Abstract
Introduction
It is important to understand which biological processes change with aging, and how such changes are associated with increased Alzheimer's disease (AD) risk. We studied how cerebrospinal fluid (CSF) proteomics changed with age and tested if associations depended on amyloid status, sex, and apolipoprotein E Ɛ4 genotype.
Methods
We included 277 cognitively intact individuals aged 46 to 89 years from Alzheimer's Disease Neuroimaging Initiative, European Medical Information Framework for Alzheimer's Disease Multimodal Biomarker Discovery, and Metabolic Syndrome in Men. In total, 1149 proteins were measured with liquid chromatography mass spectrometry with multiple reaction monitoring/Rules‐Based Medicine, tandem mass tag mass spectrometry, and SOMAscan. We tested associations between age and protein levels in linear models and tested enrichment for Reactome pathways.
Results
Levels of 252 proteins increased with age independently of amyloid status. These proteins were associated with immune and signaling processes. Levels of 21 proteins decreased with older age exclusively in amyloid abnormal participants and these were enriched for extracellular matrix organization.
Discussion
We found amyloid‐independent and ‐dependent CSF proteome changes with older age, perhaps representing physiological aging and early AD pathology.
1. INTRODUCTION
With older age, the prevalence of Alzheimer's disease (AD) rises steeply. 1 , 2 Amyloid plaques and tau tangles in the brain are considered the pathological hallmarks of AD, and the prevalence of abnormal biomarkers levels reflective of these hallmarks increases in cognitively normal (CN) individuals with older age, preceding prevalence of dementia up to 20 years. 3 , 4 Possibly, other molecular processes also may become disrupted during this time. Better characterization of age effects, and the influence of AD pathology on such relationships, would give more insight into processes that increase the brain's vulnerability to neurodegeneration during aging. Proteomic analysis of cerebrospinal fluid (CSF) measures many proteins simultaneously, and can show which molecular processes may change with age and AD in vivo. 5
Previous CSF proteomic studies found that approximately one tenth (30 of 248) 6 to one third (82 of 312) 7 of analyzed proteins show altered levels with older age in CN individuals. Those proteins showed enrichment for inflammation, response to injury, and the complement system. 7 However, those studies did not address the influence of amyloid status on the CSF proteome, while an estimated 19% of 65‐year‐old individuals with normal cognition show abnormal amyloid and are at risk to develop dementia. 3 It remains unclear to what extent observed age‐related alterations were influenced by the presence of AD pathology. Furthermore, sex and apolipoprotein E (APOE) ε4 carriership are known risk factors for AD that have been shown to either influence the CSF proteome 8 (APOE ε4), or to influence specific CSF proteins, for example, tau (sex 9 , 10 and APOE ε4 10 ). We hypothesized that associations between protein levels and age may differ depending on amyloid status, sex, and APOE ε4 carriership.
The aim of this cross‐sectional study was to investigate the relationship of age‐ and amyloid‐related changes in the CSF proteome of cognitively intact individuals spanning an age range of 46 to 89 years, including potential effects of APOE ε4 carriership and sex.
2. MATERIAL AND METHODS
2.1. Study cohorts
Three independent cohorts were included in our analysis: Alzheimer's Disease Neuroimaging Initiative (ADNI), 11 European Medical Information Framework for Alzheimer's Disease Multimodal Biomarker Discovery (EMIF‐AD MBD) 12 and Metabolic Syndrome in Men (METSIM). 13 All participants provided informed consent to participate in these studies. From these cohorts we selected CN individuals with available CSF proteomic data (Table 1).
TABLE 1.
Demographics of study cohorts and combined data
| EMIF‐AD MBD | ADNI | METSIM | Combined | |||||
|---|---|---|---|---|---|---|---|---|
| Normal amyloid | Abnormal amyloid | Normal amyloid | Abnormal amyloid | Normal amyloid | Abnormal amyloid | Normal amyloid | Abnormal amyloid | |
| N | 87 | 74 | 54 | 32 | 27 | 3 | 168 | 109 |
| Age, years, mean ± SD | 65.9 ± 7.8ab | 66.9 ± 8.3a | 75.5 ± 5.6ad | 76.1 ± 5.6a | 61.7 ± 4.5bd | 63.7 ± 6.1 | 68.3 ± 8.4 | 69.5 ± 8.6 |
| Sex, Female (%) | 43 (49%)b | 39 (53%) | 27 (50%)d | 15 (47%) | n.s. | n.s. | 70 (42%) | 54 (50%) |
| APOE ε4, 1‐2 alleles (%) | 22 (25%)a | 44 (59%)c | 5 (9.3%)ad | 16 (50%)c | 11 (41%)d | 2 (67%) | 38 (23%)c | 62 (57%)c |
| MMSE, mean ± SD | 28.9 ± 1.2 | 28.6 ± 1.2a | 29 ± 1 | 29.2 ± 1a | 28.4 ± 1.3 | 29.3 ± 0.58 | 28.8 ± 1.2 | 28.8 ± 1.2 |
| CSF amyloid 1‐42 Z‐score, mean ± SD | 0 ± 1 | –1.4 ± 0.6ac | 0 ± 1 | –3.5 ± 0.9adc | 0 ± 1 | –1.9 ± 0.15 cd | 0 ± 0.99c | –2.1 ± 1.2c |
| CSF t‐tau Z‐score, mean ± SD | 0 ± 1 | 0.74 ± 2.4 | 0±1 | 1.1 ± 1.6c | 0 ± 1 | 0.76 ± 3.2 | 0±0.99c | 0.87 ± 2.2c |
| CSF p‐tau Z‐score, mean ± SD | 0 ± 1 | 0.26 ± 1.5a | 0 ± 1 | 1.1 ± 1.7ac | 0 ± 1 | 1.2 ± 2.7 | 0 ± 0.99c | 0.56 ± 1.6c |
| CSF t‐tau status, abnormal (%) | 13 (15%) | 28 (38%)c | 5 (9.3%) | 12 (38%)c | 3 (11%) | 1 (33%) | 21 (12%)c | 41 (38%)c |
| CSF p‐tau status, abnormal (%) | 12 (14%) | 26 (35%)ac | 8 (15%) | 19 (59%)ac | 3 (11%) | 1 (33%) | 23 (14%)c | 46 (42%)c |
Abbreviations: ADNI, Alzheimer's Disease Neuroimaging Initiative; APOE, apolipoprotein E; CSF, cerebrospinal fluid; EMIF‐AD MBD, European Medical Information Framework for Alzheimer's Disease Multimodal Biomarker Discovery; METSIM, Metabolic Syndrome in Men; MMSE, Mini‐Mental State Examination; n.s., not included in study; p‐tau, phosphorylated tau; SD, standard deviation; t‐tau, total tau.
Statistical differences were tested with chi‐square test, Mann‐Whitney U test or Student's t‐test as appropriate. Amyloid, t‐tau and p‐tau Z‐scores were calculated relative to CN amyloid normal controls. a is P < .05 comparing ADNI and EMIF‐AD MBD participants, b is P < .05 comparing EMIF‐AD MBD and METSIM, c is P < .05 comparing amyloid abnormal and normal participants, d is P < .05 comparing ADNI and METSIM participants.
2.1.1. ADNI
The ADNI (adni.loni.usc.edu) was launched in 2003 as a public–private partnership, led by Principal Investigator Michael W. Weiner, MD. 11 The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD.
RESEARCH IN CONTEXT
Systematic review: We reviewed the literature on effects of age on the cerebrospinal fluid (CSF) proteome. Two studies, referenced in the text, have addressed age effects on the CSF proteome. Novel in our approach is a concomitant analysis of age and amyloid status effects on the proteome, which allows separating age‐ and Alzheimer's disease (AD)‐related effects.
Interpretation: Our finding that extracellular matrix organization overlaps between proteins with amyloid‐independent and amyloid‐dependent associations with age could indicate this process contributes to the higher AD risk with older age.
Future directions: While our results in cross‐sectional data suggest that general aging‐ and early AD‐related proteome changes overlap, further longitudinal studies of CSF protein levels in normal cognition are needed to better separate general aging‐ and AD‐related effects.
In ADNI, normal cognition was defined as documented in the ADNI General Procedures Manual, 14 as (1) absence of memory complaints, (2) normal education‐adjusted memory function measured with Logical Memory II subscale (from Wechsler Memory Scaled—Revised), (3) Mini‐Mental State Examination 15 (MMSE) score of 24 to 30, (4) Clinical Dementia Rating (CDR) 16 and Memory Box score of 0, and (5) absence of significant impairment in cognitive functions or activities of daily living. Both men and women were enrolled. The participants had a median age of 75 years (range 62 to 89 years).
2.1.2. EMIF‐AD MBD
The EMIF‐AD MBD study is a multicenter retrospective study to discover novel diagnostic and prognostic biomarkers for AD. 12 Normal cognition was defined as neuropsychological assessment within 1.5 standard deviations of age‐, sex‐, and education‐specific mean test scores. Both men and women were enrolled. The participants had a median age of 66 years (range 46 to 84 years).
2.1.3. METSIM
METSIM is an observational study on the development of diabetes. 17 We selected participants without insulin resistance from METSIM (n = 30). 13 Normal cognition was defined as living independently, absence of memory complaints, MMSE score of at least 25, and Functional Activities Questionnaire score 18 < 9. Participants were excluded if they had current (or a history of) neurological disease, significant systemic diseases, or used specific medications as described in more detail elsewhere. 13 Only men were enrolled in the study. The participants had a median age of 62 years (range 55 to 69 years).
2.2. CSF proteomics
CSF was collected and processed as described elsewhere (see ADNI 1 General Procedures Manual, 14 and study‐specific references 12 , 19 ). ADNI proteomics data were generated using a liquid chromatography mass spectrometry with multiple reaction monitoring panel 20 (311 peptides, from which we analyzed the normalized intensity data), Rules‐Based Medicine 21 multiplex (83 proteins), and enzyme‐linked immunosorbent assays (four proteins). Peptides and proteins targeting the same protein were Z‐scored relative to controls and averaged if they showed a correlation of at least 0.5. EMIF‐AD MBD CSF proteomic data was generated with tandem mass tag mass spectrometry 12 , 22 resulting in 2535 proteins (deposited at the ProteomeXchange Consortium via the PRIDE partner repository, dataset identifier: https://doi.org/10.6019/PXD019910). CSF proteomic data of 4006 proteins in METSIM was measured on the SOMAscan platform (SomaLogic). 19 Total signal intensities for the samples were median normalized by SomaLogic.
As we were interested in studying age effects, we pooled proteomic data across studies to ensure a wide age range. First, we harmonized natural log‐transformed proteomic data while anchoring age effects, by Z‐transformation relative to CN controls aged 60 to 75 years with normal amyloid, total tau (t‐tau), and phosphorylated tau (p‐tau; see schematic overview in Figure 1). Next, we averaged peptides or proteins measuring the same UniProt identifier or highly correlating proteins (r at least 0.9) with different UniProt identifiers if they were translated from the same or homologous genes into protein scores and pooled the data (5039 proteins, see Table S1 in supporting information for different peptides and proteins per protein score). We included proteins measured in at least 25% of individuals and in at least 50 individuals with abnormal amyloid, resulting in a final dataset of 1149 proteins, of which 115 were measured in all three datasets, 538 proteins were measured in both EMIF‐AD MBD and METSIM, 42 proteins in both ADNI and METSIM, and 454 proteins in EMIF‐AD MBD only.
FIGURE 1.

Overview of methods and data analysis. Data processing strategy. The combined dataset was generated by separate pre‐processing and normalization of proteomic datasets, followed by data pooling and selection of proteins measured in at least 25% of participants, of whom at least 50 were amyloid abnormal. For detailed description of the data processing strategy, see Methods; for detailed information of which peptides or protein measurements in each study were used for the pooled protein scores, see Table S1.
2.3. AD biomarker status
Amyloid status was based on center‐specific cut‐offs of CSF amyloid beta (Aβ)42 derived with Gaussian mixture modeling (EMIF‐AD MBD), 23 on CSF Aβ42 values on xMAP Luminex at a cut‐off indicating abnormality of <192 pg/mL (ADNI) 24 and on INNOTEST at <350 pg/mL (METSIM). 13 T‐tau status was based on CSF t‐tau levels using center‐specific cut‐offs (EMIF‐AD MBD), 12 with a cut‐off indicating abnormality of >93 pg/mL on xMAP Luminex (ADNI), 24 or >350 pg/mL on INNOTEST (METSIM). 13 P‐tau status was based on CSF levels using center‐specific cut‐offs 12 (EMIF‐AD MBD), >23 pg/mL on xMAP Luminex (ADNI), 24 or >60 pg/mL on INNOTEST (METSIM). 13
2.4. Determination of APOE genotypes
In ADNI, APOE genotypes were determined by polymerase chain reaction amplification and enzymatic digestion as described in ADNI research plans 25 and the resulting data were obtained from adni.loni.usc.edu. In METSIM, APOE genotypes were determined using TaqMan Allelic Discrimination Assays (Applied Biosystems). 13 In EMIF‐AD MBD, APOE genotypes were measured using the Global Screening Array (Illumina Inc.) and TaqMan assays. 26
2.5. Statistics
In all models, main effects of age, amyloid status, APOE ε4 carriership, and sex were included. In all analyses, P‐values < .05 were considered significant without multiple testing correction, as we aimed to show patterns of age‐, amyloid‐, APOE ε4‐, and sex‐related changes in the proteome rather than identifying novel biomarkers. Interaction terms were tested using analysis of variance. Main effects of age, amyloid status, APOE ε4 carriership, and sex were reported in proteins that had no significant two‐ or three‐way interactions with any of the other variables, and regression coefficients for each of these were corrected for the other variables. Effect sizes were reported with estimated marginal slopes for age or estimated marginal mean differences for amyloid status, APOE ε4, and sex.
Age effects were group‐stratified for proteins showing a significant interaction between (1) age and amyloid status, (2) age and sex, or (3) age and APOE ε4 status. Finally, we also tested three‐way interactions between age and (1) amyloid status and APOE ε4 or (2) amyloid status and sex, and when significant, stratified analyses for amyloid and APOE or amyloid and sex subgroups. Proteins included in each of the analyses are documented in Table S2 in supporting information. Finally, for each of the above models, when at least 10 proteins showed an association with the outcome, we performed pathway analyses with Reactome 27 (Reactome Pathway Gene Set version 69 downloaded from Reactome, https://reactome.org/ 28 ), using all Reactome categories except “Disease” (1779 pathways). RITAN 1.10.0 was used to test pathway enrichment with all protein‐coding genes as a background and false discovery rate correction at a q threshold of 0.05, and we report significant pathways if at least three proteins overlapped with the pathway. All analyses were run in R version 3.6.1 “Action of the Toes.” Emmeans v.1.4.2 was used to estimate effect sizes, and volcano plots were generated with EnhancedVolcano 1.4.0. Shiny v.1.4.0 was used to create a user interface allowing querying of the age associations in the CSF proteome (available at https://kwesenhagen.shinyapps.io/Aging_proteomics/).
3. RESULTS
3.1. Sample description
In total, 277 CN participants aged 46 to 89 years were included. Of these participants, 109 (39%) had abnormal amyloid levels in CSF (Table 1). Relative to participants with normal amyloid, participants with abnormal amyloid were older, more often had an APOE ε4 genotype, and more often had abnormal t‐tau and p‐tau in CSF. The cohorts were largely comparable in demographics, but ADNI participants were on average older. The METSIM study included only men, and APOE ε4 carriership was more common than in the other two studies. In addition, in EMIF‐AD MBD compared to ADNI, participants with abnormal amyloid had lower MMSE scores and less often had abnormal CSF p‐tau.
3.2. Main effects of age, amyloid status, APOE ε4 carriership, and sex
We first investigated the main effects of age, amyloid status, APOE ε4, and sex as predictors of CSF protein levels (see web interface at https://kwesenhagen.shinyapps.io/Aging_proteomics/ showing the effects for all included proteins). We tested the main effect of age in 911 of the 1149 proteins that showed no interactions between age and amyloid status, sex, and APOE ε4. Two hundred fifty‐two proteins (28%) had altered levels with older age, of which 194 (77% of 252) proteins had higher levels with older age and were enriched for immune system and signal transduction processes, and processes related to cellular responses to external stimuli, among others (Figures 2A, 2E, and 3A; Tables S3 and S4 in supporting information). Subsets of these processes were uniquely enriched among these proteins compared to the other analyses. Fifty‐eight proteins (23% of all 252 age‐related proteins) had lower levels with older age, and did not show enrichment for specific biological processes (Figures 2A and 2E; Tables S3 and S4).
FIGURE 2.
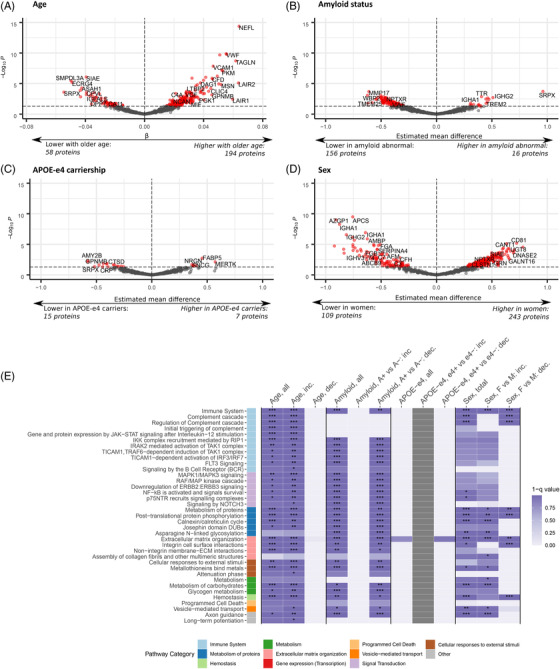
Effects of age, amyloid status, apolipoprotein E (APOE) ε4 carriership, and sex separately on the proteome. Main effect estimates from linear models including main effects of age, amyloid state, APOE ε4 carriership, and sex. Proteins were selected for the analyses using the specified selection criteria: main effect of age (no interaction between age and amyloid status, APOE ε4, or sex), main effect of amyloid status (no interaction between amyloid status and age, APOE ε4, or sex), and similarly for APOE ε4 and sex. A‐D, Effect sizes and statistical significance of protein level changes depending on the investigated factors: (A), age, (B) amyloid status, (C), APOE ε4 carriership, and (D), sex. E, Top pathway categories enriched in proteins with a significant effect of these factors. Full effect sizes and statistical significance of the investigated factors are provided in Tables S3 and S5‐7. For full overview of enriched processes, see Table S4. Inc., increased; dec., decreased; A+, amyloid abnormal; A–, amyloid normal; e4+, APOE ε4 carrier; e4–, APOE ε4 non‐carrier; F, female; M, male. *, P < .05; **, P < .01; ***, P < .001
FIGURE 3.
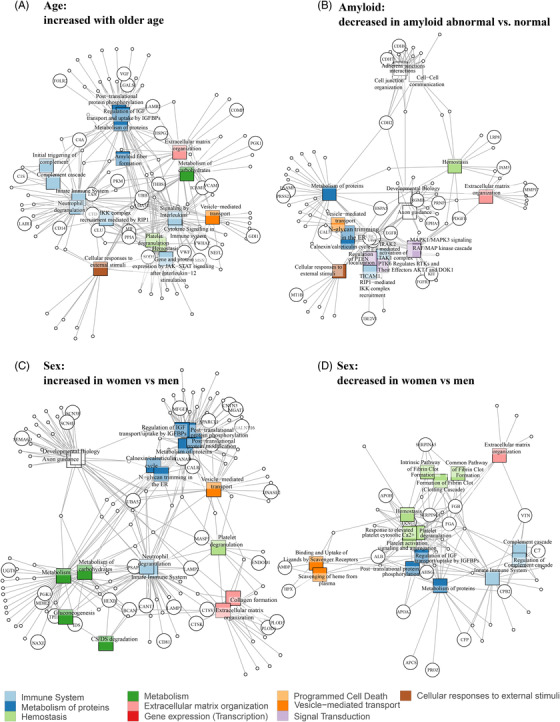
Overview of pathways enriched in proteins related to age, amyloid status and sex. Network graph showing connections between pathways (squares) and proteins found in each analysis (circles), with edges indicating that proteins are part of a pathway. An overview of top pathways and proteins is shown for each analysis; for full pathway results, see Table S4. Pathway results for (A), proteins that showed increased levels with older age, (B) proteins that showed decreased levels in amyloid abnormal versus normal participants, (C), proteins that had increased levels in women versus men, and (D), proteins that had decreased levels in women versus men.
Next, we tested the main effect of amyloid status in 864 proteins without interactions between amyloid status and age, sex, and APOE ε4. Levels of 172 proteins (20%) differed between individuals with normal and abnormal amyloid. Of the 172 proteins, 156 proteins (91%) were decreased in individuals with abnormal amyloid and showed enrichment for signal transduction, immune system, and cell cycle processes, among others (Figures 2B, 2E, and 3B; Tables S4 and S5 in supporting information). Sixteen proteins (9%) had higher levels in amyloid‐abnormal individuals (Figure 2B, Table S5); this group did not show enrichment for biological processes (Figure 2E, Table S4).
For APOE ε4 carriership, 22 out of 1016 analyzed proteins (2.2%) differed in APOE ε4 carriers and non‐carriers. Fifteen of these proteins (68% of 22 proteins) had higher levels in APOE ε4 non‐carriers (Figure 2C and 2E; Table S6 in supporting information), without enrichment of biological processes, and seven of the proteins (32% of 22 proteins) had higher levels in APOE ε4 carriers.
Finally, we tested the main effect of sex in 835 proteins without interactions between sex and age, amyloid status, and APOE ε4. Of these 835 proteins, 352 proteins (42%) had different levels in women compared to men, of which 243 (69% of 352) proteins were increased in women (Figure 2D; Table S7 in supporting information) and were enriched for metabolism, metabolism of proteins, and extracellular matrix organization–related processes, among others (Figure 2E and 3C; Table S4). The remaining 109 (31% of 352) proteins showed higher levels in men (Figure 2D; Table S7) and were enriched for metabolism of proteins, hemostasis, and immune system–related processes, among others (Figure 2E and 3D; Table S4). Because the METSIM study consisted of only men, which potentially may have biased the results, we repeated our analyses excluding the METSIM dataset. Excluding the METSIM dataset did not influence the demographics of the combined cohort (Table S8 in supporting information), and the majority of proteins and pathways related to sex remained significant after excluding METSIM (342 of 352 proteins [97%] and 63 of 65 [97%] pathways; Tables S4 and S7).
3.3. Age‐related proteome changes depending on amyloid status
Next, we looked at 39 (3% of 1149) proteins for which the association with age depended on amyloid status (Figure 4A; Table S9 in supporting information). In 24 (62% of 39) of these proteins, age effects were restricted to amyloid‐positive participants, of which 7 proteins including t‐tau and p‐tau increased with age, and the remaining 17 proteins showed decreased levels with older age. Proteins that decreased with age in amyloid abnormal individuals were enriched for extracellular matrix organization, a process that was also enriched in proteins with a main effect of age. In the amyloid normal group, seven proteins were associated with age (18% of 39). Eight proteins (21% of 39 proteins) had a significant interaction between age and amyloid status but no significant age relation in either amyloid subgroup.
FIGURE 4.
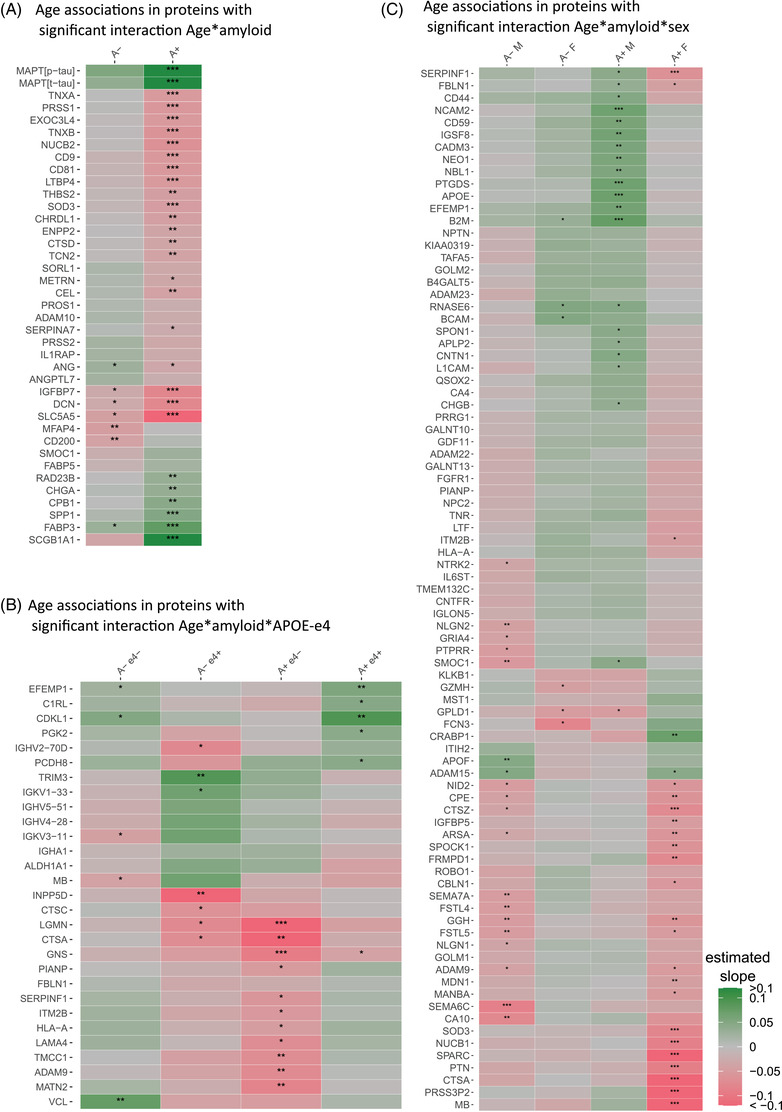
Amyloid‐dependent age effects on the cerebrospinal fluid proteome. Heatmap of regression coefficients for age: age effects in (A), proteins with a two‐way interaction between age and amyloid status, stratified for amyloid status; (B), proteins with a three‐way interaction between age, amyloid status, and apolipoprotein E (APOE) ε4, stratified for amyloid status and APOE ε4 carriership; and (C), proteins with a three‐way interaction between age, amyloid status, and sex, stratified for amyloid status and sex. A–, amyloid normal; A+, amyloid abnormal; e4+, APOE ε4 carrier; e4–, APOE ε4 non‐carrier; F, female; M, male. P < .05; **, P < .01; ***, P < .001. Full age effect estimates are provided in Tables S9‐11.
We continued to analyze proteins with age associations with three‐way interactions between age, amyloid status, and APOE ε4 carriership or sex. Twenty‐nine proteins showed age‐related effects with a three‐way interaction between age, amyloid status, and APOE ε4 (Figure 4B; Table S10 in supporting information). None of these proteins were observed in the previous models of age effects. After stratification on amyloid and APOE ε4 status, we found most age associations in amyloid abnormal APOE ε4 non‐carriers, in whom 11 out of the 29 proteins (38%) were associated with age, all with lower levels with older age and without enrichment of specific biological processes. In amyloid abnormal APOE ε4 carriers, six of the proteins showed an effect of age, all but one with increased levels with older age. Five proteins (17% of 29 proteins) had a significant three‐way interaction, but no significant age effect in any amyloid status and APOE ε4 subgroup (see Table S10 for other amyloid status and APOE ε4 subgroups).
Finally, 85 (7% of 1149) proteins showed a three‐way interaction between age, amyloid status, and sex (Figure 4C; Table S11 in supporting information). Most associations with age were found in amyloid abnormal women (25 proteins, 29%): two proteins showed higher levels with older age, and 23 proteins decreased with older age without enrichment for specific biological processes. In amyloid abnormal men, 21 proteins (25%) showed age associations, all but one with higher levels with older age, and showed no enrichment for biological processes. Twenty‐eight proteins (33% of 85 proteins) had a significant three‐way interaction, but no significant age association in any amyloid and sex subgroup (see Table S11 for other amyloid status and sex subgroups). We tested if the results remained similar when excluding the METSIM dataset, to test if this male‐only dataset drove the three‐way interaction between amyloid and sex. After excluding METSIM data, the age effects remained the same for the majority of proteins (56 of 85 proteins, 66%; Figure S1 in supporting information, Table S11).
4. DISCUSSION
In this study, we investigated in cognitively intact individuals how the CSF proteome changes with older age, and whether such changes depend on AD pathology (measured by amyloid status), APOE ε4, and sex. We found that 252 (22% of 1149) proteins showed age‐related changes independent of amyloid status, APOE ε4 carriership, and sex, of which 194 proteins showed higher levels with older age and were related to processes of the immune system, signal transduction, and cellular responses to external stimuli. Another group of 39 proteins showed age‐related changes that depended on amyloid status, with enrichment of extracellular matrix organization. These results show that the CSF proteome changes substantially with older age, and that a part may reflect early AD pathology. We found that some enriched biological processes in proteins with amyloid‐dependent and amyloid‐independent age associations overlapped, suggesting these processes may contribute to the brain's higher vulnerability to neurodegeneration with aging.
Several of the proteins that were associated with age independent of amyloid status have been previously reported to increase with older age, including NEFL, 29 YKL‐40, 30 , 31 A2M, 32 CLU, 33 C4 34 , 35 , and SNCA. 36 Two previous studies analyzed age effects on the CSF proteome, 6 , 7 which reported 30 6 and 77 7 age‐related proteins. For 31 of the 94 age‐related proteins found in these two studies, we investigated the main effect of age in our analyses. We reproduced age associations for 10 of these proteins. The other proteins were not related to age in our study (17 proteins), or were age‐related but in opposite directions in our study (three proteins). These discrepancies could be due to methodological differences, as our results were adjusted for amyloid, APOE, and sex, which previous studies did not do. 6 , 7 Potentially, the predominantly increased protein levels with older age could be due to lower CSF turnover with older age leading to more concentrated CSF. 37 , 38 However, we also see groups of proteins that decreased with older age or showed interactions between age and amyloid, APOE ε4, or sex. This makes it unlikely that CSF turnover explains age‐related proteome changes. Both age‐related and amyloid status‐related proteins showed specific associations with various immune system processes. This involvement of the immune system both with aging 39 , 40 and AD pathology 41 , 42 has been documented before. We also found overlap between age and amyloid‐related processes for pathways related to the extracellular matrix and insulin‐like growth factor (IGF) transport and uptake, in line with previous studies showing different levels of extracellular matrix proteins 5 , 43 and IGF metabolism proteins 44 , 45 in AD patients compared to controls. Perhaps, these processes may contribute to the development of AD pathology in very early phases of the disease.
We found relatively few effects of APOE ε4 carriership on the CSF proteome (22 proteins). We think that APOE ε4 effects are probably more common than we measured here. Previous studies have shown that APOE ε4 is part of a network of co‐expressed genes, 46 and that AD risk factors may differ in APOE ε4 carriers versus non‐carriers. 47 Perhaps the low number of APOE ε4 effects is due to heterogeneity in APOE ε4 effects: a previous study that analyzed differences between APOE ε4 carriers and non‐carriers in EMIF‐AD MBD and ADNI observed a relatively low correspondence between the cohorts (58%). 8 Nonetheless, we found 29 proteins that had a three‐way interaction between age, amyloid status, and APOE ε4, suggesting that the association of a proportion of CSF proteins with AD may differ in APOE ε4 carriers and non‐carriers.
Our study population contained a group of amyloid normal individuals with abnormal t‐tau or p‐tau, which may reflect suspected non‐Alzheimer's pathophysiology (SNAP). We found that in amyloid abnormal individuals, and at a trend level in amyloid normal individuals (P‐value < 0.1), levels of t‐tau and p‐tau increased with age, as has been observed previously. 48 , 49 If tau levels indeed change with age also in amyloid normal individuals, tau levels could be another factor influencing the proteome, which should be investigated in future studies.
We found different levels of many proteins between women and men (352 proteins). So far, to our knowledge, no studies have investigated sex effects on the CSF proteome in later adult life. We found that proteins that differed between men and women were enriched for processes related to metabolism of proteins, in line with a study of sex effects on the plasma proteome. 50 Overall, it is important to conduct more studies of both the effects of APOE ε4 and sex on the CSF proteome, because therapeutic targets may be different in men and women, and perhaps also depending on APOE ε4, which has important implications for clinical trials and drug discovery studies.
A potential limitation of our study is that proteomics was measured on different platforms, which may have introduced noise in the data. Still, we think such effects on our results are small, since we observed more proteins than previous studies. In addition, we used cross‐sectional data to assess age effects on the CSF proteome, and it is therefore possible that the age effects we observed here were confounded by additional unknown factors that differed between older and younger individuals in our data. Further longitudinal studies are needed to study aging effects within individuals over time. As we analyzed patterns of proteome changes without multiple testing, caution is warranted when interpreting results for single proteins. The proteins we found should therefore be reproduced also in future studies.
Strong aspects of our study are the large CSF proteomic dataset spanning a wide age range, and simultaneous analysis of the changes in the CSF proteome depending on age, amyloid status, APOE ε4 carriership, and sex. Effects of these factors on the CSF proteome can be queried through a web interface to aid biomarker and fundamental studies into AD (https://kwesenhagen.shinyapps.io/Aging_proteomics/).
In conclusion, we provide an overview of biological processes that are generally altered in aging, versus those that depended on amyloid status, sex, and/or APOE genotype. The amyloid‐dependent age associations in CSF proteins we reported here, of which many had not been previously reported as related to AD, provide more insight in the biological processes that are altered in an early phase of AD, and may ultimately help to identify novel therapeutic targets.
CONFLICTS OF INTEREST
Hilkka Soininen has served as an advisory board member of ACImmune. Prof. dr. Scheltens has acquired grants for the institution from GE Healthcare and Piramal and received consultancy/speaker fees paid to the institution from Novartis, Probiodrug, Biogen, Roche, and EIP Pharma, LLC in the past 2 years. Prof. dr. Henrik Zetterberg has served on scientific advisory boards for Eisai, Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, Nervgen, AZTherapies, and CogRx; has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, and Biogen; and is a co‐founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). Prof. dr. Teunissen received grants from the European Commission, the Dutch Research Council (ZonMW), Association of Frontotemporal Dementia/Alzheimer's Drug Discovery Foundation, The Weston Brain Institute, Alzheimer Netherlands. Prof. dr. Teunissen has functioned on advisory boards of Roche, received non‐financial support in the form of research consumables from ADxNeurosciences and Euroimmun, performed contract research or received grants from Probiodrug, Biogen, Esai, Toyama, Janssen Prevention Center, Boehringer, AxonNeurosciences, EIP farma, PeopleBio, Roche. Henrik Zetterberg has served at scientific advisory boards for Eisai, Denali, Roche Diagnostics, Wave, Samumed, Siemens Healthineers, Pinteon Therapeutics, Nervgen, AZTherapies, and CogRx; has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure, and Biogen; and is a co‐founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). The other authors reported no conflicts of interest.
AUTHOR CONTRIBUTIONS
Pieter Jelle Visser, Betty M. Tijms, Charlotte E. Teunissen and Kirsten E.J. Wesenhagen were involved in designing the study. Johan Gobom, Isabelle Bos, Stephanie J.B. Vos, Pablo Martinez‐Lage, Julius Popp, Magda Tsolaki, Rik Vandenberghe, Yvonne Freund‐Levi, Frans Verhey, Simon Lovestone, Johannes Streffer, Valerija Dobricic, Lars Bertram, Kaj Blennow, and Henrik Zetterberg were involved in coordinating the local centers of the EMIF‐AD MBD study. Johan Gobom, Kaj Blennow, and Henrik Zetterberg performed proteomic measurements in EMIF‐AD MBD. Hilkka Soininen, Maria Pikkarainen, Merja Hallikainen, Johanna Kuusisto, and Markku Laakso were involved in coordinating the METSIM study and collecting data within this study. All co‐authors provided feedback on the manuscript.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
This work has been supported by ZonMW Memorabel grant programme #733050824 (KW, BMT, and PJV) and by the Innovative Medicines Initiative Joint Undertaking under EMIF‐AD MBD grant agreement #115372. HZ is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018‐02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG‐720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809‐2016862), the AD Strategic Fund and the Alzheimer's Association (#ADSF‐21‐831376‐C, #ADSF‐21‐831381‐C and #ADSF‐21‐831377‐C), the Olav Thon Foundation, the Erling‐Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019‐0228), the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska‐Curie grant agreement No 860197 (MIRIADE), and the UK Dementia Research Institute at UCL. Alzheimerfonden (AF‐930934) and Stiftelsen för Gamla Tjänarinnor.
Data was used for this project of which collection and sharing was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH‐12‐2‐0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol‐Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann‐La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Wesenhagen KEJ, Gobom J, Bos I, et al. Effects of age, amyloid, sex, and APOE ε4 on the CSF proteome in normal cognition. Alzheimer's Dement. 2022;14:e12286. 10.1002/dad2.12286
*Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp‐content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
REFERENCES
- 1. Alzheimer's Disease International. Policy Brief: The Global Impact of Dementia 2013‐2050. n.d.
- 2. WHO . Global Action Plan on the Public Health Response to Dementia 2017 ‐ 2025. World Health Organization; 2017. [Google Scholar]
- 3. Jansen WJ, Ossenkoppele R, Knol DL, et al. Prevalence of cerebral amyloid pathology in persons without dementia. JAMA. 2015;313:1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jack CR, Therneau TM, Weigand SD, et al. Prevalence of biologically vs clinically defined Alzheimer's spectrum entities using the National institute on aging‐Alzheimer's association research framework. JAMA Neurol. 2019;76:1174‐1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wesenhagen KEJ, Teunissen CE, Visser PJ, Tijms BM. Cerebrospinal fluid proteomics and biological heterogeneity in Alzheimer's disease: a literature review. Crit Rev Clin Lab Sci. 2020;57:86‐98. [DOI] [PubMed] [Google Scholar]
- 6. Zhang J, Goodlett DR, Peskind ER, et al. Quantitative proteomic analysis of age‐related changes in human cerebrospinal fluid. Neurobiol Aging. 2005;26:207‐227. [DOI] [PubMed] [Google Scholar]
- 7. Baird GS, Nelson SK, Keeney TR, et al. Age‐dependent changes in the cerebrospinal fluid proteome by slow off‐rate modified aptamer array. Am J Pathol. 2012;180:446‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Konijnenberg E, Tijms BM, Gobom J, et al. APOE ϵ4 genotype‐dependent cerebrospinal fluid proteomic signatures in Alzheimer's disease. Alzheimer's Res Ther. 2020;12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hohman TJ, Dumitrescu L, Barnes LL, et al. Sex‐specific association of apolipoprotein e with cerebrospinal fluid levels of tau. JAMA Neurol. 2018;75:989‐998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Babapour Mofrad R, Tijms BM, Scheltens P, et al. Sex differences in CSF biomarkers vary by Alzheimer's disease stage and APOE ε4 genotype. Neurology. 2020;95:e2378‐e2388. [DOI] [PubMed] [Google Scholar]
- 11. Mueller SG, Weiner MW, Thal LJ, et al. Ways toward an early diagnosis in Alzheimer's disease: the Alzheimer's disease neuroimaging initiative (ADNI). Alzheimer's Dement. 2005;1:55‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bos I, Vos S, Vandenberghe R, et al. The EMIF‐AD multimodal biomarker discovery study: design, methods and cohort characteristics. Alzheimers Res Ther. 2018;10:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westwood S, Liu B, Baird AL, et al. The influence of insulin resistance on cerebrospinal fluid and plasma biomarkers of Alzheimer's pathology. Alzheimers Res Ther. 2017;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. ADNI . ADNI General Procedures Manual n.d. http://adni.loni.usc.edu/wp‐content/uploads/2010/09/ADNI_GeneralProceduresManual.pdf (Accessed November 14, 2021).
- 15. Folstein MF, Folstein SE, McHugh PR. Mini‐mental state. J Psychiatr Res. 1975;12:189‐198. [DOI] [PubMed] [Google Scholar]
- 16. Hughes CP, Berg L, Danziger W, Coben LA, Martin RL. A new clinical scale for the staging of dementia. Br J Psychiatry. 1982;140:566‐572. [DOI] [PubMed] [Google Scholar]
- 17. Laakso M, Kuusisto J, Stančáková A, et al. The metabolic syndrome in men study: a resource for studies of metabolic & cardiovascular diseases. J Lipid Res. 2017;58:481‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfeffer RI, Kurosaki TT, Harrah CH, Chance JM, Filos S. Measurement of functional activities in older adults in the community. J Gerontol. 1982;37:323‐329. [DOI] [PubMed] [Google Scholar]
- 19. Rohloff JC, Gelinas AD, Jarvis TC, et al. Nucleic acid ligands with protein‐like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014;3:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MRM Assays : Caprion n.d. http://www.caprion.com/en/technology/mrm‐assays.php (Accessed August 27, 2019).
- 21. Myriad RBM Operational Procedures White Paper ‐ Myriad RBM n.d. https://myriadrbm.com/scientific‐media/myriad‐rbm‐operational‐procedures‐white‐paper/ (Accessed August 27, 2019).
- 22. Tijms BM, Gobom J, Reus L, et al. Pathophysiological subtypes of Alzheimer's disease based on cerebrospinal fluid proteomics. Brain. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tijms BM, Willemse EAJ, Zwan MD, et al. Unbiased approach to counteract upward drift in cerebrospinal fluid amyloid‐β 1‐42 analysis results. Clin Chem. 2018;64:576‐585. [DOI] [PubMed] [Google Scholar]
- 24. Shaw LM, Vanderstichele H, Knapik‐Czajka M, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saykin AJ, Shen L, Foroud TM, et al. Alzheimer's disease neuroimaging initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimer's. Dement. 2010;6:265‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bos I, Vos S, Verhey F, et al. Cerebrospinal fluid biomarkers of neurodegeneration, synaptic integrity, and astroglial activation across the clinical Alzheimer's disease spectrum. Alzheimer's. Dement. 2019;15:644‐654. [DOI] [PubMed] [Google Scholar]
- 27. Fabregat A, Jupe S, Matthews L, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:D649‐D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reactome Pathway Database n.d. https://reactome.org/ (accessed November 14, 2021).
- 29. Bridel C, Van Wieringen WN, Zetterberg H, et al. Diagnostic value of cerebrospinal fluid neurofilament light protein in neurology: a systematic review and meta‐analysis. JAMA Neurol. 2019;76:1035‐1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kušnierová P, Zeman D, Hradílek P, Zapletalová O, Stejskal D. Determination of chitinase 3‐like 1 in cerebrospinal fluid in multiple sclerosis and other neurological diseases. PLoS One. 2020;15:e0233519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bonneh‐Barkay D, Wang G, Starkey A, Hamilton RL, Wiley CA. In vivo CHI3L1 (YKL‐40) expression in astrocytes in acute and chronic neurological diseases. J Neuroinflammation. 2010;7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Šunderić M, Križáková M, Malenković V, Ćujić D, Katrlík J, Nedić O. Changes due to ageing in the glycan structure of alpha‐2‐macroglobulin and its reactivity with ligands. Protein J. 2019;38:23‐29. [DOI] [PubMed] [Google Scholar]
- 33. Slot RER, Kester MI, Van Harten AC, et al. ApoE and clusterin CSF levels influence associations between APOE genotype and changes in CSF tau, but not CSF Aβ42, levels in non‐demented elderly. Neurobiol Aging. 2019;79:101‐109. [DOI] [PubMed] [Google Scholar]
- 34. Kamitaki N, Sekar A, Handsaker RE, et al. Complement genes contribute sex‐biased vulnerability in diverse disorders. Nature. 2020;582:577‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallego JA, Blanco EA, Morell C, Lencz T, Malhotra AK. Complement component C4 levels in the cerebrospinal fluid and plasma of patients with schizophrenia. Neuropsychopharmacology. 2020:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Compta Y, Valente T, Saura J, et al. Correlates of cerebrospinal fluid levels of oligomeric‐ and total‐α‐synuclein in premotor, motor and dementia stages of Parkinson's disease. J Neurol. 2015;262:294‐306. [DOI] [PubMed] [Google Scholar]
- 37. Chiu C, Miller MC, Caralopoulos IN, et al. Temporal course of cerebrospinal fluid dynamics and amyloid accumulation in the aging rat brain from three to thirty months. Fluids Barriers CNS. 2012;9:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen CPC, Chen RL, Preston JE. The influence of ageing in the cerebrospinal fluid concentrations of proteins that are derived from the choroid plexus, brain, and plasma. Exp Gerontol. 2012;47:323‐328. [DOI] [PubMed] [Google Scholar]
- 39. Moskalev A, Stambler I, Caruso C. Innate and adaptive immunity in aging and longevity: the foundation of resilience. Aging Dis. 2020;11:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos‐Lozano A, Valenzuela PL, Llavero F, et al. Successful aging: insights from proteome analyses of healthy centenarians. Aging (Albany NY). 2020;12:3502‐3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jevtic S, Sengar AS, Salter MW, McLaurin JA. The role of the immune system in Alzheimer's disease: etiology and treatment. Ageing Res Rev. 2017;40:84‐94. [DOI] [PubMed] [Google Scholar]
- 42. Nichols MR, St‐Pierre M, Wendeln A, et al. Inflammatory mechanisms in neurodegeneration. J Neurochem. 2019;149:562‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma J, Ma C, Li J, Sun Y, Ye F, Liu K, et al. Extracellular matrix proteins involved in Alzheimer's disease. Chem – A Eur J. 2020;26:12101‐12110. [DOI] [PubMed] [Google Scholar]
- 44. McGrath ER, Himali JJ, Levy D, et al. Circulating IGFBP‐2: a novel biomarker for incident dementia. Ann Clin Transl Neurol. 2019;6:1659‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hertze J, Nägga K, Minthon L, Hansson O. Changes in cerebrospinal fluid and blood plasma levels of IGF‐II and its binding proteins in Alzheimer's disease: an observational study. BMC Neurol. 2014;14:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang S, Tang L, Zhao N, Yang W, Qiu Y, Chen HZ. A systems view of the differences between APOE ε4 carriers and non‐carriers in Alzheimer's disease. Front Aging Neurosci. 2016;8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ma Y, Jun GR, Zhang X, et al. Analysis of whole‐exome sequencing data for Alzheimer's disease stratified by APOE genotype. JAMA Neurol. 2019;76:1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baldacci F, Lista S, Manca ML, et al. Age and sex impact plasma NFL and t‐Tau trajectories in individuals with subjective memory complaints: a 3‐year follow‐up study. Alzheimer's Res Ther. 2020;12:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sutphen CL, Jasielec MS, Shah AR, et al. Longitudinal ; cerebrospinal fluid biomarker changes in preclinical Alzheimer's disease during middle age. JAMA Neurol. 2015;72:1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lehallier B, Gate D, Schaum N, et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med. 2019;25:1843‐1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.


