Abstract
Post-traumatic osteoarthritis (PTOA) associated with joint injury triggers a degenerative cycle of matrix destruction and inflammatory signaling, leading to pain and loss of function. Here, prolonged RNA interference (RNAi) of matrix metalloproteinase 13 (MMP13) is tested as a PTOA disease modifying therapy. MMP13 is upregulated in PTOA and degrades the key cartilage structural protein type II collagen. Short interfering RNA (siRNA) loaded nanoparticles (siNPs) were encapsulated in shape-defined poly(lactic-co-glycolic acid) (PLGA) based microPlates (μPLs) to formulate siNP-μPLs that maintained siNPs in the joint significantly longer than delivery of free siNPs. Treatment with siNP-μPLs against MMP13 (siMMP13-μPLs) in a mechanical load-induced mouse model of PTOA maintained potent (65–75%) MMP13 gene expression knockdown and reduced MMP13 protein production in joint tissues throughout a 28-day study. MMP13 silencing reduced PTOA articular cartilage degradation/fibrillation, meniscal deterioration, synovial hyperplasia, osteophytes, and pro-inflammatory gene expression, supporting the therapeutic potential of long-lasting siMMP13-μPL therapy for PTOA.
Keywords: disease modifying osteoarthritis drug, MMP13, osteoarthritis, post-traumatic osteoarthritis, siRNA, nano-in-micro, PLGA
Graphical Abstract
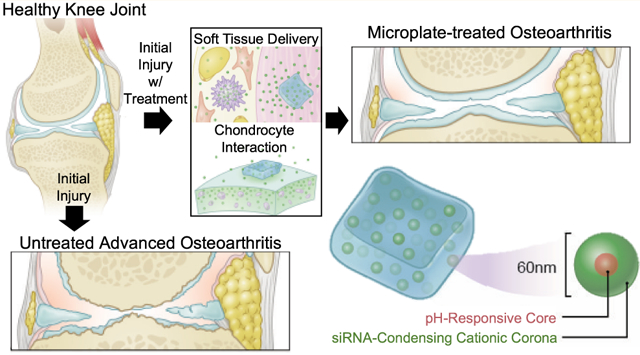
INTRODUCTION
Osteoarthritis (OA) is a chronic degenerative disease affecting joints and their surrounding tissues that affects more than 25% of the U.S. population over the age of 45.1,2 OA is classified as primary, originating from unknown causes, or as post-traumatic osteoarthritis (PTOA), secondary to injury. PTOA is induced by joint mechanical injury (e.g., anterior cruciate ligament tear), often affecting younger and highly active individuals and requiring surgical intervention on average 7–9 years earlier than typically required for primary OA.3,4 PTOA accounts for ~12% of U.S. OA cases, yet comes at a higher societal cost in terms of lost quality life years due to affecting patients of younger age and its accelerated progression.5
Current PTOA treatments are palliative, and there are no clinically approved disease modifying osteoarthritis drugs (DMOADs). Nonsteroidal anti-inflammatory drugs (NSAIDs) are typically the first line of treatment but are only marginally effective at pain relief, do not affect underlying pathology, do not slow cartilage deterioration, and can cause gastrointestinal complications.6 One challenge of systemic therapies is the lack of joint biodistribution to avascular cartilage, making local injection (intra-articular) an attractive option for increasing joint bioavailability and decreasing systemic exposure.7 For symptomatic knee OA, the Osteoarthritis Research Society International (OARSI) and the American College of Rheumatology (ACR) recommend intra-articular injection of corticosteroids.8,9 Steroids provide patients temporary pain relief but do not target the underlying cause of disease and are not recommended for long-term management.9 Recent studies indicate that patients treated with corticosteroids have increased rates of cartilage thinning (when given 4 times per year for 2 years)10 and increased chance of undergoing joint replacement surgery.11 Another challenge is that intra-articular treatments can be rapidly cleared through the synovial vasculature (small molecules) or lymphatic drainage (macromolecules);7,12 half-lives range from less than 1 h for small molecules, 1–4 h for NSAIDs/steroids, and ~1 day for large molecular weight HA.13
There is an unmet need to develop new precision medicines that target the underlying causes of OA/PTOA, have a prolonged mechanism of action allowing infrequent injections, and can modify the trajectory of joint degeneration.14,15 Many of the same inflammatory mediators and pathways are shared by both PTOA and OA.16 In the early stages, aberrant mechanical stress and disturbance of the extracellular matrix (ECM) stimulate synoviocytes and chondrocytes to produce inflammatory cytokines and matrix metalloproteinases (MMPs).17 Compromise of the intact articular surface increases exposure of type II collagen (CII) fibrils18 to MMPs,19 leading to erosion into the deeper cartilage layers, resulting in chondrocyte loss and eventual subchondral bone exposure. Released CII degradation fragments also signal to activate synovial macrophages, perpetuating PTOA/OA.20 Multiple upstream biochemical mediators of arthritis, including interleukin-1β (IL-1β), Wnt, c-Jun N-terminal kinase, and reactive oxygen species, converge to further increase expression of MMPs, suggesting that MMP inhibition may be an effective and broadly useful DMOAD.21–23
Matrix metalloproteinase 13 (MMP13) is a key proteolytic driver of cartilage loss in OA as indicated by reduced progression of surgically induced OA in MMP13 knockout mice and in wild type mice treated with the selective MMP13 inhibitor CL82198.24 Promisingly, intraperitoneal injection of CL82198 decelerated MLI-induced OA progression, increased type II collagen and proteoglycan levels, and inhibited chondrocyte apoptosis compared to saline treatment. Unfortunately, clinical trials on MMP small molecule inhibitors have been suspended due to pain associated with musculoskeletal syndrome (MSS). Patient MSS is believed to be linked to systemic delivery of small molecules that non-selectively inhibit multiple MMPs, some of which (MMP2, −3, −4, −7, and −9) are heavily involved in normal tissue homeostasis.25–27 Production of selective small molecule inhibitors is complicated by shared domains of the collagenases and the homology of the catalytic site.28 Along with the potential for side effects, small molecule inhibitors often require frequent dosing. To this point, CL82198 was injected at a high dose (10 mg/kg) intraperitoneally every other day in order to provide a therapeutic benefit.24 Systemic and high dose treatment increase the risks for off-target effects, especially in clearance organs like liver and kidney. For example, another tested MMP13 “specific” inhibitor PF152 reduced lesion severity in a canine PTOA model29 but caused nephrotoxicity thought to be associated with off-target effects on the human organic anion transporter 3.
As its mechanism is predicated on complementarity base-pairing with messenger ribonucleic acid (mRNA), RNA interference (RNAi) provides a unique opportunity to selectively inhibit conventionally undruggable targets with reduced off-target effects. MMP13 siRNA has previously been developed and tested in murine animal models of OA;30,31 unfortunately, in these studies, the observed in vivo MMP13 knockdown was not durable (only tested 2 days after injection; with indirect fluorescence imaging lasting 1 week). We previously circumvented these issues by applying matrix-anchoring as a method to increase the efficacy of siRNA targeting MMP13 with fewer doses and less off-target effects.32,33 CRISPR/cas9 technology has also been used to ablate NGF, MMP13, and Il-1β in the knee joint with adeno-associated vectors injected intra-articularly. Though viral gene editing comes with safety concerns, this promising study demonstrated that permanent MMP13 gene ablation improved knee joint structure, while combined ablation of MMP13, NGF, and Il-1β had comprehensive effects on joint structure and OA pain.34
Herein, we sought to pursue a long-lasting, joint-localized, and MMP13-selective PTOA therapy based on RNAi (Figure S1). Biomaterial depots offer a reliable approach to improve drug pharmacokinetics, particularly for treating chronic diseases.35,36 As an example, Flexion Therapeutics developed and recently achieved FDA approval for the sustained release of triamcinolone acetonide from poly(d,l-lactide-coglycolide) acid (PLGA) microparticles.37 While we and others have developed effective short interfering RNA nanoparticles (siNPs), these systems are limited in terms of long-lasting effect.32,33 Thus, we sought to engineer a nano-in-micro carrier system that leverages both the benefits of nanoscale particles (i.e., ability to be cell internalized and escape endosomes within chondrocytes/synoviocytes for intracellular cargo delivery), and micrometer-scaled polymeric particles (i.e., ability to be retained within the joint space and slowly release their loaded cargo over time). Nano-in-micro systems have recently been applied to OA,38,39 mostly comprising small molecule cargo such as the potent p38α/β MAPK inhibitor PH-797804 and kartogenin, but none utilizing siRNA therapeutic cargo.
Our design comprises endosome-escaping, siRNA-complexing nanoparticles (siNP) embedded within poly(lactic-co-glycolic acid) (PLGA) microPlates (μPLs), yielding a siNP-μPL nano-in-micro system for potent, sustained siRNA delivery (Figure S1). The siNP is a micelleplex comprising the diblock copolymer poly[DMAEMA67-bl-(DMAEMA29-co-PAA40-co-BMA75)] (DDPB);40 the DDPB polymer self-assembles into micelles with a cationic, siRNA-condensing poly(DMAEMA) (2-(dimethylamino)ethyl methacrylate) surface and the random terpolymer block of DMAEMA, PAA (2-propylacrylic acid), and BMA (N-butyl methacrylate) in the core. The core destabilizes and promotes endosome escape at acidic pH due to a shift from an approximately charge neutral to net cationic state as a result of protonation of the DMAEMA and PAA monomers, which are each approximately 50% ionized (cationic and anionic, respectively) at neutral pH. This system has been explored for siRNA delivery and more recently delivery of gene editing ribonucleoproteins.41 Controlled release of this siNP has been developed from tissue engineered scaffolds and hydrogels for wound and bone fracture healing applications.42–45 Here, we extend this concept to create an injectable format that enables sustained intra-articular siNP delivery using PLGA microPlates (μPLs).46–48 Previously, we synthesized dexamethasone loaded microPlates (DEX-μPLs) that showed sustained release, anti-inflammatory effects, and promise as a DMOAD.48 The μPLs are created by a top-down template-based strategy that enables production of size- and shape-defined cuiboidal particles with controlled drug loading. This siNP-μPL system is herein fabricated, characterized, and therapeutically tested for targeting MMP13 in PTOA knee joints.
RESULTS AND DISCUSSION
Fabrication and Characterization of siNPs and siNP-μPLs.
Polymeric μPLs were synthesized following a multistep, top-down fabrication process,46,47 embedding siRNA-loaded, endosomolytic nanoparticle micelleplexes (siNPs) (Figure S1) made of poly[DMAEMA67-b-(DMAEMA29-co-PAA40-co-BMA75)] (DDPB, Figure S2). Different siNPs (empty-NPs, Cy5-siNPs, siLUC-NPs, and siMMP13-NPs) were characterized for size and size distribution using DLS (Figure S3A). Samples showed a narrow size distribution (PDI < 0.15) with average hydrodynamic diameters of approximately 60 nm. The siRNA encapsulation efficiency (EE%) into siNPs was evaluated using a Quant-iT Ribogreen assay and was approximately 90% (Figure S3A) at a N+:P− ratio of 4 (ratio of ionized amines on the polymer to negatively charged phosphodiesters in the nucleic acid). The siNPs were cytocompatible in an active concentration range and were stable over time in solution, with siMMP13-NP-loaded μPL releasates collected at 1, 3, and 5 weeks also showing presence of siNPs similar in size to freshly formed siNPs (Figure S3B–D). Before integrating siNPs into μPLs, their stability was evaluated in the presence of acetonitrile, a solvent used in μPL fabrication. A dynamic light scattering (DLS) analysis of siNPs was performed before and after exposure to acetonitrile (1.25 nmol of siRNA in 25 μL of PBS added into 100 μL of acetonitrile to match μPL fabrication conditions). As reported in Figure 1A, acetonitrile did not significantly alter size distribution.
Figure 1.
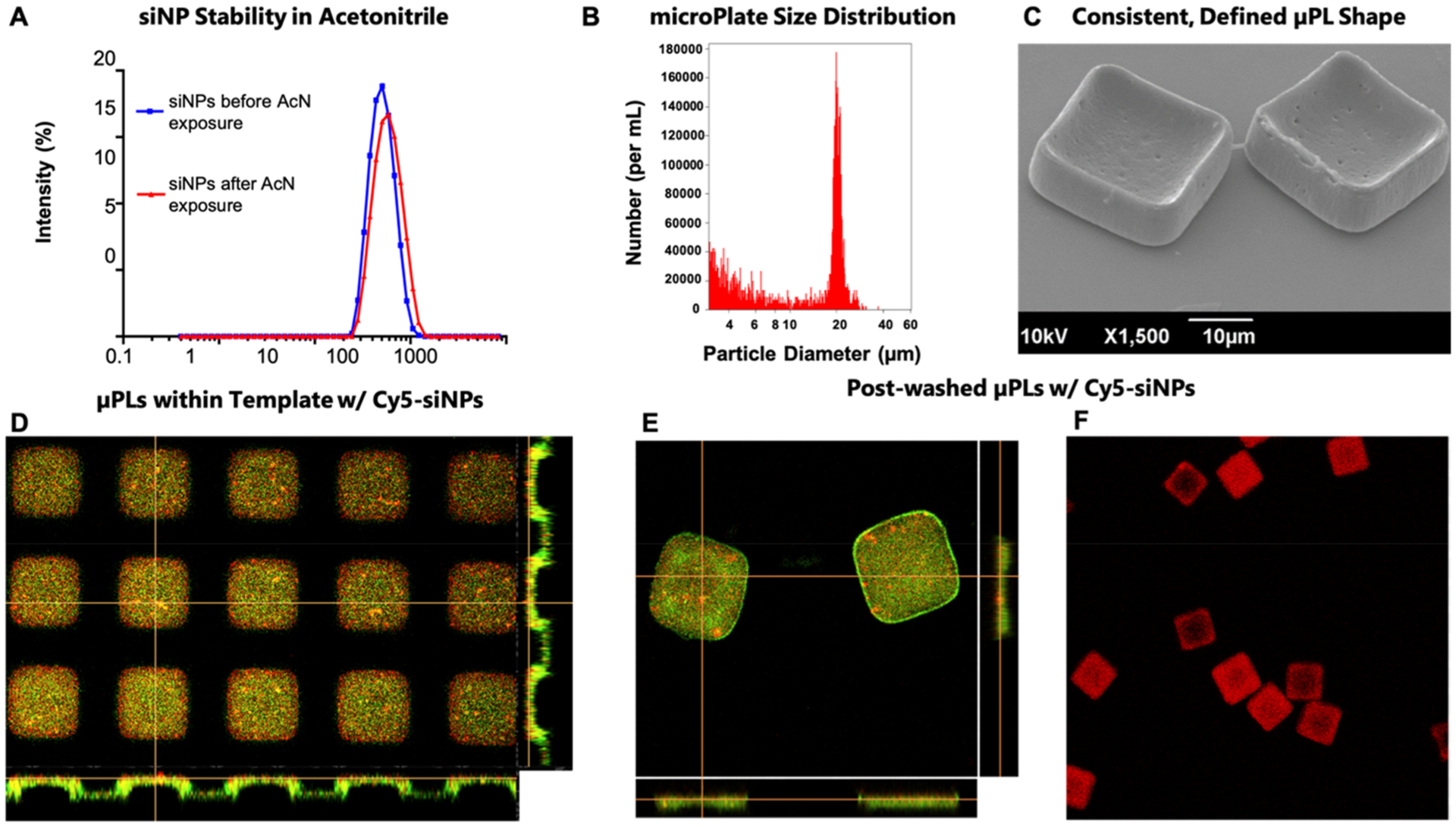
Characterization of siNP-μPLs. (A) DLS size distribution of Cy5-siNPs before (blue line) and after acetonitrile (AcN) exposure (red line). (B) Size characterization of μPLs via Multisizer analysis. (C) A 30° tilted SEM image of μPLs loaded with Cy5-siNPs. (D) Confocal microscopy image of the PVA template containing Cy5-siNPs (red) dispersed within PLGA paste (curcumin, CURC, only added for these images to enable PLGA visualization as yellow-green fluorescence). (E) Confocal microscopy image of Cy5-siNP loaded CURC-μPLs after harvest. (F) Confocal microscopy image of Cy5-siNP-μPLs (red), without CURC, utilized for releasate and in vivo pharmacokinetic studies.
The siNPs in PBS/acetonitrile solution (1.25 nmol of nucleic acid loaded onto 500 μg of DDPB polymer) were mixed into the polymeric paste (15 mg PLGA) and spread over the sacrificial poly(vinyl alcohol) (PVA) template. Following drying and harvesting of the μPLs by PVA dissolution, the resultant siNP-μPLs were characterized via Multisizer Coulter Counter and SEM (Figure 1B,C), showing production of a homogeneous population of cuboidally shaped particles. For pharmacokinetics measurements (intravital, ex vivo, and histological), Cy5-siNPs were loaded into the μPLs. Fluorescent microscopy confirmed distribution of the Cy5-siNPs throughout the μPLs both within the PVA template (Figure 1D) and following μPL isolation (Figure 1E). Note that the μPL green appearance is derived from inclusion of curcumin (CURC), used only for particle microscopy visualization purposes. Cy5-siNPs (without CURC) that were used for subsequent releasate and in vivo pharmacokinetic studies were also visualized (Figure 1F). Fluorescence microscopy images were also taken that demonstrate efficient and dispersed loading of the nucleic acid cargo within the PLGA microPlate (Figure S4). The formulation was also characterized for siRNA loading and was found to contain 1.17 μg nucleic/mg PLGA with the free siNPs loaded into the microPlates having an encapsulation efficiency of 93.8% and 88%, for siLUC and siMMP13 nucleic acids, respectively (Figure S3A).
Most PLGA microparticle (MP) systems for nucleic acid delivery published to date utilize the water-in-oil-in-water (w/o/w) emulsion method for loading the hydrophilic cargo (typically siRNA). Direct PLGA MP encapsulation of noncomplexed nucleic acids prolongs the release of the nucleic acids as expected, but the loading process is inefficient, and the released cargo has poor intracellular delivery; for example, w/o/w PLGA MP loading of siRNA against the glucocorticoid receptor yielded a loading efficiency of 53.6% with a final composition of 0.12 μg of siRNA per mg of PLGA.49 Similar w/o/w approaches achieved encapsulation of siRNA against RANK of 0.67 μg of siRNA/mg of PLGA with an encapsulation efficiency of 80.8%50 and loading of siRNA against FcγRIII precomplexed with bPEI with an encapsulation efficiency of 85–91%.51 We previously achieved some of the best encapsulation results to date in a cancer application, in which we loaded endosomolytic nanoparticles by the w/o/w method and achieved approximately 1.8 μg of nucleic acid per mg of PLGA with an encapsulation efficiency of 75%.52 Top-down fabrication approaches provide synthesis benefits over the w/o/w strategy by enabling controlled fabrication of homogeneous microparticle populations with content defined by the polymeric slurry used during fabrication while reducing aqueous exposure in production. Top-down PLGA microparticle fabrication was recently used to load 10 μg of an oligonucleotide STING agonist per microparticle (per mg PLGA varied depending on the formulation), with release properties tuned by altering microparticle polymer molecular weight.53 A survey of the literature suggests that the endosomolytic nanoparticles used here and used in our previous cancer-focused work achieved high siRNA cargo loading. Though we did not test conditions for maximizing siRNA loading using the top-down fabrication method, due to the relative ease in doping the siRNA nanocarrier into the PLGA slurry during microparticle fabrication, we anticipate that higher wt siRNA/wt PLGA than that achieved herein is attainable and that additional microparticle shape engineering could be utilized to tune drug loading efficiency and release kinetics.
In Vitro Release Kinetics and Gene Silencing Activity of siNP-μPLs.
In vitro siNP release kinetics (Figure S5) from μPLs in PBS (0.75 mg of μPLs in 1.5 mL) were quantified by both a Quant-iT Ribogreen assay (siMMP13-NPs) and fluorescence (Cy5-siNPs), showing sustained release over the 5-week experiment (Figure 2A,B). The bioactivity of siNPs released from μPLs was measured on releasates collected over 5 weeks from samples incubated at 37 °C on a shaker. Releasates were collected for μPLs loaded with both siLUC-NPs (siRNA against a luciferase model gene) and siNEG-NPs (a negative control siRNA). Gene silencing bioactivity of the releasates collected over time was measured in luciferase-expressing (LUC+) ATDC5 (murine chondrogenic) cells. Free siLUC NPs were incubated in PBS in parallel and collected at the same time points to assay their bioactivity over time in solution. The siLUC NPs released from μPLs (siLUC-μPLs) maintained silencing activity throughout the release study (Figure 2C). Free siLUC-NPs incubated in solution at body temperature (37 °C) before being applied to cells showed silencing only for 2 weeks and were inactive by 3 weeks. These data suggest that siNP silencing activity was better preserved when encapsulated within μPLs than siNPs exposed to a physiologic, aqueous solution. In a similar study, activity of the siMMP13-μPL releasates was assayed by qRT-PCR in chondrogenic ATDC5 cells stimulated with TNFα (20 ng/mL) to upregulate MMP13 expression. This study showed that the released siNPs maintained activity for weeks 1–4 when loaded with siRNA against the therapeutically relevant gene MMP13 (Figure 2D).
Figure 2.
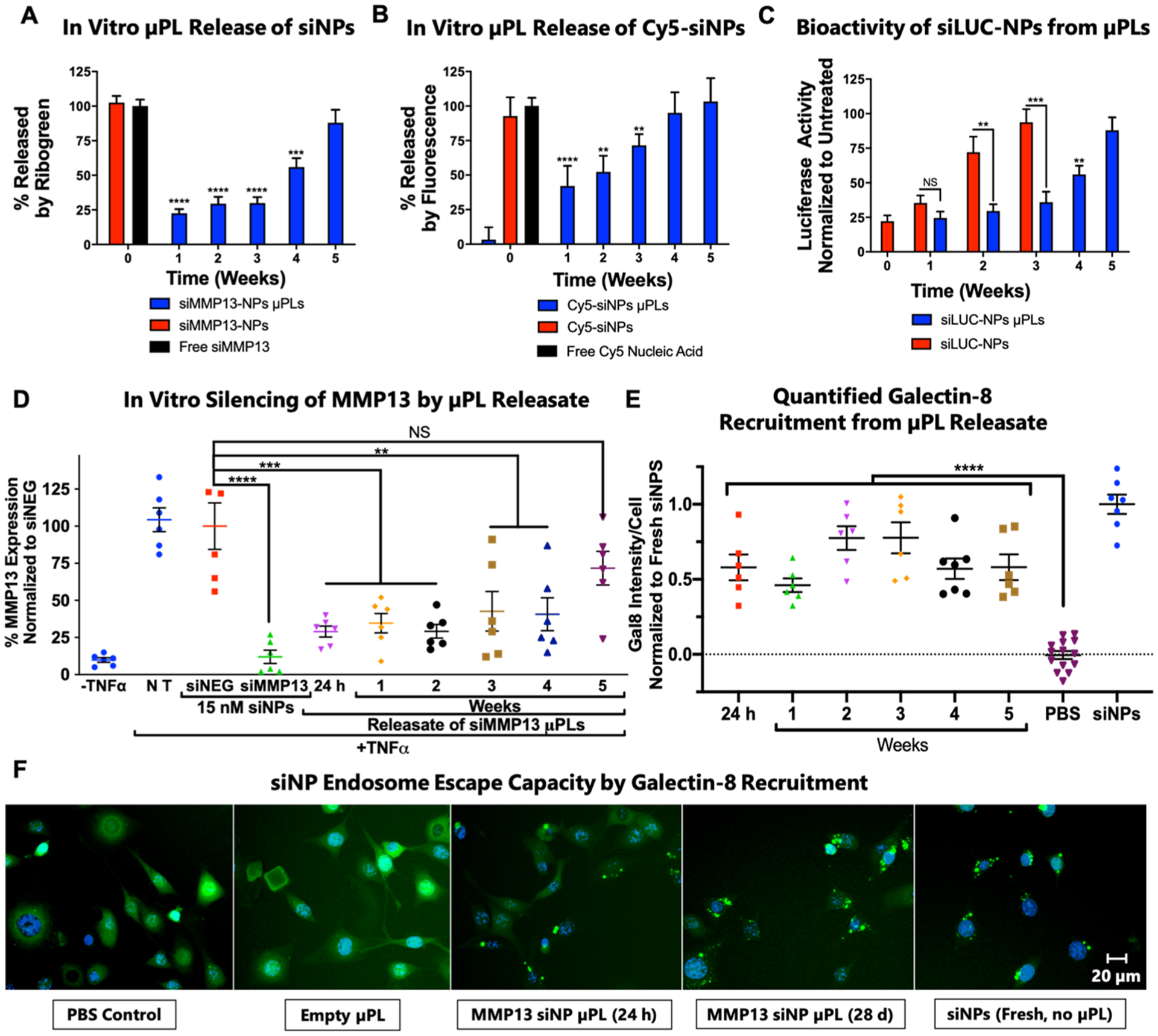
Release kinetics and bioactivity assessment of siNP-μPLs in vitro. (A) siMMP13-NP release profile from μPLs as measured by Quant-iT Ribogreen assay. (B) Cy5-siNP release kinetics from μPLs as measured by Cy5 fluorescence. (C) ATDC5 chondrogenic cell luciferase silencing by siLUC NPs collected over time from μPL releasates or incubation in PBS. (D) Silencing activity of siMMP13-μPL releasates measured by qRT-PCR on chondrogenic ATDC5 cells following stimulation with TNFα (20 ng/mL). (E) Quantification of galectin-8-YFP puncta recruitment as a marker of endosome disruption in cells treated with releasates collected at different time points. Data are normalized to recruitment in cells treated with fresh siNPs. (F) Representative images of galectin-8-YFP concentration (punctate green spots) onto disrupted endosomes (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; all error bars represent standard error).
The siNPs released from μPLs were further investigated for maintenance of cellular endocytosis and endosome escape in vitro. Reporter cells expressing galectin-8 (Gal8) fused with yellow fluorescent protein (YFP) (Gal8-YFP) show diffuse cytosolic fluorescence at baseline and punctate fluorescence upon endosome disruption. This redistribution is due to strong Gal8 binding to lectins selectively found on the inner leaflet of the endosome membrane; these lectins become exposed and available to Gal8 upon efficient disruption of the endosome. Quantification of Gal8 recruitment has been validated as a method of endosome escape analysis that is predictive of gene silencing bioactivity.54,55 The siMMP13-μPL releasates triggered robust recruitment of Gal8-YFP puncta while direct treatment with empty μPLs did not (Figure 2E,F).
Joint Pharmacokinetics of Cy5-siNP-μPLs and Cy5-siNPs.
Pharmacokinetics were assessed through a time-course of intravital imaging, ex vivo imaging, and confocal microscopy on knee joint cryosections performed following a single intra-articular injection of either free Cy5-siNPs or Cy5-siNP-μPLs (Figure 3). Mechanical loading of the mouse knee joint (9 N, 500 cycles, 5 times per week) was started immediately following injection (same as therapeutic model/study, Figures S6A,B). Confocal microscopy showed Cy5-siNPs-μPLs dispersed across the entire knee joint at 1 day after injection (Figure 3A) reaching the femoral–tibial cartilage interface, the meniscus, the infrapatellar fat pad and synovium, and the posterior joint space (Figure S7). The Cy5-siNPs-μPLs were observed to be confined to the intra-articular space and synovium and were not observed near the subchondral bone (Figures S7 and S11). Intravital imaging detected Cy5 fluorescence in mouse knees for up to 10 days, with Cy5-siNPs joint presence diminishing at a faster rate than Cy5-siNP-μPLs, yielding a higher joint area under the curve (AUC) for the Cy5-siNP-μPLs (Figure 3B–D). A nonlinear regression curve fit provided a half-life of 16.54 h for Cy5 siNPs and 176.7 h for Cy5 siNP-uPLs with Tau values of 23.86 h and 255.0 h, respectively. Although the Cy5-siNP-μPLs fluorescence signal was not detectable intravitally at 10 days, ex vivo analysis (skin off), which is more sensitive, showed signal above no treatment (NT) mice over the full time course of 25 days (Figure 3D).
Figure 3.
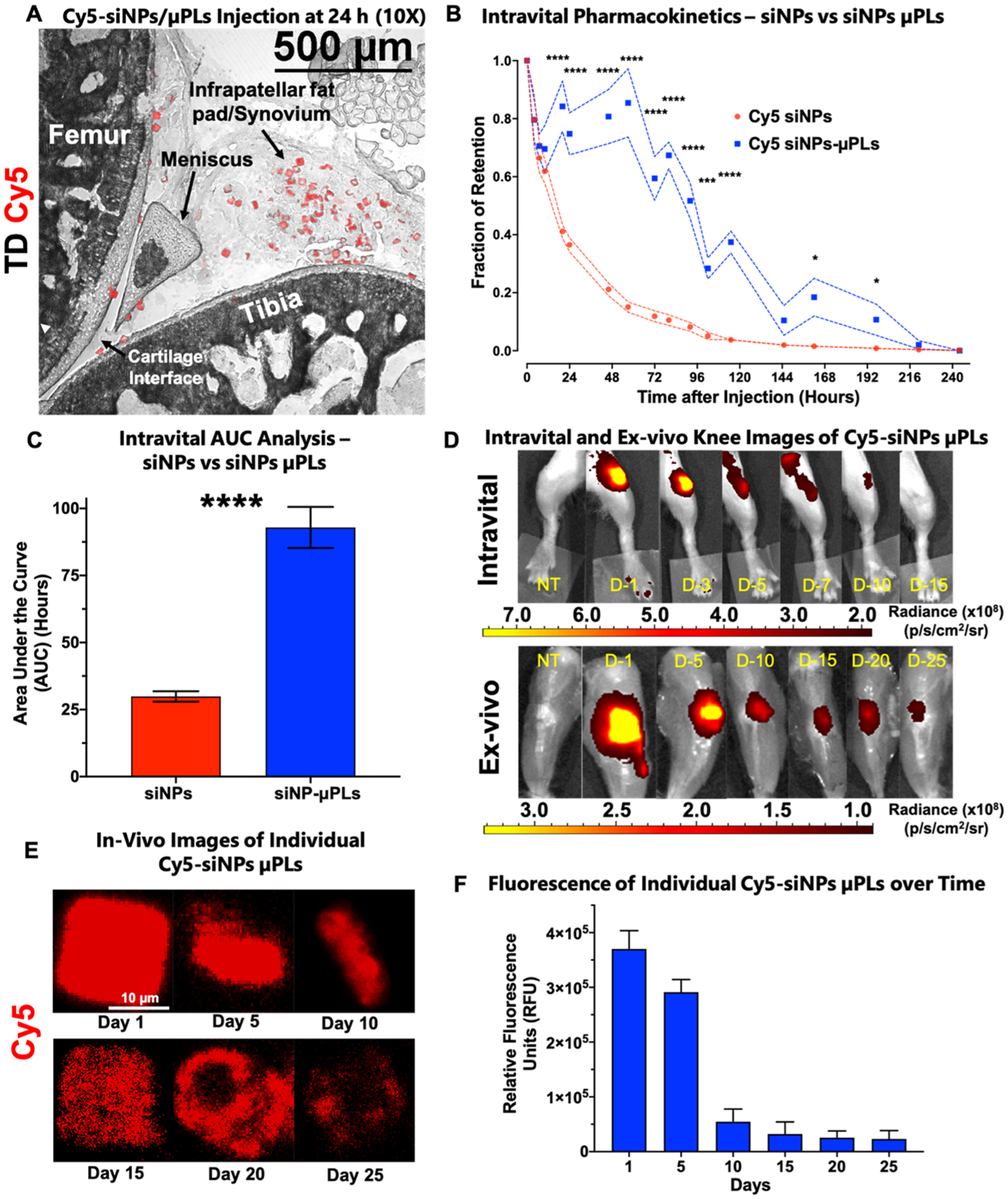
In vivo pharmacokinetic study of Cy5-siNP-μPLs and Cy5-siNPs in a mouse model of osteoarthritis. (A) Anatomically labeled sagittal cryosection from a mouse joint 1 day after intra-articular injection showing Cy5-siNP-μPLs dispersed across the joint reaching the femoral-tibial cartilage interface, the meniscus, the infrapatellar fat pad, and synovium. (B) Intravital fraction of retention over time of both Cy5-siNPs and Cy5-siNP-μPLs plotted as mean ± SEM (dotted lines). (C) Area under the curve (AUC) analysis of intravital joint pharmacokinetics data plotted as mean ± SEM. (D) Representative intravital (skin on) and ex vivo (skin off) knee images of Cy5-siNP-μPLs over time (D-no. = no. of days after injection). (E) Representative confocal microscopy images of individual Cy5-siNP-μPLs from day 1 to day 25 within the mouse joint. (F) Quantification of relative fluorescence units (RFU) of individual Cy5-siNP-μPLs in histological sections over time plotted as mean + SEM. For intravital imaging analysis, n = 4–24 limbs depending on the time point. Measurements were done on more animals at earlier time points, and the sample size at the later time points was lower because some animals were taken down throughout the course of the study for ex vivo imaging and confocal microscopy analysis. For ex vivo imaging and confocal microscopy analysis, n = 2–4 limbs per time point. TD = transmission detector. NT = no treatment. (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; all error bars represent standard error).
Confocal microscopy also confirmed the ex vivo epifluor-escence data (Figure 3D), enabling visualization of Cy5-siNPs-μPLs in the joint for the entire 25-day time course (Figure S8). Conversely, free Cy5-siNPs were only visualized on the cartilage surface and only on day 1 postinjection, likely due to initial electrostatic interactions with anionic cartilage ECM followed by clearance of free siNPs through lymphatic drainage (Figure S8). Breakdown of the μPL shape and release of the Cy5-siNP cargo over time were also apparent, as shown in the representative confocal microscopy images of Cy5-siNP-μPLs in the joints over time (Figure 3E). Relative fluorescence (RFU) of individual Cy5-siNP-μPLs was quantified over time, showing that the Cy5 signal within individual μPLs remained above baseline for the entire 25-day time course (Figure 3F). Loss of shape, structure, and fluorescence of Cy5-siNP-μPLs was also seen collectively when comparing day 1 20× images with day 25 20× images (Figures S9 and S10), showing erosion of the particles over this time frame.
Ultimately, these results support the notion that PLGA microPlate biodegradation leads to sustained release of siNPs over a time course of at least 25 days and that multiple tissues involved in joint homeostasis, including articular cartilage, synovium, infrapatellar fat pad, and the joint capsule, are directly interfacing with the μPLs and the released siNP payload. Furthermore, microPlates remain localized within the intra-articular space and are not seen outside of the joint nor in surrounding musculature or subchondral bone (Figure S11). Biodistribution analyses (Figures S12 and S13) of the Cy5 signal also showed that the cargo remained localized to the treated joint and that the liver is the likely clearance pathway for the small quantity of cargo and/or its degradation fragments that are systemically absorbed. Together, these results suggest that intra-articular use of siNP-μPLs has a low risk for off-target effects.
MMP13 Silencing from siMMP13-μPLs Treatment in a Mechanical Loading Osteoarthritis Mouse Model.
siMMP13-μPLs were investigated in a 28-day longitudinal study to assess duration of MMP13 gene silencing from a single treatment that was given at the same time as loading initiation (Figure S6A,B). This timeline approximates the clinical scenario in which treatment is initiated shortly following a known PTOA-initiating event (i.e., ligament or meniscal tear). Unlike idiopathic OA, for PTOA caused by traumatic injury, it would be feasible to initiate a prophylactic therapy shortly after the injury to reduce subsequent joint deterioration.
The 6-month-old C57BL/6 mice used in the study were subjected to a rigorous knee joint cyclic mechanical loading protocol of 9 N, 500 cycles, 5 times per week to induce PTOA-associated upregulation in MMP13 gene expression (Figure S6A,B). Gene expression measurement by qRT-PCR on combined synovial and cartilage tissues showed over 65% MMP13 gene silencing through 28 days, while gene expression measurement on isolated medial synovial tissue showed over 75% MMP13 knockdown through day 28 compared to siNEG-loaded control μPLs (Figure 4A,B).
Figure 4.
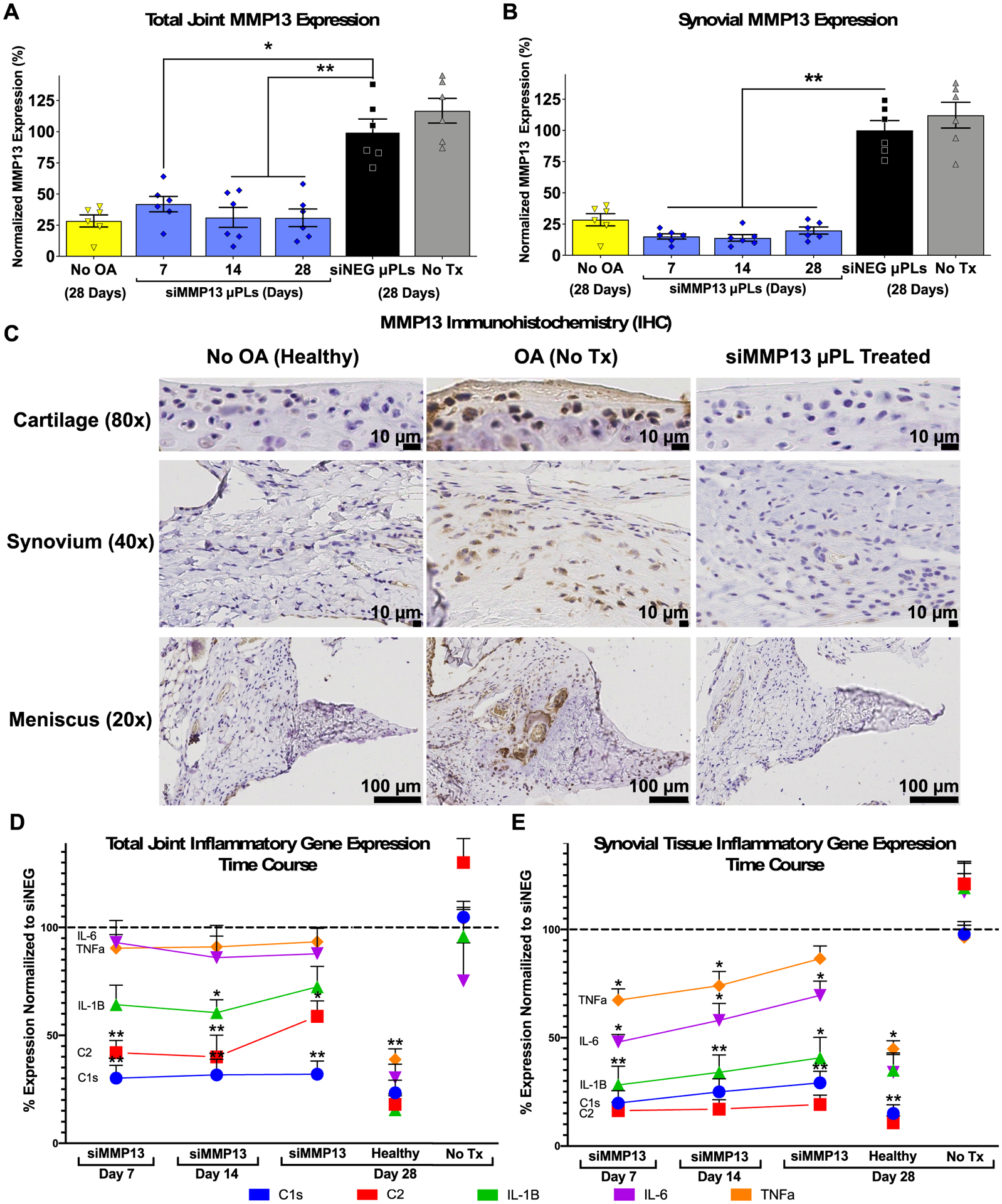
siMMP13-μPLs enable sustained in vivo inhibition of MMP13 and reduction of inflammatory gene expression in the total joint and synovium. (A and B) MMP13 gene expression in (A) combined cartilage and synovium and (B) medial synovial tissue alone over time after a single siMMP13-μPL treatment in mouse PTOA model. (C) MMP13 immunohistochemistry in the cartilage, synovial, and meniscal tissues at day 28 of loading after a single siMMP13-μPL treatment. (D and E) qRT-PCR measurement of expression of C1s, C2, IL-1β, IL-6, and TNFα in (D) the combined total joint lateral synovial and cartilage tissues and (E) medial synovial tissues. Normalization of expression for each gene was done to similar joint tissues harvested at day 28 from animals treated with siNEG-μPLs. *, p < 0.05; **, p < 0.01, compared to siNEG μPLs; all error bars represent standard error of the mean.
The 28 days of consistent target silencing achieved here was significantly extended relative to our previous cancer application of siNPs loaded into PLGA microparticles using w/o/w emulsion PLGA, which showed ~50% model gene luciferase silencing over 3 days.52 As another point of comparison, the aforementioned study on PLGA microparticles for delivery of siRNA against the glucocorticoid receptor achieved in vitro target gene mRNA level reduction for 1 week or less.49 Similarly, the previous study on RANK siRNA delivery using PEI complexes encapsulated into PLGA microparticles achieved in vitro knockdown for up to 8 days.50 Finally, intra-articular application of PLGA microparticles loaded with bPEI complexed with siRNA against FcγRIII achieved approximately 50% protein knockdown relative to controls receiving complete Freund’s adjuvant alone to induce TMJ inflammation, but the study only measured knockdown at one time point (day 9).51 The long-term gene silencing achieved in the current system is advantageous relative to prior technologies and is consistent with our prior results in which we achieved 80% silencing out to 32 days for siNP delivery from tissue engineering scaffolds.56
Immunohistochemical (IHC) staining (validated as shown in Figure S14) confirmed the MMP13 knockdown effect, revealing a treatment-associated reduction in MMP13 protein in the articular cartilage, synovial tissue, and meniscus at day 28 post-treatment (Figure 4C). Higher magnification images of the meniscus in the untreated OA group showed MMP13 expression associated with regions of calcification, pathologic vascularization, reactive synoviocytes, and potential chondroplasia/metaplasia (Figure S15), which was not seen in the tissue samples taken from healthy (unloaded, no OA) or siMMP13-μPL treated mice.
Broader Gene Expression Effects of siMMP13-μPL Treatment in a Mechanical Loading Osteoarthritis Mouse Model.
Key genes related to osteoarthritis progression were also measured by qRT-PCR in joints to verify if MMP13 inhibition was broadly impacting expression patterns in the joint of genes associated with inflammation (C1s, C2, IL-1β, IL-6, and TNFα). Serine proteases (C1S, C1RA, and C2) are components of the complement pathway known to be associated with OA progression,57,58 and these genes are assayed as a marker for influx and activation of macrophages, monocytes, and monocyte-derived dendritic cells that produce these proteases.59 Pro-inflammatory IL-6 expression was measured due to its known role in propagation of cellular stress and synovial inflammation.60,61 Similarly, IL-1β (MAPK activation cluster) and TNFα are inflammatory cytokines that are associated with OA progression by inducing nitric oxide production, MMP and aggrecanase synthesis, and suppressing proteoglycan synthesis.62 Recently published nanoString analysis in a manuscript preprint32,33 showed that PTOA joint silencing of MMP13 (from repeated administration of a shorter-acting antibody-targeted nanocarrier) was associated with reductions in expression of C1s, C2, and IL-1β. C 1s, C2, IL-1β were suppressed in the total joint qRT-PCR analysis across the full time course. There was stronger suppression of these genes in synovial tissue; all genes were suppressed at day 7, and all but TNFα remained reduced at day 14 and day 28 (Figure 4D,E).
OA progression is associated with a viscous cycle of matrix degradation, induction and propagation of inflammation through signaling properties of degradation products, and sustained inflammation driving ongoing, upregulated protease activity.63,64 It can therefore be interpreted that blocking MMP13 reduces release of inflammatory matrix byproducts and thereby more broadly suppresses inflammatory gene expression within the joint, cutting off this vicious cycle and its consequent gradual deterioration of the cartilage matrix. The reduction in inflammation associated with MMP13 silencing also provides broader protective effects that propagate throughout the joint, including reduction of hyperplasia and mineralization of soft tissues such as the meniscus and synovium. This is particularly critical to overall outcomes, as synovial health and signaling play a large role in cartilage and overall joint health.65,66
Joint Structural Protection by siMMP13-μPL Treatment.
Efficacy was next measured for siMMP13-μPLs in preventing PTOA-associated changes in articular cartilage and other joint structural features. Compared to healthy joints and those treated with siMMP13-μPLs, joints treated with siNEG-μPLs or untreated mice show reduced proteoglycan staining and structural changes such as fibrillation and loss of the articular cartilage surface (Figure 5A–C and Figures S16 and S17). The articular cartilage in coronal sections of each joint was scored by a treatment-blinded histopathologist using the Osteoarthritis Research Society International (OARSI) histopathological scoring method67,68 (Table 1). As an additional assessment of cartilage structural damage, a fluorescently labeled monoclonal antibody against collagen II (mAbCII) was systemically administered 24 h before intravital imaging (Figure 5D). The mAbCII provides a way to measure relative cartilage proteolytic damage because collagen II becomes more accessible for antibody binding as OA severity progresses.69,70
Figure 5.
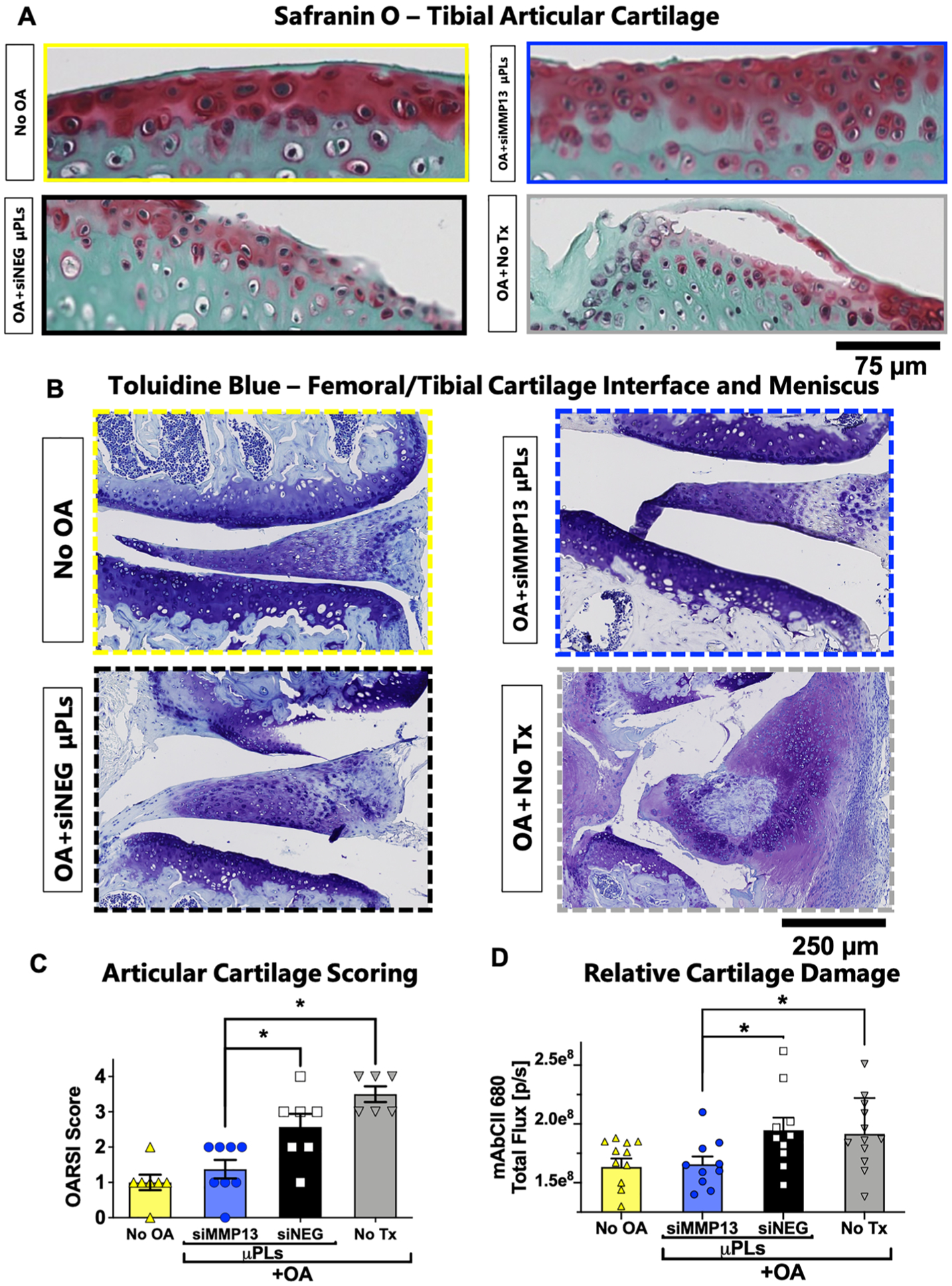
Histological and imaging biomarkers of cartilage structural changes. (A) Representative Safranin-O/Fast Green staining of the tibial plateau articular cartilage surface. (B) Representative Toluidine Blue staining of the femoral/tibial cartilage interface and meniscus. (C) Quantification of cartilage damage by the OARSI osteoarthritis cartilage histopathology assessment system. (D) Quantification of the binding of fluorescently labeled mAbCII in the joint as a marker for relative cartilage damage. (n = 6 to 8 mice knee joints; * = p < 0.05; ** = p < 0.01; all error bars represent standard error). All measurements were taken after 4 weeks of repetitive loading and/or treatment.
Table 1.
(A) Description of OARSI Scale Scoring for Safranin-O/Fast Green Stained Slides of Femoral/Tibial Cartilage Plateaus (Left) and (B) Criteria for Scoring of H&E-Stained Slides to Assess Overall Joint damage by the DJD Scale (Right)
| Degenerative Joint Disease (DJD) scale | |||
|---|---|---|---|
| Severity Score | OARSI Scale | ||
| 0 | normal | within normal limits | mild DJD as a feature of expected age-related change (e.g., attenuation of articular cartilage and proteoglycan loss) |
| 1 | loss of SO staining, articular cartilage thinning; no defects | moderate |
|
| 2 | 1 + fibrillation or pyknotic articular chondrocytes | marked |
|
| 3 | 2 + loss of articular cartilage <50% (e.g., erosion, flap, or callus) | severe |
|
| 4 | 3 + fragmentation and fissuring in 1 + articular plateau | extreme |
|
| 5 | 4 + fragmentation and fissuring of >75% cartilage surface | N/A | |
| 6 | total loss of normal articular cartilage (end-stage) | N/A | |
Both the OARSI score and mAbCII intravital binding measurement indicated that the siMMP13-μPL group articular cartilage was significantly protected relative to joints treated with siNEG-μPLs or untreated mice and was similar to mice that did not have mechanically induced PTOA (Figure 5C–D).
A more holistic joint structural appraisal was also completed by a treatment-blinded histopathologist according to the degenerative joint disease (DJD) scale based on features of H&E-stained tissue sections (Figure 6; scoring rubric, Table 1). Compared to healthy joints and those treated with siMMP13-μPLs, joints treated with siNEG-μPLs or untreated mice show hallmark soft tissue damage associated with advanced PTOA, including cartilage loss, meniscal fibrillation, osteophyte growth, ectopic mineralization, and robust synovial hyperplasia. MMP13 has long been considered a potential target for directly blocking the enzymatic degradation of collagen II to protect articular cartilage.71 However, in line with the broader gene expression changes detected, MMP13 inhibition was also found to provide more holistic joint preservation beyond protection of articular cartilage in the context of PTOA.
Figure 6.
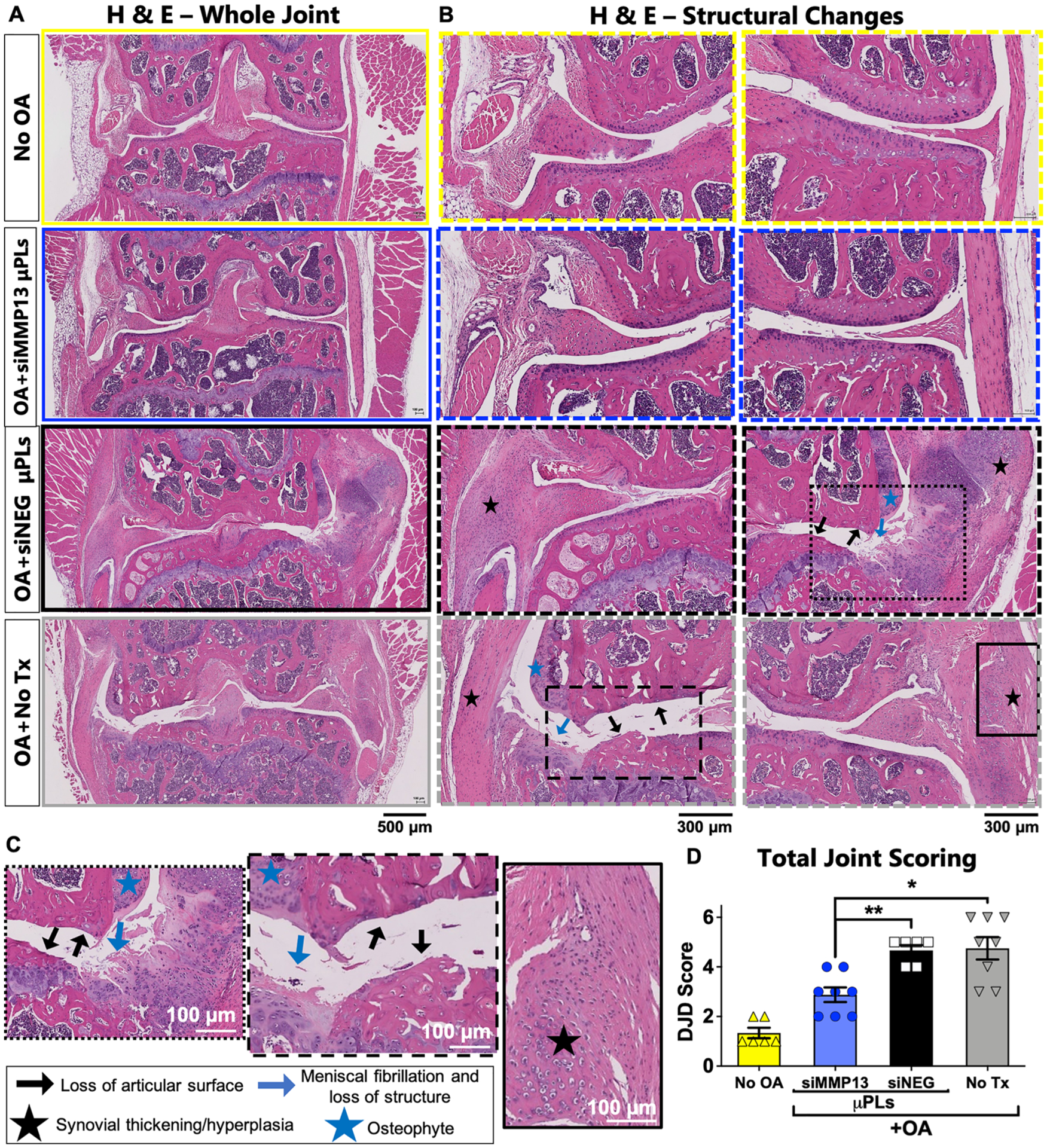
PTOA-associated joint histological changes are broadly suppressed by siMMP13-μPL treatment. (A) Representative H&E staining of whole joints. (B) Articular and synovial regions at a higher magnification. (C) High magnification inset images featuring loss of articular surface, synovial thickening/hyperplasia, meniscal fibrillation, meniscal loss of structure, and osteophyte formation from siNEG-μPLs and untreated groups. Black lines for each image represent 100 μm. (D) Degenerative Joint Disease (DJD) scoring (n = 6 to 8 mice knee joints; * = p < 0.05; ** = p < 0.01; all error bars represent standard error). All measurements were taken after 4 weeks of repetitive loading and/or treatment.
Clinical observations of osteophytes and mineralization of meniscal and synovial tissue are common in advanced OA67,72 and may be related to activation of synovial macrophages by mechanical stress.73 Osteophyte associated pain is thought to be one of the main reasons that patients with advanced OA seek total knee replacement.74 Therefore, we sought to quantitatively measure osteophytes and ectopic mineralization with micro computed tomography (microCT). Mineralization of the meniscus and synovial tissue was found to be significantly reduced in PTOA joints treated with siMMP13-μPLs (Figure 7A–C). The osteophytes emanating directly from the bone were also measured, showing reduction of osteophyte size in PTOA joints treated with siMMP13-μPLs (Figure 7D). Interestingly, the reduction of IL-1β gene expression observed with siMMP13-μPL treatment could also be related to reduced ectopic mineralization, as activation of inflammasomes by presence of ectopic calcium phosphate is known to activate expression of IL-1β.65 By reducing pathological mineralization of the joint, siMMP13-μPLs show promise in resolving the primary source of acute pain and providing global protection of cartilage, meniscus, synovium, and bone tissues of the joint. Subchondral bone density as a ratio of bone volume over total volume was also measured, and a statistically significant preservation of bone density in siMMP13-μPLs treated versus untreated groups was observed (Figure S18).
Figure 7.
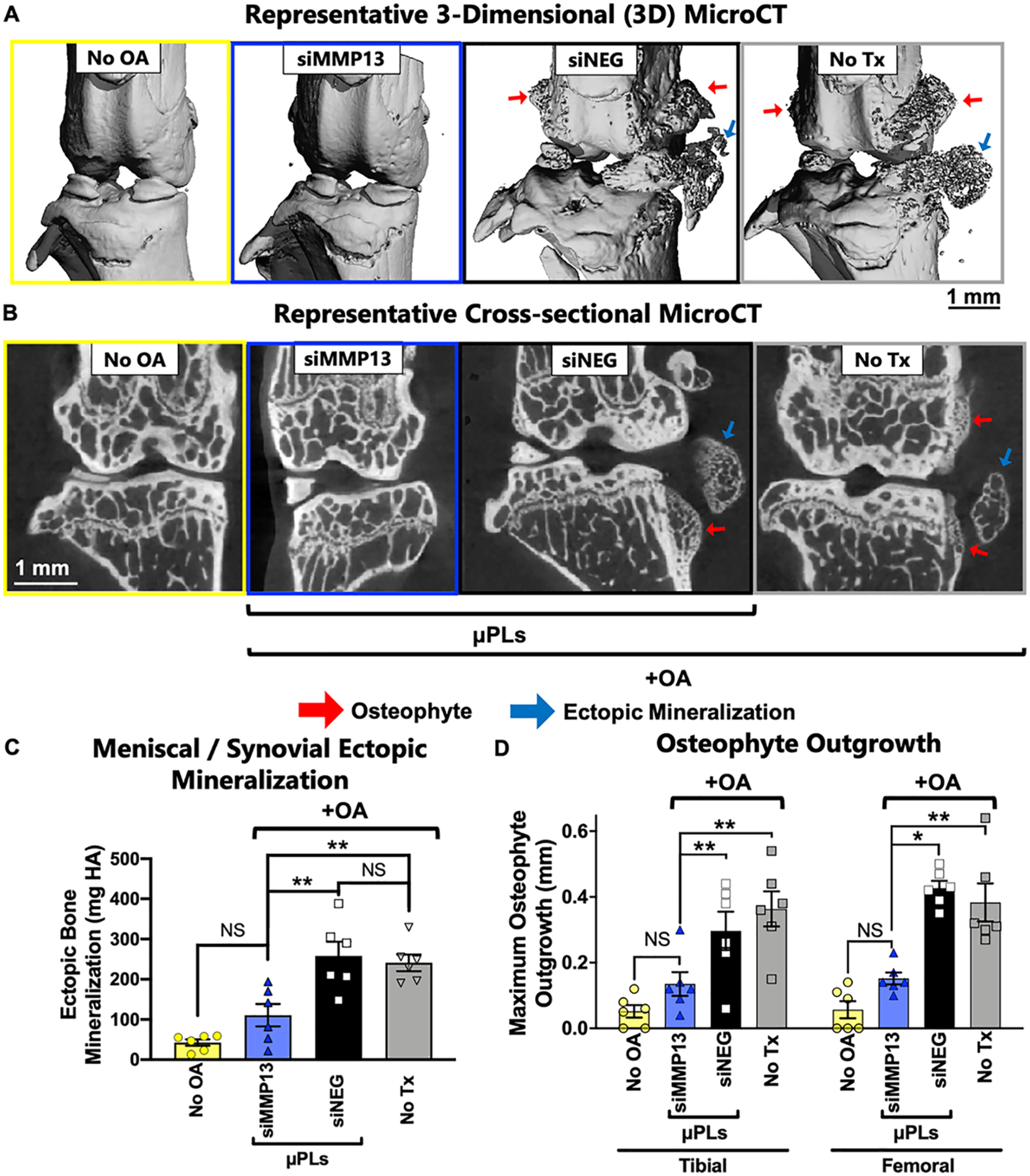
MicroCT analysis of ectopic mineralization and osteophyte outgrowth at 28 days after siMMP13-μPL treatment. (A) Representative 3-dimensional (3D) microCT renderings of mineralized tissue, highlighting osteophytes and ectopic mineralization. (B) Representative 2-dimensional (2D) cross-sectional images featuring osteophyte outgrowth and ectopic mineralization. (C) Quantified mineralization of joint soft tissues from 3-dimensional (3D) microCT analysis. (D) Maximum osteophyte outgrowth of femoral and tibial bones (mm) from 2-dimensional microCT analysis (n = 6; * = p < 0.05; ** = p < 0.01; All error bars represent standard error). All data are from animals after 4 weeks of repetitive loading and treatment.
CONCLUSION
In this study, a nano-in-micro carrier system was constructed and shows promise as a long-acting disease modifying osteoarthritis drug (DMOAD). This system was fabricated using a top-down strategy for the creation of size- and shape-defined, monodispersed PLGA μPLs loaded with endosome-escaping siNPs. The resultant formulation (siNPs-μPLs) was loaded at 1.17 μg nucleic acid/mg PLGA and was able to protect and release intact, bioactive siNPs for up to 5 weeks; released siNPs maintained chemicophysical properties, endosome-escaping ability, and gene silencing activity for the duration of the study (5 weeks). The siNPs-μPLs can be successfully dispersed within the knee joint by intra-articular injection reaching the femoral-tibial cartilage interface, the meniscus, and the infrapatellar fat pad and synovium. A single intra-articular injection of siNPs-μPLs provides sustained retention within the joint for up to at least 25 days, meanwhile free siNPs were only visualized 1 day after injection. Longitudinal in vivo studies utilizing a rigorous PTOA model demonstrated that a single intra-articular injection of siMMP13-μPLs achieved 65–75% gene knockdown of MMP13 over a 28-day study with consequently reduced MMP13 protein production. MMP13 inhibition also caused downstream reduction in the expression of a broad set of inflammatory markers implicated in osteoarthritis pathophysiology (C 1s, C2, IL-1β, IL-6, and TNFα). A single intra-articular injection of siMMP13-μPLs protected articular cartilage integrity, improved total joint histopathology, reduced ectopic mineralization, and lessened osteophyte formation through 4 weeks of rigorous mechanical overloading of mouse knee joints. In sum, this work provides a proof of concept for the utility of PLGA microPlates to extend the longevity of action and overall efficacy of siRNA nanocarriers and show that siMMP13-μPLs are a promising experimental DMOAD capable of delaying PTOA progression.
METHODS SECTION
Materials.
Polydimethylsiloxane (PDMS) (Sylgard 184) and elastomer were acquired from Dow Corning (Midland, MI). Poly(vinyl alcohol) (PVA, MW 31 000–50 000), 2-(dimethylamino)-ethyl methacrylate (DMAEMA), 2,2′-azobis(2-methylpropionitrile) (AIBN), n-butyl methacrylate (BMA), and poly(d,l-lactide-co-glycolide (PLGA, lactide:glycolide 50:50, MW 38 000–54 000) were obtained from Millipore-Sigma (Saint Louis, MO). The 2-propylacrylic acid (2-PAA) monomer was synthesized as described.75 The RAFT chain transfer agent 4-cyano-4[[(ethylsulfanyl)-carbonthioyl]sulfanyl]pentanoic acid (ECT) was synthesized following protocols previously described by Convertine et al.76 adapted from Moad et al.77 Curcumin (CURC) was purchased from Alfa Aesar (Haverhill, MA). Quant-iT Ribogreen assay kit was obtained from ThermoFisher Scientific (Waltham, MA). Syringe filters (200 nm cutoff) were procured from Corning (Corning, NY). All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). All reagents and solvents were used as received.
Synthesis and Characterization of Poly(DMAEMA) Macro-CTA.
In a 25 mL round-bottomed flask, ECT (0.2 mmol, 0.052 g), 2-(dimethylamino)ethyl methacrylate (DMAEMA) (25.2 mmol, 3.99 g, 4.28 mL), and AIBN (0.02 mmol, 0.0034 mg) were dissolved in 1,4-dioxane (5 mL) (Figure S2A). The reaction mixture was degassed by purging nitrogen through the solution for 30 min and submerged into a preheated oil bath at 70 °C for 24 h. After 24 h, the polymerization mixture was precipitated into a 10-fold excess of cold pentane and dried under vacuum overnight. The structure of PDMAEMA was characterized by 1H nuclear magnetic resonance spectroscopy (Figure S2B) NMR, 400 Bruker Spectrophotometer) in CDCl3 solvent. The molecular weight and polydispersity of the polymer was characterized by gel permeation chromatography (GPC, Agilent Technologies, Santa Clara, CA) in dimethylformamide (DMF) + 0.1 M LiBr with inline Agilent refractive index and Wyatt miniDAWN TREOS light scattering detectors (Wyatt Technology Corp., Santa Barbara, CA). Mn,GPC = 10 500, PDI = 1.15 (Figure S2C).
Synthesis and Characterization of the Diblock Copolymer of PDMAEMA67-b-P(DMAEMA29-co-BMA75-co-PAA40) (DDPB).
PDMAEMA macro-CTA was used for the synthesis of the diblock copolymer by RAFT polymerization as reported previously.43 Briefly, PDMAEMA-ECT (0.04 mmol, 0.42g), DMAEMA (2.92 mmol. 0.459 g, 0.49 mL), BMA (5.76 mmol, 0.819 g, 0.915 mL), PAA (2.92 mmol, 0.33 g), and AIBN (0.004 mmol, 0.00067 g) were dissolved in a mixture of dioxane and DMF solvent (3:1 ratio, 6 mL; Figure S2A). The polymerization solution was degassed by purging nitrogen through the solution for 30 min before submerging into a preheated oil bath at 70 °C for 24 h. After 24 h, the polymerization mixture was precipitated into a cold 50:50 mixture of ether and n-pentane, redissolved in dichloromethane, precipitated into cold pentane twice, and dried under vacuum overnight. The polymers were verified by H1 NMR and GPC (Mn,GPC = 30 500, PDI = 1.32; Figure S2B,C).
Formulation of siRNA Polymeric Nanopolyplexes (siNPs).
The siNPs were formulated by dissolving DDPB in EtOH (20 mg/mL) and then diluting to 1 mg/mL by slow addition of PBS buffer (pH 7.40, 1×) to promote spontaneous micelle formation. The final suspension was passed through a 200 nm cutoff syringe filter. Then 1.25 nanomoles of fluorescent Cyanine-5 labeled DNA or siRNA were mixed in PBS (−/−) buffer (pH 7.40, 1×) with DDPB polymer to achieve an N+:P− ratio of 4 (nucleic acid sequences provided in Table S1). The siNPs were concentrated using an Amicon Ultra 10 kDa centrifuge filter device (Millipore Corporation, Billerica, Massachusetts, USA) to a volume of 25 μL. The efficiency of siRNA encapsulation at different N+:P− ratios was evaluated using a Quant-iT Ribogreen assay kit (ThermoFisher Scientific, Waltham, MA). siLUC μPLs, siNEG μPLs, siMMP13 μPLs, and Cy5-siNP-μPLs is the nomenclature for μPLs loaded with luciferase siRNA (siLUC), negative control siRNA (siNEG), anti-MMP13 siRNA (siMMP13), and Cy5-DNA, respectively. The siNPs were evaluated using dynamic light scattering (DLS) for hydrodynamic diameter and size distribution.
Synthesis of siNP-Loaded microPlates (μPLs) (siNP-μPLs).
μPLs were synthesized in a multistep process, as previously described.46,47 Briefly, direct laser writing (DLW) was used to develop the silicon master template with a specific pattern of square wells with a length (≈20 μm) and a height (≈10 μm), separated by a 3 μm gap. Then, a PDMS template was obtained by covering the silicon master template with a mixture of PDMS and curing agent (60 °C for 8 h). The PDMS was then used to create a PVA template, which comprised the same features as the original silicon master. PLGA (in 100 μL of acetonitrile) was mixed with a solution of siNPs (initially dissolved in 25 μL of PBS) and then applied to the PVA template. Each siNP-μPL batch was synthesized using 15 mg of PLGA containing dispersed siNPs comprising 1.25 nmol of nucleic acid (17.5 μg if taking an average estimated MW of 14000 g/mol per nucleic acid) and 500 μg of DDPB polymer (1.17 μg nucleic acid/mg PLGA). Lastly, siNP-μPLs were collected by dissolution of PVA templates in deionized water at room temperature in an ultrasonic bath, followed by sequential centrifugation steps (2823g for 5 min). The resultant siNP-μPLs were stored at +4 °C if being used immediately or −80 °C until their use.
Visualizing Geometry of siNP-μPLs.
The geometric properties of siNP-μPLs were characterized using different techniques, as previously reported.46,47 Scanning electron microscopy (SEM, Elios Nanolab 650, FEI) was used to evaluate their size and shape. Briefly, a drop of sample was placed on a silicon template, sputtered with 10 nm of gold, and analyzed by an SEM operating with an acceleration voltage of 5–15 keV. siNP-μPL concentration and size distribution were also measured using a Multisizer 4 COULTER particle counter (Beckman Coulter, USA). Morphology and homogeneity of siNP distribution within μPLs was examined by a Nikon A1 confocal microscope. For the purpose of μPL confocal and fluorescent microscopy visualization, a separate template of siNP-μPLs was produced containing curcumin (0.5 mg) mixed into the PLGA; curcumin was not added into μPLs used in any of the cell or animal studies.
Encapsulation Efficiency of siNPs into μPLs.
Fluorescent Cy5-siNPs were used to quantify nucleic acid loading into μPLs. The μPLs were dissolved in an acetonitrile and deionized water (1:1, v/v) mixture and then centrifuged at 18 213g for 15 min to promote PLGA precipitation. The supernatant, containing Cy5-siNPs, was collected and concentrated using an Amicon Ultra 10 kDa centrifuge filter device (Millipore Corporation, Billerica, Massachusets, USA). Cy5-siNPs were recovered and lyophilized. Then, the lyophilized sample was dissolved in EtOH, dried, resuspended in water and centrifuged. The supernatant was collected and analyzed, relative to a Cy5-DNA standard curve, with a microPlate spectrophotometer (Tecan, Männedorf, Switzerland) at λex and λemis at 630 and 660 nm, respectively.
Characterization of In Vitro siNP Release from μPLs.
To assess release of siNPs from μPLs, siNP-μPLs were incubated in PBS at 37 °C on a shaker. Each sample was done in triplicate and comprised 6 mg of μPLs in 1.5 mL of PBS buffer, creating a 600 nM concentration of nucleic acid. The releasates were collected weekly by centrifugation of microPlates after which the full buffer was removed and replaced. Parallel samples were analyzed for concentration of released siNPs by either ribogreen assay (unlabeled siRNA loaded siNPs) or fluorescence measurement for Cy5-siNPs. The siNPs were diluted and applied to cells at a 20 nM dose to assay the potency of the released siNPs during different periods of release (see luciferase gene silencing assay section).
Size Characterization of siNPs Released from μPLs.
After spinning down the μPLs in PBS solution, the supernatant (1.5 mL) was aspirated and replaced with additional PBS. The aspirated samples used for DLS measurement were diluted to 0.16 mg/mL siNPs with isotonic double distilled water (1:10 v/v) to avoid multiscattering phenomena and analyzed at 25 °C with a Zetasizer Nano (Malvern, U.K.).
Cell Culture and Cell Viability.
Immortalized mouse chondrogenic ATDC5 cells procured from ATCC were cultured in DMEM/F-12, GlutaMAX medium with 10% serum and 1% penicillin/streptomycin. Cells were incubated at 37 °C in 5% carbon dioxide. All cellular experiments were performed at 80% confluency, except for endosome disruption galectin-8 recruitment studies with MDA-MB-231 Gal8-YFP cells54 that were performed at 40% confluency. The CellTiter-Glo (Promega Corporation, Madison, WI) assay was used to assess cytotoxicity in accordance with the manufacturer’s standard protocol.
Luciferase Gene Silencing Assay.
To measure knockdown bioactivity of the siNPs postrelease from μPLs, releasates collected form siLUC-μPLs and siNEG-μPLs were applied to ATDC5 cells lentivirally transduced to constitutively express luciferase, as in previous studies.32,33,78 The Quant-iT Ribogreen assay kit (ThermoFisher Scientific, Waltham, MA) was used to quantify RNA in the releasates, which were diluted to a concentration of 20 nM siRNA for in vitro treatments used to measure releasate silencing activity. After seeding (96-well black plates, 2000 cells per well), cells were allowed 24 h for adherence (DMEM/F12, 10% FBS, 1% penicillin/streptomycin, 1% Glutamax media) before introducing the treatment solution (releasate in Opti-MEM media at 20 nM siRNA concentration) for another 24 h. Treatments were aspirated after 24 h, and a luciferin solution (150 μg/mL luciferin) was added to the cells prior to measuring bioluminescence using an IVIS Lumina III intravital imaging system (Caliper Life Sciences, Hopkinton, MA). The bioluminescent signal form siLUC-siNP treated cells was normalized to cells treated with analogous releasates containing siNEG-siNPs. Cell viability was further gauged by comparing luciferase signal from cells treated with siNEG-siNPs releasates to untreated control cells.
Post-traumatic Osteoarthritis (PTOA) Mechanical Loading Model and In Vivo Therapeutic Study.
A mechanical overload model was used whereby the knee joints of 6-month-old C57BL/6 mice were cyclically loaded in compression using an ElectroForce 3100 test frame (TA Instruments; New Castle, DE). The mice were anaesthetized, and the knee joints were positioned in flexion at 140° with the tibia approximately vertical and placed directly under the loading point.79 The cyclic loading regimen utilized 500 cycles at 9 N force and was done 5 times per week, from the time immediately following intra-articular injection until tissues were harvested (Figure S6A,B).
Mechanical loading for both the therapeutic and pharmacokinetics studies was initiated immediately after treatment was administered. This tests the treatment in a prophylactic scenario and ensured consistent intra-articular injections in mice, which become technically more difficult after accrual of joint mechanical damage. Intra-articular injection of siNEG-μPLs and siMMP13-μPLs in PBS was done at a per knee dose of 1.875 nmol of siRNA (26.25 μg if taking an estimated average MW of 14 000 g/mol per nucleic acid) and 22.5 mg PLGA (loading capacity of 1.17 μg of nucleic acid/mg of PLGA).
RNA Harvest, cDNA Synthesis, and qRT-PCR.
After animal euthanasia, joint tissues were collected using a scalpel under a dissection microscope and placed in RNAlater solution (ThermoFisher Scientific; Waltham, MA) prior to RNA isolation with a RNeasy kit (Qiagen; Hilden, Germany). Cartilage from tibial and femoral articular surfaces were removed from the bone, and synovial tissues were separated from the menisci before processing. For total joint measurements, cartilage tissue was mixed with synovial tissue in equal mass. Synovial tissue was also separately processed. The cDNA for each sample was produced using the iScript cDNA synthesis kit (Bio-Rad; Hercules, CA), and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with TaqMan primers (below) using reagents procured from Thermofisher Scientific (Waltham, MA) according to the manufacturer’s instructions.
| GAPDH: Mm99999915_g1 | ACTB: Mm02619580_g1 |
| MMP13: Mm00439491_m1 | IL-1β: Mm00434228_m1 |
| IL-6: Mm00446190_m1 | C2: Mm00442726_m1 |
| C 1s: Mm00663210_mH | COX2: Mm03294838_g1 |
| TNFα: Mm00443258_m1 |
Histology.
All knee joints used for histology were fixed in 10% formalin for 2 days, and microCT analysis was performed in 70% ethanol after fixation. Samples were then decalcified in 20% tetrasodium EDTA for 4 days (exchanging once at 2 days) or until soft to the touch, rinsed with water, and then placed in 70% ethanol at 4 °C until paraffin-block embedded and sectioned. Embedding of tissues for coronal sectioning was aided by manually slicing the patella away such that the anterior aspect of the joint could be positioned facing down in the embedding tray, i.e., “stifled”. Sections were cut 15–20 μm thick in the coronal orientation, starting posterior to the patella and sectioning fully through the joint. At least two sections in the loading plane were chosen from each joint for Hematoxylin and Eosin, Safranin-O/Fast Green, and Toluidine Blue staining that were used in subsequent OA severity scoring. Positive staining controls of the mouse skin containing mast cells and the mouse ear were done for Toluidine Blue and Safranin-O/Fast Green, respectively (Figure S17).
MicroCT.
Formalin-fixed samples of the proximal tibia and femoral head were placed in 70% ethanol and scanned at a 15 μm isotropic voxel size, 70 kVp, 114 mA, and 200 ms integration time using a Scanco μCT50 (Scanco Medical, Basserdorf, Switzerland). Contouring was performed first to encompass all mineralized components of the joints (Figure 7A,B). Contouring for 3D analysis was done a second time to segment out and quantify mineralization in the soft tissues surrounding the cortical bone, presented as milligrams of hydroxyapatite (Figure 7C). 3D renderings (sigma, 1.5; support, 3; threshold, 388) were also generated using the Scanco software. Vertical 2D cross-sectional images were identified for each sample with the maximal osteophyte length orthogonal to tibial and femoral cortical bone, which was visualized along with soft tissue ectopic mineralization in a similar plane across all treatment groups (Figure 7B). Osteophyte length was measured on each sample, defined as maximal osteophyte length (Figure 7D). Subchondral bone volume/total volume (BV/TV) was also measured (Figure S18).
Immunohistochemistry.
Immunohistochemical staining was performed on a Leica Bond-Max autostainer (Leica Biosystems Inc., Buffalo Grove, IL). All steps besides dehydration, clearing, and coverslipping were performed on the Bond-Max where all the slides were deparaffinized. Heat-induced antigen retrieval was performed on the Bond-Max using Epitope Retrieval 1 solution (Leica Biosystems Inc.) for 20 min. Slides were incubated with anti-MMP13 (cat. no. Ab39012, Abcam, Cambridge, MA) for 1 h at a 1:750 dilution. The Bond Polymer Refine detection system (Leica Biosystems Inc.) was used for visualization. Slides were then dehydrated, cleared, and coverslipped. Antibody validation was performed on various mouse organs and positive (anti-MMP13 primary) and negative (isotype primary) controls were performed on mouse stomach (Figure S11). Slides were imaged using a Leica SCN400 Slide Scanner (Leica Biosystems Inc., Buffalo Grove, IL) and 20×, 40×, and 80× images were taken of the cartilage surface, synovium, and meniscus for each slide in each treatment group.
Joint Histological Scoring.
Coronal sections were stained with Safranin-O/Fast green for scoring by the Osteoarthritis Research Society International (OARSI) and Degenerative Joint Disease (DJD) scales in at least 2 midfrontal sections per joint.68 Scoring of deidentified sections was interpreted by a board-certified veterinary pathologist. OARSI scores (0–6 semiquantitative scale; Table 1A) were assigned from assessment of the medial and lateral plateaus of the tibia and femur.67 A degenerative joint disease (DJD) severity score (0–4 semiquantitative scale) was determined from H&E stained sections using a semiquantitative index (Table 1B) based on cartilage erosion, subchondral osteosclerosis, synovial/meniscal metaplasia, subchondral osteosclerosis, inflammation, and the growth of osteophytes and meniscal ectopic mineral deposits (Table 1B).80
In Vivo Pharmacokinetic Study.
An in vivo pharmacokinetic study was performed following the same loading and injection regimen as the PTOA model described above. Cy5-siNP-μPLs or free Cy5-siNPs were injected intra-articularly (same dose as the therapeutic study) and imaged intravitally for Cy5 fluorescence over time using an IVIS Lumina III intravital imaging system (Caliper Life Sciences, Hopkinton, MA). For IVIS image analysis, fixed regions of interest (ROIs) were drawn around both the right and the left knee joints. A preinjection (blank) image was taken followed by a time 0 (T0) image directly after intra-articular injection. The blank reading was used for background correction of all images at all time points, and dividing the radiance reading by the T0 radiance reading was used to calculate a fraction of retention for all later time points. Animals were euthanized at days 1, 5, 10, 15, and 25 postinjection for both the Cy5-siNP-μPLs and the Cy5-siNP groups. The skin was then removed from the legs (which provides better sensitivity than intravital imaging), and the knee joints were end point imaged ex vivo for remaining Cy5 fluorescence. A no treatment (NT) control was also used as an additional comparison. Legs were then snap frozen in liquid nitrogen and stored at −80 °C until processing for cryosectioning. A pharmacokinetic area under the curve (AUC) analysis and a nonlinear regression fit was performed using GraphPad Prism version 8.0.
Histological Cryosectioning and Confocal Microscopy.
Hindlimbs with intra-articular injection with Cy5-siNP-μPLs or free Cy5-siNPs were harvested at days 1, 5, 10, 15, and 25 postinjection, stored at −80 °C, and embedded into OCT freezing compound. Samples were serial sectioned in sagittal orientation until an adequate depth of the joint was reached for optimal visualization. Cryosections at various depths along the joint were then sectioned sagitally at 20 μm thick, captured utilizing a commercially available polyvinylidene chloride film coated with synthetic rubber cement (http://section-lab.jp/), and placed on a slide. Slides were then fixed in 10% neutral buffered formalin for 5 min (some were stained with DAPI), coverslipped with an aqua mount, and imaged on a Nikon Eclipse Ti inverted confocal microscope.
Imaging settings were kept constant for imaging of all microPlate-containing joint samples at each time point, but imaging settings are not equal across different days in Cy5-siNP-μPLs vs Cy5-siNPs (Figure S8) in the interest of being able to detect residual fluorescence over time without signal saturation in samples from early time points. Whole joint imaging was performed by stitching 3 × 3 10× images. here, 10× and 20× images were taken for each sample at each postinjection time point. TD = transmission detector.
Relative Fluorescence Units (RFU) Quantification.
To quantify individual microPlate (Cy5-siNP-μPLs) fluorescence over time, RFU analysis was performed on imageJ (http://rsbweb.nih.gov/ij/download.html) using the NDi6D plug-in and the following protocol (https://theolb.readthedocs.io/en/latest/imaging/measuring-cell-fluorescence-using-imagej.html). Briefly, regions-of-interests (ROIs) were drawn around microPlates (N = 3–6 per image) and background ROIs were set randomly distributed across the image (N = 3–6 per image) using the imageJ drawing/selection tools. From the Analyze menu “set measurements” were chosen to have “area integrated intensity” and “mean gray value” selected. RFU was determined through the following formula:
Statistical Methods.
Data are displayed as mean plus standard error (unless stated otherwise). Statistical tests employed either oneway ANOVA with multiple comparisons test or two-tailed Student’s t test between only two groups with α = 0.05.
Animal Ethics Statement.
All animal experiments described herein were carried out according to protocols approved by Vanderbilt University’s Institutional Animal Care and Use Committee (IACUC), and all studies followed the National Institutes of Health’s guidelines for the care and use of laboratory animals.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the assistance of the Vanderbilt Translational Pathology Shared Resource (TPSR), which is supported by NCI/NIH Cancer Center Support Grant 2P30 CA068485-14. Dynamic light scattering was conducted at the Vanderbilt Institute of Nanoscale Sciences and Engineering. Bone analysis by microCT was supported in part by the NIH (S10RR027631-01). The authors acknowledge Lauren Hug-dahl for assistance with the TOC graphic and Figure S1. The authors acknowledge R. Brock Fletcher for assistance with the Galectin-8 (Gal8) endosome escape imaging assay. We are grateful to the DOD (DOD CDMRP OR130302), NIH (NIH R01 CA224241, NIH R01 EB019409, NIH T32GM007347), Canadian Natural Sciences and Engineering Research Council (NSERC), the Rheumatology Research Foundation (RRF), the VA Merit Award (BX004151), the National Science Foundation Graduate Research Fellowship Program (NSF GRF #2016212929), the European Research Council, under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC Grant Agreement No. 616695, and by the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Sklodowska-Curie Grant Agreement No. 754490 (COFUND-2018 “MINDED”) and Grant Agreement No. 872648 (RISE-2019 “MEPHOS”) for funding support.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c04005.
siNP chemical synthesis; chemical and physical characterization of siNP-μPLs; in vitro and in vivo fluorescence and confocal microscopy images of siNP-μPLs and siNPs; mechanical loading regimen for the PTOA model; schematic of in vitro siNP-μPLs releasate study; biodistribution of Cy5-siNP-μPLs signal; in-depth knee joint histology and histology controls; subchondral bone density (BV/TV) analysis (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.1c04005
The content in this report is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
The authors declare no competing financial interest.
Contributor Information
Sean K Bedingfield, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States.
Juan M. Colazo, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States; Vanderbilt University School of Medicine, Vanderbilt University, Nashville, Tennessee 37232, United States; Medical Scientist Training Program, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, United States.
Martina Di Francesco, Laboratory of Nanotechnology for Precision Medicine, Istituto Italiano di Tecnologia, Genoa 16163, Italy.
Fang Yu, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States.
Danielle D. Liu, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States; Vanderbilt University School of Medicine, Vanderbilt University, Nashville, Tennessee 37232, United States; Medical Scientist Training Program, Vanderbilt University School of Medicine, Nashville, Tennessee 37232, United States
Valentina Di Francesco, Laboratory of Nanotechnology for Precision Medicine, Istituto Italiano di Tecnologia, Genoa 16163, Italy.
Lauren E. Himmel, Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, United States
Mukesh K. Gupta, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States
Hongsik Cho, Department of Orthopaedic Surgery and Biomedical Engineering, University of Tennessee Health Science Center-Campbell Clinic, Memphis, Tennessee 38104, United States; Research 151, VA Medical Center, Memphis, Tennessee 38104, United States.
Karen A. Hasty, Department of Orthopaedic Surgery and Biomedical Engineering, University of Tennessee Health Science Center-Campbell Clinic, Memphis, Tennessee 38104, United States; Research 151, VA Medical Center, Memphis, Tennessee 38104, United States
Paolo Decuzzi, Laboratory of Nanotechnology for Precision Medicine, Istituto Italiano di Tecnologia, Genoa 16163, Italy.
Craig L. Duvall, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37232, United States.
REFERENCES
- (1).Menon J; Mishra P Health Care Resource Use, Health Care Expenditures and Absenteeism Costs Associated with Osteoarthritis in US Healthcare System. Osteoarthritis Cartilage 2018, 26 (4), 480–484. [DOI] [PubMed] [Google Scholar]
- (2).Turkiewicz A; Petersson IF; Björk J; Hawker G; Dahlberg LE; Lohmander LS; Englund M Current and Future Impact of Osteoarthritis on Health Care: A Population-Based Study with Projections to Year 2032. Osteoarthritis and Cartilage 2014, 22 (11), 1826–1832. [DOI] [PubMed] [Google Scholar]
- (3).Wang LJ; Zeng N; Yan ZP; Li JT; Ni GX Post-Traumatic Osteoarthritis Following ACL Injury. Arthritis Res. Ther 2020, 22 (1), 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Brophy RH; Gray BL; Nunley RM; Barrack RL; Clohisy JC Total Knee Arthroplasty after Previous Knee Surgery. J. Bone Joint Surg.-Am 2014, 96A (10), 801–805. [DOI] [PubMed] [Google Scholar]
- (5).Brown TD; Johnston RC; Saltzman CL; Marsh JL; Buckwalter JA Posttraumatic Osteoarthritis: A First Estimate of Incidence, Prevalence, and Burden of Disease. J. Orthop Trauma 2006, 20 (10), 739–744. [DOI] [PubMed] [Google Scholar]
- (6).Bradley JD; Brandt KD; Katz BP; Kalasinski LA; Ryan SI Comparison of an Antiinflammatory Dose of Ibuprofen, an Analgesic Dose of Ibuprofen, and Acetaminophen in the Treatment of Patients with Osteoarthritis of the Knee. N. Engl. J. Med 1991, 325 (2), 87–91. [DOI] [PubMed] [Google Scholar]
- (7).Evans CH; Kraus VB; Setton LA Progress in Intra-Articular Therapy. Nat. Rev. Rheumatol 2014, 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Hochberg MC; Altman RD; April KT; Benkhalti M; Guyatt G; McGowan J; Towheed T; Welch V; Wells G; Tugwell P American College of Rheumatology 2012 Recommendations for the Use of Nonpharmacologic and Pharmacologic Therapies in Osteoarthritis of the Hand, Hip, and Knee. Arthritis Care Res. 2012, 64 (4), 465–74. [DOI] [PubMed] [Google Scholar]
- (9).McAlindon TE; Bannuru RR; Sullivan MC; Arden NK; Berenbaum F; Bierma-Zeinstra SM; Hawker GA; Henrotin Y; Hunter DJ; Kawaguchi H; Kwoh K; Lohmander S; Rannou F; Roos EM; Underwood M OARSI Guidelines for the Non-Surgical Management of Knee Osteoarthritis. Osteoarthritis Cartilage 2014, 22 (3), 363–88. [DOI] [PubMed] [Google Scholar]
- (10).McAlindon TE; LaValley MP; Harvey WF; Price LL; Driban JB; Zhang M; Ward RJ Effect of Intra-Articular Triamcinolone vs Saline on Knee Cartilage Volume and Pain in Patients with Knee Osteoarthritis: A Randomized Clinical Trial. JAMA 2017, 317 (19), 1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wijn SRW; Rovers MM; van Tienen TG; Hannink G Intra-Articular Corticosteroid Injections Increase the Risk of Requiring Knee Arthroplasty. Bone Joint J. 2020, 102-B (5), 586–592. [DOI] [PubMed] [Google Scholar]
- (12).Simkin PA Synovial Perfusion and Synovial Fluid Solutes. Ann. Rheum. Dis 1995, 54 (5), 424–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Larsen C; Østergaard J; Larsen SW; Jensen H; Jacobsen S; Lindegaard C; Andersen PH Intra-Articular Depot Formulation Principles: Role in the Management of Postoperative Pain and Arthritic Disorders. J. Pharm. Sci 2008, 97 (11), 4622–4654. [DOI] [PubMed] [Google Scholar]
- (14).Bracho-Sanchez E; Rocha F; Bedingfield S; Partain BD; Brusko MA; Colazo JM; Fettis MM; Farhadi SA; Helm E; Koenders K; Kwiatkowski AJ; Macias SL; Restuccia A; Wanchoo A; Avram D; Allen KD; Duvall CL; Wallet SM; Hudalla GA; Keselowsky BG Galectin-Anchored Indoleamine 2,3-Dioxygenase Suppresses Local Inflammation. bioRxiv; 2021; https://www.biorxiv.org/content/10.1101/2021.05.07.443161v1 (accessed July 15, 2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Colazo JM; Evans BC; Farinas AF; Al-Kassis S; Duvall CL; Thayer WP Applied Bioengineering in Tissue Reconstruction, Replacement, and Regeneration. Tissue Eng., Part B 2019, 25 (4), 259–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bolduc JA; Collins JA; Loeser RF Reactive Oxygen Species, Aging and Articular Cartilage Homeostasis. Free Radical Biol. Med 2019, 132, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Young IC; Chuang ST; Gefen A; Kuo WT; Yang CT; Hsu CH; Lin FH A Novel Compressive Stress-Based Osteoarthritis-Like Chondrocyte System. Exp. Biol. Med. (London, U. K.) 2017, 242 (10), 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jasin HE; Noyori K; Takagi T; Taurog JD Characteristics of Anti-Type II Collagen Antibody Binding to Articular Cartilage. Arthritis Rheum. 1993, 36 (5), 651–9. [DOI] [PubMed] [Google Scholar]
- (19).Hollander AP; Pidoux I; Reiner A; Rorabeck C; Bourne R; Poole AR Damage to Type II Collagen in Aging and Osteoarthritis Starts at the Articular Surface, Originates around Chondrocytes, and Extends into the Cartilage with Progressive Degeneration. J. Clin. Invest 1995, 96 (6), 2859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Loeser RF Integrins and Chondrocyte-Matrix Interactions in Articular Cartilage. Matrix Biol. 2014, 39, 11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lee AS; Ellman MB; Yan D; Kroin JS; Cole BJ; van Wijnen AJ; Im H-J A Current Review of Molecular Mechanisms Regarding Osteoarthritis and Pain. Gene 2013, 527 (2), 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Loeser RF Osteoarthritis Year in Review 2013: Biology. Osteoarthritis and Cartilage 2013, 21 (10), 1436–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhang Y; Jordan JM Epidemiology of Osteoarthritis. Clinics in geriatric medicine 2010, 26 (3), 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wang M; Sampson ER; Jin H; Li J; Ke QH; Im HJ; Chen D MMP13 Is a Critical Target Gene During the Progression of Osteoarthritis. Arthritis Res. Ther 2013, 15 (1), R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Krzeski P; Buckland-Wright C; Bálint G; Cline GA; Stoner K; Lyon R; Beary J; Aronstein WS; Spector TD Development of Musculoskeletal Toxicity without Clear Benefit after Administration of PG-116800, a Matrix Metalloproteinase Inhibitor, to Patients with Knee Osteoarthritis: A Randomized, 12-Month, Double-Blind, Placebo-Controlled Study. Arthritis Res. Ther 2007, 9 (5), R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Molina JR; Reid JM; Erlichman C; Sloan JA; Furth A; Safgren SL; Lathia CD; Alberts SR A Phase I and Pharmacokinetic Study of the Selective, Non-Peptidic Inhibitor of Matrix Metalloproteinase Bay 12–9566 in Combination with Etoposide and Carboplatin. Anti-Cancer Drugs 2005, 16 (9), 997–1002. [DOI] [PubMed] [Google Scholar]
- (27).Clutterbuck AL; Asplin KE; Harris P; Allaway D; Mobasheri A Targeting Matrix Metalloproteinases in Inflammatory Conditions. Curr. Drug Targets 2009, 10 (12), 1245–54. [DOI] [PubMed] [Google Scholar]
- (28).Liu J; Khalil RA Matrix Metalloproteinase Inhibitors as Investigational and Therapeutic Tools in Unrestrained Tissue Remodeling and Pathological Disorders. Progress in molecular biology and translational science 2017, 148, 355–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Settle S; Vickery L; Nemirovskiy O; Vidmar T; Bendele A; Messing D; Ruminski P; Schnute M; Sunyer T Cartilage Degradation Biomarkers Predict Efficacy of a Novel, Highly Selective Matrix Metalloproteinase 13 Inhibitor in a Dog Model of Osteoarthritis: Confirmation by Multivariate Analysis That Modulation of Type II Collagen and Aggrecan Degradation Peptides Parallels Pathologic Changes. Arthritis Rheum. 2010, 62 (10), 3006–15. [DOI] [PubMed] [Google Scholar]
- (30).Akagi R; Sasho T; Saito M; Endo J; Yamaguchi S; Muramatsu Y; Mukoyama S; Akatsu Y; Katsuragi J; Fukawa T; Takahashi K Effective Knock Down of Matrix Metalloproteinase-13 by an Intra-Articular Injection of Small Interfering RNA (SiRNA) in a Murine Surgically-Induced Osteoarthritis Model. J. Orthop. Res 2014, 32 (9), 1175–80. [DOI] [PubMed] [Google Scholar]
- (31).Nakagawa R; Akagi R; Yamaguchi S; Enomoto T; Sato Y; Kimura S; Ogawa Y; Sadamasu A; Ohtori S; Sasho T Single vs. Repeated Matrix Metalloproteinase-13 Knockdown with Intra-Articular Short Interfering RNA Administration in a Murine Osteoarthritis Model. Connect. Tissue Res 2019, 60 (4), 335–343. [DOI] [PubMed] [Google Scholar]
- (32).Bedingfield SK; Yu F; Liu DD; Jackson MA; Himmel LE; Cho H; Colazo JM; Crofford LJ; Hasty KA; Duvall CL Matrix-Targeted Nanoparticles for MMP13 RNA Interference Blocks Post-Traumatic Osteoarthritis. bioRxiv 2020; https://www.biorxiv.org/content/10.1101/2020.01.30.925321v1 (accessed July 15, 2021). [Google Scholar]
- (33).Bedingfield SK; Colazo JM; Yu F; Liu DD; Jackson MA; Himmel LE; Cho H; Crofford LJ; Hasty KA; Duvall CL Amelioration of post-traumatic osteoarthritis via nanoparticle depots delivering small interfering RNA to damaged cartilage. Nat. Biomed. Eng 2021, DOI: 10.1038/s41551-021-00780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhao L; Huang J; Fan Y; Li J; You T; He S; Xiao G; Chen D Exploration of CRISPR/Cas9-Based Gene Editing as Therapy for Osteoarthritis. Ann. Rheum. Dis 2019, 78 (5), 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wu P; Grainger DW Drug/Device Combinations for Local Drug Therapies and Infection Prophylaxis. Biomaterials 2006, 27 (11), 2450–2467. [DOI] [PubMed] [Google Scholar]
- (36).Gao W; Chen Y; Zhang Y; Zhang Q; Zhang L Nanoparticle-Based Local Antimicrobial Drug Delivery. Adv. Drug Delivery Rev 2018, 127, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bodick N; Lufkin J; Willwerth C; Kumar A; Bolognese J; Schoonmaker C; Ballal R; Hunter D; Clayman M An Intra-Articular, Extended-Release Formulation of Triamcinolone Acetonide Prolongs and Amplifies Analgesic Effect in Patients with Osteoarthritis of the Knee: A Randomized Clinical Trial. J. Bone Joint Surg Am 2015, 97 (11), 877–88. [DOI] [PubMed] [Google Scholar]
- (38).Maudens P; Seemayer CA; Pfefferle F; Jordan O; Allemann E Nanocrystals of a Potent P38 Mapk Inhibitor Embedded in Microparticles: Therapeutic Effects in Inflammatory and Mechanistic Murine Models of Osteoarthritis. J. Controlled Release 2018, 276, 102–112. [DOI] [PubMed] [Google Scholar]
- (39).Maudens P; Seemayer CA; Thauvin C; Gabay C; Jordan O; Allemann E Nanocrystal-Polymer Particles: Extended Delivery Carriers for Osteoarthritis Treatment. Small 2018, 14 (8), 1703108. [DOI] [PubMed] [Google Scholar]
- (40).Convertine AJ; Benoit DS; Duvall CL; Hoffman AS; Stayton PS Development of a Novel Endosomolytic Diblock Copolymer for SiRNA Delivery. J. Controlled Release 2009, 133 (3), 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Evans BC; Hocking KM; Kilchrist KV; Wise ES; Brophy CM; Duvall CL Endosomolytic Nano-Polyplex Platform Technology for Cytosolic Peptide Delivery to Inhibit Pathological Vasoconstriction. ACS Nano 2015, 9 (6), 5893–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Nelson CE; Gupta MK; Adolph EJ; Guelcher SA; Duvall CL siRNA Delivery from an Injectable Scaffold for Wound Therapy. Adv. Wound Care (New Rochelle) 2013, 2 (3), 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Nelson CE; Gupta MK; Adolph EJ; Shannon JM; Guelcher SA; Duvall CL Sustained Local Delivery of siRNA from an Injectable Scaffold. Biomaterials 2012, 33 (4), 1154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Martin JR; Nelson CE; Gupta MK; Yu F; Sarett SM; Hocking KM; Pollins AC; Nanney LB; Davidson JM; Guelcher SA; Duvall CL Local Delivery of PHD2 siRNA from Ros-Degradable Scaffolds to Promote Diabetic Wound Healing. Adv. Healthcare Mater 2016, 5 (21), 2751–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nelson CE; Kim AJ; Adolph EJ; Gupta MK; Yu F; Hocking KM; Davidson JM; Guelcher SA; Duvall CL Tunable Delivery of siRNA from a Biodegradable Scaffold to Promote Angiogenesis in Vivo. Adv. Mater 2014, 26 (4), 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Di Francesco M; Primavera R; Summa M; Pannuzzo M; Di Francesco V; Di Mascolo D; Bertorelli R; Decuzzi P Engineering Shape-Defined Plga Microplates for the Sustained Release of Anti-Inflammatory Molecules. J. Controlled Release 2020, 319, 201. [DOI] [PubMed] [Google Scholar]
- (47).Di Francesco M; Primavera R; Romanelli D; Palomba R; Pereira RC; Catelani T; Celia C; Di Marzio L; Fresta M; Di Mascolo D; et al. Hierarchical Microplates as Drug Depots with Controlled Geometry, Rigidity, and Therapeutic Efficacy. ACS Appl. Mater. Interfaces 2018, 10 (11), 9280–9289. [DOI] [PubMed] [Google Scholar]
- (48).Di Francesco M; Bedingfield SK; Di Francesco V; Colazo JM; Yu F; Ceseracciu L; Bellotti E; Di Mascolo D; Ferreira M; Himmel LE; Duvall C; Decuzzi P Shape-Defined Microplates for the Sustained Intra-Articular Release of Dexamethasone in the Management of Overload-Induced Osteoarthritis. ACS Appl. Mater. Interfaces 2021, 13, 31379–31392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Hong L; Wei N; Joshi V; Yu Y; Kim N; Krishnamachari Y; Zhang Q; Salem AK Effects of Glucocorticoid Receptor Small Interfering RNA Delivered Using Poly Lactic-Co-Glycolic Acid Microparticles on Proliferation and Differentiation Capabilities of Human Mesenchymal Stromal Cells. Tissue Eng., Part A 2012, 18 (7–8), 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wang Y; Tran KK; Shen H; Grainger DW Selective Local Delivery of Rank siRNA to Bone Phagocytes Using Bone Augmentation Biomaterials. Biomaterials 2012, 33 (33), 8540–8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Mountziaris PM; Tzouanas SN; Sing DC; Kramer PR; Kasper FK; Mikos AG Intra-Articular Controlled Release of Anti-Inflammatory siRNA with Biodegradable Polymer Microparticles Ameliorates Temporomandibular Joint Inflammation. Acta Biomater. 2012, 8 (10), 3552–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Garland KM; Sevimli S; Kilchrist KV; Duvall CL; Cook RS; Wilson JT Microparticle Depots for Controlled and Sustained Release of Endosomolytic Nanoparticles. Cell. Mol. Bioeng 2019, 12 (5), 429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Lu X; Miao L; Gao W; Chen Z; McHugh KJ; Sun Y; Tochka Z; Tomasic S; Sadtler K; Hyacinthe A; Huang Y; Graf T; Hu Q; Sarmadi M; Langer R; Anderson DG; Jaklenec A Engineered PLGA Microparticles for Long-Term, Pulsatile Release of STING Agonist for Cancer Immunotherapy. Sci. Transl. Med 2020, 12 (556), No. eaaz6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kilchrist KV; Dimobi SC; Jackson MA; Evans BC; Werfel TA; Dailing EA; Bedingfield SK; Kelly IB; Duvall CL Gal8 Visualization of Endosome Disruption Predicts Carrier-Mediated Biologic Drug Intracellular Bioavailability. ACS Nano 2019, 13 (2), 1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Kilchrist KV; Tierney JW; Duvall CL Genetically Encoded Split-Luciferase Biosensors to Measure Endosome Disruption Rapidly in Live Cells. ACS Sensors 2020, 5 (7), 1929–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Nelson CE; Kim AJ; Adolph EJ; Gupta MK; Yu F; Hocking KM; Davidson JM; Guelcher SA; Duvall CL Tunable Delivery of siRNA from a Biodegradable Scaffold to Promote Angiogenesis in Vivo. Adv. Mater 2014, 26 (4), 607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).John T; Stahel PF; Morgan SJ; Schulze-Tanzil G Impact of the Complement Cascade on Posttraumatic Cartilage Inflammation and Degradation. Histol Histopathol 2007, 22 (7), 781–790. [DOI] [PubMed] [Google Scholar]
- (58).Silawal S; Triebel J; Bertsch T; Schulze-Tanzil G Osteoarthritis and the Complement Cascade. Clin. Med. Insights: Arthritis Musculoskeletal Disord 2018, 11, 117954411775143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Lubbers R; van Essen MF; van Kooten C; Trouw LA Production of Complement Components by Cells of the Immune System. Clin. Exp. Immunol 2017, 188 (2), 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Zhai G; Doré J; Rahman P TGF-B Signal Transduction Pathways and Osteoarthritis. Rheumatol. Int 2015, 35 (8), 1283–92. [DOI] [PubMed] [Google Scholar]
- (61).Shen J; Li S; Chen D TGF-B Signaling and the Development of Osteoarthritis. Bone Res. 2014, 2, 14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Takahashi N; Rieneck K; van der Kraan PM; van Beuningen HM; Vitters EL; Bendtzen K; van den Berg WB Elucidation of Il-1/TGF-Beta Interactions in Mouse Chondrocyte Cell Line by Genome-Wide Gene Expression. Osteoarthritis Cartilage 2005, 13 (5), 426–38. [DOI] [PubMed] [Google Scholar]
- (63).Klatt AR; Paul-Klausch B; Klinger G; Kuhn G; Renno JH; Banerjee M; Malchau G; Wielckens K A Critical Role for Collagen II in Cartilage Matrix Degradation: Collagen II Induces Pro-Inflammatory Cytokines and MMPs in Primary Human Chondrocytes. J. Orthop. Res 2009, 27 (1), 65–70. [DOI] [PubMed] [Google Scholar]
- (64).Yasuda T Cartilage Destruction by Matrix Degradation Products. Mod. Rheumatol 2006, 16 (4), 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Hügle T; Geurts J What Drives Osteoarthritis?—Synovial versus Subchondral Bone Pathology. Rheumatology 2016, 56 (9), kew389. [DOI] [PubMed] [Google Scholar]
- (66).Chou CH; Jain V; Gibson J; Attarian DE; Haraden CA; Yohn CB; Laberge RM; Gregory S; Kraus VB Synovial Cell Cross-Talk with Cartilage Plays a Major Role in the Pathogenesis of Osteoarthritis. Sci. Rep 2020, 10 (1), 10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Glasson SS; Chambers MG; Van Den Berg WB; Little CB The OARSI Histopathology Initiative - Recommendations for Histological Assessments of Osteoarthritis in the Mouse. Osteoarthritis Cartilage 2010, 18, S17–S23. [DOI] [PubMed] [Google Scholar]
- (68).Bolon B; Stolina M; King C; Middleton S; Gasser J; Zack D; Feige U Rodent Preclinical Models for Developing Novel Antiarthritic Molecules: Comparative Biology and Preferred Methods for Evaluating Efficacy. J. Biomed. Biotechnol 2011, 2011, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Cho H; Kim BJ; Park SH; Hasty KA; Min BH Noninvasive Visualization of Early Osteoarthritic Cartilage Using Targeted Nanosomes in a Destabilization of the Medial Meniscus Mouse Model. Int. J. Nanomed 2018, 13, 1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Cho H; Pinkhassik E; David V; Stuart JM; Hasty KA Detection of Early Cartilage Damage Using Targeted Nanosomes in a Post-Traumatic Osteoarthritis Mouse Model. Nanomedicine 2015, 11 (4), 939–46. [DOI] [PubMed] [Google Scholar]
- (71).Li H; Wang D; Yuan Y; Min J New Insights on the MMP13 Regulatory Network in the Pathogenesis of Early Osteoarthritis. Arthritis Res. Ther 2017, 19 (1), 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sun Y; Mauerhan DR Meniscal Calcification, Pathogenesis and Implications. Curr. Opin. Rheumatol 2012, 24 (2), 152–157. [DOI] [PubMed] [Google Scholar]
- (73).Blom AB; van Lent PL; Libregts S; Holthuysen AE; van der Kraan PM; van Rooijen N; van den Berg WB Crucial Role of Macrophages in Matrix Metalloproteinase-Mediated Cartilage Destruction During Experimental Osteoarthritis: Involvement of Matrix Metalloproteinase 3. Arthritis Rheum. 2007, 56 (1), 147–57. [DOI] [PubMed] [Google Scholar]
- (74).Goldring SR Alterations in Periarticular Bone and Cross Talk between Subchondral Bone and Articular Cartilage in Osteoarthritis. Ther. Adv. Musculoskeletal Dis 2012, 4 (4), 249–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Evans BC; Hocking KM; Osgood MJ; Voskresensky I; Dmowska J; Kilchrist KV; Brophy CM; Duvall CL MK2 Inhibitory Peptide Delivered in Nanopolyplexes Prevents Vascular Graft Intimal Hyperplasia. Sci. Transl. Med 2015, 7 (291), 291ra95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Convertine A; Benoit D; Duvall C; Hoffman A; Stayton P Development of a Novel Endosomolytic Diblock Copolymer for siRNA Delivery. J. Controlled Release 2009, 133 (3), 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Moad G; Chong YK; Postma A; Rizzardo E; Thang SH Advances in Raft Polymerization: The Synthesis of Polymers with Defined End-Groups. Polymer 2005, 46 (19), 8458–8468. [Google Scholar]
- (78).Jackson MA; Werfel TA; Curvino EJ; Yu F; Kavanaugh TE; Sarett SM; Dockery MD; Kilchrist KV; Jackson AN; Giorgio TD; Duvall CL Zwitterionic Nanocarrier Surface Chemistry Improves siRNA Tumor Delivery and Silencing Activity Relative to Polyethylene Glycol. ACS Nano 2017, 11, 5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Poulet B; Hamilton RW; Shefelbine S; Pitsillides AA Characterizing a Novel and Adjustable Noninvasive Murine Joint Loading Model. Arthritis Rheum. 2011, 63 (1), 137–47. [DOI] [PubMed] [Google Scholar]
- (80).Aigner T; Söder S Histopathologische Begutachtung Der Gelenkdegeneration. Der Pathologe 2006, 27 (6), 431–438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


