Abstract
While population genetics of Schistosoma haematobium have been investigated in West Africa, only scant data are available from Côte d’Ivoire. The purpose of this study was to analyze both genetic variability and genetic structure among S. haematobium populations and to quantify the frequency of S. haematobium × S. bovis hybrids in school-aged children in different parts of Côte d’Ivoire. Urine samples were subjected to a filtration method and examined microscopically for Schistosoma eggs in four sites in the western and southern parts of Côte d’Ivoire. A total of 2692 miracidia were collected individually and stored on Whatman® FTA cards. Of these, 2561 miracidia were successfully genotyped for species and hybrid identification using rapid diagnostic multiplex mitochondrial cox1 PCR and PCR Restriction Fragment Length Polymorphism (PCR-RFLP) analysis of the nuclear ITS2 region. From 2164 miracidia, 1966 (90.9%) were successfully genotyped using at least 10 nuclear microsatellite loci to investigate genetic diversity and population structure. Significant differences were found between sites in all genetic diversity indices and genotypic differentiation was observed between the site in the West and the three sites in the East. Analysis at the infrapopulation level revealed clustering of parasite genotypes within individual children, particularly in Duekoué (West) and Sikensi (East). Of the six possible cox1-ITS2 genetic profiles obtained from miracidia, S. bovis cox1 × S. haematobium ITS2 (42.0%) was the most commonly observed in the populations. We identified only 15 miracidia (0.7%) with an S. bovis cox1 × S. bovis ITS2 genotype. Our study provides new insights into the population genetics of S. haematobium and S. haematobium × S. bovis hybrids in humans in Côte d’Ivoire and we advocate for researching hybrid schistosomes in animals such as rodents and cattle in Côte d’Ivoire.
Keywords: Côte d’Ivoire, Microsatellites, Population genetics, Schistosoma haematobium, Schistosomiasis
Abstract
Alors que la génétique des populations de Schistosoma haematobium a été étudiée en Afrique de l’Ouest, seules quelques données sont disponibles pour la Côte d’Ivoire. Le but de cette étude était d’analyser à la fois la variabilité génétique et la structure génétique des populations de S. haematobium et de quantifier la fréquence des hybrides S. haematobium × S. bovis chez les enfants d’âge scolaire dans différentes régions de la Côte d’Ivoire. Des échantillons d’urine ont été soumis à une méthode de filtration et examinés au microscope pour les œufs de Schistosoma dans quatre sites de l’ouest et du sud de la Côte d’Ivoire. Au total, 2 692 miracidia ont été collectés individuellement et stockés sur des cartes Whatman® FTA. Parmi ceux-ci, 2 561 miracidia ont été génotypés avec succès pour l’identification des espèces et des hybrides à l’aide de la PCR multiplex de diagnostic rapide du cox1 mitochondrial et d’une analyse du polymorphisme de longueur des fragments de restriction de PCR (PCR-RFLP) de la région ITS2 de l’ADN nucléaire. Sur 2 164 miracidia, 1 966 (90,9 %) ont été génotypés avec succès en utilisant au moins 10 loci microsatellites nucléaires pour étudier la diversité génétique et la structure de la population. Des différences significatives ont été trouvées entre les sites dans tous les indices de diversité génétique et une différenciation génotypique a été observée entre le site de l’Ouest et les trois sites de l’Est. L’analyse au niveau de l’infrapopulation a révélé un regroupement des génotypes de parasites au sein de chaque enfant, en particulier à Duekoué (Ouest) et Sikensi (Est). Parmi les six profils génétiques cox1-ITS2 possibles obtenus à partir de miracidia, S. bovis cox1 × S. haematobium ITS2 (42,0 %) était le plus fréquemment observé dans les populations. Nous avons identifié seulement 15 miracidia (0,7 %) avec un génotype S. bovis cox1 × S. bovis ITS2. Notre étude apporte de nouvelles connaissances sur la génétique des populations de S. haematobium et des hybrides S. haematobium × S. bovis chez l’homme en Côte d’Ivoire et nous plaidons pour la recherche de schistosomes hybrides chez les animaux (rongeurs et bovins) en Côte d’Ivoire.
Introduction
Schistosomiasis is a chronic neglected tropical disease caused by trematodes belonging to the genus Schistosoma [14, 40]. The disease affects both humans and animals and is of considerable public health and veterinary concern, particularly in tropical and subtropical zones. According to the World Health Organization (WHO), schistosomiasis is transmitted in over 78 countries and territories throughout a wide belt of the tropics and subtropics [68]. More than 250 million people are infected, mostly in Africa [31], and the global burden of schistosomiasis was estimated at 1.4 million disability-adjusted life years (DALYs) in 2017 [26]. Six species of schistosomes can infect humans: S. guineensis, S. haematobium, S. intercalatum and S. mansoni in Africa and the Arabian Peninsula [60], and S. mekongi and S. japonicum in Asia; S. mansoni mainly occurs in Africa, but is also found in Brazil and some Caribbean islands [51]. Schistosoma haematobium causes the urogenital form of the disease. Classified as a group I carcinogen, urogenital schistosomiasis can lead to squamous-cell carcinoma of the bladder [35]. In most countries, while human schistosomiasis is well documented, little is known about the prevalence and transmission dynamics of animal schistosomiasis.
In Côte d’Ivoire, both S. haematobium and S. mansoni are endemic, causing urogenital and intestinal schistosomiasis, respectively [13]. The former is predominant in the central and southern parts [15, 56], while the latter is mainly found in the western parts of Côte d’Ivoire [7, 47, 59]. We recently reported high prevalence of S. haematobium in school-aged children in South Côte d’Ivoire [5]. Limited data are available on animal-infecting schistosomes, such as S. bovis, a parasite of livestock and rodents. A previous study on post-mortem examinations of cattle in North Côte d’Ivoire reported a prevalence of 35% [1]. These findings were confirmed in a recent cross-sectional survey conducted in different parts along a transect from North to South Côte d’Ivoire, with the highest prevalence of S. bovis found in cattle in the northern parts of the country [39].
Habitat change and migration of hosts, which is also linked to climate change, can influence the epidemiology and distribution of schistosomiasis and enhance the occurrence of interspecies hybridization between human and animal schistosomes [38]. Schistosoma haematobium × S. bovis hybrids are well documented in West Africa, particularly in Benin, Mali, Niger and Senegal [34, 42], with occurrences also reported in Malawi [66] and Corsica, France [9, 10]. More generally, genomic studies have highlighted the fact that several natural strains that were initially identified as “pure” S. haematobium are in fact introgressed to some extent with S. bovis [44, 48]. Recently, hybrids from miracidia (although not thought to be viable) have been identified in Côte d’Ivoire between S. haematobium and S. mansoni excreted by humans [17, 41], and between S. haematobium and S. bovis being transmitted by freshwater snails of the genus Bulinus [58] and also from humans [6].
Population genetic structure and genetic diversity varies by Schistosoma species [49]. Among the African species, S. mansoni is the most widely studied. Several investigations have shown high genetic diversity and strong genetic structure in several countries, including Ethiopia, Kenya, Senegal and Uganda [2–4, 57, 61, 63]. In contrast, studies on S. haematobium have shown both less genetic diversity and less population structure compared to S. mansoni [30, 46]. This difference in genetic structuration between S. mansoni and S. haematobium is also apparent at the continental scale [30, 62, 63]. Population genetics data for S. bovis are even more limited. One study has investigated the population genetic patterns of S. bovis in Cameroon revealing an intermediate pattern with high genetic diversity (i.e. like in S. mansoni), but no genetic structuration (i.e. like in S. haematobium) at the country scale [18]. At the continental scale, a comparative genomic study revealed a significantly higher genomic diversity between S. bovis lineages than between S. haematobium lineages [48]. More recently, a study analyzed the genetic patterns of parasites collected from humans and animals in North Senegal and molecularly characterized both microsatellite markers and hybrid genotyping [11]. The authors demonstrated (i) clear genetic separation between parasites recovered from animals compared to those from humans; (ii) no genetic structuration between hybrids and pure parasites from human hosts; and (iii) significant genetic differentiation between different villages of the Senegal River basin. These authors concluded that, in Senegal, animals do not represent a real reservoir for human schistosomiasis and also, there is no cross-over of transmission between humans and animals. However, this conclusion contradicts recent observations from Benin, where S. haematobium × S. bovis hybrids were found in cows [54] and rodents [55].
The purpose of this study was to investigate the population genetic structure and genetic diversity of S. haematobium and S. haematobium × S. bovis hybrids that were obtained from human urine samples, in four sampling sites of Côte d’Ivoire. We molecularly characterized individual miracidia, using cox1 and ITS2 markers to identify S. haematobium and S. haematobium × S. bovis hybrids within the different populations, alongside microsatellite markers [65] to analyze population genetic diversity and structure.
Materials and methods
Ethics statement
Ethical clearance was obtained from the Ministère de la Santé et de l’Hygiène Publique en Côte d’Ivoire (reference no. 003–18/MSHP/CNER-kp). School authorities, teachers, parents/guardians and children were informed about the objectives, procedures and potential risks and benefits of the study. Written informed consent was obtained from children’s parents or legal guardians, while children provided oral assent. After sampling, a praziquantel treatment (40 mg/kg) was offered to children found with a Schistosoma infection.
Study sites and collection of miracidia
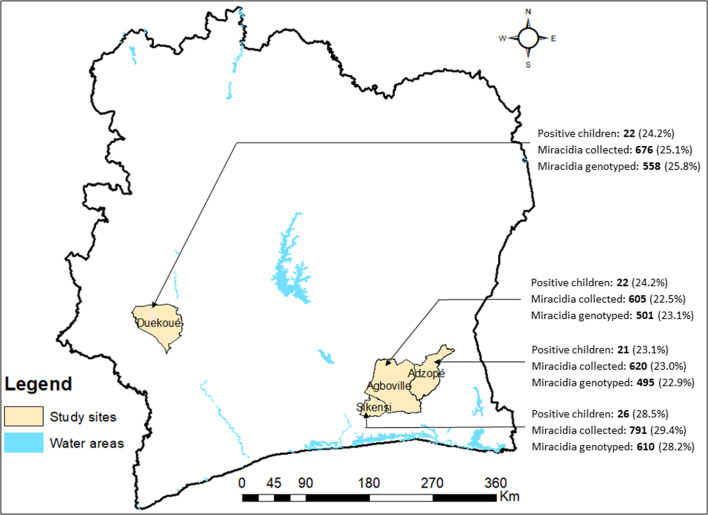
This study was carried out in four sampling sites in Côte d’Ivoire: (i) Agboville (geographical coordinates 5° 55′ 41″ N latitude, 4° 13′ 01″ W longitude) and (ii) Adzopé (6° 06′ 25″ N, 3° 51′ 36″ W) in the south-eastern part; (iii) Sikensi (5°40′34″ N, 4°34′ 33″ W) in the south-central part; and (iv) Duekoué (6° 44′ 00″ N, 7° 21′ 00″ W) in the western part of Côte d’Ivoire (Fig. 1). The study was integrated into a cross-sectional survey of the prevalence of schistosomiasis among school-aged children from January to April 2018 [5]. Schistosoma miracidia from children aged 5–14 years were collected, after egg hatching, and individually stored on Whatman® FTA cards (GE Healthcare Life Sciences; Amersham, UK) as previously described [10]. FTA cards were dried for 1 h at room temperature before being stored in a sealed plastic bag and then transferred to the “Interactions Hôtes-Pathogènes-Environnements” (IHPE) laboratory in Perpignan, France, for molecular analysis.
Figure 1.
Sites in the southern and western parts of Côte d’Ivoire and number (percentage of total sample size) of Schistosoma-infected children, miracidia collected, and miracidia genotyped.
DNA extraction for miracidia genotyping
Genomic DNA from individual miracidia was extracted using Chelex® beads (Bio-Rad; Hercules, CA, USA) from 2.0 mm discs containing sample that had been punched out of the FTA card using a Harris-Micro-Punch (VWR; London, UK [37].
Cox1 and ITS analysis for species and hybrid identification
A cox1 multiplex PCR was performed to identify the species-specific mitotype of each sample, as described in our previous work [6]. For the analysis of the nuclear internal transcribed spacer 2 (ITS2) region, we used a restriction fragment length polymorphism (RFLP) approach ([19] and Supplementary Fig. S1), following amplification using the forward primer Sc_ITS_F: 5′–GGC TGC AGC GTT AAC CAT TA–3′ and reverse primer Sc_ITS_R: 5′–ACA CAC ACC ATC GGT AC AAA–3′, which targets 505 bp of the ITS2 [19]. We performed PCRs in a total reaction volume of 25 μL, comprising 2 μL of gDNA, 5 μL of Green GoTaq flexi buffer 5×, 1.5 μL of 25 mM MgCl2, 0.5 μL of 10 mM dNTP mix, 1 μL of each 10 μM primer and 1 U of GoTaq Hot Start Polymerase (Promega; Madison, WI, USA). The reaction conditions included an activation step of 95 °C for 3 min, followed by 45 cycles of 95 °C for 40 s, 58 °C for 40 s and 72 °C for 40 s, and a final extension at 72 °C for 6 min. In the subsequent step, 5 μL of PCR products were then digested at 37 °C using 0.5 μL of restriction enzyme MboI (Thermo Fisher Scientific; Waltham, MA, USA), 2.5 μL of CutSmart buffer, and 17 μL of molecular water for 15 min, followed by inactivation at 60 °C for 20 min for a total reaction volume of 25 μL. The cut sites of the MboI enzyme for species specific SNPs were based on the following sequences positions: ↓GATC and CTAG↑. The PCR products were not cleaned or visualized before enzyme digestion. We used only samples that were positive with the cox1 multiplex PCR method. This enzymatic digestion cuts the 505 bp ITS2 fragment into four bands for S. haematobium (281 bp, 82, bp, 98 bp and 44 bp) and three bands for S. bovis (379 bp, 82 bp and 44 bp). All digested ITS2 PCR products were visualized using 2% agarose electrophoresis gels stained with GelRedTM (Biotium Inc.; Darmstadt, Germany). The nuclear ITS2 and mitochondrial cox1 identities for each individual miracidia were combined to produce a mito-nuclear genetic profile of S. haematobium (Shcox1 + ShITS2: Sh × ShSh), S. bovis (Shcox1 + ShITS2: Sb × SbSb) or S. haematobium-bovis hybrid genotypes, the latter of which is recognized by mito-nuclear discordance (Shcox1 + SbITS2: Sh × SbSb), (Shcox1 + Sb/ShITS2: Sb × SbSh), (Sbcox1 + ShITS2: Sb × ShSh) or (Sbcox1 + Sh/SbITS2: Sb × ShSb)).
Microsatellite analysis
Individual miracidia were further genotyped using a set of 18 microsatellite markers divided into two panels developed by Webster and colleagues [65]. The forward primers were fluorescently labelled with 6-FAM, VIC, NED and PET dyes (Applied Biosystems; Foster City, CA, USA) to enable identification within the multiplex PCR, as previously described [65]. Microsatellite PCRs were performed, using a Microsatellite PCR Kit (Qiagen; Hilden, Germany), in a final volume of 10 μL, including 4 μL of the DNA template, 5 μL of the 2× microsatellite PCR Buffer Kit (Qiagen; Hilden, Germany) and 1 μL of 10× microsatellite primer mix in two PCR panels of nine per multiplex. Thermal cycling was performed with an initial hot-start activation of 15 min at 95 °C, followed by 45 cycles of 94 °C for 30 s, 56 °C for 90 s and 72 °C for 60 s, with a final extension at 60 °C for 30 min. The PCR products were sent to Genoscreen (Lille, France) for genotyping. All microsatellite loci were visually peak called, using GS500Liz size standard (Applied Biosystems), and GeneMarker software. Only 16 loci were subsequently used for analysis as allelic dropout of two loci (C131 and Sh8) occurred in at least 20% of the samples (Supplementary Tab. S1). Errors due to large allele dropout or stutter bands and evidence of the presence of null alleles at each locus were checked using Micro-checker, version 2.2.3 [43].
Genetic diversity
The number and percentages of the different cox1/ITS2 profiles obtained from each miracidium were calculated for each site and any differences using a χ2 test was observed. We also calculated the number of S. bovis and S. haematobium haplotypes for cox1 or alleles for ITS2 among sites. The mitochondrial cox1 haplotype is unique as it is inherited from the maternal line, while the ITS2 was scored as one for heterozygous (Sb/Sh) and two for homozygous (SbSb or ShSh). Differences in the relative frequencies of S. bovis vs. S. haematobium haplotypes or alleles between sites were tested using a binomial test [32].
For the microsatellite data, tests for deviation from Hardy–Weinberg equilibrium per locus and site were carried out using Genepop, version 4.0 [53]. The genotypic disequilibrium test for pairs of loci overall and the adjusted p-value for 5% nominal level was performed using FSTAT, version 2.9.4 [28, 67]. Genetic diversity was compared between sites and between cox1/ITS2 genetic profiles. Expected heterozygosity (He), number of alleles (A), allelic richness (Ar), and the inbreeding coefficient (FIS) of each microsatellite locus were computed per sampling site, using FSTAT version 2.9.4 [28]. He and Ar were compared between the populations using the pairwise Friedman rank test, followed by the Nemeyi multiple comparison test implemented in the R studio PCMP plus package [36].
Genotypic differentiation and population structure
Genotypic differentiation between sampling sites was assessed using pairwise FST values [63], calculated in FSTAT, version 2.9.4 [28, 67] with a threshold of significance adjusted for multiple tests using Bonferroni’s standard correction [50]. Pairwise FST values were also calculated between samples that exhibited the different cox1/ITS2 profiles. Principal component analysis (PCA) was performed to compare either the four sites or the six cox1/ITS2 profiles, using Genetix, version 4.05 [8].
The uppermost level of genetic structure for all individuals was determined by a Bayesian clustering approach using Markov Chain Monte Carlo (MCMC) permutation analysis, implemented in Structure, version 2.3 [45], using data of children from whom we collected at least 15 miracidia. The length of the burn-in period was 250,000 with the number of MCMC replicates after burn-in at 1,000,000 and K from 1 to 8 using an admixture model. The log likelihood for each K was averaged over three runs with the CorrSieve package in R (Vienna, Austria) and the delta K-values were then computed to determine the most likely number among the K values tested [21]. For the most likely number of genetic clusters, an additional 10 runs were computed with the same initial parameters as those described. The probability of each miracidium belonging to each cluster was averaged over the 10 runs and graphically represented using Clumpp, version 1.1.2 [23] and Distruct, version 1.1 [52].
Analysis of molecular variance (AMOVA) was performed to evaluate the partitioning of the overall genetic variance to the hierarchical levels at infrapopulation “within miracidia”, “among miracidia within individual children”, “between miracidia from individual children within each site” and “between sites” using Arlequin, version 3.5 [22]. The level “within miracidia” reflects the differences between microsatellite loci and has no population genetic meaning. Hence, we report the residual variance of the remaining three levels and normalize their relative contributions to 100%.
Population genetic structure between parasite infrapopulation was assessed independently for each sampling site using Structure, version 2.3 with the same procedure as described above. If no best K was found, K was set as the number of children at each sampling site (16 for Adzopé, 18 for Agboville, 18 for Duekoué and 22 for Sikensi). We first calculated the mean probabilities assignations of the miracidia from a given child to a given cluster. Then repartitions of the miracidia among the different clusters were tested in each participant using χ2 under the null hypothesis of random repartition (1/K).
Results
Schistosoma haematobium and hybrid genotype identification
Of the 2692 miracidia collected from the urine samples from 91 Schistosoma egg-positive children at four sampling sites, 2561 were genetically analysed. cox1 and ITS2 profiles were obtained from 2164 individual miracidia (495 for Adzopé, 501 for Agboville, 610 for Sikensi and 558 for Duekoué) (Fig. 1). Of the 91 positive children, 89 (97.8%) were producing miracidia with a hybrid genotype. Among all possible hybrid profiles, the S. bovis cox1 × S. haematobium ITS2 (Sb × ShSh) genotype was the most frequently found in Adzopé (64.6%), Duekoué (56.3%) and Sikensi (30.2%), while the S. haematobium cox1 × S. haematobium/S. bovis ITS2 (Sh × ShSb) was the most common hybrid genotype in Agboville (27.7%). We also identified 15 miracidia (0.7%), from seven children, with the S. bovis cox1 × S. bovis ITS2 (Sb × SbSb) genetic profile. Overall, 791 miracidia (36.6%) presented an S. haematobium cox1 × S. haematobium ITS2 (Sh × ShSh) genetic profile. The χ2 test showed a significant difference in the distribution of miracidia with the different cox1-ITS2 profiles between sites (Table 1). The frequency of the S. bovis cox1 (50.7%) haplotype was similar to that of S. haematobium (49.3%; p = 0.27). The frequency of the S. bovis ITS2 allele (12.1%) was statistically lower than the S. bovis × S. haematobium allele (87.9%; p < 0.001). Analysis of the multiplicity of infection showed that 52 children (71.1%) were infected by worm-laying miracidia with at least three different cox1-ITS2 genetic profiles. One child (Id: AG122) from Agboville was infected by adult worms laying miracidia with all six possible cox1-ITS2 genetic profiles (Supplementary Tab. S2).
Table 1.
Total number (n) and percentage (%) of the six possible cox1-ITS2 genetic profiles identified using the haploid mitochondrial cox1 gene (first two letters) and the diploid nuclear ITS2 region (last four letters), i.e. “pure” Schistosoma haematobium (S. haematobium cox1 × S. haematobium ITS2: Sh × ShSh), S. bovis genetic signature (S. bovis cox1 × S. bovis ITS2: Sb × SbSb) and four types of hybrid (S. bovis cox1 × S. haematobium ITS2_S. bovis ITS2: Sb × ShSb; S. bovis cox1 × S. haematobium ITS2: Sb × ShSh; S. haematobium cox1 × S. bovis ITS2: Sh × SbSb; S. haematobium cox1 × S. bovis ITS2_ S. haematobium ITS2: Sh × ShSb) per area. Total number (percentage in parentheses) of S. bovis (Sb) and S. haematobium (Sh) cox1 haplotype and ITS2 alleles per site.
| Sites | Sb × SbSb |
Sh × ShSh |
Sb × ShSb |
Sb × ShSh |
Sh × SbSb |
Sh × ShSb |
Total hybrids |
All total |
Cox1 haplotypes |
ITS2 alleles |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | Sb | Sh | Sb | Sh | ||
| Adzopé | 3 (0.6) | 101 (20.4) | 47 (9.5) | 320 (64.7) | 4 (0.8) | 20 (4.0) | 391 (79.0) | 495 | 370 (74.7) | 125 (25.3) | 81 (8.2) | 909 (91.8) |
| Agboville | 9 (1.8) | 170 (33.9) | 59 (11.8) | 90 (18.0) | 34 (6.8) | 139 (27.7) | 322 (64.3) | 501 | 158 (31.5) | 343 (68.5) | 284 (28.4) | 718 (71.6) |
| Sikensi | 3 (0.49) | 311 (51.0) | 49 (8.0) | 184 (30.2) | 2 (0.33) | 61 (10.0) | 296 (48.5) | 610 | 236 (46.3) | 374 (53.7) | 120 (9.8) | 1100 (90.2) |
| Duekoué | 0 | 209 (37.5) | 19 (3.4) | 314 (56.3) | 3 (0.54) | 13 (2.33) | 349 (62.5) | 558 | 333 (59.7) | 225 (40.3) | 38 (3.4) | 1078 (96.6) |
| Total | 15 (0.7) | 791 (36.6) | 174 (8.0) | 908 (42.0) | 43 (2.0) | 233 (10.8) | 1358 (62.7) | 2 164 | 1097 (50.7) | 1067 (49.3) | 523 (12.1) | 3805 (87.9) |
χ2 test of difference of relative frequencies between sampling sites: χ2 = 555.9; df = 15; p < 0.0001. Binomial test of S. bovis vs. S. haematobium allele’s equipartition: Cox1, p = 0.27; ITS2, p < 0.00001.
Genetic diversity
Of the 2164 miracidia from which cox1/ITS2 profiles were obtained, 1966 (90.8%) also produced reliable allele data for at least 10 microsatellite loci for subsequent analyses. We found a significant genotypic disequilibrium (adjusted p-value at 5% level < 0.001) for 120 pairwise locus combinations across the whole dataset. Genetic diversity indices (He, A, Ar and FIS) and the probability of deviation from Hardy–Weinberg equilibrium (PHWE) for each microsatellite locus by sampling site are shown in Table 2. Most loci showed a deviation from Hardy–Weinberg equilibrium. All loci were highly polymorphic and the number of alleles and the allelic richness ranged from 7 to 17 and 7.0 to 15.8, respectively per locus in the whole dataset. Inter-population comparisons showed that heterozygosity (He) was relatively stable (Friedman test, χ2 = 1.33, degree of freedom (df = 3, p = 0.72) and that allelic richness was more variable ranging between 8.8 and 10.5 for Sikensi and Adzopé, respectively. A significant difference was observed for this last parameter (Friedman test, χ2 = 15.33, df = 3, p = 0.002), allelic richness from Duekoué being statistically higher compared to the remaining three sites (Nemeyi test, p < 0.05). No differences in genetic diversity indices were observed according to the different cox1/ITS2 genetic profiles.
Table 2.
Genetic diversity indices. Mean expected heterozygosity (He), total number of alleles detected (A), allelic richness (Ar) rarefied to 362 diploid individuals per population, mean inbreeding coefficient (FIS), and the probability of deviation from Hardy–Weinberg equilibrium (PHWE) for each microsatellite locus per site. n, number of miracidia genotyped per site.
| Locus | Adzopé (n = 440) |
Agboville (n = 465) |
Duekoué (n = 527) |
Sikensi (n = 534) |
Total (n = 1966) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| He | A | Ar | F IS | P HWE | He | A | Ar | F IS | P HWE | He | A | Ar | F IS | P HWE | He | A | Ar | F IS | P HWE | He | A | Ar | |
| Sh9 | 0.72 | 9 | 8.86 | 0.37 | <0.001 | 0.58 | 11 | 10.49 | 0.41 | <0.001 | 0.78 | 12 | 11.67 | 0.50 | <0.001 | 0.54 | 10 | 8.68 | 0.33 | <0.001 | 0.66 | 14 | 10.98 |
| Sh3 | 0.83 | 14 | 13.96 | 0.20 | <0.001 | 0.78 | 15 | 14.60 | 0.11 | <0.001 | 0.88 | 16 | 15.53 | 0.08 | <0.001 | 0.83 | 14 | 13.47 | 0.01 | 0.836 | 0.83 | 16 | 15.00 |
| C102 | 0.44 | 4 | 3.84 | 0.10 | 0.220 | 0.46 | 6 | 5.76 | −0.06 | <0.001 | 0.43 | 7 | 6.70 | 0.08 | 0.819 | 0.50 | 5 | 4.66 | −0.06 | <0.001 | 0.46 | 7 | 6.05 |
| Sh1 | 0.77 | 9 | 8.88 | 0.01 | <0.001 | 0.68 | 9 | 8.55 | −0.01 | <0.001 | 0.75 | 12 | 11.97 | 0.03 | <0.001 | 0.74 | 8 | 7.99 | −0.03 | <0.001 | 0.74 | 13 | 11.85 |
| Sh14 | 0.81 | 14 | 14.00 | 0.04 | <0.001 | 0.83 | 14 | 13.84 | 0.03 | <0.001 | 0.83 | 15 | 14.63 | 0.00 | <0.05 | 0.82 | 16 | 15.56 | −0.01 | <0.001 | 0.82 | 17 | 15.78 |
| Sh6 | 0.53 | 7 | 6.98 | 0.24 | <0.001 | 0.58 | 7 | 6.99 | 0.07 | <0.001 | 0.48 | 7 | 7.00 | −0.01 | <0.001 | 0.60 | 7 | 6.99 | −0.11 | <0.001 | 0.55 | 7 | 7.00 |
| C111 | 0.55 | 4 | 4.00 | 0.18 | <0.001 | 0.44 | 5 | 5.00 | 0.11 | <0.001 | 0.65 | 8 | 7.59 | 0.03 | 0.276 | 0.56 | 8 | 7.44 | 0.01 | 0.688 | 0.55 | 9 | 7.12 |
| Sh7 | 0.68 | 7 | 6.83 | 0.26 | <0.001 | 0.74 | 5 | 5.00 | 0.32 | <0.001 | 0.64 | 7 | 6.99 | 0.31 | <0.001 | 0.64 | 5 | 4.74 | 0.32 | <0.001 | 0.68 | 7 | 6.69 |
| Sh13 | 0.66 | 15 | 14.67 | 0.01 | <0.001 | 0.51 | 14 | 13.37 | 0.01 | <0.001 | 0.69 | 15 | 14.52 | −0.02 | <0.001 | 0.71 | 14 | 13.64 | −0.03 | 0.408 | 0.64 | 17 | 15.68 |
| Sh4 | 0.74 | 9 | 9.00 | 0.05 | <0.01 | 0.73 | 9 | 8.92 | 0.08 | <0.001 | 0.80 | 11 | 10.82 | 0.04 | <0.001 | 0.85 | 9 | 9.00 | 0.07 | <0.001 | 0.78 | 11 | 10.22 |
| Sh11 | 0.51 | 7 | 6.62 | −0.01 | <0.001 | 0.68 | 6 | 5.81 | 0.12 | <0.001 | 0.46 | 8 | 7.54 | 0.13 | <0.001 | 0.70 | 8 | 7.66 | 0.09 | <0.001 | 0.59 | 10 | 7.80 |
| Sh15 | 0.49 | 7 | 7.00 | 0.16 | <0.001 | 0.54 | 6 | 6.00 | 0.17 | <0.001 | 0.50 | 6 | 5.97 | 0.13 | <0.001 | 0.46 | 6 | 6.00 | −0.02 | <0.001 | 0.50 | 7 | 7.00 |
| Sh2 | 0.78 | 13 | 13.00 | 0.35 | <0.001 | 0.80 | 12 | 12.00 | 0.32 | <0.001 | 0.89 | 14 | 13.96 | 0.45 | <0.001 | 0.86 | 12 | 12.00 | 0.35 | <0.001 | 0.83 | 15 | 14.64 |
| Sh5 | 0.73 | 10 | 9.90 | 0.44 | <0.001 | 0.47 | 13 | 12.34 | 0.26 | <0.001 | 0.80 | 15 | 14.70 | 0.10 | <0.001 | 0.64 | 9 | 8.73 | 0.16 | <0.001 | 0.66 | 16 | 13.95 |
| Sh10 | 0.48 | 8 | 7.85 | 0.51 | <0.001 | 0.53 | 6 | 5.79 | 0.35 | <0.001 | 0.48 | 8 | 7.88 | 0.31 | <0.001 | 0.33 | 7 | 6.64 | 0.10 | <0.001 | 0.46 | 8 | 7.56 |
| Sh12 | 0.10 | 4 | 3.83 | −0.04 | 0.972 | 0.29 | 6 | 5.95 | 0.03 | <0.001 | 0.07 | 7 | 6.06 | −0.03 | 1.0 | 0.23 | 8 | 7.58 | −0.06 | <0.001 | 0.17 | 10 | 7.68 |
| Mean | 0.61 | 8.81 | 8.70 | 0.18 | − | 0.60 | 9.00 | 8.78 | 0.15 | − | 0.63 | 10.50 | 10.22 | 0.13 | − | 0.63 | 9.13 | 8.80 | 0.07 | − | 0.62 | 11.50 | 10.31 |
| SE | 0.19 | 3.60 | 3.61 | 0.17 | − | 0.15 | 3.61 | 3.47 | 0.14 | − | 0.22 | 3.61 | 3.59 | 0.17 | − | 0.18 | 3.28 | 3.22 | 0.15 | − | 0.18 | 3.88 | 3.65 |
SE: Standard error.
Population genotypic differentiation
The pairwise FST values between sites ranged from 4.1% to 6.9% with Duekoué being consistently most differentiated from the other sites (Table 3). The FST values between the samples that presented these cox1/ITS2 profiles ranged from 0.3% to 2.7% and thus showed that the S. bovis cox1-ITS2 profiles (Sb × SbSb) were not different from the other cox1/ITS2 profiles (Table 4). The miracidia with the S. bovis cox1 × S. bovis ITS2 (Sb × SbSb) genetic profile came from three sites (7 from Adzopé, 4 from Agboville, and 4 from Sikensi) (Supplementary Tab. S2).
Table 3.
Pairwise estimates of FST (below the diagonal) and significance (above diagonal) between parasite populations of the four sites based on 16 microsatellite loci.
| Sites | Adzopé | Agboville | Duekoué | Sikensi |
|---|---|---|---|---|
| Adzopé | – | ** | ** | ** |
| Agboville | 0.049 | – | ** | ** |
| Duekoué | 0.069 | 0.065 | – | ** |
| Sikensi | 0.047 | 0.041 | 0.056 | – |
**Significant values at p < 0.01.
Table 4.
Pairwise estimates of FST (below the diagonal) and significance (above diagonal) between cox1-ITS2 profiles of the four sampling sites based on 16 microsatellite loci.
| Genotype | Sb_SbSb | Sb_ShSb | Sb_ShSh | Sh_SbSb | Sh_ShSb | Sh_ShSh |
|---|---|---|---|---|---|---|
| Sb_SbSb | – | NS | * | * | NS | NS |
| Sb_ShSb | 0.003 | – | * | * | * | * |
| Sb_ShSh | 0.011 | 0.0071 | – | * | * | * |
| Sh_SbSb | 0.015 | 0.023 | 0.027 | – | * | * |
| Sh_ShSb | 0.006 | 0.005 | 0.014 | 0.009 | – | * |
| Sh_ShSh | 0.004 | 0.003 | 0.006 | 0.021 | 0.008 | – |
NS: not statistically significant; *Statistically significant at the p < 0.05 level.
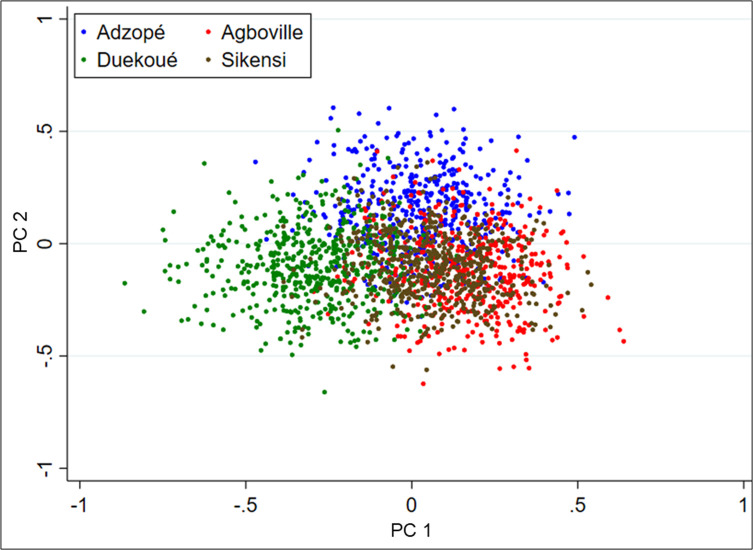
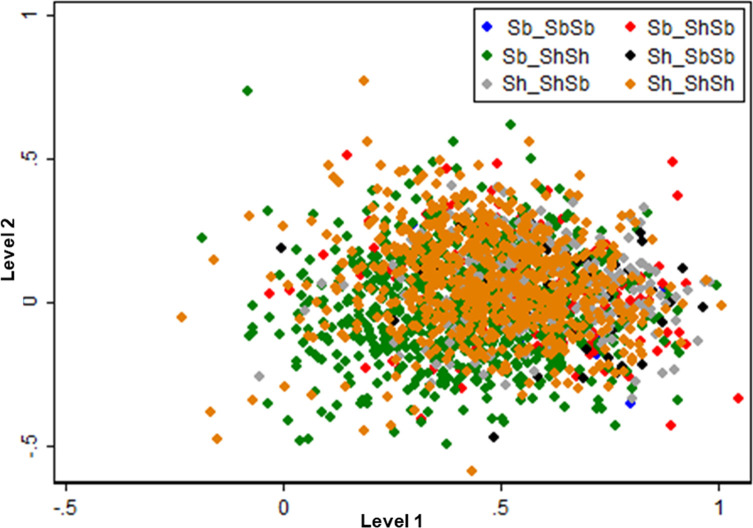
The PCA revealed only weak structuration among the four sites with only Duekoué being partially separated from the other three sites (Fig. 2). Comparing the different genetic profile, the PCA showed no genetic structuring according to the six different genetic profiles (Fig. 3).
Figure 2.
Principal component analysis (PCA) of microsatellite data by sites. Each miracidium is represented by one dot. The first two principal components (PCs) explain 41.1% and 33.7% of total inertia of the data set, respectively.
Figure 3.
Principal component analysis (PCA) of the microsatellite data with each miracidium assigned its cox1-ITS2 genotype. Each miracidium is represented by a single dot. The first two principal components (PCs) explain 61.6% and 21.0% of total inertia of the data set, respectively.
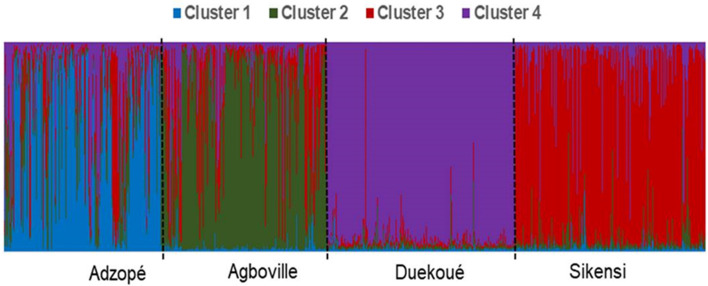
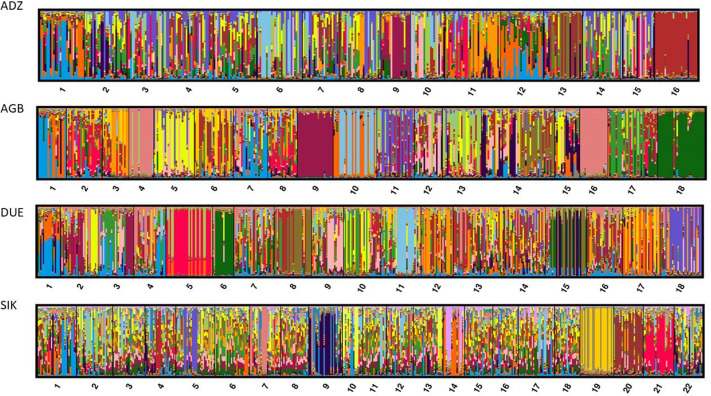
Population genetic structure was assessed both at the population level (between sampling sites) and at the infrapopulation level (between children within each sampling site – see below). K = 4 displayed maximal delta K at the population level suggesting that all miracidia were grouped into four genetic clusters. The Structure analysis showed a relatively strong genetic structure between sampling sites (Fig. 4). At the population level, an average of 51% of the genotypes were assigned to the respective dominant cluster in Adzopé, 63% in Agboville, 69% in Sikensi, and 83% in Duekoué.
Figure 4.
Bar plot depicting the genetic population structure of 1966 Schistosoma miracidia from the four sampling sites in Côte d’Ivoire produced by Structure for K = 4. Each column represents one miracidium, with colors indicating the proportional contribution of each of the four identified genetic clusters.
Infrapopulation genotypic differentiation
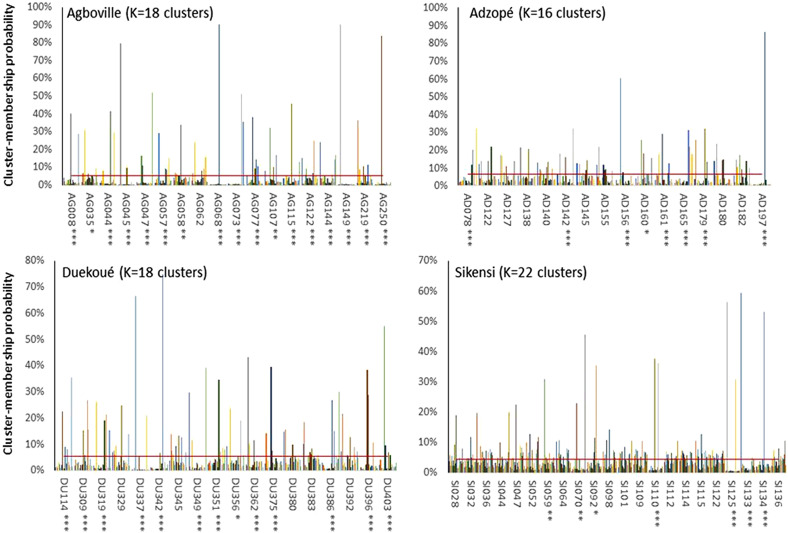
The analysis at the infrapopulation level revealed 16–22 clusters at all sites; reflecting the individual children sampled. The Structure analysis for each population with K = number of patients showed that no best K was found for all sampling sites. However, the clusters were not uniformly distributed among children and some miracidia were assigned to a restricted number of clusters (Fig. 5). This non-random repartition pattern was significant for 60.8% of the infrapopulations analysis (13/18 for Duekoué, 7/22 for Sikensi, 17/18 for Agboville and 8/16 for Adzopé). Supplementary Fig. S2 shows bar plots depicting the probability for each miracidium to belong to each cluster by child, using only those children who hosted at least 15 miracidia.
Figure 5.
Genetic clustering produced by Structure software for each population with K = number of patients. The patient codes are in abscises. *, ** and *** represent statistical difference from random repartition among the clusters at the 5%, 1% and 0.1% levels, respectively. The red line represents the null hypothesis of random repartition (1/K).
The analysis of molecular variance (AMOVA) showed that most of the miracidial genetic variance was found within sites (57.8% of the variance). There is more variance among miracidia by site rather than by host (10.4%) (Table 5).
Table 5.
Analysis of molecular variance (AMOVA) partitioning the total residual variance (after deducting the non-informative variance between microsatellite loci) observed at 16 microsatellite loci in 1966 miracidia between four hierarchical levels.
| Hierarchical level | df | Sum of squares | Variance component | Percentage of residual variation |
|---|---|---|---|---|
| Among sites | 3 | 25,659 | 7.83 | 31.8 |
| Among children within sites | 87 | 68,119 | 14.26 | 57.8 |
| Among miracidia within child | 1875 | 320,706 | 2.78 | 10.4 |
| Within miracidia (between microsatellite loci) | 1966 | 326,144 | 165.89 | – |
| Total | 3931 | 740,628 | 190.55 | – |
df: degrees of freedom.
Discussion
Our study showed a high proportion of S. haematobium × S. bovis hybrids among school-aged children in different parts of Côte d’Ivoire. Similar results were also reported from a large study in Senegal, where most children (88%) excreted hybrid miracidia [64]. Notably, virtually all children (97.8%) were producing miracidia with a hybrid genotype (i.e. a discrepancy between the mitochondrial and the nuclear signature and/or heterozygous nuclear profiles). Moreover, we found no genetic differentiation of miracidia presenting S. haematobium, S. bovis or hybrid genetic profiles, which indicates that all genotyped parasites most likely belong to a single genetic entity. This finding is in line with a recent genomic study indicating that most, if not all S. haematobium lineages across Africa, are likely to be introgressed to some extent with S. bovis [48].
We assigned all miracidia to six distinct cox1-ITS2 profiles, of which four were unambiguous hybrid profiles. Hybrids with a mitochondrial cox1 haplotype from S. bovis and homozygous ITS2 from S. haematobium (Sb × ShSh) were the most commonly identified type at 41.9%. Such hybrids have sometimes been documented in Senegal and ascribed to bidirectional introgressive hybridization [33, 63]. Beside this general pattern, significant variation was observed between sampling sites with the highest frequency of hybrids (reaching 79%) identified in Adzopé. Interestingly, the genetic contribution of the S. bovis genetic profile to the genetic make-up of schistosome populations is much more evident at the mitochondrial level than at the nuclear level. The frequency of miracidia with a heterozygous ITS2, i.e. carrying both S. bovis and S. haematobium ITS alleles, was relatively low, which could be explained by the concerted evolution of ITS2 gene region [25].
The S. bovis cox1 × S. bovis ITS2 genetic profile (SbSb) has also been reported in miracidia recovered from humans in Corsica [10]. We only detect 0.7% of this profile in our study. Importantly, this genetic profile using only two gene genotyping methods does not mean that these parasites are “pure” S. bovis parasites at the whole genome level. The genetic signature can only represent a certain introgression level between both species, and can explain some discrepancy in parasite phenotype such us the route of excretion expected to be in the feces for the S. bovis parasite.
Data on the genetic structure of S. haematobium populations are sparse [29, 30]. Previous findings based on a phylogeographic approach, using cox1 data, at the African continental scale suggested that S. haematobium is less structured than S. mansoni [63] and S. bovis [48]. In contrast, the significant differentiation between sampling sites and even within individual hosts reported here, suggests a genetic structure of miracidia at small geographical scales. The large geographical distance separating Duekoué in the western part of Côte d’Ivoire to the other three sites is likely to limit mixing between these populations and lead to genetic structuring between the schistosome populations. More surprising is the genetic structure observed at the individual level. In the majority of cases, each child hosted an infrapopulation different from the other children. Our finding does not corroborate the “genetic mixing bowl hypothesis” [16]. The authors reported that the long-living definitive hosts cumulate genotypes coming from several short-living infected intermediate host snails, which can lead to homogeneity of infrapopulations. The pattern found in our study could be explained by recent treatment of children by praziquantel one year before sample collection. In this scenario, children have only recently started to harbour parasites again and there was insufficient time for “mixing” after praziquantel treatment. This would homogenize parasite populations among children rather than promoting structuration. Children would therefore be infected with only one or a few pairs of parasites and they could also be highly exposed and have several infections. This clustering may be amplified by the genotypic specific immune responses. In a mouse model, it was found that a trickle infection (repeated light infections) can protect mice against a Schistosoma challenge infection [20]. Subsequently, the same authors have shown that this level of protection is genotype-dependent and correlated with genetic dissimilarity between the immunizing and the challenging infection [20]. The smaller the genetic distance between immunizing and challenging clones, the lower the infectivity rate of the challenging clones. This mechanism may, in turn, increase genetic diversity in each host and differentiate the parasite infrapopulation between hosts. No matter what the reasons are for this pattern, it emphasises the importance of investigating the parasites from as many different patients (or hosts in general), when investigating population genetics between different sampling sites. Increasing the number of hosts sampled, rather than the number of miracidia per host, has already been proposed after a stochastic re-sampling approach [24].
No genetic differentiation was observed according to the genetic profile. This result suggests that there are no genetic mating restrictions between the six different genetic profiles, and hence, all recombinations are possible [11, 27]. Similar findings have recently been reported from Senegal [11]. So, our results together with recent genomic studies [44, 48], call into question S. haematobium and whether forms of hybrids detected in humans thus far are truly different genomic entities.
The current study has some limitations, related to the reliability of the RFLP analysis, the absence of samples from animals, or the hybrid genotyping method. First, in our study we used an RFLP method based on a short 505 bp ITS2 fragment. Even though the results we obtained are robust, the RFLP method is known to be sensitive to different factors such as enzyme used, and the time or temperature of digestion. A new method for single-nucleotide polymorphism detection adapted to large scale screening needs to be developed. Second, miracidia from livestock or rodents were not collected. This is an important point to infer the possible zoonotic capacity of these hybrid pathogens. The situations are contrasted depending on the countries without current evidence of zoonotic transmission in Senegal [12] compared to rodent and cow reservoir hosts in Benin [54]. Finding a high frequency of hybrids in Côte d’Ivoire does not confirm zoonotic transmission and we advocate looking for the presence of hybrid schistosomes in rodents and livestock. Third, mito-nuclear genotyping methods are well adapted for large population screening; however, as previously mentioned, the whole genome cannot be resumed to these two gene assignations, and genomic studies using more genetic markers are desirable.
Conclusions
Our study investigated the genetic diversity and population structuring of schistosomes at the population level. Each child harboured a genetic cluster of schistosomes at the infrapopulation level, which could lead to putative resistant parasite selection due to drug pressure by mass drug administration (MDA). The high frequency of hybrids between S. haematobium × S. bovis observed can lead to difficulties in accurately diagnosing schistosome hybrids by conventional techniques using light microscopy. This could also have adverse consequences on schistosomiasis control towards elimination and negatively impact disease transmission. More studies are need on population genetics of schistosomes at the human and animal interface to evaluate the parasite’s gene flow.
Acknowledgments
We are grateful to the district health and village authorities for their support, to Dr. Jan Hattendorf for statistical support, and to the children and their parents/guardians for their commitment throughout the study. This work was supported financially by the Agence National de Recherche (ANR), which supports the HySWARM project (ANR-18-CE35-0001) within the framework of the “Laboratoire d’Excellence (LABEX)” TULIP program (ANR-10-LABX-41), and by the Swiss National Science Foundation (grant no. 31003A_170113 to JU, JZ and OB). EKA is a recipient of a Swiss Government Excellence Scholarship (ESKAS, grant no. 2017-0746) for which he is deeply grateful.
Cite this article as: Angora EK, Vangraefschepe A, Allienne J-F, Menan H, Coulibaly JT, Meïté A, Raso G,Winkler M, Yavo W, Toue AO, N’Goran EK, Zinsstag J, Utzinger J, Balmer O & Boissier J. 2022. Population genetic structure of Schistosoma haematobium and Schistosoma haematobium × Schistosoma bovis hybrids among school-aged children in Côte d’Ivoire. Parasite 29, 23.
Contributions of authors
EKA, HM, OB and JB conceived and designed the study. EKA, AV and JFA performed the molecular analyses. EKA, and JB performed statistical analysis. EKA wrote the first draft of the manuscript. JTC, WY, AOT, EKN, JZ, JU, OB and JB revised the manuscript. MW helped with figure resolution. All authors read and approved the final manuscript before submission and resubmission.
Conflict of interest
The authors declare no conflict of interest.
Supplementary materials
The Supplementary material of this article is available at https://www.parasite-journal.org/10.1051/parasite/2022023/olm.
Supplementary Tab. S1: Microsatellite dataset.
Supplementary Tab. S2: Total number of miracidia analyzed (total) and number (n) and percentage (%) of miracidia assigned to the six possible cox1/ITS2 profiles defined by cox1-PCR and ITS2-RFLP analysis per child and site.
Supplementary Fig. S1:
S1a: Diagram of the 505 bp fragment and the Mbo1 cut sites for both species, S1b: Banding patterns of the restriction fragment length polymorphism (RFLP) analysis by MboI of the nuclear ITS2 region of Schistosoma. Schistosoma haematobium exhibits four bands at 281, 98, 82 and 44 bp. Schistosoma bovis exhibits three bands at 379, 82 and 44 bp. Hybrids between S. haematobium × S. bovis exhibit bands at 379, 281, 98, 82 and 44 bp.
Supplementary Fig. S2:
Bar plots depicting the percentage for each miracidium belonging to each cluster by child, using children hosting at least 15 miracidia in Adzopé (ADZ), Agboville (AGB), Duekoué (DUE), and Sikensi (SIK). This was done in Structure and each site K was set equal to the number of children.
References
- 1.Achi YL, Zinsstag J, Yéo N, Dea V, Dorchies PH. 2003. Les nématodes gastro-intestinaux des bovins de la région des savanes de la Côte-d’Ivoire : enquête d’abattoir. Revue de Médecine Vétérinaire, 154, 105–112. [Google Scholar]
- 2.Aemero M, Boissier J, Climent D, Moné H, Mouahid G, Berhe N, Erko B. 2015. Genetic diversity, multiplicity of infection and population structure of Schistosoma mansoni isolates from human hosts in Ethiopia. BMC Genetics, 16, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agola LE, Steinauer ML, Mburu DN, Mungai BN, Mwangi IN, Magoma GN, Loker ES, Mkoji GM. 2009. Genetic diversity and population structure of Schistosoma mansoni within human infrapopulations in Mwea, central Kenya assessed by microsatellite markers. Acta Tropica, 111, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson TJC, LoVerde PT, Le Clec’h W, Chevalier FD. 2018. Genetic crosses and linkage mapping in schistosome parasites. Trends in Parasitology, 34, 982–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angora EK, Boissier J, Menan H, Rey O, Tuo K, Touré AO, Coulibaly JT, Méité A, Raso G, N’Goran EK, Utzinger J, Balmer O. 2019. Prevalence and risk factors for schistosomiasis among schoolchildren in two settings of Côte d’Ivoire. Tropical Medicine and Infectious Disease, 4, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angora EK, Allienne J-F, Rey O, Menan H, Touré AO, Coulibaly JT, Raso G, Yavo W, N’Goran EK, Utzinger J, Balmer O, Boissier J. 2020. High prevalence of Schistosoma haematobium × Schistosoma bovis hybrids in schoolchildren in Côte d’Ivoire. Parasitology, 147, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Assaré RK, Lai Y-S, Yapi A, Tian-Bi Y-NT, Ouattara M, Yao PK, Knopp S, Vounatsou P, Utzinger J, N’Goran EK. 2015. The spatial distribution of Schistosoma mansoni infection in four regions of western Côte d’Ivoire. Geospatial Health, 10, 345. [DOI] [PubMed] [Google Scholar]
- 8.Belkhir K. 2004. GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. http://www.genetix.univ-montp2.fr/genetix/genetix.htm [Google Scholar]
- 9.Berry A, Paris L, Boissier J, Caumes E. 2016. Schistosomiasis screening of travelers to Corsica, France. Emerging Infectious Diseases, 22, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boissier J, Grech-Angelini S, Webster BL, Allienne J-F, Huyse T, Mas-Coma S, Toulza E, Barré-Cardi H, Rollinson D, Kincaid-Smith J, Oleaga A, Galinier R, Foata J, Rognon A, Berry A, Mouahid G, Henneron R, Moné H, Noel H, Mitta G. 2016. Outbreak of urogenital schistosomiasis in Corsica (France): an epidemiological case study. Lancet Infectious Diseases, 16, 971–979. [DOI] [PubMed] [Google Scholar]
- 11.Boon NAM, Mbow M, Paredis L, Moris P, Sy I, Maes T, Webster BL, Sacko M, Volckaert FAM, Polman K, Huyse T. 2019. No barrier breakdown between human and cattle schistosome species in the Senegal River Basin in the face of hybridisation. International Journal for Parasitology, 49, 1039–1048. [DOI] [PubMed] [Google Scholar]
- 12.Borlase A, Rudge JW, Léger E, Diouf ND, Fall CB, Diop SD, Catalano S, Sène M, Webster JP. 2021. Spillover, hybridization, and persistence in schistosome transmission dynamics at the human–animal interface. Proceedings of the National Academy of Sciences of the United States of America, 118, e2110711118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chammartin F, Houngbedji CA, Hürlimann E, Yapi RB, Silué KD, Soro G, Kouamé FN, N’Goran EK, Utzinger J, Raso G, Vounatsou P. 2014. Bayesian risk mapping and model-based estimation of Schistosoma haematobium-Schistosoma mansoni co-distribution in Côte d’Ivoire. PLoS Neglected Tropical Diseases, 8, e3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colley DG, Bustinduy AL, Secor WE, King CH. 2014. Human schistosomiasis. Lancet, 383, 2253–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulibaly JT, N’Gbesso YK, N’Guessan NA, Winkler MS, Utzinger J, N’Goran EK. 2013. Epidemiology of schistosomiasis in two high-risk communities of south Côte d’Ivoire with particular emphasis on pre-school–aged children. American Journal of Tropical Medicine and Hygiene, 89, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curtis J, Minchella DJ. 2000. Schistosome population genetic structure: when clumping worms is not just splitting hairs. Parasitology Today, 16, 68–71. [DOI] [PubMed] [Google Scholar]
- 17.Depaquit J, Akhoundi M, Haouchine D, Mantelet S, Izri A. 2019. No limit in interspecific hybridization in schistosomes: observation from a case report. Parasite, 26, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djuikwo-Teukeng FF, Kouam Simo A, Allienne J-F, Rey O, Njayou Ngapagna A, Tchuem-Tchuenté LA, Boissier J. 2019. Population genetic structure of Schistosoma bovis in Cameroon. Parasites & Vectors, 12, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djuikwo Teukeng FF, Biln M, Bech N, Reguera-Gomez M, Zein-Eddine R, Kouamé-Simo AM, Allienne J-F, Tchuem-Tchuenté LA, Boissier J. 2022. Hybridization increases genetic diversity in Schistosoma haematobium populations infecting humans in Cameroon. Infectious Diseases of Poverty, 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumont M, Moné H, Mouahid G, Idris MA, Shaban M, Boissier J. 2007. Influence of pattern of exposure, parasite genetic diversity and sex on the degree of protection against reinfection with Schistosoma mansoni. Parasitology Research, 101, 247–252. [DOI] [PubMed] [Google Scholar]
- 21.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- 22.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, 10, 564–567. [DOI] [PubMed] [Google Scholar]
- 23.Francis RM. 2017. Pophelper: an R package and web app to analyse and visualize population structure. Molecular Ecology Resources, 17, 27–32. [DOI] [PubMed] [Google Scholar]
- 24.French MD, Churcher TS, Basáñez M-G, Norton AJ, Lwambo NJS, Webster JP. 2013. Reductions in genetic diversity of Schistosoma mansoni populations under chemotherapeutic pressure: the effect of sampling approach and parasite population definition. Acta Tropica, 128, 196–205. [DOI] [PubMed] [Google Scholar]
- 25.Ganley ARD, Kobayashi T. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Research, 17, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GBD 2017 DALYs, HALE Collaborators. 2018. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet, 392, 1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorton MJ, Kasl EL, Detwiler JT, Criscione CD. 2012. Testing local-scale panmixia provides insights into the cryptic ecology, evolution, and epidemiology of metazoan animal parasites. Parasitology, 139, 981–997. [DOI] [PubMed] [Google Scholar]
- 28.Goudet J, Perrin N, Waser P. 2002. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology, 11, 1103–1114. [DOI] [PubMed] [Google Scholar]
- 29.Gower CM, Gabrielli AF, Sacko M, Dembelé R, Golan R, Emery AM, Rollinson D, Webster JP. 2011. Population genetics of Schistosoma haematobium: development of novel microsatellite markers and their application to schistosomiasis control in Mali. Parasitology, 138, 978–994. [DOI] [PubMed] [Google Scholar]
- 30.Gower CM, Gouvras AN, Lamberton PHL, Deol A, Shrivastava J, Mutombo PN, Mbuh JV, Norton AJ, Webster BL, Stothard JR, Garba A, Lamine MS, Kariuki C, Lange CN, Mkoji GM, Kabatereine NB, Gabrielli AF, Rudge JW, Fenwick A, Sacko M, Dembelé R, Lwambo NJS, Tchuem Tchuenté L-A, Rollinson D, Webster JP. 2013. Population genetic structure of Schistosoma mansoni and Schistosoma haematobium from across six sub-Saharan African countries: implications for epidemiology, evolution and control. Acta Tropica, 128, 261–274. [DOI] [PubMed] [Google Scholar]
- 31.Hotez PJ, Alvarado M, Basáñez M-G, Bolliger I, Bourne R, Boussinesq M, Brooker SJ, Brown AS, Buckle G, Budke CM, Carabin H, Coffeng LE, Fèvre EM, Fürst T, Halasa YA, Jasrasaria R, Johns NE, Keiser J, King CH, Lozano R, Murdoch ME, O’Hanlon S, Pion SDS, Pullan RL, Ramaiah KD, Roberts T, Shepard DS, Smith JL, Stolk WA, Undurraga EA, Utzinger J, Wang M, Murray CJL, Naghavi M. 2014. The Global Burden of Disease Study 2010: interpretation and implications for the neglected tropical diseases. PLoS Neglected Tropical Diseases, 8, e2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell DC. 2007. Statistical methods for psychology, 6th edn. Australia, Belmont, CA: Thomson/Wadsworth. [Google Scholar]
- 33.Huyse T, Webster BL, Geldof S, Stothard JR, Diaw OT, Polman K, Rollinson D. 2009. Bidirectional introgressive hybridization between a cattle and human schistosome species. PLoS Pathogens, 5, e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huyse T, Van den Broeck F, Hellemans B, Volckaert FAM, Polman K. 2013. Hybridisation between the two major African schistosome species of humans. International Journal for Parasitology, 49, 687–689. [DOI] [PubMed] [Google Scholar]
- 35.IARC. 2012. Review of human carcinogens: biological agents. International Agency for Research on Cancer, World Health Organization: Geneva. [Google Scholar]
- 36.Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. [DOI] [PubMed] [Google Scholar]
- 37.Kebede T, Bech N, Allienne J-F, Olivier R, Erko B, Boissier J. 2020. Genetic evidence for the role of non-human primates as reservoir hosts for human schistosomiasis. PLoS Neglected Tropical Diseases, 14, e0008538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King KC, Stelkens RB, Webster JP, Smith DF, Brockhurst MA. 2015. Hybridization in parasites: consequences for adaptive evolution, pathogenesis, and public health in a changing world. PLoS Pathogens, 11, e1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouadio JN, Giovanoli Evack J, Achi LY, Fritsche D, Ouattara M, Silué KD, Bonfoh B, Hattendorf J, Utzinger J, Zinsstag J, Balmer O, N’Goran EK. 2020. Prevalence and distribution of livestock schistosomiasis and fascioliasis in Côte d’Ivoire: results from a cross-sectional survey. BMC Veterinary Research, 16, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai Y-S, Biedermann P, Ekpo UF, Garba A, Mathieu E, Midzi N, Mwinzi P, N’Goran EK, Raso G, Assaré RK, Sacko M, Schur N, Talla I, Tchuem-Tchuenté LA, Touré S, Winkler MS, Utzinger J, Vounatsou P. 2015. Spatial distribution of schistosomiasis and treatment needs in sub-Saharan Africa: a systematic review and geostatistical analysis. Lancet Infectious Diseases, 15, 927–940. [DOI] [PubMed] [Google Scholar]
- 41.Le Govic Y, Kincaid-Smith J, Allienne J-F, Rey O, de Gentile L, Boissier J. 2019. Schistosoma haematobium-Schistosoma mansoni hybrid parasite in migrant boy, France, 2017. Emerging Infectious Diseases, 25, 365–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leger E, Webster JP. 2017. Hybridizations within the genus Schistosoma: implications for evolution, epidemiology and control. Parasitology, 144, 65–80. [DOI] [PubMed] [Google Scholar]
- 43.Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P. 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4, 535–538. [Google Scholar]
- 44.Platt RN, McDew-White M, Clec’h WL, Chevalier FD, Allan F, Emery AM, Garba A, Hamidou AA, Ame SM, Webster JP, Rollinson D, Webster BL, Anderson TJC. 2019. Ancient hybridization and adaptive introgression of an invadolysin gene in schistosome parasites. Molecular Biology and Evolution, 36, 2127–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quan J-H, Choi I-W, Ismail HAHA, Mohamed AS, Jeong H-G, Lee J-S, Hong S-T, Yong T-S, Cha G-H, Lee Y-H. 2015. Genetic diversity of Schistosoma haematobium eggs isolated from human urine in Sudan. Korean Journal of Parasitology, 53, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raso G, Matthys B, N’Goran EK, Tanner M, Vounatsou P, Utzinger J. 2005. Spatial risk prediction and mapping of Schistosoma mansoni infections among schoolchildren living in western Côte d’Ivoire. Parasitology, 131, 97–108. [DOI] [PubMed] [Google Scholar]
- 48.Rey O, Toulza E, Chaparro C, Allienne J-F, Kincaid-Smith J, Mathieu-Begné E, Allan F, Rollinson D, Webster BL, Boissier J. 2021. Diverging patterns of introgression from Schistosoma bovis across S. haematobium African lineages. PLoS Pathogens, 17, e1009313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rey O, Webster BL, Huyse T, Rollinson D, Van den Broeck F, Kincaid-Smith J, Onyekwere A, Boissier J. 2021. Population genetics of African Schistosoma species. Infection, Genetics and Evolution, 89, 104727. [DOI] [PubMed] [Google Scholar]
- 50.Rice WR. 1989. Analyzing tables of statistical tests. Evolution, 43, 223–225. [DOI] [PubMed] [Google Scholar]
- 51.Rollinson D, Kaukas A, Johnston DA, Simpson AJG, Tanaka M. 1997. Some molecular insights into schistosome evolution. International Journal for Parasitology, 27, 11–28. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg NA. 2004. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138. [Google Scholar]
- 53.Rousset F. 2008. A complete re-implementation of the GENEPOP software for software for teaching and research. Molecular Ecolology Resources, 8, 103–106. [DOI] [PubMed] [Google Scholar]
- 54.Savassi BAES, Mouahid G, Lasica C, Mahaman S-DK, Garcia A, Courtin D, Allienne J-F, Ibikounlé M, Moné H. 2020. Cattle as natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 × Schistosoma bovis Sonsino, 1876 interactions, with new cercarial emergence and genetic patterns. Parasitology Research, 119, 2189–2205. [DOI] [PubMed] [Google Scholar]
- 55.Savassi BAES, Dobigny G, Etougbétché JR, Avocegan TT, Quinsou FT, Gauthier P, Ibikounlé M, Moné H, Mouahid G. 2021. Mastomys natalensis (Smith, 1834) as a natural host for Schistosoma haematobium (Bilharz, 1852) Weinland, 1858 × Schistosoma bovis Sonsino, 1876 introgressive hybrids. Parasitology Research, 120, 1755–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soumahoro MK, Bosson-Vanga AH, Coulibaly KJ, Abbes S, Angora E, Kouadio K, N’Douba AK, Sissoko D, Dosso M. 2014. Investigation d’un foyer épidémique de bilharziose urinaire dans l’école primaire du village de Guébo 2, Abidjan, Côte d’Ivoire. Bulletin de la Societé de Pathologie Exotique, 107, 185–187. [DOI] [PubMed] [Google Scholar]
- 57.Stothard JR, Kabatereine NB, Tukahebwa EM, Kazibwe F, Rollinson D, Mathieson W, Webster JP, Fenwick A. 2006. Use of circulating cathodic antigen (CCA) dipsticks for detection of intestinal and urinary schistosomiasis. Acta Tropica, 97, 219–228. [DOI] [PubMed] [Google Scholar]
- 58.Tian-Bi Y-NT, Webster B, Konan CK, Allan F, Diakité NR, Ouattara M, Salia D, Koné A, Kakou AK, Rabone M, Coulibaly JT, Knopp S, Meïté A, Utzinger J, N’Goran EK, Rollinson D. 2019. Molecular characterization and distribution of Schistosoma cercariae collected from naturally infected bulinid snails in northern and central Côte d’Ivoire. Parasites & Vectors, 12, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Utzinger J, N’Goran EK, Tanner M, Lengeler C. 2000. Simple anamnestic questions and recalled water-contact patterns for self-diagnosis of Schistosoma mansoni infection among schoolchildren in western Côte d’Ivoire. American Journal of Tropical Medicine and Hygiene, 62, 649–655. [DOI] [PubMed] [Google Scholar]
- 60.Utzinger J, Becker SL, van Lieshout L, van Dam GJ, Knopp S. 2015. New diagnostic tools in schistosomiasis. Clinical Microbiology and Infection, 21, 529–542. [DOI] [PubMed] [Google Scholar]
- 61.Van den Broeck F, Geldof S, Polman K, Volckaert FAM, Huyse T. 2011. Optimal sample storage and extraction procotols for reliable multilocus genotyping of the human parasite Schistosoma mansoni. Infection, Genetics and Evolution, 11, 1413–1418. [DOI] [PubMed] [Google Scholar]
- 62.Webster BL, Emery AM, Webster JP, Gouvras A, Garba A, Diaw O, Seye MM, Tchuenté LAT, Simoonga C, Mwanga J, Lange C, Kariuki C, Mohammed KA, Stothard JR, Rollinson D. 2012. Genetic diversity within Schistosoma haematobium: DNA barcoding reveals two distinct groups. PLoS Neglected Tropical Diseases, 6, e1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webster BL, Culverwell CL, Khamis IS, Mohammed KA, Rollinson D, Stothard JR. 2013. DNA barcoding of Schistosoma haematobium on Zanzibar reveals substantial genetic diversity and two major phylogenetic groups. Acta Tropica, 128, 206–217. [DOI] [PubMed] [Google Scholar]
- 64.Webster BL, Diaw OT, Seye MM, Webster JP, Rollinson D. 2013. Introgressive hybridization of Schistosoma haematobium group species in Senegal: species barrier break down between ruminant and human schistosomes. PLoS Neglected Tropical Diseases, 7, e2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Webster BL, Rabone M, Pennance T, Emery AM, Allan F, Gouvras A, Knopp S, Garba A, Hamidou AA, Mohammed KA, Ame SM, Rollinson D, Webster JP. 2015. Development of novel multiplex microsatellite polymerase chain reactions to enable high-throughput population genetic studies of Schistosoma haematobium. Parasites & Vectors, 8, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster BL, Alharbi MH, Kayuni S, Makaula P, Halstead F, Christiansen R, Juziwelo L, Stanton MC, LaCourse EJ, Rollinson D, Kalua K, Stothard JR. 2019. Schistosome interactions within the Schistosoma haematobium group, Malawi. Emerging Infectious Diseases, 25, 1245–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38, 1358–1370. [DOI] [PubMed] [Google Scholar]
- 68.World Health Organization. 2022. Schistosomiasis fact sheet. https://www.who.int/news-room/fact-sheets/detail/schistosomiasis.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary material of this article is available at https://www.parasite-journal.org/10.1051/parasite/2022023/olm.
Supplementary Tab. S1: Microsatellite dataset.
Supplementary Tab. S2: Total number of miracidia analyzed (total) and number (n) and percentage (%) of miracidia assigned to the six possible cox1/ITS2 profiles defined by cox1-PCR and ITS2-RFLP analysis per child and site.
Supplementary Fig. S1:
S1a: Diagram of the 505 bp fragment and the Mbo1 cut sites for both species, S1b: Banding patterns of the restriction fragment length polymorphism (RFLP) analysis by MboI of the nuclear ITS2 region of Schistosoma. Schistosoma haematobium exhibits four bands at 281, 98, 82 and 44 bp. Schistosoma bovis exhibits three bands at 379, 82 and 44 bp. Hybrids between S. haematobium × S. bovis exhibit bands at 379, 281, 98, 82 and 44 bp.
Supplementary Fig. S2:
Bar plots depicting the percentage for each miracidium belonging to each cluster by child, using children hosting at least 15 miracidia in Adzopé (ADZ), Agboville (AGB), Duekoué (DUE), and Sikensi (SIK). This was done in Structure and each site K was set equal to the number of children.