Abstract
Background
Colorectal cancer (CRC) is the third leading cause of cancer in the United States, and inflammatory bowel disease patients have an increased risk of developing CRC due to chronic intestinal inflammation with it being the cause of death in 10% to 15% of inflammatory bowel disease patients. TIPE2 (TNF-alpha-induced protein 8-like 2) is a phospholipid transporter that is highly expressed in immune cells and is an important regulator of immune cell function.
Methods
The azoxymethane/dextran sulfate sodium murine model of colitis-associated colon cancer (CAC) was employed in Tipe2 –/– and wild-type mice, along with colonoid studies, to determine the role of TIPE2 in CAC.
Results
Early on, loss of TIPE2 led to significantly less numbers of visible tumors, which was in line with its previously described role in myeloid-derived suppressor cells. However, as time went on, loss of TIPE2 promoted tumor progression, with larger tumors appearing in Tipe2 –/– mice. This was associated with increased interleukin-22/STAT3 phosphorylation signaling. Similar effects were also observed in primary colonoid cultures, together demonstrating that TIPE2 also directly regulated colonocytes in addition to immune cells.
Conclusions
This work demonstrates that TIPE2 has dual effects in CAC. In the colonocytes, it works as a tumor suppressor. However, in the immune system, TIPE2 may promote tumorigenesis through suppressor cells or inhibit it through IL-22 secretion. Going forward, this work suggests that targeting TIPE2 for CRC therapy requires cell- and pathway-specific approaches and serves as a cautionary tale for immunotherapy approaches in general in terms of colon cancer, as intestinal inflammation can both promote and inhibit cancer.
Keywords: colitis-associated colon cancer, tumor suppressor, TIPE2, IL-22
Introduction
Colorectal cancer (CRC) is the third leading cause of cancer in the United States, responsible for 8% of all cancer deaths.1 Inflammatory bowel disease (IBD) patients have an increased risk of developing CRC due to chronic intestinal inflammation; CRC is the cause of death in 10% to 15% of IBD patients.2 Spontaneous CRC and colitis-associated colon cancer (CAC) share significant pathophysiology, including the same set of mutations (ie, p53, K-Ras, and APC), although the mutation order is different in CAC.3 Murine CAC has high histological and genetic homology with both human CAC and spontaneous CRC3; thus, murine CAC models are widely used to study CRC.4 Like other cancers, CRC can evade immune system killing by upregulating inhibitory immune checkpoints, leading to an immunosuppressive microenvironment.5 Recent clinical successes have overcome these escape mechanisms, such as the use of PD-1 checkpoint inhibitors (including in CRC),5,6 leading to a focus identifying immune cell populations that support this immunosuppressive environment and the regulation of these immune cell populations, notably including myeloid-derived suppressor cells (MDSCs)7 and regulatory T cells8 in CRC.
TIPE2 (TNF-alpha-induced protein 8-like 2 or TNFAIP8L2) is a phospholipid transporter9-11 that is highly expressed in immune cells.12 TIPE2 is a complex regulator of immune cell function; its loss results in increased inflammatory cytokine production and T cell proliferation, differentiation, and activation12 and its knockout enhances macrophage phagocytosis and oxidative burst.13 Its loss also results in MDSC dysfunction.14 TIPE2 is also required for proper regulatory T cell function.15,16 In the Sv129 background, this even leads to spontaneous inflammation as the mice age.12 Perhaps most critically, loss of TIPE2 results in deficits in immune cell chemotaxis,10,11 leading to diminished murine acute dextran sodium sulfate (DSS) colitis17 and experimental autoimmune encephalitis,11,12 but with an increased susceptibility to infection.18,19 Together, TIPE2 is viewed as a critical regulator of immune cell function, with both inhibitory and excitatory effects, depending on the cell type and function being studied.20
Perhaps unsurprisingly, given its critical role in the immune system, TIPE2 has been associated with multiple types of cancer.20 In the gut, RNA levels of TIPE2 are increased in human CRC tissue, and expression correlates with disease severity,21 but its loss protects against acute DSS colitis,17 suggesting that the association with CRC is immune driven. More globally, TIPE2 has been identified in genome-wide association studies as being associated with IBD, likely through its immunoregulatory functions.22 Here, we utilize the azoxymethane (AOM)/DSS model of murine CAC (also known as AOM/DSS CAC) to uncover the role of TIPE2 in the initiation and progression of CAC.
Methods
Animals
Wild-type (WT) C57BL/6 (B6) and Tipe2–/– B6 mice were bred in house. Tipe2–/– mice were previously generated, as described elsewhere.12 All mice used in this study were housed under pathogen-free conditions in the University of Pennsylvania Animal Care Facilities. All animal protocols used were preapproved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Colitis-Associated Colon Cancer
To induce colon tumors, 8- to 12-week-old, age-matched, female23 mice were first given 10mg/kg AOM (Sigma-Aldrich, St. Louis, MO) intraperitoneally. One week later, experimental colitis was induced by adding Affymetrix brand 2.5% DSS (Thermo Fisher Scientific, Waltham, MA) drinking water for 7 days, followed by 14 days of water. Two or 3 more cycles of 7 days of DSS followed by 14 days of water were then administered. DSS concentration was 2.5%, or reduced to 2% if the weight loss of the mice became excessive enough to jeopardize their health (defined as >15% initial body weight loss). If DSS concentration was reduced, it was reduced for all mice in that study, regardless of genotype or cage that they were housed in. Weight, presence of bloody stool, and body posturing were tracked to assess for the progression of disease. At the end of the experiment, mice were sacrificed and tissues were processed for subsequent experiments. Distal colonic samples were snap frozen in dry ice for subsequent RNA and protein analyses or collected as described subsequently for other analyses.
Histopathological Scoring
Colon samples were Swiss-rolled, fixed in 10% neutral buffered formalin (Thermo Fisher Scientific) overnight at 4°C, and transferred to 70% ethanol before paraffin embedding and subsequent staining. Damage severity was evaluated using hematoxylin and eosin–stained sections by blinded investigators using a scoring system from 0 to 40 that accounts for epithelial damage and immune infiltration, as previously described.24 For evaluation of neoplasia, blinded scoring was done in house and verified with the PennVet Comparative Pathology Score. In both cases, proliferative lesions classification was done according to the original criteria of the Mouse Models of Human Cancers Consortium25 with subsequent updates of nomenclature and grading scheme.26
RNA Isolation and Real-Time Polymerase Chain Reaction
RNA was isolated using TRIzol (Thermo Fisher Scientific), followed by a Qiagen RNeasy kit (Qiagen, Valencia, CA), per manufacturer protocol, and reverse transcribed to complementary DNA with a high-capacity reverse transcription kit (Applied Biosystems, Foster City, CA). Real-time polymerase chain reaction (PCR) was performed using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) or a Quantiflex 12X Real-Time PCR system (Thermo Fisher Scientific). Relative fold changes were determined using the ΔΔCT calculation method. Values were normalized to the internal control, 18s ribosomal RNA, that was amplified using QuantiTect Primers (Qiagen). Primers that were manufactured by Integrated DNA Technologies (Coralville, Iowa) are as follows: Il6 (5ʹ-CGGAGGCTTAATTACACATGTT-3ʹ and 5ʹ-CTGGCTTTGTCTTTCTTGTTATC-3ʹ), Il1b (5ʹ-GCCCATCCTCTGTGACTCAT-3ʹ and 5ʹ-AGGCCACAGGTATTTTGTCG-3ʹ), TNF (5ʹ-ATGAGCACAGAAAGCATGATC-3ʹ and 5ʹ- TACAGGCTTGTCACTCGAATT-3ʹ), and Il22 (5ʹ-AGAAGGCTGAAGGAGACAGT-3ʹ and 5ʹ-GACATAAACAGCAGGTCCAGTT-3ʹ). Additional QuantiTect Primers include Itgae, Il23r, Il23, Tnfaip8l2, and Ptprc.
For these studies, distal colonic tissue samples were collected at the time of animal sacrifice and flash frozen in dry ice. For lamina propria and enterocyte tissue and tumor fractions, cells were isolated as described in the flow cytometry protocol, and then immediately placed in TRIzol.
Colonocytes were isolated from freshly harvested colons as previously described.27 For tumor tissue isolation, individual tumors were micro-dissected before the enterocyte preparation was performed.
Serum Cytokines
Serum cytokine levels were analyzed by Luminex Assay (Thermo Fisher Scientific) by the University of Pennsylvania’s Human Immunology Core according to manufacturer’s protocol, or via sandwich enzyme-linked immunosorbent assay (ELISA). For the sandwich ELISA, serum samples were collected and stored at -80°C before analysis. Purified and biotinylated anti-mouse interleukin (IL)-6 (Cat# 504504, 504601), IL-22 (Cat# 516402, 516407), and tumor necrosis factor α (Cat# 506302, 506311) antibodies used in ELISA were purchased from BioLegend (San Diego, CA). Quantitative ELISA was performed according to the manufacturer’s instruction. Cytokine content was presented as pg/mL.
Colonic Explant Cultures
Colonic explant studies were performed as previously described.28 Briefly, colon segments from mice at the 70-day point of our AOM/DSS protocol were sacrificed and their colons were collected and flushed with phosphate-buffered saline (PBS) to remove fecal contents, opened lengthwise, and shaken vigorously for 30 minutes in PBS. Tissue was then apportioned to wells (50-100mg of tissue per well) of a 24-well tissue culture plate (Corning, Corning, NY) and cultured in 1mL of complete RPMI 1640 medium containing 5% heat-inactivated fetal bovine serum, penicillin, streptomycin, and amphotericin B (all from Thermo Fisher Scientific/Invitrogen [Carlsbad, CA]). Tissues were incubated at 37 °C for 18 hours, and supernatants were collected and stored at -20°C until being assayed. Excreted cytokines were evaluated by sandwich ELISA, as described previously.
Paraffin-Embedded Immunohistochemistry and Immunofluorescence
Paraffin-embedded tissue sections were prepared according to the manufacturer’s protocol for the corresponding primary antibody, and primary antibody concentrations were per the manufacturer’s datasheet. An appropriate Vectakit Elite ABC (anti-rabbit or anti-mouse; Vector Laboratories, Burlingame, CA) was then used per manufacturer’s protocol. DAB reagent was from Dako (Agilent, Santa Clara, CA). Slides were then counterstained with hematoxylin (Gill No. 3 strength; Thermo Fisher Scientific). Slides were washed and mounted with VECTASHIELD Hardset Antifade Mounting Medium (Vector Laboratories).
Primary immunohistochemical antibodies were anti-pSTAT3Y705 (9145; Cell Signaling Technology, Danvers, MA) and anti-Ki67 (ab16667; Abcam, Cambridge, England).
For quantification, 3 random images or slides were always quantified. For STAT3 phosphorylation (pSTAT3) staining, the number of crypt units with >50% of enterocytes staining positive was quantified per 10 crypt lengths. For Ki-67 staining, the length of the proliferative zone was quantified for each crypt unit for all longitudinally sectioned crypts within each image.
Flow Cytometry
Colons were dissected from mice subjected to 91 days of the AOM/DSS CAC protocol. Tumors were then micro-dissected out from the tissue and pooled. Epithelial cells were sloughed off as previously described,27 with 45 minutes of shaking in Hank’s balanced salt solution buffer containing 10mM EDTA at 4 °C, followed by multiple rounds of agitation with a vortexer. Afterward, the epithelial cells were collected and the remaining tissue was cut into small pieces and digested with 100 U/mL Collagenase D (Roche/Sigma-Aldrich, St. Louis, MO), 0.1mg/mL DNase (STEMCELL Technologies, Vancouver, Canada), and 0.5mg/mL Dispase (STEMCELL Technologies) for 1 hour in RPMI 1640 media with 5% fetal bovine serum (Thermo Fisher Scientific). The resulting slurry was strained and then washed twice with PBS, before being resuspended in fluorescence-activated cell sorting (FACS) buffer (2% bovine serum albumin in PBS). These single-cell suspensions were then incubated for 20 minutes with fluorescence-labeled antibodies with the different combinations of the following antibodies: CD45.2, CD11b, CD3, CD8a, CD8b, CD11c, and Gr1 (BioLegend) and CD4 and Live/Dead Aqua (Invitrogen). Flow cytometry was conducted using FACS CantoE (BD Biosciences, Franklin Lakes, NJ). All data were analyzed with FlowJo software (version 10.0.7; FlowJo, Ashland, OR). The gating strategy is shown in Figure S3, with initial size-gating to select lymphocytes, followed by live-dead die selection, CD45-selection, and then gating on the other markers as listed previously.
RNAScope
smFISH (RNAScope; ACDBio, Newark, CA) was performed according to the manufacturer’s recommendations, using their premade probes to Tnfaip8l2 and Ptrpc. Signal was quantified using automated calculation of local maxima in FIJI (version 1.51w; National Institutes of Health, Bethesda, MD). Images were captured on a Zeiss 810 (Carl Zeiss, Oberkochen, Germany). To overcome image heterogeneity, 2 or more images per mouse were quantified and averaged (5 images per mouse on average, unless sectioning limitations dictated otherwise).
Mutant Colonoid Cultures
The colonic crypts from a C57BL/6J mouse (The Jackson Laboratory, Bar Harbor, ME; stock no: 000664) were extracted and cultured as previously described in Sato et al.29 The medium is Advanced Dulbecco’s modified Eagle medium/F12 (Thermo Fisher Scientific; 12634-010), Glutamax (Gibco/Thermo Fisher Scientific; 35050-061), 100 U/mL penicillin/streptomycin (Invitrogen/Thermo Fisher Scientific; 15140-122), 1mM N-acetyl cysteine (Sigma Aldrich; A9165-5g), B27 supplement (Invitrogen/Thermo Fisher Scientific; 17504-044), N2 supplement (Invitrogen/Thermo Fisher Scientific; 17502-048), 50ng/mL mouse epidermal growth factor (PeproTech, Cranbury, NJ; 315-09), 100ng/mL mouse Noggin (PeproTech; 250-38), and 1 µg/mL mouse R-spondin-1 (PeproTech, 315-32), 30ng/mL Recombinant Mouse Wnt-3a Protein (R&D Systems, Minneapolis, MN; 1324-WN).
The APC, P53 mutations were introduced by CRISPR-Cas9 editing. First, we generated CRISPR-Cas9 single guide RNAs (sgRNAs) targeting the APC and P53 tumor suppressor genes. Specifically, sgRNAs of APC, P53 were cloned into PX330 plasmid (Addgene, Watertown, MA; #42230) and transiently transfected separately or together into normal organoid. Two days after the transient transfection, the organoid with APC mutations were selected by removing Wnt3a and R-spondin1 from the medium, while the organoid with p53 mutations was selected by adding Nutlin-3 (10 µM/mL; Cayman Chemical Company, Ann Arbor, MI; 10004372-1). Ten subclones were picked from the engineered bulk tumoroids, and conditional PCR and Sanger sequencing (provided by the University of Pennsylvania sequencing core) were used to verify the mutations in each subclone. The subclones with verified APC or P53 mutations were pooled and used for downstream experiments. The sequences of the sgRNAs used for CRISPR-Cas9 editing and the sequences of mutated genes are presented in Figure S2.
Lentiviral Vector Construction and Infection
The shCtrl (5ʹCAACAAGATGAAGAGCACCAA-3ʹ) and shTipe2 sequence (5ʹ-GCTCAAAGAGTCTGGCACTAC-3ʹ) were inserted into the AgeI/EcoR I sites in the pLKO_TRC005 vector and were verified by nucleotide sequencing (University of Pennsylvania sequencing core). short hairpin RNA selection was performed using the RNAIDesigner tool from Thermo Fisher Scientific. Lentivirus packaging and infection were performed according to standard protocols as recommended by the manufacturer (Zhang Lab, Broad Institute, Cambridge, MA). ∆A and ∆AP colonoids were infected with 1×107 lentivirus transducing units in the presence of 5 µg/mL polybrene (Sigma-Aldrich). To stably select infected cells, puromycin (10 µg/mL; Invitrogen) were added 72 hours after transfection, for 48 hours. The expression level of TIPE2 was identified by Western blot (see the following and Figure S2F for results).
Western Blot
Cells were lysed in ice-cold lysis buffer (Thermo Fisher Scientific) with protease and phosphatase inhibitor (Roche). Equal amounts of total cell lysate (50 µg) were run on 4% to 12% gradient sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. The membranes were probed overnight with primary antibody. TNFAIP8L2 was detected using anti-TNFAIP8L2 (15940-1-AP; Proteintech Group, Chicago, IL) with anti-β-actin (3700; Cell Signaling Technology) used for loading control. After incubation, membranes were washed with Tris-buffered saline with 0.1% Tween 20 solution and incubated with the appropriate horseradish peroxidase–conjugated secondary antibody (Sigma-Aldrich) at 1:3000 dilution. Membranes were analyzed with Femto ECL reagent (Thermo Fisher Scientific) using a LI-COR Odyssey imaging system (LI-COR Biosciences, Lincoln, NE).
Colonoid Proliferation Assays
∆A and ∆AP colonoids transfected with either shCtrl or ShTipe2 were digested into single cells using TrypLE Express Enzyme (1X) (Thermo Fisher Scientific; 12605010) and then seeded into 24-well plates at a density of 4000 cells/well, with each cell type seeded in triplicate. Colonoids were cultures in media (Dulbecco’s modified Eagle medium/F12+1×B27+1×N2+ 1mM N-acetyl cysteine + 10mM HEPES+P/S [1:100] +2mM glutamax + 50ng/mL epidermal growth factor + 100ng/mL Noggin) for 4 days, with media changes every other day. EdU (Life Technologies/Thermo Fisher Scientific) was added to the culture media for 2 hours at the concentration of 10 μM and mixed well. EdU staining was performed with a Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies/Thermo Fisher Scientific; 10634), according to the manufacturer’s protocol. Stained cells were run on a FACSVerse flow cytometer (BD Biosciences). The data were analyzed using CellQuest software (BD Biosciences).
Colonoid Quantification Assay
∆A and ∆AP colonoids transfected with either shCtrl or ShTipe2 were digested into single cells (as previous) and then seeded into 24-well plates at a density of 500 cells/well. Each cell type was seeded in triple. After culture for 5 days, image the organoids were acquired on a dissecting microscope DMi8 (Leica, Wetzlar, Germany) and manually counted.
Results
Loss of Tipe2 Results in Decreased Tumor Burden
We used the well-studied AOM/DSS CAC model4,30 to study the role of TIPE2 in colon cancer initiation and progression. We first performed studies with 3 cycles of DSS administration, with each cycle compromised of 1 week of DSS followed by 2 weeks of water. At the end of the 70 days, mice were sacrificed and analyzed for both colitis and tumor burden. Studies were performed in young WT and Tipe2–/– mice on the C57BL/6 background, which as we have previously reported,17 had no baseline differences in weight or intestinal histopathology.
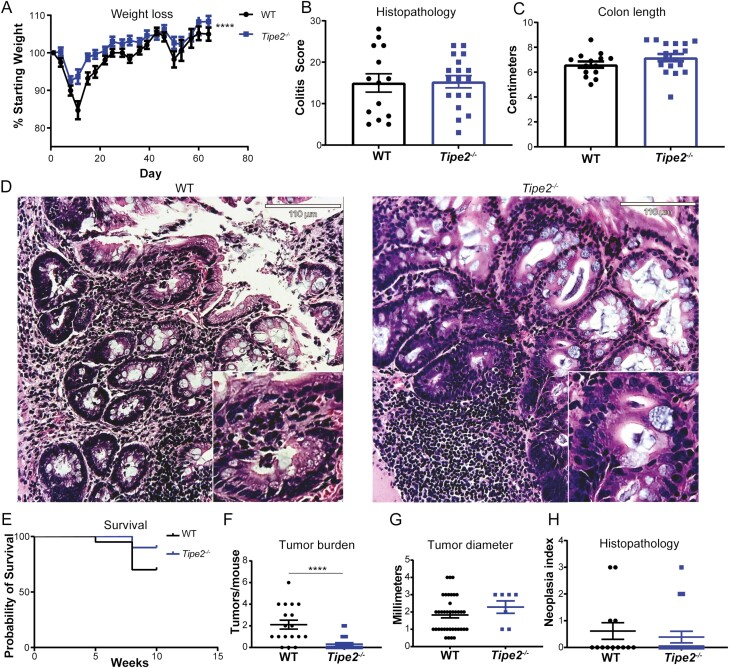
As measured by weight loss, Tipe2–/– mice were initially protected from DSS colitis (Figure 1A), as we have previously reported.17 However, over time this protection waned and by the end of the 70 days, WT and Tipe2–/– mice had similar levels of weight loss (Figure 1A), tissue damage (Figure 1B and D), changes in colon length (Figure 1C), and survival (Figure 1E), similar to what we had previously seen with experimental autoimmune encephalitis. Despite little differences in colitis between genotypes, Tipe2–/– mice did have significantly fewer tumors than WT mice (Figure 1F and G). Histopathological analysis did not show any differences in severity between the WT and Tipe2–/– tumors (Figure 1H).
Figure 1.
Colitis and tumor development in mice after azoxymethane and dextran sulfate sodium administration. A, Weight change over time; n = 17 for wild-type (WT) and 19 for Tipe2–/– mice, pooled across 3 independent experiments. Statistical analysis by 2-way analysis of variance; ∗∗∗∗P < .0001 for comparisons across time and group. B-D, Histolopathology score assessing colitis, colon length, and representative histological images at 70 days of colitis-associated colon cancer (CAC); n = 14 for WT and 18 for Tipe2–/– mice, pooled across 3 independent experiments. Scale bars=110 µm; inserts are 2× magnified. E, Survival curve over 70 days of CAC; n = 20 per group, pooled across 2 independent experiments. P = .1105 by Mantel-Cox test. F, Tumor burden per mouse; n = 18 for WT and 23 for Tipe2–/– mice pooled across 2 independent experiments. ∗∗∗∗P < .0001. G, Individual tumor diameter; n = 39 for WT and 7 for Tipe2–/– mice pooled across 2 independent experiments. H, Neoplasia index (histopathological analysis) at 70 days of CAC; n = 14 for WT and 18 for Tipe2–/– mice, pooled across 3 independent experiments. All graphs display mean±SEM unless otherwise indicated. Statistical analyses by Mann-Whitney nonparametric t test unless otherwise indicated.
Tipe2 –/– Mice Have Elevated Levels of IL-22 and pSTAT3 and Proliferative Signaling
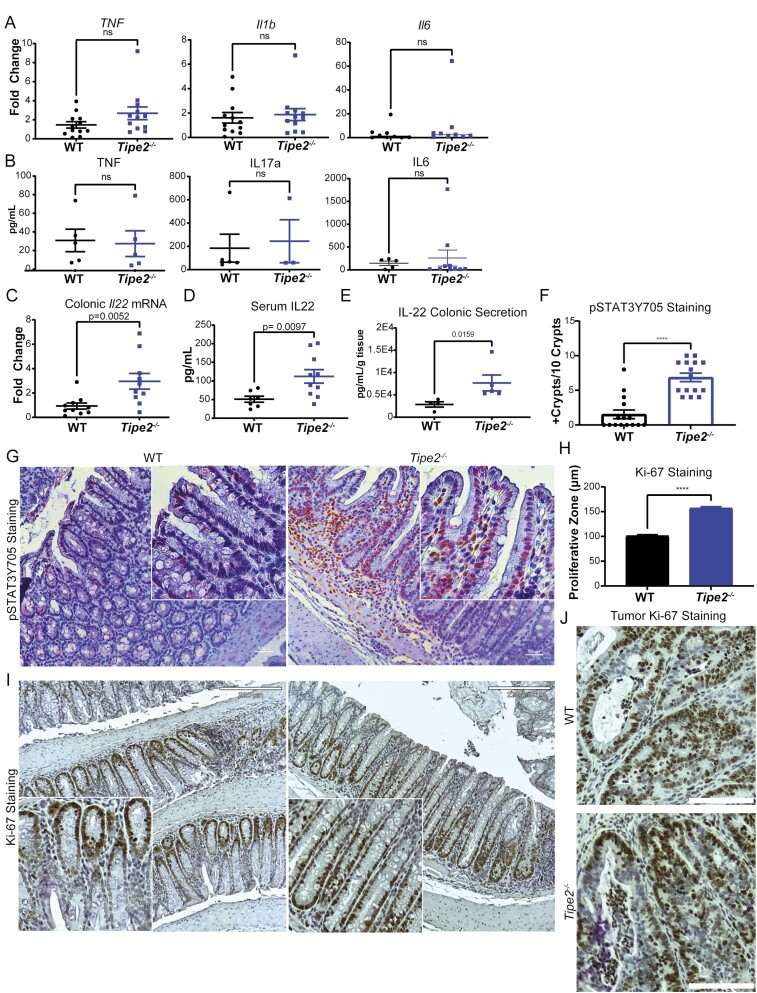
Because Tipe2–/– mice are known to have elevated levels of inflammatory cytokines at baseline,12 we assessed whether there were also differences after 70 days of CAC. Interestingly, there were no significant differences seen in colonic messenger RNA (mRNA) (Figure 2A) or serum (Figure 2B) levels of Tnfα, IL-1b, or IL-6, suggesting that the disease model overwhelmed any intrinsic differences. However, Tipe2–/– mice had elevated levels of IL-22, as measured by colonic mRNA (Figure 2C) and serum Luminex assays (Figure 2D), which we confirmed by colonic secretion ELISA studies of 70-day CAC colons (Figure 2E). IL-22 induces downstream STAT3 phosphorylation and is known to promote colon cancer tumor growth,31–34 so we next measured pSTAT3 levels in CAC intestinal tissues and found they were indeed elevated in the Tipe2–/– mice (Figure 2F and G), but staining levels were minimal and not different between genotypes in healthy colon tissue (Figure S1A). This was associated with increased levels of proliferation, as marked by Ki-67 staining, in the Tipe2–/– mice (Figure 2H-J).
Figure 2.
Tipe2 –/– mice have similar levels of inflammatory cytokines to those of wild-type (WT) mice, but increased levels of interleukin (IL)-22 and associated signaling. A, Real-time polymerase chain reaction for tumor necrosis factor (Tnf), Il1b, and Il1b in distal colonic tissue taken from azoxymethane (AOM)/dextran sulfate sodium (DSS) mice after 70 days of treatment. n = 12 per group from Tnf and Il1b, and n = 9 per group for Il6. Results were pooled across 2 independent experiments. B, Luminex assay for serum cytokines in AOM/DSS mice after 70 days of treatment. n = 5 per group for TNF, n = 5 for WT, n = 3 for Tipe2 –/– for IL17a, n = 5 for WT and n = 10 for IL6 (samples without detectable levels were excluded from analysis, leading to the variable sample numbers). Samples were pooled across 2 independent experiments. C, Real-time polymerase chain reaction for Il22 in in distal colonic tissue taken from AOM/DSS mice after 70 days of treatment; n = 10 per group, pooled across 2 independent experiments. D, Luminex assay for serum IL22 in AOM/DSS mice after 70 days of treatment; n = 7 for WT and 10 for Tipe2 –/–. Samples were pooled across 2 independent experiments. E, IL-22 excretion by AOM/DSS colonic explants harvested at 70 days of treatment, measured by sandwich enzyme-linked immunosorbent assay. n = 4 mice for WT, 5 mice for Tipe2 –/–. Representative of 2 independent experiments. F and G, pSTAT3Y705 staining in AOM/DSS mice after 70 days of treatment; results pooled from 2 independent experiments. F, Quantification done by counting 3 random sections per slide from 5 different mice per group; n = 15 per group. G, Representative micrographs: bars=50 µm, inserts 2× multiplied. H-J, Proliferation as measured by Ki-67 staining. H Quantification by measuring length of proliferative zone in all longitudinal crypts from 3 images per slide. A total of 7 WT and 8 Tipe2 –/– slides were analyzed; total n = 183 for WT and 207 for Tipe2 –/–. G, Representative micrographs: bars=220 µm, inserts 2× multiplied. J, Representative tumor-specific micrographs; bars=110 µm. All graphs display mean±SEM unless otherwise indicated. Statistical analyses by Mann-Whitney nonparametric t test unless otherwise indicated. P as indicated; ∗∗∗∗P < .0001.
At Later Stages of Colon Carcinogenesis, Loss of TIPE2 Results in Bigger Tumors
Given that the Tipe2–/– mice had increased proliferative signaling and tumors that were trending to be larger than those of WT mice, even if they were less frequent, we decided to extend our CAC model through one more 3-week cycle of DSS and water to determine if we simply needed to wait longer to observe more of the Tipe2–/– phenotype. Interestingly, by extending out the model for another cycle (91 days), we saw slightly worse colitis in the Tipe2–/– mice, with a trend toward increased weight loss, although colon length was not significantly changed (Figure 3A-D). Given that the increase in colitis score was mostly due to increased immune infiltration, it made sense that the colon lengths were not significantly different, as that is driven mostly by epithelial damage and subsequent healing with contraction. Tipe2–/– mice also had decreased survival (Figure 3F), in contrast to the 70-day time point. Interestingly, the differences in tumor burden between genotype vanished at this later time point (Figure 3G), and Tipe2–/– mice now had larger tumors (Figure 3E and H), although the tumors were not significantly more advanced when assessed by pathology (Figure 3I). When looking at serum cytokines, we again found that IL-22 was elevated in the Tipe2–/– mice, and that there were otherwise no significant differences between both genotypes (Figure 3J). However, in addition to Il22 transcripts, we did find elevated levels of Tnf and Il6 (but not Il1b) colonic mRNA, suggesting that local, if not systemic inflammation, is worse in the Tipe2–/– mice at 91 days (Figure 3K), which correlates with the increased tissue damage and decreased survival we observed.
Figure 3.
Loss of TIPE2 results in bigger tumors as disease progresses. A, Weight change over time. n = 9 for wild-type (WT) and 8 for Tipe2 –/– mice, pooled across 2 independent experiments. Statistical analysis by 2-way analysis of variance. No significant differences between groups were found. B-D, Histolopathology score assessing colitis, colon length, and representative histological images at 91 days of colitis-associated colon cancer (CAC). For histology, n = 8 for WT and 7 for Tipe2 –/–, pooled across 2 independent experiments; for colon length, n = 22 for Wt and 24 for Tipe2 –/–, pooled across 3 independent experiments. Scale bars=220 µm; inserts are 2× magnified, and statistical analyses were by Student’s t test. E, Representative (n = 5 independent experiments) whole mount images of colons after 91 days of CAC. Arrows point to individual tumors. F, Survival curve over 70 days of CAC. n = 20 per group, pooled across 2 independent experiments. ∗∗P = .006 by Mantel-Cox test. G, Tumor burden per mouse; n = 23 for WT and 26 for Tipe2 –/– mice pooled across 5 independent experiments. H, Individual tumor diameter; n = 55 for WT, and 41 for Tipe2–/– mice pooled across 5 independent experiments; ∗∗∗∗P < .0001. I, Neoplasia index (histopathological analysis) at 91 days of CAC. n = 8 for WT and 7 for Tipe2–/– mice, pooled across 2 independent experiments. J, Serum cytokines in azoxymethane (AOM)/dextran sulfate sodium (DSS) mice after 91 days of treatment, measured by sandwich enzyme-linked immunosorbent assay. n = 7 for WT and 10 for Tipe2 –/– mice, except for IL-22, where n = 9 for Tipe2– /– mice. Samples were pooled across 2 independent experiments; statistical analyses by Student’s t test. K, Real-time polymerase chain reaction of colonic tissue at 91 days for Tnf, Il6, Il22, and Il1b. n = 20 per group for Tnf, 17 per group for Il6, 18 per group for Il22, and 20 per group for Il1b, with individual replicates pooled across 2 individual experiments comprising ≥8 mice per genotype. Analysis was by Mann-Whitney nonparametric t test unless otherwise described, with significance as indicated on each graph or as described. All graphs display mean±SEM unless otherwise indicated.
Because survival and colitis was now worse in the tumor-bearing Tipe2–/– mice, compared with previous reports in which loss of TIPE2 was acutely protective against DSS colitis,17 we next performed a study to assess if Tipe2–/– mice became more susceptible to DSS colitis if the length of treatment was extended. This proved not to be the case, as Tipe2–/– mice were still significantly protected against 2 weeks of acute DSS colitis, as assessed by weight loss, survival, and histopathology (Figure S1A-C).
Loss of TIPE2 Results in Increased Numbers of CD11c+CD8a+ T Cells and Increased IL23r Expression, Suggesting an Increase in Group 3 Innate Lymphoid Cells
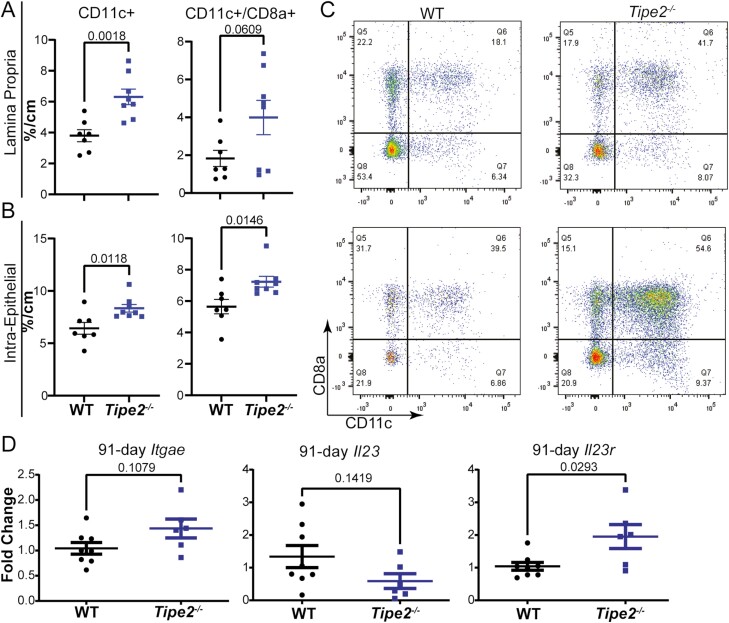
We next performed flow cytometry to assess if loss of TIPE2 led to a difference in the immunological tumor microenvironment that could explain the increased IL-22 and downstream increases in tumor size we saw over time. We performed multicolor flow cytometry for CD45, CD3, CD4, CD8a, CD8b, CD11c, and Gr1 on lamina propria and epithelial or intraepithelial cell fractions that had been subjected to our extended, 91 days of AOM/DSS CAC. Interestingly, only CD11c+, and specifically CD11c+CD8a+, cells were elevated in both the lamina propria and intraepithelial CD45+ cell fractions (although statistical significance was barely not reached for lamina propria CD11c+CD8a+ cells (Figure 4A-C); extended gating strategy shown in Figure S3).
Figure 4.
Tipe2 –/– mice with colitis-associated colon cancer have increased numbers of CD11c+CD8a+ T cells. A-C, Flow cytometry for CD11c+ and CD11c+CD8a+ cells isolated from the lamina propria and intraepithelial colonic compartments of mice subjected to 91 days of colitis-associated colon cancer; n = 7 for wild-type (WT) and 8 for Tipe2–/– mice, pooled between 2 independent experiments. C, Sample flow gates. Gating is on lymphocytes, live cells, and CD45, and sample gates are percentages of CD45+ cells (see Figure S3). D, Real-time polymerase chain reaction of colonic tissue at 91 days for Itgae, Il23, and Il23r; n = 8 for WT and 6 for Tipe2–/– mice, pooled between 2 independent experiments. Analysis by Mann-Whitney nonparametric t test, with significance as indicated on each graph. For all panels, all graphs display mean±SEM unless otherwise indicated. Statistical analyses by Student’s t test unless otherwise specified with significance as indicated on each graph.
Given limited sample availability, we then performed PCR to identify if these cells were dendritic cells or T cells, by measuring levels of CD103 (Itgae) and Il23, and to identify if group 3 innate lymphoid cells (ILC3s) could be responsible for the elevated levels of IL-22 we saw, by measuring Il23r. Neither Itgae levels nor Il23 levels were elevated in the isolated tissues (Figure 4D) suggesting that the CD11c+CD8a+ cells were T cells, not dendritic cells. However, Il23r levels were elevated (Figure 4D), suggesting that ILC3s could be the downstream generator of the increased amounts of IL-22 that we observed.
TIPE2 is an Intrinsic Colonocyte Tumor Suppressor
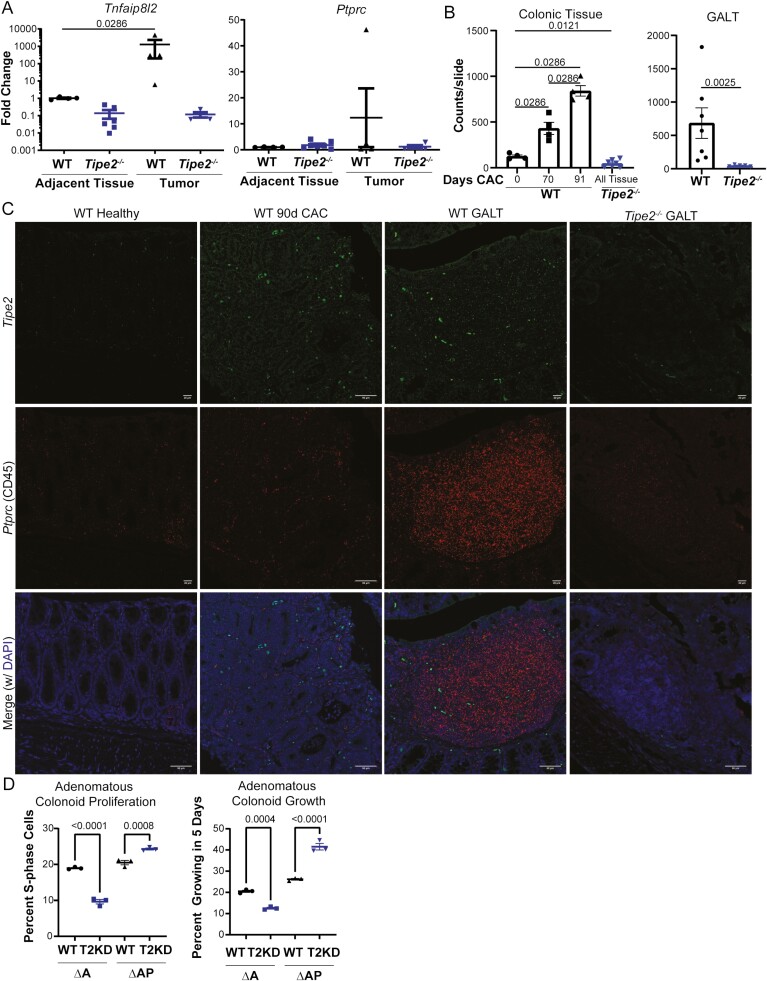
TIPE2 is not normally expressed at significant levels in the colonic epithelium,12,20 although increased expression in tumor biopsies was associated with increased colon cancer disease severity.21 To assess if TIPE2 was being induced during carcinogenesis, we performed PCR on tumor colonic epithelia and isolated lamina propria immune cells to evaluate the levels of Tipe2 gene expression, using Ptprc (CD45) gene expression as a control for immune cell levels, as TIPE2 is expressed in immune cells.12,20 Surprisingly, Tipe2 mRNA transcripts were elevated in WT tumor colonocytes, in contrast to colonocytes from adjacent tissue, and far in excess of any differences in Ptprc levels (Figure 5A). To confirm this, we then performed RNAScope, also known as in situ hybridization. We found that tumor tissue did indeed have elevated levels of Tipe2, with a more than 5-fold increase in transcripts when comparing healthy WT tissue with 91-day AOM/DSS CAC tissue (Figure 5B and C). Thus, in the context of colon cancer, colonocyte TIPE2 may function as a tumor suppressor, which puts the previous reports of elevations in TIPE2 mRNA correlating with disease severity in human CRC into a different light.21 Instead of being a driver of disease severity, these data suggest its induction is a protective, tumor-suppressive response that escalates as cancer severity increases.
Figure 5.
TIPE2 is a colonic tumor suppressor. A, Real-time polymerase chain reaction of isolated tumor and adjacent tumor colonocytes at 91 days of colitis-associated colon cancer (CAC), for Tnfaip8l2 and Ptprc. Each data point represents 2 to 3 pooled mice, and results are pooled across 3 independent experiments. n = 4 all groups, except Tipe2 –/– mice where n = 6 for Tnfaip8l2 and n = 7 for Ptprc. Analysis by Mann-Whitney nonparametric t test, with significance as indicated on each graph. B and C, RNAScope for Tipe2 and Ptprc (CD45). B and C, Quantification, using automated image analysis to measure the number of positive RNA probe signals per slide, with representative photomicrographs. For colonic tissue, n = 4 mice per group, except for Tipe2 –/–, where n = 7. For gut-associated lymphoid tissue (GALT), n = 7 for wild-type (WT) and 5 for Tipe2 –/– mice; bars=50 µm. D and E, Proliferation assay and organoid growth assessment € in shCtrl (Ctrl) and short hairpin RNA–mediated Tipe2-knockdown (T2KD) APC-mutant (∆A) and APC/p53-mutant (∆AP) murine colonoids that represent progressively advanced stages of colonic adenoma or carcinoma. n = 3 per group, representative of 2 independent experiments, and analyzed by parametric t test. All graphs display mean±SEM unless otherwise indicated. Statistical analyses by Mann-Whitney nonparametric t test unless otherwise indicated. P as indicated; ∗∗∗∗P < .0001.
CRISPR-Cas9 mutagenesis was employed as demonstrated in Figure S2A-D and as detailed in the Methods to generate murine mutant colonoid lines with loss-of-function mutations in either Apc (∆A) or Apc and Tp53 (ie, p53 [∆AP]) to use as a tool to probe the role of TIPE2 in CRC. We saw that in ∆A colonoids, decreased TIPE2 expression led to diminished proliferation of the colonoids, while knockdown of TIPE2 in ∆AP colonoids led to increased proliferation, as measured by the number of cells in S-phase (Figure 5D, Figure S2D) and overall proliferating colonoids (Figure 5E).
Discussion
Our data demonstrate that TIPE2 plays a dual role in the initiation and progression of CAC. Early on, loss of TIPE2 appears to inhibit tumor initiation; in our studies, loss of TIPE2 led to significantly less numbers of visible tumors. However, the tumors that did exist were of similar size to WT mice, suggesting that progression was not affected. Given that Tipe2–/– mice are known to have MDSC14 and regulatory T cell dysfunction,15,16 we hypothesize that loss of TIPE2 led to decreased immune cell evasion of early hyperplastic and neoplastic tumors, leading to increased tumor killing and decreased tumor burden.
However, as time went on, we saw that loss of TIPE2 promoted increased tumor progression, with larger tumors appearing in Tipe2–/– mice with an additional 21 days on the AOM/DSS protocol, and with loss of decreased tumor burden phenotype seen early on. Our data suggest that this enhanced tumor progression might occur through 2 distinct mechanisms. First, loss of TIPE2 resulted in increased IL-22 production at all time points we studied, with corresponding downstream pSTAT3. IL-22/STAT3 signaling is known to drive colon cancer tumor progression,35,36 and so this could have explained what we saw. Second, TIPE2 was induced in colonocytes as cancer progressed and might serve as a tumor suppressor. Knocking down TIPE2 in early colon cancer tumoroids (∆A) hindered tumoroid growth; however, in more advanced tumoroids (∆AP), knockdown of TIPE2 increased tumoroid growth. Interestingly, TIPE2 could not easily be detected in shCtrl ∆AP organoids (Figure S2F), but an effect was still seen with suppression of the gene, suggesting that more advanced colonic adenomas repress TIPE2. Together, these data highly suggest that TIPE2 is indeed a tumor suppressor as colon cancer progresses down the mutational cascade. Previous reports have found that overexpression of TIPE2 can inhibit cancer growth in cell lines with mutant p5337 and that TIPE2 can regulate p53 expression.38 Combined with our data, it appears that the function of TIPE2 intersects with that of p53 in suppressing tumor growth.
This finding that TIPE2 is an intrinsic tumor suppressor also explains the previous report that TIPE2 expression levels correlate with disease stage in human CRC.21 Thus, loss of TIPE2 may enhance progression of colon cancer in 2 ways: (1) by enhancing IL-22 levels and (2) through loss of its function as a tumor suppressor.
Because of its key role in regulating immune cell function, we attempted to identify the immune populations altered by loss of TIPE2 in our AOM/DSS CAC model. Flow cytometry analysis revealed an increase in CD11c+CD8a+ cells. Two distinct subsets of CD11c+CD8a+ cells exist: a cytotoxic dendritic cell population39 and a regulatory, immunosuppressive T cell population.40,41 Because of limited sample availability, we performed post hoc PCR analysis to identify which of these cell populations were more likely. Based on this analysis, we found that the likely culprit was the immunosuppressive T cell population, which may explain the increase in tumor burden overtime. Similarly, Il23r levels were elevated, implicating elevations in ILC3s as a potential source of the elevated IL-22 we observed.
Conclusions
Together, this work demonstrates that TIPE2 has multiple, sometimes opposing effects in CAC. In colonocytes, it appears to function directly as a tumor suppressor, with loss resulting enhanced tumor progression. However, in the immune system, TIPE2 has opposing roles. It may promote tumorigenesis through enhancing immune suppressor cell function but inhibit it through regulating oncogenic IL-22 secretion. Going forward, this work suggests that targeting TIPE2 for colon cancer therapy requires cell- and pathway-specific approaches and serves as a cautionary tale for immunotherapy approaches in general in terms of colon cancer, as intestinal inflammation can both promote and inhibit cancer.
Supplementary Material
Acknowledgments
We thank the University of Pennsylvania Cell and Developmental Biology Microscopy Core, Molecular Pathology and Imaging Core, and Cutaneous Phenomics and Transcriptomics Core (and specifically Dr. Stephen Prouty of this core). We thank Dr Chin Nien Li for helping measure mouse weights.
Author Contributions
Z.E. designed and performed experiments and edited the manuscript; J.R.G. designed, performed experiments, and analyzed data and wrote the manuscript, M.B.-D, Y.T., R.H., N.S., M.L., and H.S. performed experiments; C.L. provided technical guidance and input on experiments and interpretation of results; S.L. and Y.L. conceived the early studies and shared the data; Y.H.C. conceived and supervised the studies, and edited the manuscript.
Supported By
This work was funded by the National Institutes of Health (R01-AI099216, R01-AI121166, and R01-AI136945 to Y.H.C.; F32-DK116528 to J.R.G.; and T32-CA009140 to Warren Pear).
Conflicts of Interest
J.R.G. is an employee of Seres Therapeutics. Y.H.C. is a member of the advisory board of Amshenn Co and Binde Co. No other authors have any conflicts of interest to disclose.
References
- 1. Centers for Disease Control and Prevention. Colorectal Cancer Statistics. Vol. 2017; Atlanta, GA: Centers for Disease Control and Prevention; 2016. [Google Scholar]
- 2. Mattar MC, Lough D, Pishvaian MJ, Charabaty A.. Current management of inflammatory bowel disease and colorectal cancer. Gastrointest Cancer Res. 2011;4:53–61. [PMC free article] [PubMed] [Google Scholar]
- 3. Itzkowitz SH, Yio X.. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. [DOI] [PubMed] [Google Scholar]
- 4. De Robertis M, Massi E, Poeta ML, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sun X, Suo J, Yan J.. Immunotherapy in human colorectal cancer: Challenges and prospective. World J Gastroenterol. 2016;22:6362–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farkona S, Diamandis EP, Blasutig IM.. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sieminska I, Baran J.. Myeloid-derived suppressor cells in colorectal cancer. Front Immunol. 2020;11:1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Betts G, J1s E, Junaid S, et al. Suppression of tumour-specific CD4+ T cells by regulatory T cells is associated with progression of human colorectal cancer. Gut. 2012;61:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fayngerts SA, Wu J, Oxley CL, et al. TIPE3 is the transfer protein of lipid second messengers that promote cancer. Cancer Cell. 2014;26:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fayngerts SA, Wang Z, Zamani A, et al. Direction of leukocyte polarization and migration by the phosphoinositide-transfer protein TIPE2. Nat Immunol. 2017;18:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun H, Lin M, Zamani A, et al. The TIPE molecular pilot that directs lymphocyte migration in health and inflammation. Sci Rep. 2020;10:6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun H, Gong S, Carmody RJ, et al. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell. 2008;133:415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Z, Fayngerts S, Wang P, et al. TIPE2 protein serves as a negative regulator of phagocytosis and oxidative burst during infection. Proc Natl Acad Sci U S A. 2012;109:15413–15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan D, Wang J, Sun H, et al. TIPE2 specifies the functional polarization of myeloid-derived suppressor cells during tumorigenesis. J Exp Med. 2020;217:e20182005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding J, Su J, Zhang L, Ma J.. Crocetin activates Foxp3 through TIPE2 in asthma-associated Treg cells. Cell Physiol Biochem. 2015;37:2425–2433. [DOI] [PubMed] [Google Scholar]
- 16. Zhao LL. TIPE2 suppresses progression and tumorigenesis of the oral tongue squamous cell carcinoma by regulating FoxP3+ regulatory T cells. J Bioenerg Biomembr. 2020;52:279–289. [DOI] [PubMed] [Google Scholar]
- 17. Lou Y, Sun H, Morrissey S, et al. Critical roles of TIPE2 protein in murine experimental colitis. J Immunol. 2014;193:1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xi W, Hu Y, Liu Y, et al. Roles of TIPE2 in hepatitis B virus-induced hepatic inflammation in humans and mice. Mol Immunol. 2011;48:1203–1208. [DOI] [PubMed] [Google Scholar]
- 19. Wu X, Kong Q, Zhan L, et al. TIPE2 ameliorates lipopolysaccharide-induced apoptosis and inflammation in acute lung injury. Inflamm Res. 2019;68:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldsmith JR, Fayngerts S, Chen YH.. Regulation of inflammation and tumorigenesis by the TIPE family of phospholipid transfer proteins. Cell Mol Immunol. 2017;14:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li XM, Su JR, Yan SP, et al. A novel inflammatory regulator TIPE2 inhibits TLR4-mediated development of colon cancer via caspase-8. Cancer Biomark. 2014;14:233–240. [DOI] [PubMed] [Google Scholar]
- 22. Peters LA, Perrigoue J, Mortha A, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. 2017;49:1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bábíčková J, Tóthová Ľ, Lengyelová E, et al. Sex differences in experimentally induced colitis in mice: a role for estrogens. Inflammation. 2015;38:1996–2006. [DOI] [PubMed] [Google Scholar]
- 24. Murthy SN, Cooper HS, Shim H, et al. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci. 1993;38:1722–1734. [DOI] [PubMed] [Google Scholar]
- 25. Boivin GP, Washington K, Yang K, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. 2003;124:762–777. [DOI] [PubMed] [Google Scholar]
- 26. Washington MK, Powell AE, Sullivan R, et al. Pathology of rodent models of intestinal cancer: progress report and recommendations. Gastroenterology. 2013;144:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldsmith JR, Spitofsky N, Zamani A, et al. TNFAIP8 controls murine intestinal stem cell homeostasis and regeneration by regulating microbiome-induced Akt signaling. Nat Commun. 2020;11:2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larmonier CB, Uno JK, Lee KM, et al. Limited effects of dietary curcumin on Th-1 driven colitis in IL-10 deficient mice suggest an IL-10-dependent mechanism of protection. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1079–G1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. [DOI] [PubMed] [Google Scholar]
- 30. Thaker AI, Shaker A, Rao MS, Ciorba MA.. Modeling colitis-associated cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS). J Vis Exp. 2012;67:4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huber S, Gagliani N, Zenewicz LA, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bollrath J, Phesse TJ, von Burstin VA, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. [DOI] [PubMed] [Google Scholar]
- 33. Jiang R, Wang H, Deng L, et al. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer. 2013;13:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kryczek I, Lin Y, Nagarsheth N, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parks OB, Pociask DA, Hodzic Z, et al. Interleukin-22 Signaling in the Regulation of Intestinal Health and Disease. Front Cell Dev Biol. 2016;3:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keir M, Yi Y, Lu T, Ghilardi N.. The role of IL-22 in intestinal health and disease. J Exp Med. 2020;217:e20192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Tao M, Wu J, et al. Adenovirus-directed expression of TIPE2 suppresses gastric cancer growth via induction of apoptosis and inhibition of AKT and ERK1/2 signaling. Cancer Gene Ther. 2016;23:98–106. [DOI] [PubMed] [Google Scholar]
- 38. Bordoloi D, Banik K, Padmavathi G, et al. TIPE2 induced the proliferation, survival, and migration of lung cancer cells through modulation of Akt/mTOR/NF-κB signaling cascade. Biomolecules. 2019;9:836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunne PJ, Moran B, Cummins RC, Mills KHG.. CD11c+CD8α+ dendritic cells promote protective immunity to respiratory infection with Bordetella pertussis. J Immunol. 2009;183:400–410. [DOI] [PubMed] [Google Scholar]
- 40. Vinay DS, Kim CH, Choi BK, Kwon BS.. Origins and functional basis of regulatory CD11c+CD8+ T cells. Eur J Immunol. 2009;39:1552–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujiwara D, Chen L, Wei B, Braun J.. Small intestine CD11c+ CD8+ T cells suppress CD4+ T cell-induced immune colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G939–G947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.