Abstract
Proteins and phenolic compounds are two types of food ingredients with distinct functionalities. In the past decade, many attempts have been made to conjugate phenolic compounds with proteins through covalent linkages. Four types of conjugation reactions including alkaline, free radical mediated grafting, enzyme catalyzed grafting and chemical coupling methods are frequently used to synthesize phenolic–protein conjugates. The synthesized phenolic–protein conjugates can be well characterized by several different instrumental methods, such as UV spectroscopy, Fourier transform infrared spectroscopy, fluorescence spectroscopy, circular dichroism, mass spectroscopy, sodium dodecyl sulfate polyacrylamide gel electrophoresis and differential scanning calorimetry. Importantly, phenolic–protein conjugates exhibit improved biological properties (e.g. antioxidant, anticancer and antimicrobial activities) as compared with native proteins. Moreover, the applications of native proteins can be greatly widened by conjugation with phenolic compounds. Phenolic–protein conjugates have been developed as antioxidant emulsions for nutraceutical delivery, edible films for food packaging, stabilizers for metal nanoparticles, and hydrogels and nanoparticles for controlled drug release. In this review, recent advances in the synthesis, characterization, biological properties and potential applications of phenolic–protein conjugates were summarized.
This review summarizes recent advances in the synthesis, characterization, biological activities and potential applications of phenolic–protein conjugates.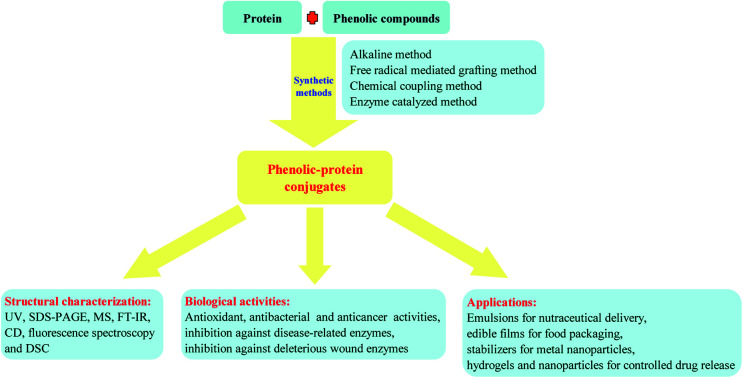
1. Introduction
Proteins are widely used in the food industry as emulsifiers, foaming agents, colloid stabilizers, biodegradable film-forming materials and microencapsulating agents.1 The functional properties of proteins are closely related to their physicochemical properties, such as solubility, swelling, foaming, emulsifying, gelling and fat binding abilities.2,3 It has been well established that the functional properties of proteins can be significantly improved by structural modification.4 Three main strategies including chemical (e.g. acetylation, succinylation, phosphorylation, lipophilization, glycosylation, thiolation and cross-linking), physical (e.g. high pressure, radiation and ultrasound) and enzymatic modifications (e.g. endopeptidases, transglutaminase and oxidase) are often selected to modify food proteins.5–9
Phenolic compounds are ubiquitous secondary metabolites in plants, which are synthesized through pentose phosphate, shikimate and phenylpropanoid pathways.10,11 Structurally, phenolic compounds comprise an aromatic ring with one or more hydroxyl substituents, ranging from simple phenolic molecules to highly polymerized compounds.12,13 Phenolic compounds are abundant micronutrients in human diet. In foods, phenolic compounds can be used as natural colorants and preservatives. Moreover, these compounds also possess many valuable biological activities, such as antioxidant, antimicrobial, antimutagenic, anticancer, antiallergenic, anti-inflammatory, antiviral, antiulcer, antidiarrheal and hepatoprotective effects.14–16 Therefore, phenolic-rich foods are generally considered to have high functionality.
In the past decade, the functionalization of proteins with phenolic compounds has gained increasing attentions. On one hand, phenolic compounds and proteins can form phenolic–protein complexes via non-covalent physical interactions (e.g. electrostatic, hydrophobic, van der Waals and hydrogen bonding).17 On the other hand, phenolic compounds can be covalently linked with proteins to form phenolic–protein conjugates.18,19 Notably, covalently linked phenolic–protein conjugates are more stable than non-covalent phenolic–protein complexes, although both physical interactions and covalent conjugation can both alter the properties of proteins.20,21 Till now, most existing studies have focused on the non-covalent interactions between phenolic compounds and proteins, which is probably because the non-covalent phenolic–protein complexes are more conveniently prepared than covalently linked phenolic–protein conjugates.17,22–24 Thus, studies on the more stable phenolic–protein conjugates should be accelerated in the next few years.
Up to now, four types of conjugation reactions including alkaline, free radical mediated grafting, enzyme catalyzed grafting and chemical coupling methods have been developed to synthesize phenolic–protein conjugates.18,19,25,26 The conjugation of phenolic compounds (especially phenolic acids and flavonoids) with proteins (mainly gelatin, whey and egg proteins) can cause significant changes in the structural, physicochemical and biological properties and widen the applications of proteins. This review summarizes recent advances on the synthesis, characterization, biological activities and potential applications of phenolic–protein conjugates.
2. Synthetic methods of phenolic–protein conjugates
2.1. Alkaline method
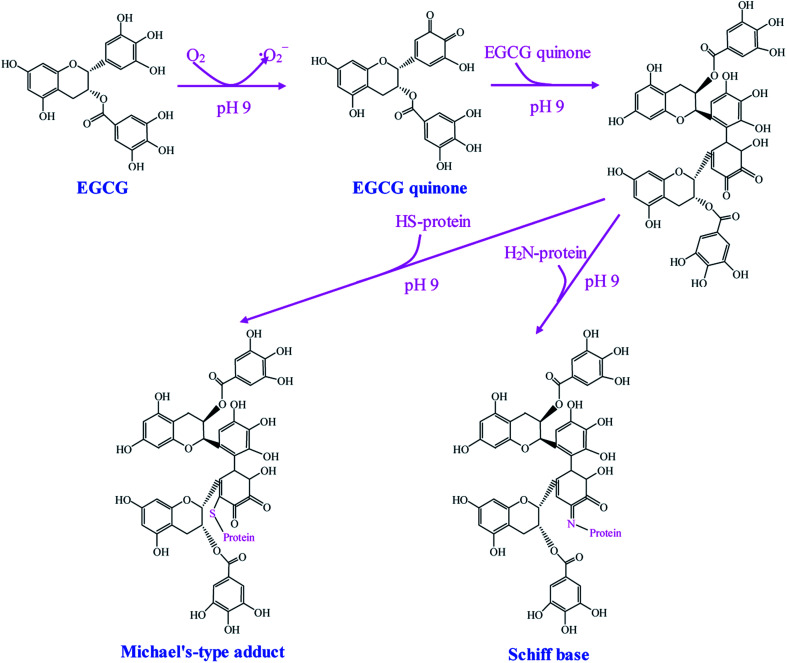
Alkaline method is a simple and effective method to synthesize phenolic–protein conjugates. The alkaline reaction is usually carried out by adjusting the pH of reaction solution to 9.0 using NaOH under atmospheric air. This method is mainly based on the oxidation of phenolic compounds.26 The synthetic mechanism of epigallocatechin-3-gallate (EGCG)–protein conjugates is proposed by Wei et al.21 As shown in Fig. 1, EGCG is oxidized to its corresponding quinone under alkaline condition with free exposure to air. Then EGCG dimerization occurs prior to the interaction with protein. Finally, the highly reactive EGCG dimer quinones can react with the nucleophilic side chains (e.g. lysine, cysteine, methionine and tryptophan) of protein.27 As a result, EGCG–protein conjugates are formed through Schiff-bases and Michael type adducts. Recently, Tao et al.28 suggest EGCG may play a bridging role to initiate a cross-linking reaction of β-lactoglobulin, with the formation of the dimerization of β-lactoglobulin after EGCG conjugation. Notably, the successful synthesis of flavonoid–protein conjugates mainly depends on the position of hydroxyl substituents on the rings B and C of flavonoids. Rawel et al.29 compare the reactivity of several flavonoids under alkaline condition and find the occurrence of catechol moiety (ortho-hydroxyl groups) on ring B of flavonoids is necessary for the covalent binding with proteins, whereas the catechol moieties on ring A of flavonoids is less reactive and unnecessary for grafting. Rawel et al.30 further suggest quercetin and rutin can react with the lysine, tryptophan and cysteine residues of whey protein. Moreover, quercetin is more reactive than rutin when conjugating with whey protein, indicating the rhamnosylglucoside at 3-O position has a great impact on the reactivity of quercetin.
Fig. 1. Schematic illustration of synthetic mechanism of EGCG–protein conjugates by alkaline method. Reproduced from ref. 21 with permission from Springer, copyright 2015.
2.2. Free radical mediated grafting method
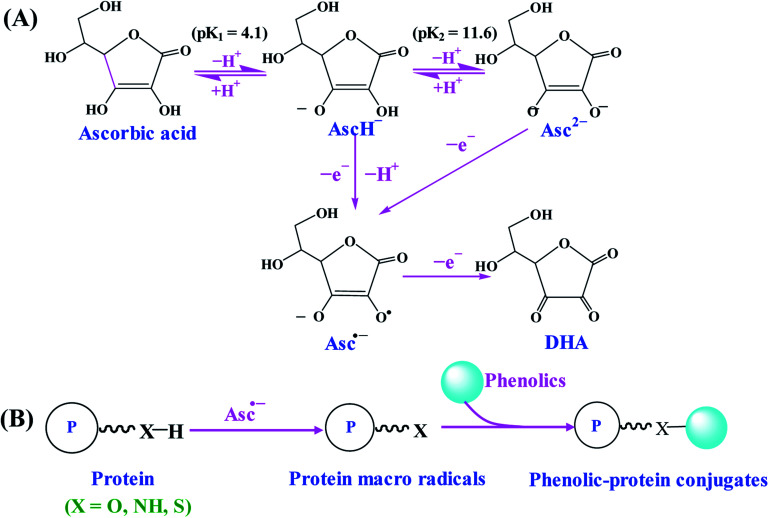
Free radical mediated grafting method, especially ascorbic acid and hydrogen peroxide (H2O2) redox pair induced grafting method has been widely used for the synthesis of phenolic–protein conjugates.19,27,31–35 The possible synthetic mechanism of phenolic–protein conjugates is recently revealed by Liu et al.36 and shown in Fig. 2. The ascorbate radicals (Asc˙−), generated by the reaction between ascorbic acid and H2O2 redox pair at pH 6, can attack the sensible residues on the side chains of protein and thus produce macro radical species on amino-acidic structure. These macro radicals can further conjugate with phenolic compounds through covalent linkages.36,37 However, the exact linkage positions between phenolic compounds and proteins are still unclear up to now. Spizzirri et al.19 suppose that the binding sites of catechin–gelatin conjugation are H-2′/H-5′ at ring B and H-6/H-8 at ring A of catechin with heteroatom-centered groups of gelatin. You et al.27 demonstrate that catechin is covalently bound to lysine (residue 327) and glutamic acid (residue 186) of ovotransferrin. Gu et al.38 first oxidize catechin by horseradish peroxidase and then conjugate the obtained catechin polymers to egg white proteins by free radical mediated grafting method, which provides a new approach to synthesize high molecular weight polyphenol–protein conjugates.
Fig. 2. Schematic illustration of synthetic mechanism of phenolic–protein conjugates by ascorbic acid/H2O2 redox initiator system. (A) The formation of Asc˙−; (B) synthesis of phenolic–protein conjugates through Asc˙−. Reproduced from ref. 36 with permission from ACS Publications, copyright 2017 and ref. 37 with permission from Elsevier, copyright 2018.
2.3. Enzyme catalyzed method
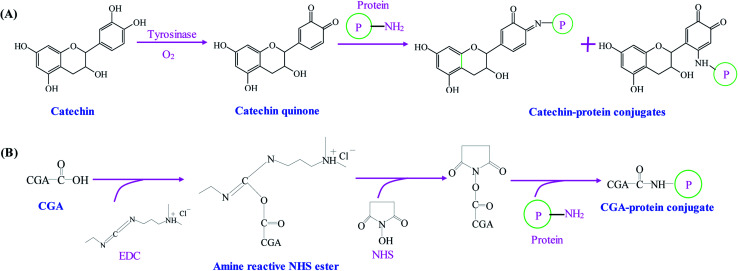
As compared with alkaline and free radical mediated grafting methods, the application of enzyme catalyzed method for the synthesis of phenolic–protein conjugates is very limited. As shown in Fig. 3A, polyphenol oxidases (e.g. laccase and tyrosinase) can catalyze phenolic compounds into electrophilic quinones. The quinones are freely diffusible and can further undergo reaction with the nucleophilic amino groups of protein through non-enzymatic reactions.39 The covalent bonds formed between phenolic compounds and proteins are Schiff-bases (C N) and Michael type adducts (C–NH).40 Thus, the synthetic mechanism of enzyme catalyzed method is much similar to that of alkaline method. However, Ali et al.41 suggest alkaline method has a higher conjugation efficiency than enzyme catalyzed method. In addition, only a few amino acids (e.g. ε-amino groups of lysine and thiol group of cysteine) are liable to modifications. Till now, phenolic compounds (especially catechin) have been successfully conjugated with different proteins through enzymatic catalysis.18,39,40,42 The optimal reaction conditions usually depends on the type of enzyme selected. The catalyzed reaction is often carried out at pH 6.0–7.5.18,39,40
Fig. 3. Schematic illustration of synthetic mechanism of (A) catechin–protein conjugates by tyrosinase catalysis. Reproduced from ref. 39 with permission from Springer, copyright 2003 and ref. 40 with permission from Springer-Link, copyright 2016; (B) chlorogenic acid (CGA)–protein conjugate through EDC and NHS coupling. Reproduced from ref. 25 with permission from Elsevier, copyright 2017.
2.4. Chemical coupling method
Phenolic compounds can be easily conjugated with proteins by chemical coupling reagents. Glutaraldehyde, a linear 5-carbon dialdehyde, is more efficient than other aldehydes in generating thermally and chemically stable cross-linkings.43 Glutaraldehyde can react with nucleophilic side chains of proteins, such as amine, thiol, phenol and imidazole. Wu et al.44 successfully synthesize EGCG–collagen conjugate through glutaraldehyde cross-linking. In another study, EGCG–gelatin conjugate is prepared by using 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium chloride and N-methylmorpholine as the cross-linking reagents in aqueous solution.45 Recently, Fu et al.25 use 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) as the coupling reagents to conjugate the carboxyl groups of chlorogenic acid with the amino groups of gelatin (Fig. 3B). El-Maksoud et al.46 prepare caffeic acid-β-lactoglobulin conjugates by EDC/NHS coupling and find the conjugation efficiency is greatly affected by the reaction pH. Notably, pH 6 is preferable to bind the maximal caffeic acid units to β-lactoglobulin, which may be due to the high activity and stability of EDC at pH 6.
Table 1 summarizes different methods used for the synthesis of phenolic–protein conjugates. Among these methods, enzyme catalyzed method is the most eco-friendly approach. However, polyphenol oxidases can oxidize the hydroxyl groups on phenolic compounds into quinone, which will eventually decrease the biological activities, e.g. antioxidant and antimicrobial activities of phenolic–protein conjugates.40 Due to similar synthetic mechanism, alkaline method also has the same limitations as enzyme catalyzed method. Although chemical coupling method is highly efficient in the synthesis of phenolic–protein conjugates, this method usually requires a large amount of chemical cross-linking reagents.25,43 These chemical cross-linking reagents are much expensive and environmentally disadvantageous, which may cause adverse impacts on human body when the products are used in food and pharmaceutical industries. By contrast, free radical mediated grafting method has several advantages. First, the grafting reagents (ascorbic acid and H2O2) used to generate free radicals are much cheaper than carbodiimide and enzymes. Moreover, ascorbic acid and H2O2 redox pair is less toxic than chemical cross-linking reagents. In addition, it is possible to perform the reaction at room temperature to avoid the degradation and oxidation of phenolic compounds. Therefore, free radical mediated grafting method has been widely used for the synthesis of phenolic–protein conjugates.19,27,31–35 Notably, the structures, biological properties and applications of phenolic–protein conjugates are closely related to the conjugation efficiency. The conjugation efficiency is affected by the type of phenolic compounds and proteins selected, the synthetic method used and the reaction conditions (e.g. reaction temperature, pH and time etc.).47 In future, the reaction conditions of each method can be optimized to achieve phenolic–protein conjugates with ideal conjugation efficiency. Meanwhile, the conjugation positions between phenolic compounds and proteins should be better elucidated.
Overview of the synthetic methods, characterization methods, biological activities and applications of phenolic–protein conjugates.
| Phenolic compounds | Proteins | Synthetic methods | Characterization methods | Biological activities | Applications | References |
|---|---|---|---|---|---|---|
| Anthocyanins | Soy protein isolate | Alkaline method | FT-IR, fluorescence | Antioxidant activity | 74 | |
| Caffeic acid | Bovine serum albumin | Free radical mediated grafting | SDS-PAGE, CD | Antioxidant activity | Nanoparticles for delivery of resveratrol | 95 |
| Caffeic acid, ferulic acid | Gelatin | Alkaline method | Edible film | 84 | ||
| Caffeic acid, tannic acid, green tea extract | Gelatin | Alkaline method | GPC, NMR | Antioxidant activity | Edible film | 56 and 86 |
| Caffeic acid | β-Lactoglobulin | EDC/NHS coupling | MALDI-TOF-MS, DSC, FT-IR, fluorescence, CD | Antioxidant activity | Emulsions for fish oil delivery | 46 |
| Catechin, quercetin | α-Casein, BSA | Enzyme catalyzed grafting | SDS-PAGE, MALDI-TOF-MS, UV, FT-IR, DSC | Antioxidant activity | 18 | |
| Catechin | Egg white protein | Free radical mediated grafting | SDS-PAGE, ESI-MS | Antioxidant activity | Emulsions for β-carotene delivery | 33 |
| Catechin | Gelatin | Enzyme catalyzed grafting | UV | Antioxidant activity | 39 | |
| Catechin, gallic acid | Gelatin | Free radical mediated grafting | UV, GPC, DSC, fluorescence | Antioxidant and anticancer activities | 19 and 49 | |
| Catechin | Keratin | Cross-linked by formaldehyde | UV, FT-IR, 1H NMR | Nanoparticles for delivery of doxorubicin | 64 | |
| Catechin, EGCG | β-Lactoglobulin, α-lactalbumin | Free radical mediated grafting | SDS-PAGE, ESI-MS, fluorescence, CD, FT-IR | Antioxidant activity | Emulsions for β-carotene delivery | 34, 35 and 55 |
| Catechin, EGC, EGCG | Ovalbumin | Free radical mediated grafting | SDS-PAGE, MALDI-TOF-MS, fluorescence, DSC, FT-IR, CD | Antioxidant activity | Emulsions for fish oil delivery | 32 |
| Catechin | Ovotransferrin | Free radical mediated grafting, alkaline method | SDS-PAGE, MS, fluorescence, LC-MS-MS | Antioxidant activity | 27 | |
| Catechin | Poly(ε-lysine) | Enzyme catalyzed grafting | Inhibition against disease-related enzymes | 42 | ||
| Catechin | Silk fibroin | Enzyme catalyzed grafting | 1H NMR, SEM, TGA | Antioxidant activity | 40 | |
| Catechin polymers | Egg white proteins | Free radical mediated grafting | LC-MS, SDS-PAGE, fluorescence | Antioxidant activity | 38 | |
| Chlorogenic acid | Bovine serum albumin | Alkaline method | CD, DSC, fluorescence | 66 | ||
| Chlorogenic acid | Bovine serum albumin, α-lactalbumin, lysozyme | Alkaline method, enzyme catalyzed grafting | MALDI-TOF-MS, GPC, SDS-PAGE, DSC | 47 | ||
| Chlorogenic acid | Gelatin | EDC/NHS coupling | 1H NMR, FT-IR | Antioxidant and antibacterial activity | 25 | |
| Chlorogenic acid | β-Lactoglobulin | Free radical mediated grafting | SDS-PAGE, CD, FT-IR | Antioxidant activity | Nanoparticles for delivery of EGCG | 31 |
| Chlorogenic acid | Myofibrillar protein | Free radical mediated grafting | Fluorescence, FT-IR, SDS-PAGE | Emulsifying and gel properties | 93 | |
| Chlorogenic acid | Whey protein isolate | SDS-PAGE, fluorescence, CD, FT-IR, DSC | Antioxidant activity | Emulsifying property | 75 | |
| Coffee-specific phenolics | Milk whey protein | Alkaline method, enzyme catalyzed grafting | CD, DSC, SDS-PAGE, MALDI-TOF-MS | Antioxidant activity | Emulsions for lutein ester delivery | 41 |
| Curcumin | Lactoferrin | EDC/NHS coupling | UV, SDS-PAGE, SEM | Anticancer activity | 76 | |
| EGCG | Collagen | Cross-linked by glutaraldehyde | Stablizer for metal nanoparticles | 44 and 87–90 | ||
| EGCG | Gelatin | Cross-linked by DMT-MM | FT-IR | Hydrogels for controlled drug release | 45 | |
| EGCG, chlorogenic acid, gallic acid | Lactoferrin | Free radical mediated grafting | SDS-PAGE, MS, FT-IR, CD, fluorescence, DSC | Antioxidant activity | 52 | |
| EGCG | Lactoferrin | Alkaline method | SDS-PAGE, CD, MALDI-TOF-MS, FT-IR, AFM, fluorescence, DSC | Emulsions for β-carotene delivery | 53, 54 and 58 | |
| EGCG | α-Lactalbumin, β-lactoglobulin, lactoferrin, sodium caseinate | Alkaline method | MALDI-TOF-MS, CD, DSC | Antioxidant activity | Emulsions for β-carotene delivery | 21 |
| EGCG | β-Lactoglobulin | Alkaline method | SDS-PAGE, CD, fluorescence | Antioxidant activity | 28 | |
| EGCG | Soy protein isolate | Alkaline method | SDS-PAGE, CD, fluorescence, surface hydrophobicity | Emulsifying property | 65 | |
| EGCG | Whey protein isolate | Free radical mediated grafting | SDS-PAGE, CD, FT-IR, ESI-MS | Antioxidant activity | Emulsions loaded with menhaden oil | 80 |
| EGCG | Zein | Alkaline method | Nanoparticles for co-delivery of curcumin and resveratrol | 94 | ||
| EGCG, quercetagetin | Zein | Alkaline method | GPC, SDS-PAGE, UV, FT-IR, CD, fluorescence, SEM | Antioxidant activity | 20 | |
| Ferulic acid, hydroxytyrosol, flaxseed polyphenols | Flaxseed protein isolate | Alkaline method | SDS-PAGE, MALDI-TOF-MS, FT-IR, CD, DSC | Antioxidant activity | 69 | |
| Gallic acid | Ovotransferrin | Alkaline method | SDS-PAGE, MALDI-TOF-MS | Emulsions for curcumin delivery | 81 | |
| Genistein, daidzein, formononetin, prunetin, biochanin A | β-Lactoglobulin | Alkaline method | MS, SDS-PAGE, CD | 29 | ||
| Phenolics from sugarcane bagasse | BSA | Free radical mediated grafting | UV, FT-IR, XRD, SEM | Anticancer activity | 37 | |
| Phenolics from Hamamelis virginiana | Gelatin | Enzyme catalyzed grafting | FT-IR, SEM | Inhibition deleterious wound enzymes and bacterial growth | Hydrogels for stimulation wound healing process | 92 |
| Pyrogallic acid | Pumpkin seed protein isolate | Alkaline method | SDS-PAGE, FT-IR, fluorescence, DSC | Antioxidant activity | 63 | |
| Quercetin | BSA | Alkaline method | Antioxidant activity | 26 | ||
| Quercetin, rutin | Whey protein, β-lactoglobulin | Alkaline method | UV, MS, SDS-PAGE, CD | 30 | ||
| Rosmarinic acid | Whey protein isolate | Alkaline method, enzyme catalyzed grafting | RP-HPLC, hydrophobicity | Antioxidant and antibacterial activity | 50 | |
| Rutin | Porcine bone protein hydrolysate | Enzyme catalyzed grafting | Surface hydrophobicity, fluorescence, CD | Emulsifying property | 79 | |
| Tannic acid | Zein | Alkaline method | Edible film | 85 |
3. Structural characterization of phenolic–protein conjugates
In general, there are two types of interactions between phenolic compounds and proteins: non-covalent and covalent interactions. The non-covalent interactions based on hydrophobic, van der Waals, hydrogen binding and ionic interactions are weaker than covalent interactions. In addition, the non-covalent interactions are mostly reversible. By contrast, the covalent interactions are generally irreversible.48 If unbound phenolic compounds are retained in the reaction systems when the reaction is over, they can certainly form non-covalent interactions with proteins and phenolic–protein conjugates. Therefore, it is necessary to remove the unbound phenolic compounds from the reaction products to avoid the formation of non-covalent interactions.33 In most cases, dialysis is often selected to remove the unbound phenolic compounds from the reaction solution, since this procedure has no negative effects on the structures and biological properties of phenolic–protein conjugates. The obtained phenolic–protein conjugates can be characterized by several instrumental methods.
3.1. UV spectroscopy
UV spectroscopy is a simple method to identify phenolic–protein conjugates. Phenolic compounds differ significantly from proteins in terms of UV spectroscopy. Phenolic acids exhibit UV absorption maxima at 270–280 nm and 305–330 nm, while flavonoids show UV absorption maxima in 270–280 nm and 310–350 nm. However, proteins exhibit UV absorption maxima at 214 nm (absorption of peptide bond) and 280 nm (absorption of aromatic amino acids).48 In general, the UV absorption maxima of proteins shift after the conjugation with phenolic compounds. The UV absorption spectra of phenolic–protein conjugates depend on the type of phenolic compounds and proteins as well as the grafting method used. For example, catechin–gelatin conjugate synthesized by laccase catalysis shows a characteristic peak at 390 nm, which is assigned to oxidized catechin moieties.39 Kim and Cavaco-Paulo18 observe a new band occurred at around 450 nm for catechin-bovine serum albumin (BSA) and catechin-α-casein conjugates, which are also synthesized by laccase catalysis. However, catechin–gelatin conjugate prepared by free radical mediated grafting method exhibits higher wavelengths at 230 and 275 nm as compared to the wavelengths of free catechin at 204 and 229 nm.49 The red shift of UV absorption band for the conjugates should be due to the formation of covalent linkages between heteroatom in the side chains of proteins and aromatic ring of phenolic compounds.19 Notably, reverse phase-high performance liquid chromatography (RP-HPLC) equipped with UV detector can be used to identified the phenolic compounds that are bounded to proteins. You et al.27 compare the RP-HPLC profile of catechin–ovotransferrin conjugate with that of ovotransferrin and catechin. They find the retention time of catechin–ovotransferrin conjugate is lower that of ovotransferrin, revealing the hydrophilicity of ovotransferrin is increased by the conjugation with catechin. The covalent interactions between phenolic compounds and proteins can change the net charge in the proteins, which in turn affect the hydrophilicity/hydrophobicity of the modified protein. In addition, RP-HPLC equipped with UV detector can be used to distinguish non-covalent and covalent interactions between phenolic compounds and proteins. This method depends on the fact that both free phenolic compounds and phenolic–protein conjugates have UV absorption but they are eluted at different retention times, whereas unmodified proteins do not show UV absorption at corresponding wavelength.50
3.2. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
Electrophoresis techniques, especially SDS-PAGE, are frequently used to determine the molecular weights of proteins and its phenolic conjugated counterparts.51 The addition of SDS can hinder non-covalent interactions between phenolic compounds and proteins. When SDS is allowed to react with proteins completely, the produced SDS–protein complexes possess similar charge densities. As a result, the mobility of the sample on SDS-PAGE will depend on the size of the samples. Therefore, the non-covalent and covalent interactions between phenolic compounds and proteins can be distinguished by SDS-PAGE. Gu et al.33 find the SDS-PAGE profile of catechin-egg white protein conjugates is considerably different to that of egg white proteins, with some of the bands diffusing and moving upwards. By contrast, the SDS-PAGE profile of catechin-egg white protein mixtures is similar to that of egg white proteins, which is because the non-covalent interactions between catechin and egg white proteins are destroyed by adding SDS. This indicates that the molecular weights of proteins are increased after conjugation with phenolic compounds. Similar phenomena are observed by many other researchers, although the grafting methods applied are totally different.18,20,27,31,35,52,53 The increase in the molecular weights of phenolic–protein conjugates should be attributed to the covalently linked phenolic moieties. Kim and Cavaco-Paulo18 suggest that phenolic molecules might act as cross-linkers to promote the formation of new dimers or polymers. By applying gel intensity analysis software, the conjugation efficiencies of phenolic–protein conjugates can be simply evaluated.35
3.3. Mass spectrometry (MS)
Apart from SDS-PAGE, MS can be also used to determine the molecular weights of phenolic–protein conjugates. Among different MS techniques, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and electrospray ionization-mass spectrometry (ESI-MS) are frequently used.18,27,35,46,52,54,55 In general, the covalent linkages between phenolic compounds and proteins slightly enhance the molecular weight of proteins. By applying MS technique, Gu et al.33 find the molecular weight of catechin-egg white protein conjugates is higher than egg white protein, which is attributed to the grafted catechin moieties. However, the mass spectrum of non-covalent interacted catechin-egg white protein mixtures was similar to that of egg white protein. Therefore, MS technique can be used to distinguish the non-covalent and covalent interactions formed between phenolic compounds and proteins. Based on the results of SDS-PAGE and MALDI-TOF-MS, You et al.27 suggest the molecular weight of catechin–ovotransferrin conjugate is related to the grafting method used. Catechin–ovotransferrin conjugate synthesized by free radical mediated method exhibits a higher molecular weight than the conjugate prepared by alkaline method. Liu et al.52 find the molecular weights of phenolic–lactoferrin conjugates synthesized by alkaline method decrease in the order of chlorogenic acid–lactoferrin > EGCG–lactoferrin > gallic acid–lactoferrin, indicating the molecular weights of conjugates are also related to the type of phenolic compound conjugated. In fact, the covalent linkage sites between phenolic compounds and proteins can be identified by LC-MS-MS.27 You et al.27 suggest that catechin is covalently bound to lysine (residues 327) and glutamic acid (residues 186) in ovotransferrin. Except for SDS-PAGE and MS, the molecular weights of phenolic–protein conjugates can be also determined by gel permeation chromatography (GPC).19,20,56
3.4. Fourier transform infrared (FT-IR) spectroscopy
FT-IR spectroscopy can provide great insights into the molecular interactions (both non-covalent and covalent interactions) between phenolic compounds and proteins. The conjugation of phenolic compounds has a complex impact on the conformation of proteins. On one hand, the conjugation of phenolic compounds can significantly reduce the functional groups in the original side chains of proteins. On the other hand, the conjugation of phenolic compounds can introduce new spatial and functional groups into protein backbones. FT-IR spectroscopy is a useful method to estimate the change in the secondary structures of proteins after conjugation with phenolic compounds by inspecting the frequencies of amide bonds. The amide band I (C O stretching, 1600–1690 cm−1), band II (N–H bending and C–N stretching, 1480–1575 cm−1), and band III (C–N stretching and N–H bending, 1229–1301 cm−1) are often employed to study the structure of proteins and phenolic–protein conjugates.57,58 In fact, amide band I is the most sensitive spectral region for the secondary structure of proteins. The major peaks in the region of amide band I can be further resolved into α-helix (1658–1650 cm−1), β-sheet (1640–1615 cm−1), β-turn (1700–1660 cm−1) and random coil (1650–1640 cm−1). Liu et al.52 apply curve fitting method to resolve amide band I region of EGCG–lactoferrin conjugates. The control lactoferrin contains 11.5% α-helix, 9.5% β-sheet, 54.1% turn and 24.9% random coil, and the EGCG–lactoferrin conjugate contains 18.1% α-helix, 15.2% β-sheet, 43.5% turn and 23.1% random coil. The same research group also find the conjugation of polyphenols prevents lactoferrin from thermal aggregation at neutral pH.58 After heat treatment, less change occurs in the FT-IR spectra (especially β-sheet) of EGCG–lactoferrin conjugate as compared to that of lactoferrin alone. By applying FT-IR spectroscopy, a major increase of β-sheet and a minor increase of α-helix are observed when tea polyphenols non-covalently interacts with β-lactoglobulin.59 Therefore, FTIR spectroscopy cannot be used to distinguish non-covalent and covalent interactions formed between phenolic compounds and proteins, since these two types of interactions can both change the conformation of proteins.
3.5. Circular dichroism (CD) spectroscopy
CD spectroscopy provides a rapid approach to determine the conformational change in the secondary structures of proteins after conjugation with phenolic compounds.34,35,46,52,53,55,58 In general, the amide chromophore of peptide bond in proteins dominates CD spectra below 250 nm. The negative bands at around 208 and 222 nm are attributed to α-helical conformation due to strong hydrogen bonding environment. CD spectroscopy of β-sheet displays a negative band at 216 nm, a positive band between 195 and 200 nm, and a negative band near 175 nm.60 Yi et al.35 observe the representative negative peak of β-lactoglobulin shifted from 216 to 206 nm after conjugation with catechin by free radical mediated method, indicating the proportion of β-sheet in β-lactoglobulin remarkably decrease. By contrast, the unordered structure of β-lactoglobulin significantly increases after conjugation with catechin, leading to the exposure of interior amino acids. Yi et al.55 find the proportion of α-helix decreases whereas β-sheet increases in α-lactalbumin after conjugation with catechin through free radical mediated method. Above studies suggest the change in the secondary structures of proteins is related to the original conformation of proteins. Recently, significant decreases in α-helix and β-sheet as well as increases in β-turn and unordered coils are observed in zein after conjugation with quercetagetin by alkaline method. However, no significant change in the secondary structure of zein is observed after conjugation with EGCG or chlorogenic acid.20 Therefore, the conformational change of phenolic–protein conjugates is also related to the type of phenolic compound conjugated. Similar with FT-IR spectroscopy, CD spectroscopy cannot be used to distinguish non-covalent and covalent interactions formed between phenolic compounds and proteins, since the two types of interactions both can change the conformation of proteins. For example, Al-Hanish et al.61 find the non-covalent interactions of EGCG and α-lactalbumin cause decrease in α-helix from 36.8% to 28.9%, while β-sheet content increased from 20.0% to 26.7% by CD spectroscopy.
3.6. Fluorescence spectroscopy
The interactions between phenolic compounds and proteins can be detected by fluorescence spectroscopy.62 On one hand, the fluorescence of proteins usually arises from the indole chromophore of the aromatic amino acids (e.g. tryptophan) and can be quenched by phenolic compounds strongly. On the other hand, some phenolic compounds also have intrinsic fluorescence properties which are exquisitely sensitive to environmental factors such as polarity and hydrogen bonding effects. A bathochromic shift is observed in the emission peak of catechin after conjugation with gelatin.19 Similar phenomena are also observed by You et al.27 and Liu et al.52 Recently, Liu et al.20 find the fluorescence intensity depends on the type of phenolic compound conjugated, which decreases in the order of EGCG–zein > quercetagetin–zein > chlorogenic acid–zein. Moreover, fluorescence spectroscopy is often used to investigate polyphenol–protein non-covalent interactions and binding affinity. For instant, β-lactoglobulin has two tryptophan residues Trp-19 and Trp-61. Trp-19 is in an apolar environment that contributes to 80% of total fluorescence, whilst Trp-61 is partly exposed to aqueous solvent and has a minor contribution to Trp fluorescence. When polyphenols interact with β-lactoglobulin, tryptophan fluorescence may change depending on the impact of non-covalent interactions on the protein conformation. On the assumption that there are n substantive binding sites for polyphenols – the quencher (Q) on protein (B), the quenching reaction can be shown as follows:59
| nQ + B ⇔ QnB | 1 |
Fluorescence quenching as a result of polyphenol–protein interactions can be described by the Stern–Volmer equation:
| F0/F = 1 + KSV[Q] | 2 |
where F0 and F are fluorescence intensities emitted by protein before and after the addition of polyphenols (quencher). KSV is the Stern–Volmer quenching constant and [Q] is the concentration of polyphenols (quencher). A linear Stern–Volmer plot indicates that one mechanism of quenching appears either static (complex formation between polyphenols and protein) or dynamic (collision of fluorophore with the quencher).48
Double logarithmic Stern–Volmer equation for the static mechanism of quenching can be applied to calculate binding constant (Ka) and the number of binding sites (n) in the protein molecule.48
| log[(F0 − F)/F] = log Ka + n × log[Q] | 3 |
By applying fluorimetric methods, the non-covalent interactions of tea polyphenols-β-lactoglobulin complexes and EGCG-α-lactalbumin complexes are determined.59,61 However, the covalent interactions between phenolic compounds and proteins have not been studied by the Stern–Volmer equation.
3.7. Differential scanning calorimetry (DSC)
DSC is a thermodynamic technique to study thermally induced transitions of proteins. The thermal behaviour of phenolic–protein conjugates is normally tested by DSC. In general, the melting endothermic peak of free phenolic compounds disappeared in the DSC thermograms of phenolic–protein conjugates. In addition, the DSC profiles of phenolic–protein conjugates usually occur at a higher temperature as compared to blank proteins, suggesting the thermal stability of proteins can be enhanced by grafting with phenolic compounds.18–21,46,52,53,63 A comparative study of non-covalent and covalent interactions between zein and polyphenols in ethanol–water solution is carried out by Liu et al.20 DSC thermogram of zein exhibits peaks corresponding to the thermal denaturation temperature and melting point of the protein. By contrast, EGCG–zein conjugate has a higher denaturation temperature than the control zein. However, EGCG–zein mixture shows a lower denaturation temperature than EGCG–zein conjugate. Except for DSC, thermogravimetric analysis (TGA) is also a promising method to evaluate the thermal property of phenolic–protein conjugates.40
Apart from aforementioned method, other methods are also useful to characterize phenolic–protein conjugates. For instant, nuclear magnetic resonance (NMR) are sometimes selected to confirm the successful conjugation of phenolic compounds with proteins.25,40,56,64 Scanning electron microscopy (SEM) and atomic force microscopy (AFM) can be used to observe the morphology of phenolic–protein conjugates.20,37,40,58 Surface hydrophobicity of phenolic–protein conjugates is determined by using a hydrophobic fluorescence probe, 1-anilino-8-naphthalene sulfonate (ANS).65 Particle size characteristics of the conjugates can be analyzed by dynamic light scattering (DLS). The zeta potential is measured to study the charges of the conjugates based on their movements in an electrical field.50 In addition, the covalent interactions between phenolic compounds and proteins can be confirmed by determining the contents of free amino and thiol groups and tryptophan. For example, the content of free amino groups is analyzed by using 2,4,6-trinitrobenzenesulfonic acid (TNBS) in the presence of 1% SDS. TNBS can bind with the nucleophilic nitrogen of the primary amino groups in the terminal amino acids and the ε-amino groups, forming adducts with yellow color. The content of free thiol groups can be determined by Ellman's reagent (5,5-dithio-bis(2-nitrobenzoic acid), DTNB). The surface and total tryptophan contents are estimated in 5 mM PBS buffer (pH 7.2) and 8 M urea using a fluorescence detector, respectively.50,66
Different structural characterization methods of phenolic–protein conjugates are summarized in Table 1. Among different methods used, SDS-PAGE and MS techniques can be used to distinguish the non-covalent and covalent interactions formed between phenolic compounds and proteins. However, other techniques, such as FT-IR, CD and fluorescence spectroscopy and DSC cannot distinguish the type of interactions.
4. Biological activities of phenolic–protein conjugates
4.1. Antioxidant activity
Reactive oxygen species (ROS) generated by NADPH oxidase during oxidative phosphorylation, are normal components of healthy cells. ROS are also mediators of the first defensive actions of cells and involved in phagocytosis, apoptosis and detoxification.67 Increasing evidence highlights that overproduction of ROS and oxygen-derived free radicals may contribute to a variety of pathological effects (e.g. DNA damages, carcinogenesis and cellular degeneration) and induce many diseases including aging, cancer, atherosclerosis, diabetes and rheumatoid arthritis.68 Therefore, it is essential to develop natural nontoxic antioxidants to protect human body from free radicals and retard the progress of many chronic diseases. Till now, studies on the biological activities of phenolic–protein conjugates are mainly focused on the antioxidant activity (Table 1). In general, the antioxidant activity of proteins could be greatly enhanced by conjugation with phenolic compounds.18–21,25,27,31,32,39,49,52 The antioxidant activity of phenolic–protein conjugates has been thoroughly evaluated by various in vitro assays, including reducing power, lipid peroxidation and low density lipoprotein oxidation inhibition assays, β-carotene-linoleic acid assay, superoxide anion, hydroxyl, 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) radicals scavenging assays and oxygen radical absorbance capacity (ORAC) assay.
The antioxidant activity of phenolic–protein conjugates is closely related to the grafting method used. You et al.27 compare the antioxidant activity of catechin–ovotransferrin conjugates synthesized by alkaline and free radical mediated methods. The conjugate obtained by free radical method has a higher antioxidant activity than that by alkaline method, which is due to different grafting mechanisms of these two methods. Ali et al.41 report the 5-caffeoylquinic acid-milk whey protein conjugate prepared under alkaline conditions has a higher conjugation efficiency than that prepared by PPO catalysis. However, the PPO catalyzed reaction produces the conjugate with a higher antioxidant activity. The same research group also finds the conjugation efficiency of rosmarinic acid with whey protein isolates under alkaline conditions is higher than that modified by PPO catalysis. In addition, the conjugate obtained under alkaline conditions exhibited higher antioxidant activity compared to that prepared by PPO catalysis.50 Above results suggest the antioxidant activity of phenolic–protein conjugates is partially influenced by the amount of phenolics covalently attached to proteins.
Moreover, the antioxidant activity of phenolic–protein conjugates is also related to the type of phenolic compound grafted. Quercetin–protein conjugates present higher (2–3 times) antioxidant activity as compared to catechin–protein conjugates synthesized by the same enzyme catalyzed method.18 Liu et al.52 find the DPPH and ABTS radical scavenging activities of phenolic–lactoferrin conjugates decrease in the order of chlorogenic acid–lactoferrin > EGCG–lactoferrin > gallic acid–lactoferrin conjugates. Recently, Pham et al.69 report that hydroxytyrosol shows a high reactivity with flaxseed protein isolate than flaxseed polyphenols and ferulic acid, which is due to the presence of more hydroxyl groups per unit in hydroxytyrosol. However, the antioxidant activity of hydroxytyrosol-flaxseed protein isolate conjugate is lower than that of flaxseed polyphenols–protein and ferulic acid–protein conjugates. Chung et al.39 reveal that catechin–gelatin conjugate shows relative lower superoxide anion radical scavenging activity whereas higher inhibition effect against human low density lipoprotein oxidation than equal amount of free catechin. This study suggests the antioxidant activity of phenolic–protein conjugates also depends on the antioxidant assay adopted. Besides, there might be some synergistic antioxidant actions between catechin moieties and gelatin backbones.
Some studies demonstrated the covalent and non-covalent interactions between phenolic compounds and proteins can reduce the antioxidant potential of polyphenols. Rohn et al.26 reveal the covalent attachment of quercetin to BSA decreases the total antioxidant activity in comparison to an equivalent amount of free quercetin. The conjugate with the highest amount of covalently bound quercetin shows only 79% of antioxidant activity in comparison with the equivalent amount of free quercetin. This is probably caused by the loss of essential amino acids (e.g. ε-amino groups of lysine and thiol groups of cysteine) and available phenolic hydroxyl groups due to covalent modification. Xiao et al.70 also report the non-covalent interactions between phenolic compounds and proteins can weaken the antioxidant potential of polyphenols. In addition, the phenolic–protein affinities are strongly influenced by the structures of dietary polyphenols. The hydrophobic force has been demonstrated to play an important role in the binding interaction between polyphenols and proteins (e.g. human plasma proteins and plasma proteins of type II diabetes).70–72 Since the non-covalent interactions between phenolic compounds and proteins is reversible, the formed polyphenols-plasma protein complex in the blood can dissociate and release the free polyphenols under normal physiological states. Thus, polyphenols and their metabolites can rapidly exchange between free and bound forms within the circulation. The reversible binding to plasma proteins may have consequences for the delivery of the polyphenols and their metabolites to cells and tissues.73
The conjugation with phenolic compounds can also alter the digestibility and allergenicity of proteins. Rawel et al.66 compare the digestibility of chlorogenic acid–BSA conjugates and unmodified BSA. They find the conjugates are partly digested even after 24 h exposure to trypsin, whereas the control BSA is digested after 5 min. This indicates the digestibility of protein is reduced by the conjugation with chlorogenic acid. Recently, black rice anthocyanins are conjugated with soy protein isolate under alkaline condition.74 The obtained conjugates shows a higher digestibility than unmodified proteins. In addition, the digestibility of the conjugates increases with increasing the amount of anthocyanins conjugated. However, transepithelial transport of peptides across Caco-2 cell monolayer is decreased in protein after conjugation with anthocyanins. Interestingly, it is also found that the anthocyanins–protein conjugates exhibit higher antioxidant capacity after gastric and intestinal digestions. Similar improvement of the antioxidant capacity is also reported for quercetin–BSA conjugate after in vitro digestion.26 Nonetheless, the antioxidant activity of the degradation products does not reach the activity of an equivalent amount of free quercetin. Recently, Xu et al.75 demonstrate the covalent conjugation with chlorogenic acid might reduce the allergenicity but improve the intestinal digestibility in vitro of whey protein isolate. Wu et al.34 further demonstrate that the allergenic capacity of β-lactoglobulin can be reduced by conjugation with EGCG and chlorogenic acid, indicating phenolic–protein conjugates can be developed as hypoallergenic foods. Further studies are needed to evaluate the allergenicity in vivo of the conjugates in order to reduce protein allergy.
Till now, only a few studies have compared the antioxidant activity of phenolic–protein conjugates with that of non-covalently interacted phenolic–protein mixtures. Gu et al.38 find the antioxidant activity of catechin polymer-egg white protein mixtures is only about 32% as compared to the corresponding conjugates. The reason associated with the low antioxidant activity of catechin polymer-egg white protein mixtures is that the interactions between catechin polymer and egg white protein are non-covalent in the physical mixtures, which results in the release and removal of some polyphenols during the dialysis step. Similarly, El-Maksoud et al.46 also find caffeic acid-β-lactoglobulin conjugates show a higher antioxidant activity than the non-covalent complex.
4.2. Anticancer activity
Cancer represents an abnormal and uncontrolled growth of cells that can disrupt normal host functions and, in some cases, spread throughout the body. The in vitro anticancer activity of phenolic–protein conjugates has been investigated by some researchers.37,49,76 A new hybrid material is developed by incorporating catechin–gelatin conjugate into carbon nanotubes. As compared to free catechin, the composite material shows enhanced anticancer activity against HeLa cells.49 Recently, phenolic compounds extracted from sugarcane bagasse are conjugated with BSA and the anticancer activity of phenolic–BSA conjugate is evaluated by in vitro cell viability assay. Notably, the viability of SW480 cells (human colon cancer cells) is significantly decreased approximately 80% by 100 μg mL−1 of conjugate whereas the free BSA or phenolics cannot affect the viability of cancer cells. This suggests phenolic–BSA conjugate has a higher anticancer activity than BSA and free phenolics. In addition, the quantity of ROS in SW480 cells is significantly increased by phenolic–BSA conjugate treatment, which is about two times as compared to free BSA and phenolics treatments. This further reveals the anticancer mechanism of phenolic–BSA conjugate is related to its ability to induce oxidative stress in cancer cells.37 A recent study reports the curcumin–lactoferrin conjugate has a considerably improved cytotoxicity of curcumin on HCT116 cells along with a higher level of cellular uptake, indicating the conjugate represents a promising chemotherapeutic agent for targeting cancerous tissue and achieving better outcome.76 In future, the in vivo anticancer activity and the related functional mechanisms of phenolic–protein conjugates can be revealed.
4.3. Antimicrobial activity
In recent years, spoilage and foodborne pathogenic microorganisms have caused serious economic losses and community-associated infections. The use of natural antimicrobial agents for food preservation is a trend that is followed by both consumers and food manufacturers. Till now, studies on the antimicrobial activity of phenolic–protein conjugates are very limited. Fu et al.25 find that chlorogenic acid–gelatin conjugate possesses antibacterial activity against Escherichia coli, Pseudomonas aeruginosa, Listeria monocytogenes and Staphylococcus aureus. The antibacterial activity of chlorogenic acid–gelatin conjugate is quite close to that of free chlorogenic acid. Recently, Ali et al.50 reveal the antimicrobial activity of rosmarinic acid-whey protein isolate conjugate obtained by PPO catalysis is higher than that obtained under alkaline conditions. However, the antimicrobial mechanisms of the conjugates are still unclear till now.
5. Potential applications of phenolic–protein conjugates
5.1. Emulsions for nutraceutical delivery
In recent years, the encapsulation of nutraceuticals into delivery systems has attracted considerable attention. The encapsulation can not only protect nutraceuticals from reacting with other food ingredients but also prevent nutraceuticals from degradation.77 Many oil-in-water nanoemulsion delivery systems encapsulated by biomacromolecules have been developed for their high stability against flocculation or aggregation, high optical clarity and improved bioavailability of lipophilic bioactive nutraceuticals.78 In general, proteins can be used as emulsifiers due to their excellent emulsification, none-toxic and biocompatible properties. It has been demonstrated that the conjugation of phenolic compounds with proteins can significantly improve the emulsification performance and antioxidant ability of native proteins, which opens up new applications of phenolic–protein conjugates in the nutraceutical and functional food ingredient fields.28 On one hand, the amphiphilic protein part anchors the conjugates to oil-water interface with the formation of coating around the oil droplets. On the other hand, the phenolic part of the conjugates possesses antioxidant ability that can protect polyunsaturated lipids from oxidation.33,79,80
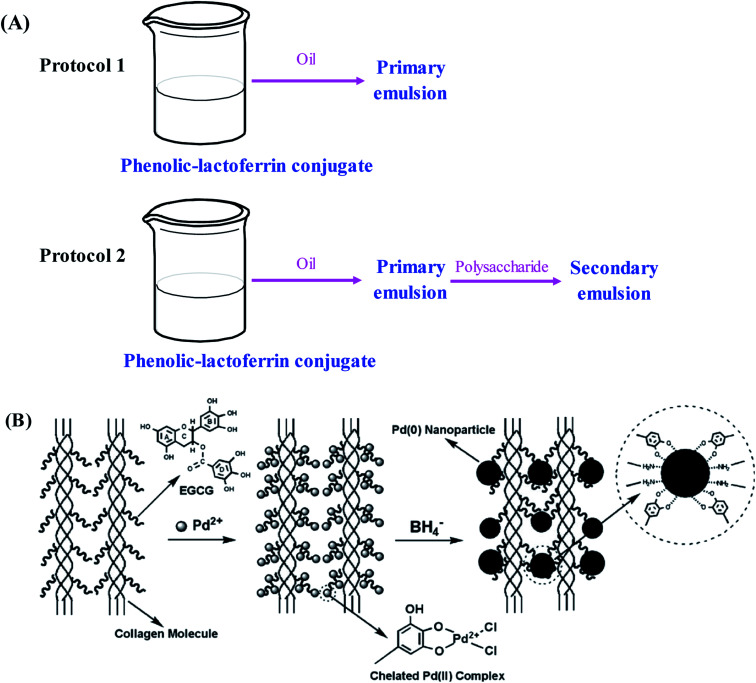
Till now, several nano-emulsion systems based on phenolic–protein conjugates have been successfully developed for nutraceutical delivery. Catechin-β-lactoglobulin conjugate nanoemulsions are prepared by Yi et al.35 as encapsulation delivery systems for β-carotene. As compared with β-lactoglobulin nano-emulsions alone, the retention of β-carotene encapsulated in conjugate nanoemulsions is significantly improved, which is probably due to the high affinity between catechin residues and β-carotene. Similarly, other phenolic–protein conjugates based nano-emulsions (e.g. catechin-egg white protein, catechin-α-lactalbumin and catechin-ovalbumin) are also developed to improve the retention of β-carotene and fish oil.32,33,55 Researchers further demonstrate the emulsifying properties of different phenolic–protein conjugates depend on the type of phenolic compound and protein used.21,52 Bilayer emulsions are developed by lactoferrin and phenolic–lactoferrin conjugates based on layer-by-layer electrostatic deposition.53 The primary and secondary emulsions stabilized by phenolic–lactoferrin conjugates exhibit improved emulsifying properties and physical stability under freeze–thaw, ionic strength and thermal treatments. In addition, the least degradation of β-carotene is observed in primary emulsion stabilized by phenolic–lactoferrin conjugate. These suggest that interfacial layers with different compositions and structures can form around lipid droplets using protein or phenolic–lactoferrin conjugates as emulsifiers. Likewise, the influence of polysaccharides (soybean soluble polysaccharides and beet pectin) on the physicochemical properties of polyphenol–lactoferrin conjugates encapsulated β-carotene emulsions is investigated by Liu et al.54 using interfacial engineering technique. The secondary emulsions stabilized by polysaccharides can effectively inhibit the deterioration of β-carotene in oil-in-water emulsions (Fig. 4A). Recently, Wei et al.81 fabricate antioxidant particle-stabilized Pickering emulsions based on gallic acid–ovotransferrin conjugate and carboxymethyldextran (CMD). The in vitro digestion study reveals that the conjugate/CMD particle-stabilized Pickering emulsions remarkably improved both the bio-accessible degree of lipolysis and curcumin. The findings of the study may have important implications for the design of nutraceutical-loaded Pickering emulsions with an excellent protective effect and delivery efficiency. In future, cellular and animal experiments can be carried out to evaluate the bio-accessibility and delivery efficiency of the conjugates.
Fig. 4. Schematic illustration of (A) preparation protocols for primary and secondary emulsions based on phenolic–lactoferrin conjugate and polysaccharide. Reproduced from ref. 54 with permission from Elsevier, copyright 2016; (B) anchoring Pd(ii) onto EGCG–collagen fibers with subsequent formation of Pd(0) nanoparticles. Reproduced from ref. 44 with permission from Elsevier, copyright 2009.
5.2. Edible films for food packaging
In recent years, the application of edible packaging films in food industry has gained increasing attentions.82 Due to the environmentally friendly and biodegradable properties, proteins are good polymeric substances to prepare edible films. However, edible films prepared from natural proteins usually exhibit low mechanical and barrier properties.83 Notably, phenolic compounds are able to act as cross-linkers between individual protein molecules and reduce the free space in the polymeric matrix, resulting in the enhancement of mechanical and barrier properties of protein-based films.56 Phenolic acid–gelatin conjugate films are prepared by mixing fish gelatin with different phenolic acids (caffeic acid and ferulic acid) at alkaline and aerobic conditions.84 Results show the solubility, oxygen and water vapor barrier properties of fish gelatin film are reduced by conjugation with caffeic acid. However, the barrier properties of gelatin film are seldom affected by conjugation with ferulic acid. This indicates the physical properties of phenolic–gelatin conjugate films are greatly influenced by the type of phenolic acid conjugated. Santos et al.85 find oxidized tannic acid–zein film has a higher tensile strength but a lower water vapor permeability than unoxidized tannic acid–zein film. The improved mechanical and water barrier property of film can be attributed to the cross-linking effect between oxidized tannic acid and zein. Recently, Choi et al.86 investigate the cross-linking effects of different oxidized phenolic substances including tannic acid, caffeic acid and green tea extract on the mechanical and barrier properties of gelatin–turmeric film. Results show the cross-linking effects of oxidized phenolic substances result in enhanced tensile strength but reduced elongation at break, water vapor permeability and water solubility of gelatin–turmeric film. Among various films, caffeic acid–gelatin–turmeric conjugate film has the highest mechanical and barrier properties. Moreover, these films possess stronger antioxidant activity, which can be used as active packaging materials to extend the shelf life of fresh ground pork. In future, it is needed to compare the physical and functional properties of phenolic–protein conjugate based films. Meanwhile, the preservation effect of the conjugate based films on different fruits and vegetables can be determined.
5.3. Stabilizers for metal nanoparticles
The synthesis of metal nanoparticles and nanocrystals with manipulated morphology and size has received increasing attentions in recent years. Phenolic–protein conjugates can supply a useful platform for metal nanoparticles to construct new materials with diverse functions.44,87–90 A novel heterogeneous palladium (Pd) nanoparticle catalyst stabilized by EGCG–collagen conjugate fibers is developed by Wu et al.44 As shown in Fig. 4C, EGCG can be easily conjugated with collagen fibers through the cross-linking of glutaraldehyde. Due to abundant hydroxyl groups in EGCG, the conjugate possesses high adsorption capacity to Pd(ii) ions through the formation of a highly stable five-member chelating ring. Afterwards, the chemical reduction of Pd(ii) ions with sodium borohydride results in the in situ formation of Pd(0) nanoparticles that are still stabilized by the conjugate. The stabilized Pd(0) nanoparticles can selectively catalyze nitrobenzene and its derivatives, and can be easily recovered and reused while maintaining high catalytic efficiency. Therefore, EGCG–collagen conjugate fibers can serve as both support and stabilizer for Pd(0) nanoparticles. Likewise, this conjugate has also been used as the stabilizer of platinum and gold nanoparticles, which can catalyze the hydrogenation of olefins and the reduction of 4-nitrophenol, respectively.89,90 In addition, silver nanoparticles stabilized by EGCG–collagen conjugate fibers show enhanced and durable antibacterial activity against E. coli.87 Due to the superior antimicrobial activity of nanoparticles, it is supposed that nanoparticle-stabilized by phenolic–protein conjugate can be used as a coating material for food products. However, the safety of the material should be evaluated before its applications.
5.4. Hydrogels and nanoparticles for controlled drug release
In general, native proteins exhibits antioxidant property due to the function of amino acids such as Trp, Tyr, Cys, Met and His, which possess phenolic, sulfhydryl and imidazole side chain groups. When phenolic compounds are conjugated with proteins, the antioxidant activity and stability of proteins can be greatly enhanced. Moreover, phenolic–protein conjugates can act as drug delivery vehicle to fight against oxidative stress.91 Phenolic–chitosan/gelatin conjugates based hydrogels are synthesized by laccase catalysis. Phenolic compounds can not only stabilize the structure of hydrogels but also stimulate wound healing process by inhibiting deleterious wound enzymes and bacterial growth.92 Honda et al.42 demonstrate the conjugation reaction can retard the degradation of EGCG–gelatin conjugate based hydrogels with a delayed release of EGCG. In vitro experiments show the conjugate based hydrogels can be adsorbed onto the outer membrane of mesenchymal stem cells and further induce osteoblastogenesis upon stem cells. Interestingly, Guo et al.93 recently demonstrated the gel property of chlorogenic acid–myofibrillar protein conjugates is closely related to the amount of phenolics covalently linked. The conjugates with high conjugation efficiency can cause excessive aggregate formation and possibly shield functional groups involved in gel formation, thereby producing a poor gel network.
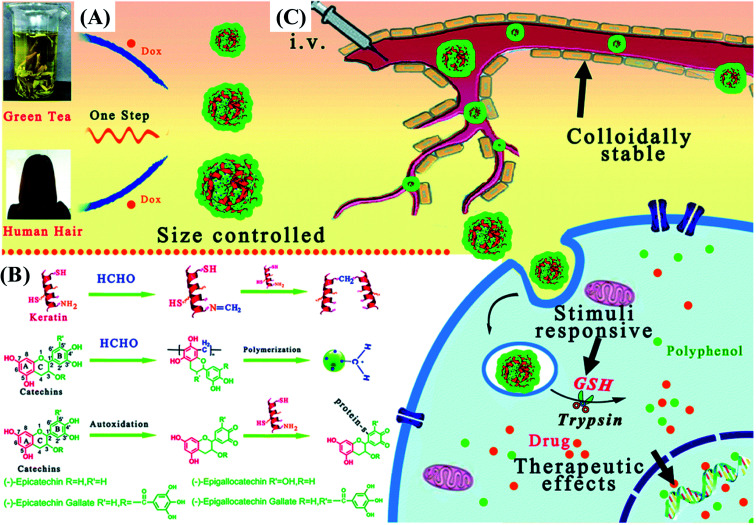
Recently, chlorogenic acid-β-lactoglobulin conjugate based nanoparticles (average particle diameters of 105–110 nm) are developed for the delivery of EGCG. The nanoparticles can not only protect EGCG from degradation but also control the release of EGCG in simulated gastrointestinal digestion fluid.31 Similarly, EGCG–zein conjugates based nanoparticles are prepared and used for the co-delivery of curcumin and resveratrol.94 Caffeic acid–BSA conjugate/zein nanoparticles are used to encapsulate resveratrol. Both thermal and UV light stability of resveratrol are significantly improved after nanoencapsulation. Cellular antioxidant assay showed resveratrol encapsulated in nanoparticles has a significantly higher antioxidant activity than resveratrol alone.95 As presented in Fig. 5, size-tunable nanoparticles (sizes ranging from 30 nm to 230 nm) based on catechin-keratin conjugate are prepared by formaldehyde cross-linking and are used as carriers of cancer drug doxorubicin. The anticancer efficiency of doxorubicin-loaded nanoparticles is higher than that of free doxorubicin.64 The potential applications of phenolic–protein conjugates are summarized in Table 1.
Fig. 5. Schematic illustration of (A) a simple strategy to fabricate catechin-keratin conjugate based nanoparticles; (B) molecular interactions of nanosphere assembly; and (C) administration of colloidally stable and glutathione (GSH)/enzyme dual stimuli-responsive nanoparticles with enhanced cancer therapy effects. Reproduced from ref. 64 with permission from Royal Society of Chemistry, copyright 2018.
6. Conclusions and future perspectives
Till now, several approaches including alkaline, free radical mediated grafting, enzyme catalysis and chemical coupling methods have been successfully developed to synthesize phenolic–protein conjugates. However, the synthetic mechanisms of phenolic–protein conjugates by alkaline and free radical mediated grafting methods are still unclear till now. Thus, more in-depth experiments are needed to reveal different synthetic mechanisms of phenolic–protein conjugates in future. Although structural changes of proteins after conjugation with phenolic compounds can be measured by many analytic methods (e.g. UV, FT-IR and fluorescence spectroscopy, CD, SDS-PAGE, MS and DSC), the linkage patters between phenolic compounds and proteins are still unrevealed. The combination of MS and NMR techniques can provide a new strategy to elucidate the primary structures of phenolic–protein conjugates. Phenolic–protein conjugates have been demonstrated to possess higher biological activities than blank proteins. However, existing studies on the biological activities of phenolic–protein conjugates are very limited and mainly focused on the antioxidant activity. In order to broaden the applications of phenolic–protein conjugates, other biological activities of phenolic–protein conjugates (e.g. anti-diabetic, anti-allergic, anti-inflammatory and antimutagenic activities etc.) are needed to be explored. Notably, existing evidences on the biological activities of phenolic–protein conjugates are based on in vitro assays. More reliable cellular and animal experiments should be carried out to fully evaluate the biological activities of phenolic–protein conjugates. Meanwhile, the structure-functionality relationships of phenolic–protein conjugates should be systematically explored. Due to the altered structures and improved biological activities of proteins, phenolic–protein conjugates will have several promising applications in food and pharmaceutical industries. Phenolic–protein conjugates can be developed as emulsions for nutraceutical delivery, edible films for food packaging, and hydrogels and nanoparticles for controlled drug release. For food and pharmaceutical applications, phenolic–protein conjugates should be carefully evaluated for their safety and toxicity to meet the regulations of government agencies. In addition, the actual applications of these useful phenolic–protein conjugates can be extended.
Funding
This work was supported by National Natural Science Foundation of China (No. 31571788), Natural Science Foundation of Jiangsu Province (No. BK20151310), Postgraduate Innovation Project of Yangzhou University (No. XKYCX19_197), and Qinglan Project of Jiangsu Province.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Material
References
- Nishinari K. Fang Y. Guo S. Phillips G. O. Food Hydrocolloids. 2014;39:301–318. doi: 10.1016/j.foodhyd.2014.01.013. [DOI] [Google Scholar]
- Kuan Y. H. Bhat R. Patras A. Karim A. A. Trends Food Sci. Technol. 2013;30:105–120. doi: 10.1016/j.tifs.2012.12.002. [DOI] [Google Scholar]
- Tian H. Guo G. Fu X. Yao Y. Yuan L. Xiang A. Int. J. Biol. Macromol. 2018;120:475–490. doi: 10.1016/j.ijbiomac.2018.08.110. [DOI] [PubMed] [Google Scholar]
- Shah N. N. Soni N. Singhal R. S. Int. J. Biol. Macromol. 2018;107:2224–2233. doi: 10.1016/j.ijbiomac.2017.10.099. [DOI] [PubMed] [Google Scholar]
- Gaspar A. L. C. de Góes-Favoni S. P. Food Chem. 2015;171:315–322. doi: 10.1016/j.foodchem.2014.09.019. [DOI] [PubMed] [Google Scholar]
- Isaschar-Ovdat S. Fishman A. Trends Food Sci. Technol. 2018;72:134–143. doi: 10.1016/j.tifs.2017.12.011. [DOI] [Google Scholar]
- Li C. P. Enomoto H. Hayashi Y. Zhao H. Aoki T. LWT--Food Sci. Technol. 2010;43:1295–1300. doi: 10.1016/j.lwt.2010.03.016. [DOI] [Google Scholar]
- Liu J. Ru Q. Ding Y. Food Res. Int. 2012;49:170–183. doi: 10.1016/j.foodres.2012.07.034. [DOI] [Google Scholar]
- O'Sullivan J. J. Park M. Beevers J. Greenwood R. W. Norton I. T. Food Hydrocolloids. 2017;71:299–310. doi: 10.1016/j.foodhyd.2016.12.037. [DOI] [Google Scholar]
- Balasundram N. Sundram K. Samman S. Food Chem. 2006;99:191–203. doi: 10.1016/j.foodchem.2005.07.042. [DOI] [Google Scholar]
- Liu J. Wang X. Yong H. Kan J. Jin C. Int. J. Biol. Macromol. 2018;116:1011–1025. doi: 10.1016/j.ijbiomac.2018.05.149. [DOI] [PubMed] [Google Scholar]
- Ignat I. Volf I. Popa V. I. Food Chem. 2011;126:1821–1835. doi: 10.1016/j.foodchem.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Liu J. Bai R. Liu Y. Zhang X. Kan J. Jin C. Int. J. Biol. Macromol. 2018;107:2242–2250. doi: 10.1016/j.ijbiomac.2017.10.097. [DOI] [PubMed] [Google Scholar]
- Cicerale S. Lucas L. J. Keast R. S. J. Curr. Opin. Biotechnol. 2012;23:129–135. doi: 10.1016/j.copbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Costa D. C. Costa H. S. Albuquerque T. G. Ramos F. Castilho M. C. Sanches-Silva A. Trends Food Sci. Technol. 2015;45:336–354. doi: 10.1016/j.tifs.2015.06.009. [DOI] [Google Scholar]
- Heleno S. A. Martins A. Queiroz M. J. R. Ferreira I. C. Food Chem. 2015;173:501–513. doi: 10.1016/j.foodchem.2014.10.057. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay P. Ghosh A. K. Ghosh C. Food Funct. 2012;3:592–605. doi: 10.1039/C2FO00006G. [DOI] [PubMed] [Google Scholar]
- Kim S. Cavaco-Paulo A. Appl. Microbiol. Biotechnol. 2012;93:585–600. doi: 10.1007/s00253-011-3524-8. [DOI] [PubMed] [Google Scholar]
- Spizzirri U. G. Iemma F. Puoci F. Cirillo G. Curcio M. Parisi O. I. Picci N. Biomacromolecules. 2009;10:1923–1930. doi: 10.1021/bm900325t. [DOI] [PubMed] [Google Scholar]
- Liu F. Ma C. McClements D. J. Gao Y. Food Hydrocolloids. 2017;63:625–634. doi: 10.1016/j.foodhyd.2016.09.041. [DOI] [Google Scholar]
- Wei Z. Yang W. Fan R. Yuan F. Gao Y. Food Hydrocolloids. 2015;45:337–350. doi: 10.1016/j.foodhyd.2014.12.008. [DOI] [Google Scholar]
- Jakobek L. Food Chem. 2015;175:556–567. doi: 10.1016/j.foodchem.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Ozdal T. Capanoglu E. Altay F. Food Res. Int. 2013;51:954–970. doi: 10.1016/j.foodres.2013.02.009. [DOI] [Google Scholar]
- Xiao J. Kai G. Crit. Rev. Food Sci. Nutr. 2012;52:85–101. doi: 10.1080/10408398.2010.499017. [DOI] [PubMed] [Google Scholar]
- Fu S. Wu C. Wu T. Yu H. Yang S. Hu Y. Food Chem. 2017;221:657–663. doi: 10.1016/j.foodchem.2016.11.123. [DOI] [PubMed] [Google Scholar]
- Rohn S. Rawel H. M. Kroll J. J. Agric. Food Chem. 2004;52:4725–4729. doi: 10.1021/jf0496797. [DOI] [PubMed] [Google Scholar]
- You J. Luo Y. Wu J. J. Agric. Food Chem. 2014;62:2581–2587. doi: 10.1021/jf405635q. [DOI] [PubMed] [Google Scholar]
- Tao F. Xiao C. Chen W. Zhang Y. Pan J. Jia Z. Int. J. Biol. Macromol. 2019;126:1186–1191. doi: 10.1016/j.ijbiomac.2019.01.017. [DOI] [PubMed] [Google Scholar]
- Rawel H. M. Ranters H. Rohn S. Kroll J. J. Agric. Food Chem. 2004;52:5263–5271. doi: 10.1021/jf0354850. [DOI] [PubMed] [Google Scholar]
- Rawel H. M. Rohn S. Kroll J. Int. J. Biol. Macromol. 2003;32:109–120. doi: 10.1016/S0141-8130(03)00044-8. [DOI] [PubMed] [Google Scholar]
- Fan Y. Zhang Y. Yokoyama W. Yi J. RSC Adv. 2017;7:21366–21374. doi: 10.1039/C6RA28462K. [DOI] [Google Scholar]
- Feng J. Cai H. Wang H. Li C. Liu S. Food Chem. 2018;241:60–69. doi: 10.1016/j.foodchem.2017.08.055. [DOI] [PubMed] [Google Scholar]
- Gu L. Su Y. Zhang M. Chang C. Li J. McClements D. J. Yang Y. Food Res. Int. 2017;96:84–93. doi: 10.1016/j.foodres.2017.03.015. [DOI] [PubMed] [Google Scholar]
- Wu X. Lu Y. Xu H. Lin D. He Z. Wu H. Liu L. Wang Z. Food Chem. 2018;256:427–434. doi: 10.1016/j.foodchem.2018.02.158. [DOI] [PubMed] [Google Scholar]
- Yi J. Zhang Y. Liang R. Zhong F. Ma J. J. Agric. Food Chem. 2015;63:297–303. doi: 10.1021/jf5056024. [DOI] [PubMed] [Google Scholar]
- Liu J. Pu H. Chen C. Liu Y. Bai R. Kan J. Jin C. J. Agric. Food Chem. 2018;66:279–289. doi: 10.1021/acs.jafc.7b05135. [DOI] [PubMed] [Google Scholar]
- Rai S. Kureel A. K. Dutta P. K. Mehrotra G. K. Int. J. Biol. Macromol. 2018;110:425–436. doi: 10.1016/j.ijbiomac.2017.11.049. [DOI] [PubMed] [Google Scholar]
- Gu L. Peng N. Chang C. McClements D. J. Su Y. Yang Y. Food Biophysics. 2017;12:198–210. doi: 10.1007/s11483-017-9476-5. [DOI] [Google Scholar]
- Chung J. E. Kurisawa M. Uyama H. Kobayashi S. Biotechnol. Lett. 2003;25:1993–1997. doi: 10.1023/B:BILE.0000004391.27564.8e. [DOI] [PubMed] [Google Scholar]
- Qi C. Wang P. Cui L. Deng C. Yu Y. Wang Q. Fan X. Appl. Microbiol. Biotechnol. 2016;100:1713–1722. doi: 10.1007/s00253-015-7064-5. [DOI] [PubMed] [Google Scholar]
- Ali M. Homann T. Khalil M. Kruse H. P. Rawel H. J. Agric. Food Chem. 2013;61:6911–6920. doi: 10.1021/jf402221m. [DOI] [PubMed] [Google Scholar]
- Ihara N. Schmitz S. Kurisawa M. Chung J. E. Uyama H. Kobayashi S. Biomacromolecules. 2004;5:1633–1636. doi: 10.1021/bm049823x. [DOI] [PubMed] [Google Scholar]
- Migneault I. Dartiguenave C. Bertrand M. J. Waldron K. C. BioTechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- Wu H. Zhuo L. He Q. Liao X. Shi B. Appl. Catal., A. 2009;366:44–56. doi: 10.1016/j.apcata.2009.06.024. [DOI] [Google Scholar]
- Honda Y. Tanaka T. Tokuda T. Kashiwagi T. Kaida K. Hieda A. Kaida K. Hieda A. Baba S. Int. J. Mol. Sci. 2015;16:14143–14157. doi: 10.3390/ijms160614143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maksoud A. A. A. El-Ghany I. H. A. El-Beltagi H. S. Anankanbil S. Banerjee C. Petersen S. V. Pérez B. Guo Z. Food Chem. 2018;241:281–289. doi: 10.1016/j.foodchem.2017.08.101. [DOI] [PubMed] [Google Scholar]
- Prigent S. V. Voragen A. G. Visser A. J. van Koningsveld G. A. Gruppen H. J. Sci. Food Agric. 2007;87:2502–2510. doi: 10.1002/jsfa.3011. [DOI] [Google Scholar]
- Czubinski J. Dwiecki K. Int. J. Food Sci. Technol. 2017;52:573–585. doi: 10.1111/ijfs.13339. [DOI] [Google Scholar]
- Cirillo G. Vittorio O. Hampel S. Spizzirri U. G. Picci N. Iemma F. Int. J. Pharm. 2013;446:176–182. doi: 10.1016/j.ijpharm.2013.02.023. [DOI] [PubMed] [Google Scholar]
- Ali M. Keppler J. K. Coenye T. Schwarz K. J. Food Sci. 2018;83:2092–2100. doi: 10.1111/1750-3841.14222. [DOI] [PubMed] [Google Scholar]
- Zhu Z. Lu J. J. Liu S. Anal. Chim. Acta. 2012;709:21–31. doi: 10.1016/j.aca.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Sun C. Yang W. Yuan F. Gao Y. RSC Adv. 2015;5:15641–15651. doi: 10.1039/C4RA10802G. [DOI] [Google Scholar]
- Liu F. Wang D. Sun C. McClements D. J. Gao Y. Food Chem. 2016;205:129–139. doi: 10.1016/j.foodchem.2016.02.155. [DOI] [PubMed] [Google Scholar]
- Liu F. Wang D. Sun C. Gao Y. Food Hydrocolloids. 2016;52:661–669. doi: 10.1016/j.foodhyd.2015.08.007. [DOI] [Google Scholar]
- Yi J. Fan Y. Zhang Y. Zhao L. Food Chem. 2016;205:73–80. doi: 10.1016/j.foodchem.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Zhang X. Do M. D. Casey P. Sulistio A. Qiao G. G. Lundin L. Lillford P. Kosaraju S. J. Agric. Food Chem. 2010;58:6809–6815. doi: 10.1021/jf1004226. [DOI] [PubMed] [Google Scholar]
- Kong J. Yu S. Acta Biochim. Biophys. Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- Liu F. Wang D. Ma C. Gao Y. Food Hydrocolloids. 2016;58:49–59. doi: 10.1016/j.foodhyd.2016.02.011. [DOI] [Google Scholar]
- Kanakis C. D. Hasni I. Bourassa P. Tarantilis P. A. Polissiou M. G. Tajmir-Riahi H. A. Food Chem. 2011;127:1046–1055. doi: 10.1016/j.foodchem.2011.01.079. [DOI] [PubMed] [Google Scholar]
- Pelton J. T. McLean L. R. Anal. Biochem. 2000;277:167–176. doi: 10.1006/abio.1999.4320. [DOI] [PubMed] [Google Scholar]
- Al-Hanish A. Stanic-Vucinic D. Mihailovic J. Prodic I. Minic S. Stojadinovic M. Radibratovic M. Milcic M. Velickovic T. C. Food Hydrocolloids. 2016;61:241–250. doi: 10.1016/j.foodhyd.2016.05.012. [DOI] [Google Scholar]
- Chaudhuri S. Sengupta B. Taylor J. Pahari B. P. Sengupta P. K. Curr. Drug Metab. 2013;14:491–503. doi: 10.2174/1389200211314040011. [DOI] [PubMed] [Google Scholar]
- Yang C. Wang B. Wang J. Xia S. Wu Y. LWT. 2019;109:443–449. doi: 10.1016/j.lwt.2019.04.034. [DOI] [Google Scholar]
- Yi Z. Sun Z. Chen G. Zhang H. Ma X. Su W. Cui X. Li X. J. Mater. Chem. B. 2018;6:1373–1386. doi: 10.1039/C7TB03293E. [DOI] [PubMed] [Google Scholar]
- Tao F. Jiang H. Chen W. Zhang Y. Pan J. Jiang J. Jia Z. J. Sci. Food Agric. 2018;98:5683–5689. doi: 10.1002/jsfa.9114. [DOI] [PubMed] [Google Scholar]
- Rawel H. M. Rohn S. Kruse H. P. Kroll J. Food Chem. 2002;78:443–455. doi: 10.1016/S0308-8146(02)00155-3. [DOI] [Google Scholar]
- Manea A. Cell Tissue Res. 2010;342:325–339. doi: 10.1007/s00441-010-1060-y. [DOI] [PubMed] [Google Scholar]
- Valko M. Jomova K. Rhodes C. J. Kuča K. Musilek K. Arch. Toxicol. 2016;90:1–37. doi: 10.1007/s00204-015-1579-5. [DOI] [PubMed] [Google Scholar]
- Pham L. B. Wang B. Zisu B. Adhikari B. Food Chem. 2019;293:463–471. doi: 10.1016/j.foodchem.2019.04.123. [DOI] [PubMed] [Google Scholar]
- Xiao J. Mao F. Yang F. Zhao Y. Zhang C. Yamamoto K. Mol. Nutr. Food Res. 2011;55:1637–1645. doi: 10.1002/mnfr.201100280. [DOI] [PubMed] [Google Scholar]
- Xiao J. Chen T. Cao H. Chen L. Yang F. Mol. Nutr. Food Res. 2011;55:310–317. doi: 10.1002/mnfr.201000208. [DOI] [PubMed] [Google Scholar]
- Xiao J. Zhao Y. Wang H. Yuan Y. Yang F. Zhang C. Kai G. Integr. Biol. 2011;3:1087–1094. doi: 10.1039/c1ib00076d. [DOI] [PubMed] [Google Scholar]
- Xie Y. Xiao J. Kai G. Chen X. Integr. Biol. 2012;4:502–507. doi: 10.1039/c2ib00185c. [DOI] [PubMed] [Google Scholar]
- Jiang L. Liu Y. Li L. Qi B. Ju M. Xu Y. Zhang Y. Sui X. Food Res. Int. 2019;120:603–609. doi: 10.1016/j.foodres.2018.11.011. [DOI] [PubMed] [Google Scholar]
- Xu H. Zhang T. Lu Y. Lin X. Hu X. Liu L. He Z. Wu X. Food Chem. 2019;298:125024. doi: 10.1016/j.foodchem.2019.125024. [DOI] [PubMed] [Google Scholar]
- Chaharband F. Kamalinia G. Atyabi F. Mortazavi S. A. Mirzaie Z. H. Dinarvand R. Artif. Cells, Nanomed., Biotechnol. 2018;46:626–636. doi: 10.1080/21691401.2017.1337020. [DOI] [PubMed] [Google Scholar]
- Donsì F. Senatore B. Huang Q. Ferrari G. J. Agric. Food Chem. 2010;58:10653–10660. doi: 10.1021/jf101804g. [DOI] [PubMed] [Google Scholar]
- McClements D. J. Li Y. Adv. Colloid Interface Sci. 2010;159:213–228. doi: 10.1016/j.cis.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Liu H. Han G. Zhang H. Liu Q. Kong B. Food Hydrocolloids. 2019;94:418–427. doi: 10.1016/j.foodhyd.2019.03.037. [DOI] [Google Scholar]
- Fan Y. Liu Y. Gao L. Zhang Y. Yi J. Food Chem. 2018;265:200–207. doi: 10.1016/j.foodchem.2018.05.098. [DOI] [PubMed] [Google Scholar]
- Wei Z. Zhang H. Huang Q. Food Funct. 2019;10:4911–4923. doi: 10.1039/C9FO01162E. [DOI] [PubMed] [Google Scholar]
- Hassan B. Chatha S. A. S. Hussain A. I. Zia K. M. Akhtar N. Int. J. Biol. Macromol. 2017;109:1095–1107. doi: 10.1016/j.ijbiomac.2017.11.097. [DOI] [PubMed] [Google Scholar]
- Wihodo M. Moraru C. I. J. Food Eng. 2013;114:292–302. doi: 10.1016/j.jfoodeng.2012.08.021. [DOI] [Google Scholar]
- Araghi M. Moslehi Z. Mohammadi Nafchi A. Mostahsan A. Salamat N. Daraei Garmakhany A. Food Sci. Nutr. 2015;3:370–375. doi: 10.1002/fsn3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T. M. Souza Filho M. D. S. M. Muniz C. R. Morais J. P. S. Kotzebue L. R. V. Pereira A. L. S. Azeredo H. M. J. Sci. Food Agric. 2017;97:4580–4587. doi: 10.1002/jsfa.8327. [DOI] [PubMed] [Google Scholar]
- Choi I. Lee S. E. Chang Y. Lacroix M. Han J. LWT--Food Sci. Technol. 2018;93:427–433. doi: 10.1016/j.lwt.2018.03.065. [DOI] [Google Scholar]
- Wu H. He L. Gao M. Gao S. Liao X. Shi B. New J. Chem. 2011;35:2902–2909. doi: 10.1039/C1NJ20674E. [DOI] [Google Scholar]
- Wu H. Huang X. Gao M. Liao X. Shi B. Green Chem. 2011;13:651–658. doi: 10.1039/C0GC00843E. [DOI] [Google Scholar]
- Wu H. Tang R. He Q. Liao X. Shi B. J. Chem. Technol. Biotechnol. 2009;84:1702–1711. doi: 10.1002/jctb.2234. [DOI] [Google Scholar]
- Wu H. Wu C. He Q. Liao X. Shi B. Mater. Sci. Eng., C. 2010;30:770–776. doi: 10.1016/j.msec.2010.03.013. [DOI] [Google Scholar]
- Raja I. S. Fathima N. N. Colloids Surf., B. 2015;128:537–543. doi: 10.1016/j.colsurfb.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Rocasalbas G. Francesko A. Touriño S. Fernández-Francos X. Guebitz G. M. Tzanov T. Carbohydr. Polym. 2013;92:989–996. doi: 10.1016/j.carbpol.2012.10.045. [DOI] [PubMed] [Google Scholar]
- Guo X. Qiu H. Deng X. Mao X. Guo X. Xu C. Zhang J. Molecules. 2019;24:3205. doi: 10.3390/molecules24173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Ma D. Luo X. Zhang Z. He L. Gao Y. McClements D. J. Food Hydrocolloids. 2018;79:450–461. doi: 10.1016/j.foodhyd.2018.01.017. [DOI] [Google Scholar]
- Fan Y. Liu Y. Gao L. Zhang Y. Yi J. Food Chem. 2018;261:283–291. doi: 10.1016/j.foodchem.2018.04.055. [DOI] [PubMed] [Google Scholar]