Abstract
Tetracycline-resistant Pasteurella aerogenes isolates obtained from the intestinal tract of swine were investigated for their tet genes by PCR analysis and hybridization experiments. In contrast to Pasteurella isolates from the respiratory tract, tet(H) genes were detected in the chromosomal DNA of only 2 of the 24 isolates, one of which also carried two copies of a tet(B) gene. All other P. aerogenes isolates carried tet(B) genes, which are the predominant tet genes among Enterobacteriaceae. A single isolate harbored a tet(B) gene as part of a truncated Tn10 element on the 4.8-kb plasmid pPAT2. Comparative analysis of the pPAT2 sequence suggested that the Tn10 relic on plasmid pPAT2 is the result of several illegitimate recombination events. The remaining 21 P. aerogenes isolates carried one or two copies of the tet(B) gene in their chromosomal DNA. In the majority of the cases, these tet(B) genes were associated with copies of Tn10 as confirmed by their SfuI and BamHI hybridization patterns. No correlation between the number of tet gene copies and the MICs of tetracycline, doxycyline and minocycline was observed.
Studies on antimicrobial resistance in bacteria presently assigned to the genus Pasteurella almost exclusively concentrate on the resistance properties of Pasteurella multocida, which represents a primary pathogen in food-producing animals, including cattle (hemorrhagic septicemia), poultry (fowl cholera), and rabbits (snuffles) (4, 34). Moreover, P. multocida strains and also bacteria of the new genus Mannheimia, which includes bacteria formerly assigned to the Pasteurella haemolytica complex (1), are also involved in multicausal respiratory diseases in swine (enzootic pneumonia and progressive atrophic rhinitis) and ruminants (enzootic bronchopneumonia in cattle, sheep, and goats), as well as in small laboratory rodents and fur-bearing animals (4, 34).
While P. multocida and those Mannheimia spp. which are involved in animal diseases are inhabitants of the mucosal surfaces of the respiratory tract, Pasteurella aerogenes represents part of the physiological flora in the intestinal tract of swine (20). Occasionally, P. aerogenes has been found to be associated with abortion and stillbirth in swine, dogs, and rabbits (20). Local wound infections in human due to P. aerogenes mainly occur in veterinarians, abattoir workers, and animal caretakers after swine bites. One report, however, also described the association of P. aerogenes with stillbirth in a woman who worked on a pig farm (39). So far, little is known about antimicrobial resistance in P. aerogenes isolates and their potential role in the diffusion of antimicrobial resistance genes among intestinal bacteria. A study on β-lactam resistance described the presence of a chromosomally located blaROB-1 gene in a single bovine P. aerogenes isolate (26), whereas a tetracycline (Tc) resistance gene of hybridization class H was detected on the 5.5-kb plasmid pPAT1 in a single porcine P. aerogenes isolate (17).
Tc resistance is a highly heterogeneous resistance property in which more than 30 different genes are involved (25, 35), many of which are located on either plasmids or transposons. Since Tcs also represent almost two-thirds of all antimicrobials used in veterinary medicine in the European Union and Switzerland (http://www.fedesa.be/eng/PublicSite/xtra/dossiers/doss9/), there is a high selective pressure under which the respective resistance genes may be exchanged. Thus, some tet genes are widely distributed among bacteria of various species and genera, whereas others are restricted to few bacterial genera living in specific habitats (35). An example for this latter case is the gene tet(H), which so far has exclusively been identified among isolates of the two closely related genera Pasteurella and Mannheimia, almost all of which were obtained from the respiratory tract of cattle, pigs, or turkeys (13, 14, 17, 18).
The aims of this study were to determine which class(es) of tet genes is present in P. aerogenes isolates obtained from the porcine intestinal tract and whether these genes are associated with plasmids and transposons. The results obtained from this study were expected to provide insight into whether enteric P. aerogenes isolates carry those tet genes predominantly seen among Enterobacteriaceae and other gram-negative enteric bacteria or those previously encountered in P. multocida and P. haemolytica isolates from the respiratory tract.
(This study was presented in part at the 40th Interscience Conference on Antimicrobial Agents and Chemotherapy, Toronto, Canada, 17 to 20 September 2000.)
MATERIALS AND METHODS
Bacterial isolates and antimicrobial susceptibility testing.
The 24 epidemiologically unrelated porcine P. aerogenes field isolates were obtained between October 1997 and April 2000 from fecal samples submitted to the Ahlemer Institute in Hannover, Germany, and were kindly provided by J. Mumme. Biochemical confirmation of the P. aerogenes isolates followed standard procedures (5, 20). The reference strain P. aerogenes DSM10153 (obtained from the national strain collection) (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) was included in these confirmatory tests. The P. aerogenes isolates were cultivated overnight at 37°C on blood agar base (Oxoid, Wesel, Germany) supplemented with 5% (vol/vol) sheep blood.
All P. aerogenes isolates were investigated for resistance to ampicillin (10 μg), chloramphenicol (30 μg), florfenicol (30 μg), gentamicin (10 μg), kanamycin (30 μg), streptomycin (10 μg), sulfamethoxazole (23.75 μg), Tc (30 μg), and trimethoprim (5 μg) by the disk diffusion test (32) on Mueller-Hinton agar (Oxoid) Zones of growth inhibition were evaluated after incubation for 16 h at 35°C according to the NCCLS standards (32) or according to the manufacturer's recommendations (Oxoid) using the following zone diameters for considering an isolate as resistant: ≤11 mm (streptomycin), ≤12 mm (chloramphenicol, gentamicin, and trimethoprim), ≤13 mm (ampicillin and kanamycin), ≤14 mm (florfenicol and Tc), and ≤16 mm (sulfamethoxazole). For a better characterization of the Tc resistance phenotype, MICs of Tc, doxycycline (Dc), and minocycline (Mc) were determined by the broth macrodilution procedure according to NCCLS document M31-A (32) using twofold dilution steps ranging from 2 to 256 μg/ml. The reference strain Escherichia coli ATCC 25922 served to control the precision and accuracy of the disk diffusion tests, whereas the reference strain Staphylococcus aureus ATCC 29213 was used as a control in the broth macrodilution experiments. Both reference strains were purchased from the Deutsche Sammlung von Mikoorganismen und Zellkulturen and were run side by side with the P. aerogenes isolates of this study. To investigate the possible influence of subinhibitory concentrations of Tc on the MICs the P. aerogenes isolates were also cultivated in Mueller-Hinton bouillon supplemented with 0.5 μg of Tc/ml prior to MIC determination. MIC determination was performed three times on independent occasions.
Identification of tet gene classes.
The identification of the tet genes was conducted by PCR as well as by Southern blot hybridization. The preparation of whole-cell DNA followed previously described protocols (17). Plasmid preparation was performed according to a previously described modification of the alkaline lysis procedure with subsequent purification by affinity chromatography on Qiagen Midi columns (17, 18). The plasmids of E. coli V517 (27) served as standards for the determination of plasmid sizes. For PCR analysis, the primers specific for the detection of tet genes of classes A to E and G (12, 13) and H (13, 18), as well as M and O (36), were used. Restriction analysis of whole-cell DNA of P. aerogenes isolates with BamHI, HindIII, or SfuI, agarose gel electrophoresis, and Southern blot hybridization were performed as described earlier (17, 18). Specific probes of the respective tet genes as described by Frech and Schwarz (12) were nonradioactively labeled by the enhanced chemiluminescence system (Amersham-Pharmacia Biotech, Freiburg, Germany). Hybridization and signal detection followed the manufacturer's recommendations. Plasmid profiles as well as endonuclease-digested whole-cell DNA served as targets for the tet gene probes. A 1,063-bp fragment amplified from Tn5706 (18) by using the sequence of the terminal 18-bp inverted repeat as PCR primer served as the specific probe for the closely related insertion elements IS1596 and IS1597. Hybridization experiments were repeated two times during the course of the study.
Transformation experiments.
Transformation of plasmids into E. coli strains JM107 and JM110 was done by either heat shock transformation into CaCl2-treated competent E. coli cells (10) or by electrotransformation into the plasmid-free and Tc-sensitive Mannheimia haemolytica strain M2000. For electrotransformation, the recipient strain was grown in brain heart infusion broth (Oxoid) until an optical density at 600 nm of 0.3 was reached. After centrifugation for 10 min at 1,650 × g, the bacterial pellet was resuspended in 100 ml of ice-cold GYT solution composed of 10% (vol/vol) glycerine, 0.125% (wt/vol) yeast extract, 0.25% (wt/vol) Caseine peptone, and and 0.02% (vol/vol) Tween 80. Centrifugation was repeated three times, and each time the bacterial pellet was resuspended in a smaller volume of GYT solution (30, 20, and 2 ml). Finally, 300 μl of the competent cells and approximately 5 μg of the plasmid DNA were mixed in a prechilled 0.2-cm cuvette and were kept on ice for 30 min. Electrotransformation was performed in a Gene Pulser II electroporation system (Bio-Rad, Munich, Germany) by applying an electric impulse (2.5 kV, 25 μFa, 800 Ω). The content of the cuvette was aseptically transferred into 1 ml of brain heart infusion broth and was incubated for 3 h at 37°C under moderate shaking (60 rpm). Subsequently, 100-μl aliquots were streaked on blood agar supplemented with 15 μg of Tc/ml. To confirm the viability of the recipients after the various concentration steps and after application of the electric impulse, aliquots of the recipient strain were streaked on nonselective blood agar plates.
Macrorestriction analysis.
Macrorestriction analysis of the P. aerogenes isolates using the enzyme SmaI (Boehringer GmbH) followed a previously described protocol (17). Since earlier experiments revealed that the SmaI fragments of P. aerogenes are mainly in a low-molecular-weight area (10 to 135 kb) as compared to those of P. multocida (17), the pulse time was increased over a 24-h period from only 2 to 5 s. The SmaI fragments of S. aureus reference strain 8325 (33) served as a size standard. The DNA fragments were separated in a CHEF DR III system (Bio-Rad) at 15 V/cm with 0.5× Tris-borate-EDTA buffer as the running buffer.
Analysis of plasmid pPAT2.
Plasmid pPAT2 was mapped using 18 different restriction endonucleases. A map of pPAT2 was constructed on the basis of the results obtained from double digests. The tetR-tet(B) gene region and its flanking areas of plasmid pPAT2 were sequenced directly on both strands using primers (Roth, Karlsruhe, Germany) designed from the conserved regions of the tetR-tet(B) sequences deposited in the databases or derived from the sequences obtained with these primers. Sequence analysis by the dideoxy chain termination method was performed using the ALF sequenator (Amersham-Pharmacia Biotech).
Nucleotide sequence accession number.
The sequence of a 2,969-bp segment of the pPAT2 sequence including the entire tetR-tet(B) region and its flanking areas has been deposited in the EMBL database under accession no. AJ278685.
RESULTS
Antimicrobial resistance and genotyping of P. aerogenes isolates.
The 24 isolates included in this study corresponded in their biochemical characteristics to those specific for isolates of the species P. aerogenes. All isolates were resistant to Tc as well as to streptomycin, and 20 of the 24 isolates exhibited additional resistances to one to five antimicrobial agents. Among them, resistance to sulfonamides (14 isolates), trimethoprim (14 isolates), and chloramphenicol (10 isolates) was observed most frequently. The MICs of Tc (MICTc) varied between 32 and >256 μg/ml, with the MICs for most of the isolates at 128 or 256 μg/ml (Table 1). The MICs of Dc varied between 16 and 64 μg/ml and those for Mc between 8 and 32 μg/ml. Preincubation of the isolates in subinhibitory concentrations of Tc occasionally increased the MICs by one- or twofold dilution.
TABLE 1.
MICs of Tc, Dc, and Mc for P. aerogenes carrying tet(H) and/or tet(B) genes
| MICs (μg/ml) of:
|
No. of tet gene copies per isolatea | No. of P. aerogenes isolates | ||
|---|---|---|---|---|
| Tc | Dc | Mc | ||
| >256 | 64 | 32 | 1 × tet(B) | 1 |
| 256 | 64 | 32 | 1 × tet(B) | 2 |
| 2 × tet(B) | 1 | |||
| 128 | 64 | 32 | 1 × tet(B) | 3 |
| 256 | 32 | 16 | 1 × tet(B) | 6 |
| 2 × tet(B) | 1 | |||
| 1 × tet(H) + 2 × tet(B) | 1 | |||
| 128 | 32 | 8 | 1 × tet(B) | 2 |
| 2 × tet(B) | 1 | |||
| tet(B) on plasmid pPAT2 | 1 | |||
| 256 | 16 | 8 | 1 × tet(B) | 1 |
| 2 × tet(B) | 1 | |||
| 128 | 16 | 8 | 1 × tet(B) | 1 |
| 32 | 16 | 8 | 1 × tet(B) | 1 |
| 64 | 16 | 8 | 1 × tet(H) | 1 |
Unless otherwise indicated, the tet genes were located in the chromosomal DNA.
Of the 24 P. aerogenes isolates, seven were plasmid free, while each of the remaining 17 isolates exhibited a different plasmid profile. These plasmid profiles comprised one to six plasmids in the size range between 1.8 and 28 kb. Most of the plasmids detected in P. aerogenes were less than 6 kb. Macrorestriction analysis identified 24 SmaI restriction patterns which differed by more than six fragments. Therefore, the corresponding isolates were considered unrelated.
Identification of tet genes.
PCR analysis of the 24 P. aerogenes isolates revealed the presence of tet genes of the two classes H and B. Single PCR products of 1,076 bp [tet(H)] and 1,170 bp [tet(B)] were detected in 1 and 22 isolates, respectively. A single P. aerogenes isolate carried both genes. The specificity of the amplicons was confirmed by BclI digestion [tet(H)] and EcoRI digestion [tet(B)]. The tet(H) amplicon yielded two BclI fragments of ca. 0.26 and 0.82 kb, whereas the tet(B) amplicon showed two EcoRI fragments of 0.56 and 0.61 kb.
Plasmid location of tet genes.
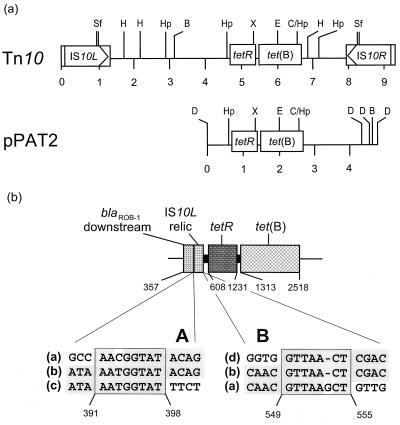
To determine the location of the tet genes on plasmids, three different experimental approaches were performed: (i) transformation into E. coli recipient strains, (ii) electrotransformation into the M. haemolytica recipient strain M2000, and (iii) hybridization of plasmid profiles. A single isolate was found to carry a tet(B) gene on a small plasmid of approximately 4.8 kb, designated pPAT2. No transformants could be obtained in repeated transformation experiments with the two E. coli recipient strains, even when selection was performed at low Tc concentrations of 5 μg/ml. However, electrotransformation into M. haemolytica recipient strain M2000 yielded transformants after selection on blood agar containing 20 μg of Tc/ml. The MICs for these transformants were 32 μg of Tc/ml and were increased to 64 μg of Tc/ml after induction of the tet(B) system. Plasmid pPAT2 did not mediate resistance properties other than Tc resistance. It revealed distinct restriction map homology to the tetR-tet(B) resistance gene area of Tn10 (Fig. 1a). Since plasmid pPAT2 was approximately half the size of Tn10, this plasmid obviously harbored a truncated copy of Tn10. Sequence analysis was conducted to determine exactly which parts of Tn10 were present in plasmid pPAT2.
FIG. 1.
(a) Comparison of the restriction maps of Tn10 (6) and plasmid pPAT2 from P. aerogenes. Restriction endonuclease cleavage sites are abbreviated as follows: B (BamHI), C (ClaI), D (DraI), E (EcoRI), H (HindIII), Hp (HpaI), Sf (SfuI), and X (XbaI). A distance scale in kilobases is given below both maps. The genes tet(B) and tetR as well as the insertion elements IS10L and IS10R are boxed. (b) Organization of the tetR-tet(B) resistance gene region and its flanking areas of plasmid pPAT2. The potential recombination sites A and B downstream of tetR are displayed as boxes. The corresponding sequences are IS10L (sequence a), pPAT2 (sequence b), noncoding sequence downstream of the blaROB-1 gene (sequence c), and Tn10-like sequence downstream of tetR (sequence d). The homologous sequences are shown on a gray background. The numbers refer to the positions of the 2,969-bp pPAT2 sequence deposited in the EMBL database under accession number AJ278685.
Within the sequenced 2,969-bp segment of pPAT2, two open reading frames for the 207-amino-acid TetR protein and the 401-amino-acid TetB protein were detected. A comparison of the tetR-tet(B) genes of pPAT2 with other tetR-tet(B) genes deposited in the databases revealed identity of the tetR genes to one another and also to pPAT2. The tet(B) gene of pPAT2, however, differed at eight positions from the nucleotide sequences of other tet(B) genes. Seven of these base pair exchanges also caused changes in the deduced amino acid sequence (Table 2), whereas the alteration at position 2389 (T→C) in the pPAT2 sequence did not change the amino acid (His-359) at the respective position.
TABLE 2.
Differences in amino acid sequences of TetB proteins
| Bacterial species | Database accession no. | Amino acid at position:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 55 | 171 | 281 | 301 | 330 | 338 | 354 | ||
| Salmonella enterica serovar Typhi | AF223162a | V | A | G | V | Q | L | A |
| Shigella flexneri | AF162223b | V | T | G | V | Q | L | A |
| E. coli | J01830c | V | T | E | D | E | L | T |
| P. aerogenes | AJ278685 | G | A | G | V | Q | F | A |
Sequence analysis of the regions flanking the tetR-tet(B) area comprised 607 bp downstream of tetR and 451 bp downstream of tet(B). In the region downstream of tetR, homology to Tn10 (positions 549 to 607) ended at an HpaI site 59 bp downstream of the translational stop codon of tetR. The region immediately downstream (positions 394 to 553) corresponded over 159 of the 160 bp to an internal segment of IS10L, the insertion element representing the left-hand end of Tn10. The sequence located further downstream (positions 357 to 398) of that IS10L relic revealed 95% identity to a noncoding region downstream of the blaROB-1 gene of P. haemolytica (GenBank database accession nos. X52872 and Z21724). The sequence of the remaining adjacent region (positions 1 to 356) revealed no significant homology to any sequences deposited in the databases. Analysis of the sequences at the junctions between the tetR-tet(B) gene region and the IS10L relic as well as between the IS10L relic and the noncoding region downstream of the blaROB-1 gene revealed in both cases areas of 8 bp which share 87.5% identity and might have served for illegitimate recombinations (Fig. 1b). In the area downstream of tet(B), homology to Tn10 was detectable over 194 bp (positions 2519 to 2712). The region located further downstream (positions 2713 to 2969) again shared no significant homology to any sequences deposited in the databases. Exactly at the junction of Tn10-related to Tn10-unrelated sequences in pPAT2, a pair of inverted repeated (IR) sequences consisting of 13 bp (IR1) and 14 bp (IR2) is located in the Tn10 sequence. Of these IR sequences, only the initial 5 bp of IR1 is left in the pPAT2 sequence.
Chromosomally located tet genes.
Negative results of transformation, electrotransformation, and hybridization of plasmid profiles suggested that the tet gene(s) might be located in the chromosomal DNA. Hybridization of HindIII-digested whole-cell DNA with the tet(H) gene probe yielded single hybridizing bands of 6.3 and >23.1 kb in the two tet(H)-carrying P. aerogenes isolates. No hybridization signals could be obtained with the probe specific for the insertion elements IS1596 and IS1597.
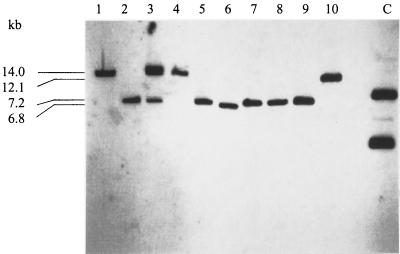
Whole-cell DNA of the tet(B)-carrying P. aerogenes isolates was digested with SfuI. Subsequent hybridization with the tet(B) gene probe revealed the presence of the following six patterns consisting of one or two hybridizing fragments: 6.8 kb (2 isolates), 7.2 kb (12 isolates), 12.1 kb (1 isolate), 14.0 kb (3 isolates), 7.2 and 14.0 kb (3 isolates), and 7.2 and 18.0 kb (1 isolate) (Fig. 2). Sixteen of the 22 P. aerogenes isolates exhibited at least one hybridizing SfuI fragment of approximately 7.2 kb, which is characteristic for the presence of a complete copy of Tn10. Since several complete copies of Tn10 present in the same isolate will also result only in a single hybridizing SfuI fragment of 7.2 kb, whole-cell DNA of all those isolates which exhibited a SfuI fragment of that size was digested with BamHI and hybridized with the tet(B) gene probe. Hybridizing BamHI fragments of >6 kb might indicate the presence of complete Tn10 copies. The four isolates which showed two hybridizing SfuI fragments also revealed two hybridizing BamHI fragments, and 10 isolates which exhibited a single 7.2-kb SfuI fragment also showed single hybridizing BamHI fragments. All but one of these hybridizing BamHI fragments were in the size range between 7.7 and >23.1 kb. However, two P. aerogenes isolates which revealed a single SfuI fragment of 7.2 kb showed two BamHI fragments of 8.2 and 15.0 kb and 10.7 and 15.0 kb. These two isolates were considered to harbor two Tn10 copies in their chromosomal DNA.
FIG. 2.
tet(B)-specific hybridization patterns of SfuI-restricted chromosomal DNA of P. aerogenes isolates (lanes 1 to 10). Sizes of the hybridizing fragments, as calculated from logarithmic plots in which HindIII-digested λ DNA (Gibco-BRL) was used as the size marker, are given in kilobases. Lane C contains the nonrestricted pCR-Blunt II-TOPO containing an internal 1,170-bp fragment of the tet(B) gene (12), which served as a positive control in these hybridization experiments.
DISCUSSION
The tet(B) gene represents the predominant gene among Enterobacteriaceae (29, 30) and has been reported to be widely distributed among other families of gram-negative bacteria (23, 35). Up to now, a tet(B) gene has only been detected once in a single bovine P. haemolytica isolate from France (Table 3). However, tet(B) genes have been found in other members of the family Pasteurellaceae, namely, isolates of Haemophilus influenzae, Haemophilus ducreyi, and Haemophilus parainfluenzae involved in infections of humans (15, 24, 28). The tet(B) gene is part of Tn10, a nonconjugative composite transposon of 9,147 bp (6, 19, 21) which often resides on large plasmids in Enterobacteriaceae (38; GenBank accession no. AP000342). Although the tet(B) gene is functionally active in a number of different hosts (11), such plasmids may be replication deficient in Pasteurella hosts; e.g., plasmids which carry the ColE1 replication system have been reported to be unable to replicate in pasteurellae (2). However, as long as these plasmids harbor complete copies of a transposon, this transposon may integrate into plasmids or the chromosomal DNA of the new host (3). After integration, the transposon can be subjected to structural alterations which also may affect those parts required for the mobility of the transposon. In this regard, the two tet(H)-carrying P. aerogenes isolates identified during the course of this study were found to harbor single copies of truncated Tn5706 elements in which both insertion elements were lost.
TABLE 3.
Comparison of tet genes detected in P. multocida and P. haemolytica from the respiratory tract and P. aerogenes isolates from intestinal tract of various animals
| Species | No. of isolates harboring tet genes of classa
|
Source or reference(s) | ||
|---|---|---|---|---|
| H | M | B | ||
| P. multocida | 20 (P) + 4 (C) | 1 (C) | 13, 14 | |
| 1 (C) | 7 | |||
| 7 (P) | 17, 18 | |||
| P. haemolytica | 2 (P) | 13 | ||
| 1 (C) | 7 | |||
| P. aerogenes | 1 (P) | 17 | ||
| 2 (C)b | 1 (P) + 22 (C)b | This study | ||
(P), located on a plasmid; (C), located in chromosomal DNA.
One isolate carried a tet(H) and two copies of a tet(B) gene.
The truncated Tn10 copy detected on the 4.8-kb tet(B)-carrying plasmid pPAT2 also lacked the insertion elements. To the best of our knowledge, plasmid pPAT2 is the first naturally occurring small multicopy plasmid carrying the tetR-tet(B) gene area of Tn10. E. coli recipient strains which harbored Tn10 or the Tn10-specific tet gene region on multicopy plasmid vectors were found to exhibit a distinctly decreased level of Tc resistance (8, 9). The lack of pPAT2-carrying E. coli transformants might confirm these findings (8, 9), since the internal BglII fragment of Tn10 which was claimed to be responsible for the reduced level of Tc resistance (8) carried the entire tetR-tet(B) gene area and was almost completely present in plasmid pPAT2. However, a general deficiency in replication of the P. aerogenes plasmid pPAT2 in E. coli recipients must also be considered since this plasmid proved to be able to replicate and confer Tc resistance in M. haemolytica strain M2000.
The tetR-tet(B) gene area of plasmid pPAT2 corresponded almost exactly to that of Tn10 (16). Assuming that a complete copy of Tn10 originally integrated into a pPAT2 precursor plasmid, truncation of Tn10 in the region downstream of tetR in pPAT2 may be explained by illegitimate recombination. In this regard, parallels between the truncated Tn10 element of pPAT2 and the truncated Tn5706 element of pPAT1, a previously described tet(H)-carrying plasmid detected in porcine P. aerogenes and P. multocida isolates (17), were observed. Although the recombination events affected the part downstream of tet(H) in pPAT1 and the part downstream of tetR in pPAT2, in both cases a small internal segment of the respective insertion element, IS1597 in pPAT1 and IS10L in pPAT2, remained to be present. Moreover, adjacent to these insertion sequence relics, sequences corresponding to those up- or downstream of blaROB-1 genes of Actinobacillus pleuropneumoniae, H. influenzae, or P. haemolytica were detected in both cases. Furthermore, the assumed recombination sites at the junctions between the region downstream of tetR and the IS10L sequence as well as between the IS10L sequence and the noncoding region downstream of blaROB-1 closely corresponded in size and nucleotide sequence identity to those recombination sites involved in the truncation of the tet(H) gene and the IS1597 element in plasmid pPAT1 (17). Thus, the left-hand portion of Tn10 (downstream of tetR) seems to be lost as a consequence of at least two independent recombination events. Loss of the right-hand portion of Tn10 [downstream of tet(B)] is difficult to explain since the sequences downstream of the Tn10-like part in pPAT2 do not exhibit homology to any sequences deposited in the databases. However, a comparison between Tn10 and pPAT2 revealed that at the junction between Tn10-homologous and non-Tn10-homologous sequences in pPAT2, an IR sequence of 13 (IR1) and 14 (IR2) bp is found in the Tn10 sequence. Of this IR sequence, only the initial 5 bp of IR1 was found to be left in pPAT2. This observation points towards another recombination event. Areas characterized by inverted repeats are considered preferential areas for illegitimate recombination events (22). If illegitimate recombination occurs at inverted repeats, these are usually destroyed in a way observed in pPAT2 (22).
Even though a tet(B) gene was previously detected in the chromosomal DNA of a single bovine P. haemolytica isolate (7), no information on the size of the hybridizing fragment and whether this tet(B) gene was associated with a complete or a truncated copy of Tn10 was given. In the present study, whole-cell DNA of tet(B)-carrying isolates was digested with SfuI, a restriction endonuclease which has two recognition sites lying closely together in each of the terminal insertion elements IS10L and IS10R but has none in the remaining part of Tn10. A complete Tn10 copy thus might be characterized by a hybridizing SfuI fragment of 7.2 kb. Of the 22 P. aerogenes isolates that harbored chromosomal tet(B) genes, 16 exhibited a SfuI fragment of that size, either alone or in addition to a second larger SfuI fragment. The larger and slightly smaller SfuI fragments observed in the P. aerogenes isolates might indicate the presence of structural alterations in the respective Tn10 elements. Tn10 elements into which other transposons (Tn1000) or insertion elements (IS911) have been inserted and thus might result in larger SfuI fragments have already been described in Pantoea agglomerans (formerly known as Enterobacter agglomerans) (37). However, complete Tn10 copies were amplified in the majority of the phylloplane bacteria studied by Schnabel and Jones (37). Complete Tn10 copies have also been detected in Haemophilus spp. (24, 28). These data as well as the data of the present study indicate that complete Tn10 copies are widely distributed among bacteria from different sources.
By using SfuI and BamHI digests, not more than two copies of the tet(B) gene could be detected in the chromosomal DNA of the P. aerogenes isolates. No correlation between the MICs of Tc, Dc, or Mc and the numbers of tet(B) gene copies was seen among the P. aerogenes isolates. For the pPAT2-harboring P. aerogenes isolate, the high MICTc (128 μg/ml) was the same as that for P. aerogenes isolates which harbored a single chromosomal copy or two chromosomal copies of the tet(B) gene (Table 1). With one exception, the MICTcs for the P. aerogenes isolates ranged between 128 and >256 μg/ml. The MICTc for the previously identified tet(B)-carrying P. haemolytica isolate was also high (256 μg/ml), while the MIC of Dc or Mc was not reported (7). The MICTcs for the P. aerogenes isolates of this study are also in good accordance with the high MICTcs reported for tet(B)-carrying enterobacterial isolates (31). While no MICs of Dc were reported for tet(B)-carrying isolates, Mendez et al. (31) showed that enterobacterial isolates carrying the tet(B) gene can be differentiated into two groups on the basis of their level of Mc resistance: those isolates for which the MIC of Mc is ≥10 μg/ml and those isolates for which the MIC of Mc is ≥5 and <10 μg/ml. This observation again corresponded closely to the situation seen among the tet(B)-carrying P. aerogenes isolates of this study (Table 1).
ACKNOWLEDGMENTS
We thank Jürgen Mumme for providing P. aerogenes isolates as well as Erika Nußbeck and Gisela Niemann for expert technical assistance. C. K. was supported by the Gesellschaft der Freunde der FAL (GdF).
REFERENCES
- 1.Angen Ø, Mutters R, Caugant D A, Olsen J E, Bisgaard M. Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb. nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov. Int J Syst Bacteriol. 1999;49:67–86. doi: 10.1099/00207713-49-1-67. [DOI] [PubMed] [Google Scholar]
- 2.Azad A K, Coote J G, Parton R. Construction of conjugative shuttle and suicide vectors for Pasteurella haemolytica and Pasteurella multocida. Gene. 1994;145:81–85. doi: 10.1016/0378-1119(94)90326-3. [DOI] [PubMed] [Google Scholar]
- 3.Bennett P M. The spread of drug resistance. In: Baumberg S, Young J P W, Wellington E M H, Saunders J R, editors. Population genetics of bacteria: 52nd symposium of the Society for General Microbiology held at the University of Leicester, January 1995. Cambridge, England: Cambridge University Press; 1995. pp. 317–344. [Google Scholar]
- 4.Biberstein E L. Pasteurella. In: Biberstein E L, Zee Y C, editors. Review of veterinary microbiology. Oxford, England: Blackwell Scientific Publications; 1990. pp. 175–180. [Google Scholar]
- 5.Carter G R. Genus I. Pasteurella Trevisan 1887. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 552–557. [Google Scholar]
- 6.Chalmers S, Sewitz R, Lipkow K, Crellin P. Complete nucleotide sequence of Tn10. J Bacteriol. 2000;182:2970–2972. doi: 10.1128/jb.182.10.2970-2972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaslus-Dancla E, Lesage-Descauses M-C, Leroy-Sétrin S, Martel J-L, Lafont J-P. Tetracycline resistance determinants, TetB and TetM, detected in Pasteurella haemolytica and Pasteurella multocida from bovine herds. J Antimicrob Chemother. 1995;36:815–819. doi: 10.1093/jac/36.5.815. [DOI] [PubMed] [Google Scholar]
- 8.Chopra I, Shales S W, Ward J M, Wallace L J. Reduced expression of Tn10-mediated tetracycline resistance in Escherichia coli containing more than one copy of the transposon. J Gen Microbiol. 1981;126:45–54. doi: 10.1099/00221287-126-1-45. [DOI] [PubMed] [Google Scholar]
- 9.Coleman D C, Foster T J. Analysis of the reduction in expression of tetracycline resistance determined by transposon Tn10 in the multicopy state. Mol Gen Genet. 1981;182:171–172. doi: 10.1007/BF00422786. [DOI] [PubMed] [Google Scholar]
- 10.Dagert M, Ehrlich S D. Prolonged incubation in calcium chloride improves the competence of E. coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 11.DeFlaun M F, Levy S B. Genes and their varied hosts. In: Levy S B, Miller R V, editors. Gene transfer in the environment. New York, N.Y: McGraw-Hill Book Co.; 1989. pp. 1–32. [Google Scholar]
- 12.Frech G, Schwarz S. Molecular analysis of tetracycline resistance in Salmonella enterica subsp. enterica serovars Typhimurium, Enteritidis, Dublin, Choleraesuis, Hadar and Saintpaul: construction and application of specific gene probes. J Appl Microbiol. 2000;89:633–641. doi: 10.1046/j.1365-2672.2000.01160.x. [DOI] [PubMed] [Google Scholar]
- 13.Hansen L M, Blanchard P C, Hirsh D C. Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob Agents Chemother. 1996;40:1558–1560. doi: 10.1128/aac.40.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen L M, McMurray L M, Levy S B, Hirsh D C. A new tetracycline resistance determinant, TetH, from Pasteurella multocida specifying active efflux of tetracycline. Antimicrob Agents Chemother. 1993;37:2699–2705. doi: 10.1128/aac.37.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuer C, Hickman R K, Curiale S M, Hillen W, Levy S B. Constitutive expression of tetracycline resistance mediated by a Tn10-like element in Haemophilus parainfluenzae results from a mutation in the repressor gene. J Bacteriol. 1987;169:990–994. doi: 10.1128/jb.169.3.990-994.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hillen W, Schollmeyer K. Nucleotide sequence of the Tn10 encoded tetracycline resistance gene. Nucleic Acids Res. 1983;11:525–539. doi: 10.1093/nar/11.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehrenberg C, Schwarz S. Identification of a truncated, but functionally active tet(H) tetracycline resistance gene in Pasteurella aerogenes and Pasteurella multocida. FEMS Microbiol Lett. 2000;188:191–195. doi: 10.1111/j.1574-6968.2000.tb09192.x. [DOI] [PubMed] [Google Scholar]
- 18.Kehrenberg C, Werckenthin C, Schwarz S. Tn5706, a transposon-like element from Pasteurella multocida mediating tetracycline resistance. Antimicrob Agents Chemother. 1998;42:2116–2118. doi: 10.1128/aac.42.8.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleckner N. Transposon Tn10. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 229–268. [Google Scholar]
- 20.Koneman E W, Allen S D, Janda W M, Schreckenberger P C, Winn W C., Jr . Color atlas and textbook of diagnostic microbiology. 5th ed. Philadelphia, Pa: Lippincott; 1997. [Google Scholar]
- 21.Lawley T D, Burland V D, Taylor D E. Analysis of the complete nucleotide sequence of the tetracycline resistance transposon Tn10. Plasmid. 2000;43:235–239. doi: 10.1006/plas.1999.1458. [DOI] [PubMed] [Google Scholar]
- 22.Leach D R F. Genetic recombination. Oxford, England: Blackwell Science Ltd.; 1996. [Google Scholar]
- 23.Levy S B. Tetracycline resistance determinants are widespread. ASM News. 1988;54:418–421. [Google Scholar]
- 24.Levy S B, Buu-Hoi A, Marshall B. Transposon Tn10-like tetracycline resistance determinants in Haemophilus parainfluenzae. J Bacteriol. 1984;160:87–94. doi: 10.1128/jb.160.1.87-94.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy S B, McMurray L M, Barbosa T M, Burdett V, Courvalin P, Hillen W, Roberts M C, Rood J I, Taylor D E. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livrelli V O, Darfeuille-Richaud A, Rich C D, Joly B H, Martel J-L. Genetic determinant of the ROB1 β-lactamase in bovine and porcine Pasteurella strains. Antimicrob Agents Chemother. 1988;32:1282–1284. doi: 10.1128/aac.32.8.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macrina F L, Kopecko D J, Jones K R, Ayers D J, McCowen S M. A multiple plasmid-containing E. coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1:417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 28.Marshall B M, Roberts M, Smith A, Levy S B. Homogeneity of transferable tetracycline resistance determinants in Haemophilus species. J Infect Dis. 1984;149:1028–1029. doi: 10.1093/infdis/149.6.1028. [DOI] [PubMed] [Google Scholar]
- 29.Marshall B M, Tachibana C, Levy S B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983;24:835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Salazar J M, Alvarez G, Gomez-Eichelmann M C. Frequency of four classes of tetracycline resistance determinants in Salmonella and Shigella spp. clinical isolates. Antimicrob Agents Chemother. 1986;30:630–631. doi: 10.1128/aac.30.4.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980;3:99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- 32.NCCLS. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard. M31-A. Wayne, Pa: NCCLS; 1999. [Google Scholar]
- 33.Pattee P A, Lee H-C, Bannantine J O. Genetic and physical mapping of the chromosome of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 41–58. [Google Scholar]
- 34.Radostits O M, Gay C C, Blood D C, Hinchcliff K W. A textbook of the diseases of cattle, sheep, pigs, goats, and horses. 9th ed. Philadelphia, Pa: W. B. Saunders Co.; 2000. [Google Scholar]
- 35.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 36.Roberts M C, Pang Y, Riley D E, Hillier S L, Berger R C, Krieger J N. Detection of Tet M and Tet O tetracycline resistance genes by polymerase chain reaction. Mol Cell Probes. 1993;7:387–393. doi: 10.1006/mcpr.1993.1057. [DOI] [PubMed] [Google Scholar]
- 37.Schnabel E L, Jones A L. Distribution of tetracycline resistance genes and transposons among phylloplane bacteria in Michigan apple orchards. Appl Environ Microbiol. 1999;65:4898–4907. doi: 10.1128/aem.65.11.4898-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherburne C K, Lawley T D, Gilmour M W, Blattner F R, Burland V, Grotbeck E, Rose D J, Taylor D E. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000;28:2177–2186. doi: 10.1093/nar/28.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorsen P, Moller B R, Apri M, Bremmelgaard A, Frederiksen W. Pasteurella aerogenes isolated from stillbirth and mother. Lancet. 1994;343:485–486. doi: 10.1016/s0140-6736(94)92736-7. [DOI] [PubMed] [Google Scholar]