Abstract
Purpose:
We develop a deep learning (DL) radiomics model and integrate it with circulating tumor cell (CTC) counts as a clinically useful prognostic marker for predicting recurrence outcomes of early-stage (ES) non-small cell lung cancer (NSCLC) patients treated with stereotactic body radiation therapy (SBRT).
Methods and Materials:
A cohort of 421 NSCLC patients was used to train a DL model for gleaning informative imaging features from computed tomography (CT) data. The learned imaging features were optimized on a cohort of 98 ES-NSCLC patients treated with SBRT for predicting individual patient recurrence risks by building DL models on CT data and clinical measures. These DL models were validated on the third cohort of 60 ES-NSCLC patients treated with SBRT to predict recurrent risks and stratify patients into subgroups with distinct outcomes in conjunction with CTC counts.
Results:
The DL model obtained a concordance-index of 0.880 (95% confidence interval, 0.879-0.881). Patient subgroups with low and high DL risk scores had significantly different recurrence outcomes (P = 3.5e-04). The integration of DL risk scores and CTC measures identified 4 subgroups of patients with significantly different risks of recurrence (χ2 = 20.11, P = 1.6e-04). Patients with positive CTC measures were associated with increased risks of recurrence that were significantly different from patients with negative CTC measures (P = 0.0447).
Conclusions:
In this first-ever study integrating DL radiomics models and CTC counts, our results suggested that this integration improves patient stratification compared with either imagining data or CTC measures alone in predicting recurrence outcomes for patients treated with SBRT for ES-NSCLC.
Introduction
Lung cancer remains the leading cause of cancer-related deaths, and the estimated number of new cases is 235,760 in the United States in 2021.1 Non-small cell lung cancer (NSCLC) accounts for approximately 87% of the diagnosed lung cancer cases and has a poor 5-year survival rate of 18%.2-4 With advances in lung cancer screening, lung cancer patients are increasingly being identified at earlier stages where treatment outcomes are substantially better, significantly improving quality of life.5,6
Stereotactic body radiation therapy (SBRT) has been increasingly used to treat early stage NSCLC (ES-NSCLC) in the medically inoperably setting due to its improved efficacy, reduced morbidity, and convenience compared with conventionally fractionated radiation therapy.7-9 Despite its high initial efficacy, 5% to 15% of patients with ES-NSCLC treated with SBRT may fail regionally and 20% to 25% fail distantly with long-term follow-up.10,11 Systemic therapy to reduce this risk in ES-NSCLC patients after surgery might be harmful to patients of stage IA,12 and it would likely have even more harmful in the medically inoperable setting. There is an opportunity to intensify treatment if we could better select which subset of patients are at high enough risk after SBRT to improve outcomes. Optimized methods to inform subsequent treatment throughout each patient’s clinical course for such purposes is thus critically needed.13-15
To predict treatment outcomes and select patients at higher risk of recurrence after SBRT, a variety of biomarkers have been investigated. Circulating tumor cell (CTC) enumeration has demonstrated promising performance as a prognostic pharmacodynamic biomarker in studies of breast cancer, prostate cancer, and lung cancer.16-19 More recently, a prospective longitudinal study of patients with ES-NSCLC treated with SBRT has revealed that higher pretreatment CTC levels and persistence of CTCs posttreatment were significantly associated with increased risk of recurrence outside the targeted treatment site.20 However, CTCs are detected in the peripheral blood of only about half of ES-NSCLC patients at diagnosis, and thus they cannot be used as a sole determinant of patients who are at high risk of recurrence after SBRT.
On the other hand, radiomics analysis has achieved promising performance in cancer research for characterizing tumor phenotypes, patient stratification, and prognosis based on quantitative imaging features.21-24 Several recent studies have also demonstrated that radiomic signatures are excellent independent biomarkers for estimating prognostic results (overall survival and metastasis) of lung cancer patients.24-29 More recently, deep learning (DL) methods have been adopted in radiomics studies and have achieved superior prediction performance compared with traditional radiomics-based machine learning methods in a variety of cancer studies,28,30-32 including lung cancer.33-35 However, most of the existing DL prognostic models formulate the prognosis as a binary classification problem of survival outcomes, and they are not equipped to effectively provide quantitative risk prediction of recurrence outcomes that could be further integrated with other biological assays to inform treatment decisions.
In this work, we investigated the integration of a DL prediction model of computed tomography (CT) imaging data and CTC measures for early prediction of treatment outcome in a large cohort of pathologically confirmed ES-NSCLC patients treated with SBRT. We hypothesize that an effective integration of the DL models and CTC measures will make accurate prognostic predictions of outcomes of ES-NSCLC patients treated with SBRT that are superior to prediction based on either imaging data or CTC measures alone.
Methods and Materials
Cohorts of NSCLC patients, imaging data, and clinical and CTC measures
This study was conducted with approval from the institutional review board. This study included 3 cohorts of lung cancer patients. The first cohort consisted of 421 lung cancer patients who were treated at MAASTRO Clinic, The Netherlands. For these patients, CT scans, manual delineations, clinical measures, and survival information are publicly available at https://wiki.cancerimagingarchive.net/display/Public/NSCLC-Radiomics.21 Median follow-up of these patients is 1.1 years. More detailed information of this data cohort is summarized in Table E1. We used this cohort as training data to train an unsupervised deep learning model for extracting informative imaging features.
The second cohort consisted of 98 consecutive patients who were treated with SBRT for ES-NSCLC (T1a, T1b, and T2a). Twelve of these patients were examined on a GE Discovery ST (GE Healthcare, Waukesha, Wisconsin), 70 were examined on a Philips Gemini/Ingenuity TF (Phillips Medical Systems, Amsterdam, The Netherlands), and the remaining 16 were examined on a Siemens Biograph 64 mCT (Siemens Healthcare, Erlangen, Germany). CT images were acquired using the following parameters: peak tube voltage of 120 kVp with the Philips scanner, 100 to 140 kVp with the Siemens scanner, and 120 to 140 kVp with the GE scanner; exposure of 30 to 300 mAs with the Philips scanner, 27 to 346 mAs with the Siemens scanner, and 4 to 21 mAs with the GE scanner; tube current of 42 to 325 mAs with the Philips scanner, 44 to 554 mAs with the Siemens scanner, and 44 to 119 mAs with the GE scanner; pixel size of 1.17 × 1.17 or 1.37 × 1.37 mm2 with the Philips scanner, 0.98 × 0.98 or 1.52 × 1.52 mm2 with the Siemens scanner, and 0.98 × 0.98 or 1.37 × 1.37 mm2 with the GE scanner; and slice thicknesses of 3 or 4 mm with the Philips scanner, 2 or 3 mm with the Siemens scanner, and 3.27 mm with the GE scanner. A standard B or sharp C filter with the Philips scanner, a flat filter with the Siemens scanner, and a body filter with the GE scanner were used for reconstruction of CT images. All the patients were treated uniformly (12.5 Gy × 4 fractions or 10 Gy × 5 fractions). The patients in this cohort had a median follow-up of 2 years that provides recurrence information of local failure, nodal failure, and distant failure. We used this cohort as training data to train a supervised DL model based on imaging features learned by the unsupervised deep learning model and for predicting recurrence risks.
For the third cohort, 5 of the 60 patients were examined on a GE Discovery ST (GE Healthcare, Waukesha, Wisconsin), 30 were examined on a Philips Gemini/Ingenuity TF (Phillips Medical Systems, Amsterdam, The Netherlands), and the remaining 25 were examined on a Siemens Biograph 64 mCT (Siemens Healthcare, Erlangen, Germany). CT images of these 60 patients were acquired using the same parameters as that of the 98-patient cohort. The CTC measures were obtained by a CTC assay validated in previous studies.20,36-38 The assay consists of an adenoviral-based probe that expresses green fluorescent protein (GFP) driven by a human telomerase reverse transcriptase promoter element in live cells with increased telomerase activity, which are then detected and enumerated by fluorescence microscopy. Increased telomerase activity is characteristic of most tumor cells (helping with forestall senescence); conversely, it is not elevated in nearly all normal cells.20 SBRT was delivered to all the subjects with a median dose of 50 Gy (range, 50-60 Gy), most commonly in 4 or 5 (88%) fractions (range, 4-20); median biologically effective dose (BED10, α/β = 10) was 100 Gy (range, 78-112.5 Gy). Seven patients (12%) received a less aggressive fractionation (>5 fractions) for larger, centrally/ultracentrally located tumors.
Table E2 provides details of the characteristics of patients in the second and third cohorts. Local failure was defined as tumor growth after initial shrinkage or progression at the local site on 2 consecutive scans confirmed by positron emission tomography images or biopsy. Nodal failure was defined as recurrence at the hilar, ipsilateral, or contralateral mediastinal, or supraclavicular lymph nodes. Distant metastasis was defined as tumor recurrence in the contralateral lung or outside the hemithorax. To differentiate metachronous primary tumors from recurrent disease, criteria from Martini and Melamed was applied.39 In all cases, pathologic confirmation of recurrent disease was obtained when possible. Additionally, date of death was determined by death certificates or institutional medical records. A flowchart of study data retrieval and exclusion of the 3 data cohorts is shown as Figure E1.
Deep learning models for predicting recurrence risks of patients in individual levels
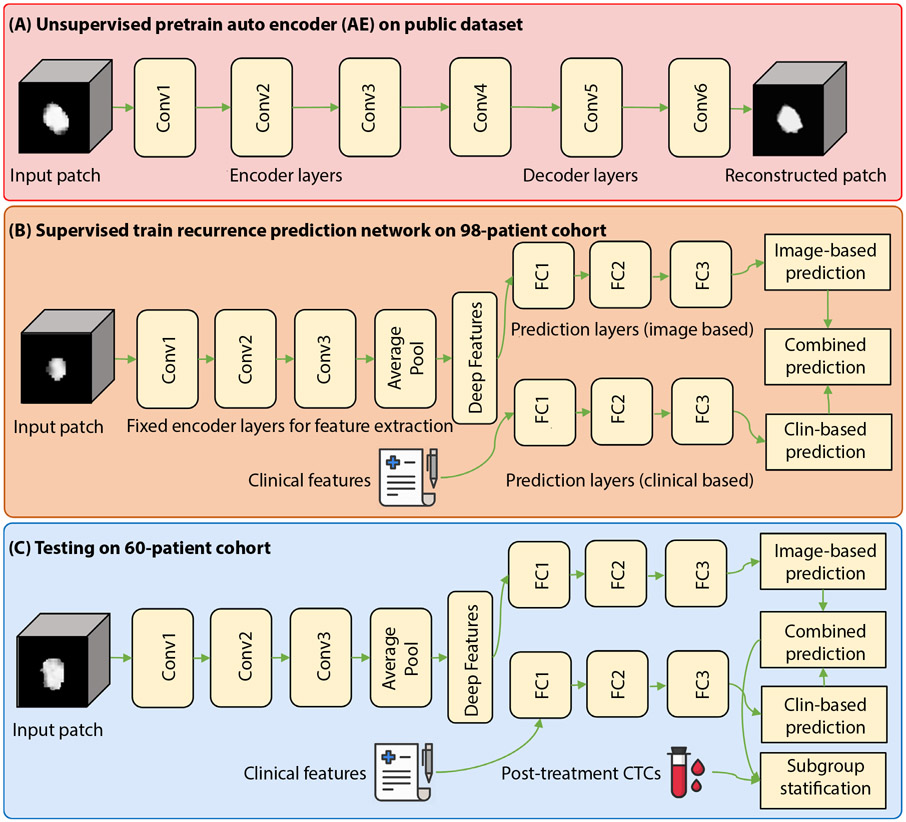
As illustrated in Figure 1, DL models were built to predict recurrence risks based on a cohort of 421 NSCLC patients and a cohort of 98 ES-NSCLC patients, and the DL models were validated on the third cohort of 60 ES-NSCLC patients. In particular, an auto-encoder (AE) DL model was trained in an unsupervised learning setting on the first cohort to extract informative imaging features from 3-dimensional (3D) CT scans of lung tumors.40 The AE model’s feature extraction component was then input to a supervised 3-layer fully connected (FC) module to predict recurrence risks in a survival analysis framework on the second cohort.28 Finally, the DL recurrence prediction model was evaluated on the third cohort.
Fig. 1.
Flowchart of the proposed recurrence prediction method. Abbreviations: AE = auto encoder; Conv = Convolution; CTC = circulating tumor cell; FC = fully connected.
Convolutional AE deep learning model
As shown in Figure 1(A), we first trained a convolutional AE model whose input and output were masked CT image patches of lung tumors defined by gross tumor target volume that were delineated in the CT scans by experienced, board-certified radiologists.21 The convolutional AE model contained 3 layers of convolutional neural networks CNNsas encoder layers (Conv1 to Conv3) to learn features from 3DCT patches of lung tumors followed by 3 convolutional decoder layers (Conv4 to Conv 6) to reconstruct the input 3DCT patches. A Sigmoid activation function was used in the convolutional AE model that was optimized to obtain representative features by minimizing reconstruction errors between its output and input CT patches. Table E3 and Figure E2 show the detailed network architecture of the convolutional AE model, the training procedures, and the training loss.
Deep learning model of recurrence risks
The proposed DL models were built on the second cohort of 98 ES-NSCLC patients in a survival analysis framework to predict recurrence risks. The DL model was built upon CNNs for survival analysis28; Table E4 and Figure E3 show the network structure. Particularly, this model shared the same encoder layers with the AE model. After these encoder layers of CNNs (Conv1 to Conv3), we used an average pooling layer to obtain the vectorized imaging features and then 3 FC layers to predict recurrence risk by optimizing a Cox loss function. We used the other stream of 3 FC layers to learn information features from clinical measures by optimizing a Cox loss function. The clinical measures included age, sex, smoking status, and body mass index. In both the streams, we used Rectified Linear Unit (ReLU) activation function in the first 2 FC layers and a Sigmoid activation function in the last FC layer. Finally, we input the risk prediction outputs of these 2 streams to a 2-factor (the image-based recurrence risks and the clinical-based recurrence risks obtained from our DL models) Cox regression model to predict recurrence risks.
In summary, we built these models to predict recurrence risk scores by optimizing the Cox-loss functions based on the second cohort. Specifically, we adopted and fixed parameters of the convolutional AE model’s encoder layers in the DL prediction models of recurrence risks. Tables E4 and E5 and Figure E3 show details of the models.
Validation of the deep learning models for predicting recurrence outcomes
We evaluated all the recurrence prediction models on the third cohort of 60 ES-NSCLC patients. Particularly, we applied the DL prediction models of recurrence risks to individuals of the third cohort for predicting risk scores. We then adopted concordance-index (c-index) of right-censored data to evaluate the prediction performance. We also compared the DL based radiomics model with a random survival forest model built on radiomic features computed from the gross tumor volume of each CT scan using pyradiomics.29,41
Survival functions of all 60 patients of the third cohort were estimated by Breslow’s estimator to dynamically describe the survival probability on individual level.42 As shown in Table E6, we also performed a cox regression analysis based on the combinations of the image-based DL risk scores, clinical measures, and CTC measures to further evaluate the recurrence prediction performance of our DL risks and other measures on the validation cohort. Table E6 also depicts the prediction performance of each of the included measure.
Patient stratification in terms of recurrence risks
The median of risk scores on the 60 patients was applied to stratify the risk scores estimated by the DL models to identify 2 subgroups of patients with high and low recurrence risks. We had previously established that high levels and persistence of CTCs after treatment defined patients at high risk of recurrence.20 For this analysis, a composite threshold of 3.9 at 3 months after SBRT was therefore chosen to identify persistent CTCs (“positive”). Based on the positive and nonpositive CTC information, patients with respectively high and low recurrence risks were further stratified into 4 subgroups with different recurrence outcomes. “False negative CTC” patients were defined as those for whom no CTCs were initially detected yet ultimately developed regional or distant recurrences.
Statistical analysis
Descriptive statistics were used to evaluate demographic characteristics, disease details, and longitudinal trends of CTC counts. Chi-square for categorical variables and 2-sample t test for continuous variables were performed to assess differences in clinicopathologic details between the demographic recordings of patients in the 2 included cohorts and stratified patient subgroups. The proportional hazards assumption of our Cox regression model was tested based on Schoenfeld residuals using proportional_hazard_test of Lifelines v0.26.3. The performance confidence interval (CI) of all prediction models was estimated with 100 runs of bootstrap sampling. A nonparametric approach was adopted to compare C-index values of prediction models based on mean of their prediction outputs of 100 runs of bootstrap sampling.43 Recurrence risks of different subgroups were assessed using log-rank test.
Results
The DL prediction model (combination of image-based and clinical-based prediction) obtained a C-index value of 0.880 (95% CI, 0.879-0.881) for predicting recurrence, better than DL prediction models built upon either clinical measures (P = .020) or imaging data alone (P = .078). In particular, the DL prediction model (Table E4) built upon the imaging data alone obtained a C-index of 0.849 (95% CI, 0.847-0.851) and the DL model (Table E5) built upon the clinical measures obtained a c-index of 0.705 (95% CI, 0.703-0.708). We also estimated the DL prediction model’s prediction performance on the third cohort’s men and women separately. The DL prediction model obtained c-index values of 0.851 (95% CI, 0.847-0.855) and 0.915 (95% CI, 0.913-0.917) for men and women, respectively.
The proportional hazards assumption testing results indicated that both the image-based recurrence risk scores and the clinical-based recurrence risk scores obtained by our DL models passed the nonproportional test, with P values of .66 and .33, respectively.
The conventional radiomics model obtained a c-index of 0.769 (95% CI, 0.759-0.779), significantly worse than the DL radiomics model (P = .048).
We also applied the attention visualization inspired by the class activation map44 to visualize the attention of our image-based DL model via calculating the contributions of locations in tumor areas to the recurrence risk results. The visualization results are shown as Figure E4. In this figure, our DL model could succeed in assigning higher recurrence risks to patients with larger and irregular-shaped tumors than the patients with smaller lesion appearance.
When using the combination of our image-based DL risk scores, clinical features, and CTC measures as input to build a cox regression model, the obtained C-index values on the 60-patient cohort was 0.870. Table E6 summarizes recurrence prediction results of imaged-based DL risks, CTC, and clinical measures. We also compared the pooled hazard rate45 with each group-specific hazard rate of all the included image-based DL risks, clinical measures, and CTC (Table E7). Specifically, our image-based DL risks and CTC measures could stratify patients into significantly different groups of recurrence outcomes (P < .05), whereas the most efficient measure (body mass index) of other included measures could just obtain stratification result that is marginally associated with the recurrence (P = .062).
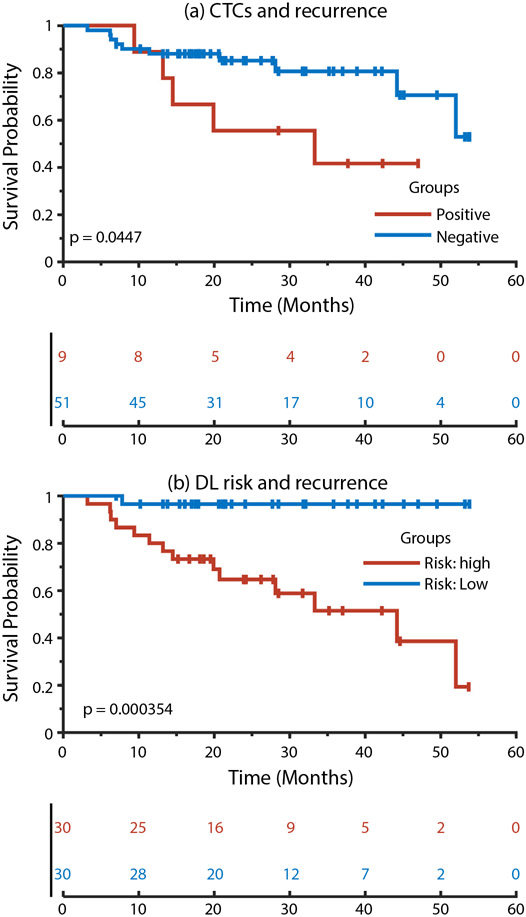
As shown in Figure 2(b), the high-risk and low-risk groups had significantly different recurrence outcomes (P = 3.5e-04, log-rank test). Significant differences were also observed in recurrence outcomes (P = .045, log-rank test) between the patients with positive and negative CTC measures, as shown in Figure 2(a).
Fig. 2.
Kaplan–Meier plots of patient subgroups with positive or negative circulating tumor cell measures and patient subgroups with high or low recurrence risks predicted by our deep-learning radiomics models. Abbreviations: CTC = circulation tumor cell; DL = deep learning.
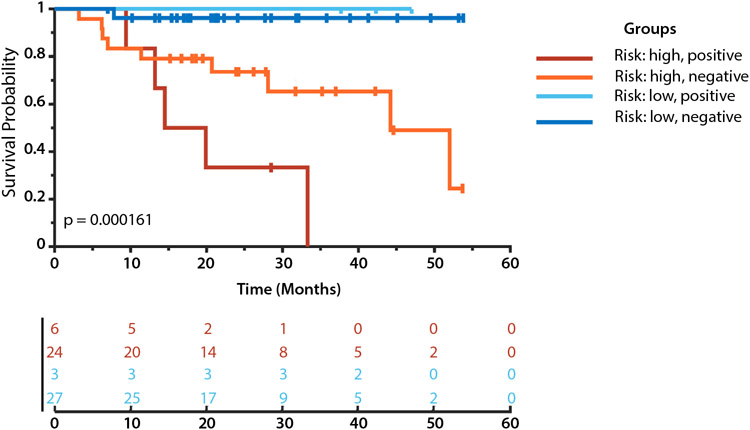
As shown in Figure 3, the integration of DL based risks and CTC positive/negative statues identified 4 subgroups with more significant differences than these 2 factors alone in their recurrence outcomes (P = 1.6e-04, log-rank test). As indicated by pairwise group difference measures summarized in Table 1 and Figure 3, the patients with high-risk estimated by the DL model and positive CTC measures had significantly increased recurrence risks in recurrence outcomes compared with those with low-risk and negative CTC measures.
Fig. 3.
Kaplan–Meier plots of patient subgroups identified jointly based on circulating tumor cell measures and our deep learning-based recurrence risks.
Table 1.
Statistical differences between patient subgroups in recurrence
| Patient subgroups | P value | χ2 |
|---|---|---|
| High-risk + positive CTC vs high-risk + negative CTC | .043 | 4.101 |
| High-risk + positive CTC vs low-risk + positive CTC | .025 | 5.002 |
| High-risk + positive CTC vs low-risk + negative CTC | 8.9e-06 | 19.729 |
| High-risk + negative CTC vs low-risk + positive CTC | .224 | 1.476 |
| High-risk + negative CTC vs low-risk + negative CTC | .006 | 7.633 |
| Low risk + positive CTC vs low-risk + negative CTC | .734 | 0.115 |
Abbreviation: CTC = circulating tumor cell.
As summarized in Table 1, for the patients with positive CTC measures, the DL based risk score was a significant risk predictor for recurrence (P = .025; log-rank test). For the patients with negative CTC measure, the DL based risk scores could also further stratify the patients into subgroups with distinct recurrence outcomes (P = .006; log-rank test). However, the CTC positivity could not further stratify the high-risk patients or low-risk patients with statistical significance.
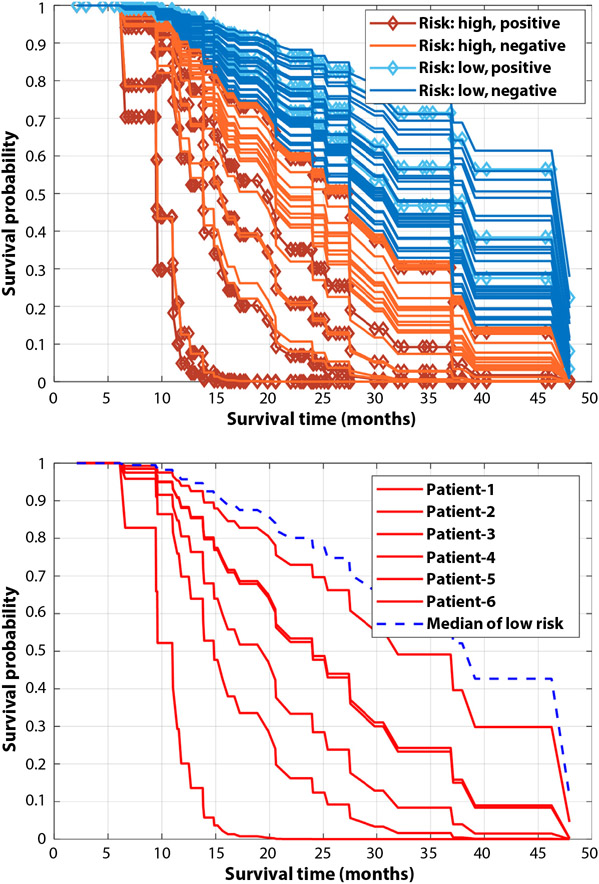
In the 60-patient cohort (9 patients with positive CTCs, 51 patients with negative CTCs), 6 patients had recurrences but with negative posttreatment CTCs (false negative CTCs). The DL prediction models predicted their recurrence. Specifically, as illustrated by their survival functions in Figure 4(b), our DL model could assign survival functions worse than the median recurrence risk patient of the low-risk group to all the 6 false negative CTCs patients. Figure 4a summarizes survival functions of all 60 patients belonging to different risk subgroups and shows that our DL-based risk prediction is efficient to stratify patients into subgroups of different posttreatment survival outcomes.
Fig. 4.
Top: Survival functions of the 60-patient data set estimated by our deep learning model, risk subgroups in this figure are the same as that in Figure 3. Bottom: Survival functions of 6 patients with negative circulating tumor cell measures and recurrence outcomes (false negative circulating tumor cells); the blue dash line represents the median survival function of patients in the low-risk group identified by our deep learning model.
Discussion
This paper presents the first study of integration of DL radiomics models and CTC measures for predicting recurrence risks of ES-NSCLC patients treated with SBRT. The objective of the present study is twofold: to evaluate if integration of radiomics and CTC counts improves stratification of ES-NSCLC patients in terms of their response to SBRT and to test if a deep learning model in a transfer learning setting can improve the performance of radiomics for prediction of outcomes of ES-NSCLC patients treated with SBRT. Our results have demonstrated that an effective integration of DL based radiomics models and CTC information could stratify ES-NSCLC with distinct recurrence risks, better than the image-based model, clinical measures-based model, and CTC measures alone. Particularly, the DL radiomics models provided complementary information to the CTC measures for predicting recurrence risks, especially for the patients with false negative CTC indictors. The prediction models may facilitate selection of patients who will benefit from intensified treatments after SBRT to improve outcomes, such as immunotherapy, chemotherapy, or additional radiation therapy.
Compared with the clinical features-based model, our image-based models built on unsupervised deep image features achieved superior performance on the recurrence prediction task when evaluated on an independent validation set of ES-NSCLC patients (the third cohort). The DL-based radiomics model performed significantly better than the conventional radiomics model that was built radiomic image features, facilitating accurate stratification of patients with high and low recurrence risks. This further demonstrated that the DL-based radiomics models could provide clinically meaningful prediction for outcomes of ES-NSCLC patients treated SBRT.
Combining the recurrence prediction results based on DL models with the CTC measures, we could further categorize the included ES-NSCLC patients into 4 subgroups with significant differences in recurrence outcomes. The refined recurrence outcome prediction combining both image-based DL models and the biologically relevant lung cancer indicator of CTCs were particularly predictive for ES-NSCLC patients treated with SBRT.
The DL-based radiomics models also achieved promising prediction performance for patients with recurrence but negative CTC measures. As illustrated in Figure 4, all these patients could be assigned with worse survival functions than the median of the low-risk group of the third cohort. These results further demonstrated that our DL-based radiomics models achieved high accuracy for predicting recurrence.
We adopted mixed unsupervised and supervised learning on separate cohorts to build prediction models for preventing overfitting. Because the public data sets do not provide follow-up information of treatment response to SBRT, it is difficult to combine them with our local cohorts. Therefore, the unsupervised learning was adopted to learn imaging features from the public data sets, and the learned unsupervised learning model was adopted to extract features and built a prediction model under a supervised learning setting. Because our local cohorts with follow-up information of treatment response were relatively small, a supervised deep learning prediction model on the second cohort had degraded performance when tested on the third cohort, with a c-index of 0.775 (95% CI, 0.751-0.798), worse than the performance obtained by the DL model built in the transfer learning setting.
A caveat of the present study is that the DL features were learned from images collected by multiple CT scanners. Though prediction models built on the DL features achieved significantly better performance than a prediction model built on conventional radiomics features, differences in the images from multiple CT scans might affect the stability of the DL features. It merits further investigation to evaluate how the difference in images from multiple CT scans/sites affect the deep learning features and deep learning models.
Because the training subjects (the second cohort) did not have CTC information, we did not directly combine the CTC information with the imaging and clinical data to build a prediction model. Instead, the CTC information was used to stratify the third cohort into 2 subgroups with negative or positive CTC. We also split the third cohort into 2 subgroups with high or low risk based on individual patients’ risk scores predicted by the combined DL prediction model built on the second cohort. These 2 different groupings were finally combined to stratify the third cohort into 4 subgroups (high-risk + positive CTC; high-risk + negative CTC; low risk + positive CTC; and low risk + negative CTC). As shown in Tables E6 and E7, statistical analyses based on Cox models on the third cohort had shown that the combination of DL radiomics risk score, clinical measures, and CTC measures obtained the best prediction performance, and both the DL radiomics risk score and CTC measure were predictors with statistical significance (P < .05). However, it merits further investigation based on large data sets to evaluate to what extent an integration of the radiomics and CTC measures can improve the prediction.
One limitation of the present study is that we did not build prediction models separately for men and women due to the small number of noncensored patients (ie, with recurrences) available for training the models (5 men and 10 women). The prediction performance of our DL prediction model was different for men and women, and the sex effect merits further investigation as well.
Conclusions
Our study suggested that integration of image- and clinical measure-based DL models with CTC measures could help distinguish patients with a high probability of cure from those at high risk of recurrence after SBRT. The integration of these novel methods merits additional study and verification and may ultimately prove to be useful tools for identifying patients with early-stage NSCLC treated with SBRT who may benefit maximally from intensified treatment, including post-SBRT immunotherapy.
Supplementary Material
Acknowledgments
This study was supported by the National Cancer Institute of the National Institutes of Health under award number CA223358.
Footnotes
Disclosures: none.
Research data for statistical analysis and the prediction model files are available at https://github.com/dumplingxxx/lung_cancer_dl_ctc.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijrobp.2021.11.006.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 2.Howlander N, Noone A, Krapcho M. SEER Cancer Statistics Review (CSR) 1975–2016. National Cancer Institute Website. 2019. Available online: https://seer.cancer.gov/csr/1975_2016/. Accessed December 15, 2019. [Google Scholar]
- 3.Ettinger DS, Akerley W, Borghaei H, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2012;10:1236–1271. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 5.International Early Lung Cancer Action Program Investigator-sHenschke CI, Yankelevitz DF, et al. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med 2006;355:1763–1771. [DOI] [PubMed] [Google Scholar]
- 6.Kovalchik SA, Tammemagi M, Berg CD, et al. Targeting of low-dose CT screening according to the risk of lung-cancer death. N Engl J Med 2013;369:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palma D, Visser O, Lagerwaard FJ, et al. Impact of introducing stereotactic lung radiotherapy for elderly patients with stage I non-small-cell lung cancer: A population-based time-trend analysis. J Clin Oncol 2010;28:5153–5159. [DOI] [PubMed] [Google Scholar]
- 8.Kapadia NS, Valle LF, George JA, et al. Patterns of treatment and outcomes for definitive therapy of early stage non-small cell lung cancer. Ann Thorac Surg 2017;104:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Videtic GM, Donington J, Giuliani M, et al. Stereotactic body radiation therapy for early-stage non-small cell lung cancer: Executive summary of an astro evidence-based guideline. Pract Radiat Oncol 2017;7:295–301. [DOI] [PubMed] [Google Scholar]
- 10.Shah JL, Loo BW. Stereotactic ablative radiotherapy for early-stage lung cancer. Sem Radiat Oncol 2017;27:218–228. [DOI] [PubMed] [Google Scholar]
- 11.Timmerman RD, Paulus R, Pass HI, et al. Stereotactic body radiation therapy for operable early-stage lung cancer: Findings from the NRG oncology RTOG 0618 trial. JAMA Oncol 2018;4:1263–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pignon J-P, Tribodet H, Scagliotti GV, et al. Lung adjuvant cisplatin evaluation: A pooled analysis by the lace collaborative group. J Clin Oncol 2008;26:3552–3559. [DOI] [PubMed] [Google Scholar]
- 13.Vachani A, Sequist LV, Spira A. AJRCCM: 100-year anniversary. The shifting landscape for lung cancer: Past, present, and future. Am J Respir Crit CareMed 2017;195:1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim C, Sekhon HS, Cutz JC, et al. Improving molecular testing and personalized medicine in non-small-cell lung cancer in Ontario. Curr Oncol 2017;24:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong FS, Hirsch FR, Machtay M. Potential future consideration for imaging and blood-based biomarkers for precision medicine in lung cancer. Transl Lung Cancer Res 2017;6:713–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non–small-cell lung cancer. J Clin Oncol 2011;29:1556–1563. [DOI] [PubMed] [Google Scholar]
- 17.Aceto N, Bardia A, Miyamoto DT, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 2007;13:7053–7058. [DOI] [PubMed] [Google Scholar]
- 19.Hou J-M, Krebs MG, Lancashire L, et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol 2012;30:525–532. [DOI] [PubMed] [Google Scholar]
- 20.Frick MA, Feigenberg SJ, Jean-Baptiste SR, et al. Circulating tumor cells are associated with recurrent disease in patients with early stage non-small cell lung cancer treated with stereotactic body radiation therapy. Clin Cancer Res 2020;26:2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Li H, Habes M, et al. Robust collaborative clustering of subjects and radiomic features for cancer prognosis. IEEE Trans Biomed Eng 2020;67:2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Galperin-Aizenberg M, Pryma D, et al. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother Oncol 2018;129:218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coroller TP, Grossmann P, Hou Y, et al. Ct-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114:345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Liu Z, He L, et al. Radiomics signature: A potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 2016;281:947–957. [DOI] [PubMed] [Google Scholar]
- 27.Lee G, Lee HY, Park H, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol 2017;86:297–307. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Boimel P, Janopaul-Naylor J, et al. Deep convolutional neural networks for imaging data based survival analysis of rectal cancer. Proc IEEE Int Symp Biomed Imaging 2019;2019:846–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao Z, Li H, Xiao Y, et al. Integration of risk survival measures estimated from pre- and posttreatment computed tomography scans improves stratification of patients with early-stage non-small cell lung cancer treated with stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2021;109:1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 2018;24:1248–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aboutalib SS, Mohamed AA, Berg WA, et al. Deep learning to distinguish recalled but benign mammography images in breast cancer screening. Clin Cancer Res 2018;24:5902–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi IL, Zhao Y, Wang R, et al. Deep learning to distinguish benign from malignant renal lesions based on routine mr imaging. Clin Cancer Res 2020;26:1944–1952. [DOI] [PubMed] [Google Scholar]
- 33.Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med 2018;15 e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Hosny A, Zeleznik R, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res 2019;25:3266–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee P, Zhou M, Lee E, et al. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat Mach Intell 2020;2:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frick MA, Kao GD, Aguarin L, et al. Circulating tumor cell assessment in presumed early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy: A prospective pilot study. Int J Radiat Oncol Biol Physics 2018;102:536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorsey JF, Kao GD, MacArthur KM, et al. Tracking viable circulating tumor cells (CTCs) in the peripheral blood of non–small cell lung cancer (NSCLC) patients undergoing definitive radiation therapy: Pilot study results. Cancer 2015;121:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacArthur KM, Kao GD, Chandrasekaran S, et al. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res 2014;74:2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606–612. [PubMed] [Google Scholar]
- 40.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 41.Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017;77:e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin D On the Breslow estimator. Lifetime Data Anal 2007;13:471–480. [DOI] [PubMed] [Google Scholar]
- 43.Kang L, Chen W, Petrick NA, Gallas BD. Comparing two correlated C indices with right-censored survival outcome: A one-shot nonparametric approach. Stat Med 2015;34:685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B, Khosla A, Lapedriza A, et al. Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on com-puter vision and pattern recognition. 2921–2929. [Google Scholar]
- 45.Fleming TR, Harrington DP. A class of hypothesis tests for one and two sample censored survival data. Commun Stat Theory Methods 1981;10:763–794. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.