Abstract
Clinical studies support a role for the protein regulated in development and DNA damage response 1 (REDD1) in ischemic retinal complications. To better understand how REDD1 contributes to retinal pathology, we examined human single-cell sequencing data sets and found specificity of REDD1 expression that was consistent with markers of retinal Müller glia. Thus, we investigated the hypothesis that REDD1 expression specifically in Müller glia contributes to diabetes-induced retinal pathology. The retina of Müller glia-specific REDD1 knockout (REDD1-mgKO) mice exhibited dramatic attenuation of REDD1 transcript and protein expression. In the retina of streptozotocin-induced diabetic control mice, REDD1 protein expression was enhanced coincident with an increase in oxidative stress. In the retina of diabetic REDD1-mgKO mice, there was no increase in REDD1 protein expression, and oxidative stress was reduced compared with diabetic control mice. In both Müller glia within the retina of diabetic mice and human Müller cell cultures exposed to hyperglycemic conditions, REDD1 was necessary for increased expression of the gliosis marker glial fibrillary acidic protein. The effect of REDD1 deletion in preventing gliosis was associated with suppression of oxidative stress and required the antioxidant transcription factor nuclear factor erythroid-2-related factor 2 (Nrf2). In contrast to diabetic control mice, diabetic REDD1-mgKO mice did not exhibit retinal thinning, increased markers of neurodegeneration within the retinal ganglion cell layer, or deficits in visual function. Overall, the findings support a key role for Müller glial REDD1 in the failed adaptive response of the retina to diabetes that includes gliosis, neurodegeneration, and impaired vision.
Introduction
Diabetic retinopathy (DR) is a common ocular complication caused by both type 1 and type 2 diabetes. Despite significant advances in therapeutic approaches to its treatment, DR remains among the most frequent causes of new cases of blindness (1). Recent clinical trials demonstrate that blockade of vascular endothelial growth factor (VEGF) signaling reduces edema and improves visual acuity in approximately half of patients (2). One of the limitations of the current single-cytokine targeted approaches is that they largely address the microvascular defects and neovascularization that occur in more advanced stages of retinal disease. Importantly, significant neuroglial deficits often precede and even predict the visible signs of microvascular disease in the retina in patients with diabetes (3,4). In the retina of patients with diabetes, glial activation likely serves as an adaptive response to mitigate tissue damage prior to clinical manifestation of DR (5,6). However, prolonged changes in expression of glial-derived factors, including VEGF, contribute to disease progression (7,8).
Müller cells are the principal retinal glia that extend radially across the entire retina to participate in both the inner and outer limiting membranes (9). Owing to this orientation, Müller glia contact virtually every other retinal cell type. Müller cells provide critical homeostatic and trophic support to both the retinal vasculature and neuronal layers. Among their many roles, Müller cells recycle neurotransmitters to prevent excitotoxicity, provide spatial buffering of ions, reabsorb fluid to prevent edema, participate in the retinoid cycle, and regulate nutrient levels (10). In addition, Müller cells release a variety of neurotrophic and angiogenic growth factors and cytokines as well as critical antioxidants such as glutathione (11). Shortly after the onset of diabetes, Müller cells become gliotic, undergoing morphological and functional alterations (12). In DR, Müller cells exhibit a range of gene and protein expression changes that contribute to retinal inflammation, microvascular defects, and neuronal dysfunction (13).
In rodent models of both type 1 and type 2 diabetes, expression of the stress response protein regulated in development and DNA damage response 1 (REDD1, also known as DDIT4/RTP801) is increased in the retina (14–17). A key role for REDD1 in DR has been demonstrated in both preclinical models and in patients with diabetes (14,18,19). The Study Evaluating Efficacy and Safety of PF-04523655 Versus Laser in Subjects with Diabetic Macular Edema (DEGAS) phase 2 clinical trial intravitreally administered stabilized siRNA targeting the REDD1 mRNA (PF-04523655) for treatment of diabetic macular edema and found a trend toward improvement in best corrected visual acuity (BCVA) when compared with focal/grid laser photocoagulation (19). In a secondary analysis of patients who completed 12-month follow-up, mean improvement in BCVA with 3 mg PF-04523655 was superior to laser (+9.1 letters vs. +3.2 letters). However, much remains unknown regarding how REDD1 contributes to retinal pathology in diabetes. Herein, we found that retinal REDD1 expression was principally localized to Müller glia. Moreover, we demonstrate the necessity of Müller glial REDD1 in development of retinal defects and visual dysfunction in diabetic mice.
Research Design and Methods
Animals
Generation of REDD1fl/fl (20) and Müller glia-specific REDD1 knockout (mgKO) mice is described in the Supplementary Materials (Supplementary Fig. 1). In REDD1-mgKO mice, Cre expression is directed to retinal Müller glia by the Pdgfra (platelet-derived growth factor receptor, α polypeptide) promoter (21). Pdgfra-Cre; HA-Rpl22 mice were generated as previously described (22). REDD1fl/fl and REDD1-mgKO littermates were administered streptozotocin (STZ) or sodium citrate buffer for 5 consecutive days to induce diabetes. As previously described (23), male and female mice were administered 50 or 75 mg/kg STZ, respectively. Diabetic phenotype was assessed with fasting blood glucose levels >250 mg/dL. All procedures were approved by the Penn State College of Medicine Institutional Animal Care and Use Committee and were in accordance with the Association for Research in Vision and Ophthalmology statement for the ethical use of animals in ophthalmic and vision research.
Müller Cell Culture
Isolation of primary Müller cells was performed using the Worthington Papain Kit (Worthington Biochemical Corp.). Briefly, mouse retinas were enucleated, dissected free of the retinal pigment epithelium, exposed to papain and DNase, and diced. Retinas were incubated at 37°C for 1 h, triturated, and spun at 300g for 5 min. The cell pellet was resuspended in an ovomucoid inhibitor-BSA solution with DNase and centrifuged at 70g for 6 min. The cell pellet was resuspended in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were transferred to culture dishes coated with 0.1% gelatin and incubated at 37°C in 5% CO2. Cultures were enriched for Müller cells in three passages. MIO-M1 Müller cell culture, REDD1 deletion by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, and stable nuclear factor erythroid-2-related factor 2 (Nrf2) knockdown were previously described (16).
RNA Isolation and Quantitative PCR
Total RNA was extracted with TRIzol (Invitrogen), reverse transcribed, and subjected to quantitative real-time PCR, as previously described (15). Primer sequences for the targets of interest are in Supplementary Table 1.
Western Blot Analysis
Retinas were flash frozen in liquid nitrogen and homogenized as previously described (14). Cell lysates and retinal homogenates were combined with Laemmli buffer, boiled, and fractionated in Criterion Precast 4–20% gels (Bio-Rad Laboratories). Proteins were transferred to a polyvinylidene fluoride membrane, blocked in 5% milk in Tris-buffered saline Tween 20, and evaluated with antibodies (Supplementary Table 2).
Immunofluorescent Microscopy
Whole eyes were excised, and corneas were punctured followed by incubation in 4% paraformaldehyde (pH 7.5) for 30 min. Eyes were washed with PBS and incubated at 4°C in 30% sucrose solution containing 0.05% sodium azide for 48 h. Eyes were embedded in optimal cutting temperature compound, flash frozen, and sectioned. Cryosections (10 µm) were permeabilized in PBS + 0.1% Triton-X-100 and blocked in 10% normal donkey serum. Sections were labeled with anti-glutamine synthetase (GS) and Alexa Fluor 488 secondary antibodies, blocked, and then labeled with anti-hemagglutinin (HA) and Alexa Fluor 647 over 2 consecutive days (Supplementary Table 2). Alternatively, sections were stained as described above, substituting anti-glial fibrillary acidic protein (GFAP) for anti-HA. Sections were also stained for cleaved caspase-3 with Alexa Fluor 488. All sections were costained with 1.6 μmol/L Hoechst. Slides were mounted with Fluoromount aqueous mounting media (Sigma-Aldrich) and imaged by confocal laser microscope (Leica SP8; Leica) with frame-stack sequential scanning.
Reactive Oxygen Species Measurement
Retinal reactive oxygen species (ROS) levels were measured as described previously (17). Retinal cryosections were fixed with 2% paraformaldehyde, stained with 1.6 μmol/L Hoechst and 10 μmol/L 2,7-dichlorofluoroscein (DCF; λex = 504 nm), and imaged by confocal laser microscopy. Alternatively, fluorescence was measured from retinal lysates exposed to 10 μmol/L DCF using a plate reader (Spectra Max M5; Molecular Devices) (excitation/emission = 504/529 nm). Additionally, nuclei were isolated from retinal tissue (24), and Nrf2 activity was assessed as previously described (16).
Spectral Domain Optical Coherence Tomography
Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) delivered by intraperitoneal injection. Pupils were dilated with topical phenylephrine HCl (0.5%). Spectral domain optical coherence tomography (SD-OCT) was performed with the Envisu R2210 (Bioptigen) mouse adapted lens with a 50° field of view, centered on the optic disk. Cross-sections from radial scan images were averaged and analyzed with the Bioptigen InVivoVue 2.2 software. Six measurements were obtained from each image, 0.287 mm on either side of the optic disk, and averaged. Autosegmentation reports were generated using Bioptigen InVivoVue Diver 3.4.4 software.
TUNEL
Cryosections were analyzed with an In Situ Cell Death Detection Kit (Millipore-Sigma). Slides were washed three times in PBS for 10 min and permeabilized in PBS with 0.1% Triton-X-100 for 2 min on ice. Sections were washed twice in PBS for 10 min and incubated with 50 µL of TUNEL reaction mixture for 60 min at 37°C. Slides were washed three times for 10 min with PBS and incubated with 1.6 μmol/L Hoechst nuclear stain and imaged by confocal laser microscopy (λex = 488 nm) with frame-stack sequential scanning.
Behavioral Assessment of Visual Function
The OptoMotry virtual optomotor system (CerebralMechanics, White Plains, NY) was used to evaluate visual function, as previously described (25). Spatial frequency (SF) threshold and contrast sensitivity (CS) were assessed using a video camera to monitor the optomotor reflex elicited in response to a vertical grating pattern rotating around the mouse on four inward-facing computer monitors. SF and CS were tested at different parameters in a random step-wise protocol (26). CS was assessed at a SF of 0.092 cycles/degree. SF was assessed at 100% contrast. The CS and SF thresholds were identified as the highest values that elicited the reflexive head movement. Thresholds were averaged for each mouse over three trials on consecutive days.
Statistical Analysis
In Fig. 2C–E, a two-sample t test was used to compare groups. In Figs. 3–6, data were analyzed overall with two-way ANOVA, and pairwise comparisons were conducted with the Dunnett or Tukey test for multiple comparisons. Significance was defined as P < 0.05 for all analyses.
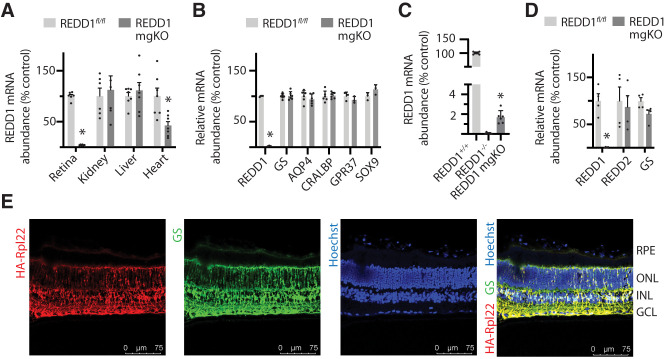
Figure 2.
Conditional deletion of REDD1 nearly eliminated retinal REDD1 expression. A: REDD1 mRNA abundance was evaluated in retina, kidney, liver, and heart of REDD1fl/fl and REDD1-mgKO mice by PCR analysis. B: REDD1, GS, AQP4, CRALBP, GRP37, and SOX9 mRNA abundances were evaluated in retinal lysates. C: REDD1 mRNA abundance was evaluated in the retina of REDD1+/+, REDD1−/−, and REDD1-mgKO mice. D: REDD1, REDD2, and GS expression in primary retinal Müller cells was evaluated. Data points represent biological replicates. Bars are means ± SD (n = 4–7). *P ≤ 0.05 vs. REDD1fl/fl in A, B, and D or REDD1−/− in C. E: Müller cell–specific Cre expression was assessed by crossing Pdgfra-Cre mice with mice homozygous for a mutation targeted to the Rpl22 locus harboring a loxP-flanked wild-type C-terminal exon 4, followed by an identical C-terminal exon 4 that is tagged with three copies of the hemagglutinin (HA) epitope before the stop codon. Whole eyes were isolated and cryo-sectioned into sagittally oriented longitudinal cross-sections. HA-Rpl22 reporter colocalization with the Müller cell–specific marker GS was evaluated in retinal cross-sections by immunofluorescence. Hoechst was used to visualize nuclei. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium.
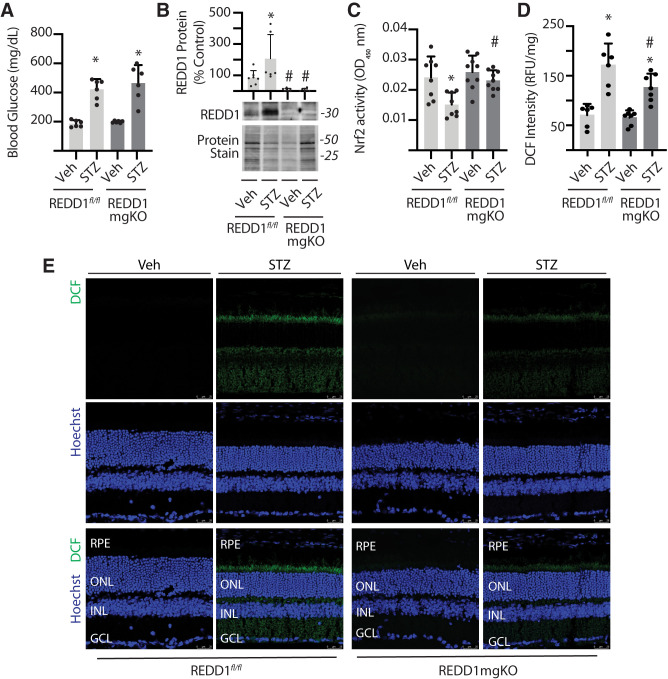
Figure 3.
Müller cell–specific REDD1 deletion prevented diabetes-induced Nrf2 suppression and attenuated oxidative stress in the retina. Diabetes was induced in REDD1fl/fl and REDD1-mgKO mice by administration of STZ. Control mice received a vehicle (Veh). All retinal analysis was performed after 6 weeks of diabetes. A: Postprandial blood glucose concentrations were measured. B: REDD1 protein expression was evaluated in retinal lysates by Western blotting. Protein loading was evaluated by reversible protein stain. Blots shown are representative of two independent experiments. Protein molecular mass (kDa) is indicated at right of blots. C: Nrf2 activity was evaluated in the nuclear fraction obtained from retinal lysates of REDD1fl/fl and REDD1-mgKO mice by DNA-binding ELISA. OD, optical density. D: ROS were quantified in the supernatants from whole retinal lysates using DCF. Values are means ± SD (n = 5–9). RFU, relative fluorescent units. Significance was defined as P < 0.05 for all analyses. *P ≤ 0.05 vs. Veh; #P ≤ 0.05 vs. REDD1fl/fl. E: ROS were visualized in whole-eye sagittal cross-sections using DCF. Hoechst was used to visualize nuclei. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium.
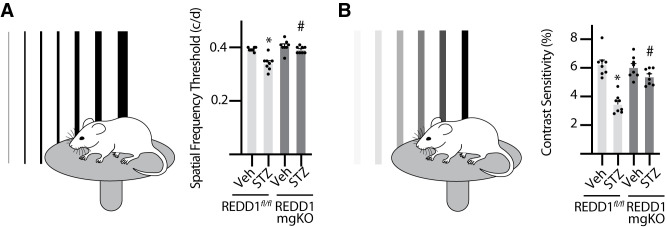
Figure 6.
Diabetes-induced visual dysfunction was absent in mice with Müller cell–specific REDD1 deletion. Diabetes was induced in REDD1fl/fl and REDD1-mgKO mice by administration of STZ. Control mice received a vehicle (Veh). All analysis was performed after 6 weeks of diabetes. Behavioral optometry was performed by displaying rotating bars on inward-facing monitors to elicit an optomotor reflex. Thresholds were averaged over three trials on consecutive days. A: SF was assessed at 100% contrast. B: CS was assessed at a spatial frequency of 0.092 cycles (c)/degree (d). CS is expressed as an inverse percentage to make data interpretation more intuitive. Bars are means ± SD (n = 8). *P ≤ 0.05 vs. Veh; #P ≤ 0.05 vs. REDD1fl/fl.
Data and Resource Availability
The data sets and resources generated during this study are available from the corresponding author upon reasonable request.
Results
REDD1 Expression Was Dominant in Müller Glia
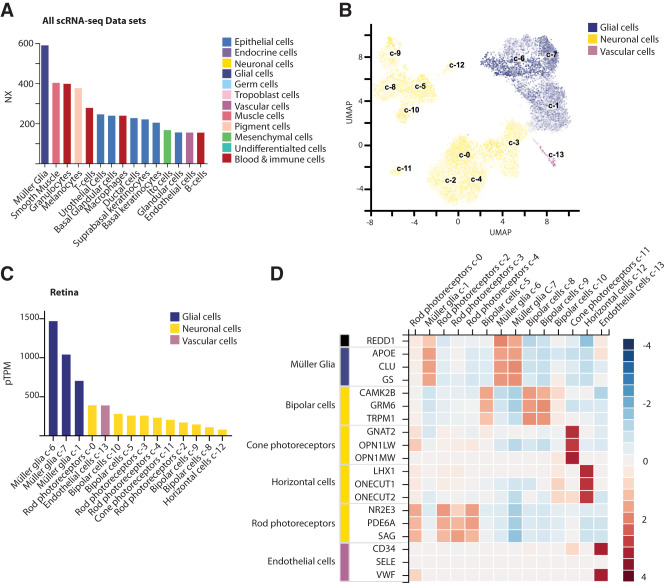
Single cell transcriptomic analysis of human retinas was previously used to identify major retinal cell types and their corresponding gene expression signatures (27,28). The specificity of REDD1 expression was examined in the single-cell RNA sequencing (scRNA-seq) data sets on the Human Protein Atlas. Remarkably, Müller glia exhibited the highest REDD1 expression of the 43 single-cell types included in the meta-analysis (Fig. 1A). Müller glia were the only cell type in which REDD1 expression was defined as enhanced, due to a normalized expression value 5.5 times the mean of all other cell types. A Uniform Manifold Approximation and Projection (UMAP) plot was used to visualize 13 single-cell clusters of retinal cells observed by scRNA-seq (Fig. 1B). Heterogeneity of Müller glia gene expression profiles was demonstrated by three unique cell clusters (c-1, c-6, and c-7). REDD1 expression in retinal cell clusters was greatest in the three Müller glia cell clusters, with c-6 Müller glia exhibiting 6.4 times the mean expression of other retinal cell types (Fig. 1C). Owing to the dominant pattern of expression in Müller glia, the specificity of REDD1 expression in retina cells was consistent with other established markers for this cell type (Fig. 1D). Examination of resource data from Cowan et al. (28) was consistent with the dominant expression in Müller glia that was observed in the meta-analysis (Supplementary Fig. 2).
Figure 1.
REDD1 expression was dominant in retinal Müller glia. REDD1 expression was evaluated in the meta-analysis of scRNA-seq data sets on the Human Protein Atlas (available from https://www.proteinatlas.org). A: Bar graph depicting top 15 normalized expression (NX) values for REDD1 in all single cell types within the meta-analysis. Color coding is based on groups of cells that share common functional features. B: Visualization of 13 single-cell clusters (c1–13) observed in the sequencing analysis of postmortem human retina by a Uniform Manifold Approximation and Projection (UMAP) plot. Each dot represents a single cell. Color intensity for individual cells is according to the percentage of maximum expression (log2(read_count + 1)/log2(max(read_count + 1) ∗ 100) in five different bins (i.e., <1%,<25%,<50%,<75%, and ≥75%). C: Bar graph illustrating REDD1 expression for each retinal cell cluster. pTPM, protein-transcripts per million. D: Heat map of Z-scores for REDD1 expression in retinal cell clusters relative to other established markers of cell type. Image credit: Human Protein Atlas (https://www.proteinatlas.org/ENSG00000168209-DDIT4/celltype).
Müller Glia-Specific REDD1 Deletion Nearly Eliminated Retinal REDD1 Expression
REDD1 mRNA abundance was dramatically attenuated in the retina of REDD1-mgKO mice compared with REDD1fl/fl mice (Fig. 2A), demonstrating that REDD1 expression in murine retina was almost exclusively localized to Pdgfra-Cre–positive cells. By contrast, REDD1 mRNA abundance was similar in the kidney and liver. A modest decrease in REDD1 mRNA was observed in the heart, supporting the presence of Pdgfra-Cre–positive cells in that tissue. The decline in REDD1 expression in the retina of REDD1-mgKO mice was not observed in association with changes in other key markers of Müller glia (Fig. 2B). Unlike whole-body REDD1−/− mice, REDD1 mRNA expression was detectable in the retina of REDD1-mgKO mice, supporting the presence of a Cre-negative REDD1-expressing retinal cell population (Fig. 2C). To evaluate REDD1 expression in Müller glia, primary Müller cell cultures were generated. Müller cells from the retina of REDD1-mgKO mice lacked REDD1 mRNA but expressed REDD2 and the Müller cell marker GS similarly to Müller cells from the retina of REDD1fl/fl mice (Fig. 2D). Specificity of PDGFRa-Cre for Müller cells was assessed by expression of an epitope-tagged Cre-reporter protein in retinal cross-sections. Consistent with the orientation of Müller glia, HA-Rpl22 expression was observed in radially oriented processes that spanned the entire thickness of the neuroretina and in branching end feet that ensheathed neurons within the ganglion cell layer (GCL) (Fig. 2D). HA-tagged Rpl22 expression in the retina strongly colocalized with GS.
Müller Cell–Specific REDD1 Deletion Prevented Diabetes-Induced REDD1 Protein Expression in the Retina
To evaluate the role of Müller cell–specific REDD1 expression in the effect of diabetes on the retina, mice were administered STZ. After 6 weeks of STZ-induced diabetes, REDD1fl/fl mice and REDD1-mgKO mice exhibited similar elevations in postprandial blood glucose concentrations (Fig. 3A). Consistent with the previous observations in wild-type mice (14,16–18), retinal REDD1 protein expression was enhanced in the retina of STZ-diabetic REDD1fl/fl mice compared with nondiabetic REDD1fl/fl controls (Fig. 3B). By contrast, REDD1 protein expression was attenuated in the retina of REDD1-mgKO mice compared with REDD1fl/fl mice, and diabetes failed to promote REDD1 expression.
Diabetes-Induced Oxidative Stress Was Attenuated in the Retina of REDD1-mgKO Mice
The principle pathways responsible for hyperglycemia-induced tissue damage are all linked to the accumulation of ROS (29). One key way cells combat elevated ROS levels is activation of the transcription factor Nrf2. We recently demonstrated that REDD1 acts, at least in part, by suppression of the Nrf2 antioxidant response (16). Nrf2 activity in nuclear isolates obtained from retinal lysates of STZ-diabetic REDD1fl/fl mice was attenuated compared with nondiabetic controls (Fig. 3C). By contrast, Nrf2 activity in the retina of STZ-diabetic REDD1-mgKO mice was enhanced compared with STZ-diabetic REDD1fl/fl mice, and the suppressive effect of diabetes was absent. To determine the impact of Müller cell–specific REDD1 expression on diabetes-induced oxidative stress, ROS were quantified in retinal supernatants from whole retina. ROS were moderately enhanced in the retina of STZ-diabetic REDD1-mgKO mice compared with nondiabetic REDD1-mgKO mice (Fig. 3D). Notably, ROS were attenuated in the retina of STZ-diabetic REDD1-mgKO mice compared with STZ-diabetic REDD1fl/fl controls (Fig. 3D and E). Visualization of ROS in retinal cross-sections suggests the decline in diabetes-induced ROS with Müller cell–specific REDD1 deletion was not localized to a particular retinal layer (Fig. 3E).
REDD1 Contributed to Development of Retinal Gliosis
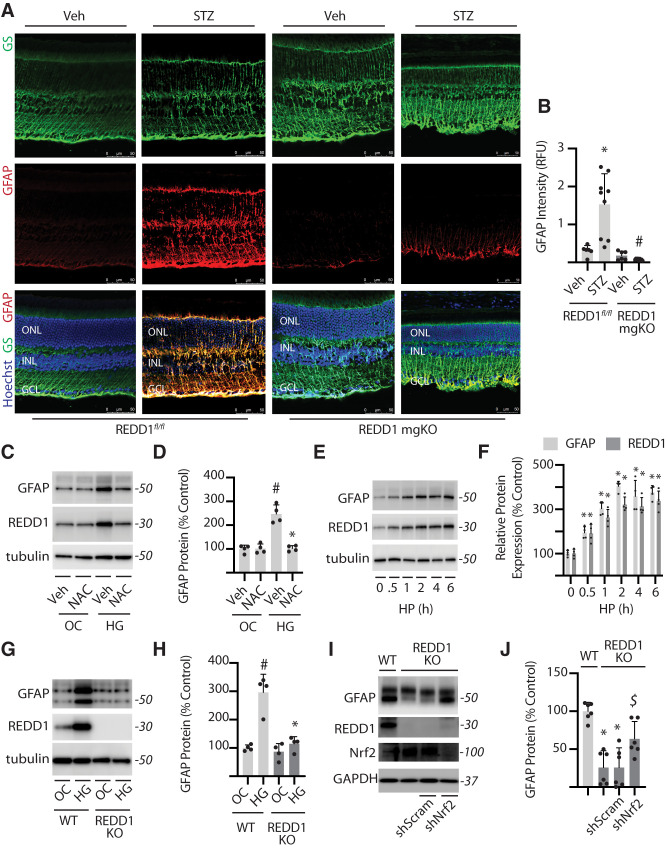
Recent studies support that chemical activation of Nrf2 is sufficient to prevent Müller cell gliosis (30–32). Diabetes promotes expression of glial fibrillary acidic protein (GFAP) in Müller cells (33), which serves as a marker of gliosis. Retinal cross-sections stained for GS demonstrated similar localization and distribution of Müller glia in the retina of REDD1-mgKO and REDD1fl/fl mice (Fig. 4A). GFAP expression in retinal cross-sections from diabetic REDD1fl/fl mice was enhanced compared with nondiabetic controls; however, a similar increase was not observed in STZ-diabetic REDD1-mgKO mice (Fig. 4A and B). To investigate the role of REDD1 and Nrf2 in development of gliosis in humans, we used a previously characterized immortalized human Müller (MIO-M1) cell line (34). Consistent with the prior report (17), REDD1 expression was enhanced in cells exposed to hyperglycemic conditions, and the ROS scavenger N-acetylcysteine (NAC) prevented the effect (Fig. 4C). Extending on the prior report, we found that GFAP expression was also increased in MIO-M1 cells exposed to hyperglycemic conditions in a manner that was prevented by NAC (Fig. 4C and D). Similarly, REDD1 and GFAP protein expression were enhanced in a coordinate manner in cells exposed to the oxidant hydrogen peroxide (Fig. 4E and F). Together, the data are consistent with a model wherein hyperglycemia-induced ROS promote REDD1 and GFAP expression. However, exposure to hyperglycemic conditions failed to promote GFAP protein content in the absence of REDD1 (Fig. 4G and H). Consistent with the prior observation (16), Nrf2 protein expression was enhanced in REDD1-deficient MIO-M1 cells exposed to hyperglycemic conditions compared with wild-type cells (Fig. 4I). In REDD1-deficient cells exposed to hyperglycemic culture conditions, Nrf2 knockdown reduced Nrf2 protein expression and increased GFAP protein expression compared with a scramble shRNA (Fig. 4I and J).
Figure 4.
REDD1 was necessary for GFAP upregulation in response to diabetes or hyperglycemic conditions. A and B: Diabetes was induced in REDD1fl/fl and REDD1-mgKO mice by administration of STZ. All analyses were performed 6 weeks after mice were administered STZ or a vehicle (Veh). Whole eyes were isolated and sectioned. GFAP and GS were evaluated in retinal cross-sections by immunofluorescence. Hoechst was used to visualize nuclei. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. B: Quantification of GFAP protein expression in retinal sections. RFU, relative fluorescent units. C: Human MIO-M1 cells were cultured in medium containing 5 mmol/L glucose and exposed to culture medium containing 30 mmol/L glucose (HG) or 5 mmol/L glucose and 25 mmol/L mannitol as an osmotic control (OC) in the presence of NAC or a vehicle (Veh) control for 4 h. Whole-cell lysates were analyzed by Western blotting. Protein molecular mass (kDa) is indicated at the right of blots. D: GFAP protein expression in C was quantified. E: MIO-M1 cells were exposed to medium supplemented with 1 mmol/L hydrogen peroxide (HP). F: GFAP and REDD1 protein expression in E were quantified. G: Wild-type (WT) and REDD1-KO MIO-M1 cells were exposed to OC or HG as described above. H: GFAP protein expression in G was quantified. I and J: Nrf2 was knocked down by stable expression of a shRNA (shNrf2). Control cells expressed a scramble shRNA (shScram). All analysis was performed 4 h after exposure to medium containing 30 mmol/L glucose. Bars are means ± SD (n = 4–9). *P ≤ 0.05 vs. Veh, time 0, or WT in B and D, F, and H, respectively; #P ≤ 0.05 vs. REDD1fl/fl or OC in B and H, respectively; $P ≤ 0.05 vs. shScram.
Diabetes-Induced Retinal Thinning Was Absent in REDD1-mgKO Mice
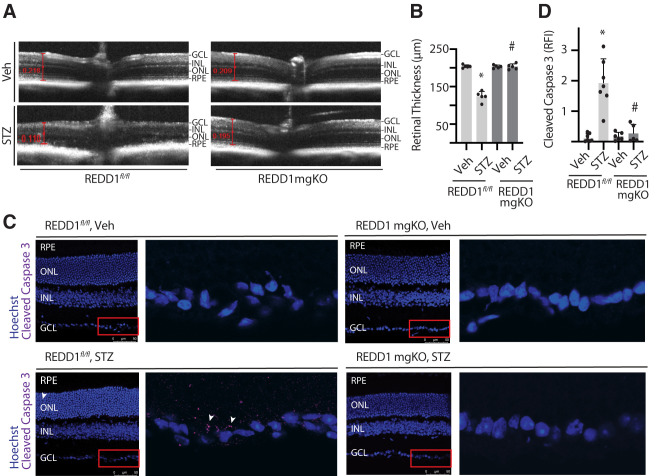
SD-OCT was used to measure the thickness of retinal layers (Fig. 5A). Neuronal retinal thickness measured from the inner limiting membrane to the photoreceptor outer segments (OS) was similar in the retina of nondiabetic REDD1fl/fl and REDD1-mgKO mice (Fig. 5B). A significant deficit in neuronal retinal thickness was observed in the retina of STZ-diabetic REDD1fl/fl mice compared with nondiabetic REDD1fl/fl controls, but a diabetes-induced deficit was not observed in REDD1-mgKO mice. Automated segmentation of OCT images was performed to further examine the impact of diabetes and REDD1 on specific retinal layers. Deficits in thickness were detected in the middle and outer retinal layers of STZ-diabetic REDD1fl/fl mice but not in the inner retinal layer (Supplementary Fig. 3A). Surprisingly, a deficit in the combined retinal nerve fiber layer (RNFL) and GCL was not observed in STZ-diabetic REDD1fl/fl mice compared with nondiabetic controls (Supplementary Fig. 3B). The most dramatic retinal thinning in STZ-diabetic REDD1fl/fl mice was localized to the outer nuclear layer (Supplementary Fig. 3C), which contains photoreceptor cell bodies. A deficit in photoreceptors was also reflected by decreased thickness in both the photoreceptor inner segments and OS (Supplementary Fig. 3D). No differences in thickness were observed in STZ-diabetic versus nondiabetic REDD1-mgKO mice.
Figure 5.
Retinal thinning was not observed in diabetic mice with Müller cell–specific REDD1 deletion. Diabetes was induced in REDD1fl/fl and REDD1-mgKO mice by administration of STZ. Control mice received a vehicle (Veh). All retinal analysis was performed after 6 weeks of diabetes. A: Retinal thickness was evaluated by SD-OCT. Representative images are shown. B: Retinal thickness from the RNFL to the photoreceptor OS was manually measured as indicated by red calipers. C: Whole eyes were isolated and cryosectioned into sagittally oriented longitudinal cross-sections, and cleaved caspase-3 was evaluated by immunohistochemistry. The red box indicates the area presented at higher magnification. White arrowheads indicate cleaved caspase-3–positive immunoreactivity. D: Cleaved caspase-3 intensity was quantified in retinal cross-sections. Bars are means ± SD (n = 5–7). *P ≤ 0.05 vs. Veh; #P ≤ 0.05 vs REDD1fl/fl. INL, inner nuclear layer; ONL, outer nuclear layer; RPE, retinal pigment epithelium; RFI, relative fluorescent intensity.
Markers of Diabetes-Induced Retinal Neurodegeneration Were Absent in REDD1-mgKO Mice
To further interrogate the cause of retinal thinning, cross-sections were analyzed for evidence of neurodegeneration. Caspase-3 is a cysteine protease that becomes active during the late stages of apoptosis. Increased cleaved caspase-3 was observed throughout the retina of STZ-diabetic REDD1fl/fl mice compared with nondiabetic controls (Fig. 5C and D) and was particularly localized to the GCL. A similar increase in cleaved caspase-3 was not observed in the retina of STZ-diabetic REDD1-mgKO mice. Increased TUNEL was also observed throughout the retina of STZ-diabetic REDD1fl/fl controls compared with nondiabetic mice, and there was a dramatic increase in TUNEL-positive nuclei localized to the GCL (Supplementary Fig. 4). By contrast, no significant difference was found in TUNEL in STZ-diabetic and nondiabetic REDD1-mgKO retinas, and only a few TUNEL-positive nuclei were observed in retinal cross-sections from STZ-diabetic REDD1-mgKO mice.
Müller Cell–Specific REDD1 Deletion Prevented Diabetes-Induced Visual Dysfunction
The effect of diabetes on functional vision was assessed by measurement of the optomotor response. STZ-diabetic REDD1fl/fl mice exhibited deficits in both SF (Fig. 6A) and CS (Fig. 6B) compared with nondiabetic controls. By comparison, neither SF nor CS was attenuated in the retina of STZ-diabetic REDD1-mgKO mice compared with nondiabetic REDD1-mgKO mice. Moreover, both thresholds were increased in STZ-diabetic mgKO mice compared with diabetic REDD1fl/fl mice.
Discussion
A mounting body of evidence supports that REDD1 plays a critical role in the development of diabetes-induced retinal dysfunction (19,35). To gain a better understanding of how REDD1 contributes to vision loss in patients with diabetes, we evaluated REDD1 expression in human scRNA-seq data sets and found a pattern of expression that was consistent with other markers of Müller glia. To manipulate REDD1 expression in Müller glia, we used Pdgfra-Cre. REDD1 deletion upon Cre-lox recombination nearly eliminated REDD1 content in whole retinal lysates and primary Müller cell cultures. An important caveat is the possibility that other retinal glia may also exhibit recombinase activity, as Pdgfra expression has been identified in astrocytes of the developing retina (36). In support of Müller glia specificity, Pdgfra-Cre–dependent recombination was observed in radially oriented processes that strongly colocalized with the Müller glia-specific marker GS. The finding is consistent with prior demonstration that Pdgfra-Cre recombinase-dependent nuclear lacZ expression in adult murine retina is exclusively localized to the inner nuclear layer (21). Cell bodies of Müller glia localize to the inner nuclear layer, whereas the cell bodies of astrocytes are more commonly found in the RNFL (37). Regardless, the studies here support that retinal REDD1 expression in murine retina is principally localized to Pdgfra-Cre–positive glia and plays a key role in their response to diabetes.
Significant neuroglial dysfunction occurs early in the progression of retinal defects that lead to DR (3,4,13). In fact, gliosis, neurodegeneration, changes in neuroretinal thickness, and deficits in visual function are hallmarks of the retinal disease caused by diabetes (38–40). In the retina of REDD1-mgKO mice, diabetes failed to promote REDD1 protein expression, and the diabetes-induced increase in Müller cell gliosis was absent. A similar role for REDD1 in the development of gliosis was recently demonstrated in a rodent model of Alzheimer disease (41). The most well-established marker of reactive gliosis is expression of the intermediate filament GFAP. In the healthy retina, GFAP is mostly observed in astrocytes, with only minimal expression in Müller cells. Shortly after the onset of diabetes, GFAP expression in astrocytes declines, and Müller cells exhibit robust upregulation of GFAP levels (38). In the current study, enhanced GFAP expression was not observed in the retina of STZ-diabetic REDD1-mgKO mice. Cleaved caspase-3 staining in the retinal GCL layer and retinal thinning were also reduced in diabetic REDD1-mgKO mice compared with diabetic controls, while visual function was preserved. Together, the observations support a key role for Müller glial REDD1 expression in the failed adaptive response of the retina to diabetes.
In the current study, spatial frequency threshold was similar in nondiabetic REDD1-mgKO and REDD1fl/fl controls, and a suppressive effect of diabetes required Müller glial REDD1 expression. A protective effect on contrast sensitivity was also observed in diabetic REDD1-mgKO mice. The observations support our prior findings that neither spatial frequency nor contrast sensitivity is impaired in the retina of diabetic REDD1−/− mice compared with diabetic controls (18). However, the protective effect of whole-body REDD1 ablation was principally observed in contrast sensitivity, which is associated with inner retinal information processing. In fact, nondiabetic REDD1−/− mice exhibit a modest deficit in the spatial frequency threshold compared with REDD1+/+ controls (18). Thus, whole-body REDD1 ablation prevents an additive deficit in spatial frequency threshold with the induction of diabetes. Reduced spatial frequency threshold with whole-body REDD1 deletion likely reflects deficits in glucose and insulin tolerance in the nondiabetic REDD1−/− mice (42). Herein, Müller glial REDD1 deletion was sufficient to prevent the diabetes-induced deficits in visual function, and there was no evidence for a change in visual thresholds in nondiabetic mice.
It is well established that thinning of inner retinal layers correlates positively with loss of visual function in patients with diabetes (43). This is consistent with modest thinning observed in the inner retinal layers of C57BL/6J mice that is observed after 6 weeks of diabetes (44). Despite the presence of significant TUNEL and cleaved caspase-3 immunoreactivity observed in the GCL of diabetic REDD1fl/fl mice, a deficit in thickness of the combined RNFL and GCL was not detected in our analysis after 6 weeks of STZ diabetes. The dramatic increase in TUNEL labeling was higher than expected based on our prior analysis of whole mounted retinas (45). Thus, widespread TUNEL labeling within the GCL may indicate over amplification of minor DNA nick events in cells undergoing early and possibly reversible stages of apoptosis, as it seems unlikely that so many retinal ganglion cells would be undergoing terminal apoptosis simultaneously. Another important caveat is the potential limitations of the layer segmentation software, as we observed obvious thinning of the total retina in response to diabetes. Surprisingly, thickness of the inner plexiform layer in STZ-diabetic REDD1fl/fl mice was attenuated compared with nondiabetic controls, which was prevented by Müller cell–specific REDD1 deletion. However, the inner nuclear layer of STZ-diabetic REDD1fl/fl mice exhibited an increase in thickness. A likely possibility is that thickness of the combined RNFL and GCL edema within the inner retinal layers of the diabetic control mice may be masking the loss of retinal cells at this relatively early stage of disease.
Müller cells surround photoreceptors from their synaptic terminals to inner segments, and adherens junctions between these two cells form the outer limiting membrane. In patients with diabetic macular edema, thickness of the photoreceptor OS is an important predictor of visual acuity (46). Thinning of the OS was observed in the retina of STZ-diabetic REDD1fl/fl controls, concomitant with the deficits in visual function. By contrast, no deficit in OS thickness was observed in the diabetic REDD1-mgKO mice compared with nondiabetic controls, and visual function was preserved. Each OS is continuously being renewed from its proximal end; thus, OS thinning may be due to a deficit in trophic support provided to photoreceptors by Müller cells that occurs as a response to REDD1. In support of this possibility, the production of the neurotrophic factors brain-derived neurotrophic factor (BDNF), nerve growth factor (NGF), and neurotrophin-3 (NT-3) by activated Müller glia is enhanced by REDD1 knockdown following optic nerve crush (47).
REDD1 mediates the cellular stress response to a number of adverse conditions by suppression of mechanistic target of rapamycin complex 1 (mTORC1) (48,49). Evidence supports that REDD1 facilitates recruitment of protein phosphatase 2A (PP2A) to dephosphorylate Akt, which in turn activates tuberous sclerosis complex 2 (TSC2) guanosine-5′-triphosphatase (GTPase) activity, leading to reduced Rheb-GTP loading, and suppressed mTORC1 activation (50). Signaling through mTORC1 plays a critical role in determining the selection of specific mRNAs for translation in part through phosphorylation of the translational repressor 4E-BP1 (51). In the retina of STZ-diabetic mice, REDD1 is required for dephosphorylation of 4E-BP1 and upregulated translation of the mRNA encoding VEGF (14). Notably, REDD1 is not required for basal VEGF expression, but rather, the REDD1-dependent increase in expression represents a stress response to diabetes and hyperglycemic conditions. In addition to its role as a repressor of mTORC1 (50), a growing body of evidence also supports a key role for REDD1 in the development of oxidative stress through both enhanced mitochondrial superoxide production and repression of the antioxidant response (35).
REDD1 acts to promote glycogen synthase kinase 3 (GSK3)-dependent Nrf2 proteasomal degradation (16). Nrf2 activity in whole retinal lysates was attenuated by diabetes in control mice, whereas REDD1 deletion in retinal Müller cells was sufficient to prevent the suppressive effect. Analysis of scRNA-seq data from the Human Protein Atlas supports that retinal Nrf2 expression is enhanced in Müller cells, with Müller cells exhibiting >2.5-fold the average expression of other retinal cell types. In our prior study (16), retinal Nrf2 activity was robustly enhanced in retinal lysates from REDD1−/− mice. This is in contrast to REDD1-mgKO mice, where no effect of Müller cell–specific REDD1 deletion was observed in the absence of diabetes. Owing to the dominant pattern of REDD1 expression in retinal Müller cells, REDD1 is essentially absent from both the retina of REDD1-mgKO and REDD1−/− mice. Thus, systemic loss of REDD1 may contribute to chronic Nrf2 hyperactivation observed in the retina of REDD1−/− mice. Regardless, the data here suggest that hyperactivation of retinal Nrf2 and absolute normalization of local ROS levels are not necessary for the benefits of REDD1 deletion. Rather, development of diabetes-induced neuroglial defects and visual dysfunction was not observed upon Müller glial REDD1 deletion, which prevented diabetes-induced suppression of Nrf2 and attenuation of retinal ROS levels.
Müller glial REDD1 deletion prevented increased expression of the gliosis marker GFAP in Müller cells of diabetic mice. A similar REDD1-dependent increase in GFAP was observed in human MIO-M1 Müller cell cultures exposed to hyperglycemic conditions, and the absence of an increase in GFAP in REDD1-deficient cells required Nrf2. Müller glia provide critical homeostatic and trophic support for both the retinal vasculature and neuronal layers. Thus, the data are consistent with a model wherein enhanced REDD1 expression leads to a failure of Müller cells to properly respond to the diabetic metabolic environment. Suppression of the Nrf2 antioxidant response in Müller glia appears to be a key player of that failed response. Overall, the findings support the pursuit of therapeutic strategies targeting REDD1 suppression and the downstream signaling events whereby this key stress response protein influences Müller cell function in diabetes.
Article Information
Acknowledgments. The authors thank Scot Kimball (Penn State College of Medicine) for critically evaluating the manuscript.
Funding. This research was supported by the American Diabetes Association Pathway to Stop Diabetes grant 1-14-INI-04, National Institutes of Health National Eye Institute grants R01 EY029702, R01 EY032879 (to M.D.D.), and F31 EY031199 (to W.P.M.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. W.P.M., A.L.T., S.S., S.A.S., and A.M.V. contributed to the investigation. W.P.M., A.L.T., S.S., S.A.S., A.M.V., and M.D.D. contributed to data curation. W.P.M., A.L.T., S.S., S.A.S., A.M.V., and M.D.D. contributed to formal analysis. W.P.M., A.L.T., D.L.W, A.J.B., and M.D.D. reviewed and edited the manuscript. W.P.M., A.L.T., and M.D.D. wrote the manuscript. W.P.M., D.L.W, A.J.B., and M.D.D. contributed to study conceptualization. W.P.M., D.L.W, A.J.B., and M.D.D. contributed to methodology. W.P.M. and M.D.D. contributed to visualization. W.P.M. and M.D.D. acquired funding. D.L.W., A.J.B., and M.D.D. provided resources. A.J.B. and M.D.D. supervised the study. M.D.D. is guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the Association for Research in Vision and Ophthalmology (ARVO) 2021 Annual Conference, virtual meeting, 2–7 May 2021.
Footnotes
W.P.M. and A.L.T. contributed equally.
This article contains supplementary material online at https://doi.org/10.2337/figshare.19149218.
References
- 1. Solomon SD, Chew E, Duh EJ, et al. Diabetic retinopathy: a position statement by the American Diabetes Association. Diabetes Care 2017;40:412–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown DM, Nguyen QD, Marcus DM, et al.; RIDE and RISE Research Group . Long-term outcomes of ranibizumab therapy for diabetic macular edema: the 36-month results from two phase III trials: RISE and RIDE. Ophthalmology 2013;120:2013–2022 [DOI] [PubMed] [Google Scholar]
- 3. Adams AJ, Bearse MA Jr. Retinal neuropathy precedes vasculopathy in diabetes: a function-based opportunity for early treatment intervention? Clin Exp Optom 2012;95:256–265 [DOI] [PubMed] [Google Scholar]
- 4. Han Y, Bearse MA Jr, Schneck ME, Barez S, Jacobsen CH, Adams AJ. Multifocal electroretinogram delays predict sites of subsequent diabetic retinopathy. Invest Ophthalmol Vis Sci 2004;45:948–954 [DOI] [PubMed] [Google Scholar]
- 5. Vujosevic S, Micera A, Bini S, Berton M, Esposito G, Midena E. Proteome analysis of retinal glia cells-related inflammatory cytokines in the aqueous humour of diabetic patients. Acta Ophthalmol 2016;94:56–64 [DOI] [PubMed] [Google Scholar]
- 6. Vujosevic S, Micera A, Bini S, Berton M, Esposito G, Midena E. Aqueous humor biomarkers of Müller cell activation in diabetic eyes. Invest Ophthalmol Vis Sci 2015;56:3913–3918 [DOI] [PubMed] [Google Scholar]
- 7. Shelton MD, Distler AM, Kern TS, Mieyal JJ. Glutaredoxin regulates autocrine and paracrine proinflammatory responses in retinal glial (Müller) cells. J Biol Chem 2009;284:4760–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, Xu X, Elliott MH, Zhu M, Le YZ. Müller cell-derived VEGF is essential for diabetes-induced retinal inflammation and vascular leakage. Diabetes 2010;59:2297–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newman E, Reichenbach A. The Müller cell: a functional element of the retina. Trends Neurosci 1996;19:307–312 [DOI] [PubMed] [Google Scholar]
- 10. Reichenbach A, Bringmann A. New functions of Müller cells. Glia 2013;61:651–678 [DOI] [PubMed] [Google Scholar]
- 11. Huster D, Reichenbach A, Reichelt W. The glutathione content of retinal Müller (glial) cells: effect of pathological conditions. Neurochem Int 2000;36:461–469 [DOI] [PubMed] [Google Scholar]
- 12. Fletcher EL, Phipps JA, Wilkinson-Berka JL. Dysfunction of retinal neurons and glia during diabetes. Clin Exp Optom 2005;88:132–145 [DOI] [PubMed] [Google Scholar]
- 13. Coughlin BA, Feenstra DJ, Mohr S. Müller cells and diabetic retinopathy. Vision Res 2017;139:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dennis MD, Kimball SR, Fort PE, Jefferson LS. Regulated in development and DNA damage 1 is necessary for hyperglycemia-induced vascular endothelial growth factor expression in the retina of diabetic rodents. J Biol Chem 2015;290:3865–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai W, Miller WP, Toro AL, et al. Deletion of the stress-response protein REDD1 promotes ceramide-induced retinal cell death and JNK activation. FASEB J 2018;32:6883–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller WP, Sunilkumar S, Giordano JF, Toro AL, Barber AJ, Dennis MD. The stress response protein REDD1 promotes diabetes-induced oxidative stress in the retina by Keap1-independent Nrf2 degradation. J Biol Chem 2020;295:7350–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller WP, Toro AL, Barber AJ, Dennis MD. REDD1 activates a ROS-generating feedback loop in the retina of diabetic mice. Invest Ophthalmol Vis Sci 2019;60:2369–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller WP, Yang C, Mihailescu ML, et al. Deletion of the Akt/mTORC1 repressor REDD1 prevents visual dysfunction in a rodent model of type 1 diabetes. Diabetes 2018;67:110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen QD, Schachar RA, Nduaka CI, et al.; DEGAS Clinical Study Group . Dose-ranging evaluation of intravitreal siRNA PF-04523655 for diabetic macular edema (the DEGAS study). Invest Ophthalmol Vis Sci 2012;53:7666–7674 [DOI] [PubMed] [Google Scholar]
- 20. Notini AJ, McClive PJ, Meachem SJ, et al. Redd1 is a novel marker of testis development but is not required for normal male reproduction. Sex Dev 2012;6:223–230 [DOI] [PubMed] [Google Scholar]
- 21. Roesch K, Jadhav AP, Trimarchi JM, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol 2008;509:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dierschke SK, Toro AL, Miller WP, Sunilkumar S, Dennis MD. Diabetes enhances translation of Cd40 mRNA in murine retinal Müller glia via a 4E-BP1/2-dependent mechanism. J Biol Chem 2020;295:10831–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saadane A, Lessieur EM, Du Y, Liu H, Kern TS. Successful induction of diabetes in mice demonstrates no gender difference in development of early diabetic retinopathy. PLoS One 2020;15:e0238727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 2010;9:457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller WP, Mihailescu ML, Yang C, et al. The translational repressor 4E-BP1 contributes to diabetes-induced visual dysfunction. Invest Ophthalmol Vis Sci 2016;57:1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci 2004;45:4611–4616 [DOI] [PubMed] [Google Scholar]
- 27. Menon M, Mohammadi S, Davila-Velderrain J, et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat Commun 2019;10:4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cowan CS, Renner M, De Gennaro M, et al. Cell types of the human retina and its organoids at single-cell resolution. Cell 2020;182:1623–1640.e1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes 2005;54:1615–1625 [DOI] [PubMed] [Google Scholar]
- 30. Cho H, Hartsock MJ, Xu Z, He M, Duh EJ. Monomethyl fumarate promotes Nrf2-dependent neuroprotection in retinal ischemia-reperfusion. J Neuroinflammation 2015;12:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Q, Zhang F, Zhang X, et al. Fenofibrate ameliorates diabetic retinopathy by modulating Nrf2 signaling and NLRP3 inflammasome activation. Mol Cell Biochem 2018;445:105–115 [DOI] [PubMed] [Google Scholar]
- 32. Deliyanti D, Alrashdi SF, Tan SM, et al. Nrf2 activation is a potential therapeutic approach to attenuate diabetic retinopathy. Invest Ophthalmol Vis Sci 2018;59:815–825 [DOI] [PubMed] [Google Scholar]
- 33. Barber AJ, Antonetti DA; The Penn State Retina Research Group . Altered expression of retinal occludin and glial fibrillary acidic protein in experimental diabetes. Invest Ophthalmol Vis Sci 2000;41:3561–3568 [PubMed] [Google Scholar]
- 34. Limb GA, Salt TE, Munro PM, Moss SE, Khaw PT. In vitro characterization of a spontaneously immortalized human Müller cell line (MIO-M1). Invest Ophthalmol Vis Sci 2002;43:864–869 [PubMed] [Google Scholar]
- 35. Miller WP, Sunilkumar S, Dennis MD. The stress response protein REDD1 as a causal factor for oxidative stress in diabetic retinopathy. Free Radic Biol Med 2021;165:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fruttiger M, Calver AR, Krüger WH, et al. PDGF mediates a neuron-astrocyte interaction in the developing retina. Neuron 1996;17:1117–1131 [DOI] [PubMed] [Google Scholar]
- 37. Reichenbach A, Bringmann A. Glia of the human retina. Glia 2020;68:768–796 [DOI] [PubMed] [Google Scholar]
- 38. Mizutani M, Gerhardinger C, Lorenzi M. Müller cell changes in human diabetic retinopathy. Diabetes 1998;47:445–449 [DOI] [PubMed] [Google Scholar]
- 39. Antonetti DA, Barber AJ, Bronson SK, et al.; JDRF Diabetic Retinopathy Center Group . Diabetic retinopathy: seeing beyond glucose-induced microvascular disease. Diabetes 2006;55:2401–2411 [DOI] [PubMed] [Google Scholar]
- 40. Jackson GR, Barber AJ. Visual dysfunction associated with diabetic retinopathy. Curr Diab Rep 2010;10:380–384 [DOI] [PubMed] [Google Scholar]
- 41. Pérez-Sisqués L, Sancho-Balsells A, Solana-Balaguer J, et al. RTP801/REDD1 contributes to neuroinflammation severity and memory impairments in Alzheimer’s disease. Cell Death Dis 2021;12:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dungan CM, Wright DC, Williamson DL. Lack of REDD1 reduces whole body glucose and insulin tolerance, and impairs skeletal muscle insulin signaling. Biochem Biophys Res Commun 2014;453:778–783 [DOI] [PubMed] [Google Scholar]
- 43. Murakami T, Nishijima K, Akagi T, et al. Segmentational analysis of retinal thickness after vitrectomy in diabetic macular edema. Invest Ophthalmol Vis Sci 2012;53:6668–6674 [DOI] [PubMed] [Google Scholar]
- 44. Sohn EH, van Dijk HW, Jiao C, et al. Retinal neurodegeneration may precede microvascular changes characteristic of diabetic retinopathy in diabetes mellitus. Proc Natl Acad Sci U S A 2016;113:E2655–E2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barber AJ, Lieth E, Khin SA, Antonetti DA, Buchanan AG, Gardner TW. Neural apoptosis in the retina during experimental and human diabetes. Early onset and effect of insulin. J Clin Invest 1998;102:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alasil T, Keane PA, Updike JF, et al. Relationship between optical coherence tomography retinal parameters and visual acuity in diabetic macular edema. Ophthalmology 2010;117:2379–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morgan-Warren PJ, O’Neill J, de Cogan F, et al. siRNA-mediated knockdown of the mTOR inhibitor RTP801 promotes retinal ganglion cell survival and axon elongation by direct and indirect mechanisms. Invest Ophthalmol Vis Sci 2016;57:429–443 [DOI] [PubMed] [Google Scholar]
- 48. Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004;18:2893–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 2005;25:5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dennis MD, Coleman CS, Berg A, Jefferson LS, Kimball SR. REDD1 enhances protein phosphatase 2A-mediated dephosphorylation of Akt to repress mTORC1 signaling. Sci Signal 2014;7:ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nandagopal N, Roux PP. Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation (Austin) 2015;3:e983402. [DOI] [PMC free article] [PubMed] [Google Scholar]