast strain construction
Deletion and mutation strains
Sequences of primers and CRISPR gene blocks are in Table S2. CRISPR-modified strains were engineered using gBlocks with ≥45 bp flanking the Cas9 recognition site, and inserted into pCRCT (Bao et al., 2015). Transformants were maintained under selection (-uracil) for 6-days, and confirmed by sequencing the genomic locus. For APL6 knockin mutations, strains were screened for loss of CRISPR-Cas9 pCRCT plasmids prior to transformation with the cargo tracking plasmids pGFP-SNA2 (-URA). For new knockout strains generated by homologous recombination, the NatMX6 cassette was amplified by PCR from p41Nat 1-F GW using 45 bp primers flanking the targeted ORF of interest, transformed, and selected on YPD agar containing 100 μg/ml nourseothricin (NAT). For new spore-derived S. cerevisiae strains, diploids were generated by mating MATa × MATalpha, sporulated, and resulting spore tetrads were dissected. Resulting spore-derived strains were analyzed for parental genetic markers by standard tetrad analysis (Teng et al., 2013) in complete minimal medium SCAUX (0.67% yeast nitrogen base without amino acids, plus amino acids required for BY4741/BY4742 auxotrophies, 2% dextrose, 20 mg/L uracil) (Teng et al., 2018).
Genetic rescue and endogenously tagged strains
Genetic rescue of Δaps3 deletion was engineered using the integrative vector pRS303 with the APS3 gene plus 133 bp 5’ UTR (distance to next gene) and C-terminal flag-tag, linearized, transformed and integrated into the his3 locus. After selection (-histidine), strains were cultured in YPD. Envy-tagged strains were engineered by capturing GFP/Envy from pFA6A-link-GFPEnvy-SpHis5 using PEP4 or VPH1-specific primers (Slubowski et al., 2015). Auxin-inducible degron (AID)-tagged strains were constructed from the TAP-tag collection (Snyder et al., 2019).
Plasmid-expressing yeast strains
N-terminally tagged GFP-SNA2 expression plasmids with native promoter pRS416 (Renard et al., 2010) were transformed into yeast, selected and maintained on synthetic complete (SC) medium minus uracil (Teng et al., 2018).
METHOD DETAILS
Genome-wide cell death screen analysis
Screen analysis and batch corrections were performed for the heat-ramp cell death dataset previously reported (Teng et al., 2011, 2013). Briefly, cell death was induced in the 4,847 knockout strains from the BY4741 YKO collection (Brachmann et al., 1998). 48 h cultures (96 format) were resuspended, diluted in YPD, quickly transferred to a programmable thermocycler, treated with a linear heat-ramp of 30°C–62°C over 20 min, and immediately plated in eight replicates (4 per dilution) on YPD. Survival was determined at 18 h after plating by semiautomated enumeration of microscopic colonies using a BioSpot reader (Teng et al., 2011, 2013). For the current study, original images of the eight replicates were reacquired from the BioSpot reader and batch corrections were applied to systematically correct for undercounting at higher colony densities. Hit cutoff was set for death-resistant strains using a boxplot (1.5 × IQR above 75th percentile of all z-scores, ~2.47). Gene ontology (GO) component analysis was performed at the Saccharomyces Genome Database (SGD https://www.yeastgenome.org/).
Heat-ramp-induced cell death
Heat-ramp assays are tunable to adjust for metabolic state and growth medium. Three different heat-ramps were applied in this study: stepped 30°C–52°C over 18 min, linear 30°C–62°C over 20 min (Figures 2A and S6 for post-diauxic Saccharomyces), and linear 30°C–52°C over 27 min (Figure 7, for Cryptococcus).
Log-phase heat-ramp assay
Throughout this study (except where noted for Figures 2A, 7A, 7B, and S6), a stepped heat-ramp of 30°C–51°C, over 18 min was applied to small volumes of log phase Saccharomyces cerevisiae in YPD using a programmable thermocycler as graphed in Figure 1A (Teng et al., 2011; Teng and Hardwick, 2013). Briefly, low density yeast cultures grown in YPD on a roller drum at 30°C overnight (~16 h), were normalized to 0.2 OD600/mL in 4–5 mL fresh YPD, allowed to continue growing ~3–4 h to log phase with adjustment to have all strains arrive at the same growth state approximately simultaneously, and equal cell numbers (100 μL of ~0.5 OD600/mL YPD) were heat-ramp treated (additional details in Figure S6).
Cryptococcus neoformans heat-ramp
Strains were grown in SAB broth for 2 days at 30°C in stationary tubes/wells (no rolling or shaking), diluted 1:10 in fresh SAB, treated with a linear heat-ramp of 30°C–52°C over 27 min, and serial dilutions were quickly spotted on SAB agar plates incubated 2 days at 30°C before imaging.
Acetic acid-induced cell death
Acetic acid-induced death was previously described (Teng and Hardwick, 2009). Overnight cultures were normalized to 0.2 or 0.25 OD600 in 4–5 mL fresh YPD and grown for 3–4 h to log phase (~0.5 OD600). Cultures were normalized to equal cell numbers (0.4–0.5 OD600/mL) in 2 mL fresh YPD and without delay, 28 or 34.5 μL glacial acetic acid was added and vortexed for a final concentration of 242 or 300 mM acetic acid, respectively. Cultures were incubated at 30°C in a roller drum for up to 3 h. Samples were serially diluted 5-fold in YPD and cell suspensions (5 μL) were spotted on YPD agar plates and imaged after 2 days at 30°C.
H2O2-induced cell death
Overnight cultures were normalized to 0.25 OD600 in 4 mL fresh YPD and grown for 3 h to log phase (~0.5 OD600). Cultures were normalized to equal cell numbers (0.4 OD600/mL) in 2 mL fresh YPD and without delay, 10.2 μL or 15.3 μL of 30% H2O2 (final 50 mM or 75 mM H2O2) was added and vortexed immediately. Cultures were incubated at 30°C on a roller drum for up to 3 h. Samples were serially diluted 5-fold in YPD and spotted (5uL) on YPD agar plates and imaged after 2 days at 30°C.
Yeast cell death quantification
For colony forming units
For colony forming units (cfu) yeast treated with a death stimulus were determined by quickly spotting 3 μL or 5 μL of 5-fold serial dilutions (in YPD for S.c. or SAB for C.n.) of samples collected before and after heat-ramp. Plates (YPD or SAB) were incubated 18–24 h at 30°C for semi-automated counting of microscopic colonies on a BioSpot Reader, or incubated 2-days at 30°C for colony visualization and imaging. Cfu/mL was calculated relative to unheated yeast samples.
Liquid phase invital dye staining
Liquid phase invital dye staining at early times points (≤2 h) post heat-ramp permits resampling of the same cultures. Aliquots of cell suspensions (100 μL undiluted cultures) were heat-ramp treated and stained with phloxine B (2 μg/mL) or propidium iodide (10 μg/mL) for ~5 min, and 10 μL stained cells were loaded on a hemocytometer and imaged within ~15 min after heat-ramp on a Zeiss Axio Imager M2 (10x Olympus objective) equipped with a Hamamatsu Orca R2 camera. Unstained, phlox-stained and PI-stained cells were serially diluted and plated in parallel to verify that the presence of dyes had no detectable effects on viability based on cfu.
Solid phase invital dye staining and video microscopy.
To avoid counting the progeny of survivors that begin to proliferate a few hours after heat-ramp, cells were stained in liquid phase and immobilized by spotting 3 μL of cells diluted in YPD directly onto YPD agar immediately after heat-ramp, and imaged on the Zeiss (e.g. 24 h and 48 h, Figure 1B). To monitor viability of the same cells over time, cells immobilized on YPD-phloxine plates were imaged using an Applied Precision DeltaVision Elite microscope system (GE Healthcare) equipped with 20xPh objective and automated stage in a 30°C incubator chamber. Images were acquired every 10 min for 16 h after heat-ramp and analyzed using ImageJ/FIJI. Initial total cell count was calculated using phase images from the first imaging time point.
Immunoblot analysis
Approximately equal cell numbers (equivalent to 2.0 OD600) were pelleted and stored frozen at −80°C. For time course experiments, cells were rapidly lysed with 192 μl 100% trichloroacetic acid (TCA), held on ice for at least 30 min, and cell pellets were washed three times with water, once with acetone, dried under vacuum for 10 min and stored frozen at 80°C (adapted from (Hughes Hallett et al., 2015). Frozen pellets from either protocol were resuspended in 200 μl sample buffer (62.5 mM Tris-HCl pH 6.8, 2% w/v SDS, 10% glycerol, 0.01% bromophenol blue, fresh 50 mM DTT, and Halt™ protease inhibitor cocktail from ThermoFisher Scientific). Lysates were vortexed 4 × 45 s with glass beads (Sigma Aldrich), maintained on ice between pulses, and heated to 95°C for 5 min before analysis by SDS-PAGE. Blots were probed with antibodies against flag-epitope (Sigma, 1:5000), PGK (Abcam, 1:5000), Pho8 (1:1000, Gregory Payne), Apm3 (1:5000, Sandra Lemmon), followed by HRP-conjugated anti-rabbit or anti-mouse (GE Healthcare, 1:5000).
Auxin inducible degron experiments
Freshly prepared 10,000x stock (500 mM) 3-indole acetic acid (IAA) dissolved in 100% ethanol was prepared for each experiment and used to treat yeast cultures (YPD) at 50 μM IAA. TAP-tagged and auxin-inducible degron TAP-AID-tagged strains were grown overnight in 5 mL YPD on a roller drum at 30°C, normalized at 0.2 OD600 in 100 mL fresh YPD, and allowed to continue growing for 4 h to log phase (~0.5–0.7 OD600). Aliquots were pre-treated with 0.5 μl ethanol control or IAA (final concentration 50 μM), and diluted for heat-ramp treatment and lysate preparation.
Imaging vacuole membrane permeabilization
Normalized overnight cultures (0.2 OD600/mL) were allowed to regrow in fresh YPD ~4 h. During the final 30 min, 1-mL aliquots were stained at 30°C with FM4–64 (10 μg/mL) and/or CellTracker™ Blue CMAC (100 μM). Stained cultures were re-normalized to equal cell numbers (0.5 OD600/mL), heat-ramp treated, and 50 μL suspensions were immobilized for 10 min on a concanavalin A-coated 8-well Ibidi μ-Slide containing 200 μL of fresh YPD with cell wall stain UVITEX2B (1 μg/mL). Medium was aspirated and replaced with 200 μL of the media type required for imaging: YPD for phloxine B (2 μg/mL), SC for FITC/GFP channels to avoid autofluorescence, SCMIN (0.67% yeast nitrogen base without amino acids, 2% dextrose, 20 mg/L uracil) for BY4709, and SCAUX for BY4741/4742 strains. Images were captured on a DeltaVision Elite microscope with a 60x (N.A. 1.42) oil immersion objective in a 30°C incubator chamber, and single image slices or 0.2 μm Z-stacks were deconvolved (Softworx, Applied Precision). Time-lapse imaging used 10 min intervals, optical axis integration (OAI, z-sweep acquisition technology), deconvolution, and ImageJ/FIJI software for analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Genome-wide screen initial hit cutoff was set at 1.5x interquartile range beyond 75th percentile of all z-scores, ~2.47. Gene ontology (GO) component enrichment analysis of the 84 screen hits compared to all knockout strains was performed at the SGD database, and p values corrected for multiple hypothesis. Statistical analyses and graphs were generated using R versions 3.6.2 and 4.1.2, Prism 9.2.1 and 9.3.1 (GraphPad Software, Inc), SQLite 3.32.3 (open source), and Excel 16.54. For comparisons of two conditions, a Student’s t-test was applied, and for comparing 3 or more conditions, ANOVA with Tukey’s HSD post-hoc test was applied to determine statistical significance as detailed in figure legends. p values < 0.05 were considered significant. Values in the text for Figure 6A are mean ± SD.
Supplementary Material
Highlights.
Yeast cells die slowly in a gene-dependent manner following stress
Deletion or inhibition of AP-3 vesicle trafficking inhibits stress-induced death
AP-3 cargo protein Yck3 kinase promotes yeast cell death
The AP-3 death pathway results in vacuole membrane permeabilization prior to death
ACKNOWLEDGMENTS
We thank G. Payne, S. Lemmon, L. Kruglyak, L. Huang, P. Morsomme, S. Michaelis, J. Boeke, and S. Emr for advice and materials listed in key resources table. We thank P. Durand and E. Koonin for manuscript comments regarding evolution of cell death. Funding sources: National Institutes of Health, United States grants AI144373 and NS127076 (J.M.H.), AI115016 and AI153414 (K.W.C.), and AI052733, AI152078, and HL059842 (A.C.); National Natural Science Foundation of China 31970550; and the Priority Academic Program Development of the Jiangsu Higher Education Institutes (X.T.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110647.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
REFERENCES
- Ameisen JC (2002). On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ 9, 367–393. 10.1038/sj/cdd/4400950. [DOI] [PubMed] [Google Scholar]
- Anand VC, Daboussi L, Lorenz TC, and Payne GS (2009). Genome-wide analysis of AP-3-dependent protein transport in yeast. Mol. Biol. Cel 20, 1592–1604. 10.1091/mbc.E08-08-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouacheria A, Cunningham KW, Hardwick JM, Palková Z, Powers T, Severin FF, and Váchová L (2018). Comment on: sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science 360, eaar6910. 10.1126/science.aar6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, and Zhao H (2015). Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol 4, 585–594. 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- Baruffini E, Ruotolo R, Bisceglie F, Montalbano S, Ottonello S, Pelosi G, Buschini A, and Lodi T (2020). Mechanistic insights on the mode of action of an antiproliferative thiosemicarbazone-nickel complex revealed by an integrated chemogenomic profiling study. Scientific Rep 10, 10524. 10.1038/s41598-020-67439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, and Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. 10.1002/(SICI)1097-0061. [DOI] [PubMed] [Google Scholar]
- Brett CL, Plemel RL, Lobingier BT, Vignali M, Fields S, and Merz AJ (2008). Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J. Cel. Biol 182, 1141–1151. 10.1083/jcb.200801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Langemeyer L, Mari M, Rethmeier R, Orban I, Perz A, Brocker C, Griffith J, Klose D, Steinhoff HJ, et al. (2010). Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cel. Biol 191, 845–859. 10.1083/jcb.201004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cap M, Stepanek L, Harant K, Vachova L, and Palkova Z (2012). Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol. Cel 46, 436–448. 10.1016/j.molcel.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Casler JC, and Glick BS (2020). A microscopy-based kinetic analysis of yeast vacuolar protein sorting. eLife 9. 10.7554/eLife.5684410.7554/eLife.56844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves SR, Rego A, Martins VM, Santos-Pereira C, Sousa MJ, and Côrte-Real M (2021). Regulation of cell death induced by acetic acid in yeasts. Front. Cell Dev. Biol 9. 10.3389/fcell.2021.642375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang G, Zhang Y, Dayhoff-Brannigan M, Diny NL, Zhao M, He G, Sing CN, Metz KA, Stolp ZD, et al. (2018). Whi2 is a conserved negative regulator of TORC1 in response to low amino acids. PLoS Genet 14, e1007592. 10.1371/journal.pgen.1007592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Aon MA, Hsu YT, Soane L, Teng X, McCaffery JM, Cheng WC, Qi B, Li H, Alavian KN, et al. (2011). Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J. Cel. Biol 195, 263–276. 10.1083/jcb.201108059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, and Hardwick JM (2008). Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ 15, 1838–1846. 10.1038/cdd.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, and Emr SD (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, and Payne GS (2012). Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat. Cel. Biol 14, 239–248. 10.1038/ncb2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T, Burd CG, and Emr SD (1998). Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J. Cel. Biol 142, 913–922. 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Mitchell PS, Sandstrom A, Vance RE, and Glass NL (2020). Molecular characterization of a fungal gasdermin-like protein. Proc. Natl. Acad. Sci. United States America 117, 18600–18607. 10.1073/pnas.2004876117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Hu J, Fan L, and Chen Q (2017). RNA-Seq-based transcriptomic and metabolomic analysis reveal stress responses and programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Scientific Rep 7, 42659. 10.1038/srep42659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand PMC (2020). Chapter 13: programmed cell death at the levels of selection. In The Evolutionary Origins of Life and Death (The University of Chicago Press; ). [Google Scholar]
- Eastwood MD, Cheung SW, Lee KY, Moffat J, and Meneghini MD (2012). Developmentally programmed nuclear destruction during yeast game-togenesis. Dev. Cel 23, 35–44. 10.1016/j.devcel.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Eastwood MD, Cheung SW, and Meneghini MD (2013). Programmed nuclear destruction in yeast: self-eating by vacuolar lysis. Autophagy 9, 263–265. 10.4161/auto.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, and Hardwick JM (2004). Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev 18, 2785–2797. 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y, Kim CH, Huganir RL, Zou S, Lindsten T, Thompson CB, Mito T, Traystman RJ, Larsen T, Griffin DE, et al. (2003). BAK alters neuronal excitability and can switch from anti- to pro-death function during postnatal development. Dev. Cel 4, 575–585. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. (2018). Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ 25, 486–541. 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Chau S, Chowdhury F, Zhou T, Hossain S, McQuibban GA, and Meneghini MD (2019). Meiotic viral attenuation through an ancestral apoptotic pathway. Proc. Natl. Acad. Sci 116, 16454–16462. 10.1073/pnas.1900751116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, and Schiestl RH (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc 2, 31–34. 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Gourlay CW, and Ayscough KR (2006). Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol 26, 6487–6501. 10.1128/MCB.00117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama R, Peli-Gulli MP, Hu Z, Jaquenoud M, Garcia Osuna GM, Sardu A, Dengjel J, and De Virgilio C (2019). Spatially distinct pools of TORC1 balance protein homeostasis. Mol. Cel 73, 325–338.e8. 10.1016/j.molcel.2018.10.040. [DOI] [PubMed] [Google Scholar]
- Heller J, Clave C, Gladieux P, Saupe SJ, and Glass NL (2018). NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc. Natl. Acad. Sci. United States America 115, E2292–E2301. 10.1073/pnas.1719705115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettenbrenner S, Maier S, Leisser C, Polgar D, Strasser S, Grusch M, and Krupitza G (2003). The evolution of cell death programs as prerequisites of multicellularity. Mutat. Res 543, 235–249. 10.1016/s1383-5742(02)00110-2. [DOI] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, and Capaldi AP (2015). Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4. 10.7554/eLife.09181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Lobkovsky AE, Wolf YI, and Koonin EV (2014). Virus-host arms race at the joint origin of multicellularity and programmed cell death. Cell cycle 13, 3083–3088. 10.4161/15384101.2014.949496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, and Hardwick JM (2005). Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cel. Biol 170, 391–399. 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarolim S, Ayer A, Pillay B, Gee AC, Phrakaysone A, Perrone GG, Breitenbach M, and Dawes IW (2013). Saccharomyces cerevisiae genes involved in survival of heat shock. G3 3, 2321–2333. 10.1534/g3.113.007971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AG, Wein T, Mayer ML, Duncan-Lowey B, Yirmiya E, Oppenheimer-Shaanan Y, Amitai G, Sorek R, and Kranzusch PJ (2022). Bacterial gasdermins reveal an ancient mechanism of cell death. Science 375, 221–225. 10.1126/science.abj8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MA, McNally EK, Samyn DR, Mattie S, and Brett CL (2018). Rab-effector-kinase interplay modulates intralumenal fragment formation during vacuole fusion. Dev. Cel 47, 80–97.e6. 10.1016/j.devcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Kornfeld OS, Lee BL, Stowe IB, O’Rourke K, Li Q, Sandoval W, Yan D, Kang J, Xu M, et al. (2021). NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136. 10.1038/s41586-021-03218-7. [DOI] [PubMed] [Google Scholar]
- Kim A, and Cunningham KW (2015). A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress. Mol. Biol. Cel 26, 4631–4645. 10.1091/mbc.E15-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim A, and Cunningham KW (2012). Vacuolar H+-ATPase (VATPase) promotes vacuolar membrane permeabilization and nonapoptotic death in stressed yeast. J. Biol. Chem 287, 19029–19039. 10.1074/jbc.M112.363390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva NA, Sokolov SS, Smirnova EA, Galkina KV, Severin FF, and Knorre DA (2021). Adaptive role of cell death in yeast communities stressed with macrolide antifungals. mSphere 6, e0074521. 10.1128/mSphere.00745-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M, Stolp ZD, and Hardwick JM (2019). Targeting intrinsic cell death pathways to control fungal pathogens. Biochem. Pharmacol 162, 71–78. 10.1016/j.bcp.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwolek-Mirek M, and Zadrag-Tecza R (2014). Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS yeast Res 14, 1068–1079. 10.1111/1567-1364.12202. [DOI] [PubMed] [Google Scholar]
- LaGrassa TJ, and Ungermann C (2005). The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex.J. Cel. Biol 168, 401–414. 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas PJ, Cunningham KW, Liu XF, Fink GR, and Culotta VC (1995). Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol. Cell. Biol 15, 1382–1388. 10.1128/MCB.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence G, Brown CC, Flood BA, Karunakaran S, Cabrera M, Nordmann M, Ungermann C, and Fratti RA (2014). Dynamic association of the PI3P-interacting Mon1-Ccz1 GEF with vacuoles is controlled through its phosphorylation by the type 1 casein kinase Yck3. Mol. Biol. Cel 25, 1608–1619. 10.1091/mbc.E13-08-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi SK, Bhattacharyya D, Strack RL, Austin JR 2nd, and Glick BS (2010). The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway. Traffic 11, 1168–1179. 10.1111/j.1600-0854.2010.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J (2016). Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158. 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinares E, Barry AO, and Andre B (2015). The AP-3 adaptor complex mediates sorting of yeast and mammalian PQ-loop-family basic amino acid transporters to the vacuolar/lysosomal membrane. Scientific Rep 5, 16665. 10.1038/srep16665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, and Williams CM (1965). Programmed cell death–I. Cytology of degeneration in the intersegmental muscles of the pernyi silkmoth. J. Insect Physiol 11, 123–133. [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, and Klionsky DJ (2010). The Cvt pathway as a model for selective autophagy. FEBS Lett 584, 1359–1366. 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar SP, Siddiqah IM, Cocca SM, and Gharakhanian E (2020). A kinase cascade on the yeast lysosomal vacuole regulates its membrane dynamics: conserved kinase Env7 is phosphorylated by casein kinase Yck3. J. Biol. Chem 295, 12262–12278. 10.1074/jbc.RA119.012346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DA, Shulga N, Undavai S, Ferrando-May E, Rexach MF, and Goldfarb DS (2005). Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS yeast Res 5, 1237–1251. 10.1016/j.femsyr.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Minina EA, Staal J, Alvarez VE, Berges JA, Berman-Frank I, Beyaert R, Bidle KD, Bornancin F, Casanova M, Cazzulo JJ, et al. (2020). Classification and nomenclature of metacaspases and paracaspases: No more confusion with caspases. Mol. Cel 77, 927–929. 10.1016/j.molcel.2019.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KL, Buffalo CZ, Sturzel CM, Heusinger E, Kirchhoff F, Ren X, and Hurley JH (2018). HIV-1 nefs are cargo-sensitive AP-1 trimerization switches in tetherin downregulation. Cell 174, 659–671.e14. 10.1016/j.cell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, and Segawa K (2021). Sensing and clearance of apoptotic cells. Curr. Opin. Immunol 68, 1–8. 10.1016/j.coi.2020.07.007. [DOI] [PubMed] [Google Scholar]
- Nie Z, Boehm M, Boja ES, Vass WC, Bonifacino JS, Fales HM, and Randazzo PA (2003). Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev. Cell 5, 513–521. 10.1016/s1534-5807(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Nishimura K, and Kanemaki MT (2014). Rapid depletion of budding yeast proteins via the fusion of an auxin-inducible degron (AID). Curr. Protoc. Cell Biol 64, 20.9.1–20.9.16. 10.1002/0471143030.cb2009s64. [DOI] [PubMed] [Google Scholar]
- Nowak MA, Tarnita CE, and Wilson EO (2010). The evolution of eusociality. Nature 466, 1057–1062. 10.1038/nature09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, Tal N, Malheiro DBA, Malitsky S, Amitai G, and Sorek R (2021). Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature 600, 116–120. 10.1038/s41586-021-04098-7. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Dell’Angelica EC, and Bonifacino JS (1998). ADP-Ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cel. Biol 142, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek HR, Stepp JD, Engle HM, Marks KM, Tan PK, Lemmon SK, and Robinson LC (1997). Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J 16, 4194–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C, Chaves S, Alves S, Salin B, Camougrand N, Manon S, Sousa MJ, and Corte-Real M (2010). Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol. Microbiol 76, 1398–1410. 10.1111/j.1365-2958.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- Pereira H, Azevedo F, Rego A, Sousa MJ, Chaves SR, and Corte-Real M (2013). The protective role of yeast cathepsin D in acetic acid-induced apoptosis depends on ANT (Aac2p) but not on the voltage-dependent channel (Por1p). FEBS Lett 587, 200–205. 10.1016/j.febslet.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Pokrzywa W, Guerriat B, Dodzian J, and Morsomme P (2009). Dual sorting of the Saccharomyces cerevisiae vacuolar protein Sna4p. Eukaryot. Cel 8, 278–286. 10.1128/EC.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniakovsky AI, Knorre DA, Markova OV, Hyman AA, Skulachev VP, and Severin FF (2005). Role of mitochondria in the pheromone- and amiodar-one-induced programmed death of yeast. J. Cel. Biol 168, 257–269. 10.1083/jcb.200408145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller TM, Denning DW, Loyse A, and Boulware DR (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis 17, 873–881. 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Farias GG, Canagarajah BJ, Bonifacino JS, and Hurley JH (2013). Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152, 755–767. 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard HF, Demaegd D, Guerriat B, and Morsomme P (2010). Efficient ER exit and vacuole targeting of yeast Sna2p require two tyrosine-based sorting motifs. Traffic 11, 931–946. 10.1111/j.1600-0854.2010.01070.x. [DOI] [PubMed] [Google Scholar]
- Robert VA, and Casadevall A (2009). Vertebrate endothermy restricts most fungi as potential pathogens. J. Infect. Dis 200, 1623–1626. 10.1086/644642. [DOI] [PubMed] [Google Scholar]
- Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, Kipps TJ, Anderson MA, Brown JR, Gressick L, et al. (2016). Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. New Engl. J. Med 374, 311–322. 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, and Emr SD (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol 8, 4936–4948. 10.1128/mcb.8.11.4936-4948.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppe J, Mari M, Yavavli E, Auffarth K, Cabrera M, Walter S, Frohlich F, and Ungermann C (2020). AP-3 vesicle uncoating occurs after HOPS-dependent vacuole tethering. EMBO J 39, e105117. 10.15252/embj.2020105117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN, Sowerby PJ, and Robinson MS (1996). Cytosolic and membrane-associated proteins involved in the recruitment of AP-1 adaptors onto the trans-Golgi network. J. Biol. Chem 271, 25446–25451. 10.1074/jbc.271.41.25446. [DOI] [PubMed] [Google Scholar]
- Segarra VA, Boettner DR, and Lemmon SK (2015). Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization. Traffic 16, 365–378. 10.1111/tra.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slubowski CJ, Funk AD, Roesner JM, Paulissen SM, and Huang LS (2015). Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins. Envy Ivy. Yeast 32, 379–387. 10.1002/yea.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder NA, Kim A, Kester L, Gale AN, Studer C, Hoepfner D, Roggo S, Helliwell SB, and Cunningham KW (2019). Auxin-inducible depletion of the essentialome suggests inhibition of TORC1 by auxins and inhibition of vrg4 by SDZ 90–215, a natural antifungal cyclopeptide. G3 9, 829–840. 10.1534/g3.118.200748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa M, Duarte AM, Fernandes TR, Chaves SR, Pacheco A, Leao C, Corte-Real M, and Sousa MJ (2013). Genome-wide identification of genes involved in the positive and negative regulation of acetic acid-induced programmed cell death in Saccharomyces cerevisiae. BMC genomics 14, 838. 10.1186/1471-2164-14-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Gaudet S, Albeck JG, Burke JM, and Sorger PK (2009). Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature 459, 428–432. 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp JD, Huang K, and Lemmon SK (1997). The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J. Cell Biol 139, 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott KE, Loyse A, Jarvis JN, Alufandika M, Harrison TS, Mwandumba HC, Day JN, Lalloo DG, Bicanic T, Perfect JR, and Hope W (2021). Cryptococcal meningoencephalitis: time for action. The Lancet Infect. Dis 10.1016/S1473-3099(20)30771-4. [DOI] [PubMed] [Google Scholar]
- Sun B, Chen L, Cao W, Roth AF, and Davis NG (2004). The yeast casein kinase Yck3p is palmitoylated, then sorted to the vacuolar membrane with AP-3-dependent recognition of a YXXPhi adaptin sorting signal. Mol. Biol. Cel 15, 1397–1406. 10.1091/mbc.E03-09-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Tang HM, Fung MC, and Hardwick JM (2015). In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Scientific reports 5, 9015. 10.1038/srep09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Cheng WC, Qi B, Yu TX, Ramachandran K, Boersma MD, Hattier T, Lehmann PV, Pineda FJ, and Hardwick JM (2011). Gene-dependent cell death in yeast. Cell Death Dis 2, e188. 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, Dayhoff-Brannigan M, Cheng WC, Gilbert CE, Sing CN, Diny NL, Wheelan SJ, Dunham MJ, Boeke JD, Pineda FJ, and Hardwick JM (2013). Genome-wide consequences of deleting any single gene. Mol. Cel 52, 485–494. 10.1016/j.molcel.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, and Hardwick JM (2009). Reliable method for detection of programmed cell death in yeast. Methods Mol. Biol 559, 335–342. 10.1007/978-1-60327-017-5_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, and Hardwick JM (2013). Quantification of genetically controlled cell death in budding yeast. Methods Mol. Biol 1004, 161–170. 10.1007/978-1-62703-383-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, and Hardwick JM (2015). Cell death in genome evolution. Semin. Cel. Dev. Biol 39, 3–11. 10.1016/j.semcdb.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X, and Hardwick JM (2019). Whi2: a new player in amino acid sensing. Curr. Genet 65, 701–709. 10.1007/s00294-018-00929-9. [DOI] [PubMed] [Google Scholar]
- Teng X, Yau E, Sing C, and Hardwick JM (2018). Whi2 signals low leucine availability to halt yeast growth and cell death. FEMS yeast Res 18. 10.1093/femsyr/foy1095. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra-Matos J, Teixeira MO, Santos-Pereira C, Noronha H, Domingues L, Sieiro C, Gerós H, Chaves SR, Sousa MJ, and Côrte-Real M (2022). Saccharomyces cerevisiae cells lacking the zinc vacuolar transporter Zrt3 display improved ethanol productivity in lignocellulosic hydrolysates.J. Fungi 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RT, and Selmecki A (2020). Expandable and reversible copy number amplification drives rapid adaptation to antifungal drugs. eLife 9. 10.7554/eLife.58349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanchurin V, Wolf YI, Katsnelson MI, and Koonin EV (2022). Toward a theory of evolution as multilevel learning. Proc. Natl. Acad. Sci 119, e2120037119. 10.1073/pnas.2120037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez R, Zamora E, Alvarez M, Alvarez ML, and Ramirez M (2016). Using mixed inocula of Saccharomyces cerevisiae killer strains to improve the quality of traditional sparkling-wine. Food Microbiol 59, 150–160. 10.1016/j.fm.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, and Talbot NJ (2006). Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 312, 580–583. 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- Vilela-Moura A, Schuller D, Mendes-Faia A, Silva RD, Chaves SR, Sousa MJ, and Corte-Real M (2011). The impact of acetate metabolism on yeast fermentative performance and wine quality: reduction of volatile acidity of grape musts and wines. Appl. Microbiol. Biotechnol 89, 271–280. 10.1007/s00253-010-2898-3. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, and Payne GS (1998). A dileucine-like sorting signal directs transport into an AP-3-dependent, clathrin-independent pathway to the yeast vacuole. EMBO J 17, 2482–2493. 10.1093/emboj/17.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, and Khaled WT (2020). Mammary development in the embryo and adult: new insights into the journey of morphogenesis and commitment. Development 147. 10.1242/dev.169862. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang W, Wen X, Bulinski PJ, Chomchai DA, Arines FM, Liu Y-Y, Sprenger S, Teis D, Klionsky DJ, and Li M (2020). TORC1 regulates vacuole membrane composition through ubiquitin- and ESCRT-dependent microautophagy. J. Cell Biol 219. 10.1083/jcb.201902127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cell death screen data are reported in Table S1 (table key on line 4559). All Original western blot images have been deposited at Mendeley Data: https://doi.org/10.17632/h8mvg73p9t. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


