Abstract
Quality control of mRNA represents an important regulatory mechanism for gene expression in eukaryotes. One component of this quality control is the nuclear retention and decay of misprocessed RNAs. Previously, we demonstrated that mature mRNAs containing a 5′ splice site (5′SS) motif, which is typically found in misprocessed RNAs such as intronic polyadenylated (IPA) transcripts, are nuclear retained and degraded. Using high-throughput sequencing of cellular fractions, we now demonstrate that IPA transcripts require the zinc finger protein ZFC3H1 for their nuclear retention and degradation. Using reporter mRNAs, we demonstrate that ZFC3H1 promotes the nuclear retention of mRNAs with intact 5′SS motifs by sequestering them into nuclear speckles. Furthermore, we find that U1-70K, a component of the spliceosomal U1 snRNP, is also required for the nuclear retention of these reporter mRNAs and likely functions in the same pathway as ZFC3H1. Finally, we show that the disassembly of nuclear speckles impairs the nuclear retention of reporter mRNAs with 5′SS motifs. Our results highlight a splicing independent role of U1 snRNP and indicate that it works in conjunction with ZFC3H1 in preventing the nuclear export of misprocessed mRNAs by sequestering them into nuclear speckles.
Keywords: intronic polyadenylated transcripts, nuclear speckles, PAXT, U1 snRNP
INTRODUCTION
Eukaryotic cells are divided into two compartments: the nucleus and cytoplasm. In the nucleus, DNA is transcribed into pre-mRNAs, which undergo a number of processing events to generate mature mRNAs. These mRNAs are then exported to the cytoplasm where they are translated into proteins. Importantly, a subset of all transcripts are misprocessed (Pickrell et al. 2010; Skandalis 2016) and, if transported to the cytoplasm, they would be translated into truncated proteins that are often toxic (Veitia 2007). To deal with these misprocessed transcripts, eukaryotic cells have evolved quality control mechanisms that prevent these mRNAs from being exported and instead target these RNAs for degradation (Lee et al. 2015). For example, transcripts that contain retained introns are sequestered in the nucleus and targeted for decay (Palazzo and Lee 2018). Such RNAs still contain intron-associated cis-elements, which are normally removed during splicing. These include the elements that specify the exon–intron boundaries (the 5′ splice site [5′SS] and the 3′ splice site [3′SS] motifs) and the branchpoint. Other misprocessed transcripts may be generated when a cryptic 3′ cleavage signal within an intron is used. These intronic polyadenylated (IPA) transcripts also include an unutilized 5′SS motif present near the 3′ end of the RNA (Tian et al. 2007). Interestingly, it has been proposed that many lncRNAs are also nuclear retained due to the fact that they have poorly spliced introns (Zuckerman and Ulitsky 2019). Moreover, other studies have found that many lncRNAs are retained by the U1 snRNP (Azam et al. 2019; Yin et al. 2020; Lubelsky et al. 2021), the component of the spliceosome that recognizes 5′SS motifs, which are likely found in lncRNAs with retained introns.
Previously, we showed that RNAs that contain a consensus 5′SS motif near their 3′ end are retained in the nucleus and degraded (Lee et al. 2015). Our findings are consistent with other lines of experiments that have demonstrated that the presence of a 5′SS motif in a mature mRNA inhibits protein expression (Boelens et al. 1993; Gunderson et al. 1998; Vagner et al. 2000; Fortes et al. 2003; Guan et al. 2007; Abad et al. 2008; Goraczniak et al. 2009; Blázquez and Fortes 2013). It remains unclear how the 5′SS motif triggers nuclear retention. One possibility is that the U1 snRNP itself may play a role. This is in line with the finding that tethering U1 components to a reporter promotes nuclear retention (Takemura et al. 2011). Interestingly, U1 snRNP is present at very high levels, about one order of magnitude higher than other snRNPs (Baserga and Steitz 1993), supporting the idea that it has additional roles beyond splicing. Indeed, the U1 snRNP components U1A and U1-70K have been shown to be required for the ability of the 5′SS motif to inhibit protein expression by repressing 3′ cleavage and polyadenylation (Boelens et al. 1993; Gunderson et al. 1998; Vagner et al. 2000). The U1 snRNP is also required for the 5′SS motif to inhibit cleavage at downstream cryptic 3′ cleavage signals in introns that produce IPA transcripts (Kaida et al. 2010; Berg et al. 2012). U1 may also help to stabilize certain transcripts. Many cryptic transcripts, especially upstream anti-sense transcripts from bidirectional promoters, are depleted in 5′SS motifs and as a consequence fail to recruit U1 and are thus rapidly cleaved and targeted for decay (Almada et al. 2013).
Previously we noted that reporter RNAs that contained 5′SS motifs accumulated in nuclear speckles (Lee et al. 2015). These liquid–liquid phase separated nuclear compartments contain splicing factors, such as SR proteins and U1 snRNPs, nuclear export factors, such as the TREX complex, and other mRNP factors, such as the exon junction complex (Huang and Spector 1992; Schmidt et al. 2006; Dias et al. 2010; Dufu et al. 2010; Spector and Lamond 2011; Daguenet et al. 2012; Akef et al. 2013; Galganski et al. 2017). It has been suggested that post-transcriptional splicing occurs in these compartments (Dias et al. 2010). Thus, the recruitment of U1 snRNP may help to target mRNAs with intact 5′SS motifs into nuclear speckles where they are either spliced (if the 5′SS is followed by a proper branch point and a 3′SS motif) or sequestered (if the 5′SS is followed by a poly(A)-tail).
Beyond the U1 snRNP, it has been unclear what other factors may be involved. One likely candidate is the PAXT complex, which is composed of MTR4, ZFC3H1, PABPN1, and other accessory proteins (Meola et al. 2016; Ogami and Manley 2017; Ogami et al. 2017; Silla et al. 2020; Wu et al. 2020). While MTR4 is found in other complexes and generally targets RNAs to the nuclear exosome (Schilders et al. 2007; Ogami et al. 2018), ZFC3H1 is thought to form the core of the PAXT complex and specifically recognize certain RNA substrates. Although it contains a zinc-finger domain and a coiled-coil region, it is largely unstructured and can promote phase-separation when expressed in cells (Wang et al. 2021). PABPN1 binds to poly(A)-tails and has been implicated in many different aspects of RNA metabolism, not only in PAXT-dependent RNA surveillance but also in nuclear export and nonsense-mediated decay (Sato and Maquat 2009; Apponi et al. 2010).
Previously, it had been shown that upon depletion of MTR4 or ZFC3H1 in HeLa cells, IPA transcripts become stabilized and then accumulate in the cytoplasm (Ogami et al. 2017). This work looked at the overall distribution of mRNAs by next-generation sequencing in MTR4-depleted cells and only specific IPA transcripts by RT-PCR in ZFC3H1-depleted cells. Depletion of PAXT components also led to the up-regulation of certain non-coding transcripts, such as a subset of enhancer RNAs (eRNAs) and promoter upstream transcripts (PROMPTs) (Meola et al. 2016; Fan et al. 2017; Silla et al. 2020; Wu et al. 2020). Interestingly, MTR4 depletion in HeLa cells also led to ZFC3H1 co-depletion (Ogami et al. 2017; Silla et al. 2018), likely by destabilizing the PAXT complex. Thus, it was not clear whether the effects observed on IPA transcripts were due to the direct depletion of MTR4, the indirect depletion of ZFC3H1, or a combination of the two. MTR4 and ZFC3H1 may directly compete with the TREX component Aly (also known as AlyRef) for RNA binding (Fan et al. 2017; Silla et al. 2018), although whether this is related to any putative U1-mediated nuclear retention remains unclear.
Here we identify factors that promote the nuclear retention of RNAs that contain a 5′SS motif, and investigate the role of nuclear speckles in this process. We find that following ZFC3H1 depletion, there is a global up-regulation of IPA transcripts, which accumulate in the cytoplasm. In addition, we observe that there is a defect in the nuclear export of mRNAs with long 3′UTRs that are generated from distal 3′ polyadenylation/cleavage sites. Using reporter mRNAs, we demonstrate that ZFC3H1, but not other PAXT components are required for the nuclear retention of mRNAs with intact 5′SS motifs in human U2OS cells. We also find that U1-70K is also required for their nuclear retention and likely acts in the same pathway as ZFC3H1. We demonstrate that ZFC3H1 and U1-70K are required for mRNAs with 5′SS motifs to be retained in nuclear speckles and that disassembly of nuclear speckles prevents nuclear retention.
RESULTS
Depletion of ZFC3H1 leads to the stabilization and cytoplasmic accumulation of intronic polyadenylated (IPA) transcripts
To investigate how ZFC3H1 affects the distribution and levels of various mRNAs, we depleted it from human osteosarcoma (U2OS) cells using lentiviral-delivered shRNA (“ZFC3H1-2”), isolated total, nuclear and cytoplasmic fractions, and purified poly(A)-selected RNAs for next-generation sequencing (Fig. 1A). The fractions had minimal cross contamination (Fig. 1B) and the depletion was almost complete (Fig. 1C). Note that ZFC3H1 depletion had no observable effects on MTR4 levels (Fig. 2A).
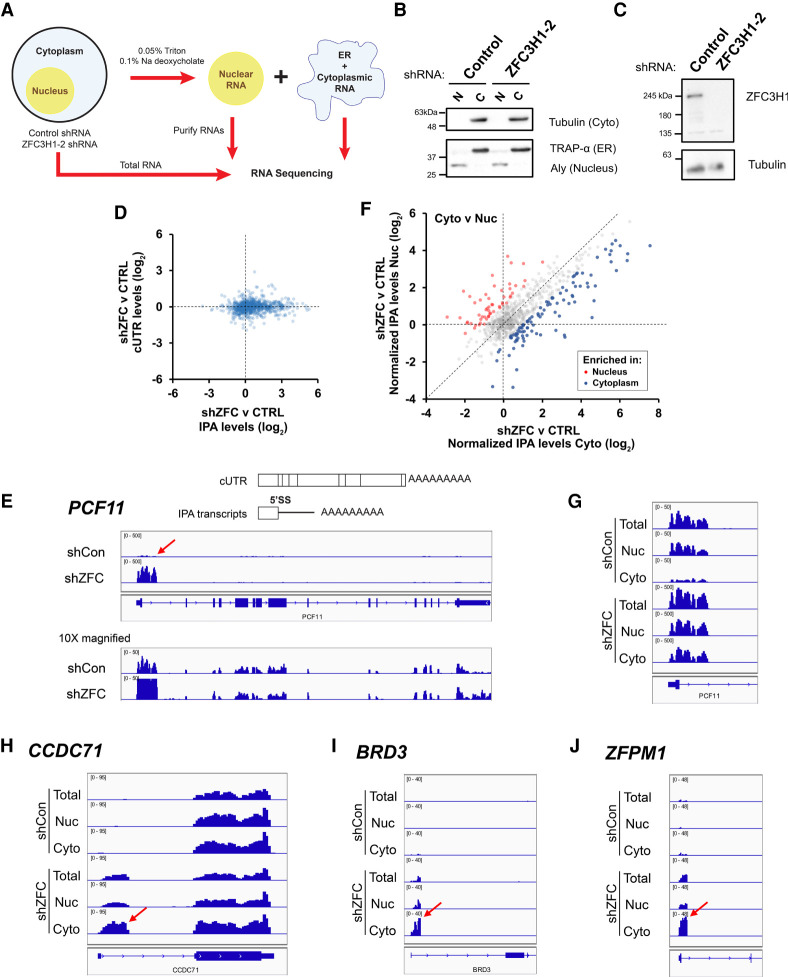
FIGURE 1.
ZFC3H1 depletion leads to cytoplasmic accumulation of endogenous 5′SS motif containing mRNAs (or intronic polyadenylated transcripts). (A) Workflow for RNA Frac-seq. ZFC3H1- or control-depleted U2OS cells were fractionated into nuclear and cytoplasmic/ER fractions (see Materials and Methods for more details). RNA was purified from these fractions and from total cell lysates, and then analyzed by Illumina sequencing. (B) Nuclear “N” and cytoplasmic “C” fractions were collected from ZFC3H1- or control-depleted U2OS cells, then separated by SDS-PAGE and analyzed by immunoblot for nuclear (Aly), ER (Trap-α), and cytoplasmic (tubulin) protein markers. (C) Lysates collected from ZFC3H1- or control-depleted U2OS cells (96 h post-transduction with lentiviral-delivered ZFC3H1-2 shRNA) were analyzed by immunoblot for ZFC3H1 and tubulin. (D) Fold change in total levels of intronic polyadenylated (IPA) transcripts (ZFC3H1 depletion vs. control depletion) (x-axis), plotted against the change in the total levels of fully processed mRNA (using cUTR reads, y-axis). Each dot corresponds to reads from one gene that is known to produce IPA transcripts (listed in Supplemental Table 1). Note that ZFC3H1 depletion leads to the up-regulation of IPA transcripts, but not fully processed mRNAs. (E) (Top) Schematic of a fully processed mRNA and IPA transcript generated from the same gene. Note that the IPA transcript is generated from a 3′ cleavage/polyadenylation signal in the first intron and contains a 5′SS motif. (Bottom) genome browser tracks of the PCF11 gene in control- “shCon” or ZFC3H1-depleted “shZFC” cells at 500× and 50× resolution. Note the large peak for the IPA transcript (intronic cleavage/polyadenylation site is denoted with a red arrow), which is up-regulated in ZFC3H1-depleted cells. Also note that the reads corresponding to the full-length transcript are unaffected by ZFC3H1 depletion. (F) Similar to D, except that the fold change of IPA transcript levels in the cytoplasmic fraction (ZFC3H1 depletion vs. control depletion, x-axis) is plotted against the fold change in the nuclear fraction (ZFC3H1 depletion vs. control depletion, y-axis). Note that ZFC3H1 depletion leads to cytoplasmic accumulation of many IPA transcripts (compare blue dots to red). To account for reads from fully processed mRNAs, the IPA transcript levels are normalized to the cUTR transcript levels of the same gene. (G) Similar to E, except the IPA peaks in the nuclear and cytoplasmic fractions are shown for PCF11. In control-depleted cells, reads from the PCF11 IPA are enriched in the nuclear, but not the cytoplasmic fractions. In ZFC3H1-depleted cells, reads from the PCF11 IPA are at comparable levels in the nuclear and cytoplasmic fractions. (H–J) Genome browser tracks of three IPA transcript-producing genes, “CCDC71,” “BRD3,” and “ZFPM1.” Note the accumulation of IPA transcripts in the cytoplasmic fractions upon ZFC3H1 depletion. The intronic 3′ cleavage/polyadenylation sites are denoted by red arrows.
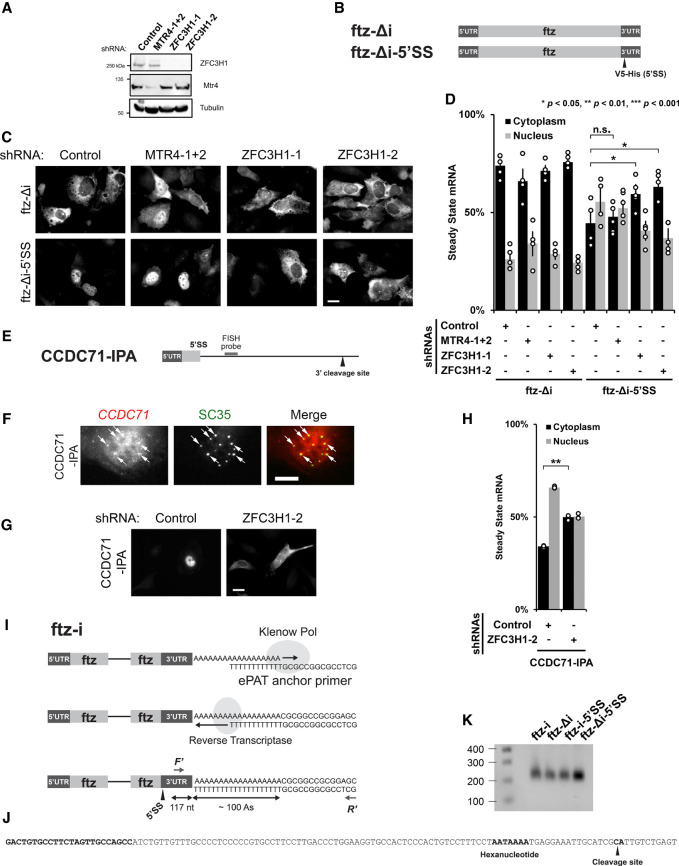
FIGURE 2.
ZFC3H1 is required for the nuclear retention of 5′SS motif containing mRNAs. (A) U2OS cells were treated with different lentivirus shRNAs against ZFC3H1 (“ZFC3H1-1” and “ZFC3H1-2”), MTR4 (“MTR4-1 + 2”) or control shRNA. Lysates were collected 96 h post-transduction, separated by SDS-PAGE and immunoprobed for ZFC3H1, MTR4, or tubulin. Note that to effectively deplete MTR4, cells were treated with lentivirus containing two shRNA plasmids. (B) Schematic of the intronless (Δi) ftz reporter (ftz-Δi) construct used in this study, with and without the V5-His element in the 3′UTR (ftz-Δi-5′SS). Note that the V5-His element contains a consensus 5′SS motif, which promotes nuclear retention. (C,D) Control-, MTR4-, and ZFC3H1-depleted cells were transfected with the intronless ftz reporter plasmid (±5′SS). Eighteen to twenty-four hours later, the cells were fixed and the mRNA was visualized by FISH. Note that depletion of ZFC3H1, but not MTR4, caused the cytoplasmic accumulation of the ftz-Δi-5′SS mRNA. Representative images are shown in C (scale bar, 10 µm) and quantification is shown in D. Each bar represents the average and standard error of at least three independent experiments, each experiment consisting of at least 30 to 60 cells. Student's t-test was performed for Figure 2D. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (E) Schematic of CCDC71-IPA reporter used in this study (see also Fig. 1H). The position of the FISH probe used to visualize the IPA RNA is marked in gray and the position of the 3′ cleavage site in the intron is as indicated. (F) U2OS cells were transfected with the CCDC71-IPA reporter and, 18 to 24 hours later, the cells were fixed. The IPA transcript was visualized by FISH and nuclear speckles were visualized by immunofluorescence against SC35. Representative images are shown with a merged overlay showing the CCDC71-IPA mRNA in red and SC35 in green. Scale bar, 10 µM. Examples of CCDC71-IPA/SC35 colocalization are indicated with arrows. (G,H) Control- and ZFC3H1-depleted cells were transfected with the CCDC71-IPA reporter and the IPA transcript was visualized by FISH. ZFC3H1 depletion increased the cytoplasmic accumulation of the CCDC71-IPA. Representative images are shown in G (scale bar, 10 µm) and quantification is shown in H. Each bar represents the average and standard error of at least three independent experiments, each experiment consisting of at least 30 to 60 cells. Student's t-test was performed for Figure 2H. (**) P < 0.01. (I–K) ePAT assay and 3′RACE were used to examine 3′ end processing. (I) Schematic of the ePAT assay as described in Janicke et al. (2012). The ftz-specific (F′) and universal (R′) primers used to amplify the ePAT amplicon are indicated. The sequence of the ePAT amplicon before the cleavage site is shown in J and is 117 nt long. (J) The sequence of the end of the 3′UTR is shown. Indicated in bold are the ftz-specific F′ primer annealing site (used in the ePAT and 3′RACE experiments), the hexanucleotide motif, and the cleavage site (as determined by 3′RACE experiments on mRNAs derived from U2OS cells transfected with either ftz-Δi or ftz-Δi-5′SS). (K) PCR products from the ePAT assay were separated on a 1% agarose gel and stained with ethidium bromide. Lane 1: Molecular weight markers with sizes in bp indicated on the left; lanes 3–6: ePAT amplicons from U2OS cells that were transfected with plasmids containing the indicated versions of the ftz reporter (without [Δi] or with [i] an intron, without or with the 5′SS motif). Note that the amplicons generated from all four reactions are the same length (∼230 nt). Since the amplified region in the 3′UTR is 117 bp long (see J), and the universal primer has a 14 nt extension (see J), the poly(A)-tail is estimated to be ∼100 nt long.
In line with what had been previously reported in MTR4-depleted cells (Ogami et al. 2017), ZFC3H1 depletion caused an up-regulation of IPA transcripts with no detectable effect on fully processed mRNAs produced from the same genes (Fig. 1D; Supplemental Table 1). When we looked at particular examples, such as PCF11, which has a well characterized IPA (Wang et al. 2019), we observed that it was up-regulated more than 10-fold after ZFC3H1 depletion (Fig. 1E, note that the 3′ end of the PCF11 IPA transcript is indicated by the red arrow), while the full-length PCF11 mRNA was unaffected (Fig. 1E, see 10× magnification). The up-regulation of IPA transcripts was also apparent for other genes (Supplemental Fig. S1A–C). Although most IPAs occur through the action of cryptic 3′ polyadenylation/cleavage sites in the first intron, a minority are generated by sites found in internal introns, for example, in the KCT13 gene, and these were also up-regulated after ZFC3H1 depletion (Supplemental Fig. S1C).
When the levels of these IPAs were compared between the nucleus and the cytoplasm, we found that their cytoplasmic levels generally increased more than their nuclear levels in the ZFC3H1-depleted cells (Fig. 1F), suggesting that ZFC3H1 was also required for their nuclear retention. This could be clearly seen in individual examples, such as the PCF11 IPA (Fig. 1G) and other IPA transcripts (Fig. 1H–J).
As documented previously, we also observed that ZFC3H1 depletion caused an up-regulation in certain PROMPTs (an example is shown in Supplemental Fig. S1D), and in many cases these tended to accumulate more rapidly in the cytoplasm than in the nucleus (an example is shown in Supplemental Fig. S1E).
From these results, we conclude that depletion of ZFC3H1 results in the up-regulation of IPA transcripts and their accumulation in the cytoplasm. These results are in line with previously published results examining MTR4-depleted cells (Ogami et al. 2017). In addition, ZFC3H1 depletion led to the up-regulation of a subset of PROMPTs, as described by others (Meola et al. 2016; Silla et al. 2020; Wu et al. 2020), and their cytoplasmic accumulation.
Depletion of ZFC3H1 leads to the inhibition of nuclear export of mRNAs generated from distal 3′ polyadenylation/cleavage sites
Although ZFC3H1 depletion did not grossly affect the nuclear/cytoplasmic distribution of most mRNAs, we did notice that for mRNAs generated from genes with several 3′ polyadenylation/cleavage signals (Supplemental Fig. S2A), the cytoplasmic/nuclear distribution of the long mRNA isoforms, generated from distal 3′ polyadenylation/cleavage sites, tended to be more nuclear in the ZFC3H1-depleted cells when compared to control cells (Supplemental Fig. S2B,C). In contrast, the overall distribution of mRNAs was generally unaffected (see “mRNAs” in Supplemental Fig. S2B). To determine whether this inhibition of export was true for short isoforms produced from the same genes, we compared the portions of the 3′UTR that are common to both short and long isoforms (cUTR, see Supplemental Fig. S2A). Reads that mapped to cUTR regions were less affected but still more skewed toward the nucleoplasmic fraction (Supplemental Fig. S2B,D). Since these reads come from both long and short isoforms, we attempted to infer the short UTR counts by subtracting out the signal in the cUTR reads that came from the long isoform (see Materials and Methods). The distribution of these inferred short UTR counts between the nucleus and cytoplasm were relatively unaffected (Supplemental Fig. S2E). To determine whether these trends were due to an overall destabilization of long isoforms, we next compared the change in cytoplasmic/nuclear distribution to the overall mRNA levels. We observed that upon ZFC3H1 depletion, the decrease in cytoplasmic/nuclear distribution was much larger than any change in the total levels for the long isoform mRNAs (Supplemental Fig. S2F). Again, this trend was less pronounced in cUTR reads (which come from both short and long isoforms), and completely disappeared in the inferred short UTR counts (Supplemental Fig. S2G,H). These trends were also seen when the levels in the cytoplasm and nucleus were directly compared with total levels (Supplemental Fig. S3).
From these results, we conclude that ZFC3H1 is likely required for the export of mRNAs generated from distal 3′ polyadenylation/cleavage sites.
ZFC3H1, but not other members of the PAXT complex, are required for the nuclear retention of reporter mRNAs containing 5′SS motifs
Previously, we documented that mRNAs that contain intact 5′SS motifs were nuclear retained. These reporters have the same configuration as IPA transcripts since both contain intact 5′SS motifs just upstream of the poly(A)-tail. In light of this, we assessed whether MTR4 or ZFC3H1 were required for the nuclear retention of reporter mRNAs containing 5′SS motifs. We treated U2OS cells with lentiviral-delivered shRNAs against MTR4 or ZFC3H1, or control shRNAs (Fig. 2A) and transfected plasmids containing the reporter mRNA fushi tarazu (ftz) with and without the 5′SS motif (Fig. 2B) as previously described (Lee et al. 2015, 2020). Note that the 5′SS motif is present in a DNA region found in many expression vectors and encodes a V5-His epitope tag; however, within the ftz reporter, this region is present downstream from the stop codon and is not translated. Additionally, this version of ftz lacks any intron (Δi). To successfully deplete MTR4, we used a pool of two separate shRNAs, as transduction with any single lentiviral-delivered shRNA gave poor depletion (data not shown). In contrast to what had been seen previously in HeLa cells (Ogami et al. 2017; Silla et al. 2018), MTR4 depletion did not lead to the co-depletion of ZFC3H1 (Fig. 2A). This allowed us to determine whether MTR4 directly affects IPAs independently of ZFC3H1. Depletion of ZFC3H1 was performed using two independent shRNAs, and this did not affect MTR4 protein levels (Fig. 2A). When we analyzed the distribution of the ftz reporter mRNA by fluorescent in situ hybridization (FISH), we found that MTR4 depletion did not affect the cytoplasmic/nuclear levels of ftz mRNA containing the 5′SS motif (ftz-Δi-5′SS) (Fig. 2C,D). In contrast, depletion of ZFC3H1 with either shRNA led to a significant inhibition in the nuclear retention of the same mRNA (Fig. 2C,D). Depletion of either MTR4 or ZFC3H1 had no drastic effect on the cytoplasmic/nuclear levels of ftz lacking the 5′SS motif (ftz-Δi), although in certain MTR4-depleted cells, this RNA became slightly more nuclear (Fig. 2C,D). Depletion of ZFC3H1, but not MTR4, also led to an increase in the total levels of both ftz and ftz-Δi-5′SS (Supplemental Fig. S4A).
We next assessed how ZFC3H1 depletion affected the distribution of two reporters, CCDC71-IPA and PCF11-IPA (Fig. 2E; Supplemental Fig. S5A), that generate IPA transcripts. Both are fragments of endogenous genes that produce natural IPA transcripts whose decay and nuclear retention require ZFC3H1 according to our sequencing results (Fig. 1E,G,H). To specifically visualize the IPA transcripts, transfected U2OS cells were probed with FISH oligonucleotides that target intronic regions upstream of the intronic 3′ cleavage/polyadenylation site (Fig. 2E; Supplemental Fig. S5A). As we had previously documented for reporter RNAs with intact 5′SS motifs, the CCDC71-IPA was enriched in nuclear speckles (Fig. 2F). Although the PCF11-IPA transcript also tended to accumulate in puncta that overlapped with speckles, the degree of colocalization was not as high (Supplemental Fig. S5B). We also observed that the depletion of ZFC3H1 significantly inhibited the nuclear retention of the CCDC71 IPA transcript (Fig. 2G,H). We observed a similar, albeit more subtle, effect on the PCF11-IPA transcript (Supplemental Fig. S5C,D). Depletion of ZFC3H1 also led to an increase in overall levels of CCDC71-IPA and PCF11IPA transcripts (Supplemental Fig. S4B,C).
Next, we analyzed the effect of PABPN1 depletion on the distribution of mRNA with 5′SS motifs. We depleted PABPN1 with two independent shRNAs in U2OS cells (Supplemental Fig. S6A) and monitored the distribution of various ftz mRNA reporters (Supplemental Fig. S6B) by FISH. We did not see a significant change in the cytoplasmic/nuclear distributions of ftz mRNA containing the 5′SS motif. This was true for intronless (ftz-Δi-5′SS) or intron-containing (ftz-i-5′SS) versions of ftz (Supplemental Fig. S6C,D). We did see a modest but significant inhibition in the nuclear export of our control reporter (ftz-Δi), in line with previous findings that PABPN1 is required for efficient mRNA nuclear export (Apponi et al. 2010). Thus, it remains formerly possible that since overall mRNA nuclear export is lower in PABPN1-depleted cells, this could have counteracted any putative inhibition of nuclear retention for ftz mRNAs containing the 5′SS motif. Previously, we showed that depletion of TPR led to the general inhibition in mRNA export and a further enhancement in the nuclear retention of ftz-Δi-5′SS mRNA (Lee et al. 2020), unlike what we see after PABPN1 depletion.
Interestingly, depletion of PABPN1 decreased the total levels of both ftz and ftz-Δi-5′SS RNAs relative to control shRNA-treated cells (Supplemental Fig. S4D). Thus, like ZFC3H1, PABPN1 appears to affect the total levels of overexpressed reporter RNAs irrespective of the presence of a 5′SS motif, although in this instance the effect is in the opposite direction.
Overall, our results indicate that ZFC3H1 is required for the nuclear retention of mRNAs that contain intact 5′SS motifs. As for other PAXT complex components, our data indicates that MTR4 is not required for this process, while for PABPN1 we are unable to make any firm assessment, as its effects on nuclear retention may be masked by its effects on mRNA export.
mRNA nuclear retention is not dependent on alterations in 3′ cleavage or poly(A)-tail length
Previously, ZFC3H1 was identified in a mass spectrometry analysis of U1 snRNP-associated factors, which also contained 3′ end processing components (So et al. 2019). Additionally, the presence of 5′SS motifs has been shown to inhibit both 3′ cleavage and polyadenylation (Boelens et al. 1993; Gunderson et al. 1998; Vagner et al. 2000). In light of these observations, ftz-Δi-5′SS mRNA might be nuclear retained due to alterations in 3′ cleavage or polyadenylation that are potentially regulated by ZFC3H1.
To determine whether the presence of a 5′SS motif disrupted normal 3′ cleavage or polyadenylation of the ftz mRNA reporter, we expressed intron lacking and containing versions of this reporter, with and without the 5′SS motif, extracted mRNA from these lysates, and assessed the 3′ cleavage by 3′ RACE and the poly(A)-tail length using the ePAT assay (Fig. 2I; Janicke et al. 2012). By 3′ RACE, both ftz-Δi and ftz-Δi-5′SS had the same 3′ cleavage site (Fig. 2J) indicating that the 5′SS did not alter the 3′ end of the transcript. We also found that the presence of the 5′SS motif did not alter the size of the ePAT amplicons (Fig. 2K), which comprises the 3′ end of the RNA and the entire poly(A)-tail, which we estimate to be ∼100 nt long (Fig. 2I). Although, it is likely that in many cases the presence of the 5′SS affects 3′ cleavage, this may depend on the strength of the 3′ cleavage/polyadenylation signal. In cases where these signals are strong, as in the ftz reporter, which contains the bovine growth hormone polyadenylation signal (Pfarr et al. 1986), cleavage and polyadenylation are likely not affected. Thus, we conclude that the ability of the 5′SS to inhibit nuclear export is functionally distinct from its ability to inhibit 3′ cleavage and polyadenylation. This finding is in agreement with the observation that the PAXT complex acts on PROMPTs after they have become polyadenylated (Wu et al. 2020).
U1-70K acts in the same pathway as ZFC3H1 in the nuclear retention of 5′SS motif containing mRNAs
As described above, ZFC3H1 was identified in a mass spectrometry analysis of U1 snRNP-interacting proteins (So et al. 2019). In addition, immunoprecipitates of ZFC3H1 contained U1-70K, a component of the U1 snRNP, as detected by mass spectrometry (Meola et al. 2016). These observations raised the possibility that 5′SS motifs are initially recognized by U1 snRNP and that in the absence of a 3′SS motif, which would normally recruit other components of the spliceosome, U1 snRNP recruits ZFC3H1 to redirect the transcript to a nuclear retention and/or decay pathway.
To assess whether U1 snRNP was required for the nuclear retention of 5′SS motif containing mRNAs, we used a mixture of four lentiviral-delivered shRNA to deplete U1-70K (Fig. 3A). U1-70K has been shown to be required for many activities of the U1 snRNP outside of splicing, including polyadenylation and 3′ cleavage (Gunderson et al. 1998; Vagner et al. 2000). We observed that U1-70K-depleted U2OS cells did not efficiently retain ftz reporters that contained a 5′SS motif (ftz-Δi-5′SS) in the nucleus (Fig. 3B,C). In contrast, U1-70K depletion had no effect on the distribution of ftz-Δi mRNA, which lacks a 5′SS (Fig. 3B,C). Note that ZFC3H1 protein levels were unaffected by U1-70K depletion.
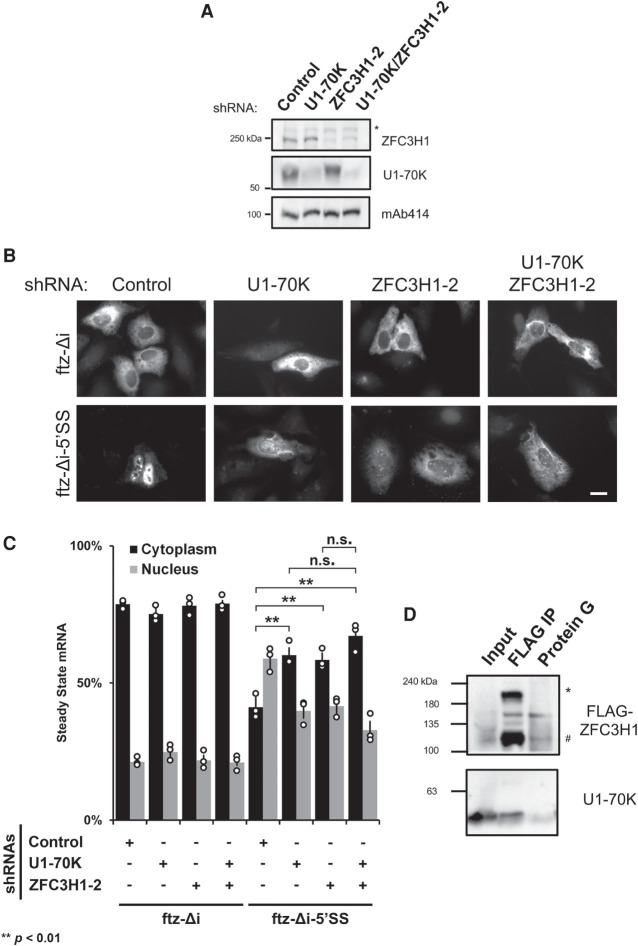
FIGURE 3.
ZFC3H1 and U1-70K function in the same pathway for the nuclear retention of 5′SS motif containing mRNAs. (A) U2OS cells were treated with lentivirus shRNA against either U1-70K, ZFC3H1, or a mixture of the two. Lysates were collected 96 h post-transduction, separated by SDS-PAGE and immunoprobed for U1-70K, ZFC3H1, and mAb414. Note that to effectively deplete U1-70K, cells were treated with lentivirus containing four shRNA plasmids. Also note that the asterisk (*) denotes a non-specific band. (B,C) Control-, U1-70K-, ZFC3H1-, or codepleted cells were transfected with the intronless ftz reporter ±5′SS as described in Figure 2. Note that the cytoplasmic/nuclear distribution of ftz-Δi-5′SS mRNA in cells codepleted of U1-70K and ZFC3H1 resembles the distribution in single depletion cells, suggesting that both proteins function in the same pathway. Representative images are shown in C (scale bar, 10 µm) and quantification is shown in D. Each bar represents the average and standard error of at least three independent experiments, each experiment consisting of at least 30 to 60 cells. Student's t-test was performed for C. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (D) HEK cells expressing carboxy-terminally tagged ZFC3H1 (ZFC3H1-FLAG) were lysed and subjected to immunoprecipitation reactions with FLAG M2 beads or mouse IgG (“Control IP”). Immunoprecipitates were separated by SDS-PAGE and immunoprobed for FLAG and U1-70K. For comparison, 1% of the input lysate was also analyzed. The full-length ZFC3H1-FLAG protein is denoted by the asterisk (*) and a shorter band, likely a degradation product, is denoted by the pound (#) sign.
To test whether these two factors functioned in the same pathway, we assessed the distribution of our reporter mRNAs in cells depleted of both ZFC3H1 and U1-70K and compared these to the single depletions (Fig. 3A). We observed that the double depleted cells had cytoplasmic/nuclear levels of ftz-Δi-5′SS that were not significantly different than in each of the single depleted cells (Fig. 3B,C). Note that the distribution of this reporter was still more nuclear than a version that lacked a 5′SS motif (compare ftz-Δi-5′SS to ftz-Δi; Fig. 3C). These data strongly indicate that nuclear retention of ftz-Δi-5′SS was only partially inhibited in the single and double knockdown cells, and that ZFC3H1 and U1-70K act in the same pathway. When the overall levels of mRNA were assessed, we found that U1-70K-depletion did not significantly affect the levels of either ftz-Δi or ftz-Δi-5′SS (Supplemental Fig. S4E). In contrast, the levels of these two reporter RNAs rose in the double depleted cells, consistent with the idea that ZFC3H1 promotes the decay of these reporters. Despite this, we did not observe a significant increase of either ftz-Δi or ftz-Δi-5′SS levels in the ZFC3H1-depleted cells in this particular set of experiments (Supplemental Fig. S4E), unlike our previous experiments (see Supplemental Fig. S4A). This difference may simply be due to the large amount of variability in these measurements (note the large error bars in all these experiments).
We next sought to confirm previous mass spectrometry data that suggested an interaction between U1 snRNP and ZFC3H1 (Meola et al. 2016; So et al. 2019). When we immunoprecipitated overexpressed ZFC3H1-FLAG from RNase-treated cell lysates, we detected endogenous U1-70K in the precipitates (Fig. 3D). These results confirm that ZFC3H1 and U1-70K are likely in a complex.
In summary, these data suggest that both ZFC3H1 and U1-70K retain 5′SS motif containing mRNAs using a common pathway, and likely by forming a complex.
ZFC3H1 and U1-70K are required for the egress of 5′SS motif containing mRNAs from nuclear speckles
Previously, we showed that 5′SS motif containing mRNAs that are retained in the nucleus also colocalize with nuclear speckles and remain in these compartments under steady state conditions (Lee et al. 2015). Since the ftz mRNA is trafficked to speckles independently of the 5′SS (Akef et al. 2013), we monitored the β-globin (βG) reporter mRNA, which requires the 5′SS motif for nuclear speckle localization (Fig. 4A), as previously described (Akef et al. 2013; Lee et al. 2015). To determine whether ZFC3H1 or U1-70K are required for targeting newly synthesized mRNAs with 5′SS motifs to nuclear speckles, we microinjected plasmids containing either the intronless β-globin gene without (βG-Δi) or with (βG-Δi-5′SS) a 5′SS motif into the nuclei of cells depleted of ZFC3H1 or U1-70K, or treated with control shRNA. We then fixed cells at various time points and monitored the colocalization of newly synthesized mRNA with the nuclear speckle marker SC35 by Pearson correlation coefficient analysis, as we have done previously (Akef et al. 2013; Lee et al. 2015). Note that microinjection allows for the generation of a large amount of mRNA in a short time span allowing one to observe events that happen only in the early stages of gene expression (Gueroussov et al. 2010).
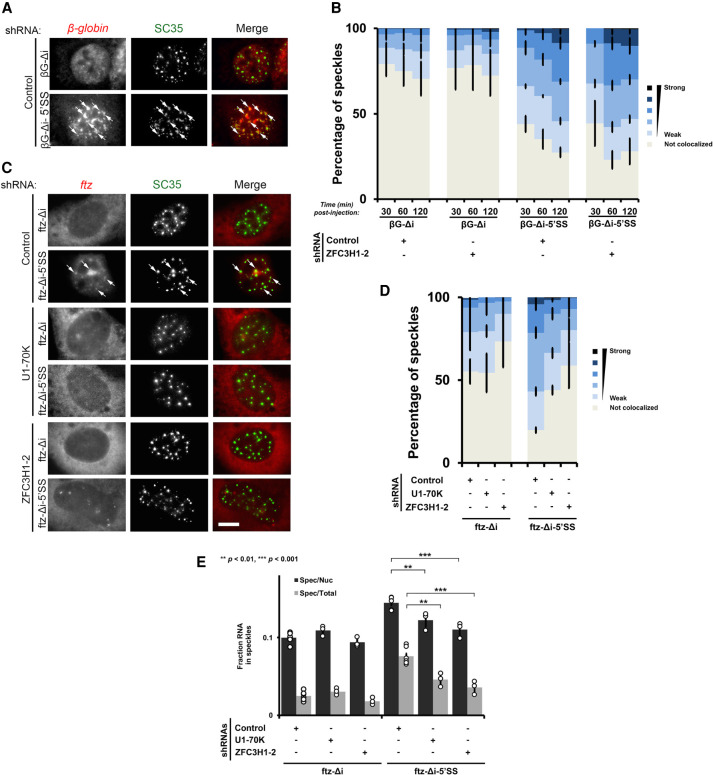
FIGURE 4.
ZFC3H1 and U1-70K are required for nuclear retention of 5′SS motif containing mRNAs in speckles. (A,B) Control or ZFC3H1-depleted U2OS cells were microinjected with plasmids containing the βG-Δi reporter ± the 5′SS motif. After the indicated times, the cells were fixed and stained for βG mRNA by FISH and for the nuclear speckle marker SC35 by immunofluorescence. (A) Example images of U2OS cells fixed 2 h post-injection with βG reporter mRNA ± the 5′SS motif, with each row representing a single field of view with white arrows pointing to examples of βG mRNA/SC35 colocalization. The merged overlayed image shows βG mRNA in red, SC35 in green. (B) Quantification of the degree of βG mRNA/SC35 colocalization in cells depleted of ZFC3H1 or control shRNA treatment by Pearson correlation coefficient analysis as previously described (Akef et al. 2013). Each bar represents the average and standard error of three independent experiments, each experiment consisting of 150 to 200 nuclear speckles from 15 to 20 cells. Note that ZFC3H1 is not required for the targeting of 5′SS motif containing mRNAs to nuclear speckles. (C–E) Control, U1-70K- or ZFC3H1-depleted U2OS cells were transfected with ftz-Δi ± 5′SS motif. Eighteen to twenty-four hours post-transfection, the cells were fixed and stained for ftz mRNA by FISH and for the nuclear speckle marker SC35 by immunofluorescence. Representative images, with each row depicting a single field of view, is shown in C with merged overlays showing ftz mRNA in red and SC35 in green. Scale bar, 10 µM. Examples of ftz/SC35 colocalization are indicated with arrows. (D) The degree of ftz/SC35 colocalization by Pearson correlation coefficient analysis was quantified as above, except that values <0.25 were counted as “not colocalized.” Note that U1-70K or ZFC3H1 depletion leads to decreased level of colocalization between ftz mRNA and SC35. Each bar represents the average and standard error of three independent experiments, each experiment consisting of 100–200 nuclear speckles from 10 to 20 cells. (E) The amount of ftz reporter mRNA present in nuclear speckles as a percentage of either the nuclear (“Spec/Nuc”) or total cellular (“Spec/Total”) mRNA levels in transfected cells. Each data point represents the average and standard error of the mean of at least three independent experiments, each experiment consisting of 10–20 cells. Student's t-test was performed, (**) P < 0.01, (***) P < 0.001.
In control shRNA-treated cells, we observed that within 30 min of expression, βG-Δi-5′SS mRNA began to accumulate in nuclear speckles and the degree of speckle localization increased over time (Fig. 4B; Supplemental Fig. S7A). In contrast, βG-Δi mRNA was not as robustly targeted to speckles during this time course. The speckle targeting βG-Δi-5′SS mRNA was not affected by depletion of ZFC3H1 or U1-70K (Fig. 4B; Supplemental Fig. S7A). We also monitored the speckle targeting of ftz-Δi without and with a 5′SS motif. Again, depletion of ZFC3H1 did not significantly impact the targeting of this mRNA to nuclear speckles (Supplemental Fig. S7B).
We next investigated whether ZFC3H1 or U1-70K was required for the egress of mRNAs containing 5′SS motifs out of nuclear speckles. To determine this, we monitored the localization of ftz without (ftz-Δi) and with (ftz-Δi-5′SS) the 5′SS motif in transfected cells, where the total amount of nuclear speckle localization reflects steady-state dynamics (i.e., both nuclear speckle targeting and egress). Note that we did not use βG-Δi-5′SS mRNA for these experiments, as the βG-Δi mRNA contains other elements that inhibit nuclear export and promote decay in a nuclear speckle-independent manner (Akef et al. 2015). Additionally, note that although newly synthesized ftz reporter mRNA transits through nuclear speckles, it is retained in nuclear speckles at later time points in a 5′SS motif-dependent manner (Lee et al. 2015). We found that depletion of either U1-70K or ZFC3H1 resulted in a decrease in nuclear speckle localized ftz-Δi-5′SS mRNA (Fig. 4C,D). When we quantified the fraction of ftz-Δi mRNA FISH signal in nuclear speckles, the 5′SS motif caused an increase, and this was reversed in cells depleted of either U1-70K or ZFC3H1 (Fig. 4E).
From these experiments, we concluded that ZFC3H1 and U1-70K help to retain 5′SS motif containing mRNAs in nuclear speckles.
Nuclear speckles are required for the efficient nuclear retention of 5′SS motif containing mRNAs
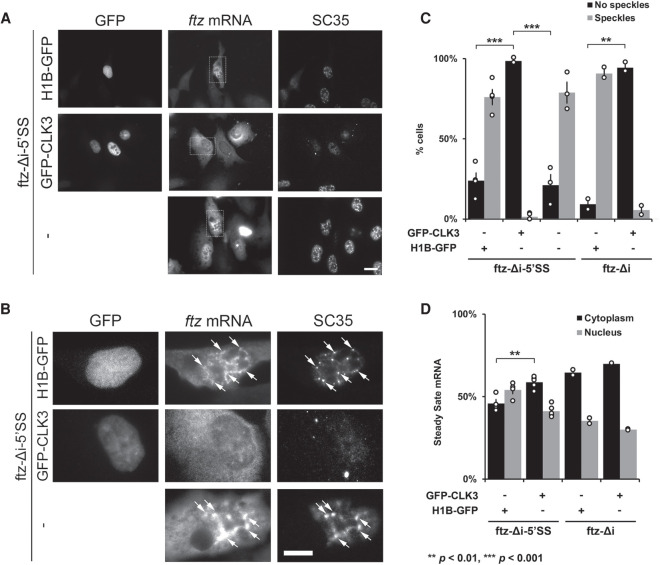
Next, we examined whether nuclear speckles are required for the nuclear retention of 5′SS motif containing mRNAs. We overexpressed GFP-CLK3 to trigger nuclear speckle disassembly, as was previously described (Wong et al. 2011), and examined the distribution of either ftz-Δi-5′SS and ftz-Δi mRNA and of the speckle marker SC35. As a control, we overexpressed Histone 1B-GFP (H1B-GFP). As was reported previously (Wong et al. 2011), expression of GFP-CLK3 resulted in the disappearance of detectable nuclear speckles (Fig. 5A–C), and in these cells we observed that ftz-Δi-5′SS mRNA was significantly enriched in the cytoplasm (Fig. 5A,D). In some cells, the nuclear speckle marker, SC35, completely disappeared, while in other cells, its levels did not significantly drop, but instead became diffuse throughout the nucleoplasm. In H1B-GFP expressing cells, ftz-Δi-5′SS mRNA was nuclear, similar to what we had seen in control U2OS cells (Fig. 5A,C). Cells expressing GFP-CLK3 still exported ftz-Δi, which lacks the 5′SS motif, indicating that expression of this kinase did not disrupt normal mRNA export (Fig. 5C,D; Supplemental Fig. S8A). GFP-CLK3 overexpression led to the increase in total ftz-Δi and ftz-Δi-5′SS RNA levels (Supplemental Fig. S4F).
FIGURE 5.
Nuclear speckles promote mRNA nuclear retention of 5′SS motif containing mRNA. (A–D) U2OS cells were cotransfected with either ftz-Δi-5′SS or ftz-Δi, and with either H1B-GFP, GFP-CLK3, or empty vector (“-”). Twenty-four hours post-transfection, cells were fixed and stained for ftz mRNA by FISH and for the speckle marker SC35 by immunofluorescence. Representative images, with each row depicting a single field of view imaged for GFP, ftz mRNA, and SC35 is shown in A. Scale bar, 10 µM. Enlarged images of the regions indicated in A are depicted in B. Examples of ftz/SC35 colocalization are indicated with arrows. Scale bar, 10 µM. Note that the overexpression of GFP-CLK3 disrupts nuclear speckles (SC35) compared to H1B-GFP or non-GFP-expressing cells. (C) The number of cells that lacked or contained nuclear speckles were quantified. Each bar represents the average and standard error of two (ftz-Δi) or four (ftz-Δi-5′SS) independent experiments, each consisting of 30–50 cells. Note that expression of GFP-CLK3 disrupts nuclear speckles compared to control cells. (**) P < 0.01, (***) P < 0.001 (Student's t-test). (D) Quantification of the cytoplasmic/nuclear FISH signal with each bar representing the average and standard error of two (ftz-Δi) or four (ftz-Δi-5′SS) independent experiments, each experiment consisting of at least 30 to 60 cells. (**) P < 0.01 (Student's t-test).
Interestingly, expression of ftz-Δi-5′SS alone caused many cells to have larger nuclear speckles than what is typically seen in untransfected cells (Fig. 5B; Supplemental Fig. S8B), perhaps because these regions grew as more mRNA became incorporated into them. Coexpression of H1B-GFP did cause some cells to have smaller nuclear speckles than what is seen in untransfected cells; however, most cells still contained these structures (Fig. 5B, also see Supplemental Fig. S8B).
In summary, our results suggest that nuclear speckles are required for the efficient nuclear retention of 5′SS motif containing mRNAs, although it remains possible that GFP-CLK3 expression disrupts mRNA nuclear retention independently of its effects on nuclear speckles.
DISCUSSION
Here we show that 5′SS motif containing mRNAs require ZFC3H1 and U1-70K for their efficient nuclear retention. Moreover, our data indicates that these two proteins act in the same pathway. It is likely that when a newly synthesized transcript has a 5′SS motif, it first recruits the U1 snRNP to initiate splicing; however, if this is followed by a strong 3′ cleavage/polyadenylation signal, ZFC3H1 is recruited instead. This leads to nuclear retention in nuclear speckles and degradation of the transcript. In support of this idea, we demonstrate that upon ZFC3H1 depletion, there is a widespread stabilization and export of endogenous IPA transcripts, which contain intact 5′SS motifs followed by poly(A)-tails. Furthermore, depletion of either ZFC3H1 or U1-70K impairs the nuclear retention of reporter mRNAs with intact 5′SSs. Although ZFC3H1 depletion also affected the stability of our reporters, this seemed independent of the presence of the 5′SS motif (Supplemental Fig. S4).
Our results indicate that ZFC3H1 has other effects on mRNA localization. ZFC3H1 promotes the nuclear retention of PROMPTs and other ncRNAs, as indicated by our RNA Frac-seq data (Supplemental Fig. S1D,E) and previous reports (Silla et al. 2018). ZFC3H1 also appears to promote the export of mRNAs generated from distal 3′ cleavage/polyadenylation sites (Supplemental Figs. S2, S3). It may be that in ZFC3H1-depleted cells, mRNAs generated from distal 3′ cleavage/polyadenylation sites are outcompeted for binding to nuclear export factors by noncoding RNAs (ncRNAs) and IPAs. Alternatively, ZFC3H1 may directly participate in their nuclear export. Further studies will be needed to test these various possibilities.
Interestingly, depletion of MTR4 and PABPN1 had minimal effects on the nuclear retention of reporter mRNAs containing 5′SS motifs in U2OS cells. This is in contrast to previous observations of IPA transcript distributions in HeLa cells (Ogami et al. 2017). Importantly, MTR4 depletion caused the co-depletion of ZFC3H1 in HeLa (Ogami et al. 2017; Silla et al. 2018) but not U2OS cells (Fig. 2A). It is also possible that other factors present in U2OS cells act redundantly with MTR4. Alternatively, nuclear retention of IPA transcripts may only require low levels of MTR4. As for PABPN1, its depletion in U2OS cells disrupted mRNA nuclear export, complicating the interpretation of our results. Previously, when mRNA export was disrupted by TPR depletion, we observed that the ftz-Δi-5′SS mRNA became even more nuclear (Lee et al. 2020), suggesting that a general block in mRNA export should increase the nuclear distribution of ftz-Δi-5′SS. However, we do not see this in PAPBN1-depleted cells. Thus, we cannot rule out the possibility that depletion of PABPN1 both inhibited export and nuclear retention, ultimately leading to no overall net change in the distribution of the ftz-Δi-5′SS mRNA.
There has been much interest in the role of nuclear speckles in promoting nuclear retention and export (Dias et al. 2010; Akef et al. 2013; Jain and Vale 2017; Palazzo and Lee 2018; Wang et al. 2018). Our new results suggest that ZFC3H1 and U1-70K prevent these mRNAs from leaving nuclear speckles (Fig. 4C,D). We still do not understand how these mRNAs are targeted to speckles. Likely this is due in part to U1 snRNP, although U1-70K does not appear to be required for this trafficking. It remains possible that U1 snRNP, which remains intact after U1-70K depletion (Rösel-Hillgärtner et al. 2013), can still target mRNAs to speckles but fails to recruit ZFC3H1 to the transcript. This is in agreement with previous results where the tethering of U1-70K to a reporter mRNA promoted nuclear retention without targeting the transcript to nuclear speckles (Takemura et al. 2011). Recently, it has been shown that the amino-terminal domain of ZFC3H1 forms phase-separated nuclear compartments, which partially overlap with nuclear speckles (Wang et al. 2021). The formation of these phase-separated compartments is thought to be required for the nuclear retention of substrate RNAs, which get trapped in these structures (Silla et al. 2018; Wang et al. 2021). It remains possible that the 5′SS motif targets mRNAs to structures that only partially overlap with speckles, and this may explain why the nuclear speckle localization of some IPA transcripts, such as the PCF11 IPA transcript (Supplemental Fig. S5B), is poor.
Intriguingly, in the fission yeast S. pombe, homologs of ZFC3H1 (Red1) and MTR4 (Mtl1) are required for both the decay and the sequestration of meiotic mRNAs and transposable element-derived transcripts into nuclear foci (Sugiyama and Sugioka-Sugiyama 2011; Shichino et al. 2018, 2020). These results suggest that the ability of ZFC3H1 to negatively regulate expression of mRNAs and ncRNAs, by promoting retention in nuclear subdomains and triggering RNA decay, is widely conserved in eukaryotes.
Previously, we showed that mRNAs with 5′SS motifs still acquired TREX components, such as UAP56, and the nuclear transport receptor NXF1 but still fail to be exported (Lee et al. 2015). Thus, it is likely that the export-promoting activity of these factors is inhibited by ZFC3H1. Indeed, it appears that UAP56-dependent ATP-hydrolysis is required for mRNAs to leave nuclear speckles (Hondele et al. 2019) and this may be directly counteracted by ZFC3H1.
In previously published results, we found that microinjected fully processed mRNAs with 5′SS motifs were efficiently exported from nuclei (Lee et al. 2015), suggesting that these mRNAs fail to recruit either U1 snRNP and/or ZFC3H1. This may indicate that these factors are recruited during transcription or processing. This idea is supported by observations that U1 snRNP directly associates with RNA polymerase II (Zhang et al. 2021). However, since microinjected mRNAs are spliced within 15 min after being injected into nuclei (Palazzo et al. 2007), it must be that microinjected mRNAs are still recognized by U1 snRNP. The fact that microinjected mRNAs with intact 5′SS motifs are exported, suggests that either ZFC3H1 or some other critical nuclear retention component may be recruited to nascent transcripts by the joint action of U1 snRNP and RNA polymerase II in a cotranscriptional manner. Further experiments are needed to validate this model.
MATERIALS AND METHODS
Plasmids constructs and antibodies
All plasmids for reporter constructs in pcDNA3.0 and H1B-GFP were previously described (Palazzo et al. 2007; Akef et al. 2013; Lee et al. 2015, 2020). Plasmid expressing GFP-CLK3 was previously described (Wong et al. 2011). Plasmid for ZFC3H1 (BC156732, IMAGE: 100062212) was obtained from the Open Freezer cDNA repository at the Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital. To generate FLAG-tagged ZFC3H1, the cDNA sequence was amplified with these primer sequences: F′ 5′-GAT ATC AAA TGG CGA CCG CAG ATA CT-3′ and R′ 5′-GCG GCC GCA AGT GAT TCT TGC TTT CTG TTT TGT T-3′ and subcloned into pcDNA 5-FRT with EcoV and NotI (NEB). To construct the CCDC71-IPA reporter, the first exon and a portion of the intron (past the intronic 3′ polyadenylation site) was amplified from HEK genomic cDNA by Phusion polymerase (NEB) based on the manufacturer's instructions using the primer sequences: F′ 5′-GGT ACC GGG CGG CCA TGT TGG AGC AG-3′ and R′ 5′-TAC AGC CCA GAT GGA GGT GAC TGA GAA C-3′. The resulting ∼1 kb PCR fragment was separated on an agarose gel, gel purified (Geneaid gel purification kit), and cloned into the pJET1.2 vector (CloneJET PCR cloning kit, Thermo Fisher). The construct was subcloned into pcDNA5-FRT/TO using KpnI and XbaI (NEB). For the PCF11-IPA reporter, a portion of the PCF11 gene (exon1–intron1–exon2) was synthesized and cloned into pJET1.2 vector (General Biosystems), and subsequently subcloned into pcDNA3.0 using NotI and XbaI (NEB). To insert the endogenous PCF11 5′UTR into this construct, the 5′UTR was first amplified from HAP1 genomic cDNA using the following primers: F′ 5′-ACG ACT CAC TAT AGG GCA CGA CGC AGC GGT TGG GAA C-3′ and R′ 5′-GAC AAC ACC CCA TGG CAT TGC GCC GCG GCC CCC-3′. The amplified 5′UTR sequence was cloned into the PCF11–pcDNA3.1 plasmid (first linearized with XhoI and KpnI) with an In-Fusion cloning kit (Takara) according to the manufacturer's protocol.
Antibodies used in this study include rabbit polyclonals against ZFC3H1 (also known as CCDC131) (Bethyl Laboratories, A301-457A), MTR4 (also known as SKIV2L2) (Bethyl Laboratories, A300-614A), PABPN1 (Bethyl Laboratories, A303-523A), U1-70K (Abcam, ab83306), Aly (Zhou et al. 2000), and TRAPα (Görlich et al. 1990) or mouse monoclonals against U1-70K (Sigma-Aldrich, clone 9C4.1), mAb414 (Sigma), SC35 (Clone SC35, Sigma), and α-tubulin (DM1A, Sigma). Immunofluorescence was performed with Alexa647-conjgated secondaries (Thermo Fisher) and HRP-conjugated secondaries (Cell Signaling). All antibodies were diluted 1:1000 for western blotting and 1:100 to 1:250 for immunofluorescence microscopy.
Cell culture, DNA transfection experiments, and lentiviral-delivered shRNA protein depletion
U2OS and HEK293T cells were grown in DMEM media (Wisent) supplemented with 10% fetal bovine serum (FBS) (Wisent) and 5% penicillin/streptomycin (Wisent). DNA transfection experiments were performed as previously described (Palazzo et al. 2007; Akef et al. 2013; Lee et al. 2015, 2020).
For all DNA transfections, U2OS cells were transfected with the appropriate amount of DNA plasmid according to the manufacturer's protocol using GenJet U2OS DNA in vitro transfection reagent (SignaGen Laboratories) for 18 to 24 h.
The lentiviral-delivered shRNA protein depletion was performed as previously described (Akef et al. 2013; Lee et al. 2015, 2020). Briefly, HEK293T were plated at 50% confluency on 60 mm dishes and transiently transfected with the gene-specific shRNA pLKO.1 plasmid (Sigma), packaging plasmid (Δ8.9), and envelope (VSVG) vectors using Lipo293T DNA in vitro transfection reagent (SignaGen Laboratories) according to the manufacturer's protocol. Forty eight hours post-transfection, viruses were harvested from the media and added to U2OS cells pretreated with 8 µg/mL hexadimethrine bromide. Cells were selected with 2 µg/mL puromycin media for at least 4 to 6 d. Western blotting was used to determine the efficiency of ZFC3H1, MTR4, PABPN1, and U1-70K depletion. The shRNAs constructs (Sigma) used in this study are as follows: ZFC3H1-1 “TRCN0000129932” 5′-CCGGGCCAAG AAGCAATCTA TCAATCTCGA GATTGATAGA TTGCTTCTTG GCTTTTTTG-3′, ZFC3H1-2 “TRCN0000432333” 5′-CCGGGACTGA TGACATCGCT AATTTCTCGA GAAATTAGCG ATGTCATCAG TCTTTTTTG-3′, MTR4-1 “TRCN0000307086” 5′-CCGGCCCAGG ATAGAAGAGT CAATACTCGA GTATTGACTC TTCTATCCTG GGTTTTTG-3′, MTR4-2 “TRCN0000296268” 5′-CCGGAGCAGG ACCACTTCGT CAAATCTCGA GATTTGACGA AGTGGTCCTG CTTTTTTG-3′, PABPN1-1 “TRCN0000000124” 5′-CCGGAGGTAG AGAAGCAGAT GAATACTCGA GTATTCATCT GCTTCTCTAC CTTTTTT-3′, PABPN1-2 “TRCN0000000120” 5′-CCGGCCCATA ACTAACTGCT GAGGACTCGA GTCCTCAGCA GTTAGTTATG GGTTTTT-3′, U1-70K-A “TRCN0000000011” 5′-CCGGCCAAGG GTAGGTGTCT CATTTCTCGA GAAATGAGAC ACCTACCCTT GGTTTTT-3′, U1-70K-B “TRCN0000349622” 5′-CCGGGACATG CACTCCGCTT ACAAACTCGA GTTTGTAAGC GGAGTGCATG TCTTTTTG-3′, U1-70K-C “TRCN0000287201” 5′-CCGGGCACCA TACATCCGAG AGTTTCTCGA GAAACTCTCG GATGTATGGT GCTTTTTG-3′, and U1-70K-D “TRCN0000287138” 5′-CCGGCGATGC CTTCAAGACT CTGTTCTCGA GAACAGAGTC TTGAAGGCAT CGTTTTTG-3′.
Microinjections, fluorescent in situ hybridization (FISH) staining, immunostaining, and nuclear speckle Pearson correlation and enrichment quantifications
Microinjections, fluorescent in situ hybridization, and immunostaining were performed as previously described (Gueroussov et al. 2010; Lee and Palazzo 2017; Lee et al. 2020). To detect intronic polyadenylated transcripts within the CCDC71-IPA and PCF11-IPA reporters, specific FISH probes were designed with the Alexa-546 conjugated to the 5′ end (IDT): CCDC71-IPA 5′-GGCCCAAGCC GTGAGGCGGG GGTCACTGAG GCAGCGAAAT GTACAGGGTG AACGGTGCTA G-3′, PCF11-IPA reporter is 5′-TTTTTAAAGG CCACTTTAAC AACAACCACC AATATATTGT TTAGAGACGG TTTCTGTCTA GTGTA-3′.
Nuclear speckle localization by Pearson correlation was performed as previously described (Akef et al. 2013; Lee et al. 2015, 2020). Briefly, for each cell, the 10 brightest nuclear speckles, as observed by SC35 staining, were selected and Pearson correlation analysis was conducted between the FISH signal and SC35 immunofluorescence signal using rectangular regions of interest 1–4 µm2 in size. Each experiment consists of an analysis of 10 cells (for a total of 100 speckles). Graphs in Figure 4 and Supplemental Figure S7 consist of the average and standard deviation of three independent experiments.
The quantification of mRNA enrichment in speckles was performed as previously described (Akef et al. 2013). Briefly, thresholds were drawn on the SC35 immunofluorescence channel so that 10% (±0.5%) of the nuclear area was selected per cell. Using this selected area, the fluorescence intensity of RNA was calculated and divided by either the total integrated mRNA signals in the nucleus (“Spec/Nuc”) or the cell body (“Spec/Total”).
3′RACE, RT-PCR, and ePAT
For 3′RACE and RT-PCR experiments, total RNA was extracted from transfected U2OS cells using TRIzol (Life Sciences). About 1 µg total RNA was used for first strand synthesized using murine MLV reverse transcriptase (Invitrogen) and oligo(dT) primer according to the manufacturer's instructions. For 3′RACE experiments, the resulting cDNA was amplified using ftz F′ primer 5′-ATG GGG TGT TGT CCC GGC TGT TGT-3′ and an oligo(dT) primer. The resulting PCR product was inserted into CloneJet (Fermentas) vector following the sticky-end cloning protocol (see manufacturer's instructions) and transformed into DH5α-competent cells. DNA was extracted from colonies using a Miniprep Kit (Geneaid) and sent for sequencing.
ePAT was performed as previously described (Janicke et al. 2012). Total RNA was extracted by TRIzol from cells transfected with the indicated ftz plasmid and expressed for 18–24 h. The ePAT anchor primer used was 5′-GCG AGC TCC GCG GCC GCG TTT TTT TTT TTT-3′, the universal primer used was 5′-GCG AGC TCC GCG GCC GCG-3′, and the ftz-specific ePAT primer used was 5′-ATG GGG TGT TGT CCC GGC TGT TGT-3′.
Co-IP experiment
Approximately 2 × 107 HEK cells transfected with ZFC3H1-FLAG plasmid were trypsinized, pelleted, and washed three times in ice-cold 1× PBS and flash frozen in liquid nitrogen. The cell pellet was lysed in ∼2 mL of IP buffer (20 mM Tris-HCl, pH8, 137 mM NaCl, 1% NP-40, 2 mM EDTA, and cOmplete Mini Protease inhibitor [Roche]) mixed for 15 min at 4°C to ensure complete lysis. To ensure that the nuclear proteins were released from the chromatin, the sample was passed through a 25¾″ needle to shear the chromatin multiple times before incubation. To release proteins from RNA, the cell lysate was treated with 0.1 mg/mL RNase A (Thermo, EN0531). The cell lysis was cleared by centrifugation at 16,100g for 10 min and ∼0.9 mL supernatant was mixed with either 50 µL FLAG M2 beads (Sigma Aldrich, A2220) or Protein G beads (NEB, #37478S), preincubated with 2% BSA overnight, and prewashed three times with IP wash buffer (same as IP buffer, except that 0.05% NP-40 was used). The cell lysate was incubated with the beads for 3 h at 4°C. Following incubation, the beads were washed five times with IP wash buffer. To elute the proteins, 50 µL of 2.5× Laemmli sample buffer was added to all the samples and boiled for 5 min. Samples were separated on an SDS-PAGE gel and transferred onto a blot for immunoblotting.
RNA Frac-seq, data processing, and analysis
RNA Frac-seq was performed as previously described (Lee et al. 2020). RNA Fraq-seq data were deposited in GEO database as GSE176144. IPA analysis was performed as described (Wang et al. 2019) using a curated list of IPA transcripts (Supplemental Table 1).
To estimate reads from short 3′UTRs, we used these calculations:
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank T. Dubric and her staff at the Donnelly Centre Sequencing Facility for performing the RNA sequencing and quality control, and S. Ihn for providing critical feedback on the manuscript. We thank A. Cochran for generously providing us with GFP-CLK3 plasmid. We thank L. Pelletier and his laboratory for accessing the cDNA repository at the Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital. This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada FN 492860.
Author contributions: E.S.L., H.W.S., and Y.E.W. performed the experiments under the guidance of A.F.P. E.S.L. and A.F.P. cowrote the paper. E.J.W. and A.G. performed the bioinformatics analysis under the guidance of A.F.P., A.E., and B.T.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079104.122.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Eliza Lee.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Eliza Lee is the first author of this paper, “ZFC3H1 and U1-70K promote the nuclear retention of mRNAs with 5′ splice site motifs within nuclear speckles.” Eliza is a postdoctoral fellow (former PhD student) in the laboratory of Alex Palazzo at the University of Toronto. She is studying mRNA nuclear export and nuclear retention as a quality control mechanism in eukaryotic cells.
What are the major results described in your paper and how do they impact this branch of the field?
The defining feature of a eukaryotic cell is that it is divided into two compartments: the nucleus, where RNA synthesis and processing occurs, and the cytoplasm, where the protein synthesis machinery is localized. This segregation allows for quality control mechanisms to act on mRNAs before they are exported into the cytoplasm and translated into protein. Normally, misprocessed mRNAs are retained in the nucleus and degraded by the nuclear surveillance machinery. It is imperative that these mRNAs are not exported into the cytoplasm as they would otherwise be translated into toxic peptides that are deleterious to the cell. Previously, I showed that the 5′ splice site (5′SS) motif promotes mRNA decay and/or nuclear retention. Within a cell, 5′SS motif containing mRNAs are formed from intronic polyadenylated (IPA) transcripts and are an example of misprocessed mRNAs. In this paper, I identified a complex, ZFC3H1-U1-70K, as required for the nuclear retention of 5′SS motif containing mRNAs. ZFC3H1 is a component of the PolyA Exosome Targeting (PAXT) complex, a nuclear surveillance complex that targets mRNAs for degradation. U1-70K is a protein component of the U1 snRNP, which is required for splicing. Interestingly, I show that the 5′SS motif targets mRNA to nuclear speckles, a liquid–liquid phase separated subcompartment of the nucleus, which contains splicing and mRNA export factors, and are sites of post-transcriptional splicing. We find that the ZFC3H1-U1-70K is required for the retention of 5′SS motif containing mRNAs, and disruption of nuclear speckles leads to partial cytoplasmic leakage of these mRNAs.
What led you to study RNA or this aspect of RNA science?
During my undergraduate studies, I became fascinated by epigenetics/chromatin biology. I was fortunate enough to work in the laboratory of Dr. Jane Mellor for my 4th year project who greatly inspired me. This led to my interest in RNA processing and RNA biology in general. I therefore jumped at the opportunity to study mRNA nuclear export when I joined the laboratory of Alex Palazzo for my PhD studies. I did not set out to study nuclear RNA surveillance: a set of interesting observations led us there, and we sort of stumbled upon this field by some happy accident. A few years later, we realized that our work supported the discoveries of many other labs (who primarily worked in the context of RNA processing, RNA degradation, or nuclear RNA surveillance) and how impactful it could be.
If you were able to give one piece of advice to your younger self, what would that be?
My advice to my younger self is that unexpected and puzzling results often give rise to new opportunities—this paper is a living example! I did not set out specifically to investigate this set of scientific questions but we have ended up with this story. It has been an interesting journey, full of twists and turns, which has been very fulfilling on a personal level.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
Kimura and Ohto proposed the “nearly neutral theory of evolution,” which has greatly influenced my work and how I (and our laboratory) approach big data and the idea of “functional” RNA. With the advent of sequencing technologies, there are a lot of hyper-adaptionalist ideas and “just so stories,” especially when we debate the “function” of noncoding RNAs. Simply put, the presence of any RNA (especially at low levels) does not immediately suggest that the RNA is “functional.” Contrary to expectations, most of evolution is shaped by drift and “neutral forces” and not necessarily by natural selection. For most mammals who have small effective population sizes (breeding population), the possibility that a slightly deleterious allele reaches fixation in the population is nonzero. These slightly deleterious mutations include transposable element replication and the generation of spurious transcriptional start sites. These ideas account for why most of the genome is “junk,” with vast quantities of spurious transcription by RNA polymerase II and a plethora of sophisticated RNA surveillance mechanisms to degrade the junk or misprocessed RNA. Importantly, the presence of a nucleus (the segregation of the splicing and protein translation machinery) has allowed for this junk RNA to persist and, over time, certain aspects of this junk RNA has been coopted by the cell and acquired a new function, which has been selected for.
REFERENCES
- Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. 2008. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res 36: 2338–2352. 10.1093/nar/gkn068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akef A, Zhang H, Masuda S, Palazzo AF. 2013. Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles. Nucleus 4: 326–340. 10.4161/nucl.26052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akef A, Lee ES, Palazzo AF. 2015. Splicing promotes the nuclear export of β-globin mRNA by overcoming nuclear retention elements. RNA 21: 1908–1920. 10.1261/rna.051987.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. 2013. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499: 360–363. 10.1038/nature12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. 2010. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum Mol Genet 19: 1058–1065. 10.1093/hmg/ddp569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam S, Hou S, Zhu B, Wang W, Hao T, Bu X, Khan M, Lei H. 2019. Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus. RNA Biol 16: 1001–1009. 10.1080/15476286.2019.1620061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga SJ, Steitz JA. 1993. The diverse world of small ribonucleoproteins. In The RNA world (ed. Gesteland R and Atkins J), pp. 359–381. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, Zhang Z, Cho S, Sherrill-Mix S, Wan L, et al. 2012. U1 snRNP determines mRNA length and regulates isoform expression. Cell 150: 53–64. 10.1016/j.cell.2012.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez L, Fortes P. 2013. U1 snRNP control of 3′-end processing and the therapeutic application of U1 inhibition combined with RNA interference. Curr Mol Med 13: 1203–1216. 10.2174/1566524011313070012 [DOI] [PubMed] [Google Scholar]
- Boelens WC, Jansen EJR, van Venrooij WJ, Stripecke R, Mattaj IW, Gunderson SI. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72: 881–892. 10.1016/0092-8674(93)90577-D [DOI] [PubMed] [Google Scholar]
- Daguenet E, Baguet A, Degot S, Schmidt U, Alpy F, Wendling C, Spiegelhalter C, Kessler P, Rio M-C, Le Hir H, et al. 2012. Perispeckles are major assembly sites for the exon junction core complex. Mol Biol Cell 23: 1765–1782. 10.1091/mbc.e12-01-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias AP, Dufu K, Lei H, Reed R. 2010. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun 1: 97. 10.1038/ncomms1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. 2010. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev 24: 2043–2053. 10.1101/gad.1898610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Kuai B, Wu G, Wu X, Chi B, Wang L, Wang K, Shi Z, Zhang H, Chen S, et al. 2017. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J 36: 2870–2886. 10.15252/embj.201696139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martínez-Chantar ML, Prieto J, Rowe D, Gunderson SI. 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci 100: 8264–8269. 10.1073/pnas.1332669100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galganski L, Urbanek MO, Krzyzosiak WJ. 2017. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45: 10350–10368. 10.1093/nar/gkx759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraczniak R, Behlke MA, Gunderson SI. 2009. Gene silencing by synthetic U1 adaptors. Nat Biotechnol 27: 257–263. 10.1038/nbt.1525 [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Herz J, Otto A, Kraft R, Wiedmann M, Knespel S, Dobberstein B, Rapoport TA. 1990. The signal sequence receptor has a second subunit and is part of a translocation complex in the endoplasmic reticulum as probed by bifunctional reagents. J Cell Biol 111: 2283–2294. 10.1083/jcb.111.6.2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan F, Caratozzolo RM, Goraczniak R, Ho ES, Gunderson SI. 2007. A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA 13: 2129–2140. 10.1261/rna.756707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueroussov S, Tarnawsky SP, Cui XA, Mahadevan K, Palazzo AF. 2010. Analysis of mRNA nuclear export kinetics in mammalian cells by microinjection. J Vis Exp 46: 2387. 10.3791/2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264. 10.1016/S1097-2765(00)80026-X [DOI] [PubMed] [Google Scholar]
- Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, Weis K. 2019. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573: 144–148. 10.1038/s41586-019-1502-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. 1992. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci 89: 305–308. 10.1073/pnas.89.1.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH. 2012. ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18: 1289–1295. 10.1261/rna.031898.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668. 10.1038/nature09479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Palazzo AF. 2017. Assessing mRNA nuclear export in mammalian cells by microinjection. Methods 126: 76–85. 10.1016/j.ymeth.2017.05.027 [DOI] [PubMed] [Google Scholar]
- Lee ES, Akef A, Mahadevan K, Palazzo AF. 2015. The consensus 5′ splice site motif inhibits mRNA nuclear export. PLoS ONE 10: e0122743. 10.1371/journal.pone.0122743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Wolf EJ, Ihn SSJ, Smith HW, Emili A, Palazzo AF. 2020. TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res 48: 11645–11663. 10.1093/nar/gkaa919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelsky Y, Zuckerman B, Ulitsky I. 2021. High-resolution mapping of function and protein binding in an RNA nuclear enrichment sequence. EMBO J 40: e106357. 10.15252/embj.2020106357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, et al. 2016. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell 64: 520–533. 10.1016/j.molcel.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Ogami K, Manley JL. 2017. Mtr4/ZFC3H1 protects polysomes through nuclear RNA surveillance. Cell Cycle 16: 1999–2000. 10.1080/15384101.2017.1377501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Richard P, Chen Y, Hoque M, Li W, Moresco JJ, Yates JR, Tian B, Manley JL. 2017. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev 31: 1257–1271. 10.1101/gad.302604.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami K, Chen Y, Manley JL. 2018. RNA surveillance by the nuclear RNA exosome: mechanisms and significance. Noncoding RNA 4: 8. 10.3390/ncrna4010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2018. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet 9: 440. 10.3389/fgene.2018.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee C-S, Dias AP, Rapoport TA. 2007. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol 5: e322. 10.1371/journal.pbio.0050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr DS, Rieser LA, Woychik RP, Rottman FM, Rosenberg M, Reff ME. 1986. Differential effects of polyadenylation regions on gene expression in mammalian cells. DNA 5: 115–122. 10.1089/dna.1986.5.115 [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pai AA, Gilad Y, Pritchard JK. 2010. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 6: e1001236. 10.1371/journal.pgen.1001236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösel-Hillgärtner TD, Hung L-H, Khrameeva E, Querrec PL, Gelfand MS, Bindereif A. 2013. A novel intra-U1 snRNP cross-regulation mechanism: alternative splicing switch links U1C and U1-70K expression. PLoS Genet 9: e1003856. 10.1371/journal.pgen.1003856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Maquat LE. 2009. Remodeling of the pioneer translation initiation complex involves translation and the karyopherin importin β. Genes Dev 23: 2537–2550. 10.1101/gad.1817109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders G, van Dijk E, Pruijn GJM. 2007. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res 35: 2564–2572. 10.1093/nar/gkm082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Richter K, Berger AB, Lichter P. 2006. In vivo BiFC analysis of Y14 and NXF1 mRNA export complexes: preferential localization within and around SC35 domains. J Cell Biol 172: 373–381. 10.1083/jcb.200503061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y, Otsubo Y, Kimori Y, Yamamoto M, Yamashita A. 2018. YTH-RNA-binding protein prevents deleterious expression of meiotic proteins by tethering their mRNAs to nuclear foci. eLife 7: e32155. 10.7554/eLife.32155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichino Y, Otsubo Y, Yamamoto M, Yamashita A. 2020. Meiotic gene silencing complex MTREC/NURS recruits the nuclear exosome to YTH-RNA-binding protein Mmi1. PLoS Genet 16: e1008598. 10.1371/journal.pgen.1008598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Karadoulama E, Mąkosa D, Lubas M, Jensen TH. 2018. The RNA exosome adaptor ZFC3H1 functionally competes with nuclear export activity to retain target transcripts. Cell Rep 23: 2199–2210. 10.1016/j.celrep.2018.04.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Schmid M, Dou Y, Garland W, Milek M, Imami K, Johnsen D, Polak P, Andersen JS, Selbach M, et al. 2020. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res 48: 2518–2530. 10.1093/nar/gkz1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skandalis A. 2016. Estimation of the minimum mRNA splicing error rate in vertebrates. Mutat Res 784–785: 34–38. 10.1016/j.mrfmmm.2016.01.002 [DOI] [PubMed] [Google Scholar]
- So BR, Di C, Cai Z, Venters CC, Guo J, Oh J-M, Arai C, Dreyfuss G. 2019. A complex of U1 snRNP with cleavage and polyadenylation factors controls telescripting, regulating mRNA transcription in human cells. Mol Cell 76: 590–599.e4. 10.1016/j.molcel.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Lamond AI. 2011. Nuclear speckles. Cold Spring Harb Perspect Biol 3: a000646. 10.1101/cshperspect.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Sugioka-Sugiyama R. 2011. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J 30: 1027–1039. 10.1038/emboj.2011.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M. 2011. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells 16: 1035–1049. 10.1111/j.1365-2443.2011.01548.x [DOI] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. 2007. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17: 156–165. 10.1101/gr.5532707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Rüegsegger U, Gunderson SI, Keller W, Mattaj IW. 2000. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA 6: 178–188. 10.1017/S1355838200991854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA. 2007. Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19: 3843–3851. 10.1105/tpc.107.055053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang L, Wang J, Chen S, Shi M, Cheng H. 2018. Intronless mRNAs transit through nuclear speckles to gain export competence. J Cell Biol 217: 3912–3929. 10.1083/jcb.201801184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Zheng D, Wei L, Ding Q, Tian B. 2019. Regulation of intronic polyadenylation by PCF11 impacts mRNA expression of long genes. Cell Rep 26: 2766–2778.e6. 10.1016/j.celrep.2019.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Fan J, Wang J, Zhu Y, Xu L, Tong D, Cheng H. 2021. ZFC3H1 prevents RNA trafficking into nuclear speckles through condensation. Nucleic Acids Res 49: 10630–10643. 10.1093/nar/gkab774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R, Balachandran A, Mao AY, Dobson W, Gray-Owen S, Cochrane A. 2011. Differential effect of CLK SR Kinases on HIV-1 gene expression: potential novel targets for therapy. Retrovirology 8: 47. 10.1186/1742-4690-8-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Schmid M, Rib L, Polak P, Meola N, Sandelin A, Jensen TH. 2020. A two-layered targeting mechanism underlies nuclear RNA sorting by the human exosome. Cell Rep 30: 2387–2401.e5. 10.1016/j.celrep.2020.01.068 [DOI] [PubMed] [Google Scholar]
- Yin Y, Lu JY, Zhang X, Shao W, Xu Y, Li P, Hong Y, Cui L, Shan G, Tian B, et al. 2020. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 580: 147–150. 10.1038/s41586-020-2105-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Aibara S, Vos SM, Agafonov DE, Lührmann R, Cramer P. 2021. Structure of a transcribing RNA polymerase II–U1 snRNP complex. Science 371: 305–309. 10.1126/science.abf1870 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. 10.1038/35030160 [DOI] [PubMed] [Google Scholar]
- Zuckerman B, Ulitsky I. 2019. Predictive models of subcellular localization of long RNAs. RNA 25: 557–572. 10.1261/rna.068288.118 [DOI] [PMC free article] [PubMed] [Google Scholar]